Background: Receptor interacting protein 3 (RIP3)-mixed lineage kinase domain-like (MLKL) interaction is essential for necroptosis.
Results: Murine RIP3 does not interact with human MLKL and vice versa due to sequence differences in and around the RIP3 phosphorylation sites.
Conclusion: Different sequences in human and mouse RIP3 control the functionally conserved RIP3-MLKL interaction.
Significance: This study provided new insights into the function of RIP3-MLKL interaction in necroptosis.
Keywords: Cell Death, Necrosis (Necrotic Death), Protein Kinases, Protein Phosphorylation, Protein-Protein Interactions, RIP, Tumor Necrosis Factor (TNF), MLKL, RIP3, Necroptosis
Abstract
Receptor interacting protein 3 (RIP3) is a protein kinase essential for TNF-induced necroptosis. Phosphorylation on Ser-227 in human RIP3 (hRIP3) is required for its interaction with human mixed lineage kinase domain-like (MLKL) in the necrosome, a signaling complex induced by TNF stimulation. RIP1 and RIP3 mediate necrosome aggregation leading to the formation of amyloid-like signaling complexes. We found that TNF induces Thr-231 and Ser-232 phosphorylation in mouse RIP3 (mRIP3) and this phosphorylation is required for mRIP3 to interact with mMLKL. Ser-232 in mRIP3 corresponds to Ser-227 in hRIP3, whereas Thr-231 is not conserved in hRIP3. Although the RIP3-MLKL interaction is required for necroptosis in both human and mouse cells, hRIP3 does not interact with mMLKL and mRIP3 cannot bind to hMLKL. The species specificity of the RIP3-MLKL interaction is primarily determined by the sequence differences in the phosphorylation sites and the flanking sequence around the phosphorylation sites in hRIP3 and mRIP3. It appears that the RIP3-MLKL interaction has been selected as an evolutionarily conserved mechanism in mediating necroptosis signaling despite that differing structural and mechanistic bases for this interaction emerged simultaneously in different organisms. In addition, we further revealed that the interaction of RIP3 with MLKL prevented massive abnormal RIP3 aggregation, and therefore should be crucial for formation of the amyloid signaling complex of necrosomes. We also found that the interaction between RIP3 and MLKL is required for the translocation of necrosomes to mitochondria-associated membranes. Our data demonstrate the importance of the RIP3-MLKL interaction in the formation of functional necrosomes and suggest that translocation of necrosomes to mitochondria-associated membranes is essential for necroptosis signaling.
Introduction
Necrosis is a type of cell death characterized by cellular organelle swelling, loss of cell membrane integrity, and subsequence leakage of cell content. It has been long neglected as a passive cell death that lacks physiological relevance. But recent studies on TNF-induced necrosis strongly support the notion that necrosis could also be a programmed cell death. This kind of programmed cell death was given the name “necroptosis” (1).
Necroptosis is involved in many physiological and pathological processes. It has been reported to protect the host from vaccinia virus and murine cytomegalovirus infection by restricting virus duplication (2–4), whereas it has been found to enhance the toxicity of Salmonella enterica serovar Typhimurium in mice (5). In addition to host defense, necroptosis is also involved in chronic inflammation and tissue injury (6), such as chronic intestinal inflammation (7, 8), liver injury (9), skin inflammation (10), retinal detachment (11, 12), hemolysis (13), and atherosclerosis (14). During embryonic development, it has been suggested that abnormal massive necroptosis is responsible for embryonic lethality in caspase-8-deficient and FADD-deficient mice (15–17).
TNF-induced necroptosis in murine fibrosarcoma L929 cells is a model system for necroptosis studies. Upon TNF stimulation, TNF receptor trimerizes and recruits a protein complex termed complex I, which contains TRADD, RIP1,3 TRAF2, and cIAP1/2 (18). Ubiquitinated RIP1 in complex I serves as an adaptor for the TAK1 complex to activate the IKK complex leading to subsequent activation of NF-κB (19). Following the deubiquitination of RIP1 (20) and internalization of the TNF receptor (18, 21), complex I switches to a RIP3-containing complex termed necrosome to initiate necroptosis (6, 18). A recent study revealed that the RIP1/RIP3 necrosome further forms amyloid aggregates and these aggregates are functional (22). In the absence of RIP3 expression, complex II (also called Death inducing signaling complex, or DISC) containing RIP1, caspase-8, TRADD, and FADD is formed instead to initiate apoptosis (18, 20). MLKL is a recently identified molecule that interacts with RIP3 in the necrosome and is required for necroptosis (23, 24). In human cells, phosphorylation of RIP3 on Ser-227 is required for its interaction with MLKL (23), which leads to MLKL phosphorylation and transduction of necroptosis signaling (23).
In an effort to understand the function of RIP3 in necroptosis, we have investigated the phosphorylation status of mRIP3 upon necroptotic stimulation and mapped the phosphorylation sites in mRIP3. We found that phosphorylation on Thr-231 and Ser-232 was induced by TNF stimulation. The phosphorylation on these two sites is required for mRIP3 to interact with mMLKL. Surprisingly, although function of the RIP3-MLKL interaction in mediating necroptosis signaling is the same in human and mouse cells, mRIP3 cannot interact with hMLKL, whereas hRIP3 cannot bind to mMLKL. Here, we have undertaken structural studies to investigate the molecular basis underlying the RIP3-MLKL interaction. As revealed here, species specificity of the RIP3-MLKL interaction is determined by the differential phosphorylation sites as well as the distinct proximal sequence surrounding the phosphorylation sites in human and mouse RIP3. Moreover, our studies further demonstrated that although the interaction between RIP3 and MLKL is not required for the formation of necrosome aggregates, its absence leads to uncontrolled, abnormal aggregation of necrosomes and therefore the RIP3-MLKL interaction is required for the formation of functional necrosome aggregates. Finally, we showed here that the RIP3-MLKL interaction is essential for proper trafficking of necrosomes to mitochondria-associated membranes during the necroptotic process.
EXPERIMENTAL PROCEDURES
Materials
Mouse TNFα and human TNFα were purchased from eBioscience (San Diego, CA). Z-VAD was obtained from Calbiochem. Myelin basic protein (MBP) and MitoTracker Deep Red were obtained from Invitrogen. Propidium iodide (PI) was from Sigma. [γ-32P]ATP was from PerkinElmer Life Sciences. Smac mimetic was kindly provided by Genentech (South San Francisco, CA). mRIP3 antibody was raised in rabbit using amino acids 287–387. FADD and MLKL antibodies were generated using Escherichia coli-expressed GST-tagged full-length protein in rabbit and guinea pig, respectively. Thr-231- and Ser-232-phosphorylated RIP3 antibody were raised in rabbits by Abmart (Shanghai, China) using polypeptides ELVDK(pT)(pS)LIRET as antigen. Mouse anti-FLAG antibodies (M2) and beads were obtained from Sigma. Rabbit anti-HA (Y-11) antibodies and mouse anti-β-actin (C4) antibodies were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Mouse anti-RIP1 antibodies were obtained from BD Biosciences (San Jose, CA).
λ-Phosphatase Treatment
The immunoprecipitate was washed first with lysis buffer (20 mm Tris-HCl, pH 7.5, 120 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1% Triton X-100, 2.5 mm sodium pyrophosphate, 1 mm β-glycerophosphate, 1 mm Na3VO4, 1 mm PMSF, 1× protease inhibitor mixture (Sigma)) 3 times followed by washing 3 times with 1× phosphatase buffer (50 mm HEPES, pH 7.5, 100 mm NaCl, 2 mm DTT, 0.01% Brij 35, 1 mm MnCl2). The immunoprecipitate was then divided into two parts. One was added with 200 units of λ-phosphatase (New England Biolabs, Ipswich, MA), whereas the other was mock treated. After incubation at 30 °C for 30 min with gentle shaking, the reactions were stopped by the addition of 2× SDS sample buffer followed by boiling for 5 min.
Identification of mRIP3 Phosphorylation Sites
FLAG-mRIP3 was immunoprecipitated using anti-FLAG beads (Sigma) followed by SDS-PAGE. The band according to RIP3 was excised from Coomassie-stained gels and subjected to in-gel digestion. Tryptic peptides were extracted from gels using 0.15% formic acid, 67% acetonitrile and dried prior to phosphopeptide enrichment. Phosphopeptides were enriched using immobilized metal ion affinity chromatography as previously described (25). Dried phosphopeptides were dissolved in 0.1% formic acid, 2% acetonitrile and separated on a fused silica capillary emitter (inner diameter, 75 μm; length, 15 cm; New Objective, Woburn, MA) packed in-house with 5-μm C18 resin (New Objective), and analyzed on AB SCIEX Triple TOF 5600 system. For information-dependent acquisition, survey scans were acquired in 250 ms and 20 product ion scans were collected in 50 ms/per scan. The acquired raw data files (.wiff) were searched with ProteinPilot software version 4.2 (AB SCIEX, Foster City, CA) against the mRIP3 protein sequence database with common contaminants included. The following search parameters were selected: sample type (identification), Cys alkylation (iodoacetamide), digestion (trypsin), instrument (TripleTOF 5600), special factors (phosphorylation emphasis), and search effort (thorough). The tolerances were specified as ±0.05 Da for peptides and ±0.05 Da for MS/MS fragments.
In Vitro Kinase Assay
Wild-type FLAG-mRIP3 protein and mutant proteins were immunoprecipitated using anti-FLAG beads from cell lysates of transfected 293T cell. After washing 3 times with kinase buffer (25 mm Tris, pH 7.5, 10 mm MgCl2, 2 mm DTT, 5 mm β-glycerophosphate, 0.1 mm Na3VO4), the beads were suspended in 20 μl of kinase buffer. 10 μg of MBP, 10 μm cold ATP, and 2 μCi of [γ-32P]ATP were added to initiate the reaction. Samples were incubated at 30 °C for 30 min, and then subjected to SDS-PAGE followed by autoradiography.
Cell Viability Assay
Cells were trypsinized and incubated in PBS containing 5 μg/ml of PI at 37 °C for 10 min. PI negative cells (living cells with integrated plasma membrane do not take up PI) were counted by flow cytometer (FACS Calibur, BD Biosciences).
Lentivirus Preparation and Infection
For lentivirus production, 293T cells were transfected with pBOB-FLAG-mRIP3 (or its mutants) and lentivirus-packing plasmids (PMDL/REV/VSVG) by the calcium phosphate precipitation method. 12 h later, cell culture medium was changed and the virus-containing medium was harvested 36 h later. For infection, 1 ml of virus-containing medium and 1.5 ml of fresh medium containing 10 μg/ml of Polybrene was added to L929 cells and NIH3T3-A cells plated in 6-well plates. The plates were centrifuged at 1500 × g for 30 min and put back into the cell incubator. Infectious medium was changed 12 h later.
Generation of RIP3 and MLKL Knock-out L929 Cells
RIP3 and MLKL knock-out L929 cells were generated by the TALEN method as described by Zhang et al. (26). The target sites for RIP3 were designed as “CTAACATTCTGCTGGA” and “TGTAGATGGACTAACC.” For MLKL, the target sites were designed as “ATCATTGGAATACCGT” and “CTTCCTGCTGCCAGGAT.” The knock-out cells were determined by sequencing of targeted loci and immunoblotting of the expression of respective proteins.
Immunoprecipitation and Triton X-100-insoluble Fraction Isolation
Cells were lysed with lysis buffer on ice for 15 min. Cell lysate was then centrifuged at 20,000 × g for 30 min. Supernatant was immunoprecipitated with anti-FLAG beads at 4 °C for 3 h or overnight. The pellet (Triton X-100 insoluble fraction) was washed with lysis buffer 3 times, dissolved in SDS sample buffer, ultrasonicated, and then boiled for 10 min.
Fluorescent Microscopy
L929 cells were infected with lentivirus encoding GFP- or RFP-tagged protein, and then plated on gelatin (0.1%)-coated glass bottom culture dishes. The cells were incubated in 37 °C, 5% CO2. Images were obtained using the Zeiss 780 confocal microscope with fluorescence optics and LSM710/780 ZEN 2010 microscopy software. Pinhole was taken as 8-μm every picture. As for staining of mitochondria, MitoTracker Deep Red was used at 5 nm concentration in cell culture medium.
Subcellular Fractionation
Crude mitochondria, pure mitochondria, mitochondria-associated membranes (MAM), and cytosol were isolated as described previously (27). Briefly, 15 plates of L929 cells (150 mm dish) were harvested with the use of a scraper and washed with twice with PBS. Cells were suspended in buffer I (225 mm mannitol, 75 mm sucrose, 0.1 mm EGTA and 30 mm Tris-HCl, pH 7.4) and homogenized with a glass homogenizer (Kontes, Vineland, NJ). Samples were centrifuged at 600 × g to remove nucleus, then centrifuged at 7,000 × g to collect the crude mitochondria (pellet). The supernatant was subjected to centrifugation at 100,000 × g, and the subsequent supernatant was collected as cytosol. For further purification, crude mitochondria were suspended in mitochondria resuspending buffer (250 mm mannitol, 5 mm HEPES, pH 7.4, and 0.5 mm EGTA) and loaded on a 20% Percoll gradient. After centrifuged at 95,000 × g for 30 min, the upper band was collected as MAM and the pellet was collected as pure mitochondria. Equivalent amounts of protein from each fraction were analyzed by SDS-PAGE and immunoblotting.
RESULTS
Identification of Phosphorylation Sites in mRIP3
Because protein phosphorylation either by itself (auto) or by other kinase(s) is often involved in the regulation of protein kinases, we sought to determine whether phosphorylation of mRIP3 is also critical for its function. The mRIP3 protein appears to be modified because Western blotting analysis revealed two protein bands corresponding to mRIP3 in both L929 cells and NIH3T3-N cells (cells of a NIH3T3 subline that express a high level of RIP3 and are subjected to necroptotic cell death in response to TNF (28)) (Fig. 1A). The band shift of mRIP3 is independent from stimulation with TNF and Z-VAD. NIH3T3-A cells are cells from another NIH3T3 subline that lack mRIP3 expression and undergo apoptosis in response to TNF (28). Because ectopic expression of FLAG-mRIP3 in NIH3T3-A cells can convert the death phenotype from apoptosis to necroptosis, it is conceivable that FLAG-mRIP3 in NIH3T3-A cells functions similarly to the endogenous RIP3 in NIH3T3-N cells in mediating necroptosis (28). As shown in Fig. 1B, ectopically expressed FLAG-mRIP3 also exhibited a band shift on SDS-PAGE. The band shift of mRIP3 resulted from protein phosphorylation because λ-phosphatase treatment completely abolished the shift (Fig. 1C). As reported earlier, HT-29 human colon cancer cell death induced by TNF plus Z-VAD and Smac mimetic treatment (29) is another cell system used in necroptosis studies. In this case, although we did not detect any band shift of hRIP3 in untreated cells, we indeed found a phosphorylation-dependent band shift after stimulation (Fig. 1D). At this stage, it is not clear whether there is any functional relevance in RIP3 phosphorylation identified in untreated cells and/or cells exposed to certain combinations of stimuli. Nevertheless, we conclude here that RIP3 is indeed present as a phosphoprotein under certain circumstances, which has been demonstrated in the presence of the protein band shift in SDS-PAGE and the disappearance of the shifted band when treated with λ-phosphatase.
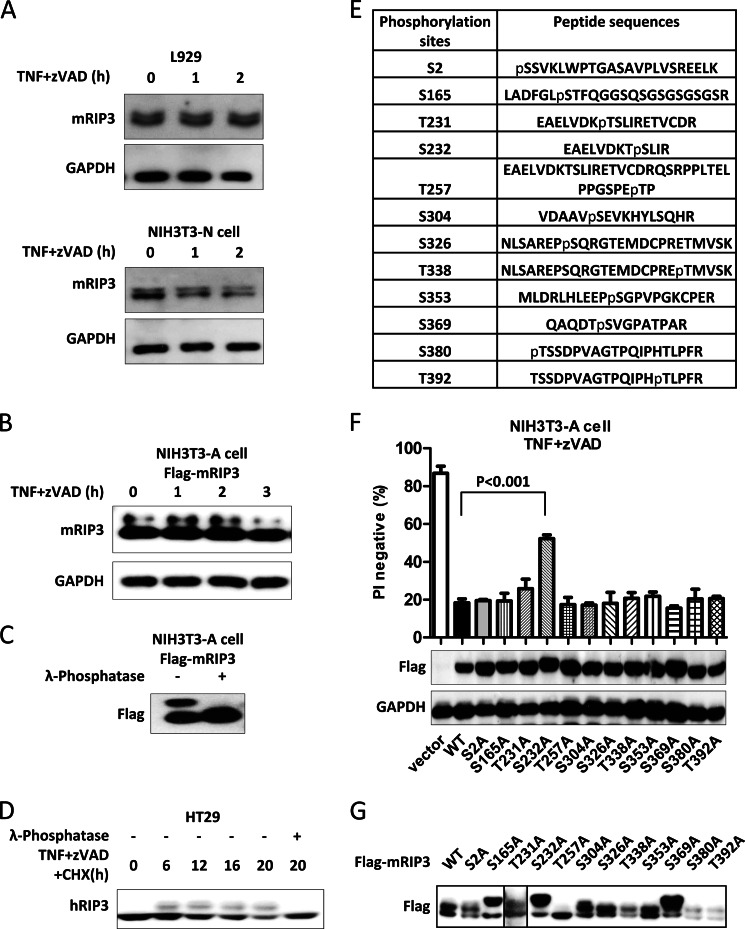
FIGURE 1.
Identification of phosphorylation sites in mRIP3. A, L929 and NIH3T3-N cells were treated with TNF (10 ng/ml) plus Z-VAD (20 μm) for the indicated time points. Cells were then harvested and the cell lysates were subjected to SDS-PAGE. Endogenous mRIP3 and GAPDH were detected by immunoblotting with mRIP3 and GAPDH antibodies. B, NIH3T3-A cells stably expressing FLAG-mRIP3 were treated with TNF (30 ng/ml) plus Z-VAD for the indicated time points. Cells were then harvested and the cell lysates were subjected to immunoblotting. C, FLAG-mRIP3-expressing NIH3T3-A cells were lysed and immunoprecipitated with anti-FLAG antibody-conjugated beads. The immunoprecipitates were treated with or without λ-phosphatase (1000 units/mg of total protein) for 30 min at 30 °C and then subjected to immunoblotting with anti-FLAG antibody. D, HT29 cells were treated with human TNF (30 nm) plus Z-VAD (20 μm) plus cycloheximide (CHX) (2 μg/ml) for the indicated time points. Cells were lysed with lysis buffer without phosphatase inhibitor, and treated with or without λ-phosphatase. Endogenous RIP3 was examined by immunoblotting. E, FLAG-mRIP3-expressing NIH3T3-A cells were treated with TNF plus Z-VAD for 2 h. Cells were then harvested and lysed before FLAG-mRIP3 was immunoprecipitated with anti-FLAG beads. After purification, FLAG-mRIP3 was digested with trypsin and the phosphopeptides were enriched by immobilized metal ion affinity chromatography, and then analyzed by mass spectrometry (see ”Experimental Procedures“). 12 potential phosphorylation sites of mRIP3 were identified. F, 12 mRIP3 mutants were created by mutating each of the 12 potential phosphorylation sites, Thr or Ser to Ala. All the mutants were introduced into NIH3T3-A cells by lentiviral vector. 48 h post-infection, cells were treated with TNF plus Z-VAD for 4 h and cell viability (PI negative) was analyzed by flow cytometer. The expression of these mutants was analyzed by immunoblotting with anti-FLAG antibody. GAPDH was analyzed as control. The data were shown as mean ± S.D. of triplicate samples. G, the 12 mRIP3 mutants were expressed in 293T cells by transient transfection. Cells were harvested and the cell lysates were then subjected to immunoblotting with anti-FLAG antibody. Samples in panels A–C and G were subjected to SDS-PAGE for a longer period time to separate shifted and non-shifted mRIP3 bands.
To identify mRIP3 phosphorylation sites, we used NIH3T3-A cells stably expressing FLAG-mRIP3. Although TNF plus Z-VAD treatment did not seem to affect the band shift, we collected the treated cells for FLAG-mRIP3 protein purification and characterization. FLAG-mRIP3 was purified from the cells, subjected to SDS-PAGE, and excised from gel. The extracted protein was digested with trypsin. Phosphopeptides were enriched by immobilized metal ion affinity chromatography and analyzed by mass spectrometry. 8 serine and 4 threonine residues were found to be phosphorylated. They are Ser-2, Ser-165, Thr-231, Ser-232, Thr-257, Ser-304, Ser-326, Thr-338, Ser-353, Ser-369, Ser-380, and Thr-392 (Fig. 1E). To evaluate the potential function of phosphorylation on these sites, each of these Ser or Thr was mutated to alanine to prevent phosphorylation. All of the mutants were introduced into NIH3T3-A cells by lentiviral vector to determine which mutant(s) could not function as wild-type mRIP3 to convert apoptotic cell death to necroptosis upon TNF plus Z-VAD stimulation. Mutation of Ser-232 to Ala partially compromised the function of mRIP3 in converting apoptosis to necroptosis, whereas other mutants behaved similarly to wild-type mRIP3 (Fig. 1F). We also analyzed the protein band shift status of these mutants on SDS-PAGE and found that the T257A mutation totally abolished the band shift, whereas S165A, S232A, and S369A mutants presented differential protein band shifting patterns and the rest of the mutants behaved in a similar manner to wild-type (Fig. 1G). Thus, it is not the phosphorylation on the sites that lead to band shifts of RIP3 but it is the phosphorylation on other site(s) such as Ser-232 that participates in the mRIP3-mediated switch of apoptosis to necroptosis upon TNF stimulation.
TNF-induced Phosphorylation of Thr-231 and Ser-232 Is Essential for mRIP3 to Mediate Necroptosis
While our research was in progress, Sun et al. (23) reported that hRIP3 was phosphorylated on Ser-227, which corresponds to Ser-232 in mRIP3. Ser-227 to Ala mutation of hRIP3 almost completely abolished the function of RIP3 in mediating necroptosis in HeLa cells (23). We then compared the effect of human S227A mutant with mouse S232A mutant in their abilities to mediate necroptosis in HeLa and NIH3T3-A cells, respectively. We found that the effect of the S227A mutation in human cells is much stronger than that of S232A in mouse cells (Fig. 2A). Sequence alignment revealed that the flanking sequence around mRIP3 Ser-232 (Ser-227 in hRIP3) is not conserved in hRIP3 (Fig. 2B). As a matter of fact, more sequence differences between hRIP3 and mRIP3 were noted in the other coding regions. This may explain why hRIP3 did not function well in murine cells (data not shown). Because it is possible that other phosphorylation sites in mRIP3 also contribute to its function, we created a mRIP3 mutant, termed mRIP3–12M with all of the 12 potential phosphorylation sites mutated to alanine. As shown in Fig. 2C, unlike the partial loss-of-function mutant, the S232A mutant, mRIP3–12M almost completely lost its function in mediating NIH3T3-A cell necroptosis, suggesting that a phosphorylation site(s) other than Ser-232 is also required for mRIP3 to execute its function in necroptosis to the full extent.
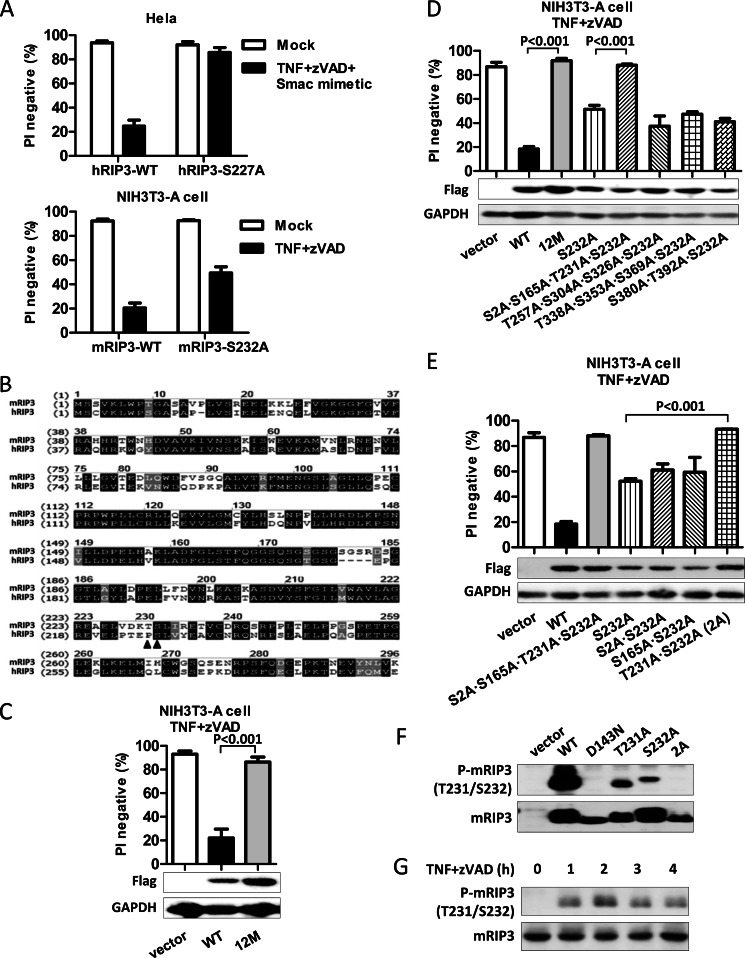
FIGURE 2.
Phosphorylation of mRIP3 on Thr-231 and Ser-232 is essential for mRIP3 to mediate necroptosis. A, HeLa cells were infected with lentivirus encoding hRIP3-WT or hRIP3-S227A. 48 h later, cells were treated with human TNF (30 ng/ml) plus Z-VAD (20 μm) plus Smac mimetic (10 nm) for 24 h. Cell viability was then analyzed. NIH3T3-A cells were infected with lentivirus encoding mRIP3-WT or mRIP3-S232A. 48 h later, cells were treated with mouse TNF plus Z-VAD for 4 h followed by cell viability analysis. S227A mutation completely abolished the function of hRIP3 in mediating cell death but the S232A mutation in mRIP3 only partly impaired its function. B, alignment of the kinase domain of mRIP3 and hRIP3. Identical amino acid residues were shown in black background. Similar amino acid residues were shown in gray background. The solid triangle denotes the phosphorylation sites. C, a mRIP3 mutant, mRIP3–12M, in which the 12 potential phosphorylation sites were mutated to Ala was introduced into NIH3T3-A cells. The TNF plus Z-VAD-induced cell death was analyzed as described in panel A. D and E, mRIP3 mutants as indicated in the figure were introduced into NIH3T3-A cells. Cell viability was analyzed as in panel A. The simultaneous mutation of Thr-231 and Ser-232 to Ala (2A mutant) has similar effects as the mutation of all 12 potential phosphorylation sites in mRIP3. The data were shown as mean ± S.D. of triplicate samples. F, expression vector encoding nothing (empty vector), mRIP3-WT, D143N (kinase-dead), T231A, S232A, or 2A was transfected into 293T cells. 24 h after transfection, cells were harvested and the cell lysates were immunoblotted with mRIP3 antibody or anti-phosphorylated mRIP3 antibody. The anti-phospho-mRIP3 antibody detects Thr-231 and Ser-232 dual phosphorylated RIP3. G, L929 cells were treated with or without TNF plus Z-VAD for different periods of time. Cells were lysed and the lysates were subjected to immunoblotting with anti-phospho-Thr-231/Ser-232-mRIP3 and anti-RIP3 antibodies.
To find out which phosphorylation site(s) functions synergistically with Ser-232, we constructed another four mRIP3 mutants, S2A/S165A/T231A/S232A, T257A/S304A/S326A/S232A, T338A/S353A/S369A/S232A, and S380A/T392A/S232A. Among these mutants, the S2A/S165A/T231A/S232A mutant behaved like the mRIP3–12M (Fig. 2D). We then further created three mRIP3 mutants, S2A/S232A, S165A/S232A, and T231A/S232A. Among the three mutants, expression of the T231A/S232A (termed 2A thereafter) mutant in NIH3T3-A cells failed to mediate TNF-induced necroptosis (Fig. 2E). Thus, it appears that phosphorylation of both Thr-231 and Ser-232 in mRIP3 is required for its function.
To measure the phosphorylation of Thr-231 and Ser-232, we generated a specific antibody against phospho-Thr-231/Ser-232-mRIP3 (Fig. 2F). The dual phosphorylation on Thr-231 and Ser-232 in mRIP3 is undetectable in resting L929 cells and TNF plus Z-VAD stimulation induces dual phosphorylation of mRIP3 (Fig. 2G).
T231A and S232A Double Mutation Does Not Affect mRIP3 Kinase Activity but Disrupts the Interaction between mRIP3 and mMLKL
Recent studies showed that MLKL interacts with RIP3 in the necroptotic process (23, 24). Because it was revealed previously that hRIP3 carrying the S227A mutation cannot interact with hMLKL when co-transfected in 293T cells (23), we tested here whether mRIP3 T231A, S232A, and 2A mutants were capable to interact with mMLKL or not. Although both T231A and S232A mutations reduced the interaction between mRIP3 and mMLKL to a certain extent, the reduction was much stronger in the case of the S232A mutant (Fig. 3A). As expected, the mRIP3–2A mutant completely lost its ability to interact with MLKL (Fig. 3A). In contrast to the differential interaction with MLKL, all these mRIP3 mutants interacted normally with RIP1 (Fig. 3B), indicating that the different structural basis in RIP3 is involved in its interaction with RIP1 when compared with MLKL.
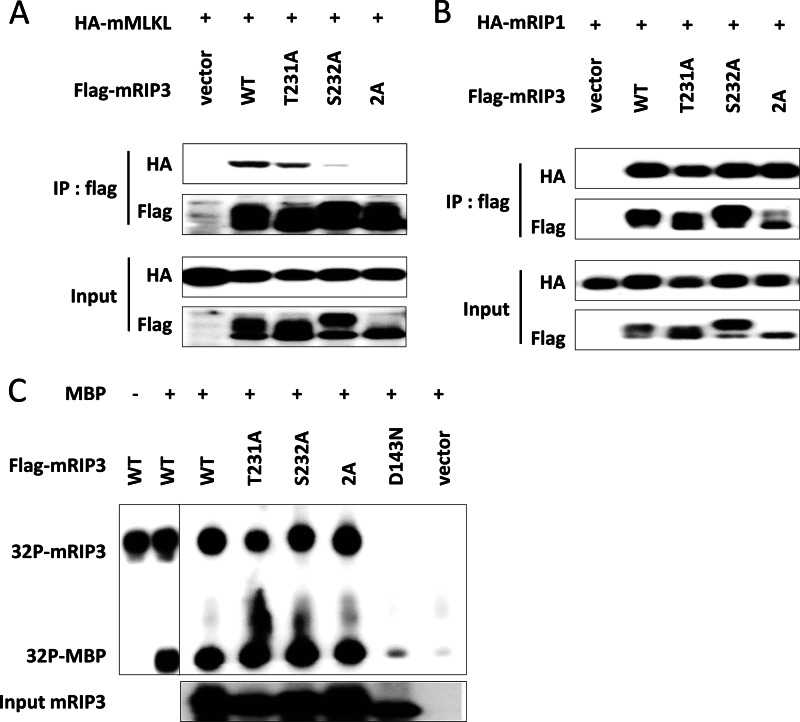
FIGURE 3.
T231A/S232A double mutation disrupts the interaction of mRIP3 with mMLKL without affecting its kinase activity. A, empty vector, FLAG-mRIP3 wild-type, or T231A, S232A, 2A mutants were co-transfected with HA-mMLKL in 293T cells, respectively. 18 h later, cells were lysed and the lysates were subjected to immunoprecipitation with anti-FLAG beads. The input and immunoprecipitates (IP) were immunoblotted with anti-HA and anti-FLAG antibodies. B, the experiments were performed as in A except that HA-mMLKL was replaced by HA-mRIP1. C, 293T cells were transfected with empty vector, FLAG-mRIP3 wild-type, or FLAG-mRIP3 mutants as indicated. 36 h later, FLAG-mRIP3 wild-type and mutants from cell lysates were immunoprecipitated with anti-FLAG beads. The immunoprecipitates were subjected to in vitro kinase assay with [γ-32P]ATP using MBP as substrate. The amount of input RIP3 protein was analyzed by immunoblotting with anti-FLAG antibody. mRIP3 kinase-dead mutant D143N was included as a negative control.
Because phosphorylation sites Thr-231 and Ser-232 are located in the mRIP3 kinase domain, we questioned whether mutation (Thr or Ser to Ala mutation) of either or both residues would affect the mRIP3 kinase activity. All 3 mutants were isolated from transfected cells and were further subjected to an in vitro kinase assay using MBP as substrate. Wild-type mRIP3 and a kinase-dead mutant mRIP3-D143N were included as positive and negative controls, respectively. Surprisingly, we found that the mutation of either or both residues (Thr-231 and Ser-232) to Ala did not affect their autophosphorylation and phosphorylation of MBP (Fig. 3C).
mRIP3 Thr-231 and Ser-232 Phosphorylation Is Essential for the Recruitment of mMLKL to the Necrosome but Not for the Necrosome Assembly
To determine the role of Thr-231 and Ser-232 phosphorylation in necrosome signaling, we analyzed the effect of Thr-231 and Ser-232 phosphorylation on necrosome formation. Wild-type and RIP3 knock-out (KO) L929 cells generated by the TALEN method were used in the experiments (Fig. 4A). As expected, RIP3 KO L929 cells were resistant to TNF plus Z-VAD-induced necroptosis. Reconstituting FLAG-mRIP3, but not the mRIP3–2A mutant, into RIP3 KO L929 cells restored necroptotic sensitivity (Fig. 4B). The same result was obtained using NIH3T3-A cells (Fig. 2E).
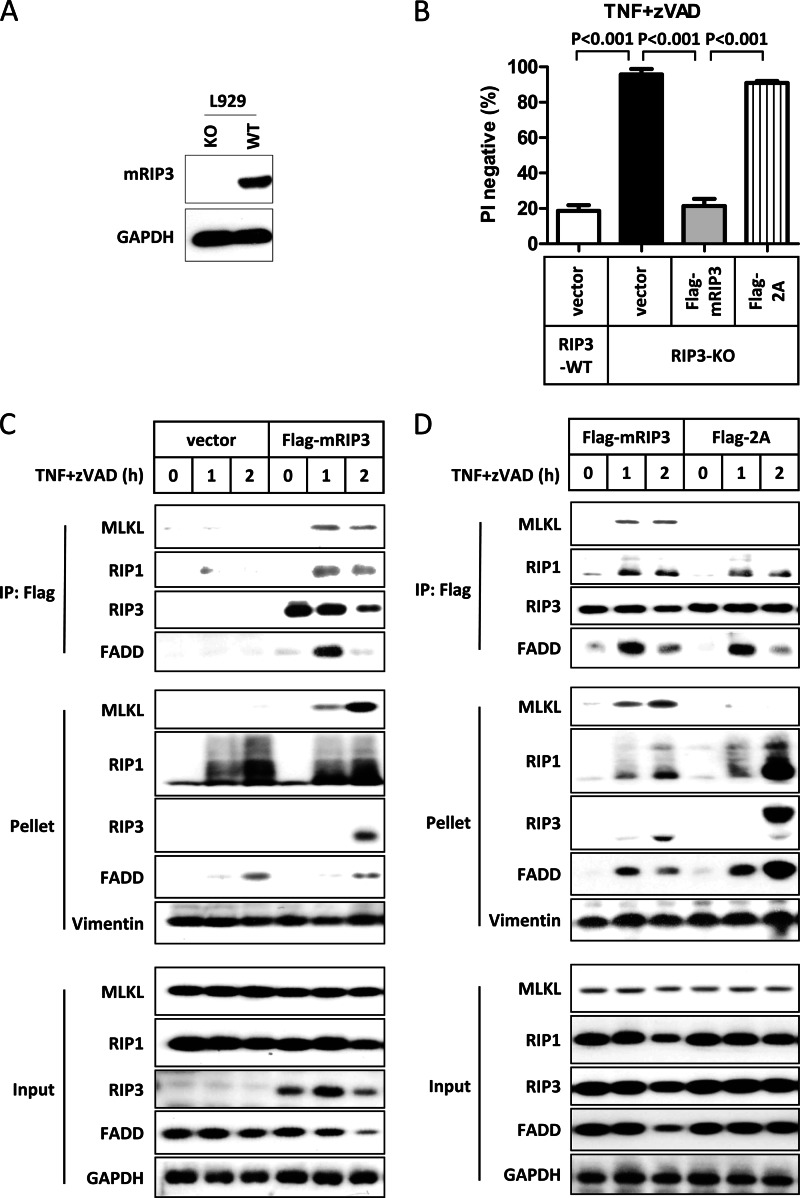
FIGURE 4.
Thr-231 and Ser-232 phosphorylation in mRIP3 is essential for the recruitment of mMLKL to necrosomes. A, cell lysates from RIP3 wild-type and RIP3 knock-out (KO) L929 cells were immunoblotted with mRIP3 antibody. GAPDH was immunoblotted as the loading control. B, L929 RIP3-KO cells were reconstituted with empty vector, FLAG-mRIP3 wild-type, and 2A mutant by lentiviral vector. 48 h later, cells were treated with TNF plus Z-VAD for 4 h and cell viability was analyzed. The data were shown by mean ± S.D. of triplicate samples. L929 parental (RIP3 wild-type) cells were included as control. C and D, RIP3 KO L929 cells stably expressing FLAG-mRIP3, 2A mutant, or empty vector were treated with TNF plus Z-VAD for the indicated time points. Cells were lysed with lysis buffer containing 1% Triton X-100. Supernatants (input) were subjected to RIP3 complex immunoprecipitation (IP) using anti-FLAG beads. The pellets (Triton X-100-insoluble fractions) were mixed with SDS sample buffer, ultrasonicated, and boiled. The immunoprecipitates, pellets, and inputs were immunoblotted with antibodies as indicated. Vimentin and GAPDH were immunoblotted as loading control of the pellet and total cell lysate, respectively.
It is known that necroptotic stimuli induce the formation of necrosomes, which contain RIP1, RIP3, and other proteins. In addition, translocation of RIP1 and RIP3 into the Triton X-100 (or Nonidet P-40) insoluble fraction upon necroptotic stimulus treatment was observed in previous studies (22, 30). However, it remains unknown whether MLKL also goes into the Triton X-100-insoluble fraction as RIP1 and RIP3 at this point. To this end, we generated two stable lines by transfecting empty vector or FLAG-mRIP3 expressing vector into RIP3 KO L929 cells and then analyzed the necrosome formation in these cells. Consistent with previous reports, TNF plus Z-VAD stimulation induced interactions of RIP3 with MLKL, RIP1, and FADD (Fig. 4C). In addition to RIP1 and RIP3, MLKL was also found in the Triton X-100-insoluble fraction after TNF plus Z-VAD stimulation. We then used RIP3 KO cells expressing the mRIP3–2A mutant to perform the same experiment. MLKL was not pulled down by the mRIP3–2A mutant and was not found in the Triton X-100-insoluble fraction (Fig. 4D), indicating that interaction with RIP3 is required for translocation of MLKL to the Triton X-100-insoluble fraction during necroptosis. In contrast, the mRIP3–2A mutant still formed a complex with RIP1 and FADD and all of them were present in the Triton X-100-insoluble fraction (Fig. 4D).
The Interaction of RIP3 and MLKL Is Species Specific
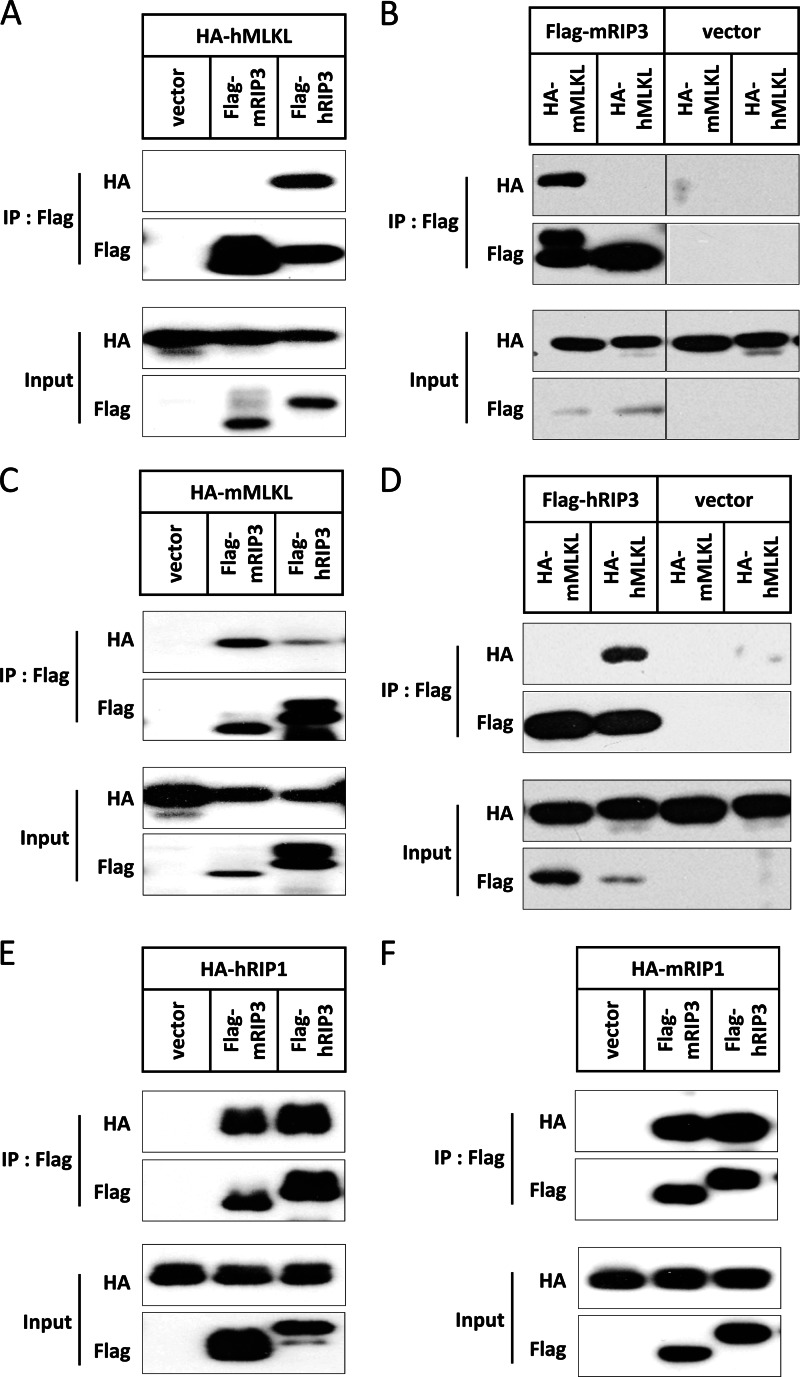
mRIP3 and hRIP3 share 60% identity and 69% similarity at the amino acid level. The kinase domain between these two species is more conserved with 70% identity and 78% similarity. It was shown that hRIP3 interacts with hMLKL through its kinase domain (23). We noticed that mRIP3 cannot interact with hMLKL (Fig. 5, A and B), whereas hRIP3 interacted with mMLKL very weakly (Fig. 5, C and D). In contrast, there is no species specificity for the interaction between RIP3 and RIP1 (Fig. 5, E and F). The fact that hMLKL cannot interact with mRIP3 was also mentioned by Sun et al. (23).
FIGURE 5.
The interaction of RIP3 and MLKL is species specific. A, empty vector, FLAG-mRIP3, and FLAG-hRIP3 were co-transfected with HA-hMLKL. 18 h later, cells were harvested and the cell lysates were then subjected to immunoprecipitation (IP) with anti-FLAG beads. Both immunoprecipitates and total cell lysates (input) were immunoblotted with anti-HA and anti-FLAG antibodies. B, HA-mMLKL and HA-hMLKL were co-transfected with FLAG-mRIP3 or empty vector. 18 h later, cells were harvested and the cell lysates were subjected to immunoprecipitation with anti-FLAG beads. Both immunoprecipitates and total cell lysate were immunoblotted with anti-HA and anti-FLAG antibodies. C, the same as in A except that HA-hMLKL was replaced by HA-mMLKL. D, the same as in B except that FLAG-mRIP3 was replaced by FLAG-hRIP3. E, the same as in A except that HA-hMLKL was replaced by HA-hRIP1. F, the same as in A except that HA-hMLKL was replaced by HA-mRIP1.
The Sequence Flanking the RIP3 Phosphorylation Sites Determines the Species-specific Interaction between RIP3 and MLKL
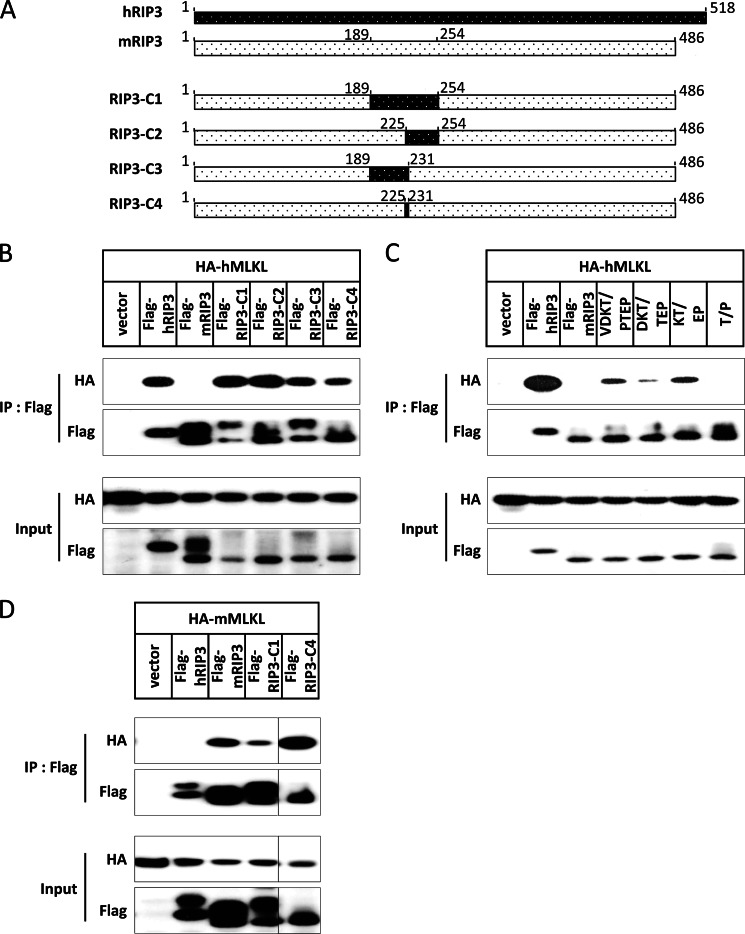
To find out the structural basis of this species-specific binding between RIP3 and MLKL, we used the Phyre server to predict three-dimensional structures of mouse and human RIP3 kinase domain (31), and found that there is a loop around the phosphorylation sites in both hRIP3 and mRIP3, whereas many amino acids around the phosphorylation sites are not conserved (Fig. 2B). To determine whether the sequence flanking phosphorylation site Ser-227 in hRIP3 is involved in species specificity of the RIP3-MLKL interaction, we generated a series of chimeric constructs using mRIP3 as backbone and replaced the amino acids in and around the loop (189–254 aa) by the corresponding hRIP3 fragment (184–249 aa) (Fig. 6A). Here, RIP3-C1 was made in the mRIP3 backbone and the amino acids 189–254 were replaced with hRIP3 amino acids 184–249. Similarly, RIP3-C2, -C3, and -C4 were constructed by replacing mRIP3 aa 225–254 with hRIP3 aa 220–249, mRIP3 aa 189–231 with hRIP3 aa 184–226, and mRIP3 aa 225–231 with hRIP3 aa 220–226, respectively. RIP3-C1 and RIP3-C2 bound to hMLKL as strongly as hRIP3 (Fig. 6B). RIP3-C3 and RIP3-C4 had reduced binding affinity to hMLKL (Fig. 6B). Therefore, the sequence that determines the selective interaction of hRIP3 with hMLKL should be primarily located within the fragment spanning amino acid residues 220–249 in hRIP3 (corresponding to 225–254 aa in mRIP3). Because 220–226 aa (VELPTEP) of hRIP3 (corresponding to 225–231 aa AELVDKT in mRIP3) is the overlapping sequence among RIP3-C2, RIP3-C3, and RIP3-C4, the weak binding of RIP3-C3 and RIP3-C4 to hMLKL is thus most likely contributed by this overlapping sequence, which is also the most diverse motif in this region. To determine which amino acid is more important in this overlapping sequence, we generated more mutations by changing VDKT, DKT, KT, and Thr in mRIP3 to the corresponding PTEP, TEP, EP, and Pro in hRIP3, respectively, and performed protein-protein interaction experiments with hMLKL. Conclusively, change of 2 amino acids, Lys and Thr, in mRIP3 to the human version, Glu and Pro, appears to be the minimal requirement for mRIP3 to interact with hMLKL (Fig. 6C). In addition, we also analyzed whether modifying mRIP3 to be able to interact with hMLKL would compromise its interaction with mMLKL. As shown in Fig. 6D, RIP3-C1 had less affinity to mMLKL in comparison with mRIP3, whereas the interaction between RIP3-C4 and mMLKL was stronger than RIP3-C1. Collectively, our data demonstrated that the sequence flanking the phosphorylation sites plays a primary role in determining the species specificity of the interaction between RIP3 and MLKL.
FIGURE 6.
The sequence flanking the phosphorylation sites determines the species-specific interaction between RIP3 and MLKL. A, schematic presentation of RIP3 chimeric constructs. RIP3-C1 was made using mRIP3 as the backbone and amino acids 189–254 were replaced with hRIP3 aa 184–249. Similarly, RIP3-C2, -C3, and -C4 were made by replacing mouse 225–254 aa with human 220–249 aa, mouse 189–231 aa with human 184–226 aa, and mouse 225–231 aa with human 220–226 aa, respectively. B, empty vector and FLAG-hRIP3, FLAG-mRIP3, RIP3-C1, -C2, -C3, and -C4 were co-transfected with HA-hMLKL into 293T cells. 18 h after transfection, cells were harvested and the cell lysates were subjected to immunoprecipitation (IP) with anti-FLAG beads. Both immunoprecipitates and total cell lysates were immunoblotted with anti-HA and anti-FLAG antibodies. C, RIP3-VDKT/PTEP, RIP3-DKT/TEP, RIP3-KT/EP, and RIP3-T/P were made by using mRIP3 as backbone and the amino acids 228VDKT231 were replaced with hRIP3 223PTEP226, 229DKT231 with hRIP3 224TEP226, 230KT231 with hRIP3 225EP226, and Thr-231 with hRIP3 Pro-226, respectively. These constructs together with FLAG-hRIP3 and FLAG-mRIP3 were co-transfected with HA-hMLKL and the results analyzed as in B. D, empty vector and FLAG-hRIP3, FLAG-mRIP3, RIP3-C1, and -C4 were co-transfected with HA-mMLKL into 293T cells and the results analyzed as in B.
Loss of Interaction with MLKL Enhances RIP3 Aggregation Upon TNF Stimulation
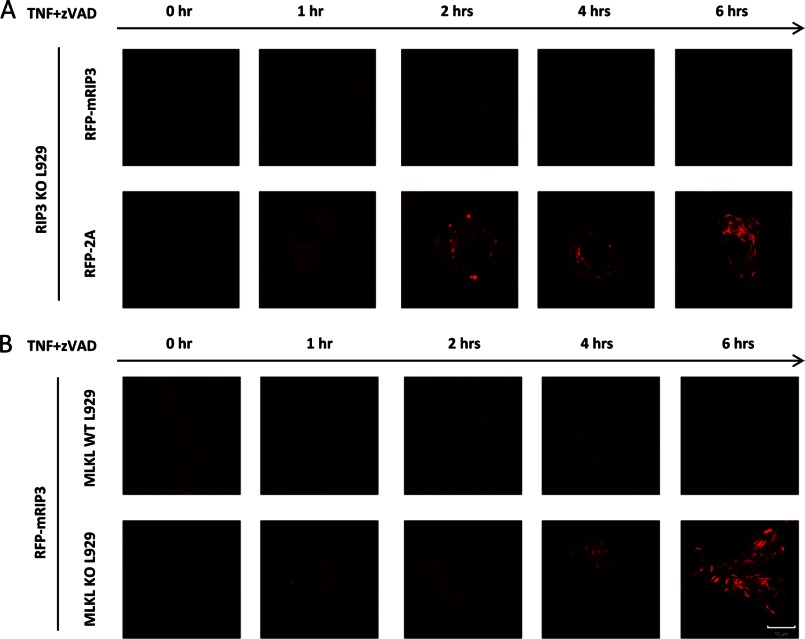
A recent study showed that RIP1/RIP3 form amyloid-like aggregates, which are required for necroptotic signaling (22). Because T231A and S232A mutations in mRIP3 abolished its interaction with MLKL, we sought to evaluate whether loss of interaction with MLKL affects the aggregation of RIP3. We transfected RIP3 KO L929 cells with the expression vectors of RFP-mRIP3 or RFP-mRIP3–2A. Similar to FLAG-mRIP3, the RFP-mRIP3 expression in RIP3 KO L929 cells restored the cells' sensitivity to TNF-induced necroptosis (data not shown). As anticipated, the RFP-mRIP3–2A mutant had no effect on TNF sensitivity when expressed in RIP3 KO cells (data not shown). We treated the RFP-mRIP3 or RFP-mRIP3–2A reconstituted RIP3 KO cells with TNF plus Z-VAD for different periods of time and examined the cells under a fluorescent microscope. RIP3 is uniformly distributed in cytosol with very tiny dots (Fig. 7A). TNF plus Z-VAD stimulation did not affect the pattern of RIP3 in most of the cells, even in detached dying cells at 6 h treatment (Fig. 7A). However, the puncta structure of RIP3, which was reported to be induced by TNF stimulation (23), was indeed observed in a small amount of cells (<7% of total) after TNF plus Z-VAD stimulation (data not shown). It is difficult to conclude whether or not the punctae of RIP3 precedes cell death based only on microscope analysis. The distribution pattern of the mRIP3–2A mutant is similar to that of mRIP3 in the resting stage (Fig. 7A). To our surprise, TNF plus Z-VAD treatment induced high-intensity punctae of the RFP-mRIP3–2A mutant in most of the cells (>80% cells) and the punctae appeared as rod-like structures at later time points (Fig. 7A). Because the expression level of the mRIP3–2A mutant was not changed by TNF plus Z-VAD treatment (Fig. 4D and data not shown), the induced RFP-mRIP3–2A dots or rods should be the results of the aggregation of necrosomes containing mRIP3–2A.
FIGURE 7.
Loss of interaction with MLKL leads to massive aggregation of RIP3 in TNF-stimulated L929 cells. A, RIP3 KO L929 cells were infected with lentivirus encoding RFP-mRIP3 or RFP-mRIP3–2A. The infected cells were then treated with TNF plus Z-VAD for different periods of time as indicated and live cell imaging was recorded. B, the same as in A except that MLKL wild-type and KO L929 cells were infected with lentivirus encoding RFP-mRIP3. Scale bar, 10 μm.
Because the mRIP3–2A mutant cannot interact with MLKL, it is possible that the massive aggregation of this RIP3 mutant is due to the loss of its binding to MLKL. To evaluate this possibility, we used a L929 line in which MLKL was knocked out using the TALEN method. Just like MLKL knockdown cells (23, 24), MLKL KO cells are resistant to TNF plus Z-VAD-induced necroptosis (data not shown). We expressed RFP-mRIP3 in MLKL KO and parental L929 cells, and then treated the cells with TNF plus Z-VAD for different periods of time. As shown in Fig. 7B, the stimulation induced an extensive aggregation of mRIP3 in MLKL KO cells but not in parental L929 cells. Thus, our data obtained from using the RIP3–2A mutant and MLKL KO cells demonstrated that without interaction with MLKL, RIP3 undergoes abnormal, massive aggregation upon TNF stimulation. This aggregation most likely occurred with the participation of RIP1 and other components of the necrosome because we observed a large amount of RIP1 in the Triton X-100-insoluble fraction with the RIP3–2A mutant in TNF plus Z-VAD-treated cells (Fig. 4D).
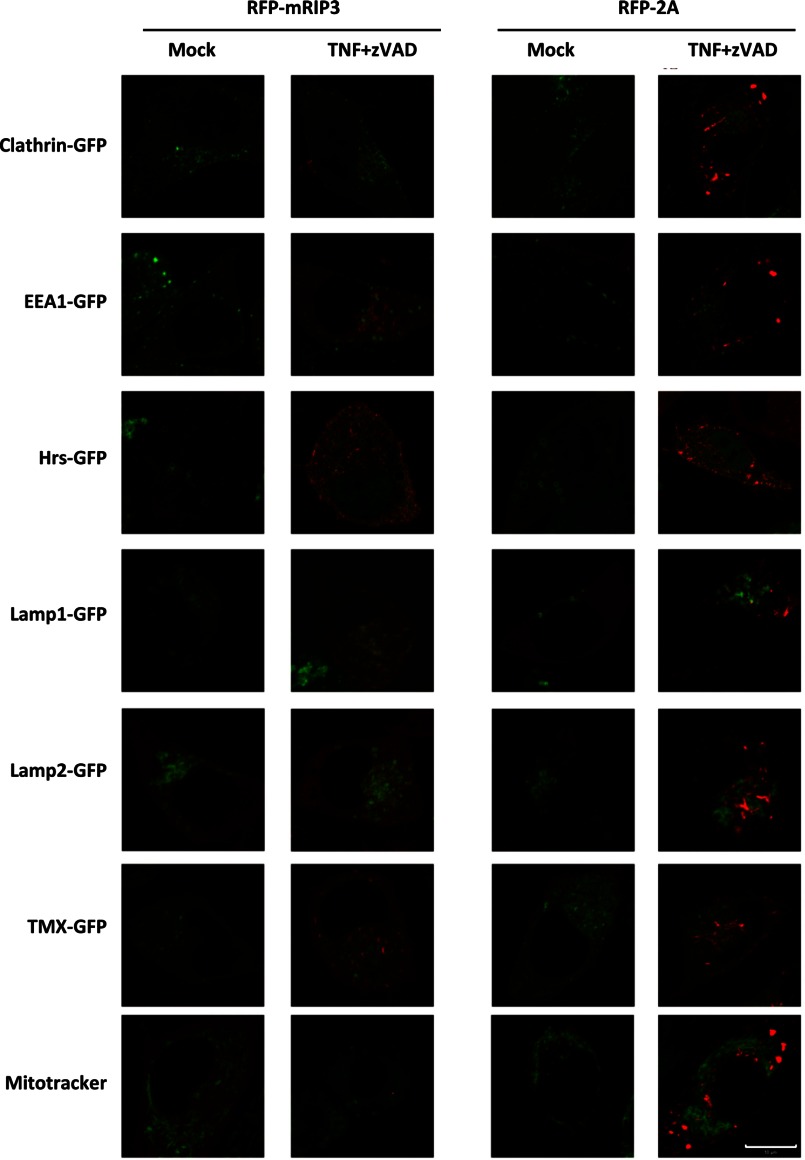
The formation of amyloid-like aggregates of necrosomes is required for necroptotic signaling (22), but it is unknown whether only aggregates of a certain size can induce necroptosis because the cells that had abnormal, massive aggregation of mRIP3–2A did not undergo cell death. Our data suggest that aggregation alone is not sufficient to trigger necroptosis. Because the aggregates of RFP-mRIP3–2A can be easily seen under a fluorescent microscope, we used a number of markers on subcellular organelles to determine the location of the mRIP3–2A aggregates. As a comparison, we included RFP-RIP3 samples and selected the cells that had RFP punctae after TNF plus Z-VAD treatment although the cells were representative of a small portion of the cells. Unfortunately, we were unable to find a precise co-localization of either the mRIP3–2A or the mRIP3 aggregates with the markers we used. Nevertheless, we can exclude clathrin-dependent endocytosis vesicles, early endosome, multivesicular bodies, lysosomes, and endoplasmic reticulum (ER) as the potential target locations for the aggregates of mRIP3 and the mRIP3–2A mutant (Fig. 8).
FIGURE 8.
RIP3 KO L929 cells were reconstituted with RFP-mRIP3 or RFP-mRIP3–2A. The cells were then either transfected with GFP-clathrin (marker of clathrin-dependent endocytosis vesicles), GFP-EEA1 (marker of early endosomes), GFP-Hrs (marker of multivesicular bodies), GFP-TMX (ER marker), GFP-Lamp1 (Lysosome marker), GFP-Lamp2 (Lysosome marker), or stained with MitoTracker. The cells were treated with or without TNF plus Z-VAD for 3 h and live cell imaging was recorded. Scale bar, 10 μm.
Translocation of Necrosomes to MAM Is Associated with the Necroptotic Process and the Interaction of RIP3 with MLKL Is Required for Correct Trafficking of Necrosomes
A previous study suggested that Triton X-100-insoluble fractions contain mitochondria. Thus, translocation of necrosomes to mitochondria could be essential for necroptotic signaling (30, 32). However, we were unable to confirm co-localization of RIP3 with mitochondrial markers in L929 cells before and after TNF stimulation using immunostaining, GFP, or RFP fusion mitochondrial marker proteins and MitoTracker (data not shown and Fig. 8). Because of the inducible presence of RIP1/RIP3 in the Triton X-100-insoluble fraction, TNF stimulation should induce not only the formation but also the translocation of necrosomes. The undetectable change of RIP3 distribution under a microscope (Fig. 7, A and B) is most likely due to the fact that only a small portion of RIP3 translocated.
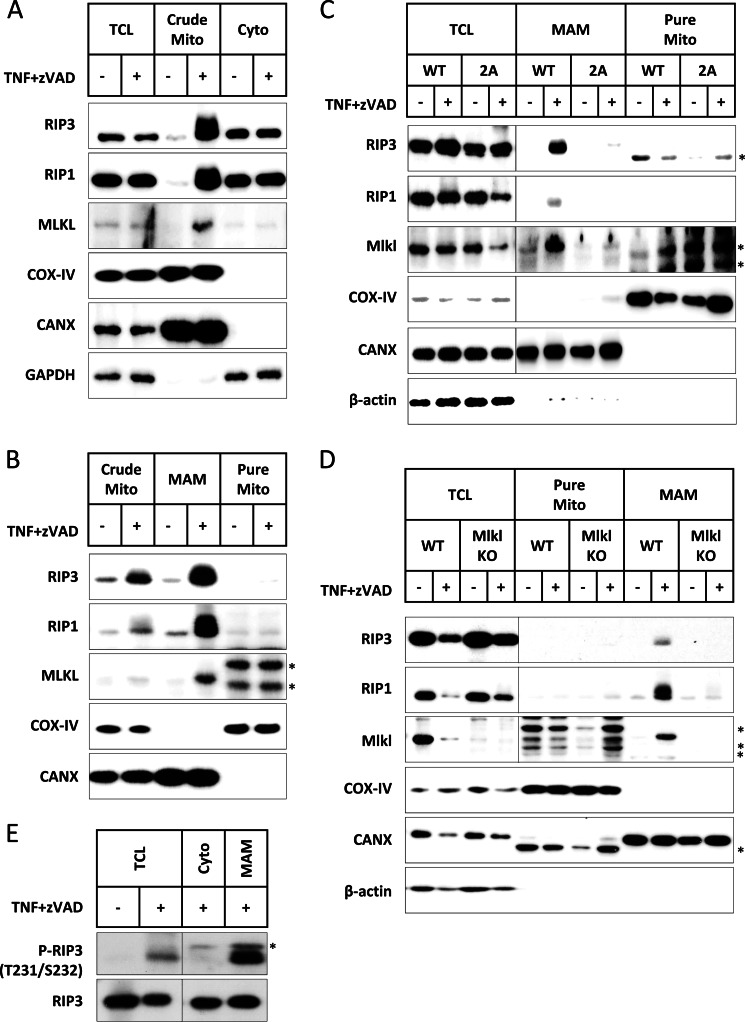
To further address where the necrosomes moved during the process of necroptosis, we fractionated L929 cells before and after TNF plus Z-VAD treatment and we indeed observed an induced increase of RIP1, RIP3, and MLKL in the crude mitochondria fraction (Fig. 9A). However, when we further fractionated the crude mitochondria fraction into pure mitochondria and MAM, we found that RIP1, RIP3, and MLKL were much more enriched in the MAM fraction (Fig. 9B). Thus, MAM is most likely the subcellular compartment that necrosomes translocate to during TNF-induced necroptosis.
FIGURE 9.
The interaction of RIP3 with MLKL leads to translocation of necrosomes to MAM. A, L929 cells were fractionated before and after 3 h of TNF plus Z-VAD treatment. The total cell lysates (TCL), crude mitochondrial (Crude Mito), and cytosolic (Cyto) fractions were immunoblotted with antibodies against RIP3, RIP1, MLKL, COX-IV (mitochondrial marker), CANX (ER marker, enriched in MAM), and GAPDH. B, the crude mitochondria-containing fraction was further fractionated into highly pure mitochondria (Pure Mito) and MAM. The crude mitochondria, MAM, and pure mitochondria were immunoblotted with antibodies against RIP3, RIP1, MLKL, COX-IV, and CANX. C, RIP3 KO L929 cells were reconstituted with wild-type RIP3 or RIP3–2A. The cells were treated with or without TNF plus Z-VAD for 3 h and the cell lysates were then fractionated. The TCL, MAM, and pure mitochondria were immunoblotted with antibodies against RIP3, RIP1, MLKL, COX-IV, CANX, and actin. D, the same as in C except that MLKL wild-type and KO L929 cells were used here. E, L929 cells were treated with or without TNF plus Z-VAD for 3 h and then fractionated. The TCL, Cyto, and MAM were immunoblotted with an antibody against Thr-231 and Ser-232 dual-phosphorylated mRIP3 (P-RIP3). Total RIP3 was immunoblotted as loading controls. Asterisks indicate nonspecific bands.
To determine whether the RIP3 and MLKL interaction plays any role in TNF-induced translocation of necrosomes to MAM, we fractionated RIP3 KO L929 cells expressing mRIP3 or mRIP3–2A mutant before and after TNF plus Z-VAD treatment. In the RIP3-reconstituted cells, TNF induced MAM translocation of RIP3, RIP1, and MLKL (Fig. 9C). Interestingly, we did not find RIP3, RIP1, and MLKL at the MAM fraction in TNF plus Z-VAD-treated RIP3 KO cells expressing RIP3–2A (Fig. 9C). It is apparent that RIP3 and RIP3–2A translocated to different subcellular locations although both can be collected in the Triton X-100-insoluble fraction. It is highly possible that the defect of RIP3–2A in mediating necroptosis is due to the fact that it cannot translocate to MAM. To further confirm that interaction with MLKL is required for RIP3 trafficking to MAM, we used MLKL KO L929 cells and indeed we found that TNF plus Z-VAD stimulation cannot induce the translocation of RIP3 and RIP1 to MAM (Fig. 9D).
To determine whether phosphorylated RIP3 is translocated to MAM after TNF stimulation, we compared the amounts of phospho-RIP3 in total cell lysate, cytosol, and MAM. As shown in Fig. 9E, phospho-RIP3 is much more enriched in the MAM fraction, consistent with the idea that the interaction of RIP3 with MLKL is required for the translocation of necrosomes. More importantly, our findings suggest that only phosphorylated RIP3 moves with necrosomes to the MAM.
DISCUSSION
Recent studies have revealed that MLKL is required for RIP3 to mediate necroptosis signaling (23, 24). The phosphorylation of Ser-227 in hRIP3 is essential for the interaction between hRIP3 and hMLKL, which is crucial for the execution of necroptotic cell death (23). In this study we identified Thr-231 and Ser-232 as the phosphorylation sites in mRIP3 and the phosphorylation of these sites is important for interaction between mRIP3 and mMLKL. The function of RIP3 and MLKL in transducing necroptosis signaling is conserved in human and mouse. However, mRIP3 cannot interact with hMLKL and vice versa. Ser-232 in mRIP3 corresponds to Ser-227 in hRIP3. But the amino acids surrounding this phosphorylation site are not conserved between human and mouse RIP3. In our studies, we found that the fragment spanning the ∼30 amino acids flanking this phosphorylation site (including the second phosphorylation site in mouse) plays a primary role in determining the species specificity of the interaction between RIP3 and MLKL. It appears that the interaction between RIP3 and MLKL is an evolutionarily conserved event in necroptosis signaling, whereas the structural basis for the interaction of these two proteins diverged somehow during evolution in different organisms, such as human and mouse as we demonstrated here. A recent study has suggested that the amyloid signaling complex of necrosomes is required for necroptosis (22). Our data demonstrated that the RIP3-MLKL interaction is required for proper formation of necrosome aggregates to mediate necroptosis because the abnormal, massive aggregation of RIP3 resulted from the loss of its interaction with MLKL does not cause cell death. Furthermore, our data suggest that not only the formation of amyloid-like aggregates of necrosomes but also the translation of the necrosome complex to MAM are required for the progression of necroptosis and that the RIP3-MLKL interaction plays a critical role in both processes.
As shown here, the band shift of mRIP3 in SDS-PAGE is phosphorylation dependent; but the phosphorylation that leads to the band shifting is not related to its function in the necroptotic signaling pathway (Fig. 1). Unlike mRIP3, there is no band shift for hRIP3 in resting HT29 cells. However, certain extracellular stimuli could indeed induce the hRIP3 protein band shift (Fig. 1D). Band shift of both hRIP3 and mRIP3 could be observed when they were overexpressed in 293T cells (Figs. 1G and 5C). The mobility shifted hRIP3 band was shown previously to correspond to Ser-199-phosphorylated hRIP3 (29). Interestingly, the S199A hRIP3 mutant was still able to mediate necroptosis despite the absence of phosphorylation on this site (data not shown). So, similar to mRIP3, hRIP3 phosphorylation that leads to the band shift is not critical for its function in necroptosis. Although it is already known that phosphorylation in other sites (other than Ser-199) regulates the interaction between RIP3 and MLKL, it is not clear yet at this stage if the biological indication of Ser-199 phosphorylation in hRIP3 and further investigation is needed to shed light on this issue.
It was proposed earlier that phosphorylation on Ser-227 in hRIP3 is autophosphorylation because the kinase-dead mutant of hRIP3 cannot interact with hMLKL (23). We found it appears to be the same case for mRIP3 because the kinase-dead mutant of mRIP3 (mRIP3-D143N) cannot interact with mMLKL either (data not shown). To demonstrate that it is the autophosphorylation but not the loss of kinase activity that diminishes the interaction between RIP3 and MLKL, we created an additional phosphorylation mimic mutation on Ser-227 in the hRIP3 kinase-dead mutant. Unlike the kinase-dead mutant (hRIP3-K50A), the phosphorylation mimic mutant (hRIP3-K50A/S227D) can still interact with hMLKL (data not shown). Ser-227 phosphorylation on hRIP3 was identified in TNF stimulated but not in unstimulated HT29 cells by mass spectrum analysis (23). Consistently, Thr-231 and Ser-232 phosphorylation of mRIP3 is induced upon TNF plus Z-VAD treatment in L929 (Figs. 2G and 8E). Because overexpressed hRIP3 and mRIP3 can both interact with their corresponding MLKL, respectively, in the absence of extracellular stimulation, the autophosphorylation on Ser-227 in hRIP3 and on Thr-231 and Ser-232 in mRIP3 can be considered to be present constitutively in this case. Based on these observations, we propose that an important signaling event in the necrosome is that TNF induced Ser-227 autophosphorylation in hRIP3 or Thr-231 and Ser-232 autophosphorylation in mRIP3 recruits the corresponding MLKL, and the recruited MLKL then plays a key role in transducing the downstream signaling.
It was reported that TNF stimulation led to the gradual formation and enlarging of discrete hRIP3 punctae along with the progression of necroptosis (23). Similarly, we also observed the induced formation and enlargement of mRIP3-containing complexes, but only in a small percentage of TNF-stimulated L929 cells. Given the fact that the non-necrotic mRIP3–2A mutant can form much more and larger punctae, it remains unclear whether the visible RIP3 punctae really function in necroptosis. In this study, the data we obtained from cellular fractionation showed that MAM is the site where the necrosome aggregates translocate and is thus likely the subcellular compartment where necrosomes initiate necroptosis. MAM is the contact site of ER and mitochondria, and is the primary compartment for Ca2+ traffic between these two organelles. Thus, MAM has been indicated to play a role in protein folding, oxidative phosphorylation, and Ca2+-mediated apoptosis (33). To date the ER Ca2+ has not been found to play any role in TNF-induced necroptosis. A recent work suggested that mitochondria fission is required for necroptosis (30). ER and mitochondria interaction is known to enhance mitochondria fission (34). However, it is unknown at this stage whether there is any linkage between necrosome translocation to MAM and mitochondria fission. What is the functional indication for translocated necrosomes in MAM is a future challenge in our study of necroptosis.
In conclusion, we demonstrated in this work that the recruitment of MLKL by RIP3 is essential for necroptotic cell death in both human and mouse. But the structural basis for the RIP3-MLKL interaction in human and mouse is not exactly the same, which is reflected as the species specificity of this interaction. Although TNF-induced autophosphorylation of RIP3 is required for both hRIP3 and mRIP3 to interact with hMLKL and mMLKL, respectively, the phosphorylation sites required for this interaction are different. The sequence flanking the required phosphorylation sites is not conserved between human and mouse and the sequence differences appear to contribute to species specificity of the RIP3-MLKL interaction. More importantly, we concluded in this study that the evolutionary conservation of the RIP3-MLKL interaction does have its functional relevance because the RIP3-MLKL interaction controls the proper formation of necrosome aggregates and is required for the translocation of necrosomes to the proper subcellular location to initiate necroptosis.
This work was supported by 973 program Grant 2009CB522201, National Science Foundation of China Grants 91029304, 31221065, 81061160512, 91229201, 863 program Grant 2012AA02A201, 111 Project B12001, Chinese National Scientific and Technological Major Project Grant 2013ZX10002002, and Open Research Fund of State Key Laboratory of Cellular Stress Biology, Xiamen University Grant SKLCSB2012KF003.
- RIP3
- receptor interacting protein 3
- MLKL
- mixed lineage kinase domain-like
- Z
- benzyloxycarbony
- MBP
- myelin basic protein
- PI
- propidium iodide
- MAM
- mitochondria-associated membranes
- aa
- amino acid(s)
- ER
- endoplasmic reticulum.
REFERENCES
- 1. Degterev A., Huang Z., Boyce M., Li Y., Jagtap P., Mizushima N., Cuny G. D., Mitchison T. J., Moskowitz M. A., Yuan J. (2005) Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat. Chem. Biol. 1, 112–119 [DOI] [PubMed] [Google Scholar]
- 2. Cho Y. S., Challa S., Moquin D., Genga R., Ray T. D., Guildford M., Chan F. K. (2009) Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell 137, 1112–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Upton J. W., Kaiser W. J., Mocarski E. S. (2010) Virus inhibition of RIP3-dependent necrosis. Cell Host Microbe 7, 302–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Upton J. W., Kaiser W. J., Mocarski E. S. (2012) DAI/ZBP1/DLM-1 complexes with RIP3 to mediate virus-induced programmed necrosis that is targeted by murine cytomegalovirus vIRA. Cell Host Microbe 11, 290–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Robinson N., McComb S., Mulligan R., Dudani R., Krishnan L., Sad S. (2012) Type I interferon induces necroptosis in macrophages during infection with Salmonella enterica serovar Typhimurium. Nat. Immunol. 13, 954–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Han J., Zhong C. Q., Zhang D. W. (2011) Programmed necrosis. Backup to and competitor with apoptosis in the immune system. Nat. Immunol. 12, 1143–1149 [DOI] [PubMed] [Google Scholar]
- 7. Welz P. S., Wullaert A., Vlantis K., Kondylis V., Fernández-Majada V., Ermolaeva M., Kirsch P., Sterner-Kock A., van Loo G., Pasparakis M. (2011) FADD prevents RIP3-mediated epithelial cell necrosis and chronic intestinal inflammation. Nature 477, 330–334 [DOI] [PubMed] [Google Scholar]
- 8. Günther C., Martini E., Wittkopf N., Amann K., Weigmann B., Neumann H., Waldner M. J., Hedrick S. M., Tenzer S., Neurath M. F., Becker C. (2011) Caspase-8 regulates TNF-α-induced epithelial necroptosis and terminal ileitis. Nature 477, 335–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liedtke C., Bangen J. M., Freimuth J., Beraza N., Lambertz D., Cubero F. J., Hatting M., Karlmark K. R., Streetz K. L., Krombach G. A., Tacke F., Gassler N., Riethmacher D., Trautwein C. (2011) Loss of caspase-8 protects mice against inflammation-related hepatocarcinogenesis but induces non-apoptotic liver injury. Gastroenterology 141, 2176–2187 [DOI] [PubMed] [Google Scholar]
- 10. Bonnet M. C., Preukschat D., Welz P. S., van Loo G., Ermolaeva M. A., Bloch W., Haase I., Pasparakis M. (2011) The adaptor protein FADD protects epidermal keratinocytes from necroptosis in vivo and prevents skin inflammation. Immunity 35, 572–582 [DOI] [PubMed] [Google Scholar]
- 11. Trichonas G., Murakami Y., Thanos A., Morizane Y., Kayama M., Debouck C. M., Hisatomi T., Miller J. W., Vavvas D. G. (2010) Receptor interacting protein kinases mediate retinal detachment-induced photoreceptor necrosis and compensate for inhibition of apoptosis. Proc. Natl. Acad. Sci. U.S.A. 107, 21695–21700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Murakami Y., Matsumoto H., Roh M., Suzuki J., Hisatomi T., Ikeda Y., Miller J. W., Vavvas D. G. (2012) Receptor interacting protein kinase mediates necrotic cone but not rod cell death in a mouse model of inherited degeneration. Proc. Natl. Acad. Sci. U.S.A. 109, 14598–14603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fortes G. B., Alves L. S., de Oliveira R., Dutra F. F., Rodrigues D., Fernandez P. L., Souto-Padron T., De Rosa M. J., Kelliher M., Golenbock D., Chan F. K., Bozza M. T. (2012) Heme induces programmed necrosis on macrophages through autocrine TNF and ROS production. Blood 119, 2368–2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lin J., Li H., Yang M., Ren J., Huang Z., Han F., Huang J., Ma J., Zhang D., Zhang Z., Wu J., Huang D., Qiao M., Jin G., Wu Q., Huang Y., Du J., Han J. (2013) A role of RIP3-mediated macrophage necrosis in atherosclerosis development. Cell Rep. 3, 200–210 [DOI] [PubMed] [Google Scholar]
- 15. Kaiser W. J., Upton J. W., Long A. B., Livingston-Rosanoff D., Daley-Bauer L. P., Hakem R., Caspary T., Mocarski E. S. (2011) RIP3 mediates the embryonic lethality of caspase-8-deficient mice. Nature 471, 368–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Oberst A., Dillon C. P., Weinlich R., McCormick L. L., Fitzgerald P., Pop C., Hakem R., Salvesen G. S., Green D. R. (2011) Catalytic activity of the caspase-8-FLIP(L) complex inhibits RIPK3-dependent necrosis. Nature 471, 363–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang H., Zhou X., McQuade T., Li J., Chan F. K., Zhang J. (2011) Functional complementation between FADD and RIP1 in embryos and lymphocytes. Nature 471, 373–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Micheau O., Tschopp J. (2003) Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell 114, 181–190 [DOI] [PubMed] [Google Scholar]
- 19. Bertrand M. J., Milutinovic S., Dickson K. M., Ho W. C., Boudreault A., Durkin J., Gillard J. W., Jaquith J. B., Morris S. J., Barker P. A. (2008) cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol. Cell 30, 689–700 [DOI] [PubMed] [Google Scholar]
- 20. Wang L., Du F., Wang X. (2008) TNF-α induces two distinct caspase-8 activation pathways. Cell 133, 693–703 [DOI] [PubMed] [Google Scholar]
- 21. Schneider-Brachert W., Tchikov V., Neumeyer J., Jakob M., Winoto-Morbach S., Held-Feindt J., Heinrich M., Merkel O., Ehrenschwender M., Adam D., Mentlein R., Kabelitz D., Schütze S. (2004) Compartmentalization of TNF receptor 1 signaling. Internalized TNF receptosomes as death signaling vesicles. Immunity 21, 415–428 [DOI] [PubMed] [Google Scholar]
- 22. Li J., McQuade T., Siemer A. B., Napetschnig J., Moriwaki K., Hsiao Y. S., Damko E., Moquin D., Walz T., McDermott A., Chan F. K., Wu H. (2012) The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell 150, 339–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sun L., Wang H., Wang Z., He S., Chen S., Liao D., Wang L., Yan J., Liu W., Lei X., Wang X. (2012) Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell 148, 213–227 [DOI] [PubMed] [Google Scholar]
- 24. Zhao J., Jitkaew S., Cai Z., Choksi S., Li Q., Luo J., Liu Z. G. (2012) Mixed lineage kinase domain-like is a key receptor interacting protein 3 downstream component of TNF-induced necrosis. Proc. Natl. Acad. Sci. U.S.A. 109, 5322–5327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wu X., Tian L., Li J., Zhang Y., Han V., Li Y., Xu X., Li H., Chen X., Chen J., Jin W., Xie Y., Han J., Zhong C. Q. (2012) Investigation of receptor interacting protein (RIP3)-dependent protein phosphorylation by quantitative phosphoproteomics. Mol. Cell Proteomics 11, 1640–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang F., Cong L., Lodato S., Kosuri S., Church G. M., Arlotta P. (2011) Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat. Biotechnol. 29, 149–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wieckowski M. R., Giorgi C., Lebiedzinska M., Duszynski J., Pinton P. (2009) Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nat. Protoc. 4, 1582–1590 [DOI] [PubMed] [Google Scholar]
- 28. Zhang D. W., Shao J., Lin J., Zhang N., Lu B. J., Lin S. C., Dong M. Q., Han J. (2009) RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science 325, 332–336 [DOI] [PubMed] [Google Scholar]
- 29. He S., Wang L., Miao L., Wang T., Du F., Zhao L., Wang X. (2009) Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-α. Cell 137, 1100–1111 [DOI] [PubMed] [Google Scholar]
- 30. Wang Z., Jiang H., Chen S., Du F., Wang X. (2012) The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell 148, 228–243 [DOI] [PubMed] [Google Scholar]
- 31. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the Web. A case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 32. Ciarlo L., Manganelli V., Garofalo T., Matarrese P., Tinari A., Misasi R., Malorni W., Sorice M. (2010) Association of fission proteins with mitochondrial raft-like domains. Cell Death Differ. 17, 1047–1058 [DOI] [PubMed] [Google Scholar]
- 33. Hayashi T., Rizzuto R., Hajnoczky G., Su T. P. (2009) MAM. More than just a housekeeper. Trends Cell Biol. 19, 81–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Friedman J. R., Lackner L. L., West M., DiBenedetto J. R., Nunnari J., Voeltz G. K. (2011) ER tubules mark sites of mitochondrial division. Science 334, 358–362 [DOI] [PMC free article] [PubMed] [Google Scholar]