Background: It is unknown if phosphatidylethanolamine (PE), the major non-bilayer-forming mitochondrial phospholipid, is involved in the biogenesis of outer membrane proteins.
Results: Depletion of PE impairs import of β-barrel proteins by the outer membrane translocase TOM.
Conclusion: PE is required for full activity but not stability of TOM.
Significance: PE plays a different role in the biogenesis of mitochondrial outer membrane proteins compared with cardiolipin.
Keywords: Membrane Biogenesis, Mitochondria, Phospholipid, Protein Targeting, Protein Translocation
Abstract
The mitochondrial outer membrane contains proteinaceous machineries for the import and assembly of proteins, including TOM (translocase of the outer membrane) and SAM (sorting and assembly machinery). It has been shown that the dimeric phospholipid cardiolipin is required for the stability of TOM and SAM complexes and thus for the efficient import and assembly of β-barrel proteins and some α-helical proteins of the outer membrane. Here, we report that mitochondria deficient in phosphatidylethanolamine (PE), the second non-bilayer-forming phospholipid, are impaired in the biogenesis of β-barrel proteins, but not of α-helical outer membrane proteins. The stability of TOM and SAM complexes is not disturbed by the lack of PE. By dissecting the import steps of β-barrel proteins, we show that an early import stage involving translocation through the TOM complex is affected. In PE-depleted mitochondria, the TOM complex binds precursor proteins with reduced efficiency. We conclude that PE is required for the proper function of the TOM complex.
Introduction
Mitochondrial outer membrane proteins fulfill essential functions in the eukaryotic cell, including transport of metabolites and communication with the cytosol and other organelles. Porin, also termed VDAC (for voltage-dependent anion channel), mediates the transport of metabolites (1). TOM (translocase of the outer membrane) forms the main entry gate for mitochondrial precursor proteins that are synthesized on cytosolic ribosomes (2–8). ERMES (endoplasmic reticulum-mitochondria encounter structure) tethers mitochondria to the endoplasmic reticulum (9), and the outer membrane harbors components important for mitochondrial fusion and fission (10–12). Additionally, the outer membrane plays an important role in apoptosis (13). Thus, the biogenesis of outer membrane proteins and their correct assembly into protein machineries are essential for mitochondrial function.
The outer membrane contains two types of integral membrane proteins: proteins with α-helical transmembrane segments and β-barrel proteins. All outer membrane proteins are synthesized on cytosolic ribosomes and are targeted to the mitochondrial surface. β-Barrel precursor proteins are transported via the protein-conducting channel Tom40 of the TOM complex (14–16). Subsequently, small TIM chaperones of the intermembrane space guide the β-barrel precursors to SAM (sorting and assembly machinery) (15–19). The SAM complex, also called the TOB (topogenesis of mitochondrial outer membrane β-barrel proteins) complex, binds to a conserved β-signal of the precursor protein and mediates the insertion into the outer membrane (15, 16, 19–23). For α-helical outer membrane proteins, a common import pathway has not been found. The receptor protein Tom70 and the mitochondrial import proteins Mim1 and Mim2 play a crucial role in the biogenesis of multispanning outer membrane proteins such as Ugo1, whereas TOM core subunits and the SAM complex are not involved (24–27). Additionally, Mim1 is also involved in the import of single-spanning outer membrane proteins such as Tom20 and small Tom proteins (25–29). A specialized SAM form containing Mdm10 (mitochondrial distribution and morphology protein 10) mediates the biogenesis of Tom22, whereas other single-spanning precursor proteins insert independently of known proteinaceous factors but can be impaired by elevated levels of ergosterol (31–38).
The outer membrane of mitochondria contains two non-bilayer-forming phospholipids, cardiolipin (CL)5 and phosphatidylethanolamine (PE) (39–41). In yeast mitochondria, Crd1 (cardiolipin synthase 1) and Psd1 (phosphatidylserine decarboxylase 1) catalyze the formation of CL and PE, respectively (42–48). Minor amounts of mitochondrial PE can also be provided by the activity of Psd2 at the vacuole/Golgi membranes, the acyltransferases Tgl3 and Ale1, and the CDP-ethanolamine pathway (49–53). Additionally, several mitochondrial proteins were reported to regulate the levels of CL and PE (9, 54–62). Lack of CL affects stability and function of various mitochondrial membrane protein complexes, including the respiratory chain of the inner membrane (55, 56, 58, 63–71). Although the outer membrane contains only small amounts of CL, lack of CL causes destabilization of TOM and SAM complexes and thus reduced precursor binding by the translocases. The biogenesis of β-barrel proteins and the assembly of some α-helical outer membrane proteins are impaired in CL-deficient mitochondria (41). A double deletion of PSD1 and CRD1 is lethal, suggesting overlapping functions of PE and CL that are essential for cell viability (72). Depletion of CL as well as of PE leads to a decreased activity of the respiratory chain, which in turn impairs preprotein import into the inner membrane and matrix due to a reduced inner membrane potential (58, 63, 73, 74). CL and PE also have overlapping functions in the fusion of mitochondria (75). However, because neither respiratory activity nor mitochondrial fusion is strictly essential for the cell viability of yeast, the effects of CL and PE on respiration and fusion cannot explain the synthetic lethality of the double deletion.
The role of PE in the biogenesis of mitochondrial outer membrane proteins has not been addressed so far, although PE is one of the major phospholipids of the outer membrane (39, 40). For this study, we analyzed the biogenesis of outer membrane proteins in PE-depleted mitochondria. Although no defect in the biogenesis of α-helical proteins was observed, the import of β-barrel proteins was impaired at the stage of translocation through the TOM complex. The stability of TOM and SAM complexes was not affected, yet the TOM complex bound precursor proteins with reduced efficiency. We conclude that PE is required for the efficient function of the TOM machinery.
EXPERIMENTAL PROCEDURES
Yeast Strains, Growth Conditions, and Isolation of Mitochondria and Outer Membrane Vesicles
The yeast strains crd1Δ, psd1Δ, and psd1Δ psd2Δ and the corresponding wild-type strain BY4741 have been described (41, 76). Cells were grown in YPLac or YPG medium (58) at 21–30 °C to an early logarithmic growth phase. Mitochondria were isolated, adjusted to a protein concentration of 10 mg/ml, aliquoted, and shock-frozen with liquid nitrogen as described (77). Outer membrane vesicles were isolated from purified mitochondria via sucrose density centrifugation as described (40).
Protein Import into Mitochondria
For import studies, 35S-labeled precursor proteins were synthesized with the TnT coupled transcription/translation kit (Promega). The import was performed as described (77). To remove non-imported precursor proteins, proteinase K was added to a final concentration of 50 μg/ml, and the samples were incubated for 15 min on ice. The activity of the protease was blocked by addition of PMSF to a final concentration of 2 mm. For blue native electrophoresis, mitochondria were solubilized with 1% (w/v) digitonin in digitonin buffer (20 mm Tris-HCl (pH 7.4), 50 mm NaCl, 0.1 mm EDTA, and 10% (v/v) glycerol) for 15 min on ice. After a clarifying spin (16,100 × g, 10 min, 4 °C), samples were loaded on a blue native gel. The blue native gel was prepared as described (77).
Co-immunoprecipitation
Mitochondria were solubilized with 1% digitonin in digitonin buffer and incubated with protein A-Sepharose (GE Healthcare) coupled to Tom5-specific antibodies or preimmune antibodies. Binding was performed for 1 h at 4 °C under constant rotation. After excessive washing, bound proteins were eluted with 0.1 m glycine (pH 2.5) and subjected to SDS-PAGE.
Determination of Mitochondrial Phospholipid Distribution
Lipids were extracted from isolated mitochondria with chloroform/methanol (2:1, v/v) as described (78). Subsequently, washing steps of the organic phase with 0.034% (w/v) MgCl2 solution, 2 n KCl/methanol (4:1, v/v), and methanol/water/chloroform (48:47:3, per volume) were performed. Phospholipids were separated by thin-layer chromatography as described (48). Phospholipids were detected by iodine vapor, scrapped off, and quantified (79).
RESULTS
Biogenesis of β-Barrel Proteins Is Impaired in PE-deficient Mitochondria
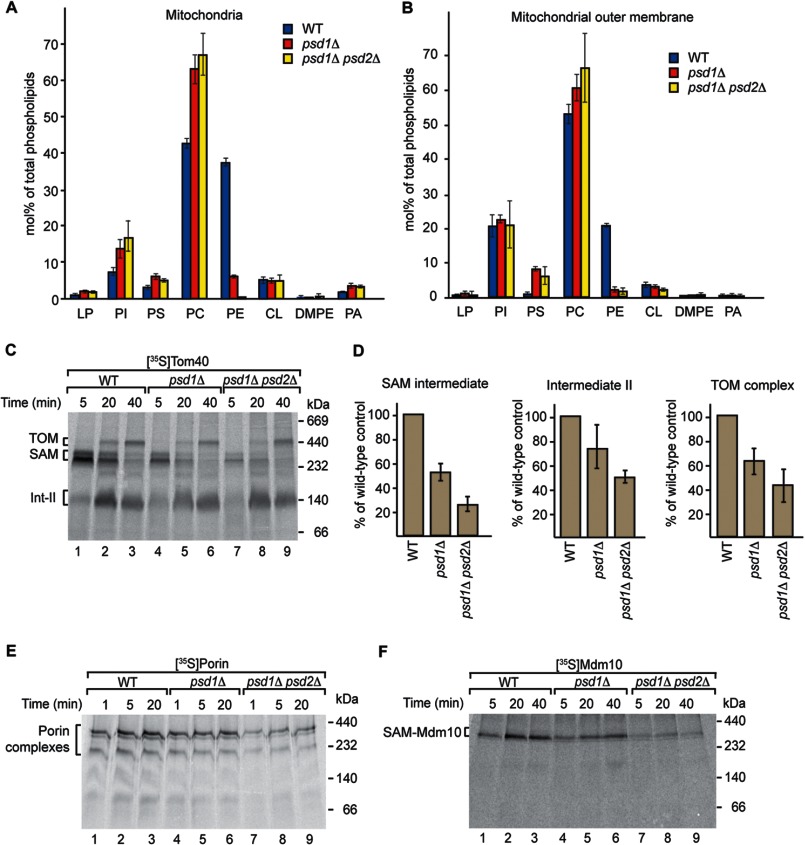
To study the role of PE in the biogenesis of mitochondrial outer membrane proteins, we isolated mitochondria from the psd1Δ yeast strain and the psd1Δ psd2Δ double deletion strain (47, 73, 76). As expected, the levels of PE were strongly reduced (Fig. 1A and B). The levels of phosphatidylcholine (PC), phosphatidylinositol, and phosphatidylserine were increased in the mutant mitochondria, whereas the level of CL was unchanged (Fig. 1A) (47). We also determined the phospholipid distribution in purified mitochondrial outer membrane vesicles. Similar to purified mitochondria, the levels of phosphatidylserine and PC were increased in the mutants, whereas the level of phosphatidylinositol remained unchanged (Fig. 1B). To study the biogenesis of β-barrel proteins, we imported the 35S-labeled precursor of Tom40 into isolated wild-type and mutant mitochondria. The import of this model β-barrel precursor allows the analysis of distinct assembly steps, which can be visualized by blue native gel electrophoresis (Fig. 1C, lanes 1–3) (14–16, 20, 21, 80–82). Upon short import times, the Tom40 precursor binds to the SAM complex. Subsequently, the precursor is released to form a second intermediate and finally assembles into the mature TOM complex of ∼450 kDa (16). In the PE-depleted mutant mitochondria, all assembly steps of Tom40 were reduced (Fig. 1, C, lanes 4–9, and D). Binding of the Tom40 precursor to the SAM complex was considerably decreased, whereas formation of intermediate II and the mature TOM complex was moderately affected (Fig. 1D). We studied the assembly of two additional β-barrel proteins, porin and Mdm10, by monitoring formation of the mature complexes by blue native electrophoresis (16, 31). The biogenesis of both proteins was impaired by the depletion of PE (Fig. 1, E and F).
FIGURE 1.
Biogenesis of β-barrel proteins is impaired in PE-depleted mitochondria. A, the phospholipid distribution of isolated WT, psd1Δ, and psd1Δ psd2Δ mitochondria was determined. Means ± S.E. (n = 4) are shown for the mitochondrial preparation. B, the phospholipid distribution of isolated WT, psd1Δ, and psd1Δ psd2Δ outer membrane vesicles was determined. The means with range from two independent experiments are depicted. LP, lysophospholipids; PI, phosphatidylinositol; PS, phosphatidylserine; DMPE, dimethylphosphatidylethanolamine; PA, phosphatidic acid. C, 35S-labeled Tom40 was imported into isolated WT, psd1Δ, or psd1Δ psd2Δ mitochondria at 25 °C for the indicated time periods. The mitochondria were lysed with digitonin and analyzed by blue native electrophoresis and digital autoradiography. SAM, Tom40 precursor bound to the SAM complex; Int-II, second intermediate of the Tom40 assembly pathway. D, quantification of the three assembly steps of Tom40 (import was performed and analyzed as described for C). Means ± S.E. (n = 5) are shown for the formation of the SAM intermediate after 5 min of import, intermediate II after 20 min of import, and the mature TOM complex after 40 min of import. E and F, porin or Mdm10, respectively, was imported into isolated WT, psd1Δ, or psd1Δ psd2Δ mitochondria at 25 °C for the indicated time periods. The mitochondria were lysed with digitonin and analyzed by blue native electrophoresis and digital autoradiography.
We also analyzed the biogenesis of three outer membrane proteins that contain α-helical transmembrane segments: Tom22, Tom20, and Ugo1. In contrast to the biogenesis defects of β-barrel precursors, the assembly of 35S-labeled Tom22 and Tom20 into the TOM complex was not inhibited in psd1Δ and psd1Δ psd2Δ mitochondria (Fig. 2, A and B). The assembly of Tom20 was mildly increased in psd1Δ psd2Δ mitochondria (Fig. 2B, lanes 7–9). Similarly, import of Ugo1, determined by formation of the mature dimer (25, 26), was not decreased in the mutant mitochondria and was even enhanced (Fig. 2C). We conclude that PE-depleted mitochondria are impaired in the import of β-barrel proteins but not in the biogenesis of several α-helical outer membrane proteins.
FIGURE 2.
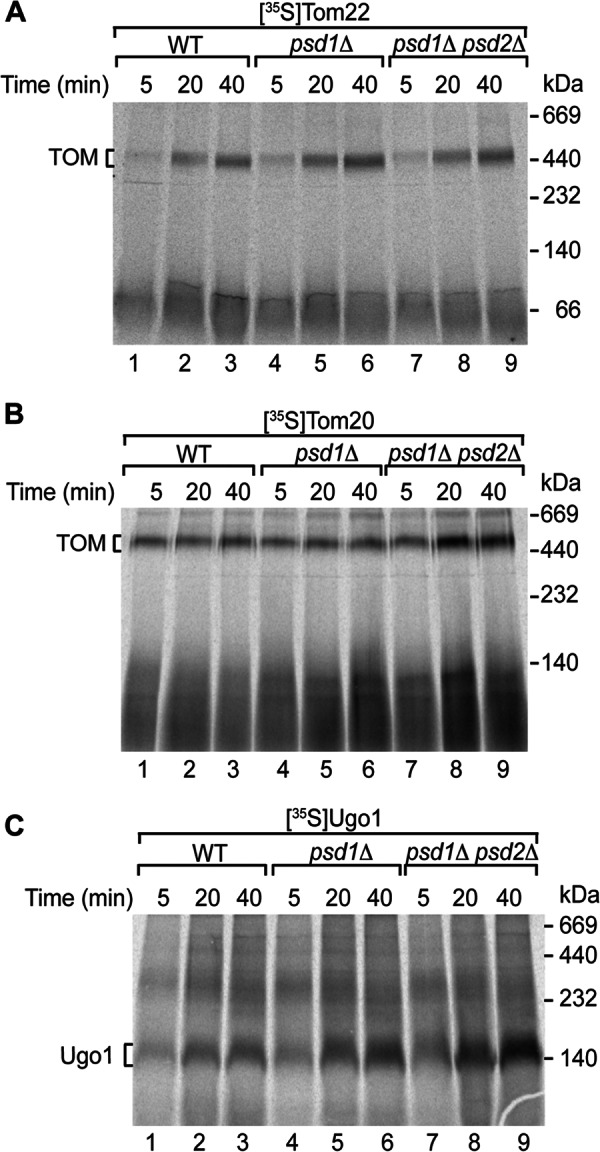
Biogenesis of α-helical outer membrane proteins is not inhibited in PE-depleted mitochondria. 35S-Labeled Tom22 (A), Tom20 (B), or Ugo1 (C) was imported into isolated WT, psd1Δ, or psd1Δ psd2Δ mitochondria at 25 °C for the indicated time periods. The mitochondria were lysed with digitonin and analyzed by blue native electrophoresis and digital autoradiography.
Transport of β-Barrel Proteins across the TOM Complex Is Impaired in PE-depleted Mitochondria
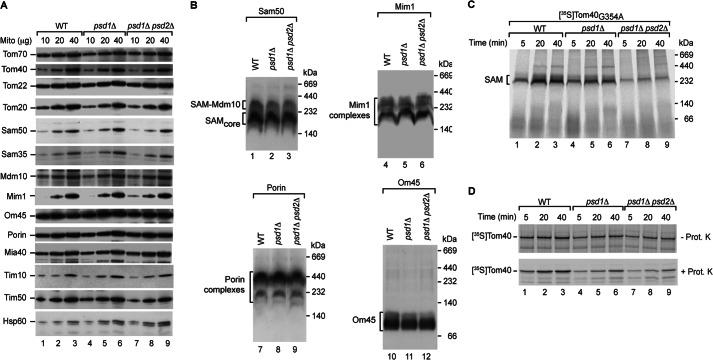
To address how PE depletion may impair β-barrel biogenesis, we first analyzed the steady-state levels of mitochondrial proteins, including TOM and SAM subunits, Mim1, and control proteins of different mitochondrial compartments. The protein levels were not markedly altered and thus could not explain the defect in β-barrel assembly (Fig. 3A).
FIGURE 3.
Impaired transport of β-barrel precursors across the outer membrane of PE-depleted mitochondria. A, proteins of isolated WT, psd1Δ, and psd1Δ psd2Δ mitochondria were analyzed by SDS-PAGE and detected by immunodecoration with the indicated antisera. B, WT, psd1Δ, and psd1Δ psd2Δ mitochondria were lysed with digitonin and separated by blue native electrophoresis. Protein complexes were detected by immunodecoration with the indicated antisera. C, 35S-labeled Tom40(G354A) was imported into WT, psd1Δ, and psd1Δ psd2Δ mitochondria. The imported proteins were analyzed by blue native electrophoresis and autoradiography. D, 35S-labeled Tom40 was imported into WT, psd1Δ, and psd1Δ psd2Δ mitochondria, followed by proteinase K (Prot. K) treatment as indicated. Proteins were separated by SDS-PAGE.
In crd1Δ mitochondria lacking CL, the assembly of Tom40 is compromised. In this case, the assembly defect is at least partially caused by a destabilized SAM complex (41). We asked if the SAM complex is affected in PE-depleted mitochondria. We lysed mitochondria with the nonionic detergent digitonin and studied outer membrane protein complexes by blue native electrophoresis (Fig. 3B). The two SAM complexes, SAMcore and SAM-Mdm10, as well as additional outer membrane protein complexes, Mim1, Om45, and porin (25, 31, 36, 83–87), were not or only moderately affected by depletion of PE (Fig. 3B).
To identify the PE-dependent stage of β-barrel biogenesis, we dissected the biogenesis of Tom40 into distinct steps. To analyze binding of the β-barrel precursor to the SAM complex, we imported a mutant form of Tom40 that is blocked in release from the SAM complex due to a single amino acid exchange in the β-signal (22). The mutant Tom40 precursor efficiently accumulated at the SAM complex of wild-type mitochondria (Fig. 3C, lanes 1–3). Binding of the Tom40 precursor to the SAM complex was impaired in psd1Δ mitochondria and strongly inhibited in psd1Δ psd2Δ mitochondria (Fig. 3C, lanes 4–9). Thus, PE is involved in an early step of the Tom40 assembly pathway, at the stage of binding to the SAM complex or at an earlier stage leading to the SAM complex.
Precursors of β-barrel proteins are initially imported by the TOM complex to the intermembrane space side (14–18, 88, 89). This initial import step cannot be resolved by blue native electrophoresis, as no blue native-stable intermediate is formed, but can be analyzed by protection of the precursor against externally added protease (15–18, 21, 81). Therefore, we imported Tom40 into the mutant mitochondria and determined the accessibility to added proteinase K. The amount of protease-protected Tom40 precursor was reduced in the PE-depleted mitochondria (Fig. 3D, lower panel). Moreover, the binding of the Tom40 precursor to the mitochondrial surface was moderately decreased (Fig. 3D, upper panel). We conclude that PE is required at an early stage of Tom40 import into mitochondria that takes place before binding to the SAM complex, i.e. for the initial translocation of the precursor across the outer membrane by the TOM complex. The analysis of Tom22 assembly supports the conclusion that the SAM complex was not generally (unspecifically) damaged by lack of PE because the biogenesis of Tom22 depends on each SAM subunit (31, 33, 36, 81, 84, 90) but is not altered in PE-depleted mitochondria (Fig. 2A).
PE Is Required for the Function but Not the Stability of the TOM Complex
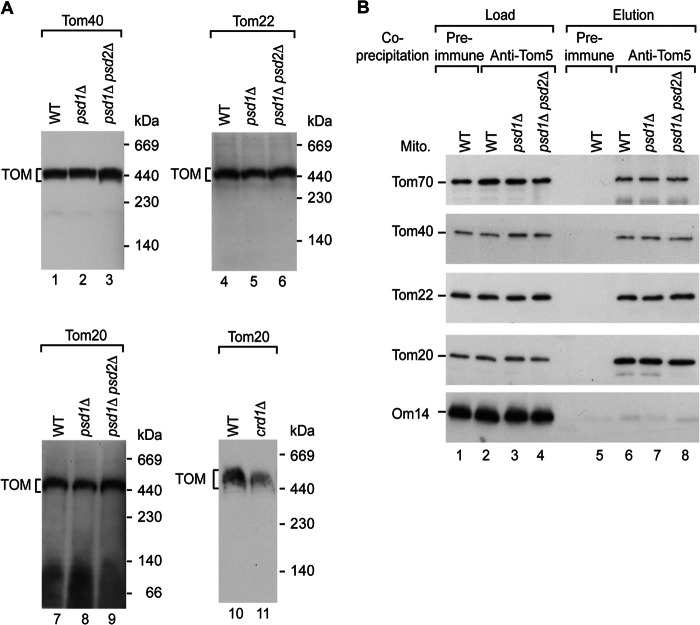
We asked if PE is required for the composition and/or function of the TOM complex. We resolved the TOM complex by blue native electrophoresis. For crd1Δ mitochondria, it has been shown that the interaction of Tom20 with the TOM complex is disturbed (Fig. 4A, lane 11) (41). In contrast, the TOM complex of psd1Δ and psd1Δ psd2Δ mitochondria was not altered on blue native electrophoresis. Tom40, Tom22, and Tom20 were present in the mature TOM complex in both mutants to a similar extent as in wild-type mitochondria (Fig. 4A, lanes 1–9). Furthermore, co-immunoprecipitation with Tom5-specific antibodies revealed that the reduced PE content did not disturb the association of the Tom20, Tom22, and Tom70 receptors with the Tom40-Tom5 core of the TOM complex (Fig. 4B) (91). Thus, the stability of the TOM translocon is not altered in psd1Δ and psd1Δ psd2Δ mitochondria.
FIGURE 4.
Depletion of PE does not affect the stability of the TOM complex. A, WT, psd1Δ, psd1Δ psd2Δ, and crd1Δ mitochondria were lysed with digitonin and separated by blue native electrophoresis. Protein complexes were detected by immunodecoration with the indicated antisera. B, WT, psd1Δ, and psd1Δ psd2Δ mitochondria (Mito.) were lysed with digitonin and subjected to co-immunoprecipitation with the indicated antisera. Proteins were eluted, separated by SDS-PAGE, and detected by immunodecoration with the indicated antisera (4% load and 100% elution).
To directly analyze the activity of the TOM complex in the interaction with preproteins, we used the presequence-carrying Oxa1 precursor that is targeted to the inner membrane. In the absence of an inner membrane potential, Oxa1 is efficiently arrested at the TOM complex of the outer membrane and forms a blue native-stable intermediate (41, 92, 93). The interaction of 35S-labeled Oxa1 with the TOM complex was strongly decreased in PE-depleted mitochondria (Fig. 5). In the psd1Δ psd2Δ mitochondria, the formation of the Oxa1-TOM intermediate was virtually blocked (Fig. 5), demonstrating that PE is required for the proper function of the TOM complex.
FIGURE 5.
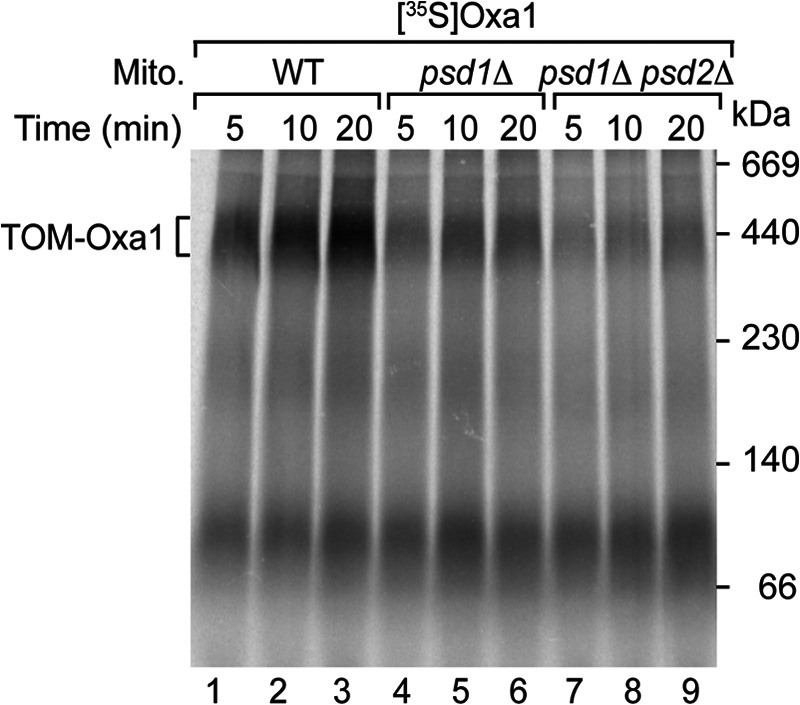
PE is required for precursor binding to the TOM complex. 35S-Labeled Oxa1 was imported into WT, psd1Δ, and psd1Δ psd2Δ mitochondria (Mito.) in the absence of a membrane potential. The samples were analyzed by blue native electrophoresis and autoradiography.
DISCUSSION
We have demonstrated that PE is required for the full activity of the main protein entry gate of mitochondria. The capability of the TOM complex to interact with precursor proteins is strongly compromised in PE-depleted mitochondria. PE is thus required for the initial translocation of β-barrel precursor proteins across the outer membrane that occurs through the TOM complex. The biogenesis of outer membrane proteins with α-helical membrane anchors, which are not transported through the TOM channel (25–30, 33), is not inhibited by depletion of PE, demonstrating that the protein import activity of the outer membrane is not generally impaired.
PE is the most abundant non-bilayer-forming phospholipid of the outer membrane (Fig. 1B) (39, 40). Several membrane proteins require non-bilayer-forming lipids for optimal activity (94, 95). Therefore, the shift to a higher level of bilayer-forming lipids such as phosphatidylserine and PC in the outer membrane might affect the activity of the TOM complex in PE-depleted mitochondria. It has been shown that PE can assist in the refolding of integral membrane proteins (96). Although the overall composition and stability of the TOM complex do not depend on the presence of PE, it is conceivable that PE may play a more specific role in the proper conformation of some TOM subunits and thus may be required for the full function of the TOM complex.
Surprisingly, the biogenesis of some α-helical outer membrane proteins such as Ugo1 and Tom20 is enhanced in PE-deficient mitochondria. These precursor proteins use the Mim1 complex on their biogenesis pathway and not the TOM channel (25–30). It will be interesting to address in future studies if PE plays an inhibitory role in the Mim1 pathway or if the increased levels of the phospholipids phosphatidylserine and PC in PE-deficient outer membranes support the activity of components of the Mim1 pathway.
PE is not required for the overall stability of the TOM and SAM complexes of the mitochondrial outer membrane, whereas lack of CL results in destabilization of both TOM and SAM (41). Thus, both non-bilayer-forming phospholipids CL and PE are required for the biogenesis of mitochondrial outer membrane proteins, but they play different roles. Whereas CL affects the stability and thus also the function of several translocase complexes (41), PE plays a selective role in the activity of the TOM complex. Because Tom40 is essential for cell viability, its biogenesis is rate-limiting for the growth of yeast (4, 5, 7, 8, 97, 98). The involvement of CL and PE in the biogenesis pathway of Tom40 provides a possible explanation for the synthetic lethality of a double deletion of PSD1 and CRD1 (72).
Acknowledgments
We thank Dr. Martin van der Laan for discussion and Nicole Zufall for expert technical assistance.
This work was supported by Deutsche Forschungsgemeinschaft Sonderforschungsbereich 746, Excellence Initiative of the German Federal and State Governments Grants EXC 294 BIOSS and GSC-4 (Spemann Graduate School), the Bundesministerium für Bildung und Forschung, and Austrian Science Fund FWF Project 21429 and DK Molecular Enzymology Grant W901-B05 (to G. D.).
- CL
- cardiolipin
- PE
- phosphatidylethanolamine
- PC
- phosphatidylcholine.
REFERENCES
- 1. Colombini M. (2012) VDAC structure, selectivity, and dynamics. Biochim. Biophys. Acta 1818, 1457–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mihara K. (2000) Targeting and insertion of nuclear-encoded preproteins into the mitochondrial outer membrane. BioEssays 22, 364–371 [DOI] [PubMed] [Google Scholar]
- 3. Koehler C. M. (2004) New developments in mitochondrial assembly. Annu. Rev. Cell Dev. Biol. 20, 309–335 [DOI] [PubMed] [Google Scholar]
- 4. Dolezal P., Likic V., Tachezy J., Lithgow T. (2006) Evolution of the molecular machines for protein import into mitochondria. Science 313, 314–318 [DOI] [PubMed] [Google Scholar]
- 5. Neupert W., Herrmann J. M. (2007) Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76, 723–749 [DOI] [PubMed] [Google Scholar]
- 6. Baker M. J., Frazier A. E., Gulbis J. M., Ryan M. T. (2007) Mitochondrial protein-import machinery: correlating structure with function. Trends Cell Biol. 17, 456–464 [DOI] [PubMed] [Google Scholar]
- 7. Endo T., Yamano K. (2009) Multiple pathways for mitochondrial protein traffic. Biol. Chem. 390, 723–730 [DOI] [PubMed] [Google Scholar]
- 8. Becker T., Böttinger L., Pfanner N. (2012) Mitochondrial protein import: from transport pathways to an integrated network. Trends Biochem. Sci. 37, 85–91 [DOI] [PubMed] [Google Scholar]
- 9. Kornmann B., Currie E., Collins S. R., Schuldiner M., Nunnari J., Weissman J. S., Walter P. (2009) An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science 325, 477–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Okamoto K., Shaw J. M. (2005) Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu. Rev. Genet. 39, 503–536 [DOI] [PubMed] [Google Scholar]
- 11. Hoppins S., Lackner L., Nunnari J. (2007) The machines that divide and fuse mitochondria. Annu. Rev. Biochem. 76, 751–780 [DOI] [PubMed] [Google Scholar]
- 12. Westermann B. (2010) Mitochondrial fusion and fission in cell life and death. Nat. Rev. Mol. Cell Biol. 11, 872–884 [DOI] [PubMed] [Google Scholar]
- 13. Taylor R. C., Cullen S. P., Martin S. J. (2008) Apoptosis: controlled demolition at the cellular level. Nat. Rev. Mol. Cell Biol. 9, 231–241 [DOI] [PubMed] [Google Scholar]
- 14. Model K., Meisinger C., Prinz T., Wiedemann N., Truscott K. N., Pfanner N., Ryan M. T. (2001) Multistep assembly of the protein import channel of the mitochondrial outer membrane. Nat. Struct. Biol. 8, 361–370 [DOI] [PubMed] [Google Scholar]
- 15. Paschen S. A., Waizenegger T., Stan T., Preuss M., Cyrklaff M., Hell K., Rapaport D., Neupert W. (2003) Evolutionary conservation of biogenesis of β-barrel membrane proteins. Nature 426, 862–866 [DOI] [PubMed] [Google Scholar]
- 16. Wiedemann N., Kozjak V., Chacinska A., Schönfisch B., Rospert S., Ryan M. T., Pfanner N., Meisinger C. (2003) Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature 424, 565–571 [DOI] [PubMed] [Google Scholar]
- 17. Wiedemann N., Truscott K. N., Pfannschmidt S., Guiard B., Meisinger C., Pfanner N. (2004) Biogenesis of the protein import channel Tom40 of the mitochondrial outer membrane. Intermembrane space components are involved in an early stage of the assembly pathway. J. Biol. Chem. 279, 18188–18194 [DOI] [PubMed] [Google Scholar]
- 18. Hoppins S. C., Nargang F. E. (2004) The Tim8-Tim13 complex of Neurospora crassa functions in the assembly of proteins into both mitochondrial membranes. J. Biol. Chem. 279, 12396–12405 [DOI] [PubMed] [Google Scholar]
- 19. Gentle I., Gabriel K., Beech P., Waller R., Lithgow T. (2004) The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J. Cell Biol. 164, 19–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ishikawa D., Yamamoto H., Tamura Y., Moritoh K., Endo T. (2004) Two novel proteins in the mitochondrial outer membrane mediate β-barrel protein assembly. J. Cell Biol. 166, 621–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chan N. C., Lithgow T. (2008) The peripheral membrane subunits of the SAM complex function codependently in mitochondrial outer membrane biogenesis. Mol. Biol. Cell 19, 126–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kutik S., Stojanovski D., Becker L., Becker T., Meinecke M., Krüger V., Prinz C., Meisinger C., Guiard B., Wagner R., Pfanner N., Wiedemann N. (2008) Dissecting membrane insertion of mitochondrial β-barrel proteins. Cell 132, 1011–1024 [DOI] [PubMed] [Google Scholar]
- 23. Walther D. M., Papic D., Bos M. P., Tommassen J., Rapaport D. (2009) Signals in bacterial β-barrel proteins are functional in eukaryotic cells for targeting to and assembly in mitochondria. Proc. Natl. Acad. Sci. U.S.A. 106, 2531–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Otera H., Taira Y., Horie C., Suzuki Y., Suzuki H., Setoguchi K., Kato H., Oka T., Mihara K. (2007) A novel insertion pathway of mitochondrial outer membrane proteins with multiple transmembrane segments. J. Cell Biol. 179, 1355–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Becker T., Wenz L. S., Krüger V., Lehmann W., Müller J. M., Goroncy L., Zufall N., Lithgow T., Guiard B., Chacinska A., Wagner R., Meisinger C., Pfanner N. (2011) The mitochondrial import protein Mim1 promotes biogenesis of multi-spanning outer membrane proteins. J. Cell Biol. 194, 387–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Papic D., Krumpe K., Dukanovic J., Dimmer K. S., Rapaport D. (2011) Multispan mitochondrial outer membrane protein Ugo1 follows a unique Mim1-dependent import pathway. J. Cell Biol. 194, 397–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dimmer K. S., Papić D., Schumann B., Sperl D., Krumpe K., Walther D. M., Rapaport D. (2012) A crucial role of Mim2 in the biogenesis of mitochondrial outer membrane proteins. J. Cell Sci. 125, 3464–3473 [DOI] [PubMed] [Google Scholar]
- 28. Becker T., Pfannschmidt S., Guiard B., Stojanovski D., Milenkovic D., Kutik S., Pfanner N., Meisinger C., Wiedemann N. (2008) Biogenesis of the mitochondrial TOM complex. Mim1 promotes insertion and assembly of signal-anchored receptors. J. Biol. Chem. 283, 120–127 [DOI] [PubMed] [Google Scholar]
- 29. Hulett J. M., Lueder F., Chan N. C., Perry A. J., Wolynec P., Likić V. A., Gooley P. R., Lithgow T. (2008) The transmembrane segment of Tom20 is recognized by Mim1 for docking to the mitochondrial TOM complex. J. Mol. Biol. 376, 694–704 [DOI] [PubMed] [Google Scholar]
- 30. Popov-Celeketić J., Waizenegger T., Rapaport D. (2008) Mim1 functions in an oligomeric form to facilitate the integration of Tom20 into the mitochondrial outer membrane. J. Mol. Biol. 376, 671–680 [DOI] [PubMed] [Google Scholar]
- 31. Meisinger C., Rissler M., Chacinska A., Sanjuán Szklarz L. K., Milenkovic D., Kozjak V., Schönfisch B., Lohaus C., Meyer H. E., Yaffe M. P., Guiard B., Wiedemann N., Pfanner N. (2004) The mitochondrial morphology protein Mdm10 functions in assembly of the preprotein translocase of the outer membrane. Dev. Cell 7, 61–71 [DOI] [PubMed] [Google Scholar]
- 32. Setoguchi K., Otera H., Mihara K. (2006) Cytosolic factor- and TOM-independent import of C-tail-anchored mitochondrial outer membrane proteins. EMBO J. 25, 5635–5647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stojanovski D., Guiard B., Kozjak-Pavlovic V., Pfanner N., Meisinger C. (2007) Alternative function for the mitochondrial SAM complex in biogenesis of α-helical TOM proteins. J. Cell Biol. 179, 881–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Meineke B., Engl G., Kemper C., Vasiljev-Neumeyer A., Paulitschke H., Rapaport D. (2008) The outer membrane form of the mitochondrial protein Mcr1 follows a TOM-independent membrane insertion pathway. FEBS Lett. 582, 855–860 [DOI] [PubMed] [Google Scholar]
- 35. Kemper C., Habib S. J., Engl G., Heckmeyer P., Dimmer K. S., Rapaport D. (2008) Integration of tail-anchored proteins into the mitochondrial outer membrane does not require any known import components. J. Cell Sci. 121, 1990–1998 [DOI] [PubMed] [Google Scholar]
- 36. Becker T., Wenz L. S., Thornton N., Stroud D., Meisinger C., Wiedemann N., Pfanner N. (2011) Biogenesis of mitochondria: dual role of Tom7 in modulating assembly of the preprotein translocase of the outer membrane. J. Mol. Biol. 405, 113–124 [DOI] [PubMed] [Google Scholar]
- 37. Krumpe K., Frumkin I., Herzig Y., Rimon N., Özbalci C., Brügger B., Rapaport D., Schuldiner M. (2012) Ergosterol content specifies targeting of tail-anchored proteins to mitochondrial outer membranes. Mol. Biol. Cell 23, 3927–3935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Merklinger E., Gofman Y., Kedrov A., Driessen A. J. M., Ben-Tal N., Shai Y., Rapaport D. (2012) Membrane integration of a mitochondrial signal-anchored protein does not require additional proteinaceous factors. Biochem. J. 442, 381–389 [DOI] [PubMed] [Google Scholar]
- 39. Sperka-Gottlieb C. D. M., Hermetter A., Paltauf F., Daum G. (1988) Lipid topology and physical properties of the outer mitochondrial membrane of the yeast, Saccharomyces cerevisiae. Biochim. Biophys. Acta 946, 227–234 [DOI] [PubMed] [Google Scholar]
- 40. Zinser E., Sperka-Gottlieb C. D. M., Fasch E. V., Kohlwein S. D., Paltauf F., Daum G. (1991) Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eukaryote Saccharomyces cerevisiae. J. Bacteriol. 173, 2026–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gebert N., Joshi A. S., Kutik S., Becker T., McKenzie M., Guan X. L., Mooga V. P., Stroud D. A., Kulkarni G., Wenk M. R., Rehling P., Meisinger C., Ryan M. T., Wiedemann N., Greenberg M. L., Pfanner N. (2009) Mitochondrial cardiolipin involved in outer-membrane protein biogenesis: implications for Barth syndrome. Curr. Biol. 19, 2133–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Clancey C. J., Chang S. C., Dowhan W. (1993) Cloning of a gene (PSD1) encoding phosphatidylserine decarboxylase from Saccharomyces cerevisiae by complementation of an Escherichia coli mutant. J. Biol. Chem. 268, 24580–24590 [PubMed] [Google Scholar]
- 43. Trotter P. J., Pedretti J., Voelker D. R. (1993) Phosphatidylserine decarboxylase from Saccharomyces cerevisiae. Isolation of mutants, cloning of the gene, and creation of a null allele. J. Biol. Chem. 268, 21416–21424 [PubMed] [Google Scholar]
- 44. Jiang F., Rizavi H. S., Greenberg M. L. (1997) Cardiolipin is not essential for the growth of Saccharomyces cerevisiae on fermentable or non-fermentable carbon sources. Mol. Microbiol. 26, 481–491 [DOI] [PubMed] [Google Scholar]
- 45. Chang S. C., Heacock P. N., Mileykovskaya E., Voelker D. R., Dowhan W. (1998) Isolation and characterization of the gene (CLS1) encoding cardiolipin synthase in Saccharomyces cerevisiae. J. Biol. Chem. 273, 14933–14941 [DOI] [PubMed] [Google Scholar]
- 46. Tuller G., Hrastnik C., Achleitner G., Schiefthaler U., Klein F., Daum G. (1998) YDL142c encodes cardiolipin synthase (Cls1p) and is non-essential for aerobic growth of Saccharomyces cerevisiae. FEBS Lett. 421, 15–18 [DOI] [PubMed] [Google Scholar]
- 47. Birner R., Bürgermeister M., Schneiter R., Daum G. (2001) Roles of phosphatidylethanolamine and its several biosynthetic pathways in Saccharomyces cerevisiae. Mol. Biol. Cell 12, 997–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Horvath S. E., Böttinger L., Vögtle F. N., Wiedemann N., Meisinger C., Becker T., Daum G. (2012) Processing and topology of the yeast mitochondrial phosphatidylserine decarboxylase 1. J. Biol. Chem. 287, 36744–36755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Trotter P. J., Voelker D. R. (1995) Identification of a non-mitochondrial phosphatidylserine decarboxylase activity (PSD2) in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 270, 6062–6070 [DOI] [PubMed] [Google Scholar]
- 50. Bürgermeister M., Birner-Grünberger R., Nebauer R., Daum G. (2004) Contribution of different pathways to the supply of phosphatidylethanolamine and phosphatidylcholine to mitochondrial membranes of the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1686, 161–168 [DOI] [PubMed] [Google Scholar]
- 51. Riekhof W. R., Voelker D. R. (2006) Uptake and utilization of lysophosphatidylethanolamine by Saccharomyces cerevisiae. J. Biol. Chem. 281, 36588–36596 [DOI] [PubMed] [Google Scholar]
- 52. Riekhof W. R., Wu J., Jones J. L., Voelker D. R. (2007) Identification and characterization of the major lysophosphatidylethanolamine acyltransferase in Saccharomyces cerevisiae. J. Biol. Chem. 282, 28344–28352 [DOI] [PubMed] [Google Scholar]
- 53. Rajakumari S., Daum G. (2010) Janus-faced enzymes yeast Tgl3p and Tgl5p catalyze lipase and acyltransferase reactions. Mol. Biol. Cell 21, 501–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Birner R., Nebauer R., Schneiter R., Daum G. (2003) Synthetic lethal interaction of the mitochondrial phosphatidylethanolamine biosynthetic machinery with the prohibitin complex of Saccharomyces cerevisiae. Mol. Biol. Cell 14, 370–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tamura Y., Harada Y., Yamano K., Watanabe K., Ishikawa D., Ohshima C., Nishikawa S., Yamamoto H., Endo T. (2006) Identification of Tam41 maintaining integrity of the TIM23 protein translocator complex in mitochondria. J. Cell Biol. 174, 631–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tamura Y., Endo T., Iijima M., Sesaki H. (2009) Ups1p and Ups2p antagonistically regulate cardiolipin metabolism in mitochondria. J. Cell Biol. 185, 1029–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tamura Y., Onguka O., Hobbs A. E., Jensen R. E., Iijima M., Claypool S. M., Sesaki H. (2012) Role of two conserved intermembrane space proteins, Ups1p and Ups2p, in intra-mitochondrial phospholipid trafficking. J. Biol. Chem. 287, 15205–15218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kutik S., Rissler M., Guan X. L., Guiard B., Shui G., Gebert N., Heacock P. N., Rehling P., Dowhan W., Wenk M. R., Pfanner N., Wiedemann N. (2008) The translocator maintenance protein Tam41 is required for mitochondrial cardiolipin biosynthesis. J. Cell Biol. 183, 1213–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Osman C., Haag M., Potting C., Rodenfels J., Dip P. V., Wieland F. T., Brügger B., Westermann B., Langer T. (2009) The genetic interactome of prohibitins: coordinated control of cardiolipin and phosphatidylethanolamine by conserved regulators in mitochondria. J. Cell Biol. 184, 583–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Osman C., Voelker D. R., Langer T. (2011) Making heads or tails of phospholipids in mitochondria. J. Cell Biol. 192, 7–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kuroda T., Tani M., Moriguchi A., Tokunaga S., Higuchi T., Kitada S., Kuge O. (2011) FMP30 is required for the maintenance of a normal cardiolipin level and mitochondrial morphology in the absence of mitochondrial phosphatidylethanolamine synthesis. Mol. Microbiol. 80, 248–265 [DOI] [PubMed] [Google Scholar]
- 62. Nguyen T. T., Lewandowska A., Choi J. Y., Markgraf D. F., Junker M., Bilgin M., Ejsing C. S., Voelker D. R., Rapoport T. A., Shaw J. M. (2012) Gem1 and ERMES do not directly affect phosphatidylserine transport from ER to mitochondria or mitochondrial inheritance. Traffic 13, 880–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jiang F., Ryan M. T., Schlame M., Zhao M., Gu Z., Klingenberg M., Pfanner N., Greenberg M. L. (2000) Absence of cardiolipin in the crd1 null mutant results in decreased mitochondrial membrane potential and reduced mitochondrial function. J. Biol. Chem. 275, 22387–22394 [DOI] [PubMed] [Google Scholar]
- 64. Pfeiffer K., Gohil V., Stuart R. A., Hunte C., Brandt U., Greenberg M. L., Schägger H. (2003) Cardiolipin stabilizes respiratory chain supercomplexes. J. Biol. Chem. 278, 52873–52880 [DOI] [PubMed] [Google Scholar]
- 65. Zhang M., Mileykovskaya E., Dowhan W. (2005) Gluing the respiratory chain together. Cardiolipin is essential for organization of complexes III and IV into a supercomplex in intact yeast mitochondria. J. Biol. Chem. 280, 29403–29408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gallas M. R., Dienhart M. K., Stuart R. A., Long R. M. (2006) Characterization of Mmp37p, a Saccharomyces cerevisiae mitochondrial matrix protein with a role in mitochondrial protein import. Mol. Biol. Cell 17, 4051–4062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. van der Laan M., Meinecke M., Dudek J., Hutu D. P., Lind M., Perschil I., Guiard B., Wagner R., Pfanner N., Rehling P. (2007) Motor-free mitochondrial presequence translocase drives membrane integration of preproteins. Nat. Cell Biol. 9, 1152–1159 [DOI] [PubMed] [Google Scholar]
- 68. Claypool S. M., Oktay Y., Boontheung P., Loo J. A., Koehler C. M. (2008) Cardiolipin defines the interactome of the major ADP/ATP carrier protein of the mitochondrial inner membrane. J. Cell Biol. 182, 937–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Gonzalvez F., Schug Z. T., Houtkooper R. H., MacKenzie E. D., Brooks D. G., Wanders R. J., Petit P. X., Vaz F. M., Gottlieb E. (2008) Cardiolipin provides an essential activating platform for caspase-8 on mitochondria. J. Cell Biol. 183, 681–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. DeVay R. M., Dominguez-Ramirez L., Lackner L. L., Hoppins S., Stahlberg H., Nunnari J. (2009) Coassembly of Mgm1 isoforms requires cardiolipin and mediates mitochondrial inner membrane fusion. J. Cell Biol. 186, 793–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mileykovskaya E., Dowhan W. (2009) Cardiolipin membrane domains in prokaryotes and eukaryotes. Biochim. Biophys. Acta 1788, 2084–2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gohil V. M., Thompson M. N., Greenberg M. L. (2005) Synthetic lethal interaction of the mitochondrial phosphatidylethanolamine and cardiolipin biosynthetic pathways in Saccharomyces cerevisiae. J. Biol. Chem. 280, 35410–35416 [DOI] [PubMed] [Google Scholar]
- 73. Böttinger L., Horvath S. E., Kleinschroth T., Hunte C., Daum G., Pfanner N., Becker T. (2012) Phosphatidylethanolamine and cardiolipin differentially affect the stability of mitochondrial respiratory chain supercomplexes. J. Mol. Biol. 423, 677–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tasseva G., Bai H. D., Davidescu M., Haromy A., Michelakis E., Vance J. E. (2013) Phosphatidylethanolamine deficiency in mammalian mitochondria impairs oxidative phosphorylation and alters mitochondrial morphology. J. Biol. Chem. 288, 4158–4173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Joshi A. S., Thompson M. N., Fei N., Hüttemann M., Greenberg M. L. (2012) Cardiolipin and mitochondrial phosphatidylethanolamine have overlapping functions in mitochondrial fusion in Saccharomyces cerevisiae. J. Biol. Chem. 287, 17589–17597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Horvath S. E., Wagner A., Steyrer E., Daum G. (2011) Metabolic link between phosphatidylethanolamine and triacylglycerol metabolism in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1811, 1030–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Stojanovski D., Pfanner N., Wiedemann N. (2007) Import of proteins into mitochondria. Methods Cell Biol. 80, 783–806 [DOI] [PubMed] [Google Scholar]
- 78. Folch J., Lees M., Sloane Stanley G. H. (1957) A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 226, 497–509 [PubMed] [Google Scholar]
- 79. Broekhuyse R. M. (1968) Phospholipids in tissues of the eye. I. Isolation, characterization and quantitative analysis by two-dimensional thin-layer chromatography of diacyl and vinyl-ether phospholipids. Biochim. Biophys. Acta 152, 307–315 [DOI] [PubMed] [Google Scholar]
- 80. Humphries A. D., Streimann I. C., Stojanovski D., Johnston A. J., Yano M., Hoogenraad N. J., Ryan M. T. (2005) Dissection of the mitochondrial import and assembly pathway of human Tom40. J. Biol. Chem. 280, 11535–11543 [DOI] [PubMed] [Google Scholar]
- 81. Dukanovic J., Dimmer K. S., Bonnefoy N., Krumpe K., Rapaport D. (2009) Genetic and functional interactions between the mitochondrial outer membrane proteins Tom6 and Sam37. Mol. Cell. Biol. 29, 5975–5988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Becker T., Guiard B., Thornton N., Zufall N., Stroud D. A., Wiedemann N., Pfanner N. (2010) Assembly of the mitochondrial protein import channel: role of Tom5 in two-stage interaction with the SAM complex. Mol. Biol. Cell 21, 3106–3113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Meisinger C., Wiedemann N., Rissler M., Strub A., Milenkovic D., Schönfisch B., Müller H., Kozjak V., Pfanner N. (2006) Mitochondrial protein sorting: differentiation of β-barrel assembly by Tom7-mediated segregation of Mdm10. J. Biol. Chem. 281, 22819–22826 [DOI] [PubMed] [Google Scholar]
- 84. Thornton N., Stroud D. A., Milenkovic D., Guiard B., Pfanner N., Becker T. (2010) Two modular forms of the mitochondrial sorting and assembly machinery are involved in biogenesis of α-helical outer membrane proteins. J. Mol. Biol. 396, 540–549 [DOI] [PubMed] [Google Scholar]
- 85. Wideman J. G., Go N. E., Klein A., Redmond E., Lackey S. W. K., Tao T., Kalbacher H., Rapaport D., Neupert W., Nargang F. E. (2010) Role of Mdm10, Tom7, Mdm12 and Mmm1 proteins in the assembly of mitochondrial outer membrane proteins in Neurospora crassa. Mol. Biol. Cell 21, 1725–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Krimmer T., Rapaport D., Ryan M. T., Meisinger C., Kassenbrock C. K., Blachly-Dyson E., Forte M., Douglas M. G., Neupert W., Nargang F. E., Pfanner N. (2001) Biogenesis of porin of the outer mitochondrial membrane involves an import pathway via receptors and the general import pore of the TOM complex. J. Cell Biol. 152, 289–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Yamano K., Tanaka-Yamano S., Endo T. (2010) Mdm10 as a dynamic constituent of the TOB/SAM complex directs coordinated assembly of Tom40. EMBO Rep. 11, 187–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Yamano K., Tanaka-Yamano S., Endo T. (2010) Tom7 regulates Mdm10-mediated assembly of the mitochondrial import channel protein Tom40. J. Biol. Chem. 285, 41222–41231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Habib S. J., Waizenegger T., Lech M., Neupert W., Rapaport D. (2005) Assembly of the TOB complex of mitochondria. J. Biol. Chem. 280, 6434–6440 [DOI] [PubMed] [Google Scholar]
- 90. Lackey S. W. K., Wideman J. G., Kennedy E. K., Go N. E., Nargang F. E. (2011) The Neurospora crassa TOB complex: analysis of the topology and function of Tob38 and Tob37. PLoS ONE 6, e25650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. van Wilpe S., Ryan M. T., Hill K., Maarse A. C., Meisinger C., Brix J., Dekker P. J. T., Moczko M., Wagner R., Meijer M., Guiard B., Hönlinger A., Pfanner N. (1999) Tom22 is a multifunctional organizer of the mitochondrial preprotein translocase. Nature 401, 485–489 [DOI] [PubMed] [Google Scholar]
- 92. Frazier A. E., Chacinska A., Truscott K. N., Guiard B., Pfanner N., Rehling P. (2003) Mitochondria use different mechanisms for transport of multispanning membrane proteins through the intermembrane space. Mol. Cell. Biol. 23, 7818–7828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Chacinska A., Lind M., Frazier A. E., Dudek J., Meisinger C., Geissler A., Sickmann A., Meyer H. E., Truscott K. N., Guiard B., Pfanner N., Rehling P. (2005) Mitochondrial presequence translocase: switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell 120, 817–829 [DOI] [PubMed] [Google Scholar]
- 94. van der Does C., Swaving J., van Klompenburg W., Driessen A. J. M. (2000) Non-bilayer lipids stimulate the activity of the reconstituted bacterial protein translocase. J. Biol. Chem. 275, 2472–2478 [DOI] [PubMed] [Google Scholar]
- 95. van den Brink-van der Laan E., Killian J. A., de Kruijff B. (2004) Nonbilayer lipids affect peripheral and integral membrane proteins via changes in the lateral pressure profile. Biochim. Biophys. Acta 1666, 275–288 [DOI] [PubMed] [Google Scholar]
- 96. Bogdanov M., Umeda M., Dowhan W. (1999) Phospholipid-assisted refolding of an integral membrane protein. Minimum structural features for phosphatidylethanolamine to act as a molecular chaperone. J. Biol. Chem. 274, 12339–12345 [DOI] [PubMed] [Google Scholar]
- 97. Chacinska A., Koehler C. M., Milenkovic D., Lithgow T., Pfanner N. (2009) Importing mitochondrial proteins: machineries and mechanisms. Cell 138, 628–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Dukanovic J., Rapaport D. (2011) Multiple pathways in the integration of proteins into the mitochondrial outer membrane. Biochim. Biophys. Acta 1808, 971–980 [DOI] [PubMed] [Google Scholar]