Background: POT1 and TPP1 protect the ends of human telomeres.
Results: The secondary structure of single-stranded DNA alters the coordinated binding of multiple POT1-TPP1 proteins.
Conclusion: Single-stranded telomere DNA is more susceptible to DNase I digestion when coated with POT1-TPP1 proteins.
Significance: By altering DNA properties, coating of telomere DNA by POT1-TPP1 proteins may have a functional role in telomere maintenance.
Keywords: DNA Binding Protein, DNA Structure, Nucleotide, Telomerase, Telomeres, G-quadruplex, POT1, TPP1
Abstract
Telomeres are macromolecular nucleoprotein complexes that protect the ends of eukaryotic chromosomes from degradation, end-to-end fusion events, and from engaging the DNA damage response. However, the assembly of this essential DNA-protein complex is poorly understood. Telomere DNA consists of the repeated double-stranded sequence 5′-TTAGGG-3′ in vertebrates, followed by a single-stranded DNA overhang with the same sequence. Both double- and single-stranded regions are coated with high specificity by telomere end-binding proteins, including POT1 and TPP1, that bind as a heterodimer to single-stranded telomeric DNA. Multiple POT1-TPP1 proteins must fully coat the single-stranded telomere DNA to form a functional telomere. To better understand the mechanism of multiple binding, we mutated or deleted the two guanosine nucleotides residing between adjacent POT1-TPP1 recognition sites in single-stranded telomere DNA that are not required for multiple POT1-TPP1 binding events. Circular dichroism demonstrated that spectra from the native telomere sequence are characteristic of a G-quadruplex secondary structure, whereas the altered telomere sequences were devoid of these signatures. The altered telomere strands, however, facilitated more cooperative loading of multiple POT1-TPP1 proteins compared with the wild-type telomere sequence. Finally, we show that a 48-nucleotide DNA with a telomere sequence is more susceptible to nuclease digestion when coated with POT1-TPP1 proteins than when it is left uncoated. Together, these data suggest that POT1-TPP1 binds telomeric DNA in a coordinated manner to facilitate assembly of the nucleoprotein complexes into a state that is more accessible to enzymatic activity.
Introduction
Telomeres are nucleoprotein complexes that cap and protect the ends of all eukaryotic chromosomes. Telomeres contain G-rich arrays of double-stranded DNA that end in single-stranded DNA (ssDNA)3 overhangs (1). A primary function of telomeres is to protect chromosomes from deleterious events such as degradation and end-to-end fusions (2, 3). Specialized telomeric proteins bind telomere DNA to prevent the induction of DNA damage response pathways (4–7). In addition, telomeres protect against the loss of coding DNA during cell division. Because of the “end replication problem,” DNA polymerases are unable to fully replicate the extreme end of the lagging strand of DNA (8). As such, the telomeres of somatic cells shorten with each cell division until they become critically short. At this point, the cell is no longer capable of dividing and enters a state of replicative senescence (9, 10). In highly proliferative cells, including germ line cells and the majority of cancer cells, telomere shortening is counterbalanced by the activation of a ribonucleoprotein called telomerase (11–13). Telomerase helps maintain telomere length by adding nucleotides (nt) to the ssDNA of the telomere at each round of cell division. It is becoming increasingly clear that telomere end-binding proteins regulate telomerase activity both positively and negatively (14–18). The combination of these diverse functions makes telomere end-binding proteins critical for proper telomere homeostasis mechanisms.
In humans, telomeres consist of tandem hexameric DNA repeats of the sequence TTAGGG, which interacts with at least six unique proteins to form a dynamic entity collectively referred to as shelterin (2, 19). The double-stranded region of the telomere is bound in a sequence-specific manner by two separate proteins called telomere repeat-binding factors 1 and 2 (TRF1 and TRF2) (20). Meanwhile, the protection of telomeres 1 (POT1) protein binds and coats the telomere ssDNA overhang (21–23). Other shelterin proteins include TIN2, RAP1, and TPP1, all of which interact with TRF1/2 and POT1 at the telomere (16, 24–26).
The single-stranded region of telomeres extends for 50–300 nucleotides in mammals and is bound with nanomolar affinity by POT1 protein (21, 22, 27, 28). Multiple splicing variants of human POT1 have been identified in vivo (21, 29). The primary products of these variants are the full-length protein and a truncated isoform that lacks a C-terminal domain (POT1-N). Although the DNA recognition sequence of the two isoforms is identical, only the full-length isoform interacts with TPP1 via its C-terminal domain (30–32). The x-ray crystal structure of the human POT1-N isoform reveals that the N terminus of POT1 is comprised of dual oligonucleotide/oligosaccharide binding folds (22). In humans, the two oligonucleotide/oligosaccharide binding folds of a single POT1 protein interact with a 10-nucleotide tract of telomeric DNA (5′-TTAGGG TTAG-3′) to coordinate binding. Although human TPP1 is not known to interact with telomeric DNA directly, the POT1-TPP1 heterodimer binds ssDNA with an affinity 10-fold greater than that of POT1 alone (14, 15). Clearly it is one function of POT1-TPP1 to protect the single-stranded region of telomeric DNA from degradation, recombination, and subsequent signaling of a DNA damage response (4, 5, 33–35). In an entirely separate role, however, TPP1 recruits the enzyme telomerase to the single-stranded region of telomeres where POT1-TPP1 acts as a processivity enhancement factor in stimulating telomerase activity (15, 16, 36).
In vitro, multiple POT1-TPP1 proteins saturate long tracts of single-stranded telomeric DNA templates to form compact structures (23). In the absence of protein, ssDNA with a G-rich telomere sequence readily forms highly stable, intramolecular G-quadruplex formations (37–39). Aside from shielding DNA, the binding of POT1 is thought to unwind G-quadruplex formations, thus altering its structure from an inaccessible state to one that is telomerase-extendable (22, 40, 41). Once accessible, telomerase binds the shortened, single-stranded telomere overhang and extends it by hundreds of nucleotides through a processive repeat addition mechanism (42, 43). Together, these findings demonstrate that telomere maintenance is a complex function regulated by members of the shelterin complex, the integrity and structural composition of telomere DNA, and the recruitment and activity of telomerase.
In this report, we investigated the role DNA secondary structure may play in the binding of multiple POT1-TPP1 proteins to long strands of telomeric DNA. First we examined the putative role of the two guanosine nucleotides residing between adjacent POT1 recognition sites in single-stranded telomeric DNA on the secondary structure in the absence of protein and then in terms of assembly of the macromolecular complexes. Although these guanosines play a direct role in G-quadruplex formation of the unbound oligonucleotide, we discovered that mutation or deletion of these two guanosines did not affect the ability of multiple POT1-TPP1 proteins to coat the DNA template. Finally, we assessed the ability of multiple POT1-TPP1 proteins to protect telomeric DNA from DNase I digestion. We show that a 48-nt, single-stranded telomeric DNA is more susceptible to DNase I digestion when coated with multiple POT1-TPP1 proteins than it is when coated with POT1 alone or when left uncoated by protein. This observation was exclusive for the native telomere DNA sequence and indicates an expanded role of POT1-TPP1 in regulating accessibility to telomere DNA. Together, our data provide insight into the organization and biochemical properties of multiple POT1-TPP1 proteins coating telomeric DNA and, thus, the contribution to telomere maintenance.
EXPERIMENTAL PROCEDURES
Circular Dichroism Spectroscopy
CD spectra were obtained using an Applied Photophysics π*-180 (Applied Photophysics Ltd, United Kingdom) spectrometer. A quartz cuvette with 1-cm path length was used to acquire CD spectra from 220–340 nm with a 2-nm bandwidth, 2-nm step size, and 2-s collection time per data point. DNA solutions also contained 50 mm HEPES (pH 8.0) and 75 mm salt, either NaCl or KCl. The DNA concentrations for all three oligonucleotides in this study were adjusted to 10 μm on the basis of absorbance readings obtained using a NanoDrop spectrometer (Thermo Fisher Scientific, Inc., Waltham, MA) and were confirmed on the AP π*-180 instrument. The DNA samples were heat-denatured at 95 °C for 5 min followed by cooling on ice for at least 5 min. Each 500-μl DNA sample was scanned three times, and the average represents the depicted data.
POT1 and POT1-TPP1 Purification
Full-length POT1 and 6× His-TPP1 were coinfected and expressed in insect cells using recombinant baculovirus as described previously (23). The TPP1 construct comprises residues 89–334, which encompasses both its POT1- and telomerase-interacting domains (15, 16, 30–32, 44, 45). Cells were lysed via sonication in buffer containing 25 mm HEPES (pH 8), 150 mm NaCl, 5 mm DTT, 5 mm benzamidine, 1 mm PMSF, and protease inhibitor mixture (Roche). Cellular debris was pelleted by ultracentrifugation, and the supernatant was loaded onto precharged nickel-nitrilotriacetic acid resin (Qiagen). Gravity binding was performed at 4 °C, after which protein was eluted with buffer containing 200 mm imidazole, 25 mm HEPES (pH 8.0), and 150 mm NaCl. Eluted fractions were further purified using size exclusion chromatography on a Superdex 200 HiLoad 16/60 column (GE Healthcare). Protein fractions were pooled and concentrated and then flash frozen and stored at −80 °C.
POT1-N was expressed in Spodoptera frugiperda 9 insect cells and the baculovirus expression system. The protein was expressed as a fusion protein with an N-terminal GST tag. After affinity pull-down, the GST tag was cleaved using Prescission protease (GE Healthcare), and POT1-N protein was purified using size exclusion chromatography. Individual aliquots of POT1-N were flash-frozen and stored at −80 °C.
Gel Shift Assays
Gel shifts were performed to quantitatively compare the ability of POT1-N and POT1-TPP1 to bind multiple recognition sequences in the various DNA substrates. Telomere oligonucleotides (Invitrogen) were 5′-radiolabeled with [γ-32P]ATP and T4 polynucleotide kinase (Roche). Protein-DNA binding reactions were performed in buffer containing 50 mm HEPES (pH 8.0), 75 mm NaCl, 5 mm DTT, and 5% glycerol. Reactions were performed using 200 nm DNA containing ∼4% 32P-labeled DNA and 0–1600 nm of recombinant POT1-N or POT1-TPP1 protein in 160 nm increments. To attempt to achieve saturation with M3 and M4 mutants, protein concentrations were increased up to 4000 nm in 400 nm increments while maintaining DNA concentration at 200 nm. Binding reactions were incubated for 15 min at 4 °C before 8 μl of the reaction was loaded onto a 4–20% Tris-borate non-denaturing gel (Invitrogen). Gels were run at 120 V for 3–5 h, dried, and scanned using a Storm 860 PhosphorImager system (GE Healthcare). Densitometry was performed using ImageQuantTL software (GE Healthcare).
To determine an apparent dissociation constant from the binding data, the fraction of DNA fully saturated with POT1-TPP1 was plotted versus concentration of the heterodimer. These data were fit to the Hill equation, F = Pn/(Pn + Kn), where F is the fraction of total DNA in the complex with four proteins bound, P is the total concentration of POT1-TPP1 added to the reaction, and K is an empirical constant representing the concentration of protein at which 50% of the DNA was shifted to the population bound by four proteins. All experiments were performed in triplicate, and the calculated Kapp values represent the average of the three replicates with percent standard deviation (supplemental Table 1).
DNase I Digestion Assays
Telomere oligonucleotides (Invitrogen) were 5′ end-labeled using T4 PNK (polynucleotide kinase) (New England Biolabs) and [γ-32P]ATP 6000Ci/mmol (Perkin Elmer LifeSciences). Briefly, 10 units of T4 PNK and 25 μm DNA were incubated in 10 μl of 1× PNK buffer for 30 min at 37 °C. The reaction was quenched with 10 mm EDTA (pH 8.0) and incubation at 65 °C for 10 min. Radiolabeled DNA was diluted to 40 μl and purified using Illustra MicroSpin G-25 columns (GE Healthcare) following the standard protocol. 3′ end labeling was performed with terminal transferase (New England Biolabs) and [α-32P]dGTP 3000Ci/mmol (Perkin Elmer LifeSciences) according to the protocol of the manufacturer.
POT1-TPP1 nucleoprotein complexes were assembled as follows: 1.2 μm POT1-TPP1 was incubated with 100 nm DNA at 4 °C for 15 min in protein buffer (25 mm Hepes (pH 8.0), 150 mm NaCl, 6% glycerol). Free DNA or POT1-TPP1-DNA complexes were then incubated in DNase I reaction buffer (40 mm Tris-HCl (pH 8.0), 10 mm MgSO4, 10 mm CaCl2) with one unit of RQ1 RNase-Free DNase I (Promega). Reactions were incubated at room temperature for the time points indicated and then quenched by the addition of 20 mm EGTA (pH 8.0) (Fisher Scientific) combined with heating to 95 °C for 10 min. These reactions were also performed by substituting NaCl with either KCl or LiCl molar equivalents in the protein buffer to determine the influence of different monovalent cations on DNase accessibility. Reactions were then resolved on a 12% acrylamide, 7 m urea, 1× Tris borate-EDTA sequencing gel. Gels were dried, and bands were quantified using a Storm 860 PhosphorImager system (GE Healthcare). Densitometry was performed using ImageQuantTL software (GE Healthcare). All experiments were performed in triplicate with error bars indicated in each figure. Note that several error bars, especially those used for hT48GG → CC, are smaller than the labels used for individual points.
RESULTS
Human Telomeric Sequence and G-quadruplex Formation
To gain insight into the potential role of the secondary structure on multiple POT1-TPP1 binding, we analyzed three slightly different telomeric DNA sequences by CD. Native telomere sequence and structure is represented by a 48-nt probe comprised of a human telomere sequence with eight hexameric repeats (hT48wt, (TTAGGGTTAGGG)4) (Fig. 1A). To disrupt the G-quadruplex structure and to limit alternative binding modes because of the presence of overlapping POT1-TPP1 binding sites, the two guanosines of the second hexameric repeat were mutated to cytidine (hT48GG→CC, (TTAGGGTTAGCC)4) (Fig. 1A). The third model DNA was a 40-nt probe in which the two guanosines of every second hexameric repeat were deleted (hT48→40ΔGG, (TTAGGGTTAG)4) (Fig. 1A) and was designed to provide information on the effect of decreased spacing between adjacent binding sites on multiple binding. All three of these sequences maintained four cognate POT1-TPP1 binding sites.
FIGURE 1.
Circular dichroism spectra of single-stranded DNA with varying sequences. A, diagram of three 48-nt DNA templates investigated. hT48wt is comprised of eight hexameric repeats of a wild-type telomere DNA sequence. The hT48GG→CC construct is comprised of eight hexameric repeats of telomeric DNA in which the last two guanosines of each double repeat are mutated to cytidines. hT48→40ΔGG is comprised of eight hexameric repeats of telomeric DNA in which the last two guanosines of each double repeat are deleted. Each construct contains four consecutive POT1-TPP1 recognition sites, as indicated by the lines above each sequence. B, the CD spectra representing the average of three readings for each DNA sequence depicted in A are shown in buffer containing 75 mm NaCl. The hT48wt spectrum is characteristic of antiparallel G-quadruplexes typical of telomeric DNA in the presence of Na+. The spectra of hT48GG→CC and hT48→40ΔGG, however, are indicative of DNA adopting B-form helices, the most common ssDNA conformation. C, CD spectra as obtained in B but in the presence of 75 mm KCl. The hT48wt displays features that are characteristic of hybrid-type G-quadruplexes consisting of both parallel and antiparallel strands, known to be the dominant conformation of longer telomere sequences in the presence of K+. The spectra for both hT48GG→CC and hT48→40ΔGG lack these features, as both present as consistent with common B-form helices.
G-rich DNA with native telomere sequence could potentially form G-quadruplexes (38, 39), whereas deletion or mutation of the guanosines in hT48→40ΔGG and hT48GG→CC is predicted to disrupt or alter the distribution of conformations containing G-quadruplexes. We employed CD spectroscopy to characterize the secondary structure of hT48wt (the wild-type control) and the two mutants, hT48GG→CC and hT48→40ΔGG.
Parallel and antiparallel G-quadruplexes have characteristic spectra in CD spectroscopy. The spectra of parallel G-quadruplexes display a positive band at 260 nm and a small negative band at 240 nm, whereas antiparallel G-quadruplexes display a peak at 290 nm and a valley at 265 nm and another positive peak at 245 nm (38, 39). As expected, the CD spectrum of the hT48wt probe with the native telomere sequence had a characteristic peak at 295 nm and a valley at 265 nm, followed by a peak at 245 nm in buffer containing NaCl. This result clearly indicates the presence of an antiparallel G-quadruplex (Fig. 1B). Conversely, the CD spectra of both hT48GG→CC and hT48→40ΔGG in the presence of NaCl revealed a single peak near 270 nm and a valley at 240 nm (Fig. 1B). These spectra indicate that both of these oligonucleotides adopt B-form helical structure or are relatively disordered and do not form G-quadruplexes characteristic of native telomere DNA (39). The CD spectrum obtained from the oligonucleotides in buffer containing KCl supports the formation of a G-quadruplex structure in the hT48wt sequence (Fig. 1C). This spectrum displays a strong positive peak at 290 nm, a shoulder peak around 250 nm, and a small negative peak at 240 nm. These features are characteristic of a mixed population containing multiple G-quadruplex conformations, which was demonstrated previously for longer telomere sequences in the presence of potassium ions (38). This distinct spectrum is absent from the data from hT48GG→CC and hT48→40ΔGG, which are again more consistent with general B-form helices (Fig. 1C).
POT1-N and POT1-TPP1 Saturate Native and Mutant DNA Templates
Next, we determined whether the mutation or deletion of the two G residues separating individual POT1 binding sites influences the ability of the protein to fully saturate the DNA. Omission of the two Gs disrupts G-quadruplex formation (Fig. 1), which may affect the mechanism of multiple binding events of POT1-TPP1 protein. Therefore, to gain insight into the mechanism of POT1-TPP1 multiple binding, we compared the binding reactions of POT1-TPP1 proteins to the three telomeric probes hT48wt, hT48GG→CC, and hT48→40ΔGG.
We first examined the binding of POT1-TPP1 to ssDNA telomere substrates exhibiting the minimal 10-nt POT1 binding sequence and flanked with two C nt on either the 3′ or 5′ end. EMSAs were used to determine that POT1-TPP1 bound the mutant substrates as effectively as native sequence, with a complete shift between 1.5 and 2 times molar excess of protein (supplemental Fig. 1). In addition, the high degree of similarity between the three shifts indicates that neither the presence nor the placement of the C nt outside of the POT1 recognition sequence effects binding of a single POT1-TPP1 heterodimer to the 12-nt oligonucleotide.
We next used the gel shift assays to examine binding properties of multiple POT1-TPP1 proteins to the longer telomere DNA substrates exhibiting G-to-C mutations or G deletions. An inherent advantage of this method is the ability to separate complexes with different numbers of proteins loaded onto the DNA. This allows the distribution of multiple bound forms to be characterized and quantified. Stoichiometric binding conditions, in which the concentrations of both DNA and protein are in excess of the Kd for an individual binding interaction, were used to most sensitively reveal the effects of changes in DNA structure on the distribution of partially saturated intermediates in the binding reaction. Binding of POT1-TPP1 to native ssDNA (hT48wt) showed four distinct bands, indicating four possible binding stoichiometries with one, two, three, or four POT1-TPP1 proteins bound per DNA (Fig. 2A). As the concentration of POT1-TPP1 is increased, the intensities of these different complexes changes, and the slowest migrating band, indicative of hT48wt bound by four POT1-TPP1 heterodimers, becomes the most prevalent species.
FIGURE 2.

Binding of multiple POT1-TPP1 proteins to wild-type and mutant telomere DNA templates. EMSA experiments of hT48wt (A), hT48GG→CC (B), and hT48→40ΔGG (C) constructs, respectively. EMSAs were performed with DNA concentration remaining constant in each lane and POT1-TPP1 concentration increases from 0 – 2× molar excess/POT1 binding site on the DNA template (in 0.2× molar excess increments from left to right). Numbers in A indicate the number of POT1-TPP1 heterodimers bound to the DNA. D, quantification of the EMSA data for POT1-TPP1 binding to the three oligonucleotides described.
However, even at the highest POT1-TPP1 concentrations, populations exist that contains only one, two, or three proteins bound to the DNA. Additionally, the concentration dependence of the individually bound species does not follow the pattern expected for a simple binding mechanism in which POT1-TPP1 loads from the DNA 3′ end and in which the affinity of the protein is the same at each site. In this model there should be no preferential accumulation of any individual species. If intermediates are observed, then there should be a stoichiometric progression of intensity proceeding through intermediates with increasing numbers of proteins bound.
In contrast to the wild-type DNA, the four binding states were not significantly populated in the binding reaction using hT48GG→CC DNA. A strong preference was shown for either a single heterodimer bound or an entirely saturated construct with four POT1-TPP1 proteins bound (Fig. 2B). The bands representing the bound states with two and three heterodimers are far less discernible. The GG to CC mutation results in a concentration dependence that is consistent with a simple cooperative loading mechanism for POT1-TPP1 multiple binding in which only the single bound and fully saturated forms accumulate. The lack of accumulation of species with only two or three proteins bound suggests that the presence of G quadruplex structure may act as a barrier to cooperative multiple binding on native telomere DNA.
In the binding reactions performed with the hT48→40ΔGG DNA, all four bound states can be distinguished by EMSA (Fig. 2C). In this case, the accumulation and depletion of individually bound forms follows a progression consistent with a simple sequential binding model. Importantly, these data indicate that four POT1-TPP1 proteins were able to saturate the available binding sites on the hT48→40ΔGG construct despite the lack of nucleotides between adjacent POT1-binding sites in the DNA. Nonetheless, relative to the hT48GG→CC DNA there is clearly a greater accumulation of partially bound forms more similar to the results observed with the wild-type telomere model. These results are consistent with the interpretation that the POT1-TPP1 heterodimer is capable of saturating four tandem binding sites, regardless of nucleotide sequence or spacing between POT1 recognition sequences. However, alteration of the G-quadruplex structure and spacing of adjacent telomeric repeats results in differences in the overall binding mechanism, resulting in the differential accumulation of partially bound forms.
To determine whether these changes in binding mechanism result in significant differences in the overall capability of POT1-TPP1 to coat the three DNA constructs, an apparent binding constant (Kapp) was calculated by empirically fitting the data to the Hill binding equation. The resulting Kapp, therefore, represents the concentration of POT1-TPP1, where 50% of the DNA oligonucleotide population is bound with four POT1-TPP1 proteins (Fig. 2D). The Kapp was very similar for the three DNA constructs, being 840 nm ± 8.3%, 760 nm ± 22%, and 670 nm± 11% for hT48wt, hT48GG→CC, and hT48→40ΔGG, respectively (supplemental Table 1). In addition, we performed similar multiple binding experiments using POT1-N in place of POT1-TPP1. Although slightly lower Kapp values were observed, as expected (supplemental Table 1), the multiple binding profile of POT1-N was similar to that of POT1-TPP1 for all three DNA constructs (supplemental Fig. 2).
Taken together, these data indicate that the two G nucleotides residing between adjacent POT1-binding sites are unnecessary for sequential binding of multiple POT1-TPP1 heterodimers onto a single-stranded telomeric DNA template. However, the differences in populations of complexes containing less than four proteins bound to the hT48wt and hT48GG→CC DNA suggests that the presence of G-quadruplexes may influence the mechanism of multiple binding events.
DNA-dependent Binding Drives Loading of Multiple POT1-TPP1 Proteins
Previous studies have shown that sequential binding of Sterkiella nova telomere end-binding proteins, TEBPα and TEBPβ (respective homologs of human POT1 and TPP1), to a G-rich telomeric substrate is cooperative (46). Conversely, recent data demonstrated that the Saccharomyces cerevisiae telomere end-binding protein Rap1p binds each telomeric recognition site independently (47). Therefore, the potential exists for bound POT1-TPP1 to influence the binding of additional proteins in a directional and cooperative manner.
A key result indicating a cooperative mechanism is the observation that binding of POT1-TPP1 to the hT48GG→CC DNA accumulates only the single bound and fully bound forms. This characteristic is not idiosyncratic to a 48-nt DNA because similar results were observed with constructs of up to 72 nt (supplemental Fig. 3). It has been demonstrated previously that POT1 has an affinity over 100-fold greater for the 3′ terminal binding site of telomeric DNA (48). It follows, then, that in a simple sequential and cooperative binding mechanism initial binding occurs first at the 3′ end, followed by progressive loading of proteins in a 3′-to-5′ direction.
To test this idea, we analyzed DNAs containing the hT48GG→CC sequence with an additional G-to-C mutation to either the third or fourth POT1-binding site (M3 and M4, respectively) (Fig. 3A). A G-to-C mutation within the POT1 recognition sequence reduces the efficiency of POT1 binding to less than 15% compared with the native telomere sequence (22). A cooperative, directional binding model predicts that POT1-TPP1 binding to the M3 construct would result in accumulation of protein bound at the most 3′ POT1-binding site. At higher concentrations of POT1-TPP1, the lower affinity at the adjacent site would be overcome, and subsequent tight binding would result in saturation of the DNA with four POT1-TPP1 proteins. In contrast to the hT48GG→CC construct, which is readily coated with four POT1-TPP1 proteins (Fig. 3B), EMSA analysis reveals accumulation of bands corresponding to the singly bound and fully saturated forms for the M3 DNA (Fig. 3C). We interpret these results as indicating that the mutation results in two dominant populations at high concentrations of protein, representing a single bound POT1-TPP1 protein and the fully saturated construct with four POT1-TPP1 proteins. Quantification of the bands reveals that, with 3–5× POT1-TPP1 proteins/POT1 binding site on the ssDNA, ∼37% is bound by a single POT1-TPP1 protein, and ∼58% is bound by four POT1-TPP1 proteins.
FIGURE 3.
Binding of POT1-TPP1 to sequential recognition sites. A, diagram of DNA sequences investigated. Solid bars above the sequence indicate individual POT1 binding sites. Dashed lines indicate a G→C (asterisk) mutation within the POT1 binding site of the DNA. Shown are electrophoretic mobility shift assays of POT1-TPP1 binding to the hT48GG→CC (B), M3 (C), and M4 (D) DNA fragments, respectively. Experiments were performed the same as in Fig. 2, except that protein concentrations were increased to 0–5× molar excess/POT1 binding site in an attempt to fully saturate the DNA with four bound POT1-TPP1 proteins. Densitometry performed on the last five lanes corresponding to 3–5× molar excess POT1-TPP1 per binding site was quantified to reveal the proportion of each bound species for the individual constructs. E, model depicting a simple mechanistic explanation for the cooperative binding observed in the presence of multiple POT1-TPP1 loading events and the process that M3 and M4 mutants obstruct. Step 1, POT1-TPP1 binds preferentially to the most 3′ binding site. Step 2, POT1-TPP1 assumes a conformational change upon binding of an appropriate DNA substrate. Step 3, the conformational change assumed in step 2 facilitates recruitment of an additional POT1-TPP1 heterodimer to the 5′ adjacent binding site. Step 4, the second bound POT1-TPP1 undergoes the DNA-binding dependent conformational change and then facilitates recruitment and binding of additional POT1-TPP1 heterodimers in a sequential 3′→5′ manner until the single-stranded telomeric DNA is fully coated.
Next we determined the effect of weakening the binding affinity of the preferential 3′ end binding site. As shown in Fig. 3D, EMSA analysis revealed primarily two populations of nucleoprotein complexes with the distribution representative of two POT1-TPP1 proteins bound and a second with four (Fig. 3D). Quantification of the individual bands reveals that, with 3–5× POT1-TPP1 proteins/POT1 binding site on the ssDNA, the relative distribution of one, two, three, or four POT1-TPP1 proteins bound to the 48-nt substrate is 7, 48, 1, and 44%, respectively.
In both reactions, the lack of accumulation of a species representing three POT1-TPP1 proteins to the DNA (∼1%) discounts the possibility of POT1-TPP1 binding only to the three native POT1-binding sites in both mutant DNA. The accumulation of singly bound POT1-TPP1 when the M3 site was mutated is consistent with a sequential, directional binding model in which POT1-TPP1 proteins must first bind properly at the 3′ end of a telomeric substrate, and in pairs, before complete coating of the DNA can occur. Interestingly, a significant portion of the population can go on to bind to saturation despite the weakened binding at the M3 site. The persistence of the singly bound form, even at apparently saturating concentrations of protein, may indicate kinetic trapping of this species.
Interestingly, mutation of the 3′ binding site does not block saturation with all four binding sites occupied. Rather, the assembly reaction “stalls,” resulting in accumulation of an intermediate bound form with only 2 POT1-TPP1 proteins bound. Assuming that the species with 2 POT1-TPP1 proteins bound is an intermediate, then weakened binding at the 3′ site, in turn, decreases the apparent affinity for binding at the adjacent internal site. This result suggests that the conformation of the DNA-protein complex with two POT1-TPP1 proteins bound is altered when the M4 site is mutated so that enhanced binding of the next POT1-TPP1 protein is not observed. The net result is likely to be that the cooperative binding of POT1-TPP1 is uncoupled at the addition of the third protein.
These results can be interpreted with a model for multiple binding in which each POT1-TPP1 binding event proceeds through two stages (Fig. 3E). Because of the 3′ end preference, the first binding event involves the recognition and binding of a single POT1-TPP1 heterodimer to the terminal 3′ binding site. Upon binding appropriately, a DNA-dependent conformational change is induced in the POT1-TPP1 heterodimer. The fully bound state then facilitates the recruitment and binding of a subsequent POT1-TPP1 protein to the adjacent 5′ binding site. This process continues to coordinate multiple binding events in a sequential manner until the native telomeric DNA is fully coated with POT1-TPP1 heterodimers.
Particular steps in this model can be impeded if a cognate binding site is altered, as in the mutant M3 and M4 constructs. In the case of the M3 mutant, the first heterodimer binds to the cognate 3′ terminal site normally, yet loading of the second protein is inhibited because of the mutated binding site. Once protein overcomes the lower affinity for the disfavored mutant site, no more impediments exist, and the only other substantially observed species is the fully coated DNA. M4, however, decreases the affinity of the preferential terminal binding site. The 3′ end preference, at more than 100-fold compared for internal sites, is much greater than the decrease in affinity introduced by the G-to-C M4 mutation within the POT1 binding site. The observation that two bound POT1-TPP1 heterodimers accumulate in the M4 sequence in the absence of one or three indicates that although the protein must bind, the reduced affinity inhibits the proper conformational change that must occur to facilitate subsequent binding events.
Telomere DNA Is More Susceptible to DNase I Digestion when Coated with POT1-TPP1 Proteins
A DNase I digestion assay was performed to elucidate the protective properties of POT1-TPP1 coated telomere DNA. The hT48wt telomere DNA was 5′ end labeled and divided into two groups. One group remained as uncoated hT48wt DNA, and a second group was coated with three times molar excess POT1-TPP1. Each population was then incubated with DNase I, and aliquots were analyzed for the following one to 24 h (Fig. 4A, left panel, and supplemental Fig. 4A). It is immediately clear that DNase I does not readily degrade the uncoated wild-type 48-nt telomere DNA. In fact, the uncoated telomere DNA is only digested partially (∼50%), even 24 h after incubating with DNase I. This relative level of DNase I protection is likely due to the G-quadruplex secondary structure exhibited by wild-type telomere DNA. The tight packing of the G-tetrads must render the DNA more inaccessible to enzymatic cleavage.
FIGURE 4.
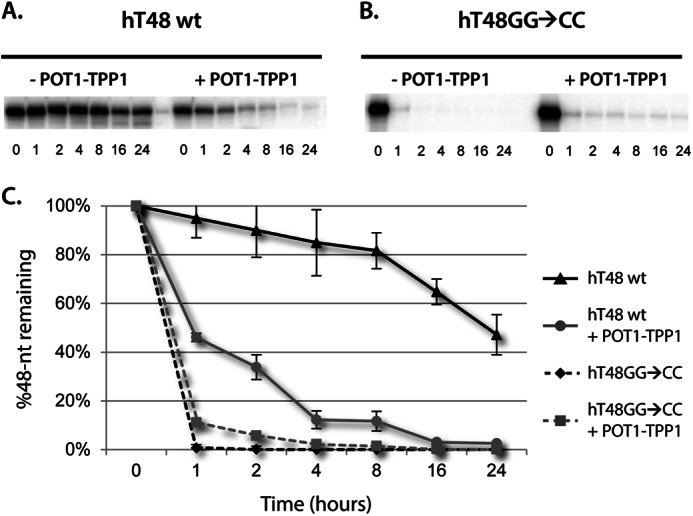
DNase I digestion of 48-nt native and mutant telomeric DNA and POT1-TPP1-DNA nucleoprotein complexes. A ssDNA probe with a native hT48wt (A) or mutant hT48GG→CC (B) telomere sequence was 5′ end-labeled and subjected to degradation by DNase I. DNA samples were either incubated with DNase I alone (-POT1-TPP1) or were first coated with POT1-TPP1 protein (+POT1-TPP1). All samples were subjected to degradation by DNase I digestion in a time course from 0–24 h, as indicated. C, the rate of degradation of each DNA 48-nt substrate with and without POT1-TPP1. The entire gels are provided in supplemental Fig. 5.
The POT1-TPP1-coated hT48wt DNA, however, was degraded readily, with only ∼10% of the full-length oligonucleotide remaining intact after 4 h and virtually all of it degraded by 16 h (Fig. 4A, right panel). The striking result that uncoated telomere DNA is better protected from DNase I digestion than POT1-TPP1-coated DNA can most likely be attributed to the tertiary structure of the formed complexes. Although POT1-TPP1-coated telomere DNA forms compact structures (23), the DNA within them is more susceptible to DNase I digestion. This is likely linked to the native function of POT1 and TPP1 to make the DNA more accessible to enzymes like telomerase, thereby allowing for telomere elongation (40).
To compare the effects of POT1 coating in the absence of TPP1, the hT48wt DNA was coated with POT1-N proteins and subjected to DNase I digestion (supplemental Fig. 4). The DNA in this complex was degraded slightly more readily than that of the DNA alone but much less than that of the POT1-TPP1-coated DNA. These data indicate a contribution of TPP1 to assist in unfolding of the DNA that is cumulatively greater than the effects of POT1 alone.
With the mutation of guanosines to cytidines, the hT48GG→CC DNA does not form G-quadruplexes, as demonstrated by circular dichroism and, as such, would be more readily digested by DNase I. To this end, we repeated the DNase I digestion assay with this mutant telomere DNA construct. As expected, the hT48GG→CC DNA was degraded rapidly (Fig. 4B, left panel, and supplemental Fig. 5). In contrast to the native hT48wt DNA construct, the hT48GG→CC construct was better protected from DNase I digestion when coated with POT1-TPP1 proteins (Fig. 4B, right panel). However, the stable moieties formed by uncoated, native hT48wt DNA were by far the most resistant to DNase I digestion (Fig. 4C). As mentioned previously, a variety of monovalent cations are able to coordinate the formation of different G-quadruplex conformations. Therefore, the DNase experiments were repeated with either potassium or lithium chloride in place of sodium chloride to determine whether different G-quadruplex conformations affected the rate of DNase digestion. These experiments demonstrated similar DNase protection in all monovalent salts tested, suggesting that the type of G-quadruplex formed does not greatly alter enzyme accessibility (supplemental Fig. 6).
DISCUSSION
The in vivo stoichiometry of human POT1-TPP1 proteins is 50–100/telomere (19). Because one POT1 protein binds essentially two hexameric DNA repeats, the amount of cellular POT1-TPP1 proteins is, therefore, more than enough to coat the 50- to 300-nt telomere overhang. In vitro, multiple POT1-TPP1 proteins do, indeed, coat long, single-stranded telomeric DNA to form compact nucleoprotein complexes (23). The crystal structure of the N-terminal, DNA-interacting domain reveals that an individual POT1 protein interacts with 10 nt of telomere DNA (22). This leaves two unbound guanosine residues between adjacent POT1 recognition sites in the ssDNA. As such, these two guanosines may serve as a linker residing between tandem bound POT1-TPP1 proteins and facilitate compaction of the assembled nucleoprotein complex. Alternatively, by contributing to G-quadruplex formation of unbound DNA, these two guanosines may help regulate multiple POT1-TPP1 binding events and, thus, govern assembly of the nucleoprotein complex. Our data expand on the intricate relationship between multiple POT1-TPP1 binding events and the secondary structure of the unbound DNA substrate. Additionally, our results relate multiple POT1-TPP1 binding with the cumulative protective properties against DNase I digestion.
Although the complete coating of four proteins onto the three DNA constructs investigated here was similar, as evidenced by an equivalent Kapp assessment, the pattern for loading of multiple proteins was altered in mutant versus native telomere DNA templates. Although the GG-to-CC mutant DNA becomes coated entirely with four POT1-TPP1 proteins, the DNA with native telomere sequence continues to display a small proportion with only one, two, or three proteins bound even at the highest protein:DNA ratios. One explanation can be attributed to putative, overlapping binding sites in the native sequence. Only DNA with a native sequence presents POT1-TPP1 proteins with an alternative binding event in which there are different registers on a given segment of telomere DNA. Conversely, the GG-to-CC mutation or the GG deletion establishes four unique and tandem POT1 recognition sites in the mutant DNA templates. This alternative binding explanation is unlikely, however, for the following reason. If POT1-TPP1 simply bound any POT1 recognition site with little or no cooperativity, then the M3 and M4 constructs should primarily present the three-protein bound state as protein concentrations are increased. This does not occur, though, with less than 1% representing the three-protein bound state for either construct (Fig. 3). Notably, the incomplete coating of hT48wt is unique to the POT1-TPP1 heterodimer because the DNA-binding domain of POT1 alone coats nearly 100% of the POT1 recognition sites on both a 48-nt (supplemental Fig. 2) and a 72-nt telomere DNA substrate (23). Together, our data indicate that a combination of native telomere DNA sequence and the inclusion of TPP1 limit the complete coating of the native telomere ssDNA in vitro.
A more plausible explanation for the differences in coating patterns is due to G-quadruplex formations that inhibit the complete coating of POT1-TPP1 proteins to the hT48wt construct. Our data show that coating of POT1-TPP1 onto an unstructured template, such as hT48GG→CC, indicates cooperative binding because four proteins saturate the template almost immediately following the first binding event. In contrast, loading onto the native template appears more gradual, presumably because subsequent binding events require further unwinding of the G-quadruplex structure.
Across many species, the protection of telomere DNA involves the high-specificity binding of multiple, specialized proteins. In all eukaryotes, the 3′ tails of telomeres end in single-stranded DNA overhangs that are recognized by POT1- or POT1-like proteins (49). Structurally, these proteins contain conserved oligonucleotide/oligosaccharide binding fold motifs that are involved in ssDNA recognition and specificity (50, 51). Recent evidence shows that full-length Schizosaccharomyces pombe POT1 molecules bind telomeric DNA repeats as a dimer (52). Taken together, these data suggest that oligomerization, potentially DNA-dependent, may facilitate cooperative binding for the telomere end-binding proteins in a range of eukaryotic organisms. Neither oligomerization nor cooperative binding, however, has been shown to occur for mammalian POT1. Nandakumar and Cech (52) hypothesize that the lack of cooperativity in human POT1 binding is due to the absence of a heterogenous DNA linker in the telomere sequence, which exists in other organisms, including budding and fission yeast. Support for this idea is observed in our data when a mutation (e.g. GG→CC) is introduced into the telomere DNA sequence to create a heterogeneous linker. Our data reveal that, when the telomeric DNA is heterogenous, the loading of multiple human POT1-TPP1 proteins onto a long ssDNA substrate is more cooperative/coordinated.
Similar to S. pombe POT1 (52), our data suggest a DNA-dependent oligomerization property of human POT1-TPP1 for loading on to a long ssDNA substrate. Human POT1 displays a more than 100-fold preference for binding sites located at the 3′ end of telomeric DNA (48). Our data show that a mutation to the penultimate POT1 binding site of a 48-nt oligonucleotide limits POT1-TPP1 binding to a single POT1-TPP1 protein, at least at lower concentrations of POT1-TPP1. At higher concentrations of protein, POT1-TPP1 eventually binds to the mutant binding sequence in DNA, which results in rapid loading of all four POT1-TPP1 proteins onto the 48-nt DNA (Fig. 3C). Loading of multiple POT1-TPP1 proteins onto a 48-nt oligonucleotide with a 3′ POT1 binding site, however, altered results in primarily two or four POT1-TPP1 proteins binding to the DNA (Fig. 3D). The maximum loading of four proteins can be explained as before. First, an initial binding event occurs at the mutant site on the 3′ end of the DNA. This binding event then facilitates immediate loading of subsequent POT1-TPP1 proteins to the oligonucleotide. We found the binding of only two POT1-TPP1 proteins to this oligonucleotide somewhat surprising. Although we cannot exclude that the two binding events occur within the DNA template and not at the 3′ mutated site, this is an unlikely explanation. If this were true, then it would be expected that three POT1-TPP1 proteins would bind the three native sites in the DNA template. Our quantitative gel shifts show that there is clearly no such population. As such, our data suggest that improper binding of a single POT1-TPP1 protein is adequate for recruitment of a second POT1-TPP1 protein. However, proper binding of two POT1-TPP1 heterodimers appears to induce coating (four POT1-TPP1 proteins) of the 48-nt DNA (Fig. 3E).
Our data show that coating of single-stranded telomere DNA with multiple POT1-TPP1 proteins accelerates degradation by DNase I. This finding was exclusive to ssDNA with a native telomere sequence, as POT1-TPP1 coating of the hT48GG→CC construct delayed its degradation by DNase I. These results indicate three primary populations of DNA: a highly stable structure consisting of native telomeric DNA in a G-quadruplex configuration, a more accessible yet ordered structure consisting of telomeric DNA (mutant or native) coated with POT1-TPP1, and a population of unstructured DNA (at least for the mutant strands) (Fig. 5). Taken together, these data support the dichotomous role of POT1-TPP1, both protecting free DNA from degradation while making it more accessible to enzymes such as DNase I and telomerase (40).
FIGURE 5.
Schematic representation of telomere complexes and their relative rates of DNase I digestion. The left panel shows a 48-nt strand of native, G-rich telomeric DNA folded into an illustrative G-quadruplex. This DNA formation is highly resistant to DNase I digestion. Both native and mutant ssDNA can be saturated with POT1-TPP1 proteins to form compact nucleoprotein complexes, as indicated in the center panel. The POT1-TPP1-coated DNA structure is degraded by DNase I more quickly than the G-quadruplex DNA but slower than that of unstructured and unbound DNA. Relatively unstructured ssDNA such as hT48GG→CC (right panel) is rapidly degraded by DNase I.
Acknowledgments
We thank Dr. Noa Noy, Dr. Bill Merrick, and Dr. Malligarjunan Rajavel for critical review of the manuscript. We also thank Dr. Elayne Podell and Dr. Tom Cech (University of Colorado at Boulder) for providing infected insect cells expressing POT1-N.
This work was supported by American Heart Association Scientist Development Grant 11SDB5580010 (to D. J. T.).

This article contains supplemental Figs. 1–6 and Table 1.
- ssDNA
- single-stranded DNA
- nt
- nucleotide(s).
REFERENCES
- 1. Moyzis R. K., Buckingham J. M., Cram L. S., Dani M., Deaven L. L., Jones M. D., Meyne J., Ratliff R. L., Wu J. R. (1988) A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl. Acad. Sci. U.S.A. 85, 6622–6626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. de Lange T. (2005) Shelterin. The protein complex that shapes and safeguards human telomeres. Genes Dev. 19, 2100–2110 [DOI] [PubMed] [Google Scholar]
- 3. de Lange T. (2009) How telomeres solve the end-protection problem. Science 326, 948–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Denchi E. L., de Lange T. (2007) Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 448, 1068–1071 [DOI] [PubMed] [Google Scholar]
- 5. Guo X., Deng Y., Lin Y., Cosme-Blanco W., Chan S., He H., Yuan G., Brown E. J., Chang S. (2007) Dysfunctional telomeres activate an ATM-ATR-dependent DNA damage response to suppress tumorigenesis. EMBO J. 26, 4709–4719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Moser B. A., Subramanian L., Khair L., Chang Y. T., Nakamura T. M. (2009) Fission yeast Tel1(ATM) and Rad3(ATR) promote telomere protection and telomerase recruitment. PLoS Genet. 5, e1000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pinto A. R., Li H., Nicholls C., Liu J. P. (2011) Telomere protein complexes and interactions with telomerase in telomere maintenance. Front. Biosci. 16, 187–207 [DOI] [PubMed] [Google Scholar]
- 8. Watson J. D. (1972) Origin of concatemeric T7 DNA. Nat. New. Biol. 239, 197–201 [DOI] [PubMed] [Google Scholar]
- 9. Harley C. B., Futcher A. B., Greider C. W. (1990) Telomeres shorten during ageing of human fibroblasts. Nature 345, 458–460 [DOI] [PubMed] [Google Scholar]
- 10. Wright W. E., Pereira-Smith O. M., Shay J. W. (1989) Reversible cellular senescence. Implications for immortalization of normal human diploid fibroblasts. Mol. Cell Biol. 9, 3088–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim N. W., Piatyszek M. A., Prowse K. R., Harley C. B., West M. D., Ho P. L., Coviello G. M., Wright W. E., Weinrich S. L., Shay J. W. (1994) Specific association of human telomerase activity with immortal cells and cancer. Science 266, 2011–2015 [DOI] [PubMed] [Google Scholar]
- 12. Wright W. E., Piatyszek M. A., Rainey W. E., Byrd W., Shay J. W. (1996) Telomerase activity in human germline and embryonic tissues and cells. Dev. Genet. 18, 173–179 [DOI] [PubMed] [Google Scholar]
- 13. Mantell L. L., Greider C. W. (1994) Telomerase activity in germline and embryonic cells of Xenopus. EMBO J. 13, 3211–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang F., Podell E. R., Zaug A. J., Yang Y., Baciu P., Cech T. R., Lei M. (2007) The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature 445, 506–510 [DOI] [PubMed] [Google Scholar]
- 15. Xin H., Liu D., Wan M., Safari A., Kim H., Sun W., O'Connor M. S., Songyang Z. (2007) TPP1 is a homologue of ciliate TEBP-β and interacts with POT1 to recruit telomerase. Nature 445, 559–562 [DOI] [PubMed] [Google Scholar]
- 16. Abreu E., Aritonovska E., Reichenbach P., Cristofari G., Culp B., Terns R. M., Lingner J., Terns M. P. (2010) TIN2-tethered TPP1 recruits human telomerase to telomeres in vivo. Mol. Cell Biol. 30, 2971–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. van Steensel B., de Lange T. (1997) Control of telomere length by the human telomeric protein TRF1. Nature 385, 740–743 [DOI] [PubMed] [Google Scholar]
- 18. Smogorzewska A., de Lange T. (2004) Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 73, 177–208 [DOI] [PubMed] [Google Scholar]
- 19. Takai K. K., Hooper S., Blackwood S., Gandhi R., de Lange T. (2010) In vivo stoichiometry of shelterin components. J. Biol. Chem. 285, 1457–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Broccoli D., Smogorzewska A., Chong L., de Lange T. (1997) Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet. 17, 231–235 [DOI] [PubMed] [Google Scholar]
- 21. Baumann P., Cech T. R. (2001) Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292, 1171–1175 [DOI] [PubMed] [Google Scholar]
- 22. Lei M., Podell E. R., Cech T. R. (2004) Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat. Struct. Mol. Biol. 11, 1223–1229 [DOI] [PubMed] [Google Scholar]
- 23. Taylor D. J., Podell E. R., Taatjes D. J., Cech T. R. (2011) Multiple POT1-TPP1 proteins coat and compact long telomeric single-stranded DNA. J. Mol. Biol. 410, 10–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. O'Connor M. S., Safari A., Xin H., Liu D., Songyang Z. (2006) A critical role for TPP1 and TIN2 interaction in high-order telomeric complex assembly. Proc. Natl. Acad. Sci. U.S.A. 103, 11874–11879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ye J. Z., Donigian J. R., van Overbeek M., Loayza D., Luo Y., Krutchinsky A. N., Chait B. T., de Lange T. (2004) TIN2 binds TRF1 and TRF2 simultaneously and stabilizes the TRF2 complex on telomeres. J. Biol. Chem. 279, 47264–47271 [DOI] [PubMed] [Google Scholar]
- 26. Choi K. H., Farrell A. S., Lakamp A. S., Ouellette M. M. (2011) Characterization of the DNA binding specificity of Shelterin complexes. Nucleic Acids Res. 39, 9206–9223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huffman K. E., Levene S. D., Tesmer V. M., Shay J. W., Wright W. E. (2000) Telomere shortening is proportional to the size of the G-rich telomeric 3′-overhang. J. Biol. Chem. 275, 19719–19722 [DOI] [PubMed] [Google Scholar]
- 28. Wright W. E., Tesmer V. M., Huffman K. E., Levene S. D., Shay J. W. (1997) Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 11, 2801–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Baumann P., Podell E., Cech T. R. (2002) Human Pot1 (protection of telomeres) protein. Cytolocalization, gene structure, and alternative splicing. Mol. Cell Biol. 22, 8079–8087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Houghtaling B. R., Cuttonaro L., Chang W., Smith S. (2004) A dynamic molecular link between the telomere length regulator TRF1 and the chromosome end protector TRF2. Curr. Biol. 14, 1621–1631 [DOI] [PubMed] [Google Scholar]
- 31. Liu D., Safari A., O'Connor M. S., Chan D. W., Laegeler A., Qin J., Songyang Z. (2004) PTOP interacts with POT1 and regulates its localization to telomeres. Nat. Cell Biol. 6, 673–680 [DOI] [PubMed] [Google Scholar]
- 32. Ye J. Z., Hockemeyer D., Krutchinsky A. N., Loayza D., Hooper S. M., Chait B. T., de Lange T. (2004) POT1-interacting protein PIP1. A telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 18, 1649–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kibe T., Osawa G. A., Keegan C. E., de Lange T. (2010) Telomere protection by TPP1 is mediated by POT1a and POT1b. Mol. Cell Biol. 30, 1059–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hockemeyer D., Palm W., Else T., Daniels J. P., Takai K. K., Ye J. Z., Keegan C. E., de Lange T., Hammer G. D. (2007) Telomere protection by mammalian Pot1 requires interaction with Tpp1. Nat. Struct. Mol. Biol. 14, 754–761 [DOI] [PubMed] [Google Scholar]
- 35. Wu L., Multani A. S., He H., Cosme-Blanco W., Deng Y., Deng J. M., Bachilo O., Pathak S., Tahara H., Bailey S. M., Deng Y., Behringer R. R., Chang S. (2006) Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell 126, 49–62 [DOI] [PubMed] [Google Scholar]
- 36. Latrick C. M., Cech T. R. (2010) POT1-TPP1 enhances telomerase processivity by slowing primer dissociation and aiding translocation. EMBO J. 29, 924–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sen D., Gilbert W. (1988) Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature 334, 364–366 [DOI] [PubMed] [Google Scholar]
- 38. Ambrus A., Chen D., Dai J., Bialis T., Jones R. A., Yang D. (2006) Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 34, 2723–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kypr J., Kejnovská I., Renciuk D., Vorlícková M. (2009) Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 37, 1713–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zaug A. J., Podell E. R., Cech T. R. (2005) Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc. Natl. Acad. Sci. U.S.A. 102, 10864–10869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang H., Nora G. J., Ghodke H., Opresko P. L. (2011) Single molecule studies of physiologically relevant telomeric tails reveal POT1 mechanism for promoting G-quadruplex unfolding. J. Biol. Chem. 286, 7479–7489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Morin G. B. (1989) The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell 59, 521–529 [DOI] [PubMed] [Google Scholar]
- 43. Greider C. W. (1991) Telomerase is processive. Mol. Cell Biol. 11, 4572–4580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nandakumar J., Bell C. F., Weidenfeld I., Zaug A. J., Leinwand L. A., Cech T. R. (2012) The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature 492, 285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhong F. L., Batista L. F., Freund A., Pech M. F., Venteicher A. S., Artandi S. E. (2012) TPP1 OB-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell 150, 481–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Buczek P., Horvath M. P. (2006) Structural reorganization and the cooperative binding of single-stranded telomere DNA in Sterkiella nova. J. Biol. Chem. 281, 40124–40134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Williams T. L., Levy D. L., Maki-Yonekura S., Yonekura K., Blackburn E. H. (2010) Characterization of the yeast telomere nucleoprotein core. Rap1 binds independently to each recognition site. J. Biol. Chem. 285, 35814–35824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Loayza D., Parsons H., Donigian J., Hoke K., de Lange T. (2004) DNA binding features of human POT1. A nonamer 5′-TAGGGTTAG-3′ minimal binding site, sequence specificity, and internal binding to multimeric sites. J. Biol. Chem. 279, 13241–13248 [DOI] [PubMed] [Google Scholar]
- 49. Baumann P., Price C. (2010) Pot1 and telomere maintenance. FEBS Lett. 584, 3779–3784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Horvath M. P. (2011) Structural anatomy of telomere OB proteins. Crit. Rev. Biochem. Mol. Biol. 46, 409–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lewis K. A., Wuttke D. S. (2012) Telomerase and telomere-associated proteins. Structural insights into mechanism and evolution. Structure 20, 28–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nandakumar J., Cech T. R. (2012) DNA-induced dimerization of the single-stranded DNA binding telomeric protein Pot1 from Schizosaccharomyces pombe. Nucleic Acids Res. 40, 235–244 [DOI] [PMC free article] [PubMed] [Google Scholar]





