Background: The bivalency of IgG and Fc fusion could cause undesired therapeutic properties.
Results: We developed a stable monomeric Fc modality by N-glycosylation engineering, enabling the generation of crystal structure.
Conclusion: The monomeric Fc prolonged the half-life of Fab domain through the interaction with neonatal Fc receptor.
Significance: The monomeric Fc will be used for pharmacokinetics enhancement of biotherapeutics that require monovalent properties.
Keywords: Antibody Engineering, Crystal Structure, Glycosylation, Pharmacokinetics, Protein Engineering, N-Glycosylation, Half-life Extension
Abstract
Human IgG is a bivalent molecule that has two identical Fab domains connected by a dimeric Fc domain. For therapeutic purposes, however, the bivalency of IgG and Fc fusion proteins could cause undesired properties. We therefore engineered the conversion of the natural dimeric Fc domain to a highly soluble monomer by introducing two Asn-linked glycans onto the hydrophobic CH3-CH3 dimer interface. The monomeric Fc (monoFc) maintained the binding affinity for neonatal Fc receptor (FcRn) in a pH-dependent manner. We solved the crystal structure of monoFc, which explains how the carbohydrates can stabilize the protein surface and provides the rationale for molecular recognition between monoFc and FcRn. The monoFc prolonged the in vivo half-life of an antibody Fab domain, and a tandem repeat of the monoFc further prolonged the half-life. This monoFc modality can be used to improve the pharmacokinetics of monomeric therapeutic proteins with an option to modulate the degree of half-life extension.
Introduction
Prolonged survival time of IgG molecules in serum is achieved through the interaction of their Fc region with the neonatal Fc receptor (FcRn)2 (1, 2). The long half-life of full-length IgG and Fc fusion proteins, which have been used as therapeutic molecules for the treatment of various diseases, allows for less frequent dosing in patients (3, 4). Human IgG is a bivalent molecule that provides avidity effect and maximizes the host defense against pathogenic bacteria and virus. For other therapeutic purposes, however, the bivalency of IgG might not be necessary and could cause undesired properties. For example, if the targets are multimeric soluble molecules, the dimeric nature of IgG can result in formation of a cross-linked network in plasma (5). Furthermore, when the targets to be antagonized are on a cell surface, the IgG may result in unwanted agonist activity (6). To overcome these issues, “one-armed” antibody and one-armed Fc fusion proteins have been recently created using Fc heterodimers for various therapeutic targets and shown to improve the biological activity, bioavailability, and pharmacokinetics of a molecule (7, 8). As an alternative approach for half-life extension of biotherapeutics, we sought a novel monomeric Fc modality that allows single polypeptide chains, simplifying production development.
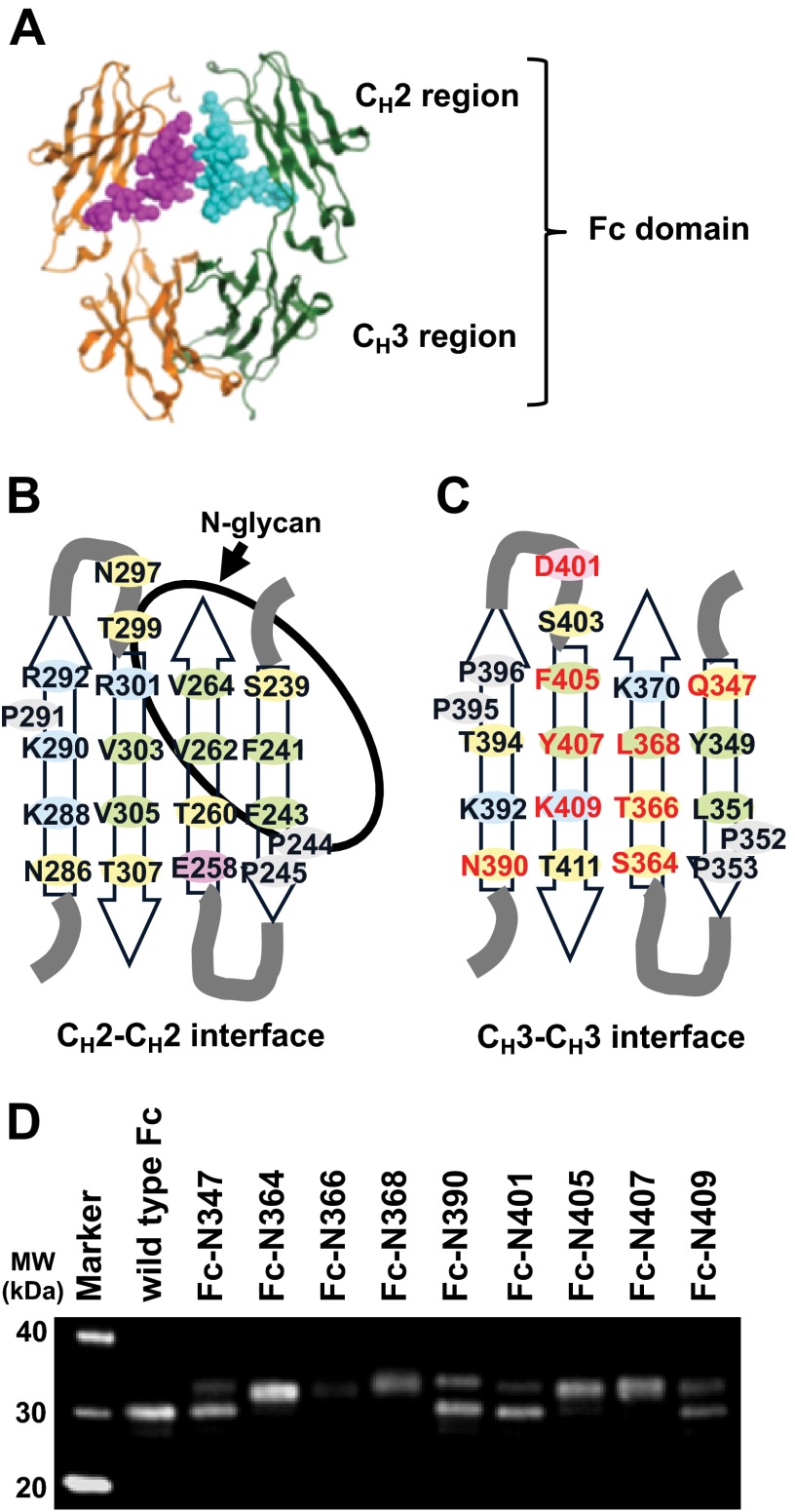
Asn-linked glycosylation (N-glycosylation) is one of the most common forms of post-translational modification of proteins in eukaryotic organisms. In general, the modification occurs at an asparagine residue in the consensus sequence of Asn-X-Ser/Thr, where X is any amino acid except proline (9). N-Glycosylation can have an impact on the protein stability, susceptibility to protease, and immunogenicity as well as in vivo bioactivity of therapeutic proteins (10, 11). Native human antibodies have an N-glycan at Asn297 on the CH2 region of the dimeric form of the Fc domain. Crystal structures of the Fc domains have revealed that the carbohydrates are packed within the internal space enclosed by the CH2 domain (see Fig. 1A). Although the CH2 domains from two polypeptide chains make no direct interactions due to the carbohydrate (see Fig. 1, A and B), the CH3 domains associate through a large hydrophobic interface (see Fig. 1, A and C). To engineer a stable monomeric form of Fc domain, we utilized the N-glycosylation engineering approach, in which the engineered carbohydrates will not only disrupt the CH3-CH3 interface, but also mask the exposed hydrophobic surface of CH3 domain.
FIGURE 1.
Structure-based design and protein expression of N-glycosylated variants of human antibody Fc domain. A, the crystal structure of the Fc domain of human IgG B12 (PDB ID: 1HZH) is shown. In this model, each polypeptide chain of the Fc dimer is shown as ribbon models (orange and green), and all the atoms of carbohydrate moieties are shown as space-filling models (magenta and cyan). All the molecular graphic figures were prepared with the program MOE (Chemical Computing Group). B and C, schematic drawing of dimer interface with amino acid residues (Eu numbering (14)) of CH2 domain (B) and CH3 domain (C) of human immunoglobulin γ1 chain. The amino acid residues that were replaced with asparagine for N-glycosylation incorporation are shown as red. Hydrophobic, hydrophilic, negatively charged, and positively charged residues are shown as green, yellow, pink, and blue. Proline residues are shown as gray. The area covered by N-glycan at Asn297 is shown as an orbit. D, Western blot analysis of mutational variants under reducing condition: Fc-N347 (Q347N/Y349T); Fc-N364 (S364N); Fc-N366 (T366N/L368T); Fc-N368 (L368N/K370T); Fc-N390 (K390N/K392T); Fc-N401 (D401N); Fc-N405 (F405N/Y407T); Fc-N407 (Y407N/K409T); Fc-N409 (K409N). The genes were transiently expressed in HEK293F cells. Expression levels of all the mutants were similar to wild type Fc domain except that Fc-N366 was expressed poorly. The Fc variants with additional N-glycosylation migrated with mobility corresponding to a molecular mass of ∼32 kDa, whereas wild type Fc molecule migrated at ∼30 kDa. The natural glycosylation of Fc may account for the difference from the theoretical molecular mass of polypeptide part of wild type Fc (25,010 Da). MW, molecular weight standard.
EXPERIMENTAL PROCEDURES
Selection of the Positions for N-Glycosylation Engineering
Four criteria were employed to determine the residues for introducing N-glycosylation mutational sites (Asn-X-Ser/Thr) at the CH3-CH3 interface. First, we identified residues located on the interface. To calculate the percentage of accessible surface area (%ASA) of each residue in both Fc dimer (native form) and one chain of Fc dimer (hypothetical Fc monomer), we used the crystal structure of the Fc domain of human IgG B12 (Protein Data Bank (PDB) ID: 1HZH), which is the high resolution structure of the entire human antibody. The residues with a higher %ASA(dimer) should be the residues that are exposed to solvent. We hypothesized that the residues with the higher %ASA(monomer) value are more likely to be either exposed to solvent or buried in the CH3-CH3 interface. Therefore, we estimated the degree of interface involvement by subtracting the %ASA of monomer from that of Fc dimer as ΔASA = %ASA(monomer) − %ASA(dimer) and selected 22 interface residues whose ΔASA values were above a threshold value of 10% (Table 1). Second, we avoided mutagenesis of proline, glycine, and cysteine residues that generally play an important role in maintaining the structural framework of proteins. Third, we only incorporated Asn-X-Ser/Thr-Y in regions where neither X nor Y is a proline residue because proline at either of these positions strongly inhibits glycosylation efficiency (9, 12). When the residue at position +2 needed to be mutated, we chose threonine over serine because it has been shown that threonine at the position tends to produce higher N-glycosylation occupancy than serine (13). Finally, we performed a manual inspection and selected nine residues where the engineered carbohydrate would have a substantial impact on disruption of the CH3-CH3 interface.
TABLE 1.
%ASA of candidate residues for mutagenesis
ΔASA = %ASA(monomer) − %ASA(dimer). % of ASA was calculated with the program MOE (Chemical Computing Group).
| Interface residues | %ASA (dimer) | %ASA (monomer) | ΔASA |
|---|---|---|---|
| Gln347a | 26.5 | 41.8 | 15.4 |
| Tyr349 | 5.1 | 41.4 | 36.3 |
| Leu351 | 3.8 | 41.9 | 38.0 |
| Ser354 | 13.6 | 60.2 | 46.6 |
| Asp356 | 47.9 | 74.4 | 26.5 |
| Glu357 | 2.8 | 26.9 | 24.1 |
| Lys360 | 42.7 | 62.9 | 20.2 |
| Ser364a | 3.8 | 18.5 | 14.8 |
| Thr366a | 0.7 | 21.2 | 20.5 |
| Leu368a | 1.4 | 15.2 | 13.8 |
| Lys370 | 17.1 | 37.3 | 20.0 |
| Asn390a | 39.9 | 55.6 | 15.7 |
| Lys392 | 42.8 | 77.6 | 34.9 |
| Thr394 | 2.5 | 42.7 | 40.2 |
| Val397 | 13.6 | 42.3 | 28.7 |
| Ser400 | 56.7 | 89.2 | 32.5 |
| Asp401a | 14.0 | 32.4 | 18.5 |
| Phe405a | 0 | 24.2 | 24.2 |
| Tyr407a | 0 | 37.3 | 37.3 |
| Lys409a | 1.5 | 50.5 | 48.9 |
| Lys439 | 27.9 | 41.3 | 13.4 |
| Ser444 | 56.8 | 68.8 | 12.0 |
a The positions selected for N-glycosylation engineering.
Protein Expression and Purification
The expression plasmid of wild type Fc fragment was constructed as an N-terminal hexahistidine tag followed by the human immunoglobulin γ1 constant region starting with Gly236 (refer to the numbering of the Eu antibody amino acid sequence in Ref. 14). The mutational constructs were generated by PCR with the primers that generate desired amino acid replacement. KLH (keyhole limpet hemocyanin protein)-derived monoclonal antibody was used as a control antibody for mouse pharmacokinetics (PK) study as it comprises no target in mouse (internal data). The expression vectors of Fab-monoFc variants were constructed by PCR amplification of the Fab fragment of the KLH-derived antibody and monoFc constructs. For initial protein production of N-glycosylated Fc variants, HEK293F cells were transiently transfected with the expression plasmids by using 293fectin reagent and grown in FreeStyle293 medium according to the manufacturer's protocol (Invitrogen). All the Fc variants were purified by using HiTrap chelating column and HiTrap protein A column (GE Healthcare). For in vivo analysis of Fab-monoFc variants, CHO cells were transfected with the expression plasmids by Lipofectamine 2000 (Invitrogen). Stable clones were selected with G418 and methotrexate for 2–3 weeks. The proteins were purified by using HiTrap protein G column followed by Superdex200 column (GE Healthcare). All the purified fractions were dialyzed against PBS and stored at −80 °C.
Size Exclusion Chromatography-Multiangle Light Scattering (SEC-MALS)
Average molar mass and oligomerization state of wild type Fc domain and N-glycosylated Fc variants were determined using SEC-MALS. Protein samples were prepared at concentrations of 4.5–7.0 mg/ml in PBS buffer. Each sample (200 μg) was injected onto an analytical Superdex 200 10/300 GL column (GE Healthcare) connected to an Agilent 1100 HPLC system (Foster City, CA). Protein peaks resolved on the sizing column were analyzed using the Wyatt Technology miniDAWN three-angle light scattering detector and Optilab-REX refractometer (Santa Barbara, CA) connected in-line to the HPLC system. Data acquisition and analysis were done using the Wyatt Technology Astra software with a Δn/Δc value of 0.185 ml/g for protein. Glycan mass contribution was determined by applying the protein conjugation template in Astra software using an approximated Δn/Δc value of 0.14 ml/g for the sugar moiety.
Differential Scanning Calorimetry (DSC)
Thermal stabilities of wild type Fc domain and N-glycosylated Fc variants were analyzed using the MicroCal capillary DSC system, VP-DSC (Northampton, MA). The protein sample at concentration of 0.02 mm in PBS buffer was placed in the sample cell. The heat capacity difference between the sample cell and reference cell was recorded and analyzed using Origin7.0 software from MicroCal.
Capillary Gel Electrophoresis
The relative percentage of glycosylated and unglycosylated species in each protein sample was measured under reducing condition using Caliper LabChip GXII (Hopkinton, MA). Deglycosylated control was prepared by incubating the protein with Glycanase F (ProZyme) for 3 h at 37 °C in PBS buffer. Samples for the Caliper assay were prepared according to the manufacturer's instructions. Protein loading, separation, staining, and destaining were performed on a quartz chip photo-etched with microchannels according to the LabChip Protein Express program. An electropherogram was generated for each sample and analyzed using LabChip GX version 3.0 software.
Solubility Assessment by Analytical SEC
Protein samples were dialyzed against HEPES buffer (50 mm HEPES, 150 mm NaCl, pH 8.0), phosphate buffer (50 mm phosphate, 150 mm NaCl, pH 7.0), histidine buffer (50 mm, histidine, 150 mm NaCl, pH 6.0), or acetate buffer (50 mm acetate, 150 mm NaCl, pH 5.0). Protein in each buffer was concentrated up to 200 mg/ml by Vivaspin ultrafiltration spin column with 10,000 molecular weight cut-off (Vivascience Ltd.) and then incubated for 7 days at room temperature. A total of 10 μl for each sample was injected onto an analytical Superdex 200 10/300 GL column (GE Healthcare) connected to an Agilent 1100 HPLC system (Foster City, CA).
FcRn Binding Assays
The FcRn binding assays were carried out using a surface plasmon resonance (SPR) biosensor, Biacore 3000 (Biacore). The SPR experiments were conducted at 25 °C in PBS buffer (pH 6.0) with 0.005% P20. Human and mouse FcRn proteins were purchased from ARVYS Proteins, Inc. (Stamford, CT). All the experiments were repeated three times.
1-to-1 Binding Assay
Immobilization of N-glycosylated Fc variants or Fab-monoFc variants on a CM5 sensor chip was conducted by the amine coupling method. The equilibrium binding was measured by injecting 150 μl of soluble human or mouse FcRn protein at flow rate of 5 μl/min. The sensor surfaces were regenerated by running PBS buffer (pH 7.2). We observed that all the binding curves of each concentration reached the plateau at the end of injection (30 min). Steady-state response units were recorded at the end of the injection, and the equilibrium dissociation constant (Kd) was calculated using the BIAevaluation software (GE Healthcare).
Avidity Assay
To assess the avidity of analyte (molecule in mobile phase), we used the SPR-based “avidity assay format” that was previously reported (15–17). Briefly, FcRn protein was immobilized on a CM5 sensor chip by the amine coupling method. The equilibrium binding was measured by injecting 30 μl of Fab-monoFc variants over the FcRn surface at flow rate of 2 μl/min. The sensor surfaces were regenerated by running 100 mm Tris-HCl, pH 8.0.
pH Switch Assay
To evaluate the efficiency of dissociation of Fab-monoFc variants from FcRn at neutral pH, we employed a “pH switch assay” modified from the method that was previously reported (18). In this assay, FcRn protein was immobilized on a CM5 sensor chip by the amine coupling method. The binding was measured by injecting 100 nm of Fab-monoFc variants in running buffer (PBS, pH 6.0) followed by injection of either running buffer (PBS, pH 6.0) or neutral buffer (PBS, pH 7.2) alone over the FcRn surface.
Crystallization and Structure Determination
Protein was concentrated to 30 mg/ml in Tris buffer (25 mm Tris-HCl, 150 mm NaCl, pH 7.5) for crystallization trials of monoFc (Fc-N364/N407).3 Crystallization was performed with the Mosquito crystallization robot (TTP LabTech) using the hanging-drop vapor diffusion method at 18 °C, with the drops containing 0.2 μl of protein solution and 0.2 μl of reservoir solution equilibrated against the reservoir solution. Crystals were cryo-protected in the presence of 20% glycerol in the mother liquor and immediately flash-cooled in liquid nitrogen. X-ray diffraction data were collected from a single crystal up to 1.9 Å resolution on the SER-CAT beamline 22-ID, Advanced Photon Source (APS), Argonne, IL. The data were indexed, integrated, and scaled with HKL2000 (the statistics are given in Table 2). The crystals belonged to the space group P3112, with cell dimensions of a = b = 64.22 Å and c = 146.94 Å. The structure was solved by molecular replacement with PHASER using the crystal structure of a mutated, antibody-dependent cell-mediated cytotoxicity-enhanced human Fc domain (PDB ID: 2QL1) (19) as a search model. After the monoFc monomer was located, the initial model was subjected to minimization with BUSTER and was further rebuilt using COOT. Several rounds of refinement alternating with rebuilding produced the final refined model corresponding to an Rcryst of 0.22 and Rfree of 0.24 (the refinement statistics are given in Table 2).
TABLE 2.
X-ray data collection and model refinement statistics
Numbers in parentheses reflect statistics for the last shell.
| Crystal 1 | |
|---|---|
| Data collection | |
| Space group | P3112 |
| Cell dimensions | |
| a, b, c (Å) | 64.22, 64.22, 146.94 |
| α, β, γ | 90.0, 90.0, 120.0 |
| Resolution (Å) | 50-1.9 (1.93-1.90) |
| Rmergea | 0.048 (0.66) |
| I/σI | 33.2 (1.12) |
| Completeness (%) | 96.8 (67.3) |
| Redundancy | 4.7 (2.2) |
| Refinement | |
| Resolution (Å) | 1.9 |
| No. of reflections | 26,861 |
| Rworkb/Rfreec (%) | 22.2/23.8 |
| No. of atoms | |
| Protein | 1,665 |
| Carbohydrate | 113 |
| Water | 161 |
| B-factors | |
| Protein | 49.35 |
| r.m.s.d deviations | |
| Bond lengths (Å) | 0.009 |
| Bond angles (°) | 1.15 |
a Rmerge = |Ih − 〈Ih〉|/Ih, where 〈Ih〉 is the average intensity over symmetry equivalents.
b Rwork = ‖Fobs| − |Fcalc‖/|Fobs|.
c Rfree is equivalent to Rwork, but calculated for a randomly chosen 5% of reflections omitted from the refinement process.
d r.m.s., root mean square.
PK Study in Mice
Male BALB/c mice (∼8-week-old males) were purchased from Charles River (Wilmington, MA). Six mice per group received a single dose of Fab-monoFc variants via intravenous route. The administered dose of 5 mg/kg was based on the most recent scheduled body weights. The test articles were prepared in PBS, and the dosing volume was 4 ml/kg. At 0 min, 10 min, 6 h, 24 h, and 2, 3, 4, 7, 14 and 21 days after dose, blood samples of 10 μl were collected from the tail vein via capillary tubes. The Pfizer Institutional Animal Care and Use Committee approved all aspects of these studies. All studies were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Study samples were quantitated using biotinylated goat anti-human antibody (Bethyl Laboratories) captured onto streptavidin-coated beads (affinity capture column of the Gyrolab CD microstructure). After being captured onto the affinity capture column, bound Fab-monoFc variants or KLH-derived antibodies were detected with Alexa Fluor 647-labeled goat anti-human antibody (Molecular Probes). Sample concentrations were determined by interpolation from a standard curve that was fit using a five-parameter logistic curve fit with 1/y2 response weighting in Watson (Version 7.4). Plasma PK parameters were calculated using noncompartmental methods with the aid of Watson (Version 7.4). Data in the terminal log-linear phase were analyzed by linear regression to estimate the terminal rate constant (k) and half-life (t½ = 0.693/k). At least the last three time points were used to calculate k. Total AUCinf was determined as the sum of AUC0-last and AUCextrap, where AUC0-last was calculated from 0 to the last time point (Tlast) with the last measurable concentration (Clast) using the linear trapezoidal rule and AUCextrap was the extrapolated portion of the area from Tlast to infinite using Clast/k. Total body clearance (CL) based on plasma concentrations was calculated as dose/AUCinf, and the volume of distribution at steady-state (Vdss) was calculated as CL × AUMC/AUC, where AUMC was the area under the first moment curve. The intersubject variability was relatively higher for the KLH-derived antibody than for other constructs.
RESULTS AND DISCUSSION
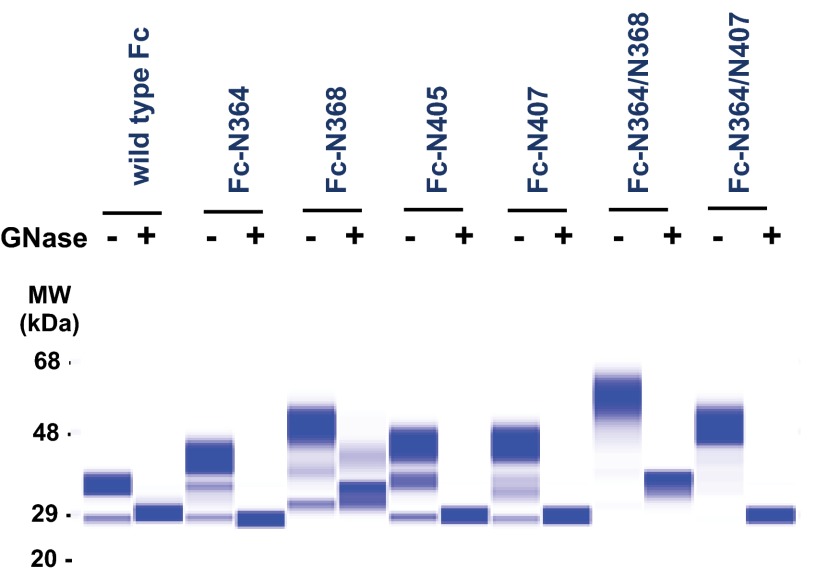
We designed mutational sites for N-glycosylation incorporation onto the CH3-CH3 interface based on the crystal structure of Fc domain (see the details under “Experimental Procedures”). Nine selected residues are illustrated in Fig. 1C. Individual N-glycosylated Fc variants were constructed using the Fc domain of human immunoglobulin γ1 without hinge region (Gly236 to Lys497) as a template. Western blot analysis showed that the mutagenesis at 364, 368, 405, and 407 resulted in efficient expression and N-glycosylation, whereas other mutants resulted in either lower expression of the protein or less than 50% incorporation of N-glycosylation (Fig. 1D). These four N-glycosylated variants (denoted as Fc-N364, Fc-N368, Fc-N405, and Fc-N407) were purified and further characterized. Capillary gel electrophoresis was used to estimate the N-linked glycosylation occupancy of the purified proteins (Fig. 2 and Table 3). Distribution of oligomeric species was analyzed by SEC-MALS. The molecular mass determined by SEC-MALS showed that Fc-N405 was mostly monomeric, whereas Fc-N364, Fc-N368, and Fc-N407 were mixtures of monomeric and dimeric forms (Table 3).
FIGURE 2.
Deglycosylation of engineered N-glycosylated Fc variants. Capillary gel electrophoresis of purified N-glycosylated variants shows that each band shift correlates with the number of incorporated N-glycosylation sequons. Capillary gel electrophoresis was also carried out with the treatment of Glycanase F (GNase), confirming that the shifting was caused by occupation of N-glycosylation in each position. The electropherogram was analyzed using LabChip GX version 3.0 software to quantitate the ratio of glycosylated and nonglycosylated species at each position (Table 3). MW, molecular weight standard.
TABLE 3.
Summary of biophysical characterizations of N-glycosylated Fc variants
| N-Glycosylated Fc variants | % of unglycosylationa | Molar massb |
Tmc | FcRn binding Kd (1-to-1)d |
||
|---|---|---|---|---|---|---|
| Monomer | Dimer | Human FcRn | Mouse FcRn | |||
| % | °C | nm | ||||
| Wild type Fc | 12 | None | 51,860 | 72/83 | 290 | 280 |
| Fc-N364 | 6 | 28,390 | 47,490 | 64 | 310 | 340 |
| Fc-N368 | 9 | 29,370 | 42,110 | 58 | 320 | 460 |
| Fc-N405 | 7 | 27,090 | None | 62 | 260 | 290 |
| Fc-N407 | 5 | 32,640/33,580e | 63 | 450 | 340 | |
| Fc-N364/N368 | <0.5 | Aggregation | 53/62 | 580 | 790 | |
| Fc-N364/N407 | <0.5 | 29,270 | None | 64 | 210 | 220 |
a Capillary gel electrophoresis was used to quantitate the yields of glycosylated and nonglycosylated species at each position. The percentage of unglycosylation was calculated as 100× [unglycosylated protein]/[glycosylated protein].
b SEC-MALS was used to estimate the molecular mass and distribution of oligomeric species of the N-glycosylated mutants.
c The melting temperature (Tm) was measured by DSC.
d The apparent equilibrium dissociation constants (Kd) for human or mouse FcRn binding were obtained from the 1-to-1 binding assay with an SPR biosensor.
e Two broad peaks were observed with average molecular mass between monomer and dimer.
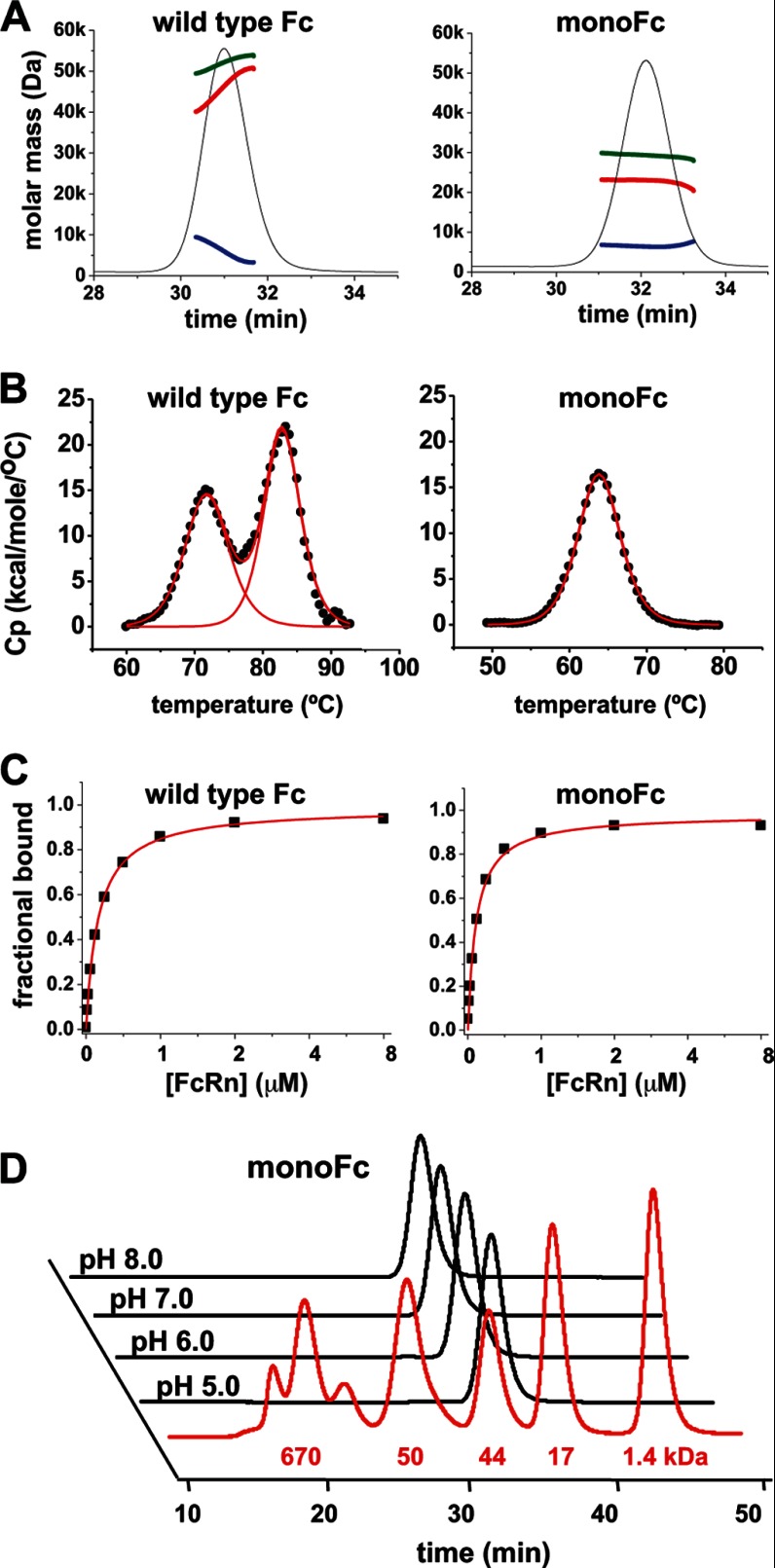
We attempted to eliminate the residual dimeric forms of these variants by combining two N-glycosylation sites. We created two double mutants, namely Fc-N364/N368 and Fc-N364/N407. Further increases in size were observed in Fc-N364/N368 and Fc-N364/N407 (Fig. 2). We found that nonglycosylated molecules of both Fc-N364/N368 and Fc-N364/N407 were decreased to an undetectable level (Fig. 2 and Table 3). SEC-MALS and DSC data show that Fc-N364/N407 was completely monomeric and the most stable among all the variants, whereas Fc-N364/N368 showed a tendency to aggregate and lower thermal stability (Fig. 3, A and B, and Table 3). The equilibrium binding assay confirmed that our monomeric Fc-N364/N407 retained binding to FcRn with almost the same affinity as wild type Fc (Fig. 3C and Table 3). Here, we demonstrated that the double N-glycosylation variant Fc-N364/N407 improved the properties of monomeric Fc while maintaining the ability to bind to FcRn, and we named this monomeric version of Fc molecule as monoFc. Two independent groups have recently reported engineering of monomeric forms of Fc domain by site-directed mutagenesis. Rose et al. (20) found that mutation of Tyr407 promoted the formation of monomeric Fc as well as sialylation of N-linked glycan at Asn297 in the CH2 domain. Ying et al. (21) also found that amino acid replacement at the positions of Pro395, Phe405, Tyr407, and Lys409 resulted in the monomeric form of Fc with FcRn binding affinity retained. We believe that N-glycosylation incorporation not only can disrupt the Fc dimer but also can improve physicochemical properties such as protein solubility and stability. We tested the solubility of monoFc at various pH levels and found that monoFc was highly soluble and remained monomeric up to 200 mg/ml (Fig. 3D). In addition, our approach may be advantageous when compared with the mutational approaches because the glycan moieties could sterically shield these residues, masking potential immune recognition or anti-drug antibody binding (22).
FIGURE 3.
Biophysical characterization of monoFc. A, SEC-MALS analysis shows the molar mass of total molecules (green), polypeptides (red), and carbohydrate moiety (blue) of wild type Fc domain (left panel) and monoFc (right panel). B, DSC analysis shows the thermograms (black dot) with model fitting (red line). The excess heat capacity (Cp) is plotted versus temperature. Wild type Fc dimer (left panel) yielded two transitions with the melting temperatures (tm) of 72 and 83 °C (right panel). In contrast, monoFc yielded a single transition with the melting temperature of 64 °C (right panel). C, SPR binding analysis shows the FcRn interaction with wild type Fc (left panel) and monoFc (right panel). The fractions at equilibrium for each injection were plotted versus the concentrations of mouse FcRn (black dots), and the data were fit to a steady-state affinity models (red line) to calculate Kd for immobilized Fc variants (Table 3) D, analytical SEC analysis of concentrated monoFc solution at various pH levels. Protein in each buffer was concentrated up to 200 mg/ml and then incubated for 7 days at room temperature. The SEC profile shows the overlay of monoFc samples in HEPES buffer (50 mm HEPES, 150 mm NaCl, pH 8.0), in phosphate buffer (50 mm phosphate, 150 mm NaCl, pH 7.0), in histidine buffer (50 mm, histidine, 150 mm NaCl, pH 6.0), and in acetate buffer (50 mm acetate, 150 mm NaCl, pH 5.0). Overlay of molecular weight (MW) standard (gel filtration standard, Bio-Rad) is shown as red line: 670 kDa (thyroglobulin), 158 kDa (γ-globulin), 44 kDa (ovalbumin), 17 kDa (myoglobin), and 1.4 kDa (vitamin B12).
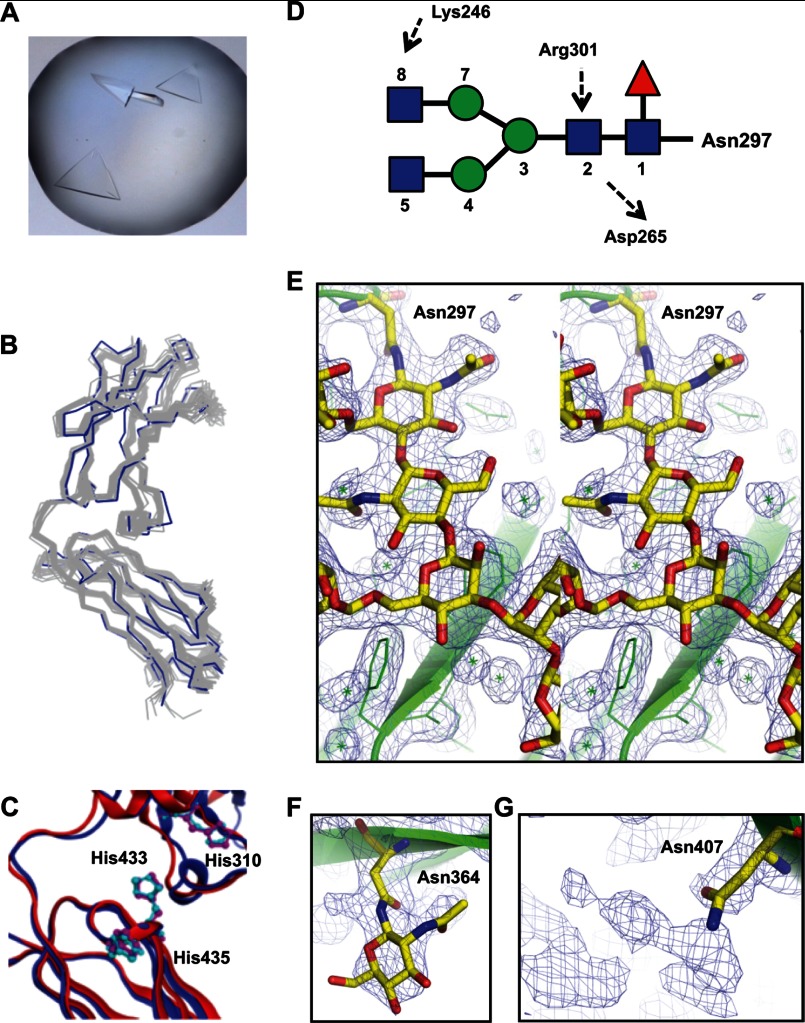
We solved the crystal structure of monoFc, which, to our knowledge is the first crystal structure of the monomeric form of the Fc domain. Crystals of monoFc were grown that diffracted to 1.9 Å (Fig. 4A and Table 2). The asymmetric unit contents of the crystal accounted for only one copy of monoFc, and the calculated electron density maps showed clear density for the entire backbone of the monomer from Gly236 to Ser444. Interestingly, the overall structure of monoFc was very similar to those of one polypeptide chain of Fc dimer (Fig. 4B), although the CH3-CH3 interface of Fc dimer is completely separated. It is also noteworthy that the spatial alignment of key residues involved in the FcRn interaction appears to have been maintained between monoFc and wild type Fc dimer (Fig. 4C). This might explain our finding that monoFc had the same FcRn binding affinity as wild type Fc (Fig. 3C and Table 3).
FIGURE 4.
Crystal structure of monoFc. A, crystals of monoFc protein. Large trigonal crystals were obtained using 2.2 m ammonium sulfate and 200 mm sodium fluoride as a precipitant. B, crystal structure of monoFc (blue) superimposed onto the structures of one polypeptide chain of 18 Fc domains (gray) available at the PDB. C, comparison of spatial alignment of His310, His433, and His435 of the monoFc (red) and Fc domain of human IgG1 (blue, PDB ID: 2DTS). D, schematic drawing of the sequence of carbohydrate at Asn297 and its interaction with amino acid residues of monoFc. Symbols are assigned for N-acetylglucosamine (blue square), mannose, (green circle), and fucose (red triangle). Dotted arrows represent the direction of hydrogen bonds from hydrogen donor to hydrogen acceptor. E, stereo representation of 2Fo − Fc electron density maps around the carbohydrate moiety connected to Asn297 of monoFc superimposed with refined molecular structures. F, clear density was seen for one sugar residue attached to Asn364. G, continuous but weak extra density was seen adjacent to Asn407.
The electron density of eight sugar residues connected to Asn297 was clearly visible on the CH2 domain of monoFc (Fig. 4, D and E), and the conformation of carbohydrate on monoFc was very similar to that of wild type Fc dimer. The internal space enclosed by the CH2 domain of the wild type Fc dimer is filled with the Asn297 carbohydrate that has multiple contacts with the inner surface of CH2 domain (Fig. 1A). In particular, Lys246, Asp265, and Arg301 are the key residues to form hydrogen bonds to sugar residues in the Fc dimer structures (23). Because the same hydrogen-bonding patterns were observed in the structure of monoFc (Fig. 4D), these hydrogen bonds most likely play an important role in maintaining the spatial orientation of Asn297 carbohydrates of monoFc. In contrast, the carbohydrates at the engineered Asn364 and Asn407 sites appeared highly mobile and therefore displayed only partial electron density of carbohydrates (Fig. 4, F and G). These observations could indicate that monoFc is stabilized by two different types of carbohydrates: the static nature of the carbohydrate at Asn297 and the dynamic nature of the carbohydrates at Asn364 and Asn407. In this study, we describe the method of N-glycosylation engineering to generate a monomeric molecule of the Fc domain that is naturally a dimer through tight hydrophobic interactions. We believe that our strategy can be applied to other systems to disrupt protein-protein interfaces and to stabilize the exposed protein surface by either the static or the dynamic nature of carbohydrates.
The Fc region of IgG is responsible for the prolonged circulation time of IgGs in serum through FcRn recycling (1, 2). To assess whether monoFc molecule can extend the serum lifetime of protein therapeutics, we studied the FcRn binding and PK of monoFc. We used the Fab fragment from the KLH-derived antibody fused to monoFc (Fab-monoFc) providing a prototype with no target in mice (internal data). To elucidate the FcRn involvement for the serum lifetime of monoFc, we also constructed the Fab-monoFc fusion proteins with FcRn-binding knock-out variant (Fab-monoFc[H310A/H433A]) (24) and the FcRn-binding enhancement variant (Fab-monoFc[M428L/N434S]) (15). First, we examined the FcRn binding of these constructs with the 1-to-1 binding assay format. As expected, Fab-monoFc[M428L/N434S] had higher binding affinity than Fab-monoFc, whereas no FcRn binding was observed for Fab-monoFc[H310A/H433A] (Table 4). Furthermore, we characterized the PK properties of these constructs in mice. The mean plasma concentration profiles following a single intravenous injection of the Fab-monoFc variants are illustrated in Fig. 5A. Fab-monoFc and Fab-monoFc[M428L/N434S] had improved PK over Fab-monoFc[H310A/H433A]. When comparing the PK parameters and FcRn binding affinity of these three Fab-monoFc variants (Tables 4 and 5), we found that the higher affinity for FcRn binding corresponded to the longer serum half-life in vivo. This correlation between FcRn binding and serum half-life suggests that the monoFc extended the plasma half-life of the fusion proteins through FcRn-mediated recycling mechanism.
TABLE 4.
FcRn binding affinity of Fab-monoFc variants
Two different SPR binding assays were used to measure the binding affinity of Fab-monoFc variants for FcRn. The 1-to-1 binding was examined by injecting either human or mouse FcRn over Fab-monoFc variants on the biosensor surface. The avidity binding was examined by injecting Fab-monoFc variants onto FcRn on the surface. Apparent equilibrium dissociation constants (Kd) were calculated by plotting the steady-state binding.
| Fab-monoFc fusions | Human FcRn binding |
Mouse FcRn binding |
||
|---|---|---|---|---|
| Kd (1-to-1) | Kd (avidity) | Kd (1-to-1) | Kd (avidity) | |
| nm | nm | |||
| Fab-monoFc | 210 | 1300 | 250 | 180 |
| Fab-monoFc[H310A/H433A] | >8,000 | >8,000 | >8,000 | >8,000 |
| Fab-monoFc[M428L/N434S] | 35 | 240 | 68 | 35 |
| Fab-monoFc-monoFc | 250 | 44 | 250 | 4.5 |
| KLH-derived antibody | 290 | 28 | 280 | 9.3 |
FIGURE 5.
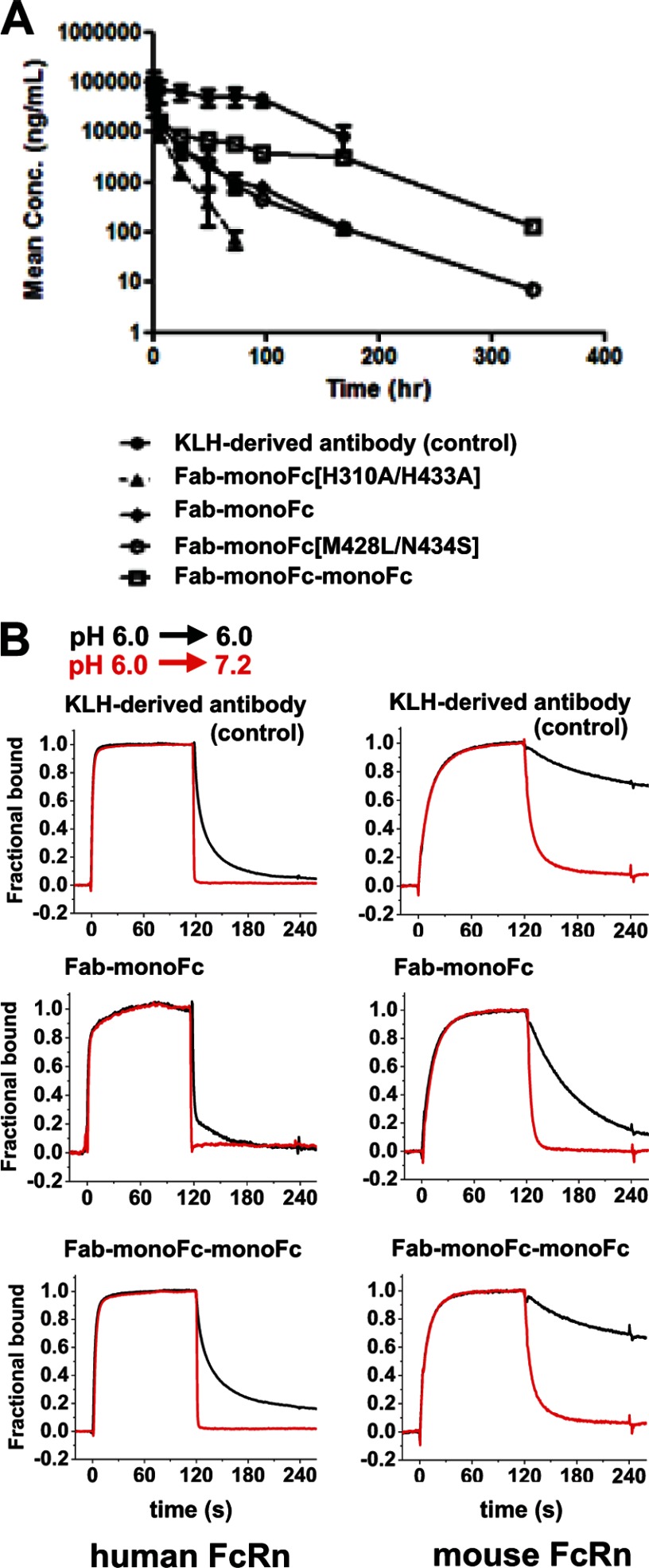
Pharmacokinetics and FcRn binding of monoFc variants fused to Fab fragment. A, BALB/c mice were intravenously administrated with 5 mg/kg of monoFc fused to the Fab fragment from the KLH-derived antibody (Fab-monoFc), FcRn-binding knock-out variant (Fab-monoFc[H310A/H433A]), FcRn-binding enhancement variant (Fab-monoFc[M428L/N434S]), and tandem monoFc fusion (Fab-monoFc-monoFc) as well as the KLH-derived antibody that is a control human antibody with no target in mice. At 0 min, 10 min, 6 h, 24 h, and 2, 3, 4, 7, 14, and 21 days after dose, blood samples of 10 μl were collected from the tail vein via capillary tubes. Each data point represents the mean ± S.D. (n = 6 each). The clearance for the KLH-derived antibody was comparable with that of typical IgGs at 0.3 ml/h/kg. The PK parameters are summarized in Table 5. Mean Conc., mean concentration. B, the pH-dependent interactions of Fab-monoFc, Fab-monoFc-monoFc, and the KLH-derived antibody with either human FcRn (left panels) or mouse FcRn (right panels). Overlays of sensorgrams show sequential injections of 100 nm of each Fab-monoFc variant in acidic buffer (pH 6.0) followed by injection of either acidic buffer (pH 6.0, black) or neutral buffer (pH 7.2, red) at 120 s. Each sensorgram was normalized for the comparison of dissociation phases (120–240 s) between pH 6.0 and pH 7.2.
TABLE 5.
Pharmacokinetic parameters of Fab-monoFc variants
Numbers are the average of six animals for intravenous administration of 5 mg/kg of proteins. PK parameters were calculated using noncompartmental methods using Watson (Version 7.4). AUC = area under the concentration curve (AUC represents AUCInf for all the constructs except for KLH-derived antibody, which used AUCLast due to the high AUCExtrap). C0 = Initial concentration extrapolated to time zero; t1/2 = terminal half-life; CL = clearance; Vdss = volume of distribution at steady-state.
| Variants | AUCInf | AUCExtrap | C0 | t1/2 | CL | Vdss |
|---|---|---|---|---|---|---|
| μg × h/ml | % | μg/ml | h | ml/h/kg | ml/kg | |
| Fab-monoFc | 579 | 1 | 70 | 32 | 9.0 | 177 |
| Fab-monoFc[H310A/H433A] | 364 | 0 | 72 | 11 | 14 | 93 |
| Fab-monoFc[M428L/N434S] | 548 | 1 | 58 | 42 | 9.2 | 213 |
| Fab-monoFc-monoFc | 1,288 | 20 | 95 | 97 | 3.0 | 323 |
| KLH-derived antibody | 5,955 | 62 | 140 | 173 | 0.3 | 78 |
The serum half-life for the Fab-monoFc molecule, however, was much shorter than that for the control antibody (Table 5). Hence, we proposed a hypothesis that avidity could improve the pharmacokinetic property of monoFc. To test the hypothesis, we constructed a tandem repeat of monoFc molecules fused to the Fab (Fab-monoFc-monoFc). The PK data showed that Fab-monoFc-monoFc had a 3-fold longer serum half-life than Fab-monoFc (Fig. 5A and Table 5). We next examined the FcRn binding in a format that permitted avid interaction (FcRn on the biosensor surface) as well as in the 1-to-1 format (monoFc on the biosensor surface). It has been known that mouse FcRn displays 2–10-fold higher affinity for human IgG than human FcRn (25–27). We observed a similar trend in which monoFc variants were bound to mouse FcRn with a higher affinity than human FcRn in the avidity format (Table 4). Although Fab-monoFc-monoFc had similar binding affinity to that of Fab-monoFc in the 1-to-1 binding format, it had ∼40- and 30-fold higher binding affinities than Fab-monoFc in the avidity format for mouse and human FcRn, respectively (Table 4). Fab-monoFc-monoFc appears to bind to FcRn on the biosensor surface with a slow dissociation rate, similar to that of control antibody (Fig. 5B). This is probably due to the avidity effect of tandem monoFc when it binds to the immobilized FcRn on the biosensor surface. The release of IgGs from FcRn at neutral pH is as important as the binding to FcRn at acidic pH for the FcRn-mediated recycling mechanism of IgGs (28). Our data demonstrated that Fab-monoFc-monoFc not only associated tightly to the immobilized FcRn at acidic pH, but also dissociated readily from the FcRn at neutral pH (Fig. 5B). These results strongly indicate that a tandem repeat of monoFc can gain avidity in a similar way to wild type Fc dimer, which results in slow dissociation from the surface FcRn at acidic pH while maintaining the fast dissociation at neutral pH. The Bjorkman group (29) has recently reported that the bivalency of Fc dimer is required for efficient cell-based FcRn-mediated recycling mechanism. Our FcRn binding and in vivo PK data further confirmed that bivalency is important to achieve the slow dissociation that is required for efficient FcRn recycling and thus longer serum half-life. The same group has also proposed a model in which a network of Fc-FcRn complex is formed between adjacent membranes (30, 31). The tandem monoFc might be able to gain the avidity for FcRn due to a close proximity of each receptor molecule on the membrane surface. In this context, avidity would slow the dissociation of tandem monoFc from FcRn in endosome at acidic pH, which in turn could prevent it from entering a degradation pathway in the lysosome (31).
In summary, we have engineered two N-glycosylation sites on the CH3 domain to stabilize the monomeric form of Fc and solved the crystal structure, revealing the stabilized protein surface by carbohydrates. We have demonstrated that monoFc can prolong the half-life of Fab domain via FcRn-mediated recycling and that the avidity through a tandem repeat of monoFc can further prolong the half-life. This monomeric Fc modality could possibly constitute an alternative approach for the PK enhancement of protein and peptide therapeutics that require monovalent properties.
Acknowledgments
We are grateful for technical assistance from Mark Krebs, Scott Gatto, and Richard Zollner. We also thank Madan Katragadda for helpful discussion.
The atomic coordinates and structure factors (code 4J12) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Throughout this study, the following mutant designations were used throughout: Fc-N364/N407, Fc[S364N/Y407N/K409T]; Fc-N364/N368, Fc[S364N/L368N/K370T]; Fc-N347, Q347N/Y349T; Fc-N364, S364N; Fc-N366, T366N/L368T; Fc-N368, L368N/K370T; Fc-N390, K390N/K392T; Fc-N401, D401N; Fc-N405, F405N/Y407T; Fc-N407, Y407N/K409T; Fc-N409, K409N.
- FcRn
- neonatal Fc receptor
- CH2
- constant region 2 of antibody heavy chain
- CH3
- constant region 3
- SEC-MALS
- size exclusion chromatography coupled with multiangle light scattering
- DSC
- differential scanning calorimetry
- SEC
- size exclusion chromatography
- PK
- pharmacokinetics
- monoFc
- monomeric Fc
- %ASA
- percentage of accessible surface area
- KLH
- keyhole limpet hemocyanin protein
- AUC
- area under the curve.
REFERENCES
- 1. Brambell F. W., Hemmings W. A., Morris I. G. (1964) A theoretical model of γ-globulin catabolism. Nature 203, 1352–1354 [DOI] [PubMed] [Google Scholar]
- 2. Raghavan M., Bonagura V. R., Morrison S. L., Bjorkman P. J. (1995) Analysis of the pH dependence of the neonatal Fc receptor/immunoglobulin G interaction using antibody and receptor variants. Biochemistry 34, 14649–14657 [DOI] [PubMed] [Google Scholar]
- 3. Morell A., Terry W. D., Waldmann T. A. (1970) Metabolic properties of IgG subclasses in man. J. Clin. Invest. 49, 673–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lobo E. D., Hansen R. J., Balthasar J. P. (2004) Antibody pharmacokinetics and pharmacodynamics. J. Pharm. Sci. 93, 2645–2668 [DOI] [PubMed] [Google Scholar]
- 5. Marrack J. (1955) The structure of antigen-antibody aggregates and complement fixation. Annu. Rev. Microbiol. 9, 369–386 [DOI] [PubMed] [Google Scholar]
- 6. Prat M., Crepaldi T., Pennacchietti S., Bussolino F., Comoglio P. M. (1998) Agonistic monoclonal antibodies against the Met receptor dissect the biological responses to HGF. J. Cell Sci. 111, 237–247 [DOI] [PubMed] [Google Scholar]
- 7. Demignot S., Pimm M. V., Baldwin R. W. (1990) Comparison of biodistribution of 791T/36 monoclonal antibody and its Fab/c fragment in BALB/c mice and nude mice bearing human tumor xenografts. Cancer Res. 50, 2936–2942 [PubMed] [Google Scholar]
- 8. Dumont J. A., Low S. C., Peters R. T., Bitonti A. J. (2006) Monomeric Fc fusions: impact on pharmacokinetic and biological activity of protein therapeutics. BioDrugs 20, 151–160 [DOI] [PubMed] [Google Scholar]
- 9. Shakin-Eshleman S. H., Spitalnik S. L., Kasturi L. (1996) The amino acid at the X position of an Asn-X-Ser sequon is an important determinant of N-linked core-glycosylation efficiency. J. Biol. Chem. 271, 6363–6366 [DOI] [PubMed] [Google Scholar]
- 10. Solá R. J., Griebenow K. (2009) Effects of glycosylation on the stability of protein pharmaceuticals. J. Pharm. Sci. 98, 1223–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Elliott S., Lorenzini T., Asher S., Aoki K., Brankow D., Buck L., Busse L., Chang D., Fuller J., Grant J., Hernday N., Hokum M., Hu S., Knudten A., Levin N., Komorowski R., Martin F., Navarro R., Osslund T., Rogers G., Rogers N., Trail G., Egrie J. (2003) Enhancement of therapeutic protein in vivo activities through glycoengineering. Nat. Biotechnol. 21, 414–421 [DOI] [PubMed] [Google Scholar]
- 12. Roitsch T., Lehle L. (1989) Structural requirements for protein N-glycosylation. Influence of acceptor peptides on cotranslational glycosylation of yeast invertase and site-directed mutagenesis around a sequon sequence. Eur. J. Biochem. 181, 525–529 [DOI] [PubMed] [Google Scholar]
- 13. Bause E., Legler G. (1981) The role of the hydroxy amino acid in the triplet sequence Asn-Xaa-Thr(Ser) for the N-glycosylation step during glycoprotein biosynthesis. Biochem. J. 195, 639–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Edelman G. M., Cunningham B. A., Gall W. E., Gottlieb P. D., Rutishauser U., Waxdal M. J. (1969) The covalent structure of an entire γG immunoglobulin molecule. Proc. Natl. Acad. Sci. U.S.A. 63, 78–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zalevsky J., Chamberlain A. K., Horton H. M., Karki S., Leung I. W., Sproule T. J., Lazar G. A., Roopenian D. C., Desjarlais J. R. (2010) Enhanced antibody half-life improves in vivo activity. Nat. Biotechnol. 28, 157–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yeung Y. A., Leabman M. K., Marvin J. S., Qiu J., Adams C. W., Lien S., Starovasnik M. A., Lowman H. B. (2009) Engineering human IgG1 affinity to human neonatal Fc receptor: impact of affinity improvement on pharmacokinetics in primates. J. Immunol. 182, 7663–7671 [DOI] [PubMed] [Google Scholar]
- 17. Suzuki T., Ishii-Watabe A., Tada M., Kobayashi T., Kanayasu-Toyoda T., Kawanishi T., Yamaguchi T. (2010) Importance of neonatal FcR in regulating the serum half-life of therapeutic proteins containing the Fc domain of human IgG1: a comparative study of the affinity of monoclonal antibodies and Fc-fusion proteins to human neonatal FcR. J. Immunol. 184, 1968–1976 [DOI] [PubMed] [Google Scholar]
- 18. Wang W., Lu P., Fang Y., Hamuro L., Pittman T., Carr B., Hochman J., Prueksaritanont T. (2011) Monoclonal antibodies with identical Fc sequences can bind to FcRn differentially with pharmacokinetic consequences. Drug Metab. Dispos. 39, 1469–1477 [DOI] [PubMed] [Google Scholar]
- 19. Oganesyan V., Damschroder M. M., Leach W., Wu H., Dall'Acqua W. F. (2008) Structural characterization of a mutated, ADCC-enhanced human Fc fragment. Mol. Immunol. 45, 1872–1882 [DOI] [PubMed] [Google Scholar]
- 20. Rose R. J., van Berkel P. H., van den Bremer E. T., Labrijn A. F., Vink T., Schuurman J., Heck A. J., Parren P. W. (2013) Mutation of Y407 in the CH3 domain dramatically alters glycosylation and structure of human IgG. mAbs 5, 219–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ying T., Chen W., Gong R., Feng Y., Dimitrov D. S. (2012) Soluble monomeric IgG1 Fc. J. Biol. Chem. 287, 19399–19408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gribben J. G., Devereux S., Thomas N. S., Keim M., Jones H. M., Goldstone A. H., Linch D. C. (1990) Development of antibodies to unprotected glycosylation sites on recombinant human GM-CSF. Lancet 335, 434–437 [DOI] [PubMed] [Google Scholar]
- 23. Krapp S., Mimura Y., Jefferis R., Huber R., Sondermann P. (2003) Structural analysis of human IgG-Fc glycoforms reveals a correlation between glycosylation and structural integrity. J. Mol. Biol. 325, 979–989 [DOI] [PubMed] [Google Scholar]
- 24. Ye L., Zeng R., Bai Y., Roopenian D. C., Zhu X. (2011) Efficient mucosal vaccination mediated by the neonatal Fc receptor. Nat. Biotechnol. 29, 158–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Datta-Mannan A., Witcher D. R., Tang Y., Watkins J., Jiang W., Wroblewski V. J. (2007) Humanized IgG1 variants with differential binding properties to the neonatal Fc receptor: relationship to pharmacokinetics in mice and primates. Drug Metab. Dispos. 35, 86–94 [DOI] [PubMed] [Google Scholar]
- 26. Deng R., Loyet K. M., Lien S., Iyer S., DeForge L. E., Theil F. P., Lowman H. B., Fielder P. J., Prabhu S. (2010) Pharmacokinetics of humanized monoclonal anti-tumor necrosis factor-α antibody and its neonatal Fc receptor variants in mice and cynomolgus monkeys. Drug Metab. Dispos. 38, 600–605 [DOI] [PubMed] [Google Scholar]
- 27. Ober R. J., Radu C. G., Ghetie V., Ward E. S. (2001) Differences in promiscuity for antibody-FcRn interactions across species: implications for therapeutic antibodies. Int. Immunol. 13, 1551–1559 [DOI] [PubMed] [Google Scholar]
- 28. Vaccaro C., Zhou J., Ober R. J., Ward E. S. (2005) Engineering the Fc region of immunoglobulin G to modulate in vivo antibody levels. Nat. Biotechnol. 23, 1283–1288 [DOI] [PubMed] [Google Scholar]
- 29. Tesar D. B., Tiangco N. E., Bjorkman P. J. (2006) Ligand valency affects transcytosis, recycling and intracellular trafficking mediated by the neonatal Fc receptor. Traffic 7, 1127–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Burmeister W. P., Huber A. H., Bjorkman P. J. (1994) Crystal structure of the complex of rat neonatal Fc receptor with Fc. Nature 372, 379–383 [DOI] [PubMed] [Google Scholar]
- 31. Raghavan M., Bjorkman P. J. (1996) Fc receptors and their interactions with immunoglobulins. Annu. Rev. Cell Dev. Biol. 12, 181–220 [DOI] [PubMed] [Google Scholar]