Background: YidC interacts with SecY during membrane protein insertion but details on this interaction are missing.
Results: YidC binds to the lateral gate of SecY and is detached by nascent membrane proteins but not by SecA.
Conclusion: Nascent membrane-induced lateral gate movements directly influence the SecY-YidC interaction.
Significance: This is the first detailed analysis of the SecY-YidC interaction.
Keywords: Membrane Biogenesis, Membrane Trafficking, Protein Targeting, Secretion, Signal Recognition Particle
Abstract
Most membrane proteins are co-translationally inserted into the lipid bilayer via the universally conserved SecY complex and they access the lipid phase presumably via a lateral gate in SecY. In bacteria, the lipid transfer of membrane proteins from the SecY channel is assisted by the SecY-associated protein YidC, but details on the SecY-YidC interaction are unknown. By employing an in vivo and in vitro site-directed cross-linking approach, we have mapped the SecY-YidC interface and found YidC in contact with all four transmembrane domains of the lateral gate. This interaction did not require the SecDFYajC complex and was not influenced by SecA binding to SecY. In contrast, ribosomes dissociated the YidC contacts to lateral gate helices 2b and 8. The major contact between YidC and the lateral gate was lost in the presence of ribosome nascent chains and new SecY-YidC contacts appeared. These data demonstrate that the SecY-YidC interaction is influenced by nascent-membrane-induced lateral gate movements.
Introduction
Approximately 30% of all proteins synthesized in the cytosol of eukaryotic or prokaryotic cells execute their functions in extra cytosolic compartments. Dedicated protein targeting pathways ensure that these proteins are specifically recognized and delivered to protein transport channels, which then facilitate their transport across or into membranes (1). The Sec translocon is particularly important for protein transport because it constitutes a universally conserved protein conducting channel that orchestrates co-translational and post-translational protein transport processes in prokaryotes and eukaryotes. The bacterial Sec translocon is a heterotrimeric membrane protein complex consisting of SecY, SecE, and SecG. SecY is the core component and consists of 10 transmembrane domains (TMs),2 which assemble around a central pore (2). The 10 TMs are arranged in a clam-shell like structure, in which the two halves (TMs 1–5 and TMs 6–10, respectively) are connected by a loop between TM 5 and TM 6. This hinge region is embraced at the back by SecE. Opposite the hinge region, TMs 2b, 3, 7, and 8 constitute the lateral gate (Fig. 1), which is thought to open during the insertion of signal sequences or during the lipid insertion of membrane proteins (2, 3). SecG the third component of the bacterial Sec translocon is not essential for protein transport in vitro and is thought to play a role during post-translational transport (4).
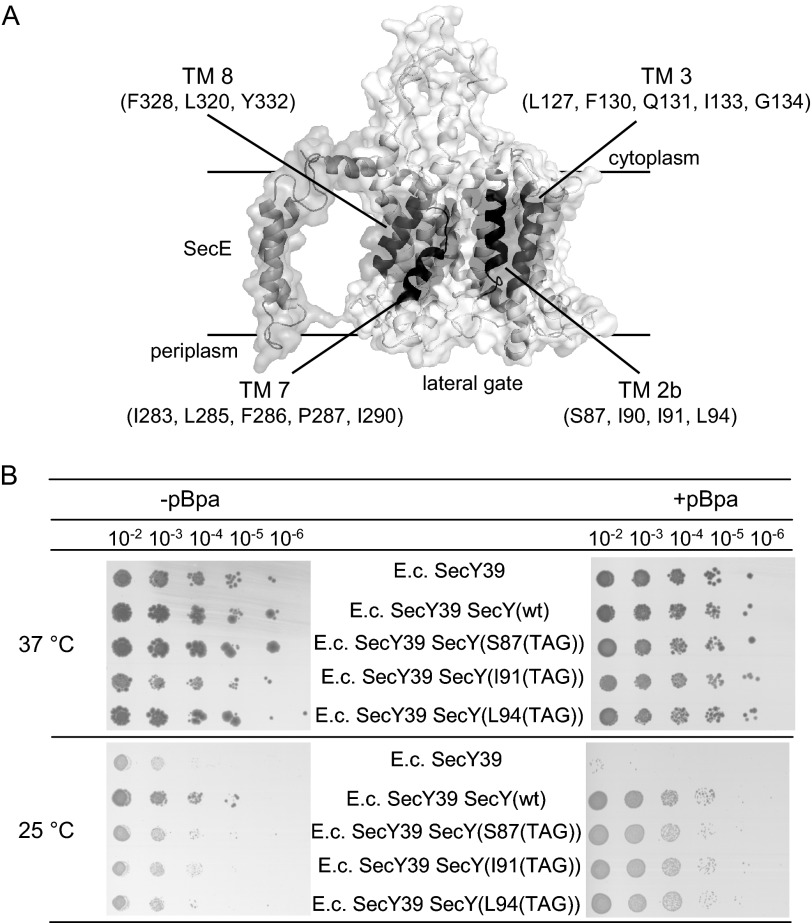
FIGURE 1.
Mapping contacts of the lateral gate of SecY by in vivo cross-linking. A, cryo-EM structure of the E. coli SecYEG complex (3) (Protein Data Bank codes 3J00 and 3J01); the four TM domains that constitute the lateral gate are highlighted and the residues where pBpa was incorporated for cross-linking in vivo and in vitro are indicated. B, functionality of pBpa-containing SecY derivatives was analyzed by expressing them in the cold-sensitive E. coli SecY39 strain that contained the orthogonal aminoacyl-tRNA synthetase/tRNACUA pair. Cells were grown overnight on LB medium, diluted, and grown for another 2 h before serial dilution. Of each dilution, 10 μl were spotted on either LB plates (−pBpa) or on LB plates containing 1 mm pBpa (+pBpa). Cells were grown overnight at either 25 or 37 °C.
The SecY channel is a passive pore and associates with soluble and membrane-bound partner proteins for facilitating protein transport (1). During post-translational protein transport, the ATPase SecA binds to cytosolic loops C4, C5, and C6 of SecY. SecA serves a dual function, it acts as a pre-protein receptor and provides the energy for translocation by ATP hydrolysis (1, 5). The SRP receptor FtsY binds to the C4 and C5 loops of SecY during co-translational protein transport and recruits the SRP-ribosome nascent chain complex (SRP-RNCs) to the Sec translocon (6–8). Translation at the ribosome is then thought to provide the major driving force for co-translational membrane protein insertion.
In addition to interactions with cytosolic partner proteins, SecY also cooperates with membrane proteins, but the physiological significance of these interactions is less defined. The SecDFYajC membrane complex associates transiently with the SecYEG complex (9), but is not essential for cell viability in Escherichia coli (10). A recent crystal structure of Thermus thermophilus SecDF shows that both SecF and SecD consist of 6 TMs and a large periplasmic domain each (11). SecDF is suggested to be involved in proton-motive force-dependent translocation and in the release of translocated secretory proteins from the periplasmic side of the membrane (11, 12). A role for SecDF in membrane protein insertion has also been proposed (13) and SecDF was found to co-purify with YidC (14), an essential protein that is specifically required for membrane protein insertion (15). This led to the hypothesis that SecDF is required for tethering YidC to the SecYEG translocon during membrane protein insertion (14). YidC is a member of the Oxa family of membrane proteins (16) and can function as an insertase for membrane proteins independently or in concert with SecYEG (17–20). Several functions have been attributed to SecYEG-associated YidC. YidC could be required for the release of TMs from the SecY channel (21), or for the assembly of multiple TMs prior to their release into the lipid phase (22). YidC has also been shown to control the topology of SecYEG-dependent membrane proteins like LacY (23). Finally, a role of YidC for the assembly of individual subunits into functional membrane protein complexes (24) and for membrane protein quality control has been proposed (25). Although YidC was found to co-purify with the Sec translocon (26) and in complex with SecYEG on Blue Native PAGE (27), the actual interaction between SecYEG and YidC has not yet been determined.
In the current study we have used an in vivo and in vitro site-directed cross-linking approach for determining the SecY-YidC interaction sites. Our data demonstrate that YidC is in contact with all four TMs of the lateral gate. These interactions dynamically respond to the binding of ribosomes or RNCs to SecY but not to the binding of SecA.
EXPERIMENTAL PROCEDURES
Strains and Plasmids, Growth Conditions
The following E. coli strains were used: DH5α (28), BL21 pSup-BpaRS-6TRN (29), C43 pSup-BpaRS-6TRN, BL325 (9), KC6(DE3) pftsQ-tnaC (a gift from R. Beckmann, Munich), and SecY39 (30). Cells were grown in LB medium at either 30 or 37 °C. TAG stop codons were incorporated at the indicated positions of pTRc99aSecY(His)EG (8) or pTrc99a-YidC (20) using the Phusion PCR Kit (NE Biolabs, Frankfurt, Germany) with 5′-phosphorylated oligonucleotides (Table 1).
TABLE 1.
Oligonucleotide primer used for generating SecY amber stop codon mutants
| Primer | |
|---|---|
| TM 2b | |
| SecY_S87_for | 5′-att tag gcg tcg atc att atc cag-3′ |
| SecY I90amb_for | 5′-att tcg gcg tcg tag att atc cag- 3′ |
| SecY_I91_for | 5′-att tcg gcg tcg atc tag atc cag-3′ |
| SecY_L94_for | 5′-att tcg gcg tcg atc att atc cag tag ctg acg-3′ |
| SecY I90amb_rev | 5′-ata cgg cat gat ccc ca gag-3′ (used also for Ser-87, Ile-91, Leu-94) |
| TM 3 | |
| SecY_L127_for | 5′-gtg tag gca ata ttc cag tcg atc-3′ |
| SecY_F130_for | 5′-gtg ctg gca ata tag cag tcg atc-3′ |
| SecY_Q131_for | 5′-gtg ctg gca ata ttc tag tcg atc-3′ |
| SecY_I133_for | 5′-gtg ctg gca ata ttc cag tcg tag ggt att-3′ |
| SecY_G134_for | 5′-gtg ctg gca ata ttc cag tcg atc tag att gct-3′ |
| SecY_L127_rev | 5′-cag agt acc gta gcg ggt gta ctg-3′ (used also for Phe-130, Gln-131, Ile-133, Gly-134) |
| TM 7 | |
| SecY I283amb_for | 5′-ttc gct tcc agt tag att ctg ttc-3′ |
| SecY_L285_for | 5′-ttc gct tcc agt att att tag ttc ccg-3′ |
| SecY_F286_for | 5′-ttc gct tcc agt att att ctg tag ccg gcg-3′ |
| SecY_P287_for | 5′-ttc gct tcc agt att att ctg ttc tag gcg acc − 3′ |
| SecY I283amb_rev | 5′-gat tgc cgg gat tac ccc-3′ (used also for Leu-285, Phe-286, Pro-287) |
| SecY_I290_for | 5′-gcg acc tag gcg tca tgg-3′ |
| SecY_I290_rev | 5′-cgg gaa cag att aat act gga-3′ |
| TM 8 | |
| L316_for | 5′-caa ccg tag tat gtg tta ctc-3′ |
| L320_for | 5′-caa ccg ctt tat gtg tta tag tat gcg-3′ |
| L316_rev | 5′-ccc agg ctg caa ata cag-3′ (used also for Leu-320) |
| SecY F328amb_for | 5′-atc atc ttc tag agt ttc ttc tac acg-3′ |
| SecY Y332amb_for | 5′-atc atc ttc ttc agt ttc ttc tag acg gcg-3′ |
| SecY F328/Y332amb_rev | 5′-tgc aga cgc ata gag taa cac ata aag-3′ |
| Hinge region | |
| V225_for | 5′-ttg gtt gca tag tta gta ttt-3′ |
| V225_rev | 5′-cag caa cac gag gaa-3′ |
| F383_for | 5′-tat att acc tag atc agc ctg atc-3′ |
| F383_rev | 5′-cag cgc acc aac cag-3′ |
Activity Assays for pBpa Containing SecY Derivatives
The functionality of the pBpa-containing SecY constructs was analyzed by expressing them in the cold-sensitive SecY mutant E. coli SecY39 pSup-BpaRS-6TRN (8, 30). TAG stop codons were incorporated at different positions in secY using pTRc99aSecY(His)EG and the plasmid-borne copies were transformed into SecY39. Growth was monitored on LB plates at 37 or 25 °C in the absence or presence of pBpa. The growth experiments were performed in the absence of isopropyl 1-thio-β-d-galactopyranoside, because the basal expression level of SecYEG was sufficient for complementation.
In Vivo and in Vitro pBpa Cross-linking
BL21 cells carrying the plasmids pSup-BpaRS-6TRN and pTrc99a-SecY(His)EG or pTrc99a-YidC were grown according to the procedure described previously at 30 °C in the presence of 1 mm pBpa (8, 31). E. coli C43 cells carrying pSup-BpaRS-6TRN and pTrc99a-SecY(His)EG containing TAG amber stop codons in TMs 7 and 8 were grown at 37 °C in LB medium in the presence of 1 mm pBpa. After reaching the log phase, cells were induced with 0.1 mm isopropyl 1-thio-β-d-galactopyranoside and grown for 5 h at 25 °C. Cells were harvested by centrifugation, washed once with 50 mm TeaOAc buffer, pH 7.5, and resuspended in 1× PBS buffer (137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.76 mm NaH2HPO4, pH 7.6). For UV exposure, cells were transferred to a 6-well microtiter plate and UV exposed for 30 min on ice. Cells were broken in a French pressure cell and membranes were prepared and solubilized with 1% dodecyl maltoside. SecY and its cross-linking products were purified via Talon® Affinity Resin (Clontech, Mountain View, CA). Purified samples were separated on SDS-PAGE and cross-linking products were identified by immune detection and mass spectrometry. For performing the in vivo cross-linking in SecDF-depleted cells, E. coli BL325 containing pSup-BpaRS-6TRN and pTrc99a-SecY(His)EG with the amber stop codon at position Ile-91 within SecY were grown overnight on LB medium containing 0.2% arabinose. After a washing step these cells were used to inoculate a 1-liter culture containing pBpa and either 0.2% arabinose (SecDF+) or 0.2% glucose (SecDF−). Cells were grown to an A600 of 1.0 and SecY(His)EG expression was induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside. Cells continued to grow up to an A600 of 2.0 and were then harvested. UV exposure and purification of cross-linking products followed the above described protocol.
For in vitro cross-linking, inner membrane vesicles (INVs) were prepared from E. coli cells expressing stop codon containing YidC or SecY derivatives in the presence of 1 mm pBpa as described previously (20). INV (4 μg/μl in INV buffer (100 mm TeaOAc, pH 7.5, 250 mm sucrose, 1 mm DTT)) were cross-linked on ice by UV irradiation for 20 min, followed by metal-affinity purification as described above. Cross-linking products were detected by Western blotting and mass spectrometry. Where indicated, puromycin was added to a final concentration of 2 mm. For in vitro cross-linking in the presence of RNCs, N terminally His-tagged RNCs carrying the first 102 amino acids of FtsQ followed by an HA tag and a TnaC stalling sequence were expressed in vivo and purified essentially as described (32). 70S ribosomes were purified via sucrose gradient centrifugation as described (20). INVs were incubated with ribosome-associated nascent chains in INV buffer adjusted to pH 8.0 and supplemented with 5 mm Mg(Ac)2 on ice and UV irradiated for 20 min. The reaction mixture was then solubilized with 1% dodecyl maltoside and SecY cross-linking products were purified via Talon affinity resin and analyzed by Western blotting or mass spectrometry.
Mass Spectrometry
Lanes of an SDS-PAGE of YidC(K249pBpa) were cut into slices, subjected to in-gel digestion using trypsin, and analyzed by nano-HPLC-ESI-MS as described (8). Analysis of the remaining samples (YidC(L540pBpa), SecY(I91pBpa), SecY(L127pBpa), and SecY(L320pBpa)) was performed using an UltiMate 3000 RSLCnano/LTQ-Orbitrap XL system (Thermo Fisher Scientific, Bremen, Germany) essentially as described (33), except for the use of a 5-min washing step on the pre-column and a linear gradient ranging from 5 to 40% solvent B (0.1% (v/v) formic acid in 86% (v/v) acetonitrile) in 30 min for the analytical HPLC. Proteins were identified by database searches against E. coli protein sequences deposited at the UniProt database (release 2011_02), scored, and quantified as described in Ref. 20. For SecY(I91pBpa), SecY(L127pBpa), and SecY(L320pBpa) INV and whole cells, the mass spectrometric data were analyzed using the MaxQuant program (34, 35) as described (33) and the organism-specific UniProt database (The UniProt Consortium, 2011, version 2012_09) for E. coli (taxonomy: 83333, keywords: 181 and 1185).
In Vitro Synthesis and Transport
Proteins were in vitro synthesized and radioactively labeled using a CTF cell extract as described (20). The [35S]methionine/[35S]cysteine labeling mix was obtained from PerkinElmer Life Sciences. Proteoliposomes (comprised of E. coli polar lipids and purified proteins at a concentration of 0.4 mg/ml) were prepared as described previously (20, 31). Transport of in vitro synthesized proteins was carried out for 30 min at 37 °C. After synthesis, the transport assay was split in two parts. One part was directly TCA precipitated, whereas the other part was digested for 25 min at 25 °C using 0.5 mg/ml of proteinase K, followed by TCA precipitation and SDS-PAGE.
RESULTS
YidC Contacts Both Sides of the Lateral Gate of SecY
To determine contacts between SecY and YidC, we used an in vivo site-directed cross-linking approach employing the UV-activated phenylalanine derivative para-benzoyl-l-phenylalanine (pBpa) (29). Amber stop codons (TAG) were incorporated at different positions in the SecY transmembrane domains that constitute the lateral gate (Fig. 1A). These constructs were then expressed in cells that carried a plasmid-borne orthogonal aminoacyl-tRNA synthetase/tRNACUA pair (29) and that were grown in pBpa-supplemented media. This resulted in the incorporation of pBpa at the amber stop codon position. The functionality of the pBpa-containing SecY derivatives was verified by testing their ability to suppress the cold-sensitive phenotype of the conditional E. coli SecY39 mutant strain (30; Fig. 1B). SecY39 cells expressing SecY derivatives carrying the TAG codon at different positions within TM2b revealed a growth defect when grown at 25 °C in the absence of pBpa, but grew like cells expressing wild type SecY when pBpa was present in the medium (Fig. 1B). This demonstrated that the incorporation of pBpa at the indicated positions of SecY did not significantly interfere with SecY activity. These assays were also performed with the other TAG-containing SecY derivatives and gave similar results (data not shown).
Wild type E. coli cells expressing the tRNA synthetase/tRNACUA pair and the amber stop codon containing SecY derivatives were then grown in the presence of pBpa and exposed to UV light. Cells were fractionated and SecY and its cross-linked partner proteins were partially purified via a C-terminal His6 tag on SecY. The purified material was analyzed by immune detection using α-YidC antibodies. The specificity of the cross-linking reaction was controlled by purifying wild type SecY lacking pBpa. For both wild type SecY and the pBpa containing SecY derivatives, we observed that YidC co-purified with SecY (Fig. 2A). Co-purification of SecY and YidC was observed before and provided the first indication for a SecY-YidC interaction (26).
FIGURE 2.
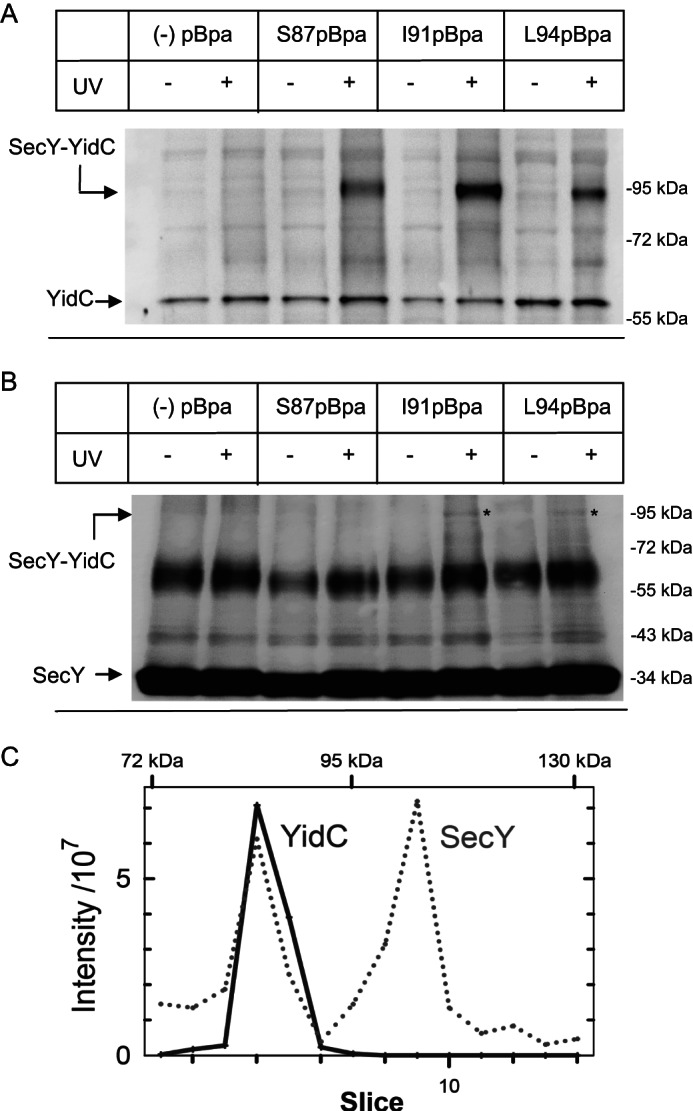
YidC is in contact with helix 2b of the lateral gate of SecY. A, E. coli BL21 cells expressing either wild type SecY (−pBpa) or SecY derivatives carrying pBpa at positions Ser-87, Ile-91, or Leu-94 in TM 2b of SecY were harvested and exposed to UV light for activating pBpa. One aliquot was not UV exposed and served as control. Subsequently, cells were fractionated and SecY/SecY cross-linking products were purified via metal affinity chromatography and separated on SDS-PAGE (approximately 100 μg of protein) followed by immune detection using α-YidC antibodies. Indicated is the SecY-YidC cross-linking product and YidC that co-purified with His-tagged SecY. B, the same material as in A was decorated with α-SecY antibodies, raised against an N-terminal SecY peptide. The putative SecY-YidC cross-linking product is indicated (*). C, a non-UV irradiated (not cross-linked, −UV) and a UV irradiated (cross-linked, +UV) sample of SecY(I91pBpa) purified from whole cells was separated on a 5–15% SDS gel and the proteins were visualized by Coomassie staining. The −UV and +UV lanes were cut into equal slices followed by in-gel trypsin digestion and mass spectrometry. Shown are intensity profiles of SecY (dotted line) and YidC (solid line) peptides found in the individual gel slices. The intensity values were corrected for the values obtained in the absence of UV treatment.
For the SecY pBpa mutants at positions 87, 91, or 94 in TM2b, we observed a UV-specific cross-linking product at approximately 90 kDa that was recognized by α-YidC antibodies (Fig. 2A). In particular incorporating pBpa at position Ile-91 of SecY resulted in a very strong cross-linking product. The 90-kDa band was strictly UV-dependent and only observed with pBpa containing SecY derivatives and not detectable in cells expressing wild type SecY (Fig. 2A). The mass of the cross-linking product would be consistent with the predicted mass of a cross-link between SecY (migrating at approximately 34 kDa on SDS-PAGE, despite its predicted molecular mass of 48 kDa) and YidC (migrating at approximately 60 kDa). α-SecY antibodies detected the non-cross-linked SecY at approximately 34 kDa but also a cross-reacting UV-independent band at approximately 65 kDa (Fig. 2B). However, the putative 90-kDa SecY-YidC cross-linking product was only weakly detectable by α-SecY antibodies in the I91pBpa and L94pBpa samples (Fig. 2B (*)) and was undetectable in the S87pBpa sample. The α-SecY antibodies were raised against an N-terminal SecY peptide and the weak detection of the cross-linking product could reflect a partial shielding of this SecY epitope in the cross-linking product. We therefore performed mass spectrometry (MS) for obtaining independent proof that the 90-kDa band corresponded to a SecY-YidC cross-linking product. Purified SecY(I91pBpa) from UV-treated and untreated cells was separated on SDS-PAGE and individual gel slices were subjected to MS. Intensity profiles of the gel area above the 72-kDa marker band showed that SecY peptides were found in two prominent peaks with maximum intensities corresponding to molecular masses of approximately 85 and 105 kDa (Fig. 2C, dotted line; Table 2). A third minor peak was detectable at approximately 115 kDa. YidC peptides were found in a single peak with a maximum intensity also at approximately 85 kDa (Fig. 2C, solid line; Table 2), confirming that the approximate 90-kDa band detected by α-YidC antibodies on SDS-PAGE corresponded to a specific UV-dependent SecY-YidC cross-linking product. The 105-kDa peak contained 32 peptides of the 68-kDa membrane-bound chaperone PpiD, resulting in sequence coverage of more than 66% (Fig. 2C, Table 2). A SecY-PpiD cross-linking product at approximately 105 kDa was confirmed by Western blotting using α-PpiD antibodies (data not shown). Calculating the relative intensities (−UV/+UV) of the SecY-YidC and SecY-PpiD cross-linking products showed that they were strictly UV-dependent (Table 2). However, the significance of the SecY-PpiD interaction was not further analyzed in this study. The observation that one SecY position can be cross-linked to two different proteins in vivo has been observed before (5, 8, 20) and probably reflects the heterogeneity of the SecYEG populations present in E. coli cells. The minor peak at 115 kDa contained peptides of several proteins like SecG, SlyD, RhlE, or CusS. The SecY peptides found in this band were not strictly UV-dependent, because their relative intensity was above “0” and therefore the possible roles of the identified proteins were not further analyzed. A complete list of proteins that either co-purified with SecY in these assays or cross-linked to SecY(I91pBpa) can be found in supplemental Table S1.
TABLE 2.
Mass spectrometric analyses of the SecY(I91pBpa) cross-linking products
A non-UV irradiated (not cross-linked, −UV) and a UV-irradiated (cross-linked, +UV) sample of SecY(I91pBpa) purified from whole cells was separated on 5–15% SDS gels and the proteins were visualized by Coomassie staining. The−UV and +UV lanes were cut into equal slices followed by in-gel trypsin digestion and mass spectrometry. Shown are the quantification for SecY, YidC, and the membrane-bound chaperone PpiD.
| Proteina | Molecular massb | Gel molecular massc | Relative intensity (−UV/+UV)d | Coveragee | Peptidesf |
|---|---|---|---|---|---|
| kDa | |||||
| YidC | 61.5 | 85 | 0.00 | 24.8 | 13 |
| PpiD | 68.1 | 103.6 | 0.00 | 66.5 | 32 |
| SecY | 48.5 | 85 | 0.04 | 23.5 | 11 |
| 105 | 0.02 | 18.3 | 8 | ||
| 115 | 0.26 | 16.7 | 7 | ||
a Protein identified.
b Calculated molecular mass.
c Molecular mass of gel slice determined by extrapolation.
d Relative intensity observed in gel slices from the control lane (−UV) compared to the +UV lane.
e Sequence coverage of total sequence by detected peptides.
f Number of peptides detected.
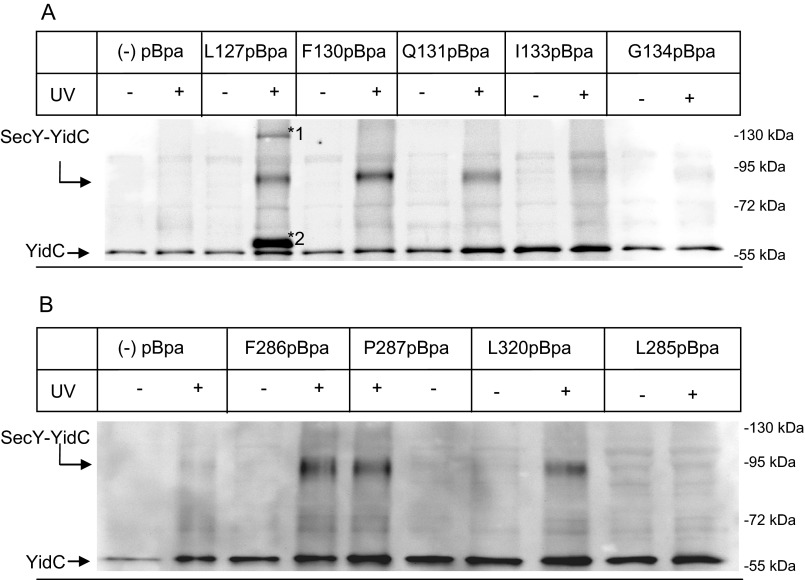
To determine whether YidC also contacts TM 3 located on the same side of the lateral gate as TM 2b, we analyzed five residues within this TM and found the 90-kDa cross-linking product for positions 127, 130, and 131 (Fig. 3A). Positions 133 and 134 gave only a very faint SecY-YidC cross-linking product (Fig. 3A). Surprisingly, for position 127, two additional UV-dependent bands at approximately 65 and 130 kDa were detected by α-YidC antibodies (Fig. 3A, *1 and *2)). Mass spectrometry confirmed the presence of UV-dependent YidC peptides at 65, 92, and 130 kDa (Table 3). In addition, we observed UV-independent YidC peptides at 55 kDa, which corresponds to YidC that co-purified with SecY (Table 3). The 130-kDa band could correspond to a (SecY)2-YidC complex and the 65-kDa band to a proteolytically degraded SecY-YidC cross-linking product. However, as both bands were only observed for position 127, this was not further investigated in the current study.
FIGURE 3.
YidC is also in contact with TM 3, 7, and 8 of the lateral gate of SecY. A, cells expressing either wild type SecY (−pBpa) or derivatives carrying pBpa at different positions within TM 3 were harvested, exposed to UV light, and further treated as described in the legend to Fig. 2A. Two additional UV-dependent bands were recognized by α-YidC antibodies for SecY(L127pBpa) and are indicated (*1 and *2). B, as in A but different residues within TM 7 (Phe-286, Pro-287, and Leu-285) and one residue within TM 8 (Leu-320) were analyzed. Note that due to their low expression in E. coli BL21, TM 7, and TM 8 SecY(pBpa) derivatives were expressed in E. coli C43.
TABLE 3.
Mass spectrometric analyses of the SecY(L127pBpa) cross-linking products
A non-UV irradiated (not cross-linked, −UV) and a UV-irradiated (cross-linked, +UV) sample of SecY(L127pBpa) purified from whole cells was separated on 5–15% SDS gels and the proteins were visualized by Coomassie staining. Gel areas around the bands that were detected by α-YidC antibodies (Fig. 3A) were cut out from the −UV and +UV lanes. Following in-gel trypsin digestion, the samples were analyzed by mass spectrometry. Shown is the quantification for YidC peptides.
| Proteina | Molecular massb | Gel molecular massc | Relative intensity (−UV/+UV)d | Coveragee | Peptidesf |
|---|---|---|---|---|---|
| kDa | |||||
| YidC | 61.5 | 55 | 0.60 | 46.7 | 17 |
| 65 | 0.01 | 50.2 | 22 | ||
| 92 | 0.00 | 34.1 | 11 | ||
| 130 | 0.00 | 27.6 | 12 | ||
a Protein identified.
b Calculated molecular mass.
c Molecular mass of gel slice determined by extrapolation.
d Relative intensity observed in gel slices from the control lane (−UV) compared to the +UV lane.
e Sequence coverage of total sequence by detected peptides.
f Number of peptides detected.
TM 7 and TM 8 constitute the other side of the lateral gate and the 90-kDa SecY-YidC cross-linking products were observed for positions 286 and 287 within TM 7 (Fig. 3B) and for position 320 within TM 8 (Fig. 3B). Additional positions like 285 within TM 7 (Fig. 3B) were tested but showed no or only very weak SecY-YidC cross-links (Table 4). The specificity of the SecY-YidC interaction was further tested by incorporating pBpa in the loop that connects TM 5 and TM 6 at the back of SecY. The positions (Phe-383 and Val-225) were selected because they were not shielded by SecE and therefore accessible to YidC if it were located at the back of SecY. However, no cross-links to YidC were observed for either position (Table 4). This indicates that YidC specifically contacts the lateral gate of SecY. In summary, we demonstrate that YidC is located in close proximity to the front of the SecYEG translocon where it makes multiple contacts to all four TMs that constitute the lateral gate of SecY (Table 4).
TABLE 4.
Summary of SecY positions that cross-link to YidC as determined by Western blotting using α-YidC antibodies
“+” indicates a strong cross-linking product and “−“ that no cross-link was observed. “(+)” indicates a weak cross-linking product.
| SecY region | pBpa position | SecY-YidC cross-link |
|---|---|---|
| TM 2b | Ser-87 | + |
| Ile-90 | (+) | |
| Ile-91 | + | |
| Leu-94 | + | |
| TM 3 | Leu-127 | + |
| Phe-130 | + | |
| Gln-131 | + | |
| Ile-133 | (+) | |
| Gly-134 | (+) | |
| TM 7 | Leu-283 | − |
| Leu-285 | − | |
| Phe-286 | + | |
| Pro-287 | + | |
| Ile-290 | − | |
| TM 8 | Tyr-332 | − |
| Phe-328 | − | |
| Leu-320 | + | |
| Hinge region | Val-225 | − |
| Phe-383 | − |
YidC Interacts with the Lateral Gate of SecY Independently of the SecDFYajC Complex
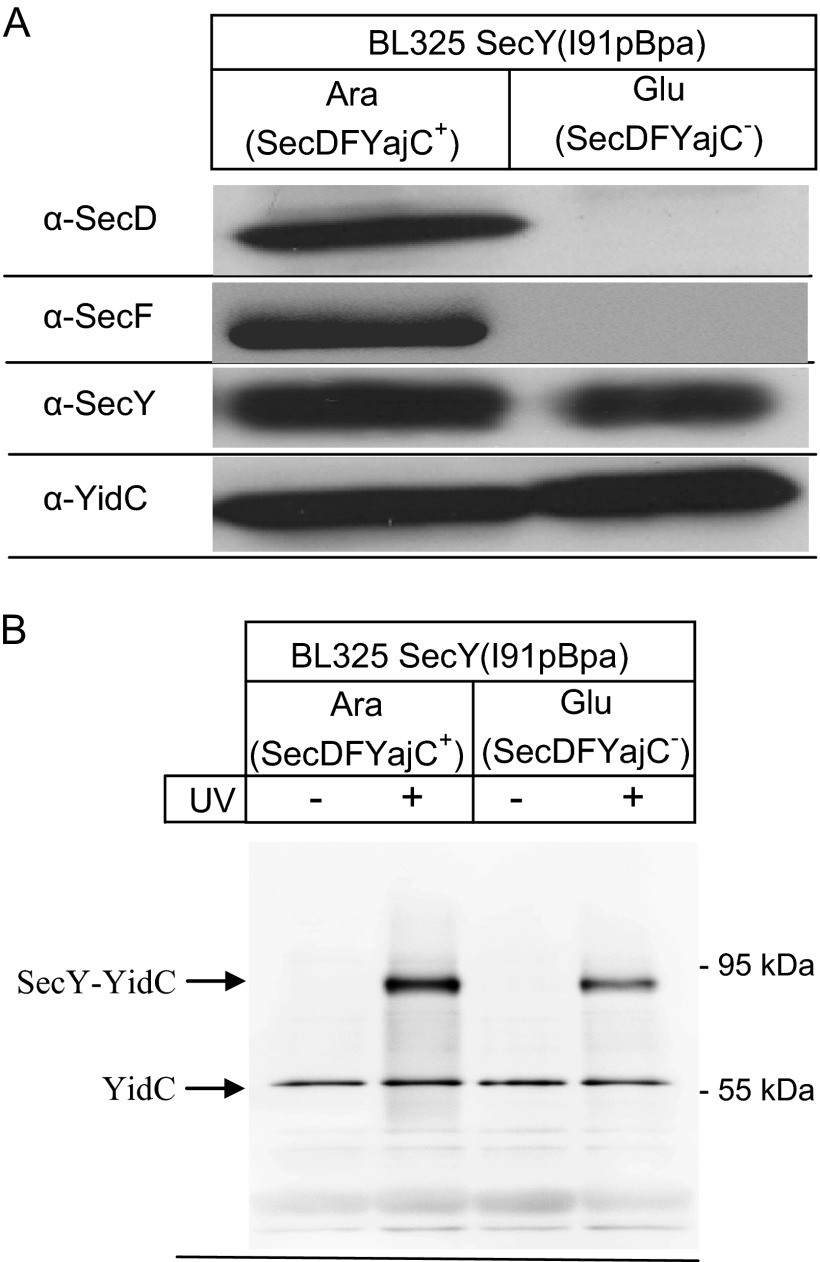
Previous studies have suggested that YidC interacts with SecY via the SecDFYajC membrane protein complex (13, 14, 36). We therefore analyzed the cross-linking pattern in the conditional SecDF depletion strain BL325, which contains secDF under control of an arabinose inducible promoter (9). BL325 cells carrying the orthogonal aminoacyl-tRNA synthetase/tRNACUA pair and expressing SecY(I91pBpa) were grown in arabinose or glucose containing media and the SecDF content in membranes was analyzed by Western blotting. SecDF, SecY, and YidC were detectable in membranes of arabinose-grown cells, but SecDF was undetectable in membranes of glucose-grown BL325 cells (Fig. 4A). The steady-state amounts of SecY or YidC were not significantly influenced by SecDF depletion (Fig. 4A). SecDF-containing and SecDF-depleted BL325 cells were then UV exposed for activating pBpa and SecY and its cross-linking products were subsequently purified. The 90-kDa SecY-YidC cross-linking product was detectable both in the presence of SecDF and in its absence. These data demonstrate that YidC can interact with the lateral gate of SecY independently of the SecDFYajC complex. Nevertheless, the 90-kDa band was slightly weaker in the absence of SecDF (Fig. 4B). This could indicate that SecDFYajC stabilizes or enhances the SecY-YidC interaction without being permanently associated with the assembled SecYEG-YidC complex. This would also be in agreement with previous Blue Native PAGE analyses (27).
FIGURE 4.
The YidC contact to the lateral gate is maintained in the absence of SecDFYajC. A, the conditional SecDFYajC depletion strain E. coli BL325 expressing SecY(I91pBpa) was grown either in the presence of arabinose for inducing SecDFYajC expression or in the presence of glucose for SecDFYajC depletion. After reaching an A600 of 2.0, cells were harvested and INV were isolated by differential centrifugation. INV of both cultures were then separated on SDS-PAGE (approximately 10 μg of protein) and after Western transfer decorated with antibodies against SecD, SecF, SecY, and YidC. B, SecDFYajC containing and SecDFYajC-depleted BL325 cells, which expressed SecY(I91pBpa) were grown as described in A and subsequently kept either in the dark or UV exposed. After cell breakage, SecY was purified as described in the legend to Fig. 2. Approximately 100 μg of protein was loaded on SDS-PAGE and decorated with α-YidC antibodies after Western transfer.
The large periplasmic loop (P1) of YidC has been proposed to interact with the SecDFYajC complex and residues 215–265 within the periplasmic loop were suggested to bind to the SecF subunit (36). We therefore incorporated pBpa into position 249 of the periplasmic loop of YidC and purified YidC(249pBpa) after UV exposure of whole cells. Mass spectrometry revealed cross-links to YajC, SecD, and also to SecG and signal peptidase I (Lep) (Table 5). The relative intensity of the SecD peptides indicated that their presence in the 151-kDa band was not strictly UV-dependent (Table 5), but at least partially also the result of co-purification. Although these data confirmed that YidC is in close proximity to the YajC and SecD subunits of the SecDFYajC complex, we did not find cross-links to SecF. This might be related to the low abundance of the SecDFYajC complex in E. coli cells, which is estimated to exist in only approximately 40 copies per cell (10). We therefore switched to an in vitro cross-linking approach using INV from cells expressing YidC(L540pBpa) carrying pBpa at its C terminus. By using these INV we previously succeeded in detecting cross-links to ribosomal proteins, which we did not detect by the in vivo cross-linking method (20). When this approach was applied and YidC(L540pBpa) purified after UV exposure, cross-links between YidC(L540pBpa) and SecF, YajC, and signal peptidase I were identified by mass spectrometry in two independent experiments (Table 5). The presence of SecF peptides in the 84-kDa band was not strictly UV-dependent (Table 5), but partially also related to co-purification. These data confirm a YidC-SecDFYajC interaction, although this interaction appears not to be essential in tethering YidC to the lateral gate of SecY (Fig. 3B). The MS data also identified a cross-link between YidC(L540pBpa) and SecY (Table 5). This provides additional proof for the YidC-SecY interaction and demonstrates that the C terminus of YidC interacts with SecY.
TABLE 5.
Mass spectrometric analyses of the YidC-SecDFYajC interaction
For in vivo cross-linking, a non-UV irradiated (not cross-linked, −UV) and a UV irradiated (cross-linked, +UV) sample of YidC(K249pBpa), purified from whole cells was separated on a 5–15% SDS gels and the proteins were visualized by Coomassie staining. The −UV and +UV lanes were cut into equal slices followed by in-gel trypsin digestion and mass spectrometry. Only those proteins are listed that showed a UV-dependent mass shift, indicative of specific cross-linking. For in vitro cross-linking, INV from cells expressing YidC(L540pBpa) were isolated via sucrose gradient centrifugation and subsequently UV-exposed. Non-UV irradiated INV served as control.
| Proteina | Molecular massb | Gel molecular massc | Relative Intensity (−UV/+UV)d | Coveragee | Peptidesf |
|---|---|---|---|---|---|
| kDa | |||||
| YidC(K249pBpa) in vivo cross-links | |||||
| YajC | 11.9 | 65.5 | 0.00 | 11.8 | 1 |
| SecD | 66.6 | 151 | 0.19 | 23.3 | 11 |
| SecG | 11.4 | 67.4 | 0.00 | 16.4 | 1 |
| Lep | 35.9 | 125.4 | 0.00 | 34.6 | 12 |
| YidC(L540pBpa) in vitro cross-links | |||||
| YajC | 11.9 | 69.9 | 0.00 | 46.4 | 4 |
| SecF | 35.4 | 83.6 | 0.24 | 24.5 | 9 |
| SecY | 48.5 | 91.9 | 0.04 | 15.6 | 7 |
| Lep | 35.9 | 91.9 | 0.04 | 16.7 | 5 |
a Protein identified.
b Calculated molecular mass of the identified protein.
c Molecular mass of the cross-linking product as determined by extrapolation.
d Relative intensity observed in gel slices from the control lane (−UV) compared to the +UV lane.
e Sequence coverage of total sequence by detected peptides.
f Number of peptides detected.
The SecY-YidC Interaction Does Not Require the Presence of a Substrate
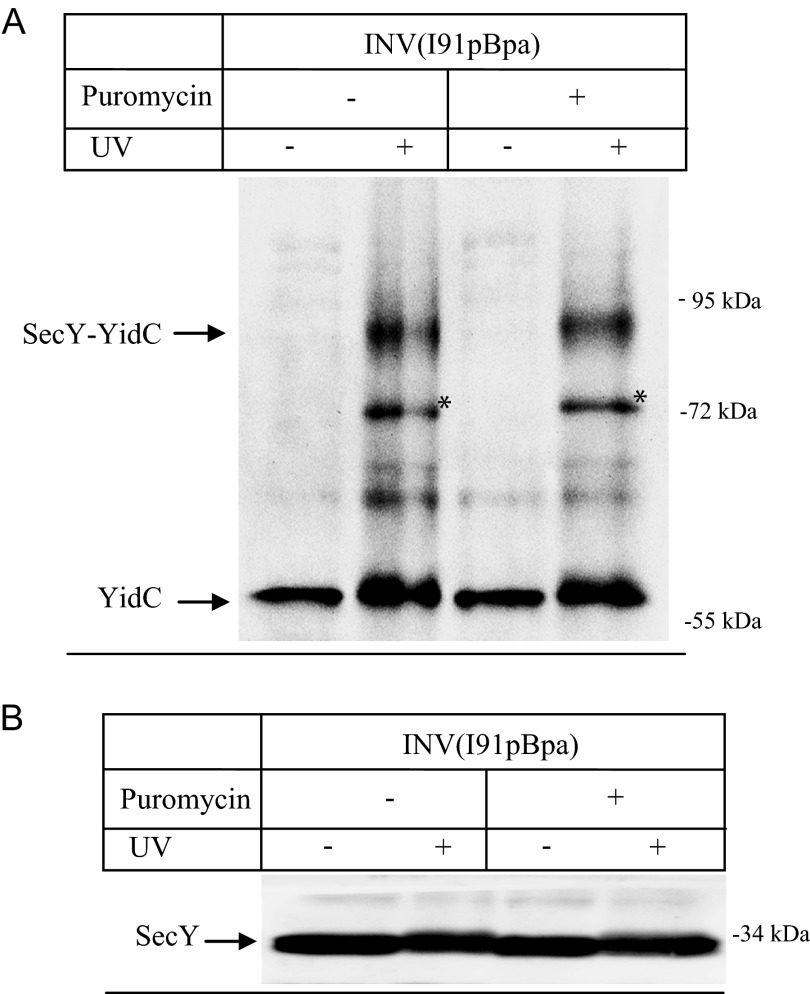
The insertion of YidC into the inner membrane of E. coli is SecY dependent (37) and the in vivo SecY-YidC cross-link could therefore correspond to YidC that is in the process of being co-translationally inserted via SecY. To exclude this possibility, we performed the cross-linking in vitro, which allowed us to perform cross-linking with actively transporting or with resting SecYEG translocons. INV of E. coli cells expressing SecY(I91pBpa) were isolated and purified via sucrose gradient centrifugation. These INV were subsequently UV exposed for activating pBpa and SecY and its cross-linking partner was purified and separated on SDS-PAGE. After Western transfer, the 90-kDa SecY-YidC cross-linking product was detected (Fig. 5A), indicating that the SecY-YidC interaction is maintained in purified INV. Although we considered it unlikely that actively translating ribosomes were still attached to these INV, we also treated them with the antibiotic puromycin before UV exposure. The tRNA analog puromycin dissociates translating ribosomes (7, 38), but this treatment had no significant effect on the occurrence of the SecY-YidC cross-linking product (Fig. 5A). Probing these samples with α-SecY antibodies demonstrated that equal amounts of INV were loaded in these experiments (Fig. 5B). α-YidC antibodies recognized an additional UV-dependent band at approximately 72 kDa, which was also not influenced by puromycin treatment. This band was also detectable in SecY(F130pBpa) and SecY(L320pBpa) INV (cf. Figs. 6 and 7). MS analysis on SecY(L320pBpa) INV that were either UV exposed or kept in the dark confirmed that the 72-kDa band also corresponded to a YidC-SecY cross-link (Table 6). In summary, these data indicate that the SecY-YidC cross-linking product does not represent YidC that is being inserted via SecY. These experiments also show that YidC is not specifically recruited to the lateral gate of actively transporting SecYEG translocons but instead it binds also to SecY in the absence of a substrate.
FIGURE 5.
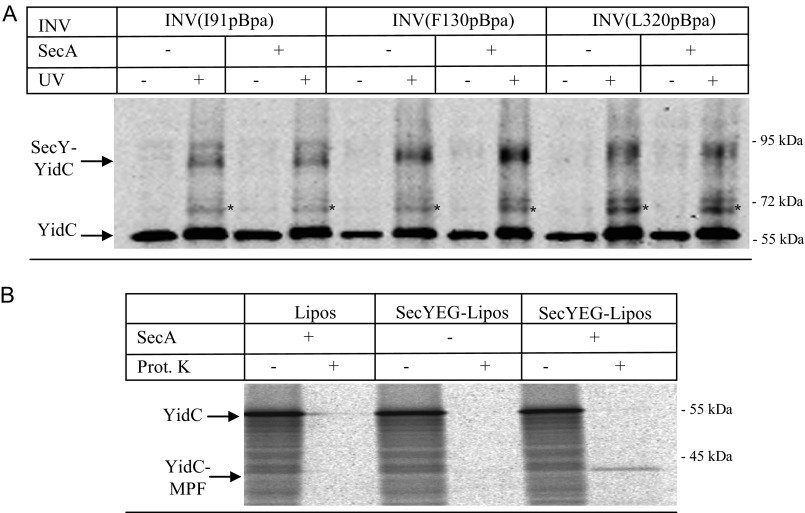
The SecY-YidC interaction does not require the presence of a substrate. A, INVs from BL21 SecY(I91pBpa) cells were isolated by sucrose gradient centrifugation and either UV exposed or kept in the dark. When indicated, puromycin (final concentration 2 mm) was added to dissociate translating ribosomes and the INV were incubated for 15 min at 37 °C before UV treatment. Subsequently, SecY was purified as described in the legend to Fig. 2. Samples were separated on a 5–20% SDS-PAGE, and after Western transfer, the blot was decorated with α-YidC antibodies. Asterisk (*) indicates a second UV-dependent SecY-YidC cross-linking product at approximately 72 kDa that was only observed when the cross-linking was performed in vitro (Table 6). B, the same material as in A was probed with antibodies against SecY to control that equal amounts of protein were loaded.
FIGURE 6.
The SecY-YidC interaction is not influenced by SecA-binding to SecY. A, INVs from BL21 or C43 cells expressing SecY derivatives with pBpa incorporated at the indicated positions were isolated by sucrose gradient centrifugation and adjusted to a final SecY concentration of approximately 6 μm. These INV were incubated with either 6 μm purified SecA or with buffer (−) and UV exposed or kept in the dark. Subsequently, SecY and its cross-linking products were purified via metal affinity chromatography. INVs were separated on SDS-PAGE and after Western transfer decorated with α-YidC antibodies. Asterisk (*) indicates the second UV-dependent SecY-YidC cross-linking product at 72 kDa. B, the functionality of SecA was tested with reconstituted proteoliposomes. YidC was in vitro synthesized in the presence of either liposomes (Lipos) or liposomes reconstituted with SecYEG. When indicated, SecA was present during in vitro synthesis and transport. After 30 min of synthesis at 37 °C, the samples were split in half and one part was directly precipitated with trichloroacetic acid (TCA) (−). The other half was first treated for 20 min at 25 °C with 0.5 mg/ml of proteinase K (Prot. K) and only then TCA precipitated. YidC and its membrane-protected fragment (YidC-MPF) are indicated. The YidC-MPF corresponds to the first two transmembrane domains of YidC plus the connecting 325-amino acid long periplasmic loop (37).
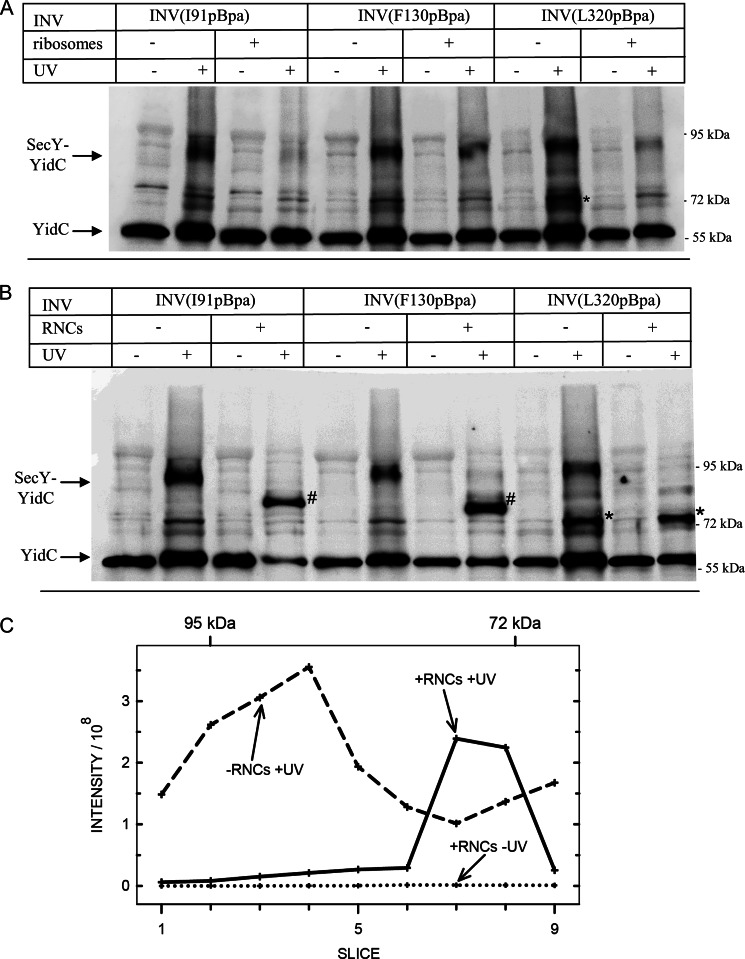
FIGURE 7.
Ribosomes and ribosome nascent chains induce conformational changes at the SecY-YidC interface. A, the INVs described in the legend to Fig. 6 (final SecY concentration 6 μm) were incubated with 6 μm sucrose gradient-purified E. coli ribosomes. After UV exposure, SecY and its cross-linking products were purified via metal affinity chromatography and the purified material was separated on SDS-PAGE, blotted, and decorated with α-YidC antibodies. The 72-kDa band labeled with an asterisk (*) indicates the second UV-dependent SecY-YidC cross-linking product at 72 kDa. B, INVs as in A were incubated with 6 μm affinity purified FtsQ-RNCs of 102 amino acids in length and UV exposed. Indicated is the 92-kDa SecY-YidC cross-linking product and the co-purifying YidC. The bands labeled with number signs (#) correspond to SecY-YidC cross-linking products, which specifically appear in the presence of RNCs. The band labeled with an asterisk (*) corresponds to the 72-kDa SecY-YidC cross-linking product. C, integrated MS intensity of YidC plotted against the position of the gel slice of a SDS-PAGE in the molecular mass range between approximately 100 and 70 kDa. Purified SecY(I91pBpa) UV treated in the presence (solid line, +RNCs + UV) or absence (dashed line, -RNCs + UV) of RNCs. As control, the analysis of SecY(I91pBpa) INV, which were incubated with RNCs but not UV-treated (dotted line, +RNCs −UV), is shown.
TABLE 6.
Mass spectrometric analyses of the SecY(L320pBpa) cross-linking products
A non-UV irradiated (not cross-linked, −UV) and a UV-irradiated (cross-linked, +UV) sample of SecY(L320pBpa) INV were purified, separated on 5–15% SDS gels and the proteins were visualized by Coomassie staining. Gel areas around the bands that were detected by α-YidC antibodies (Fig. 7B) were cut out from the −UV and the +UV lanes. Following in-gel trypsin digestion, the samples were analyzed by mass spectrometry. Shown is the quantification for YidC peptides.
| Proteina | Molecular massb | Gel molecular massc | Relative intensity (−UV/+UV)d | Coveragee | Peptidesf |
|---|---|---|---|---|---|
| kDa | |||||
| YidC | 61.5 | 72 | 0.02 | 21.2 | 7 |
| 92 | 0.07 | 21.7 | 7 | ||
a Protein identified.
b Calculated molecular mass.
c Molecular mass of gel slice determined by extrapolation.
d Relative intensity observed in gel slices from the control lane (−UV) compared to the +UV lane.
e Sequence coverage of total sequence by detected peptides.
f Number of peptides detected.
The SecY-YidC Interaction Is Not Influenced by SecA Binding to SecY
SecA, the motor protein of the post-translational protein transport is suggested to induce an opening of the lateral gate upon binding to SecY (39). We therefore analyzed whether the addition of SecA would influence the SecY-YidC interaction. INV containing SecY(I91pBpa) at a concentration of 6 μm were incubated with equimolar concentrations of SecA and subsequently UV exposed. Buffer-treated INV served as control. INV were then solubilized and SecY and its cross-linking products were purified by metal affinity chromatography. The 90-kDa SecY-YidC cross-linking product was detectable both in the presence and absence of SecA (Fig. 6A), indicating that the interaction between TM 2b of the lateral gate and YidC is not significantly influenced when SecA is bound to SecYEG. The addition of SecA also did not significantly influence the interaction of YidC with TM 3 (SecY(F130pBpa) or TM 8 (SecY(L320pBpa) (Fig. 6A). Independently of the SecA addition, the weaker 72-kDa cross-linking product was also recognized in all samples.
For excluding the possibility that the SecA used in these assays was non-functional, we performed in vitro transport assays using reconstituted SecYEG-proteoliposomes. YidC was chosen as substrate for in vitro protein transport because the translocation of its large periplasmic loop requires a functional SecA-SecY interaction (37). When YidC was synthesized in the presence of SecA and SecYEG proteoliposomes, a 42-kDa membrane-protected fragment of YidC (YidC-MPF) was observed after proteinase K treatment (Fig. 6B). This fragment corresponds to the first two transmembrane domains of YidC and the connecting periplasmic loop and is indicative for complete membrane insertion (37). In contrast, no protease protection of YidC was observed when SecA was omitted or when SecA was added to liposomes lacking SecYEG (Fig. 6B). Thus, the SecA used in these studies is active and our data therefore demonstrate that binding of SecA to SecYEG and the associated lateral gate movements (39, 40) do not significantly influence the SecY-YidC interaction.
Differential Effects of Ribosomes and Ribosome Nascent Chains on the SecY-YidC Interaction
Lateral gate movements have also been suggested to occur during ribosome or RNC binding to SecYEG (3, 41). These movements are probably required for allowing a transmembrane domain to exit into the lipid phase. When SecY(I91pBpa) INV were incubated with 6 μm sucrose gradient-purified 70 S ribosomes, the 90-kDa SecY-YidC cross-linking product almost completely disappeared (Fig. 7A). Further analyses using SecY(F130pBpa) within TM 3 and SecY(L320pBpa) within TM 8 revealed that for these two TMs the SecY-YidC interaction became significantly weaker in the presence of ribosomes. However, the contacts of YidC to TM 3 and TM 8 of SecY did not completely vanish upon ribosome binding (Fig. 7A). The UV-dependent band at 72 kDa was strongest for positions Leu-320, but became significantly weaker in the presence of ribosomes and thus showed the same pattern as the 90-kDa cross-linking band.
In conclusion, our data show that binding of non-translating ribosomes to the SecYEG translocon induces a significant weakening of the SecY-YidC interaction. In particular, TM 2b loses contact to YidC in the presence of ribosomes.
We next tested the influence of FtsQ RNCs on the SecY-YidC interaction. FtsQ is a single spanning membrane protein that is targeted by the SRP pathway to SecY and has been shown to contact YidC during insertion (21). The strongest YidC interaction was observed for RNCs of a length between 97 and 108 amino acids (21). We therefore used purified FtsQ-RNCs of 102 amino acid length and incubated them with INV at a RNC:SecY ratio of 1:1. The addition of RNCs to SecY(I91pBpa) INV resulted in a complete loss of the 90-kDa cross-linking product (Fig. 7B). However, in the presence of FtsQ-RNCs an additional band at approximately 80 kDa became visible (Fig. 7B). The disappearance of the 90-kDa SecY-YidC cross-link upon RNC addition and the appearance of the 80-kDa band was also observed for position Phe-130 (Fig. 7B). These additional bands were not detected in the presence of non-translating ribosomes or SecA, suggesting that they specifically resulted from RNC binding to SecY. Adding RNCs to SecY(L320pBpa) INV also induced the loss of the 90-kDa cross-linking product, but no additional band emerged (Fig. 7B). Finally, the 72-kDa YidC cross-linking product that disappeared in the presence of non-translating ribosomes was retained in the presence of RNCs (Fig. 7B).
The strong recognition of the additional bands at 80 and 72 kDa by α-YidC antibodies and their UV dependence suggested that they also corresponded to a SecY-YidC cross-linking product. This was further verified by mass spectrometry. SecY(I91pBpa) INV were separated on SDS-PAGE after UV treatment in the presence or absence of FtsQ-RNCs. Subsequently, the gel region between approximately 100 and 70 kDa was cut into slices, each of which was subjected to in-gel digestion using trypsin followed by MS analysis. Fig. 7C shows intensity profiles of YidC after UV-induced cross-linking in the presence (continuous line) or absence of FtsQ-RNCs (dashed line). As an additional control, the intensity profile of SecY(I91pBpa) INV incubated with FtsQ-RNCs but UV untreated is included (dotted line). This latter profile shows no visible intensity in the entire mass range, demonstrating that all YidC peptides monitored in our analyses originated from YidC that was cross-linked to SecY. In the absence of RNCs, the SecY-YidC cross-link was mainly detected in the 95–85-kDa range, corresponding to the 90-kDa SecY-YidC band that is detectable by Western blotting. In addition, we observed slightly increased intensities between 70 and 72 kDa, which likely reflect the UV-dependent band at 72 kDa and is also weakly detectable in SecY(I91pBpa) INV (Fig. 7B). Upon addition of RNCs, YidC peptides were exclusively detected in a new peak at approximately 80 kDa. The peak at approximately 90 kDa and the increased intensity at approximately 72 kDa completely disappeared in the presence of RNCs, which is in agreement with the immune detection. These analyses verified that the 80-kDa band that is observed upon addition of RNCs also correspond to SecY-YidC cross-linking products. MS did not provide any indication that the faster migrating SecY-YidC cross-linking products corresponded to degraded YidC. We identified identical YidC peptides in both samples (±RNC), resulting in a sequence coverage for YidC of 47% in each case. Thus, it is likely that SecY-YidC cross-linking products exhibit position-dependent migration on SDS-PAGE. This has also been observed for SecY-SecA and SecY-FtsY cross-linking products (6, 8).
In summary, our data demonstrate a highly dynamic SecY-YidC interface that specifically responds to the addition of ribosomes or RNCs but not to the addition of SecA. These differences probably mirror the specific activation of the passive SecYEG channel by cytosolic ligands.
DISCUSSION
The SecYEG translocon is thought to facilitate membrane protein insertion by allowing hydrophobic TMs to exit the SecY channel via a lateral gate (42, 43). The transfer of a TM from SecY into the lipids is assisted by YidC (21, 22, 26, 44), but how YidC interacts with the SecYEG translocon is largely unknown.
Our data demonstrate that YidC is localized in front of the SecYEG translocon where it makes multiple contacts to all four TMs of the lateral gate. This position of YidC relative to SecY is significant because it provides the explanation for the length-dependent interaction of nascent membrane proteins with SecY and YidC that has been shown in previous studies (21, 22). It also provides strong support for the concept of TMs exiting the SecY channel via the lateral gate (2). Finally, the presence of YidC with its largely hydrophobic surface close to the lateral gate explains why multiple transmembrane domains can be retained close to the SecY translocon before their cooperative exit into the lipid phase (22, 45).
Structural information on YidC is so far limited to a medium-resolution projection structure of a YidC dimer (46) and high-resolution structure of the large periplasmic loop (47, 48). It was therefore unknown how YidC would interact with the Sec machinery. Previous data suggested that YidC might be tethered to SecYEG via the SecDFYajC complex (14, 36). However, our data show that the interaction between YidC and the lateral gate of SecY does not require SecDFYajC. Still, in support of a possible involvement of SecDFYajC in SecY-YidC interaction, we found a slightly reduced cross-linking efficiency in the absence of SecDFYajC. Considering the low abundance of the SecDFYajC complex in E. coli (approximately 40 copies/cell; 10) it is likely that SecDFYajC assists the YidC-SecYEG interaction without being permanently associated with the SecYEG-YidC complex. The periplasmic loop of YidC was proposed to interact with the SecDFYajC complex, which we confirm here by cross-linking position 249 within the periplasmic loop of YidC to SecD and YajC. In addition, we show that the C terminus of YidC is in contact with SecF and YajC. Importantly, SecY also cross-links to the C terminus of YidC. In previous studies it was already shown that the C terminus of YidC binds to SRP, FtsY (20), and ribosomal subunits (20, 49). The short C terminus of E. coli YidC apparently constitutes a major contact site for cytosolic and membrane integral partner proteins during protein transport.
Binding of SecA to SecY is thought to induce a pre-open state of the translocon, which results in TM 2b being separated from TM 7 and TM 8 (39, 50, 51). This leads to the formation of a hydrophobic crack through which signal sequences or transmembrane domains could exit the SecY channel. This separation appears to be crucial because protein translocation is blocked when TM 2b and TM 7 are fixed in the closed state (40). However, whether these lateral gate movements influence the SecY interaction with partner proteins was unknown. Our study shows that the SecY-YidC interaction is preserved in the SecA-induced pre-open state. In contrast, non-translating ribosomes induce a significant re-orientation of the SecY-YidC interface, resulting in the loss of the TM2b-YidC interaction and in a weakening of the YidC contacts to TM 3 and 8. Movements of helices 2A and 2B upon ribosome binding is in agreement with molecular dynamics simulations (52, 53). On the other hand, no major lateral gate movements were detected in the cryo-EM analyses of the ribosome-bound eukaryotic Sec61 complex (41). However, this cryo-EM structure could represent a post-insertion state and therefore it does not exclude ribosome-induced lateral gate movements. The different effects of ribosome or SecA binding on the SecY-YidC interface suggest that the ribosome-induced pre-activation of the SecYEG complex is different from the SecA-induced pre-open state. This is consistent with a recent study showing that the plug domain (helix 2A) of SecY responds differently to SecA-dependent secretory proteins and inner membrane proteins (54). However, considering the large binding surface of ribosomes, it is also possible that ribosome binding to a SecYEG-YidC complex induces conformational changes within YidC that lead to the loss of the SecY-YidC interaction.
The 90-kDa cross-link between YidC and TMs 2b, 3, and 8 of SecY were also lost upon RNCs binding to SecY. This provides further support for the substrate-induced lateral gate movements that were proposed based on x-ray crystallography (55) and cryo-EM studies (2, 50). However, in comparison to non-translating ribosomes, two major differences were visible in the cross-link pattern: 1) an additional cross-linking species at approximately 80 kDa appeared for TM 2b and TM 3, and 2) the 72-kDa cross-linking product of SecY(L320 pBpa) was not influenced by RNCs. These differences likely are caused by the presence of the nascent FtsQ chain, which is long enough to contact YidC (21) and could therefore retain YidC in close proximity to the SecY channel. This is probably crucial for the proposed role of YidC in folding and quality control of SecY-inserted membrane proteins (43).
The exact nature of the 72- and 80-kDa bands are currently unknown. MS analyses clearly identified them as SecY-YidC cross-links and identified the same SecY and YidC peptides as for the 90-kDa cross-linking product. It therefore seems unlikely that both bands result from proteolytic degradation of the 90-kDa SecY-YidC cross-linking product, although this cannot entirely be excluded. Previous studies have shown that SecY-SecA and SecY-FtsY cross-links exhibit a position-dependent mobility on SDS-PAGE (5, 8, 56). SecY-FtsY cross-links migrate at approximately 130 kDa when SecY is cross-linked to the NG-domain of FtsY and at approximately 160 kDa when SecY cross-links to the A-domain (8). The cross-link position most likely influences the radius of gyration, resulting in faster and slower migrating species on SDS-PAGE. The three different SecY-YidC cross-link species we observe (90, 80, and 72 kDa) could also result from this position-dependent mobility. This would further support the conclusion that the SecY-YidC interface dynamically responds to ribosome or RNC binding. With the possible exception of L127pBpa, we did not detect differently migrating SecY-YidC cross-linking species in the in vivo approach, although a fraction of SecY should be in contact with ribosomes/RNCs in living E. coli cells. Due to the unsynchronized nature and the presence of unstalled ribosomes in the in vivo method, abundance of the 72- and 80-kDa cross-link species is probably too low to be detected by Western blotting.
Our data suggest that the sequential interaction of SecY with the ribosome and the signal anchor sequence of RNCs cause a massive reorientation at the SecY-YidC interface that is likely to allow the exit of a membrane protein into the lipid phase of the membrane. Coordination of lateral gate movements with conformational changes within YidC could be a key event during lipid insertion of transmembrane domains. Importantly, these reorientations do not completely compromise the SecY-YidC interaction, consistent with the observation that RNCs of different lengths have been found to make sequential contacts with SecY and YidC (21) and that the SecY-YidC interaction appears to be more stable on Blue Native PAGE in the presence of RNCs (27).
The finding that YidC is located at the lateral gate is also of importance for the proposed orientation of a possible SecYEG dimer. In the “back to back” orientation, the dimer interface would mainly cover the hinge region and one TM of SecE (57–59), whereas in the “front to front” orientation, the lateral gate would constitute the contact site of the two protomers (60). This latter orientation is difficult to imagine in light of our findings. Nevertheless, in a recent in vivo study it was proposed that resting SecYEG translocons can exist in both orientations (61). Whether YidC interacts with both dimer populations and if so, which SecY surface is used in the front to front orientation requires further analyses.
Acknowledgments
We gratefully acknowledge Roland Beckman and Eli van der Sluis, Ludwig-Maximilians-Universität Munich, for providing the FtsQ plasmid and Matthias Müller and Michaela Fürst for providing α-PpiD antibodies.
This work was supported by the Deutsche Forschungsgemeinschaft Grants FOR929, FOR 967, and GRK1478 (to H. G. K.), grants from the Excellence Initiative of the German Federal and State Governments EXC 294 BIOSS Centre for Biological Signaling Studies (to B. W.) and GSC-4 Spemann Graduate School for Biology and Medicine (to H. G. K.), a FF-Nord Foundation Fellowship (to T. W.), and an Erasmus Fellowship of the European Union (to N. P.).

This article contains supplemental Table S1.
- TM
- transmembrane
- INV
- inverted inner membrane vesicle
- MPF
- membrane-protected fragment
- pBpa
- para-benzoyl-l-phenylalanine
- RNC
- ribosome-associated nascent chain
- SRP
- signal recognition particle
- TeaOAc
- triethanolamine acetate.
REFERENCES
- 1. Park E., Rapoport T. A. (2012) Mechanisms of Sec61/SecY-mediated protein translocation across membranes. Annu. Rev. Biophys. 41, 21–40 [DOI] [PubMed] [Google Scholar]
- 2. Van den Berg B., Clemons W. M., Jr., Collinson I., Modis Y., Hartmann E., Harrison S. C., Rapoport T. A. (2004) X-ray structure of a protein-conducting channel. Nature 427, 36–44 [DOI] [PubMed] [Google Scholar]
- 3. Frauenfeld J., Gumbart J., Sluis E. O., Funes S., Gartmann M., Beatrix B., Mielke T., Berninghausen O., Becker T., Schulten K., Beckmann R. (2011) Cryo-EM structure of the ribosome-SecYE complex in the membrane environment. Nat. Struct. Mol. Biol. 18, 614–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nishiyama K., Suzuki T., Tokuda H. (1996) Inversion of the membrane topology of SecG coupled with SecA-dependent preprotein translocation. Cell 85, 71–81 [DOI] [PubMed] [Google Scholar]
- 5. Mori H., Ito K. (2006) Different modes of SecY–SecA interactions revealed by site-directed in vivo photo-cross-linking. Proc. Natl. Acad. Sci. U.S.A. 103, 16159–16164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Angelini S., Boy D., Schiltz E., Koch H. G. (2006) Membrane binding of the bacterial signal recognition particle receptor involves two distinct binding sites. J. Cell Biol. 174, 715–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Angelini S., Deitermann S., Koch H. G. (2005) FtsY, the bacterial signal-recognition particle receptor, interacts functionally and physically with the SecYEG translocon. EMBO Rep. 6, 476–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kuhn P., Weiche B., Sturm L., Sommer E., Drepper F., Warscheid B., Sourjik V., Koch H. G. (2011) The bacterial SRP receptor, SecA and the ribosome use overlapping binding sites on the SecY translocon. Traffic 12, 563–578 [DOI] [PubMed] [Google Scholar]
- 9. Duong F., Wickner W. (1997) The SecDFyajC domain of preprotein translocase controls preprotein movement by regulating SecA membrane cycling. EMBO J. 16, 4871–4879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pogliano J. A., Beckwith J. (1994) SecD and SecF facilitate protein export in Escherichia coli. EMBO J. 13, 554–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tsukazaki T., Mori H., Echizen Y., Ishitani R., Fukai S., Tanaka T., Perederina A., Vassylyev D. G., Kohno T., Maturana A. D., Ito K., Nureki O. (2011) Structure and function of a membrane component SecDF that enhances protein export. Nature 474, 235–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Matsuyama S., Fujita Y., Mizushima S. (1993) SecD is involved in the release of translocated secretory proteins from the cytoplasmic membrane of Escherichia coli. EMBO J. 12, 265–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen M., Xie K., Yuan J., Yi L., Facey S. J., Pradel N., Wu L. F., Kuhn A., Dalbey R. E. (2005) Involvement of SecDF and YidC in the membrane insertion of M13 procoat mutants. Biochemistry 44, 10741–10749 [DOI] [PubMed] [Google Scholar]
- 14. Nouwen N., Driessen A. J. (2002) SecDFyajC forms a heterotetrameric complex with YidC. Mol. Microbiol. 44, 1397–1405 [DOI] [PubMed] [Google Scholar]
- 15. Stiegler N., Dalbey R. E., Kuhn A. (2011) M13 procoat protein insertion into YidC and SecYEG proteoliposomes and liposomes. J. Mol. Biol. 406, 362–370 [DOI] [PubMed] [Google Scholar]
- 16. Funes S., Hasona A., Bauerschmitt H., Grubbauer C., Kauff F., Collins R., Crowley P. J., Palmer S. R., Brady L. J., Herrmann J. M. (2009) Independent gene duplications of the YidC/Oxa/Alb3 family enabled a specialized cotranslational function. Proc. Natl. Acad. Sci. U.S.A. 106, 6656–6661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Samuelson J. C., Chen M., Jiang F., Möller I., Wiedmann M., Kuhn A., Phillips G. J., Dalbey R. E. (2000) YidC mediates membrane protein insertion in bacteria. Nature 406, 637–641 [DOI] [PubMed] [Google Scholar]
- 18. van der Laan M., Bechtluft P., Kol S., Nouwen N., Driessen A. J. (2004) F1F0 ATP synthase subunit c is a substrate of the novel YidC pathway for membrane protein biogenesis. J. Cell Biol. 165, 213–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Serek J., Bauer-Manz G., Struhalla G., van den Berg L., Kiefer D., Dalbey R., Kuhn A. (2004) Escherichia coli YidC is a membrane insertase for Sec-independent proteins. EMBO J. 23, 294–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Welte T., Kudva R., Kuhn P., Sturm L., Braig D., Müller M., Warscheid B., Drepper F., Koch H. G. (2012) Promiscuous targeting of polytopic membrane proteins to SecYEG or YidC by the Escherichia coli signal recognition particle. Mol. Biol. Cell 23, 464–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Urbanus M. L., Scotti P. A., Froderberg L., Saaf A., de Gier J. W., Brunner J., Samuelson J. C., Dalbey R. E., Oudega B., Luirink J. (2001) Sec-dependent membrane protein insertion. Sequential interaction of nascent FtsQ with SecY and YidC. EMBO Rep. 2, 524–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Beck K., Eisner G., Trescher D., Dalbey R. E., Brunner J., Müller M. (2001) YidC, an assembly site for polytopic Escherichia coli membrane proteins located in immediate proximity to the SecYE translocon and lipids. EMBO Rep. 2, 709–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nagamori S., Smirnova I. N., Kaback H. R. (2004) Role of YidC in folding of polytopic membrane proteins. J. Cell Biol. 165, 53–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wagner S., Pop O. I., Haan G. J., Baars L., Koningstein G., Klepsch M. M., Genevaux P., Luirink J., de Gier J. W. (2008) Biogenesis of MalF and the MalFGK2 maltose transport complex in Escherichia coli requires YidC. J. Biol. Chem. 283, 17881–17890 [DOI] [PubMed] [Google Scholar]
- 25. van Bloois E., Dekker H. L., Fröderberg L., Houben E. N., Urbanus M. L., de Koster C. G., de Gier J. W., Luirink J. (2008) Detection of cross-links between FtsH, YidC, HflK/C suggests a linked role for these proteins in quality control upon insertion of bacterial inner membrane proteins. FEBS Lett. 582, 1419–1424 [DOI] [PubMed] [Google Scholar]
- 26. Scotti P. A., Urbanus M. L., Brunner J., de Gier J. W., von Heijne G., van der Does C., Driessen A. J., Oudega B., Luirink J. (2000) YidC, the Escherichia coli homologue of mitochondrial Oxa1p, is a component of the Sec translocase. EMBO J. 19, 542–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Boy D., Koch H. G. (2009) Visualization of distinct entities of the SecYEG translocon during translocation and integration of bacterial proteins. Mol. Biol. Cell 20, 1804–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hanahan D. (1983) Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580 [DOI] [PubMed] [Google Scholar]
- 29. Ryu Y., Schultz P. G. (2006) Efficient incorporation of unnatural amino acids into proteins in Escherichia coli. Nat. Methods 3, 263–265 [DOI] [PubMed] [Google Scholar]
- 30. Baba T., Jacq A., Brickman E., Beckwith J., Taura T., Ueguchi C., Akiyama Y., Ito K. (1990) Characterization of cold-sensitive secY mutants of Escherichia coli. J. Bacteriol. 172, 7005–7010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Braig D., Bär C., Thumfart J. O., Koch H. G. (2009) Two cooperating helices constitute the lipid-binding domain of the bacterial SRP receptor. J. Mol. Biol. 390, 401–413 [DOI] [PubMed] [Google Scholar]
- 32. Seidelt B., Innis C. A., Wilson D. N., Gartmann M., Armache J. P., Villa E., Trabuco L. G., Becker T., Mielke T., Schulten K., Steitz T. A., Beckmann R. (2009) Structural insight into nascent polypeptide chain-mediated translational stalling. Science 326, 1412–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Niemann M., Wiese S., Mani J., Chanfon A., Jackson C., Meisinger C., Warscheid B., Schneider A. (2013) Mitochondrial outer membrane proteome of Trypanosoma brucei reveals novel factors required to maintain mitochondrial morphology. Mol. Cell. Proteomics 12, 515–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 35. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., Mann M. (2011) Andromeda. A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 36. Xie K., Kiefer D., Nagler G., Dalbey R. E., Kuhn A. (2006) Different regions of the nonconserved large periplasmic domain of Escherichia coli YidC are involved in the SecF interaction and membrane insertase activity. Biochemistry 45, 13401–13408 [DOI] [PubMed] [Google Scholar]
- 37. Koch H. G., Moser M., Schimz K. L., Muller M. (2002) The integration of YidC into the cytoplasmic membrane of Escherichia coli requires the signal recognition particle, SecA and SecYEG. J. Biol. Chem. 277, 5715–5718 [DOI] [PubMed] [Google Scholar]
- 38. Darken M. A. (1964) Puromycin inhibition of protein synthesis. Pharmacol. Rev. 16, 223–243 [PubMed] [Google Scholar]
- 39. Zimmer J., Nam Y., Rapoport T. A. (2008) Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature 455, 936–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. du Plessis D. J., Berrelkamp G., Nouwen N., Driessen A. J. (2009) The lateral gate of SecYEG opens during protein translocation. J. Biol. Chem. 284, 15805–15814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Becker T., Bhushan S., Jarasch A., Armache J. P., Funes S., Jossinet F., Gumbart J., Mielke T., Berninghausen O., Schulten K., Westhof E., Gilmore R., Mandon E. C., Beckmann R. (2009) Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome. Science 326, 1369–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Luirink J., von Heijne G., Houben E., de Gier J. W. (2005) Biogenesis of inner membrane proteins in Escherichia coli. Annu. Rev. Microbiol. 59, 329–355 [DOI] [PubMed] [Google Scholar]
- 43. Dalbey R. E., Wang P., Kuhn A. (2011) Assembly of bacterial inner membrane proteins. Annu. Rev. Biochem. 80, 161–187 [DOI] [PubMed] [Google Scholar]
- 44. Houben E. N., ten Hagen-Jongman C. M., Brunner J., Oudega B., Luirink J. (2004) The two membrane segments of leader peptidase partition one by one into the lipid bilayer via a Sec/YidC interface. EMBO Rep. 5, 970–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hou B., Lin P.-J., Johnson A. E. (2012) Membrane protein TM segments are retained at the translocon during integration until the nascent chain cues FRET-detected release into bulk lipid. Mol. Cell 48, 398–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lotz M., Haase W., Kühlbrandt W., Collinson I. (2008) Projection structure of yidC. A conserved mediator of membrane protein assembly. J. Mol. Biol. 375, 901–907 [DOI] [PubMed] [Google Scholar]
- 47. Ravaud S., Wild K., Sinning I. (2008) Purification, crystallization and preliminary structural characterization of the periplasmic domain P1 of the Escherichia coli membrane-protein insertase YidC. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 64, 144–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Oliver D. C., Paetzel M. (2008) Crystal structure of the major periplasmic domain of the bacterial membrane protein assembly facilitator YidC. J. Biol. Chem. 283, 5208–5216 [DOI] [PubMed] [Google Scholar]
- 49. Kohler R., Boehringer D., Greber B., Bingel-Erlenmeyer R., Collinson I., Schaffitzel C., Ban N. (2009) YidC and Oxa1 form dimeric insertion pores on the translating ribosome. Mol. Cell. 34, 344–353 [DOI] [PubMed] [Google Scholar]
- 50. Hizlan D., Robson A., Whitehouse S., Gold V. A., Vonck J., Mills D., Kühlbrandt W., Collinson I. (2012) Structure of the SecY complex unlocked by a preprotein mimic. Cell Rep. 1, 21–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tsukazaki T., Mori H., Fukai S., Ishitani R., Mori T., Dohmae N., Perederina A., Sugita Y., Vassylyev D. G., Ito K., Nureki O. (2008) Conformational transition of Sec machinery inferred from bacterial SecYE structures. Nature 455, 988–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gumbart J., Trabuco L. G., Schreiner E., Villa E., Schulten K. (2009) Regulation of the protein-conducting channel by a bound ribosome. Structure 17, 1453–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Haider S., Hall B. A., Sansom M. S. (2006) Simulations of a protein translocation pore. SecY. Biochemistry 45, 13018–13024 [DOI] [PubMed] [Google Scholar]
- 54. Lycklama a Nijeholt J. A., Wu Z. C., Driessen A. J. (2011) Conformational dynamics of the plug domain of the SecYEG protein-conducting channel. J. Biol. Chem. 286, 43881–43890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Egea P. F., Stroud R. M. (2010) Lateral opening of a translocon upon entry of protein suggests the mechanism of insertion into membranes. Proc. Natl. Acad. Sci. U.S.A. 107, 17182–17187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Das S., Oliver D. B. (2011) Mapping of the SecA·SecY and SecA·SecG interfaces by site-directed in vivo photocross-linking. J. Biol. Chem. 286, 12371–12380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kaufmann A., Manting E. H., Veenendaal A. K., Driessen A. J., van der Does C. (1999) Cysteine-directed cross-linking demonstrates that helix 3 of SecE is close to helix 2 of SecY and helix 3 of a neighboring SecE. Biochemistry 38, 9115–9125 [DOI] [PubMed] [Google Scholar]
- 58. Dalal K., Chan C. S., Sligar S. G., Duong F. (2012) Two copies of the SecY channel and acidic lipids are necessary to activate the SecA translocation ATPase. Proc. Natl. Acad. Sci. U.S.A. 109, 4104–4109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Breyton C., Haase W., Rapoport T. A., Kühlbrandt W., Collinson I. (2002) Three-dimensional structure of the bacterial protein-translocation complex SecYEG. Nature 418, 662–665 [DOI] [PubMed] [Google Scholar]
- 60. Mitra K., Schaffitzel C., Shaikh T., Tama F., Jenni S., Brooks C. L., 3rd, Ban N., Frank J. (2005) Structure of the E. coli protein-conducting channel bound to a translating ribosome. Nature 438, 318–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Park E., Rapoport T. A. (2012) Bacterial protein translocation requires only one copy of the SecY complex in vivo. J. Cell Biol. 198, 881–893 [DOI] [PMC free article] [PubMed] [Google Scholar]