Abstract
The DNA damage-binding protein 2 (DDB2) is an adapter protein that can direct a modular Cul4-DDB1-RING E3 Ligase complex to sites of ultraviolet light-induced DNA damage to ubiquitinate substrates during nucleotide excision repair. The DDB2 transcript is ultraviolet-inducible; therefore, its regulation is likely important for its function. Curiously, the DDB2 mRNA is reportedly short-lived, but the transcript does not contain any previously characterized cis-acting determinants of mRNA stability in its 3′ untranslated region (3′UTR). Here, we used a tetracycline regulated d2EGFP reporter construct containing specific 3′UTR sequences from DDB2 to identify novel cis-acting elements that regulate mRNA stability. Synthetic 3′UTRs corresponding to sequences as short as 25 nucleotides from the central region of the 3′UTR of DDB2 were sufficient to accelerate decay of the heterologous reporter mRNA. Conversely, these same 3′UTRs led to more rapid induction of the reporter mRNA, export of the message to the cytoplasm and the subsequent accumulation of the encoded reporter protein, indicating that this newly identified cis-acting element affects transcriptional and post-transciptional processes. These results provide clear evidence that nuclear and cytoplasmic processing of the DDB2 mRNA is inextricably linked.
INTRODUCTION
Damage-specific DNA binding protein 2 (DDB2) was originally identified in association with DDB1 as part of an ultraviolet (UV)-induced protein complex with DNA damage-binding activity (1,2). Mutations in the DDB2 gene are associated with xeroderma pigmentosum group E, a rare genetic disorder characterized by DNA repair defects, sun-sensitivity and a predisposition to skin cancer. Fibroblasts derived from xeroderma pigmentosum group E patients have modest UV sensitivity with a measurable defect in the global genomic subpathway of nucleotide excision repair (GG-NER) (3–5). Although its precise role in GG-NER remains somewhat enigmatic, it is thought that DDB2 facilitates GG-NER through early damage recognition (6–9). It is now recognized that the DDB1 protein is part of a modular Cul4-DDB1-RING E3 Ligase complex (CRL4) (2,10,11) and that this complex can be directed to a wide array of substrates through adapter proteins like DDB2 (CRL4DDB2) (2). Thus, DDB2 can direct CRL4 to UV lesions where it can locally ubiquitinate a variety of proteins to affect local chromatin structure and facilitate the assembly of a GG-NER complex (2,11–15). In addition to its role in GG-NER, CRL4DDB2 can also regulate cell cycle progression and senescence through the ubiquitination of other substrates (16–24). These various functions of DDB2 indicate that this protein plays several critical anti-neoplastic roles; therefore, understanding its regulation is critical to understand its role in GG-NER and cancer.
DDB2 expression and activity is tightly controlled through a variety of transcriptional and post-translational mechanisms (11,25–29). Transcription of the DDB2 gene is regulated, in large part, by the p53 tumour suppressor, a transcription factor that positively regulates the expression of genes encoding proteins involved in DNA repair, cell cycle checkpoints and apoptosis (27,30). Recently, we used a temperature sensitive variant of p53 that permits rapid reversible regulation of p53 activity to monitor the induction and subsequent decay of p53-induced gene expression using oligonucleotide microarrays (29). The p53 transcriptional response was readily reversible, and it was dominated by short-lived mRNAs, including DDB2 mRNA (29). Although most of the unstable p53-induced mRNAs contained a variety of sequence motifs associated with rapid turnover of mRNAs, the DDB2 mRNA did not contain any consensus destabilizing elements in its 3′UTR (29). Therefore, the primary sequence of the DDB2 mRNA did not predict in an obvious way its rapid decay following transient p53 activation.
Here, we report the characterization of the 3′UTR of DDB2 using tetracycline-regulated heterologous d2EGFP-3′UTR reporter constructs, expressing a variant of green flourescent protein (GFP). Using this system, we were able to identify a short region within the 3′UTR of DDB2 that affects transcription and decay of the reporter transcript. Our results suggest that this 3′UTR sequence is recognized co-transcriptionally linking transcriptional and post-transcriptional regulatory pathways. The present work suggests that the 3′UTR of DDB2 plays an important role in regulating DDB2 expression and that this likely contributes to its anti-neoplastic activity. Furthermore, similar sequences were identified in the 3′UTRs of unrelated transcripts, suggesting that expression of their encoded proteins may be regulated similarly.
MATERIALS AND METHODS
Cell culture and complementary DNA synthesis
HeLa Tet-Off® cells were purchased from Clontech Laboratories (Mountain View, CA), whereas HT29-tsp53 cells were obtained from Mats Ljungman (University of Michigan) (31). Cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum (Thermo Fisher Scientific, Waltham, MA) and 200 µg/ml G418 antibiotic (Sigma-Aldrich, St. Louis, MO) at 37°C in a humidified atmosphere with 5% CO2.
Generation of d2EGFP 3′UTR reporter constructs
The 3′UTR of DDB2 and defined regions of this 3′UTR were cloned using a reverse transcriptase polymerase chain reaction (RT-PCR)-based strategy with complementary DNA (cDNA) derived from colorectal cancer cells (HT29-tsp53 cells) (29). Briefly, cells were detached with trypsin, and harvested, and total RNA was collected from cells using the RNeasy RNA isolation kit (QIAGEN). Five micrograms of RNA were reverse-transcribed using oligo-dT primers and the First Strand cDNA Synthesis Kit (MBI Fermentas, Burlington, ON). Primers containing either EcoRI (5′) or XbaI (3′) linkers were used to amplify specific regions of the 3′UTR of DDB2 by RT-PCR (Supplementary Table S1). A variety of smaller synthetic 3′UTRs corresponding to mutant versions spanning nucleotides 149–198 of the DDB2 3′UTR were synthesized as complementary oligonucleotides (Sigma-Aldrich Canada Ltd, Oakville, ON, Canada) with the same 5′ and 3′ linker sequences (Supplementary Table S2). These oligonucleotides were subsequently annealed in vitro in Oligo Annealing Buffer (Promega, Madison, WI) according to manufacturer’s instructions.
Heterologous d2EGFP-DDB2 3′UTR reporter constructs were generated by subcloning the corresponding dsDNA into EcoRI and XbaI restriction sites located between the d2EGFP open reading frame and the SV40 polyadenylation signal of the pTRE-d2EGFP vector (PT3228-5, Clontech laboratories, Mountain View, CA) (32). The identity of all constructs was confirmed by DNA sequencing (StemCore Laboratories, Ottawa, ON).
Transfections and fluorescence-activated cell sorting
HeLa Tet-Off® cells or HEK293 Tet-On® cells were seeded at 50% confluence and 24 h later were transfected with pTRE-d2EGFP or heterologous reporter constructs using GeneJuice Transfection Reagent (EMD Chemicals, Gibbstown, NJ). In transient transfection experiments, doxycycline was added 24 h following transfection. To generate stable HeLa Tet-Off®-derived d2EGFP-expressing populations, d2EGFP positive cells were identified by fluorescence-activated cell sorting (FACS) using a DakoCytomation MoFlo flow cytometer (Dako, Denmark). The fluorescent cells were expanded and further enriched by two additional rounds of FACS yielding a highly enriched d2EGFP-positive population. To generate stable HEK293 Tet-On®-derived d2EGFP-expressing populations, FACS was performed similarly except that doxycycline was added 24 h before each sort to express d2EGFP.
For quantitative analysis of green fluorescence, d2EGFP-positive cells were detached with trypsin, washed and collected in phosphate buffered saline (PBS). Flow cytometry was carried out using a Beckman Coulter Epics XL flow cytometer (Beckman Coulter, Mississauga, ON). The d2EGFP fluorescence was analysed using FCS Express 3.0 software package (De Nova Software, Los Angeles, CA).
Quantitative RT-PCR
The cDNAs of interest were generated as described earlier in the text. Quantitative RT-PCR (qRT-PCR) analysis was performed using the Sybr green fluorescent DNA stain (Invitrogen, Carlsbad, CA) and the LightCycler 2 thermo cycler machine with Light-Cycler software version 3 (Roche Diagnostics, Switzerland). The expression of the d2EGFP mRNA was determined using the following primer pairs: CGACGGCAACTACAAGACC and CCATCATCCTGCTCCTCCAC. Expression of d2EGFP mRNA was normalized to the expression of β-actin (GGGCATGGGTCAGAAGGAT and GTGGCCATCTCTTGCTCGA) and/or Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (AACAGCGACACCCACTCCTC and GGAGGGGAGATTCAGTGTGGT) mRNA levels, with similar results.
Polysome isolation
Polysomes were isolated as previously described (33). Briefly, cells from two 15-cm diameter plates were incubated in 100 µg/ml cycloheximide (CHX) for 3 min at 37°C. Cells were washed with ice-cold PBS containing CHX, scraped and collected by centrifugation at 200× g. The cells were lysed in 500 µl of ice-cold polysome lysis buffer [15 mM Tris–HCl (ph 7.4), 15 mM MgCl2, 300 mM NaCl, 1% (v/v) Triton X-100, CHX (0.1 mg/ml), Rnasin (100 U/ml)], transferred to microcentrifuge tubes and incubated on ice for 10 min. Nuclei were pelleted at 5000 rpm for 5 min at 4°C. Supernatants were transferred to fresh tubes while the nuclear fractions were stored at −80°C. The non-nuclear fractions were centrifuged for 5 min at 14 000 rpm at 4°C to remove debris. These supernatants were transferred to fresh tubes, and they were stored at −80°C. Lysates containing equal units of optical density at 254 nm were placed onto linear sucrose gradients (10–50%) and centrifuged at 39 000 rpm for 90 min at 4°C. Fractions from each gradient were sequentially collected from the top of the gradient using a Teledyne ISCO R1/R2 fraction collector coupled with a UA6 Absorbance detector (Teledyne, Lincoln, NE). Fractions were stored at −80°C. RNA from each fraction, including nuclear pellets, was isolated by phenol-chloroform extraction followed by ethanol precipitation. Equal volumes of RNA were used to generate cDNA and analysed by quantitative RT-PCR, as described earlier in the text.
Immunoblot analysis
Immunoblotting was performed as previously described (34). Briefly, 20 µg of total protein derived from whole-cell lysates were resolved by gel electrophoresis using NuPAGE 3–8% gradient polyacrylamide gels (Invitrogen). Proteins were transferred to Hybond-C nitrocellulose (GE Healthcare, Baie d’Urfé, QC), and blots were stained with Ponceau S Red (5 mg/ml Ponceau S Red, 2% glacial acetic acid) to visualize total transferred proteins. Blots were then blocked in TBSMT [50 mM Tris, 150 mM NaCl (pH 7.6) (TBS), 5% non-fat milk powder, 0.05% Tween 20]. Proteins were detected using HRP-conjugated antibodies against GFP (clone B2, 1:200, Santa Cruz) diluted in TBSM (TBS, 0.5% nonfat milk powder) and were visualized using SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific) in combination with X-ray film (Kodak, Rochester, NY). The anti-GFP antibodies were removed using Restore Western Blot Stripping Buffer (Thermo Fisher Scientific) and reprobed with anti-β-actin (clone AC-74, 1:4000, Sigma-Aldrich Canada Ltd, Oakville, ON) followed by horse radish peroxidase (HRP)–conjugated goat-anti-mouse secondary antibodies and visualized similarly.
Statistical analysis
Statistical analysis was performed using the Prism 3.02 software package (GraphPad, San Diego, CA). The statistical comparison of means was performed by one-way analysis of variance (ANOVA) followed by Tukey’s multple comparisons test. Statistical comparison of time course experiments was performed by two-way ANOVA. Unless stated otherwise, statistical significance was inferred using a threshold of P ≤ 0.05.
RESULTS
The central region from 3′UTR of DDB2 acts in cis to accelerate the decay of heterologous RNA
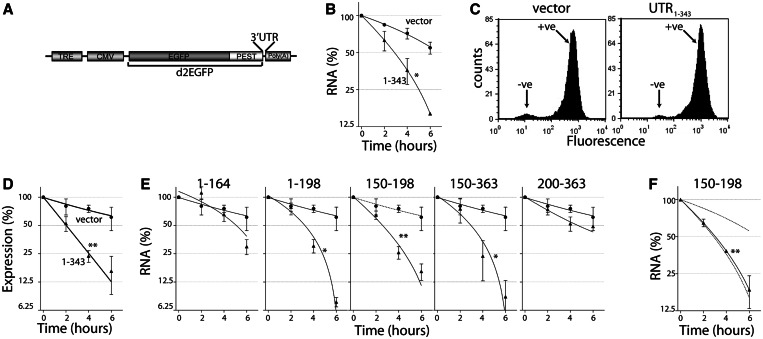
The DDB2 mRNA was reported to have a short half-life; yet, the 3′UTR contained no identifiable destabilizing elements (29); therefore, we sought to identify novel cis-acting elements in its 3′UTR. To do this, we first subcloned various regions of the 3′UTR of DDB2 into the pTRE-d2EGFP plasmid between the d2EGFP open reading frame and the polyadenylation sequence (Figure 1A). These constructs placed the heterologous reporter cDNA under tetracycline-repressible control in HeLa Tet-Off® cells (29). Transient transfection of HeLa Tet-Off® cells with the d2EGFP-UTR1–343 construct (containing nucleotides 1–343 of the 3′UTR of DDB2) led to readily detectable d2EGFP mRNA that could be depleted significantly more rapidly than vector control-encoded d2EGFP mRNA lacking this 3′UTR sequence following addition of doxycycline (Figure 1B).
Figure 1.
The 3′UTR of DDB2 confers instability to a heterologous reporter gene. (A) Schematic representation of the pTRE-d2EGFP vector used to generate 3′UTR reporter constructs. The location of the tetracycline response element (TRE), minimal cytomegalovirus promoter (CMV), EGFP with a C-terminal PEST sequence (d2EGFP), 3′UTR cloning site and the polyadenylation signal are indicated. (B) HeLa Tet-Off® cells were transfected with the pTRE-d2EGFP vector or vectors containing a 3′UTR corresponding to the indicated nucleotides from the 3′UTR of DDB2. In (B) and (F), doxycycline (1 µg/ml) was added 24 h after transfection and total RNA was collected at the indicated times to monitor loss of d2EGFP expression by qRT-PCR. (C) Stable pools of d2EGFP positive cells were generated through multiple rounds of fluorescence activated cell sorting. (D and E) In all, 1 µg/ml of doxycycline was added to stable cell lines expression d2EGFP with the indicated 3′UTR sequence. At the indicated time, total RNA was collected for qRT-PCR analysis of d2EGFP expression. Each value in (B), (D), (E) and (F) represents the mean (±SEM) determined in several independent experiments. The asterisk and double asterisk indicate the curve is significantly different from the vector control as determined by two-way ANOVA at P < 0.05 and P < 0.0001, respectively.
To facilitate quantitative analysis of d2EGFP mRNA and protein expression, we generated stable pools of d2EGFP-expressing cells by transfecting HeLa Tet-Off® cells with these pTRE-d2EGFP constructs. The transfected cells were subjected to three rounds of FACS and expansion of the d2EGFP-positive cells to generate homogeneous stable populations expressing the d2EGFP gene (Figure 1C). Addition of doxycycline to vector control and d2EGFP-UTR1–343 cells led to a reduction in d2EGFP mRNA expression with the 3′UTR of DDB2 accelerating the decay of the reporter mRNA (Figure 1D). Importantly, the rate of decay of these mRNAs was comparable with that observed in transient transfection experiments with the same constructs (compare Figure 1B and D). Therefore, the 3′UTR of DDB2 functions in cis to regulate mRNA decay.
To localize cis-acting determinants of mRNA stability in the 3′UTR of the DDB2 transcript, the decay of d2EGFP mRNA was monitored in other pooled cell lines expressing heterologous d2EGFP mRNAs with different fragments of the 3′UTR of DDB2. The addition of doxycycline led to decreased expression of all reporter genes tested (Figure 1E). Although the d2EGFP-UTR1–164 and d2EGFP-UTR200–363 mRNAs decayed at a rate similar to vector controls, the d2EGFP-UTR1–198, d2EGFP-UTR150–198 and d2EGFP-UTR150–363 mRNAs decayed significantly faster that the control mRNA, and his was similar to the mRNAs containing the full-length 3′UTR of DDB2 (Figure 1E). These unstable mRNAs were similarly short-lived in transient transfection experiments (Figure 1F and data not shown). Therefore, a 49-nt sequence common to these three heterologous reporter mRNAs, corresponding to nucleotides 150–198 was sufficient to accelerate the decay of the reporter construct.
Discordance between d2EGFP mRNA and protein levels
The d2EGFP protein is a fusion of eGFP with a C-terminal PEST sequence derived from ornithine decarboxylase; therefore, this fluorescent protein is turned over rapidly in a proteasome-dependent manner with a half-life of 2 h (32). We chose this short-lived reporter protein because unstable proteins require ongoing translation; therefore, their levels were expected to correlate with the amount of template mRNA available for translation. However, the relationship between mRNA and protein levels was more complex than anticipated.
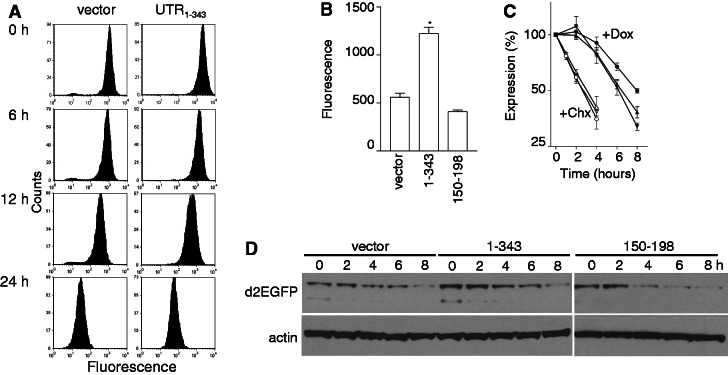
Flow cytometric analysis of d2EGFP expression indicated that the addition of doxycycline to vector control or d2EGFP-UTR1–343 cells resulted in a decrease in green fluorescence without any remarkable qualitative differences between cell lines (Figure 2A). Quantitative analysis of d2EGFP, d2EGFP-UTR1–343 and d2EGFP-UTR150–198 cells indicated that fluorescence was highest in d2EGFP-UTR1–343 cells, despite the instability of the encoding mRNA (Figure 2B). In time course experiments, green fluorescence decreased marginally faster in the d2EGFP-UTR1–343 and d2EGFP-UTR150–198 cells compared with control cells (Figure 2C). The initial difference in d2EGFP expression and the similar rate of decrease in d2EGFP expression was confirmed by immunoblot analysis (Figure 2D). Although the d2EGFP expressed from short-lived mRNAs disappeared more rapidly in the presence of doxycycline (Figure 2C), the large difference in mRNA expression (Figure 1B and D–F) was not quantitatively reflected by the small difference in d2EGFP protein expression. This disparity between protein and mRNA expression did not result from differences in the rate of d2EGFP protein turnover because the d2EGFP protein expressed from these vectors exhibited similar half-lives in the presence of CHX (Figure 2C). Taken together, the 3′UTR of DDB2 had a dramatic effect on d2EGFP mRNA decay with a much smaller effect on the expression of the protein.
Figure 2.
The effect of the 3′UTR on d2EGFP protein expression. (A) Stable cell lines were exposed to doxycycline to shut off reporter gene expression and at the indicated times fluorescence was assessed by flow cytometry. (B) Fluorescence was determined by flow cytometry of actively growing cultures expressing d2EGFP from the control, UTR1–343 and UTR150–198 vectors before the addition of doxycycline. (C) Control (circles), d2EGFP-UTR1–343 (triangles) and d2EGFP-UTR150–198 (inverted triangles) cells were either treated with 1 µg/ml of doxycycline (+Dox, closed symbols) or 10 µg/ml of cycloheximide (+CHX, open symbols). Fluorescence was determined at the indicated time by flow cytometry and is expressed as a percentage of untreated controls. (D) Cells were treated as described in (A) except that whole-cell lysates were collected for western blot analysis of d2EGFP expression. Each value in (B) and (C) represents the mean (±SEM) determined from several independent experiments. The asterisk indicates the value is significantly different from control as determined by one-way ANOVA (P < 0.0001) followed by a Tukey’s multiple comparisons test (P < 0.001).
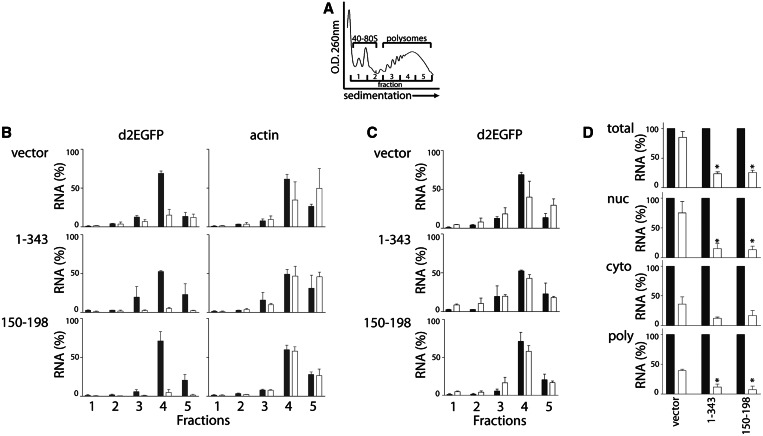
To address the disparity between mRNA and protein levels, similar biological experiments were performed except that cytoplasmic RNA was isolated and separated by sucrose gradient sedimentation to capture ribosome-associated RNAs (polysomes). For reference, a representative OD260 sedimentation profile is presented in Figure 3A. The expression of the d2EGFP and actin mRNAs was highest in fraction 4, regardless of the 3′UTR present in the reporter construct, suggesting that all of these mRNAs were translated at a similar rate (Figure 3B). Addition of doxycycline led to a decrease in d2EGFP expression; however, the distribution of remaining d2EGFP transcripts across these fractions remained largely unchanged (Figure 3B–C). Quantitative analysis indicated that there was a significant decrease in d2EGFP expression in polysomes (fractions 3–5) in all cell lines (Figure 3D, P < 0.001, single sample t-test). A similar decrease in d2EGFP mRNA expression was detected in the cytoplasmic fraction collected before sucrose-gradient sedimentation (Figure 3D). This modest difference in the amount of cytoplasmic and polysome-associated d2EGFP mRNA among cell lines correlated with the small differences in d2EGFP protein expression, reported in Figure 2.
Figure 3.
Subcellular localization of d2EGFP mRNA. HeLa Tet-Off®-derived cells were treated with either 0 or 1 µg/ml of doxycycline (shaded and open bars, respectively) to inhibit reporter gene expression and 4 h later these cells were collected. Total cellular, nuclear, cytoplasmic and polysomal RNA fractions were isolated. (A) A representative sucrose gradient sedimentation profile of the polysome preparation is provided for reference. Polysomal mRNAs are contained in fractions 3–5, as indicated. (B) The relative expression of d2EGFP and beta actin across these gradient fractions was determined by qRT-PCR. RNA levels are expressed relative to total input RNA level from no doxycycline control samples. (C) For comparison, the expression of d2EGFP mRNA following doxycycline treatment in (B) is expressed as a percentage of its respective total input RNA. (D) The expression of d2EGFP in several distinct cellular compartments was determined. Each value represents the mean (±SEM) from several independent experiments. The asterisk indicates the value is significantly different from controls in the presence of doxycycline, as determined by one-way ANOVA followed by a Tukey’s multiple comparisons test (P < 0.05).
Remarkably, the control d2EGFP mRNA expression was significantly higher than the d2EGFP-UTR1–343 and d2EGFP-UTR150–198 mRNAs in the nuclear RNA fraction collected before polysome fractionation, and this was comparable with d2EGFP mRNA in total cellular RNA (Figure 3D). In contrast, the unstable d2EGFP-UTR1–343 and d2EGFP-UTR150–198 transcripts disappeared as rapidly from this pool of RNA as they did from the polysome and cytoplasmic fractions (Figure 3D). Therefore, the highly stable d2EGFP mRNA encoded by the control vector resides primarily in the nucleus and does not contribute to nascent translation, explaining the initial disparity detected between d2EGFP mRNA and protein levels. The persistence of the control mRNA in the nucleus indicates that this transcript is not degraded efficiently in the nucleus nor is it exported efficiently to the cytoplasm. Conversely, our results suggest that sequences within the 150–198 region of the 3′UTR of DDB2 affect the export of the heterologous mRNAs and/or its degradation in the nucleus.
The induction of d2EGFP
The potential effect of 3′UTR sequences on nuclear export motivated us to re-express d2EGFP by washing out doxycycline. We hypothesized that differences in the rate of mRNA export could profoundly influence d2EGFP protein without greatly affecting mRNA expression. Vector control, d2EGFP-UTR1–343 and d2EGFP-UTR150–198 cells were treated with a low concentration of doxycycline for 48 h to block reporter gene expression, and then cells were thoroughly washed in PBS, and fresh doxycycline-free media was replaced. The induction of d2EGFP was apparent within 4 h following the removal of doxycycline in d2EGFP-UTR150–198 and d2EGFP-UTR1–343-expressing cells by flow cytometry, whereas re-expression of d2EGFP was delayed in vector control cells (Figure 4A and B). The d2EGFP-UTR150–198 cells recovered d2EGFP protein to pretreatment levels fastest, restoring a normal flow cytometric histogram within 16 h (Figure 4A and B). Similar results were obtained through immunoblot analysis (Supplementary Figure S1). Unexpectedly, d2EGFP mRNA levels were also induced faster in d2EGFP-UTR1–343 and d2EGFP-UTR150–198 compared with vector control cells, indicating that the DDB2 derived 3′UTRs influence d2EGFP mRNA synthesis (Figure 4C). The more rapid recovery of d2EGFP protein but not mRNA in the d2EGFP-UTR150–198 compared with the d2EGFP-UTR1–343 cells suggests that this small RNA sequence is more effective at stimulating reporter expression post-transcriptionally. Therefore, the UTR150–198 sequence facilitates the recovery of reporter gene expression, whereas other sequences in the 3′UTR of DDB2 likely have opposing effects on RNA metabolism as well.
Figure 4.
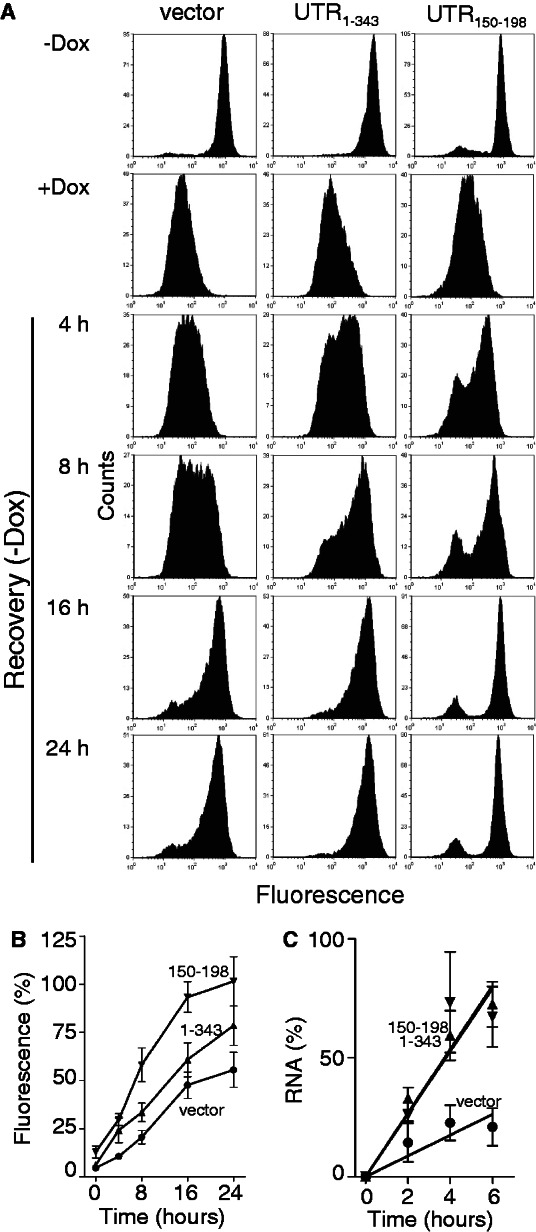
Re-expression of d2EGFP expression in stable cell lines. Stable HeLa Tet-Off®-derived cell lines expressing the indicated reporter constructs were treated with a 0.1 ng/ml doxycycline for 2 days to inhibit reporter gene expression (+Dox). Doxycycline was washed out, and the cells were replaced with fresh medium for the indicated time. (A) Cells were collected and analysed by flow cytometry. Similar results were obtained in three independent experiments. (B) The recovery of green fluorescence from experiments in (A) are expressed as a percentage of the untreated control. (C) Total cellular RNA was collected from similarly treated cells and the recovery of d2EGFP mRNA expression is presented. Each value in (B) and (C) represents the mean (±SEM) from three independent experiments. The curves in B and C were significantly different by two-way ANOVA (P < 0.0001).
To ensure that these effects were not unique to this particular cell line, HEK293 Tet-On® cells were transfected with these heterologous constructs, and d2EGFP positive cells were isolated by FACS. Addition of doxycycline, in these cell lines, led to increased expression of d2EGFP. Once again, the UTR150–198-expressing cells accumulated d2EGFP protein more rapidly (Figure 5A, B and C). Taken together, nucleotides 150–198 from the 3′UTR of DDB2 positively influence reporter protein expression through a combination of increased synthesis, export and/or translation of the reporter mRNAs.
Figure 5.
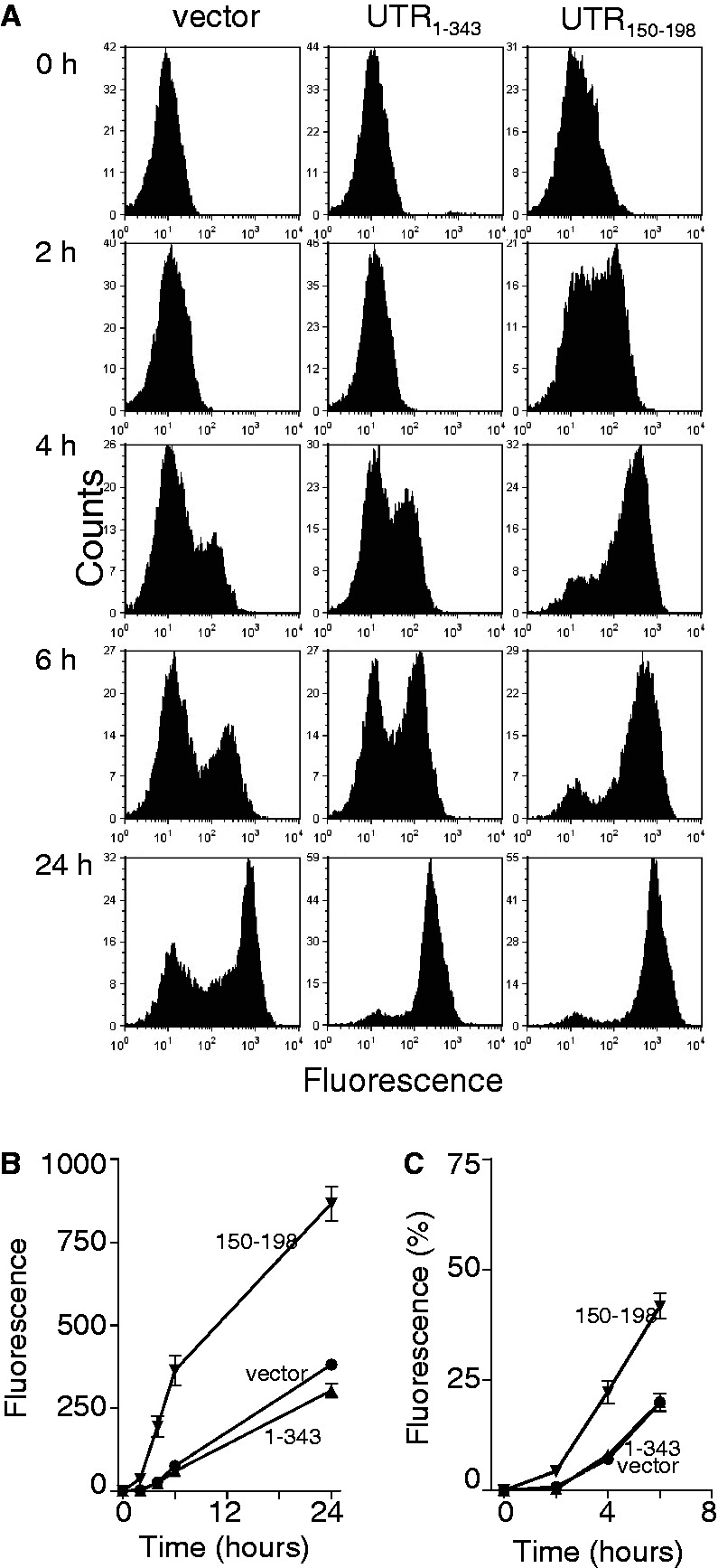
Inducible expression of d2EGFP. Stable HEK293 Tet-On® -derived cells expressing the indicated reporter constructs were treated with a 1 µg/ml doxycycline to induce reporter gene expression. (A) Cells were collected at the indicated for flow cytometric analysis. Similar results were obtained in three independent experiments. (B) The mean increase in green florescence at each time point, determined from experiments in (A), is presented. (C) The results in (B) are also presented as a percentage of fluorescence detected by 24 h. Each value in (B) and (C) represents the mean (±SEM) from three independent experiments. The curves in panels (B) and (C) were significantly different by two-way ANOVA (P < 0.0001).
Inverted repeat and U-rich sequences in the 3′UTR of DDB2
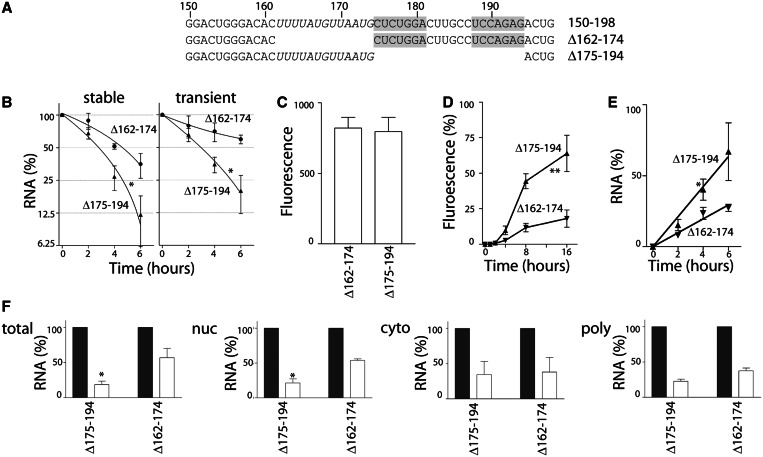
The most obvious feature within the 49 nt sequence was a 7-nt inverted repeat sequence with a 6-nt intervening sequence, corresponding to nucleotides 175–194 of the 3′UTR of DDB2 (Figure 6A). To determine whether this inverted repeat sequence may be essential for accelerated mRNA decay, stable cell lines were generated expressing a heterologous d2EGFP mRNA with the Δ175–194 3′UTR (Figure 6A). The d2EGFP-UTRΔ175–194 reporter mRNA decayed as rapidly as the d2EGFP-UTR150–198 transcript (Figure 6B). Similarly, the d2EGFP-UTRΔ175–194 mRNA was unstable in transiently transfected HeLa Tet-Off® cells (Figure 6B). Therefore, the inverted repeat sequence was not essential for accelerated decay of this mRNA, and the minimal destabilizing region falls within a 25-nt region corresponding to nucleotides 150–174.
Figure 6.
A short U-rich sequence in the 3′UTR of DDB2 regulates the induction, export and decay of the reporter mRNA (A) Synthetic 3′UTRs corresponding to nucleotide 150–198 with deletions of the inverted repeat (Δ175–194) and U-rich rich regions (Δ162–174) were used to generate additional heterologous reporter constructs. The inverted repeats are shaded, and the U-rich region is italicized for clarity. (B) Stable pools of d2EGFP-positive cells and transiently transfected HeLa Tet-Off® cells were exposed to 1 µg/ml doxycycline, and d2EGFP expression was monitored by qRT-PCR. (C) The indicated stable cell lines were collected, and green fluorescence was quantitatively determined by flow cytometry. (D and E) The expression of d2EGFP was blocked with a low concentration of doxycycline then washed out to permit re-expression of d2EGFP, as described in Figure 4. The recovery of green fluorescence (D) and d2EGFP mRNA (E) is expressed as the percentage of their respective no doxycycline controls. (F) The remaining d2EGFP mRNA in the indicated pool of RNA 4 h following the addition of doxycycline is presented. Each value represents the mean (±SEM) from several independent experiments. In (B), (D) and (E), the asterisk and double asterisk indicate the UTRΔ175–194 curve is significantly different from the UTR Δ162–174, as determined by two-way ANOVA at P < 0.05 and P < 0.0001, respectively. In (F), the asterisk indicates the residual expression of d2EGFP mRNA following doxycycline treatment is significantly different in UTRΔ175–194 compared with UTRΔ175–194 cells, as determined by student t-test (P < 0.05).
The region immediately to the 5′ side of the inverted repeat was rich in uridine (8 of 13 residues) and was devoid of cytosine (Figure 6A). We similarly generated a synthetic 3′UTR containing the Δ162–174 sequence (Figure 6A). The resulting heterologous d2EGFP-UTRΔ162–174 mRNA was significantly more stable than either the d2EGFP-UTR150–198 or the d2EGFP-UTRΔ175–194 mRNAs in both stable cell lines and transiently transfected HeLa Tet-Off® cells (Figure 6B and C). Taken together, the inverted repeat sequence (nucleotides 175–194) is dispensable for accelerated mRNA decay while the adjacent U-rich region (nucleotides 162–174) appears to be important for rapid turnover.
Next, we sought to determine whether the induction of reporter gene expression or subcellular localization of the reporter mRNAs was affected in the d2EGFP-UTRΔ162–174 and d2EGFP-UTRΔ175–194 cell lines. Reporter gene expression recovered more rapidly at the protein and mRNA level in the d2EGFP-UTRΔ175–194 cells compared with the d2EGFP-UTRΔ162–174-expressing cells while basal levels of d2EGFP were similar (Figure 6C–E). Consistent with the data presented in Figure 3, the stable d2EGFP-UTRΔ162–174 reporter mRNA was retained primarily in the nucleus, whereas the expression of this mRNA decreased in the polysomal fraction within 4 h. Taken together, all of the heterologous reporter mRNAs containing the U-rich region from the 3′UTR of DDB2 (UTR1–343, UTR150–198 and UTRΔ175–194) exhibited accelerated d2EGFP mRNA induction, mRNA export and mRNA decay, whereas the stable vector control and d2EGFP-UTRΔ162–174 mRNAs were induced more slowly and tended to be retained in the nucleus (recall Figures 1–6). Our results indicate that the 3′UTR of DDB2 contains cis-acting elements that profoundly influence gene expression through a combination of transcriptional and post-transcriptional mechanisms.
DISCUSSION
The 3′UTR of DDB2 contains a novel cis-acting determinant of mRNA stability
The abundance of any given transcript is a function of its rates of both production and turnover. It has become clear that although transcription plays a dominant role in determining the expression of mRNAs, transcriptional regulation is insufficient to fully explain RNA abundance (35). The half-lives of mRNAs are often highly conserved and transcripts encoding functionally related proteins show coordinated changes in mRNA stability, indicating that this level of gene regulation contributes to gene expression (36–38). Therefore, although transcription has historically been the focus of efforts to understand the regulation of gene expression, mRNA decay has emerged as another important level of regulation (36,39–42).
The mRNA decay machinery receives instruction from cis-acting elements that are typically located in the 3′UTR of targeted transcripts. The best characterized elements are adenylate-uridylate-rich elements (ARE) and guanidylate-uridylate-rich elements (43–47). Transacting RNA-binding proteins bind these elements and direct transcripts for decay through de-adenylation followed by exosome-mediated decay (3′–5′) and/or 5′decapping with subsequent Xrn1-dependent exonucleolytic degradation (5′–3′) (45,46). These destabilizing elements, when inserted into the 3′UTR of otherwise stable reporter mRNA, accelerate decay of heterologous mRNAs (48,49). These reporter systems provide a straight forward way to characterize sequences that target specific mRNAs for accelerated decay (48,49). Here, we used this strategy to localize a functional destabilizing element to a 25-nt region that contained a 13-nt U-rich sequence that when deleted prevented the accelerated decay of the heterologous reporter mRNA.
BlastN analysis was performed using the 25-nt sequence corresponding to nucleotides 150–174 from the 3′UTR of DDB2 to query the human RNA sequence database (RefSeq RNA at http://blast.ncbi.nlm.nih.gov/) (50). Four additional transcripts (ZNF493, SS18, MAK and XPNPEP3) were identified that contained highly similar sequences that were all localized to 3′UTRs in the sense orientation (Supplementary Table S3). No similarity to either coding or antisense UTR sequences was identified through this approach, strongly suggesting that these sequences represent a novel cis-acting element. Through a survey of other 3′UTRs studied in our laboratories, we also identified a similar sequence in the sense orientation in the 3′UTR of baculoviral inhibitor of apoptosis (IAP) repeat containing 2 (BIRC2) mRNA (Supplementary Table S3). This sequence had previously been cloned into a chloramphenicol acetyltransferase (CAT) 3′UTR reporter vector and destabilized the reporter mRNA (51). Taken together, the 3′UTR of DDB2 contains a short U-rich sequence that is capable of destabilizing heterologous RNAs, and this element appears to be present and may be similarly functional in other 3′UTRs.
The 3′UTR of DDB2 affects the distribution of heterologous reporter mRNAs
Conceptually similar reporter constructs have been generated from the same pTRE-d2EGFP vector to study the role of 3′UTRs from p16(INK4a) and MKP1 on mRNA decay (48,49). One of the benefits of this system that had yet to be used is the fact that the d2EGFP protein is short-lived owing to the presence of a proteasome-targeting sequence at its C-terminus (32). Therefore, d2EGFP protein and mRNA expression were expected to correlate in this model system. Consistent with this rationale, insertion of the 3′UTR of IL-1β containing an ARE (52) led to decreased protein expression (Supplementary Figure S2). In contrast, the 3′UTR of DDB2 unexpectedly increased basal d2EGFP protein levels, consistent with increased translation from the heterologous d2EGFP-UTR1–343 mRNA. To study the fraction of d2EGFP mRNA immediately available for translation, ribosome-associated RNA was isolated. The amount of polysome-associated RNA correlated much better than total mRNA with d2EGFP protein levels. Meanwhile, the stable d2EGFP mRNAs (vector control and d2EGFP-UTRΔ162–174) were detected in the nuclear fraction, whereas the unstable heterologous mRNAs (UTR1–343, UTR150–198 and UTRΔ175–194) were depleted equally well from all fractions. Therefore, the bulk of the stable d2EGFP messages were in the nucleus and unavailable for translation. The most parsimonious explanation is the 3′UTR of DDB2 facilitates both nuclear export and cytoplasmic decay of the heterologous reporter mRNAs.
The 3′UTR of DDB2 and inducible gene expression
Comparing nascent transcription to RNA abundance suggests that changes in gene expression cannot be explained by changes in transcription rate alone, and that changes in mRNA stability are integral to regulation of gene expression (35,53,54). Genome-wide analysis of mRNA decay indicates that transcripts encoding housekeeping proteins, metabolic enzymes and structural proteins tend to have longer half-lives, whereas those encoding transcription factors, cell signaling proteins and cell cycle regulators tend to be short-lived (36,39–42). Short-lived mRNAs tend to be transcriptionally responsive, whereas the longer-lived transcripts are typically constitutively expressed (36,39–42). Metabolic labelling experiments in bacterial lipopolysaccharide-treated dendritic cells indicate that mRNA abundance was predominantly determined by NFκB-dependent transcription, whereas changes in mRNA stability permitted transcripts to reach new equilibrium levels and subsequently recover faster, giving rise to a ‘peaked’ response (55). Our own analysis of the p53 response indicated that most p53-responsive mRNAs, including DDB2, were labile and exhibited a similar peaked response (29). Taken together, mRNA decay appears to have a pronounced effect on shaping expression patterns with short-lived mRNAs exhibiting rapid peaked responses and long-lived mRNAs exhibiting slower and comparatively sustained responses (55,56).
Recent evidence in Saccharomyces cerevisiae suggests a direct and causal relationship between transcription and mRNA decay (57–59). The fate of mRNA in the cytoplasm is determined, in part, through promoter sequences (57–59). It is thought that the binding of specific transcription factors to promoter elements affects the co-transcriptional assembly of the mRNA ribonucleoprotein complex, leaving a lasting mark on the nascent transcript that directs it to a particular fate (60). The Rbp4/7 heterodimer and Ccr4-Not complex represent two examples of factors that associate with nascent transcripts and link transcription, mRNA export and mRNA decay (59–63). The term ‘synthegradase’ has been coined to describe the contribution of proteins to the synthesis and decay of mRNAs (64).
Reporter genes have been powerful tools to study gene induction and mRNA decay, but these processes have usually been studied independently. Here, we developed heterologous reporter constructs under control of a tetracycline-regulated promoter that permitted us to monitor both the induction and decay of transcripts uniquely differing in 3′UTR sequences. Heterologous reporter constructs containing the 3′UTR of DDB2, or portions of this 3′UTR, were capable of impacting the synthesis, subcellular localization and decay of the reporter mRNAs. These multiple effects were entirely unexpected and provided strong evidence that transcription, nuclear export and cytoplasmic decay are also tightly coupled in human cells. Although the precise mechanism remains to be defined, it is plausible that these sequences identified in the 3′UTR of DDB2 are recognized co-transcriptionally such that the mRNA ribonucleoprotein complex differs among reporter mRNAs. Whether the orthologous Rbp4/7 and/or Ccr4-Not complexes are involved in determining the fate of these mRNAs is still a matter of speculation.
Opposing effects of the 3′UTR on mRNA stability and protein expression
Heterologous reporter constructs containing the 3′UTR of DDB2 expressed higher steady state levels and permitted more rapid induction of d2EGFP expression. Similarly, the 3′UTR of BIRC2 was able to destabilize a separate reporter mRNA (CAT) while positively influencing CAT protein expression (Supplementary Figure S3). Therefore, two distinct heterologous mRNAs (CAT and D2EGFP) exhibited a similar inverse relationship between mRNA stability and protein expression when the 3′UTRs of BIRC2 and DDB2, containing similar sequences in their 3′UTRs were present. This was not typical of unstable reporter mRNAs in either reporter system. For example, the presence of 3′UTRs with ARE sequences (TNFα and IL1β) decreased mRNA and protein levels concordantly (Supplementary Figures S2 and S3). Taken together, the 3′UTR of DDB2 contains a previously uncharacterized cis-acting element that confers mRNA instability but increased protein expression. This element appears to be present in the sense orientation in other 3′UTRs (Supplementary Table S3), and thus it may be similarly involved in regulating the expression of these mRNAs and proteins.
In summary, nucleotides 150–174 derived from the 3′UTR of DDB2 function confer a variety of interesting features on heterologous 3′UTR reporter mRNAs. The mRNAs containing this sequence were unstable, but these unstable transcripts were induced better than a variety of mRNAs lacking this sequence. The positive effect on gene expression was associated with increase induction, nuclear export and protein expression. This synthetic 3′UTR facilitated shut-off of the same reporter gene, suggesting that the modified vector represents a significant improvement over current commercially available tetracycline-regulated expression vectors and likely other vectors as well. Not only does the present work provide novel insight into DDB2 regulation but the positive influence of this sequence on reporter gene expression may facilitate conditional expression of transgenes in experimental models and may even have uses in gene therapy.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3 and Supplementary Figures 1–3.
ACKNOWLEDGEMENTS
The authors would like to acknowledge the assistance of the Flow Cytometry and DNA sequence facilities at StemCore Laboratories at the Ottawa Hospital Research Institute.
FUNDING
Canadian Institutes of Health Research (to B.C.M.) and National Science and Engineering Research Council (to M.H.). They also thank the Ottawa Regional Cancer Foundation for seed funding. B.D.M. held an Ontario Graduate Scholarship in Science and Technology. M.H. is the CHEO Volunteer Association Endowed Scholar. B.C.M. was supported with a Research Scientist Award from the National Cancer Institute of Canada supported with funds provided by the Canadian Cancer Society. Funding for open access charge: University of Ottawa Open Access Author Fund.
Conflict of interest statement. None declared.
REFERENCES
- 1.Dualan R, Brody T, Keeney S, Nichols AF, Admon A, Linn S. Chromosomal localization and cDNA cloning of the genes (DDB1 and DDB2) for the p127 and p48 subunits of a human damage-specific DNA binding protein. Genomics. 1995;29:62–69. doi: 10.1006/geno.1995.1215. [DOI] [PubMed] [Google Scholar]
- 2.Scrima A, Fischer ES, Lingaraju GM, Bohm K, Cavadini S, Thoma NH. Detecting UV-lesions in the genome: the modular CRL4 ubiquitin ligase does it best! FEBS Lett. 2011;585:2818–2825. doi: 10.1016/j.febslet.2011.04.064. [DOI] [PubMed] [Google Scholar]
- 3.Fujiwara Y, Masutani C, Mizukoshi T, Kondo J, Hanaoka F, Iwai S. Characterization of DNA recognition by the human UV-damaged DNA-binding protein. J. Biol. Chem. 1999;274:20027–20033. doi: 10.1074/jbc.274.28.20027. [DOI] [PubMed] [Google Scholar]
- 4.Tang J, Chu G. Xeroderma pigmentosum complementation group E and UV-damaged DNA-binding protein. DNA Repair. 2002;1:601–616. doi: 10.1016/s1568-7864(02)00052-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nichols AF, Itoh T, Graham JA, Liu W, Yamaizumi M, Linn S. Human damage-specific DNA-binding protein p48. Characterization of XPE mutations and regulation following UV irradiation. J. Biol. Chem. 2000;275:21422–21428. doi: 10.1074/jbc.M000960200. [DOI] [PubMed] [Google Scholar]
- 6.Fitch ME, Cross IV, Ford JM. p53 responsive nucleotide excision repair gene products p48 and XPC, but not p53, localize to sites of UV-irradiation-induced DNA damage, in vivo. Carcinogenesis. 2003;24:843–850. doi: 10.1093/carcin/bgg031. [DOI] [PubMed] [Google Scholar]
- 7.Fitch ME, Cross IV, Turner SJ, Adimoolam S, Lin CX, Williams KG, Ford JM. The DDB2 nucleotide excision repair gene product p48 enhances global genomic repair in p53 deficient human fibroblasts. DNA Repair. 2003;2:819–826. doi: 10.1016/s1568-7864(03)00066-1. [DOI] [PubMed] [Google Scholar]
- 8.Wang QE, Zhu Q, Wani G, Chen J, Wani AA. UV radiation-induced XPC translocation within chromatin is mediated by damaged-DNA binding protein, DDB2. Carcinogenesis. 2004;25:1033–1043. doi: 10.1093/carcin/bgh085. [DOI] [PubMed] [Google Scholar]
- 9.Moser J, Volker M, Kool H, Alekseev S, Vrieling H, Yasui A, van Zeeland AA, Mullenders LH. The UV-damaged DNA binding protein mediates efficient targeting of the nucleotide excision repair complex to UV-induced photo lesions. DNA Repair. 2005;4:571–582. doi: 10.1016/j.dnarep.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 10.Jackson S, Xiong Y. CRL4s: the CUL4-RING E3 ubiquitin ligases. Trends Biochem. Sci. 2009;34:562–570. doi: 10.1016/j.tibs.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer ES, Scrima A, Bohm K, Matsumoto S, Lingaraju GM, Faty M, Yasuda T, Cavadini S, Wakasugi M, Hanaoka F, et al. The molecular basis of CRL4DDB2/CSA ubiquitin ligase architecture, targeting, and activation. Cell. 2011;147:1024–1039. doi: 10.1016/j.cell.2011.10.035. [DOI] [PubMed] [Google Scholar]
- 12.Rapic-Otrin V, McLenigan MP, Bisi DC, Gonzalez M, Levine AS. Sequential binding of UV DNA damage binding factor and degradation of the p48 subunit as early events after UV irradiation. Nucleic Acids Res. 2002;30:2588–2598. doi: 10.1093/nar/30.11.2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kapetanaki MG, Guerrero-Santoro J, Bisi DC, Hsieh CL, Rapic-Otrin V, Levine AS. The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc. Natl Acad. Sci. USA. 2006;103:2588–2593. doi: 10.1073/pnas.0511160103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lan L, Nakajima S, Kapetanaki MG, Hsieh CL, Fagerburg M, Thickman K, Rodriguez-Collazo P, Leuba SH, Levine AS, Rapic-Otrin V. Monoubiquitinated H2A destabilizes photolesion-containing nucleosomes with the concomitant release of the UV-damaged DNA-binding protein E3 ligase. J. Biol. Chem. 2012;287:12039–12049. doi: 10.1074/jbc.M111.307058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luijsterburg MS, Lindh M, Acs K, Vrouwe MG, Pines A, van Attikum H, Mullenders LH, Dantuma NP. DDB2 promotes chromatin decondensation at UV-induced DNA damage. J. Cell Biol. 2012;197:267–281. doi: 10.1083/jcb.201106074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sugasawa K. UV-DDB: a molecular machine linking DNA repair with ubiquitination. DNA Repair. 2009;8:969–972. doi: 10.1016/j.dnarep.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 17.Stoyanova T, Roy N, Kopanja D, Raychaudhuri P, Bagchi S. DDB2 (damaged-DNA binding protein 2) in nucleotide excision repair and DNA damage response. Cell Cycle. 2009;8:4067–4071. doi: 10.4161/cc.8.24.10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stoyanova T, Roy N, Kopanja D, Bagchi S, Raychaudhuri P. DDB2 decides cell fate following DNA damage. Proc. Natl Acad. Sci. USA. 2009;106:10690–10695. doi: 10.1073/pnas.0812254106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stoyanova T, Roy N, Bhattacharjee S, Kopanja D, Valli T, Bagchi S, Raychaudhuri P. p21 cooperates with DDB2 protein in suppression of ultraviolet ray-induced skin malignancies. J. Biol. Chem. 2012;287:3019–3028. doi: 10.1074/jbc.M111.295816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roy N, Stoyanova T, Dominguez-Brauer C, Park HJ, Bagchi S, Raychaudhuri P. DDB2, an essential mediator of premature senescence. Mol. Cell. Biol. 2010;30:2681–2692. doi: 10.1128/MCB.01480-09. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Bagchi S, Raychaudhuri P. Damaged-DNA binding protein-2 drives apoptosis following DNA damage. Cell Div. 2010;5:3. doi: 10.1186/1747-1028-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan Y, Zhang X, Legerski RJ. Artemis interacts with the Cul4A-DDB1DDB2 ubiquitin E3 ligase and regulates degradation of the CDK inhibitor p27. Cell Cycle. 2011;10:4098–4109. doi: 10.4161/cc.10.23.18227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castells E, Molinier J, Benvenuto G, Bourbousse C, Zabulon G, Zalc A, Cazzaniga S, Genschik P, Barneche F, Bowler C. The conserved factor DE-ETIOLATED 1 cooperates with CUL4-DDB1DDB2 to maintain genome integrity upon UV stress. EMBO J. 2011;30:1162–1172. doi: 10.1038/emboj.2011.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oh KS, Emmert S, Tamura D, DiGiovanna JJ, Kraemer KH. Multiple skin cancers in adults with mutations in the XP-E (DDB2) DNA repair gene. J. Invest. Dermatol. 2011;131:785–788. doi: 10.1038/jid.2010.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Itoh T, O'Shea C, Linn S. Impaired regulation of tumor suppressor p53 caused by mutations in the xeroderma pigmentosum DDB2 gene: mutual regulatory interactions between p48(DDB2) and p53. Mol. Cell. Biol. 2003;23:7540–7553. doi: 10.1128/MCB.23.21.7540-7553.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu W, Nichols AF, Graham JA, Dualan R, Abbas A, Linn S. Nuclear transport of human DDB protein induced by ultraviolet light. J. Biol. Chem. 2000;275:21429–21434. doi: 10.1074/jbc.M000961200. [DOI] [PubMed] [Google Scholar]
- 27.Hwang BJ, Ford JM, Hanawalt PC, Chu G. Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc. Natl Acad. Sci. USA. 1999;96:424–428. doi: 10.1073/pnas.96.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shiyanov P, Hayes SA, Donepudi M, Nichols AF, Linn S, Slagle BL, Raychaudhuri P. The naturally occurring mutants of DDB are impaired in stimulating nuclear import of the p125 subunit and E2F1-activated transcription. Mol. Cell. Biol. 1999;19:4935–4943. doi: 10.1128/mcb.19.7.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melanson BD, Bose R, Hamill JD, Marcellus KA, Pan EF, McKay BC. The role of mRNA decay in p53-induced gene expression. RNA. 2011;17:2222–2234. doi: 10.1261/rna.030122.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brady CA, Attardi LD. p53 at a glance. J. Cell Sci. 2010;123:2527–2532. doi: 10.1242/jcs.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKay BC, Chen F, Perumalswami CR, Zhang F, Ljungman M. The tumor suppressor p53 can both stimulate and inhibit ultraviolet light-induced apoptosis. Mol. Biol. Cell. 2000;11:2543–2551. doi: 10.1091/mbc.11.8.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X, Zhao X, Fang Y, Jiang X, Duong T, Fan C, Huang CC, Kain SR. Generation of destabilized green fluorescent protein as a transcription reporter. J. Biol. Chem. 1998;273:34970–34975. doi: 10.1074/jbc.273.52.34970. [DOI] [PubMed] [Google Scholar]
- 33.Liwak U, Thakor N, Jordan LE, Roy R, Lewis SM, Pardo OE, Seckl M, Holcik M. Tumor suppressor PDCD4 represses internal ribosome entry site-mediated translation of antiapoptotic proteins and is regulated by S6 kinase 2. Mol. Cell. Biol. 2012;32:1818–1829. doi: 10.1128/MCB.06317-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mackinnon-Roy C, Stubbert LJ, McKay BC. RNA interference against transcription elongation factor SII does not support its role in transcription-coupled nucleotide excision repair. Mut. Res. 2011;706:53–58. doi: 10.1016/j.mrfmmm.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 35.Fan J, Yang X, Wang W, Wood WH, III, Becker KG, Gorospe M. Global analysis of stress-regulated mRNA turnover by using cDNA arrays. Proc. Natl Acad. Sci. USA. 2002;99:10611–10616. doi: 10.1073/pnas.162212399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Friedel CC, Dolken L, Ruzsics Z, Koszinowski UH, Zimmer R. Conserved principles of mammalian transcriptional regulation revealed by RNA half-life. Nucleic Acids Res. 2009;37:e115. doi: 10.1093/nar/gkp542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Munchel SE, Shultzaberger RK, Takizawa N, Weis K. Dynamic profiling of mRNA turnover reveals gene-specific and system-wide regulation of mRNA decay. Mol. Biol. Cell. 2011;22:2787–2795. doi: 10.1091/mbc.E11-01-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keene JD. The global dynamics of RNA stability orchestrates responses to cellular activation. BMC Biol. 2010;8:95. doi: 10.1186/1741-7007-8-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang E, van Nimwegen E, Zavolan M, Rajewsky N, Schroeder M, Magnasco M, Darnell JE., Jr Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes. Genome Res. 2003;13:1863–1872. doi: 10.1101/gr.1272403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raghavan A, Bohjanen PR. Microarray-based analyses of mRNA decay in the regulation of mammalian gene expression. Brief. Funct. Genomics Proteomics. 2004;3:112–124. doi: 10.1093/bfgp/3.2.112. [DOI] [PubMed] [Google Scholar]
- 41.Mata J, Marguerat S, Bahler J. Post-transcriptional control of gene expression: a genome-wide perspective. Trends Biochem. Sci. 2005;30:506–514. doi: 10.1016/j.tibs.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 42.Sharova LV, Sharov AA, Nedorezov T, Piao Y, Shaik N, Ko MS. Database for mRNA half-life of 19 977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells. DNA Res. 2009;16:45–58. doi: 10.1093/dnares/dsn030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee JE, Lee JY, Wilusz J, Tian B, Wilusz CJ. Systematic analysis of cis-elements in unstable mRNAs demonstrates that CUGBP1 is a key regulator of mRNA decay in muscle cells. PloS One. 2010;5:e11201. doi: 10.1371/journal.pone.0011201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vlasova IA, Tahoe NM, Fan D, Larsson O, Rattenbacher B, Sternjohn JR, Vasdewani J, Karypis G, Reilly CS, Bitterman PB, et al. Conserved GU-rich elements mediate mRNA decay by binding to CUG-binding protein 1. Mol. Cell. 2008;29:263–270. doi: 10.1016/j.molcel.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hau HH, Walsh RJ, Ogilvie RL, Williams DA, Reilly CS, Bohjanen PR. Tristetraprolin recruits functional mRNA decay complexes to ARE sequences. J. Cell. Biochem. 2007;100:1477–1492. doi: 10.1002/jcb.21130. [DOI] [PubMed] [Google Scholar]
- 46.Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 47.Chen CY, Xu N, Shyu AB. mRNA decay mediated by two distinct AU-rich elements from c-fos and granulocyte-macrophage colony-stimulating factor transcripts: different deadenylation kinetics and uncoupling from translation. Mol. Cell. Biol. 1995;15:5777–5788. doi: 10.1128/mcb.15.10.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuwano Y, Kim HH, Abdelmohsen K, Pullmann R, Jr, Martindale JL, Yang X, Gorospe M. MKP-1 mRNA stabilization and translational control by RNA-binding proteins HuR and NF90. Mol. Cell. Biol. 2008;28:4562–4575. doi: 10.1128/MCB.00165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang W, Martindale JL, Yang X, Chrest FJ, Gorospe M. Increased stability of the p16 mRNA with replicative senescence. EMBO Rep. 2005;6:158–164. doi: 10.1038/sj.embor.7400346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao TT, Graber TE, Jordan LE, Cloutier M, Lewis SM, Goulet I, Cote J, Holcik M. hnRNP A1 regulates UV-induced NF-kappaB signalling through destabilization of cIAP1 mRNA. Cell Death Diff. 2009;16:244–252. doi: 10.1038/cdd.2008.146. [DOI] [PubMed] [Google Scholar]
- 52.Kastelic T, Schnyder J, Leutwiler A, Traber R, Streit B, Niggli H, MacKenzie A, Cheneval D. Induction of rapid IL-1 beta mRNA degradation in THP-1 cells mediated through the AU-rich region in the 3′UTR by a radicicol analogue. Cytokine. 1996;8:751–761. doi: 10.1006/cyto.1996.0100. [DOI] [PubMed] [Google Scholar]
- 53.Garcia-Martinez J, Aranda A, Perez-Ortin JE. Genomic run-on evaluates transcription rates for all yeast genes and identifies gene regulatory mechanisms. Mol. Cell. 2004;15:303–313. doi: 10.1016/j.molcel.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 54.Cheadle C, Fan J, Cho-Chung YS, Werner T, Ray J, Do L, Gorospe M, Becker KG. Control of gene expression during T cell activation: alternate regulation of mRNA transcription and mRNA stability. BMC Genomics. 2005;6:75. doi: 10.1186/1471-2164-6-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rabani M, Levin JZ, Fan L, Adiconis X, Raychowdhury R, Garber M, Gnirke A, Nusbaum C, Hacohen N, Friedman N, et al. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat. Biotechnol. 2011;29:436–442. doi: 10.1038/nbt.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Elkon R, Zlotorynski E, Zeller KI, Agami R. Major role for mRNA stability in shaping the kinetics of gene induction. BMC Genomics. 2010;11:259. doi: 10.1186/1471-2164-11-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bregman A, Avraham-Kelbert M, Barkai O, Duek L, Guterman A, Choder M. Promoter elements regulate cytoplasmic mRNA decay. Cell. 2011;147:1473–1483. doi: 10.1016/j.cell.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 58.Trcek T, Larson DR, Moldon A, Query CC, Singer RH. Single-molecule mRNA decay measurements reveal promoter- regulated mRNA stability in yeast. Cell. 2011;147:1484–1497. doi: 10.1016/j.cell.2011.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dori-Bachash M, Shema E, Tirosh I. Coupled evolution of transcription and mRNA degradation. PLoS Biol. 2011;9:e1001106. doi: 10.1371/journal.pbio.1001106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choder M. mRNA imprinting: additional level in the regulation of gene expression. Cell. Logist. 2011;1:37–40. doi: 10.4161/cl.1.1.14465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goler-Baron V, Selitrennik M, Barkai O, Haimovich G, Lotan R, Choder M. Transcription in the nucleus and mRNA decay in the cytoplasm are coupled processes. Genes Dev. 2008;22:2022–2027. doi: 10.1101/gad.473608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harel-Sharvit L, Eldad N, Haimovich G, Barkai O, Duek L, Choder M. RNA polymerase II subunits link transcription and mRNA decay to translation. Cell. 2010;143:552–563. doi: 10.1016/j.cell.2010.10.033. [DOI] [PubMed] [Google Scholar]
- 63.Shalem O, Groisman B, Choder M, Dahan O, Pilpel Y. Transcriptome kinetics is governed by a genome-wide coupling of mRNA production and degradation: a role for RNA Pol II. PLoS Genet. 2011;7:e1002273. doi: 10.1371/journal.pgen.1002273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dahan N, Choder M. The eukaryotic transcriptional machinery regulates mRNA translation and decay in the cytoplasm. Biochim. Biophys. Acta. 2013;1829:169–173. doi: 10.1016/j.bbagrm.2012.08.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.