Background: GDP-fucose and other sugar nucleotide biosynthetic pathways are conserved in the P. falciparum genome.
Results: These pathways are active in the intraerythrocytic life cycle of the parasite.
Conclusion: The parasite biosynthesizes GDP-fucose and other sugar nucleotides not related to the glycosylphosphatidylinositol structures
Significance: Their presence strongly suggests that they are involved in the biosynthesis of glycans not yet characterized.
Keywords: Glycobiology, Malaria, Metabolism, Parasite Metabolism, Plasmodium, FS, Fucose, GMD, Sugar Nucleotides
Abstract
Carbohydrate structures play important roles in many biological processes, including cell adhesion, cell-cell communication, and host-pathogen interactions. Sugar nucleotides are activated forms of sugars used by the cell as donors for most glycosylation reactions. Using a liquid chromatography-tandem mass spectrometry-based method, we identified and quantified the pools of UDP-glucose, UDP-galactose, UDP-N-acetylglucosamine, GDP-mannose, and GDP-fucose in Plasmodium falciparum intraerythrocytic life stages. We assembled these data with the in silico functional reconstruction of the parasite metabolic pathways obtained from the P. falciparum annotated genome, exposing new active biosynthetic routes crucial for further glycosylation reactions. Fucose is a sugar present in glycoconjugates often associated with recognition and adhesion events. Thus, the GDP-fucose precursor is essential in a wide variety of organisms. P. falciparum presents homologues of GDP-mannose 4,6-dehydratase and GDP-l-fucose synthase enzymes that are active in vitro, indicating that most GDP-fucose is formed by a de novo pathway that involves the bioconversion of GDP-mannose. Homologues for enzymes involved in a fucose salvage pathway are apparently absent in the P. falciparum genome. This is in agreement with in vivo metabolic labeling experiments showing that fucose is not significantly incorporated by the parasite. Fluorescence microscopy of epitope-tagged versions of P. falciparum GDP-mannose 4,6-dehydratase and GDP-l-fucose synthase expressed in transgenic 3D7 parasites shows that these enzymes localize in the cytoplasm of P. falciparum during the intraerythrocytic developmental cycle. Although the function of fucose in the parasite is not known, the presence of GDP-fucose suggests that the metabolite may be used for further fucosylation reactions.
Introduction
Malaria is a global health problem caused by protozoan parasites of the genus Plasmodium and transmitted by the Anopheles mosquito. Approximately half of the world's population is at risk of infection, and there are around 200 million cases annually, leading to more than half a million deaths each year (1). Most of the people who die are children living in resource-poor countries in sub-Saharan Africa. Among the five Plasmodium species that cause malaria in humans (Plasmodium falciparum, Plasmodium vivax, Plasmodium malariae, Plasmodium ovale, and Plasmodium knowlesi), P. falciparum is the most deadly and is responsible for the majority of the malaria-linked deaths in sub-Saharan Africa (1). Chemotherapy is one of the central strategies for malaria treatment, but unfortunately, drug resistance to commonly used antimalarial drugs has spread very rapidly (2, 3). The implementation of new approaches to prevent and fight malaria has greatly benefited from the sequencing of the parasite genome (4) and the development of improved tools for functional genomics (5–8). However, this area of research remains greatly limited by our incomplete knowledge of parasite biology (9). Therefore, it is critical to promote research efforts that probe the basic biochemistry and cell biology of Plasmodium with the aim of characterizing essential proteins, metabolic pathways, and other processes that could be suitable for intervention and to validate new candidate drug targets.
Glycosylphosphatidylinositol (GPI)4 anchors (10) represent the major carbohydrate modification described in P. falciparum cell surface proteins (11). Several of these GPI-anchored glycoproteins are essential for parasite invasion and virulence (12), and GPI anchors are generated via a complex synthetic pathway (13). Recently, Bushkin et al. (14) presented new evidence demonstrating the existence of functional N-glycosylation in the intraerythrocytic stages of P. falciparum. In general, protozoan parasite N-glycosylation patterns are atypical of those described in higher eukaryotes (15, 16). In P. falciparum, the secondary loss of enzymes related to the biosynthesis of N-glycosylation precursors and the quality control of glycoprotein folding in the endoplasmic reticulum (17, 18) result in a very unusual N-glycosylation (14) that, if essential, could be therapeutically exploitable. To our knowledge, despite some controversy (19–24), O-glycans have never been unequivocally described in P. falciparum. Thus, although there have been significant efforts to understand the glycobiology of the parasite, several questions on this topic remain unanswered.
Sugar nucleotides, the essential intermediates in carbohydrate metabolism and glycoconjugate biosynthesis, are activated forms of sugars produced by the cell as donor precursors for most of the glycosylation reactions. They are formed in two main ways: by a salvage pathway involving “activation” of the sugar using a kinase and a pyrophosphorylase or by a de novo pathway involving the bioconversion of an existing sugar or sugar nucleotide. Specific P. falciparum metabolic databases (25), based on the parasite genome sequence, predict the conservation of the de novo biosynthetic pathways for UDP-N-acetyl-glucosamine, GDP-mannose, GDP-fucose, and UDP-glucose. Most of the predicted open reading frames (ORFs) that encode for the enzymes involved in these pathways are also present in other Plasmodium species (P. vivax, P. knowlesi, Plasmodium chabaudi, Plasmodium yoelii, and Plasmodium berghei) (26), suggesting that they are conserved and encode for proteins playing important roles in the parasite. Indeed, UDP-N-acetyl-glucosamine and GDP-mannose (through its product dolichol-phosphomannose) are essential donor substrates for the biosynthesis of GPI structures (27). Because sugar nucleotides are the “basic building blocks” of glycoconjugates, the conservation of their biosynthetic metabolic routes strongly suggests the existence of further downstream glycosylation reactions in which these metabolites are involved (i.e. glycan biosynthesis).
In this work, we identify and quantify the sugar nucleotides present in different stages of the intraerythrocytic life cycle of P. falciparum using a liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based assay, and we present the results in the context of the functional metabolic pathways reconstructed from the genome of the parasite. We demonstrate that P. falciparum is unable to take up significant amounts of tritiated fucose from the culture media. Furthermore, we show that the putative GDP-fucose biosynthesis enzymes are functional in vitro, by expressing them and detecting GDP-fucose production. Endogenous forms of these enzymes localize to the cytosol of P. falciparum. We show that the genes encoding these enzymes are expressed at low abundance, and their levels are modulated throughout the red blood cell stages of the parasite. Finally, we also demonstrate that the parasite expresses a protein o-fucosyltransferase homolog, and schizont extracts incorporate tritiated GDP-fucose.
EXPERIMENTAL PROCEDURES
Parasites and Parasite Culture
P. falciparum 3D7 (obtained from MR4-ATCC) parasites were cultured with human erythrocytes (3–4% hematocrit) in RPMI medium (Sigma) supplemented with 10% AB+ human serum, incubated at 37 °C in an atmosphere of 92% N2, 3% O2, and 5% CO2 using standard methods (28). For sugar nucleotide analysis and labeling experiments, parasites were grown in RPMI medium supplemented with 0.5% Albumax II (Invitrogen) to avoid possible variations due to the uses of different human serum batches. Parasite growth was monitored by counting the infected erythrocytes in Giemsa-stained thin blood smears under light microscopy.
Sugar Nucleotide Analysis
Osmotic lysis of red blood cells of sorbitol-synchronized cultures at different stages of the intraerythrocytic life cycle (rings, trophozoites, and schizonts, approximately 18, 35, and 44 h postinvasion, respectively) and 10–12% parasitemia was performed by resuspending erythrocyte pellet twice in 60 volumes of cold erythrocyte lysis buffer (10× stock solution: 0.15 m NH4Cl, 0.1 m KHCO3, 0.01 m EDTA). Non-infected red blood cells were lysed and included as controls in every analysis to discard noise due to the detection of sugar nucleotides carried away from the erythrocytes. The suspension was incubated on ice until lysis was completed (∼10 min) (29). Pellets were washed three times with cold phosphate-buffered saline (PBS), and sugar nucleotide analysis was performed as described elsewhere (30). Briefly, parasite pellets were lysed in 70% ethanol in the presence of 20 pmol of GDP-glucose internal standard, and sugar nucleotides were extracted using Envi-Carb columns (31). Sugar nucleotides were then analyzed using LC-MS/MS, using multiple-reaction monitoring for detection. HPLC conditions were adapted from Ref. 31, and acetonitrile was added postcolumn to produce stable electrospray ionization. The peak areas for each sugar nucleotide, along with their empirically determined molar relative response factors and the known amount of internal GDP-glucose, were used to quantify sugar nucleotides. Analyses were performed on three different sugar nucleotide extracts.
Metabolic Labeling of Parasite with Tritiated Sugars
Parasite cultures at 10–12% parasitemia were synchronized to the ring state and then washed and resuspended in RPMI medium supplemented with Albumax II. Cells were incubated at 3–4% hematocrit for 16–18 h, until the trophozoite stage. The parasites were then washed with glucose-free RPMI supplemented with Albumax II and 20 mm fructose, and they were metabolically labeled with 3H-sugars (50 μCi/ml) for 1 or 4 h, adapting the conditions used in previous work (23, 32, 33). Triplicates of each condition were included, and controls were “mock-labeled” for a few seconds using medium at 4 °C supplemented with an excess (100-fold) of the unlabeled (cold) sugar. Labeled cultures and controls were washed three times with PBS at 4 °C and treated with saponin to release parasites. Free parasites were washed again three times with PBS at 4 °C and lysed, and the radioactivity was measured by liquid scintillation counting.
To calculate the amount of sugar incorporated into the glycoprotein and glycolipid compartments, after the labeling, free parasites were further lysed, and the proteins were precipitated with 10% trichloroacetic acid and filtered (glycoproteins), or the glycolipids were extracted with organic solvents (glycolipids) (34).
Expression and Purification of GDP-fucose Biosynthesis Genes in Escherichia coli
Codon harmonization was used for the production of heterologously expressed P. falciparum GDP-mannose 4,6-dehydratase (PfGMD, gene ID PF3D7_0813800) and GDP-l-fucose synthase (PfFS, gene ID PF3D7_1014000) in E. coli (35). Codon-harmonized genes (Genscript) were cloned into the pGEX 6P expression vector (GE Healthcare) and expressed in BL21 (DE3) E. coli cells. 3-Liter cultures were grown to an A600 nm of 0.6 and induced overnight at 18 °C with 100 μm isopropyl-β-d-thiogalactopyranoside. Cells were harvested and lysed in PBS including 0.1 mg/ml lysozyme in the presence of Complete protease inhibitor mixture (Roche Applied Science). Lysis was ensured by sonication (Misonix Microson XL 2000). 0.5% Triton X-100 was added, and, after a 30-min incubation at 4 °C, the lysate was clarified by centrifugation at 12,000 × g for 20 min, and the supernatant was added to 1 ml of PBS-washed GST beads (GE Healthcare) and incubated overnight at 4 °C. The beads were collected in chromatography columns, washed with >10 bead volumes of PBS before three elutions of 1 ml with 10 mm glutathione in 50 mm Tris-HCl, pH 8. The eluates were pooled, diafiltered with PBS, and concentrated in 10,000 nominal molecular weight limit centrifugal filter units (Amicon, Millipore).
GDP-fucose Biosynthesis Assay
Assay conditions were adapted from elsewhere (36–38). Reactions were performed in 25 μl (final volume) of 100 mm MOPS, pH 7.0, 100 mm NaCl, 10 mm dithiothreitol, 5 mm EDTA, 1 mm GDP-mannose (Sigma), 0.4 mm NADPH, and 1 mm NADP. The reaction was started by adding 1.5 mg/ml of the recombinant PfGMD enzyme, and the reaction was left to proceed for 3 h at 37 °C. Recombinant PfFS (1.5 mg/ml) was then added, and the concentration of NADPH was adjusted to 1.5 mm. For experiments without the PfFS enzyme, only NADPH was added at this point. Reactions were stopped after 2 h at 37 °C by heating to 100 °C for 2 min, and then the samples were filtered to remove insoluble material, and sugar nucleotides were analyzed by HPLC (31), including standards for every HPLC series of experiments.
For LC-MS/MS analysis, HPLC conditions were adapted from Ref. 39, using a porous graphitic carbon column (Hypercarb, 100 × 2.1 mm, 5-μm particle size, Thermo Scientific) and MS-compatible mobile phases. Starting buffer was 0.1% formic acid, brought to pH 9.0 with ammonia, followed by a 36-min gradient from 10 to 50% acetonitrile at a flow rate of 100 μl/min. Detection was performed by negative mode electrospray ionization-MS on an API3000 triple quadrupole LC-MS/MS mass spectrometer (PE-Sciex) with a declustering potential of −50 V, focusing potential of −300 V, collision energy of 30 V, and source temperature of 375 °C.
Cloning and Expression of Epitope-tagged Versions of PfGMD and PfFS in P. falciparum
Gene PfGMD was amplified from P. falciparum genomic DNA using primer pARL1F-GMD(KpnI) (CGCGGTACCATGCGAGTTGCTTTAATC) and pARL1R-GMD(PstI) (CGCCTGCAGTTGCTTTTTACCCATTT). PfFS was amplified from P. falciparum cDNA using primer pARL1F-FS(KpnI) (CGCGGTACCATGACACGAATTTGTCTTGTAACTG) and pARL1R-FS(PstI) (CGCCTGCAGTTTTCTTACATTTTTGTATTCGTCAATAAACCA).P. falciparum genes were cloned in the KpnI-PstI site of transfection vector pARL1a-3HA (40) under the control of the pfcrt promoter region (41). P. falciparum were transfected as described previously (41). Briefly, 150 μg of each plasmid was used to electroporate (310 V, 950 millifarads) 200 μl of infected red blood cells at >5% parasitemia, synchronized for ring stage parasites. Transfected parasites were selected on 2 nm of WR99210 drug, and resistant parasites appeared in culture from 25 to 35 days after drug application.
Indirect Immunofluorescence Assays
Cultured P. falciparum transgenic lines were washed in PBS and then fixed with 4% EM grade paraformaldehyde and 0.075% EM grade glutaraldehyde in PBS (42). Fixed cells were permeabilized with 0.1% Triton X-100 in PBS and blocked for 1 h at room temperature in 3% PBS-bovine serum albumin (PBS-BSA). Samples were incubated overnight with primary antibody (rat anti-HA (1:10; Roche Applied Science) or rabbit anti-HSP70 (1:50; StressMarq)) (43) diluted in 3% PBS-BSA, followed by a 1-h incubation with secondary antibody (anti-rat conjugated with Alexa Fluor 488 or anti-rabbit conjugated with Alexa Fluor 594 (1:200, Invitrogen)) diluted in 3% PBS-BSA. Nuclei were stained for 1 h with 4′,6-diaminido-2-phenylindole (DAPI; 2 mg/ml diluted in PBS) during the secondary antibody incubation. Confocal microscopy was performed using a laser scanning confocal microscope (TCS-SP5; Leica Microsystems) at the microscopy scientific and technical services facility of the Universitat de Barcelona.
RNA Preparation and Quantitative Real-time PCR
Tight synchronization of parasites was achieved by Percoll purification of schizonts followed by sorbitol lysis 5 h later, to obtain a population of a defined age window of 0–5 h postinvasion. RNA was purified using the TRIzol method at the ring, trophozoite, and schizont stage, and cDNA was synthesized by reverse transcription performed using random hexaprimers and SuperScript-III reverse transcriptase (RT; Invitrogen) according to the manufacturer's instructions and including controls without RT. All quantitative PCRs were performed using PowerSYBR Green Master Mix (Invitrogen), and expression values were calculated using the relative standard curve method. Results are expressed in arbitrary units, relative to a genomic DNA standard curve, and were normalized against seryl-tRNA synthetase (PF3D7_0717700). The primers used were GGTGATTGCTCAAAGGCAAAA and TTCATAAACGAGCTGGGTAAATGTAT for PfGMD; CTTGTATTTTCCCTGTAAATTGTTCTCTAC and GATGGTATAACGTGCGCATTTTC for PfFS; TTTTTTAAAGTGATAGACAGGGTTAT and AAAAAAGTCCCCTAAATATACATCCT for PfPoFUT2; and AAGTAGCAGGTCATCGTGGTT and TTCGGCACATTCTTCCATAA for seryl-tRNA synthetase.
Preparation of P. falciparum Cell Extracts and [3H]GDP-fucose Incorporation Assays
Percoll-purified infected red blood cells (36–42 h postinvasion) grown at 10–12% parasitemia were subjected to saponin lysis and washed three times with PBS. The cell pellet was placed on ice and homogenized for 1 h (at 109 cells/ml) with 1% Nonidet P-40, 10 mm Tris-HCl, pH 7.2, 150 mm NaCl, 10 mm MnCl2, 5 mm MgCl2, 1× protease inhibitor mixture (Roche Applied Science), 1 mm phenylmethylsulfonyl fluoride (PMSF), and 100 μg/ml Nα-p-tosyl-l-lysine chloromethyl ketone. Inactivated extracts were boiled for 20 min with 3 m urea, 0.1 m dithiothreitol, and 5 mm EDTA. 2 μCi of [3H]GDP-fucose (American Radiochemical Chemicals, Inc.) was added to the extract, and the reaction was incubated at 37 °C for 1 h. Proteins were TCA-precipitated and washed three times with cold 100% acetone, and tritium incorporated into the TCA pellets was measured by scintillation counting.
RESULTS
Sugar Nucleotide Biosynthetic Pathways Predicted in P. falciparum
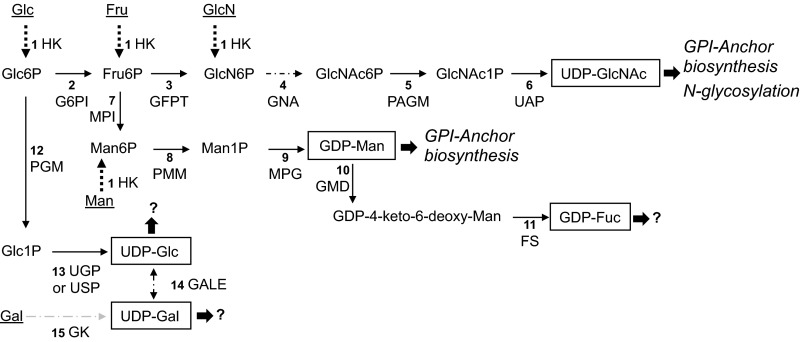
Based on the current knowledge of P. falciparum sugar biochemistry (11, 14, 23, 32) and the predicted metabolic pathways (25), we have reconstructed the sugar nucleotide biosynthetic routes present in P. falciparum (Fig. 1). The enzymes and genes involved in these metabolic routes are shown in Table 1. Bioinformatic analysis suggests that sugar nucleotides are made by conventional eukaryotic de novo routes from glucose 6-phosphate, although other salvage pathways also exist (Fig. 1). Conserved pathways for the biosynthesis of UDP-GlcNAc and GDP-mannose agree well with the monosaccharide content of known P. falciparum glycoconjugates (i.e. GPI anchors and N-glycans). However, the conservation of UDP-glucose and GDP-fucose biosynthetic pathways (Fig. 1) does not correspond with any previous description of fucose and/or glucose-containing glycans in the parasite. Because sugar nucleotides are obligate donors for glycosylation reactions mediated by glycosyltransferases, the presence of these metabolic routes in the parasite genome prompted us to identify the sugar nucleotide pools present in the blood stages of the parasite to assess the activation state of these pathways.
FIGURE 1.
Sugar nucleotide biosynthetic pathways identified in the genome of P. falciparum. The numbers refer to the enzymes and known or candidate genes described in Table 1. Predicted known fates of sugar nucleotide donors according to the glycoconjugates described in P. falciparum are in italic type (or marked with a question mark if the fate is unknown). Sugar nucleotides identified in this study are boxed. Dotted lines indicate confirmed salvage pathways, and sugars, taken up from the medium, are underlined. The discontinuous arrow (step 4) represents the glucosamine-phosphate N-acetyltransferase activity (EC 2.3.1.4) for which a candidate gene is not yet identified. HK, hexokinase; G6PI, glucose-6-phosphate isomerase; GFPT, glucosamine-fructose-6-phosphate aminotransferase; GNA, glucosamine-phosphate N-acetyltransferase; PAGM, phosphoacetylglucosamine mutase; UAP, UDP-N-acetylglucosamine pyrophosphorylase; MPI, mannose-1-phosphate isomerase; PMM, phosphomannomutase; MPG, mannose-1-phosphate guanyltransferase; GMD, GDP-mannose 4,6-dehydratase; FS, GDP-l-fucose synthase; PGM, phosphoglucomutase; UGP, UTP-glucose-1-phosphate uridylyltransferase; GALE, UDP-glucose 4-epimerase; GK, galactokinase. The suggested pathway for the biosynthesis of UDP-Gal through the activity of the UDP-sugar pyrophosphorylase enzyme is indicated with a gray discontinuous arrow (see “Discussion”).
TABLE 1.
Enzymes and genes involved in P. falciparum sugar nucleotide biosynthesis
| Step no.a | Enzyme name | Enzyme no. | Putative P. falciparum homologuesb |
|---|---|---|---|
| 1 | Hexokinase (HK) | EC 2.7.1.1 | PF3D7_0624000 |
| 2 | Glucose-6-phosphate isomerase (G6PI) | EC 5.3.1.9 | PF3D7_1436000c |
| 3 | Glucosamine-fructose-6-phosphate aminotransferase (GFPT) | EC 2.6.1.16 | PF3D7_1025100 |
| 4 | Glucosamine-phosphate N-acetyltransferase (GNA) | EC 2.3.1.4 | No gene identifiedd |
| 5 | Phosphoacetylglucosamine mutase (PAGM) | EC 5.4.2.3 | PF3D7_1130000 |
| 6 | UDP-N-acetylglucosamine pyrophosphorylase (UAP) | EC 2.7.7.23 | PF3D7_1343600 |
| 7 | Mannose-6-phosphate isomerase (MPI) | EC 5.3.1.8 | PF3D7_0801800 |
| 8 | Phosphomannomutase (PMM) | EC 5.4.2.8 | PF3D7_1017400 |
| 9 | Mannose-1-phosphate guanyltransferase (MPG) | EC 2.7.7.13 | PF3D7_1420900 |
| 10 | GDP-mannose 4,6-dehydratase (GMD) | EC 4.2.1.47 | PF3D7_0813800 |
| 11 | GDP-l-fucose synthase (FS) | EC 1.1.1.271 | PF3D7_1014000 |
| 12 | Phosphoglucomutase (PGM) | EC 5.4.2.2 | PF3D7_1012500 |
| 13 | UTP-glucose-1-phosphate uridylyltransferase (UGP) or UDP-sugar pyrophosphorylase (USP) | EC 2.7.7.9 or EC 2.7.7.64 | PF3D7_0517500 |
| 14 | UDP-glucose 4-epimerase (GALE) | EC 5.1.3.2 | No gene identified |
| 15 | Galactokinase (GK) | EC 2.7.1.6 | No gene identified |
a Step numbers refer to those shown in Fig. 1.
b Gene ID numbers underlined are as identified and annotated in the P. falciparum genome (25, 46), and those in boldface type and underlined have been functionally characterized in this paper.
c Glucose-6-phosphate isomerase (EC 5.3.1.9) enzyme, PF3D7_1436000 has been crystallized, but to our knowledge its activity has not been published (77).
Identification and Quantification of Sugar Nucleotide Pools in P. falciparum Blood Stages
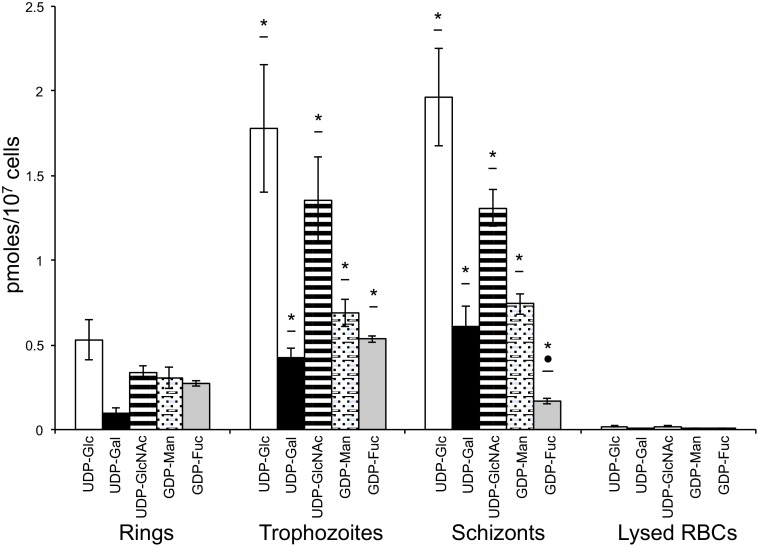
Commercially available sugar nucleotides were used as standards to identify and quantify the products of the conserved sugar nucleotide metabolic pathways (25) using the 3D7 strain of P. falciparum. Variable levels of UDP-galactopyranose, UDP-Glc, UDP-GlcNAc, GDP-mannose (GDP-Man), and GDP-fucose (GDP-Fuc) pools were detected through the different blood stages (i.e. rings, trophozoites, and schizonts) of the parasite (Fig. 2 and Table 2). The sugar nucleotide levels were, in general, 10–100-fold lower than those of other extracellular protozoan parasites. However, in the case of GDP-fucose, levels detected were comparable (Table 2) (30). To exclude the possibility of contamination from host cell material, non-infected osmotically lysed erythrocyte ghosts were analyzed and included as negative controls. The results were at least 1 order of magnitude lower than in the parasite analysis for every sugar nucleotide (Fig. 2 and Table 2). The sugar nucleotides detected in the parasite agree well with the monosaccharide contents of known glycoconjugates in P. falciparum consisting of glucosamine (derived from GlcNAc) and Man residues present in GPI anchors and N-glycans (11, 14). Furthermore, the detection of UDP-GlcNAc, UDP-Glc, GDP-Man, and GDP-Fuc is consistent with the presence of known or candidate sugar nucleotide biosynthetic enzymes encoded in the genome of P. falciparum (25). We were surprised to find that pools of UDP-galactopyranose can be identified at the different blood stages of the parasites, although no apparent candidates for UDP-glucose-4′-epimerase can be detected in the genome of the parasite. The possible metabolic route for the biosynthesis of this sugar nucleotide is discussed below.
FIGURE 2.
Specific sugar nucleotide levels in different blood stages of P. falciparum. Values of UDP-Glc (open bars), UDP-Gal (black solid bars), UDP-GlcNAc (striped bars), GDP-Man (dotted bars), and GDP-Fuc (gray solid bars) indicate the average of three different extractions. Error bars, S.D. The last set of bars represent the levels of sugar nucleotides measured on osmotically lysed non-infected red blood cells (RBCs; used as control). Values are indicated in pmol/107 cells. Analyses were performed in triplicate, and mean values ± S.D. are shown. Statistically significant differences (one-way analysis of variance Tukey's post-test) between stages are shown with an asterisk (*, p < 0.05, rings versus trophozoites or schizonts) or a dot (●, p < 0.05, trophozoites versus schizonts).
TABLE 2.
Sugar nucleotide levels in blood stages of P. falciparum
| Sugar nucleotide | Stage |
Lysed red blood cellsa,b | ||
|---|---|---|---|---|
| Ringsa | Trophozoitesa | Schizontsa | ||
| UDP-Glc | 0.53 ± 0.12 | 1.78 ± 0.38 | 1.96 ± 0.29 | 0.02 |
| UDP-Gal | 0.09 ± 0.04 | 0.42 ± 0.06 | 0.61 ± 0.12 | <0.01 |
| UDP-GlcNAc | 0.34 ± 0.04 | 1.35 ± 0.26 | 1.31 ± 0.11 | 0.02 |
| GDP-Man | 0.31 ± 0.06 | 0.69 ± 0.08 | 0.74 ± 0.06 | <0.01 |
| GDP-Fuc | 0.27 ± 0.01 | 0.53 ± 0.02 | 0.17 ± 0.02 | <0.01 |
a Amounts are indicated in pmol/107 cells.
b S.D. values are at least 1 order of magnitude smaller than the calculated amount and are not included.
Metabolic Labeling of Parasites with Tritiated Sugars
To assess the presence of carbohydrate salvage routes in the parasite P. falciparum-infected erythrocytes, cultures were metabolically labeled with tritiated Man or Fuc in medium containing 20 mm d-fructose (32, 34, 44, 45). Whereas [3H]Man was incorporated as described previously (23), [3H]Fuc was not significantly taken up by the parasite (Fig. 3A), contributing to less than 15% of the average pool amount, and we did not detect a significant incorporation into its glycoproteins or glycolipids (Fig. 3B). This indicates that the main source of GDP-Fuc for P. falciparum intraerythrocytic life stages is through the bioconversion of an existing sugar/sugar nucleotide, which is consistent with the apparent absence of genes encoding enzymes involved in the GDP-fucose salvage pathway in the P. falciparum genome (25). Thus, the de novo GDP-Fuc synthetic route from GDP-Man, which depends on GMD and FS (Fig. 1 and Table 1), is the most feasible pathway for the biosynthesis of this sugar nucleotide in P. falciparum.
FIGURE 3.
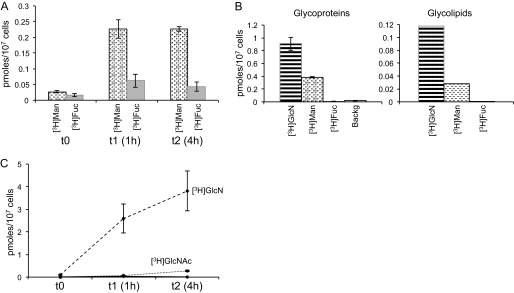
[3H]Sugars incorporated by P. falciparum trophozoites. A, amounts of mannose (dotted bars) and fucose (gray solid bars) indicate the average of three different determinations at different times. Error bars, S.D. Values are indicated in pmol of sugar/107 cells. t0 controls were mock-labeled for a few seconds using medium at 4 °C supplemented with an excess (100-fold) of the unlabeled (cold) sugar. B, amount of sugar incorporated to the glycoproteins (left; determinations performed in triplicate; error bars, S.D.) or glycolipids (right) of P. falciparum. 3H incorporated is indicated in pmol of sugar/107 cells. C, [3H]GlcN (dashed line) and [3H]GlcNAc (dotted line) incorporated by P. falciparum trophozoites. l-Glc, which is not incorporated by the parasite, is included as a negative control of incorporation (solid line). Determinations were performed in triplicate at three different times, and mean values ± S.D. are shown. t0 controls were generated as in A.
Similarly, metabolic labeling with [3H]GlcNAc showed that parasites were unable to take up this sugar from the media (Fig. 3C). This, together with the labeling of the parasite with [3H]GlcN, strongly suggests that the sequence of intermediates in the P. falciparum UDP-GlcNAc biosynthetic pathway is GlcN6P → GlcNAc6P → GlcNAc1P → UDP-GlcNAc.
P. falciparum GDP-Fuc Biosynthetic Genes Are Active in Vitro
Both PfGMD and PfFS genes are identified and annotated in the P. falciparum genome (25, 46). The predicted PfGMD (PF3D7_0813800) protein product is assigned to EC 4.2.1.47 and presents 48.1% sequence identity and 61.4% similarity to its human counterpart. PfFS (PF3D7_1014000) encodes a protein assigned to EC 1.1.1.271 that has a 35.7% sequence identity and 57.1% similarity to its human homolog.
The presence of putative PfGMD and PfFS genes, together with the detection of GDP-Fuc in P. falciparum intraerythrocytic stages, suggests that the protein products of these genes would be active GDP-Fuc biosynthetic enzymes. To assess this, we expressed both PfGMD and PfFS genes in E. coli with the addition of N-terminal GST tags. In both cases, soluble proteins could be purified by GST affinity chromatography (Fig. 4A). The enzymes were assayed as described under “Experimental Procedures,” and the sugar nucleotides produced were analyzed either by LC-MS/MS or by reverse phase ion pairing chromatography. When both enzymes were added to the reaction, GDP-Fuc was readily detected (Fig. 4B, top panels). Both enzymes are required for the production of GDP-Fuc, because in the presence of PfFS only, there was no turnover of GDP-Man (not shown), and with only PfGMD, a novel GDP-deoxyhexose with a different retention time was generated (Fig. 4B, middle panels). The production of a novel GDP-deoxyhexose by a GMD enzyme in the absence of an FS enzyme has been reported before (36, 47). In addition, in the presence of PfGMD alone, we detected also a product at 586 m/z, which coincides with the molecular weight of GDP-4-keto-6-deoxy mannose. Although there are no commercial standards available to analyze the transition of GDP-4-keto-6-deoxy mannose for identification, the fragmentation of this novel product produced a major product ion at m/z 442 corresponding to a [GDP-H]−, strongly resembling the fragmentation of GDP-deoxyhexoses (Fig. 4B, bottom panels) (30, 36).
FIGURE 4.
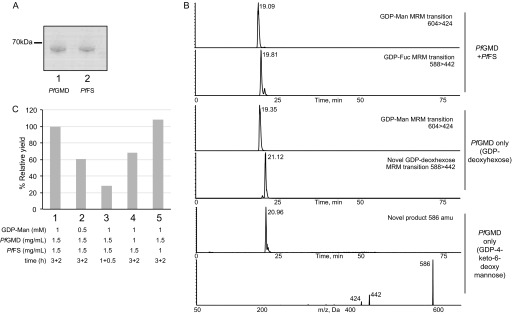
Activity of expressed PfGMD and PfFS. A, Coomassie Blue-stained SDS-polyacrylamide gel of purified, GST-tagged, recombinantly expressed PfGMD (lane 1) and PfFS (lane 2) proteins. B, the purified recombinant enzymes were assayed using GDP-Man as a substrate. Sugar nucleotides were analyzed by multiple-reaction monitoring LC-MS/MS. Multiple-reaction monitoring (MRM) transitions are indicated in the panels. C, effect of time and GDP-Man and enzyme concentration on the yield of GDP-Fuc by PfGMD and PfFS. Conditions tested are: 1 mm GDP-Man, 3 h of 1.5 mg/ml PfGMD, and 2 h of 1.5 mg/ml PfFS (1) (control); 0.5 mm GDP-Man, 3 h of 1.5 mg/ml PfGMD, and 2 h of 1.5 mg/ml PfFS (2); 1 mm GDP-Man, 1 h of 1.5 mg/ml PfGMD, and 0.5 h of 1.5 mg/ml PfFS (3); 1 mm GDP-Man, 3 h of 1 mg/ml PfGMD, and 2 h of 1 mg/ml PfFS (4); and 1 mm GDP-Man, 3 h of 1.5 mg/ml PfGMD, and 2 h of 1 mg/ml PfFS (5).
To assess the effect of time and GDP-Man and PfGMD/PfFS concentration on the production of GDP-Fuc, we performed different activity assays where sugar nucleotides were measured by reverse phase HPLC. GDP-Fuc production was dependent on time and the initial concentration of GDP-Man (Fig. 4C). Interestingly, at the PfGMD and GDP-Man concentrations tested (1.5 mg/ml and 1 mm, respectively), PfFS did not become rate-limiting at 1 mg/ml, whereas decreasing the initial concentration of PfGMD to 1 mg/ml reduced the relative yield of GDP-Fuc to a 70%.
PfGMD and PfFS Are Expressed during P. falciparum Intraerythrocytic Life Cycle and Their Protein Products Localize in the Cytosol of the Parasite
Real-time quantitative RT-PCR analysis of PfGMD and PfFS RNA transcripts was performed at the different asexual stages of P. falciparum (rings, trophozoites, and schizonts), and expression values were calculated using the relative standard curve method. In both cases, the highest expression levels were found at the schizont phase (Fig. 5), in agreement with what had been observed previously in other studies and with what is annotated in P. falciparum metabolic databases (25, 48, 49). Nevertheless, transcription of both genes through the whole intraerythrocytic cycle is ∼10-fold lower than that of the seryl-tRNA synthetase housekeeping gene expression taken as a reference (48), with much lower PfFS levels in the ring stage (Fig. 5).
FIGURE 5.
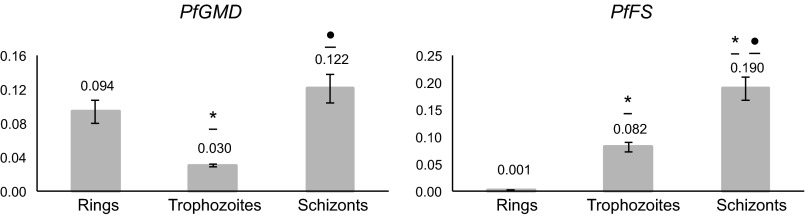
Real-time quantitative RT-PCR analysis of PfGMD (left) and PfFS (right) at different stages of P. falciparum intraerythrocytic cycle. Results are expressed in arbitrary units and normalized against seryl-tRNA synthetase. Data are representative of two independent RNA extractions per stage, assayed in triplicate. Statistically significant differences (one-way analysis of variance Tukey's post-test) between stages are shown with an asterisk (*, p < 0.05, rings versus trophozoites or schizonts) or a dot (●, p < 0.05, trophozoites versus schizonts).
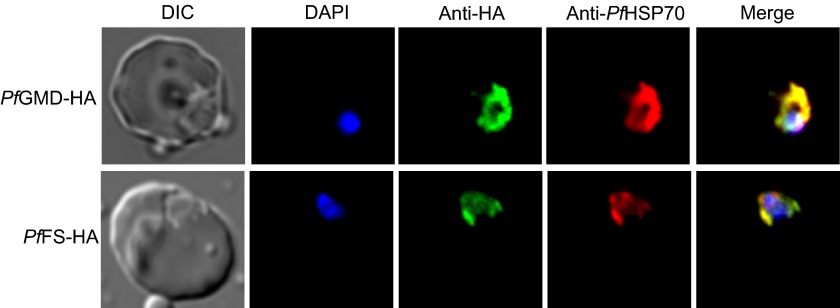
To assess the subcellular location of PfGMD and PfFS proteins, HA-tagged versions of the genes were transiently expressed under the control of the crt promoter using the pARL1a-3HA expression vector (41). Immunofluorescence microscopy using anti-HA antibodies produced a typical cytosolic distribution in the different stages of the intraerythrocytic life cycle (Fig. 6). To confirm this, we also stained the cells using anti-HSP70 antibody, a commonly used cytosolic marker (43). The merged image shows co-localization, indicating that the expressed HA-tagged PfGMD and PfFS are localized in the cytosol of P. falciparum through the different stages of the parasite asexual life cycle.
FIGURE 6.
Subcellular localization of PfGMD and PfFS expressed in P. falciparum. PfGMD-HA-expressing and PfFS-HA-expressing trophozoites were labeled with anti-HA (green), anti-PfHSP70 (red), and DAPI for nuclear staining (blue). The first column represents the differential interference contrast (DIC), and last column represents the merge of HA and PfHSP70 signals. Similar co-localization was observed in other stages of the intraerythrocytic life cycle (not shown).
P. falciparum Schizonts Express a Protein o-Fucosyltransferase (PoFUT2) Homolog and Incorporate [3H]GDP-Fuc in a Cell-free Assay
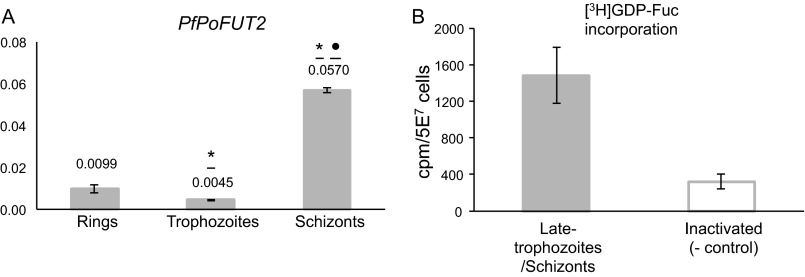
The P. falciparum genome presents a homolog of PoFUT2 that, in other organisms, is involved in the O-fucosylation of thrombospondin type 1 repeat (TSR) domains (50). Because this gene (PF3D7_0909200) is the most suitable candidate to encode for a fucosyltransferase activity, we performed real-time quantitative RT-PCR analysis of PfPoFUT2 at the different blood stages of P. falciparum. Despite the low levels, a peak of expression was observed at the schizont phase of the parasite (Fig. 7A) coinciding with the maximum expression of PfGMD and PfFS genes. Furthermore, P. falciparum schizont extracts incorporated [3H]GDP-Fuc, strongly suggesting that fucosylation processes are active in the late stages of the asexual life cycle of the parasite (Fig. 7B).
FIGURE 7.
P. falciparum late blood stages express PfPoFUT2 and incorporate [3H] GDP-fucose. A, real-time quantitative RT-PCR results are expressed in arbitrary units and normalized against seryl-tRNA synthetase. Data are representative of two independent RNA extractions per stage, assayed in triplicate. Statistically significant differences (one-way analysis of variance Tukey's post-test) between stages are shown with an asterisk (*, p < 0.05, rings versus trophozoites or schizonts) or a dot (●, p < 0.05, trophozoites versus schizonts). B, detergent homogenates of late trophozoite/schizont stage parasites were incubated with [3H]GDP-fucose before (gray solid bar) and after inactivation (open bar). The experiment was repeated two times on different days, and [3H]GDP-fucose incorporation was assayed in triplicate. Data are expressed as mean values ± S.D. (error bars).
DISCUSSION
Carbohydrate structures that decorate the surface of cells play important roles in the biology of host-pathogen interactions. In the particular case of the malaria parasite, glycan structures associated with the parasite itself appear to be limited to GPI anchors (11, 51) and the recently described N-glycans (14). This seems to confirm the general trend of Plasmodium parasites to reduce several metabolic and biosynthetic pathways and reflects the process of evolving toward a parasitic niche (52). However, the parasite genome encodes orthologues for enzymes involved in the biosynthesis of sugar nucleotides not related to GPI anchor or N-glycan structures, which prompted us to survey the sugar nucleotide pools present in the asexual life stages of P. falciparum. Qualitatively, the sugar nucleotide content of P. falciparum obtained by LC-electrospray ionization-MS/MS agrees well with the presence of known or candidate sugar nucleotide biosynthesis enzymes (Fig. 1) (25), with the exception of the presence of a significant pool of UDP-Gal (see below). Quantitatively, the sugar nucleotide amounts observed are consistent with the increased metabolic activity of mature stage parasites (53, 54). The analysis performed, together with the information available in annotated P. falciparum metabolic databases (25), enabled us to thoroughly verify the presence of active sugar nucleotide metabolic routes during the parasite blood stages and, in some cases, add new evidence to support the annotations of genes likely to be involved in sugar nucleotide biosynthesis. The presence and conservation of these sugar nucleotide metabolic pathways in several Plasmodium genomes (46), together with the detection of their final products, are strong arguments to presume the existence of further downstream reactions in which these metabolites are involved and expose new active metabolic pathways accessible for the exploration of their biological function.
The identification of UDP-Gal is somewhat puzzling. The P. falciparum genome lacks candidate genes for a UDP-glucose 4-epimerase (EC 5.1.3.2) that can produce UDP-galactose via the epimerization of UDP-glucose or a galactose-1-phosphate uridylyl transferase activity (EC 2.7.7.12) (25). Thus, two main activities can be proposed for the production of this sugar nucleotide. Either an enzyme with UTP-glucose-1-phosphate uridylyltransferase activity (EC 2.7.7.9) presents a weak galactose-1-phosphate uridylyltransferase activity (EC 2.7.7.10), as described in mammals (55, 56), or a broad substrate range UDP-sugar pyrophosphorylase (EC 2.7.7.64) is present in the genome of P. falciparum, as described in plants or Leishmania major (57–61). UDP-sugar pyrophosphorylase is an enzyme that can nonspecifically utilize UTP and glucose 1-phosphate or galactose 1-phosphate to produce UDP-glucose or UDP-galactose and pyrophosphate. Interestingly, searches with functionally characterized UDP-sugar pyrophosphorylase orthologues from plants and L. major identify a match, PF3D7_0517500, with more than 30% similarity and an e value of <10−55. However, although galactose competes for P. falciparum PfHT1 hexose permease (45) and its incorporation into the parasite galactolipids has been reported (62, 63), there is not a clear galactokinase candidate in the P. falciparum genome, and tritiated galactose is not significantly incorporated into the parasite proteins (23) or into the α-galactose moieties of digalactosyl diglycerides (64).
Bioinformatic analysis (25) and our own work suggest that UDP-GlcNAc, the direct donor for all GlcNAc transferases, is made in P. falciparum by the conventional eukaryotic de novo route from glucose 6-phosphate (Fig. 1), although a salvage pathway also exists, via the action of hexokinase (GlcN → GlcN-6-P) (22, 23). With more than 30% similarity and e values of <10−20 to plant and human UDP-GlcNAc diphosphorylases (EC 2.7.7.23), PF3D7_1343600 is the most suitable candidate gene to encode this activity in the P. falciparum genome (Fig. 1 and Table 1), although redundant activity from PF3D7_0517500 might be expected. The lack of a candidate for a gene encoding for UDP-glucose 4-epimerase (EC 5.1.3.2) that could present a UDP-GlcNAc 4-epimerase activity (EC 5.1.3.7) to convert UDP-GlcNAc into UDP-GalNAc indicates that the parasite probably does not produce UDP-N-acetyl galactosamine, in agreement with the inability of the parasite to synthesize GalNAc and the absence of mucin-type O-glycosylation in P. falciparum (63, 65).
The main question regarding the UDP-GlcNAc biosynthetic pathway is the identity of a gene encoding for the glucosamine-phosphate N-acetyltransferase activity (EC 2.3.1.4 (25)). The identification of pools of UDP-GlcNAc in the parasite (Fig. 2), along with the labeling of GPI-anchored proteins using tritiated glucosamine (Fig. 3C) (23, 63), strongly suggests that the UDP-GlcNAc metabolic route (25) (Fig. 1) is active in P. falciparum. Most likely the UDP-GlcNAc de novo pathway from glucose is the most important in vivo for the parasite, because glucosamine is not an abundant free sugar in the mammalian or insect host. Because the glucosamine moiety, derived from GlcNAc by de-N-acetylation (27), is present in GPI structures that serve as membrane anchors for many important surface antigens of the parasite invasive stages (12, 66), the inability to identify a candidate gene encoding for the glucosamine-phosphate N-acetyltransferase activity is very intriguing because this pathway is potentially targetable for selective anti-malarial drug design.
The presence of the fucose donor GDP-Fuc in the intraerythrocytic stages of P. falciparum is not surprising, because the parasite contains homologues of the enzymes involved in the biosynthesis of this precursor from GDP-Man. We have shown that PfGMD and PfFS are active in vitro (Fig. 4), and they are expressed through the parasite's asexual life cycle (Fig. 5) (67). The GDP-Fuc pools in P. falciparum are, on average (Table 2), comparable with the pools of other protozoan parasites, such as trypanosomatids (30), including T. brucei, for which this metabolite is essential (36). The apparent lack of agreement with the amount of the GDP-Fuc pool in the schizont stage of the parasite when the PfFS gene is more expressed may be due to the specific demand of the metabolite at that stage. Because sugar nucleotides are donors for glycosylation reactions, the rates of turnover and the amount of the pools detected may change, reflecting their utilization. The localization of both enzymes in the cytoplasmic compartment (Fig. 6), as aldolase and other enzymes involved in the metabolism of carbohydrates (68), agrees well with the identification of a putative GDP-Fuc transporter in the P. falciparum endoplasmic reticulum/Golgi apparatus (69), where N- and O-glycosylation processes occur. The location and function of a putative fucose-containing glycan in P. falciparum remain enigmatic, and biosynthetic labeling with tritiated fucose is not feasible, because there is no prominent GDP-Fuc salvage pathway in the parasite, and/or hexose transporters do not efficiently take up fucose (Fig. 3). However, the labeling of parasite extracts after incubation with modified precursors opens a door to the identification of putative fucose-containing glycoconjugates (Fig. 7B). Interestingly, the C-terminal region of P. falciparum circumsporozoite surface protein (CS), in which the RTS,S malaria vaccine is based (70), and other proteins of the parasite (71) contain TSR domains. TSR domains are generally O-fucosylated in higher eukaryotes by PoFUT2, and the fucose residue can be further modified by the addition of β1–3 glucose (72–74). The C-terminal region of CS, containing the TSR domain, was recently expressed in HEK293T cells and structurally analyzed, showing the presence of a fucose and a glucose residue (75, 76). The expression of PoFUT2 in P. falciparum (Fig. 7A) and its conservation in other Plasmodium species raise the possibility of the presence of a mechanism of O-fucosylation of CS and other TSR-containing proteins in the parasite.
In summary, in this work, we have reported the first evidence of the presence of sugar nucleotides in the blood stages of P. falciparum, and we have described the active metabolic routes involved in their biosynthesis. In addition, we have characterized the de novo route of GDP-Fuc, a metabolite that may probably be involved in the biosynthesis of novel fucosylated glycans not yet described in the malaria parasite.
Acknowledgments
We thank Hernando A. del Portillo for support, reagents, and helpful scientific discussions throughout this project. We thank Mike Ferguson for helpful discussions and for access to the LC-MS/MS system to measure sugar nucleotides. We are grateful to I. H. Hallyburton, J. A. Nunes, M. Ramírez, and CCiTUB (Scientific and Technological Centers Universitat de Barcelona) for technical support, advice, and assistance. We also thank L. Martín-Jaular, A. Cortés, N. Rovira-Graells, and M. Llinás for useful suggestions and comments on the manuscript.
This work was supported by Spanish Ministry of Economy and Competitiveness Grant SAF2010-21069 (to L. I.).
- GPI
- glycosylphosphatidylinositol
- Man
- mannose
- Fuc
- fucose
- TSR
- thrombospondin type 1 repeat.
REFERENCES
- 1. World Health Organization (2011) World Malaria Report 2011, World Health Organization, Geneva [Google Scholar]
- 2. Price R. N., Douglas N. M., Anstey N. M. (2009) New developments in Plasmodium vivax malaria. Severe disease and the rise of chloroquine resistance. Curr. Opin. Infect. Dis. 22, 430–435 [DOI] [PubMed] [Google Scholar]
- 3. Guinovart C., Navia M. M., Tanner M., Alonso P. L. (2006) Malaria. Burden of disease. Curr. Mol. Med. 6, 137–140 [DOI] [PubMed] [Google Scholar]
- 4. Gardner M. J., Hall N., Fung E., White O., Berriman M., Hyman R. W., Carlton J. M., Pain A., Nelson K. E., Bowman S., Paulsen I. T., James K., Eisen J. A., Rutherford K., Salzberg S. L., Craig A., Kyes S., Chan M. S., Nene V., Shallom S. J., Suh B., Peterson J., Angiuoli S., Pertea M., Allen J., Selengut J., Haft D., Mather M. W., Vaidya A. B., Martin D. M., Fairlamb A. H., Fraunholz M. J., Roos D. S., Ralph S. A., McFadden G. I., Cummings L. M., Subramanian G. M., Mungall C., Venter J. C., Carucci D. J., Hoffman S. L., Newbold C., Davis R. W., Fraser C. M., Barrell B. (2002) Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419, 498–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sakata T., Winzeler E. A. (2007) Genomics, systems biology, and drug development for infectious diseases. Mol. Biosyst. 3, 841–848 [DOI] [PubMed] [Google Scholar]
- 6. Meissner M., Agop-Nersesian C., Sullivan W. J., Jr. (2007) Molecular tools for analysis of gene function in parasitic microorganisms. Appl. Microbiol. Biotechnol. 75, 963–975 [DOI] [PubMed] [Google Scholar]
- 7. Ekland E. H., Fidock D. A. (2007) Advances in understanding the genetic basis of antimalarial drug resistance. Curr. Opin. Microbiol. 10, 363–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kooij T. W., Janse C. J., Waters A. P. (2006) Plasmodium post-genomics. Better the bug you know? Nat. Rev. Microbiol. 4, 344–357 [DOI] [PubMed] [Google Scholar]
- 9. Greenwood B. M., Fidock D. A., Kyle D. E., Kappe S. H., Alonso P. L., Collins F. H., Duffy P. E. (2008) Malaria. Progress, perils, and prospects for eradication. J. Clin. Invest. 118, 1266–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ferguson M. A., Homans S. W., Dwek R. A., Rademacher T. W. (1988) Glycosyl-phosphatidylinositol moiety that anchors Trypanosoma brucei variant surface glycoprotein to the membrane. Science 239, 753–759 [DOI] [PubMed] [Google Scholar]
- 11. von Itzstein M., Plebanski M., Cooke B. M., Coppel R. L. (2008) Hot, sweet and sticky. The glycobiology of Plasmodium falciparum. Trends Parasitol. 24, 210–218 [DOI] [PubMed] [Google Scholar]
- 12. Sanders P. R., Kats L. M., Drew D. R., O'Donnell R. A., O'Neill M., Maier A. G., Coppel R. L., Crabb B. S. (2006) A set of glycosylphosphatidyl inositol-anchored membrane proteins of Plasmodium falciparum is refractory to genetic deletion. Infect. Immun. 74, 4330–4338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Delorenzi M., Sexton A., Shams-Eldin H., Schwarz R. T., Speed T., Schofield L. (2002) Genes for glycosylphosphatidylinositol toxin biosynthesis in Plasmodium falciparum. Infect. Immun. 70, 4510–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bushkin G. G., Ratner D. M., Cui J., Banerjee S., Duraisingh M. T., Jennings C. V., Dvorin J. D., Gubbels M. J., Robertson S. D., Steffen M., O'Keefe B. R., Robbins P. W., Samuelson J. (2010) Suggestive evidence for Darwinian selection against asparagine-linked glycans of Plasmodium falciparum and Toxoplasma gondii. Eukaryot. Cell 9, 228–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Izquierdo L., Atrih A., Rodrigues J. A., Jones D. C., Ferguson M. A. (2009) Trypanosoma brucei UDP-glucose:glycoprotein glucosyltransferase has unusual substrate specificity and protects the parasite from stress. Eukaryot. Cell 8, 230–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Izquierdo L., Schulz B. L., Rodrigues J. A., Güther M. L., Procter J. B., Barton G. J., Aebi M., Ferguson M. A. (2009) Distinct donor and acceptor specificities of Trypanosoma brucei oligosaccharyltransferases. EMBO J. 28, 2650–2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Banerjee S., Vishwanath P., Cui J., Kelleher D. J., Gilmore R., Robbins P. W., Samuelson J. (2007) The evolution of N-glycan-dependent endoplasmic reticulum quality control factors for glycoprotein folding and degradation. Proc. Natl. Acad. Sci. U.S.A. 104, 11676–11681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Samuelson J., Banerjee S., Magnelli P., Cui J., Kelleher D. J., Gilmore R., Robbins P. W. (2005) The diversity of dolichol-linked precursors to Asn-linked glycans probably results from secondary loss of sets of glycosyltransferases. Proc. Natl. Acad. Sci. U.S.A. 102, 1548–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dayal-Drager R., Hoessli D. C., Decrind C., Del Guidice G., Lambert P. H., Nasir-ud-Din (1991) Presence of O-glycosylated glycoproteins in the Plasmodium falciparum parasite. Carbohydr. Res. 209, c5–c8 [DOI] [PubMed] [Google Scholar]
- 20. Nasir-ud-Din, Drager-Dayal R., Decrind C., Hu B. H., Del Giudice G., Hoessli D. (1992) Plasmodium falciparum synthesizes O-glycosylated glycoproteins containing O-linked N-acetylglucosamine. Biochem. Int. 27, 55–64 [PubMed] [Google Scholar]
- 21. Nasir-un-Din, Hassan M., Qazi M. H., Fayyazuddin, Sinaldi G., Hoessli D., Walker-Nasir E. (1992) Plasmodium falciparum synthesizes 43000 daltons protein containing O-linked glucosamine. Biochem. Soc. Trans. 20, 388S. [DOI] [PubMed] [Google Scholar]
- 22. Dieckmann-Schuppert A., Bender S., Odenthal-Schnittler M., Bause E., Schwarz R. T. (1992) Apparent lack of N-glycosylation in the asexual intraerythrocytic stage of Plasmodium falciparum. Eur. J. Biochem. 205, 815–825 [DOI] [PubMed] [Google Scholar]
- 23. Gowda D. C., Gupta P., Davidson E. A. (1997) Glycosylphosphatidylinositol anchors represent the major carbohydrate modification in proteins of intraerythrocytic stage Plasmodium falciparum. J. Biol. Chem. 272, 6428–6439 [DOI] [PubMed] [Google Scholar]
- 24. Kimura E. A., Couto A. S., Peres V. J., Casal O. L., Katzin A. M. (1996) N-Linked glycoproteins are related to schizogony of the intraerythrocytic stage in Plasmodium falciparum. J. Biol. Chem. 271, 14452–14461 [DOI] [PubMed] [Google Scholar]
- 25. Ginsburg H. (2006) Progress in in silico functional genomics. The malaria metabolic pathways database. Trends Parasitol. 22, 238–240 [DOI] [PubMed] [Google Scholar]
- 26. Stoeckert C. J., Jr., Fischer S., Kissinger J. C., Heiges M., Aurrecoechea C., Gajria B., Roos D. S. (2006) PlasmoDB v5. New looks, new genomes. Trends Parasitol. 22, 543–546 [DOI] [PubMed] [Google Scholar]
- 27. Ferguson M. A. (1999) The structure, biosynthesis and functions of glycosylphosphatidylinositol anchors, and the contributions of trypanosome research. J. Cell Sci. 112, 2799–2809 [DOI] [PubMed] [Google Scholar]
- 28. Trager W., Jensen J. B. (1976) Human malaria parasites in continuous culture. Science 193, 673–675 [DOI] [PubMed] [Google Scholar]
- 29. Di Girolamo F., Raggi C., Birago C., Pizzi E., Lalle M., Picci L., Pace T., Bachi A., de Jong J., Janse C. J., Waters A. P., Sargiacomo M., Ponzi M. (2008) Plasmodium lipid rafts contain proteins implicated in vesicular trafficking and signalling as well as members of the PIR superfamily, potentially implicated in host immune system interactions. Proteomics 8, 2500–2513 [DOI] [PubMed] [Google Scholar]
- 30. Turnock D. C., Ferguson M. A. (2007) Sugar nucleotide pools of Trypanosoma brucei, Trypanosoma cruzi, and Leishmania major. Eukaryot. Cell 6, 1450–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Räbinä J., Mäki M., Savilahti E. M., Järvinen N., Penttilä L., Renkonen R. (2001) Analysis of nucleotide sugars from cell lysates by ion-pair solid-phase extraction and reversed-phase high-performance liquid chromatography. Glycoconj. J. 18, 799–805 [DOI] [PubMed] [Google Scholar]
- 32. Gerold P., Dieckmann-Schuppert A., Schwarz R. T. (1994) Glycosylphosphatidylinositols synthesized by asexual erythrocytic stages of the malarial parasite, Plasmodium falciparum. Candidates for plasmodial glycosylphosphatidylinositol membrane anchor precursors and pathogenicity factors. J. Biol. Chem. 269, 2597–2606 [PubMed] [Google Scholar]
- 33. Azzouz N., de Macedo C. S., Ferguson M. A., Smith T. K., Schwarz R. T. (2005) Mannosamine can replace glucosamine in glycosylphosphatidylinositols of Plasmodium falciparum in vitro. Mol. Biochem. Parasitol. 142, 12–24 [DOI] [PubMed] [Google Scholar]
- 34. Azzouz N., Gerold P., Schwarz R. T. (2008) Metabolic labeling and structural analysis of glycosylphosphatidylinositols from parasitic protozoa. Methods Mol. Biol. 446, 183–198 [DOI] [PubMed] [Google Scholar]
- 35. Williams M., Sprenger J., Human E., Al-Karadaghi S., Persson L., Louw A. I., Birkholtz L. M. (2011) Biochemical characterisation and novel classification of monofunctional S-adenosylmethionine decarboxylase of Plasmodium falciparum. Mol. Biochem. Parasitol. 180, 17–26 [DOI] [PubMed] [Google Scholar]
- 36. Turnock D. C., Izquierdo L., Ferguson M. A. (2007) The de novo synthesis of GDP-fucose is essential for flagellar adhesion and cell growth in Trypanosoma brucei. J. Biol. Chem. 282, 28853–28863 [DOI] [PubMed] [Google Scholar]
- 37. Rhomberg S., Fuchsluger C., Rendić D., Paschinger K., Jantsch V., Kosma P., Wilson I. B. (2006) Reconstitution in vitro of the GDP-fucose biosynthetic pathways of Caenorhabditis elegans and Drosophila melanogaster. FEBS J. 273, 2244–2256 [DOI] [PubMed] [Google Scholar]
- 38. Sullivan F. X., Kumar R., Kriz R., Stahl M., Xu G. Y., Rouse J., Chang X. J., Boodhoo A., Potvin B., Cumming D. A. (1998) Molecular cloning of human GDP-mannose 4,6-dehydratase and reconstitution of GDP-fucose biosynthesis in vitro. J. Biol. Chem. 273, 8193–8202 [DOI] [PubMed] [Google Scholar]
- 39. Pabst M., Grass J., Fischl R., Léonard R., Jin C., Hinterkörner G., Borth N., Altmann F. (2010) Nucleotide and nucleotide sugar analysis by liquid chromatography-electrospray ionization-mass spectrometry on surface-conditioned porous graphitic carbon. Anal. Chem. 82, 9782–9788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bernabeu M., Lopez F. J., Ferrer M., Martin-Jaular L., Razaname A., Corradin G., Maier A. G., Del Portillo H. A., Fernandez-Becerra C. (2012) Functional analysis of Plasmodium vivax VIR proteins reveals different subcellular localizations and cytoadherence to the ICAM-1 endothelial receptor. Cell Microbiol. 14, 386–400 [DOI] [PubMed] [Google Scholar]
- 41. Crabb B. S., Rug M., Gilberger T. W., Thompson J. K., Triglia T., Maier A. G., Cowman A. F. (2004) Transfection of the human malaria parasite Plasmodium falciparum. Methods Mol. Biol. 270, 263–276 [DOI] [PubMed] [Google Scholar]
- 42. Tonkin C. J., van Dooren G. G., Spurck T. P., Struck N. S., Good R. T., Handman E., Cowman A. F., McFadden G. I. (2004) Localization of organellar proteins in Plasmodium falciparum using a novel set of transfection vectors and a new immunofluorescence fixation method. Mol. Biochem. Parasitol. 137, 13–21 [DOI] [PubMed] [Google Scholar]
- 43. Pesce E. R., Acharya P., Tatu U., Nicoll W. S., Shonhai A., Hoppe H. C., Blatch G. L. (2008) The Plasmodium falciparum heat shock protein 40, Pfj4, associates with heat shock protein 70 and shows similar heat induction and localisation patterns. Int. J. Biochem. Cell Biol. 40, 2914–2926 [DOI] [PubMed] [Google Scholar]
- 44. Geary T. G., Divo A. A., Bonanni L. C., Jensen J. B. (1985) Nutritional requirements of Plasmodium falciparum in culture. III. Further observations on essential nutrients and antimetabolites. J. Protozool. 32, 608–613 [DOI] [PubMed] [Google Scholar]
- 45. Woodrow C. J., Burchmore R. J., Krishna S. (2000) Hexose permeation pathways in Plasmodium falciparum-infected erythrocytes. Proc. Natl. Acad. Sci. U.S.A. 97, 9931–9936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Aurrecoechea C., Brestelli J., Brunk B. P., Dommer J., Fischer S., Gajria B., Gao X., Gingle A., Grant G., Harb O. S., Heiges M., Innamorato F., Iodice J., Kissinger J. C., Kraemer E., Li W., Miller J. A., Nayak V., Pennington C., Pinney D. F., Roos D. S., Ross C., Stoeckert C. J., Jr., Treatman C., Wang H. (2009) PlasmoDB. A functional genomic database for malaria parasites. Nucleic Acids Res. 37, D539–D543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tonetti M., Zanardi D., Gurnon J. R., Fruscione F., Armirotti A., Damonte G., Sturla L., De Flora A., Van Etten J. L. (2003) Paramecium bursaria Chlorella virus 1 encodes two enzymes involved in the biosynthesis of GDP-l-fucose and GDP-d-rhamnose. J. Biol. Chem. 278, 21559–21565 [DOI] [PubMed] [Google Scholar]
- 48. Bozdech Z., Llinás M., Pulliam B. L., Wong E. D., Zhu J., DeRisi J. L. (2003) The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 1, E5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rovira-Graells N., Gupta A. P., Planet E., Crowley V. M., Mok S., Ribas de Pouplana L., Preiser P. R., Bozdech Z., Cortés A. (2012) Transcriptional variation in the malaria parasite Plasmodium falciparum. Genome Res. 22, 925–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Luo Y., Koles K., Vorndam W., Haltiwanger R. S., Panin V. M. (2006) Protein O-fucosyltransferase 2 adds O-fucose to thrombospondin type 1 repeats. J. Biol. Chem. 281, 9393–9399 [DOI] [PubMed] [Google Scholar]
- 51. Mendonça-Previato L., Todeschini A. R., Heise N., Previato J. O. (2005) Protozoan parasite-specific carbohydrate structures. Curr. Opin. Struct. Biol. 15, 499–505 [DOI] [PubMed] [Google Scholar]
- 52. Olszewski K. L., Llinás M. (2011) Central carbon metabolism of Plasmodium parasites. Mol. Biochem. Parasitol. 175, 95–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rosenthal P. J. (2002) Hydrolysis of erythrocyte proteins by proteases of malaria parasites. Curr. Opin. Hematol. 9, 140–145 [DOI] [PubMed] [Google Scholar]
- 54. Roth E., Jr. (1990) Plasmodium falciparum carbohydrate metabolism. A connection between host cell and parasite. Blood Cells 16, 453–460; discussion 461–456 [PubMed] [Google Scholar]
- 55. Isselbacher K. J. (1958) A mammalian uridinediphosphate galactose pyrophosphorylase. J. Biol. Chem. 232, 429–444 [PubMed] [Google Scholar]
- 56. Leslie N., Yager C., Reynolds R., Segal S. (2005) UDP-galactose pyrophosphorylase in mice with galactose-1-phosphate uridyltransferase deficiency. Mol. Genet. Metab. 85, 21–27 [DOI] [PubMed] [Google Scholar]
- 57. Damerow S., Lamerz A. C., Haselhorst T., Führing J., Zarnovican P., von Itzstein M., Routier F. H. (2010) Leishmania UDP-sugar pyrophosphorylase. The missing link in galactose salvage? J. Biol. Chem. 285, 878–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dickmanns A., Damerow S., Neumann P., Schulz E. C., Lamerz A. C., Routier F. H., Ficner R. (2011) Structural basis for the broad substrate range of the UDP-sugar pyrophosphorylase from Leishmania major. J. Mol. Biol. 405, 461–478 [DOI] [PubMed] [Google Scholar]
- 59. Lamerz A. C., Damerow S., Kleczka B., Wiese M., van Zandbergen G., Lamerz J., Wenzel A., Hsu F. F., Turk J., Beverley S. M., Routier F. H. (2010) Deletion of UDP-glucose pyrophosphorylase reveals a UDP-glucose independent UDP-galactose salvage pathway in Leishmania major. Glycobiology 20, 872–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Litterer L. A., Schnurr J. A., Plaisance K. L., Storey K. K., Gronwald J. W., Somers D. A. (2006) Characterization and expression of Arabidopsis UDP-sugar pyrophosphorylase. Plant Physiol. Biochem. 44, 171–180 [DOI] [PubMed] [Google Scholar]
- 61. Kotake T., Hojo S., Yamaguchi D., Aohara T., Konishi T., Tsumuraya Y. (2007) Properties and physiological functions of UDP-sugar pyrophosphorylase in Arabidopsis. Biosci. Biotechnol. Biochem. 71, 761–771 [DOI] [PubMed] [Google Scholar]
- 62. Maréchal E., Azzouz N., de Macedo C. S., Block M. A., Feagin J. E., Schwarz R. T., Joyard J. (2002) Synthesis of chloroplast galactolipids in apicomplexan parasites. Eukaryot. Cell 1, 653–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Dieckmann-Schuppert A., Bender S., Holder A. A., Haldar K., Schwarz R. T. (1992) Labeling and initial characterization of polar lipids in cultures of Plasmodium falciparum. Parasitol. Res. 78, 416–422 [DOI] [PubMed] [Google Scholar]
- 64. Ramasamy R., Field M. C. (2012) Terminal galactosylation of glycoconjugates in Plasmodium falciparum asexual blood stages and Trypanosoma brucei bloodstream trypomastigotes. Exp. Parasitol. 130, 314–320 [DOI] [PubMed] [Google Scholar]
- 65. Dieckmann-Schuppert A., Bause E., Schwarz R. T. (1993) Studies on O-glycans of Plasmodium falciparum-infected human erythrocytes. Evidence for O-GlcNAc and O-GlcNAc-transferase in malaria parasites. Eur. J. Biochem. 216, 779–788 [DOI] [PubMed] [Google Scholar]
- 66. Gilson P. R., Nebl T., Vukcevic D., Moritz R. L., Sargeant T., Speed T. P., Schofield L., Crabb B. S. (2006) Identification and stoichiometry of glycosylphosphatidylinositol-anchored membrane proteins of the human malaria parasite Plasmodium falciparum. Mol. Cell Proteomics 5, 1286–1299 [DOI] [PubMed] [Google Scholar]
- 67. Otto T. D., Wilinski D., Assefa S., Keane T. M., Sarry L. R., Böhme U., Lemieux J., Barrell B., Pain A., Berriman M., Newbold C., Llinás M. (2010) New insights into the blood-stage transcriptome of Plasmodium falciparum using RNA-Seq. Mol. Microbiol. 76, 12–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bhowmick I. P., Kumar N., Sharma S., Coppens I., Jarori G. K. (2009) Plasmodium falciparum enolase. Stage-specific expression and sub-cellular localization. Malaria J. 8, 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Martin R. E., Ginsburg H., Kirk K. (2009) Membrane transport proteins of the malaria parasite. Mol. Microbiol. 74, 519–528 [DOI] [PubMed] [Google Scholar]
- 70. Agnandji S. T., Lell B., Soulanoudjingar S. S., Fernandes J. F., Abossolo B. P., Conzelmann C., Methogo B. G., Doucka Y., Flamen A., Mordmüller B., Issifou S., Kremsner P. G., Sacarlal J., Aide P., Lanaspa M., Aponte J. J., Nhamuave A., Quelhas D., Bassat Q., Mandjate S., Macete E., Alonso P., Abdulla S., Salim N., Juma O., Shomari M., Shubis K., Machera F., Hamad A. S., Minja R., Mtoro A., Sykes A., Ahmed S., Urassa A. M., Ali A. M., Mwangoka G., Tanner M., Tinto H., D'Alessandro U., Sorgho H., Valea I., Tahita M. C., Kaboré W., Ouédraogo S., Sandrine Y., Guiguemdé R. T., Oud́raogo J. B., Hamel M. J., Kariuki S., Odero C., Oneko M., Otieno K., Awino N., Omoto J., Williamson J., Muturi-Kioi V., Laserson K. F., Slutsker L., Otieno W., Otieno L., Nekoye O., Gondi S., Otieno A., Ogutu B., Wasuna R., Owira V., Jones D., Onyango A. A., Njuguna P., Chilengi R., Akoo P., Kerubo C., Gitaka J., Maingi C., Lang T., Olotu A., Tsofa B., Bejon P., Peshu N., Marsh K., Owusu-Agyei S., Asante K. P., Osei-Kwakye K., Boahen O., Ayamba S., Kayan K., Owusu-Ofori R., Dosoo D., Asante I., Adjei G., Chandramohan D., Greenwood B., Lusingu J., Gesase S., Malabeja A., Abdul O., Kilavo H., Mahende C., Liheluka E., Lemnge M., Theander T., Drakeley C., Ansong D., Agbenyega T., Adjei S., Boateng H. O., Rettig T., Bawa J., Sylverken J., Sambian D., Agyekum A., Owusu L., Martinson F., Hoffman I., Mvalo T., Kamthunzi P., Nkomo R., Msika A., Jumbe A., Chome N., Nyakuipa D., Chintedza J., Ballou W. R., Bruls M., Cohen J., Guerra Y., Jongert E., Lapierre D., Leach A., Lievens M., Ofori-Anyinam O., Vekemans J., Carter T., Leboulleux D., Loucq C., Radford A., Savarese B., Schellenberg D., Sillman M., Vansadia P., and RTS,S Clinical Trials Partnership (2011) First results of phase 3 trial of RTS,S/AS01 malaria vaccine in African children. N. Engl. J. Med. 365, 1863–1875 [DOI] [PubMed] [Google Scholar]
- 71. Morahan B. J., Wang L., Coppel R. L. (2009) No TRAP, no invasion. Trends Parasitol. 25, 77–84 [DOI] [PubMed] [Google Scholar]
- 72. Tan K., Duquette M., Liu J. H., Dong Y., Zhang R., Joachimiak A., Lawler J., Wang J. H. (2002) Crystal structure of the TSP-1 type 1 repeats. A novel layered fold and its biological implication. J. Cell Biol. 159, 373–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hofsteenge J., Huwiler K. G., Macek B., Hess D., Lawler J., Mosher D. F., Peter-Katalinic J. (2001) C-Mannosylation and O-fucosylation of the thrombospondin type 1 module. J. Biol. Chem. 276, 6485–6498 [DOI] [PubMed] [Google Scholar]
- 74. Kozma K., Keusch J. J., Hegemann B., Luther K. B., Klein D., Hess D., Haltiwanger R. S., Hofsteenge J. (2006) Identification and characterization of abeta1,3-glucosyltransferase that synthesizes the Glc-β1,3-Fuc disaccharide on thrombospondin type 1 repeats. J. Biol. Chem. 281, 36742–36751 [DOI] [PubMed] [Google Scholar]
- 75. Doud M. B., Koksal A. C., Mi L. Z., Song G., Lu C., Springer T. A. (2012) Unexpected fold in the circumsporozoite protein target of malaria vaccines. Proc. Natl. Acad. Sci. U.S.A. 109, 7817–7822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Song G., Koksal A. C., Lu C., Springer T. A. (2012) Shape change in the receptor for gliding motility in Plasmodium sporozoites. Proc. Natl. Acad. Sci. U.S.A. 109, 21420–21425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Aoki K., Tanaka N., Kusakabe Y., Fukumi C., Haga A., Nakanishi M., Kitade Y., Nakamura K. T. (2010) Crystallization and preliminary x-ray crystallographic study of phosphoglucose isomerase from Plasmodium falciparum. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66, 333–336 [DOI] [PMC free article] [PubMed] [Google Scholar]