Background: SNX10 and SNX11 exhibit antagonistic activity in regulating endosome vacuolation.
Results: Two additional α-helices downstream of the conventional PX domain were identified in the SNX11 crystal structure.
Conclusion: SNX11 contains an extended PX (PXe) domain critical for the inhibition of vacuolation induced by SNX10.
Significance: This study reports a novel PXe domain that may represent a new subgroup of the PX domain.
Keywords: Cell Sorting, Phosphatidylinositol, Protein Sorting, Protein Structure, Sorting, PX Domain, PXe Domain, SNX10, SNX11, Endosome
Abstract
Sorting nexins are phox homology (PX) domain-containing proteins involved in diverse intracellular endosomal trafficking pathways. The PX domain binds to certain phosphatidylinositols and is recruited to vesicles rich in these lipids. The structure of the PX domain is highly conserved, containing a three-stranded β-sheet, followed by three α-helices. Here, we report the crystal structures of truncated human SNX11 (sorting nexin 11). The structures reveal that SNX11 contains a novel PX domain, hereby named the extended PX (PXe) domain, with two additional α-helices at the C terminus. We demonstrate that these α-helices are indispensible for the in vitro functions of SNX11. We propose that this PXe domain is present in SNX10 and is responsible for the vacuolation activity of SNX10. Thus, this novel PXe domain constitutes a structurally and functionally important PX domain subfamily.
Introduction
Endocytosis and intracellular protein trafficking are well regulated biological processes (1, 2). The endosomal system is critical for endocytosis and intracellular protein trafficking. Research has shown that different components of the endosomal system contain distinct species of phosphatidylinositols (PIs)2 that are able to recruit certain sorting proteins to the specific sites of the endosome membrane (3, 4). Several PI-binding domains have been identified, such as the pleckstrin homology domain, the FYVE domain, the epsin N-terminal homologue domain, and the phox homology (PX) domain (3).
The PX domain is found in yeast and mammalian proteins and contains three N-terminal β-strands, followed by three α-helices (3). In addition to their capability to interact with PIs, previous research showed that some PX domains are also involved in protein-protein interaction (5), but the mechanism for PX domain interaction with other proteins remains unclear.
Sorting nexins (SNXs), which are proteins that contain a PX domain (3, 4, 6), are involved in several protein trafficking processes. To this day, 33 SNXs have been identified. In addition to the PX domain, most SNXs contain other recognizable domains and have been named SNXPX-BAR or SNXPX-other. A small number of SNXs contain only the PX domain and have been named SNXPX, including SNX3, SNX10, SNX11, SNX12, SNX22, and SNX24 (3).
Among the SNX family proteins, the roles of SNXPX-BAR proteins (including SNX1, SNX2, SNX5, and SNX6) in protein trafficking are best characterized (7–13). Recently, several SNXPX proteins have been investigated. SNX3 was reported to be part of a novel retromer required for the recycling of Wntless from the membrane to the trans-Golgi network (14, 15). SNX10, which has been reported to be involved in the homeostasis regulation of endosomes and required for primary cilial formation (16, 17), is another SNXPX protein. Recently, SNX10 has been reported to regulate osteoclast formation and resorption activity (18) and has been related to osteopetrosis disease (19). However, the structural basis for these SNXPX activities remains largely unknown.
SNX11 is a member of the SNXPX family and shares the highest sequence homology with SNX10 among the SNXPX proteins. This similarity in sequence may indicate that their functions are related. In a related study, we reported that overexpression of SNX11 inhibited SNX10-induced vacuolation.3 Here, we report the crystal structure of SNX11 in two forms. Surprisingly, we found that SNX11 contains a novel PX domain with two additional α-helices next to the conventional PX domain. We further demonstrate that these two additional α-helices are critical for SNX11 to inhibit SNX10-induced vacuolation in vitro. We named this functional extended PX domain the “PXe domain.” Sequence alignment indicated that SNX10 may also contain this novel PXe domain.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Human SNX11 (hSNX11) was cloned into pCR3.1-GFP-FLAG vectors for expression in mammalian cells. Two hSNX11 fragments (hSNX11-142C with residues 7–142 and hSNX11-170C with residues 7–170) were constructed for the structural study. These two cDNA fragments were inserted into pET-21a and verified by sequencing. Proteins were expressed in Rosetta 2(DE3) cells grown in LB medium. hSNX11-142C was expressed at 16 °C for 22 h after induction with 0.3 mm isopropyl β-d-thiogalactopyranoside at A600 = 0.6–0.8, whereas hSNX11-170C was expressed at 25 °C for 14 h after induction with 0.3 mm isopropyl β-d-thiogalactopyranoside at A600 = 0.6–0.8. Bacteria were collected by centrifugation, washed, and resuspended in lysis buffer (20 mm Tris-HCl (pH 8.0), 300 mm NaCl, and 7 mm β-mercaptoethanol) at 4 °C. Cells were lysed by three cycles of sonication. hSNX11-142C was purified using a Ni2+ affinity column (GE Healthcare) following standard protocols. Eluted protein was further purified by gel filtration using a Superdex 75 column (GE Healthcare) equilibrated against buffer containing 20 mm MES (pH 6.5), 100 mm NaCl, and 5 mm DTT. hSNX11-170C was purified similarly, except it was subjected to reductive methylation (20) after the Ni2+ affinity column purification.
Crystallization, Data Collection, and Structure Determination
hSNX11-142C (6.5 mg/ml) and methylated hSNX11-170C (5.5 mg/ml) were crystallized at 293 K by the sitting-drop vapor diffusion method using the sparse matrix crystallization kit from Hampton Research. Crystals of hSNX11-142C were grown from 0.2 m ammonium sulfate, 0.1 m sodium cacodylate trihydrate (pH 6.5), and 30% PEG 8000. Crystals of methylated hSNX11-170C appeared in 0.1 m HEPES (pH 7.5) and 20% PEG 8000. Prior to data collection, crystals were cryo-cooled using the mother liquor with 30% PEG 400 added as the cryoprotectant. Diffraction data were collected at beamline BL17U1 of the Shanghai Synchrotron Radiation Facility at 100 K with wavelength of 0.9792 Å. Data sets were processed using the program Mosflm (21). The structure of hSNX11-142C was solved by molecular replacement using Grd19p (Protein Data Bank code 1OCS) as the search model. The initial phase was improved with DM (22). ARP/wARP (23) was used to complete the model building. The crystal structure of methylated hSNX11-170C was solved by molecular replacement using the hSNX11-142C model. Buccaneer (24) was used for model building for the hSNX11-170C structure. Structure refinement was conducted using REFMAC5 (25). All main chain torsion angles fall in the most favored region of the Ramachandran plot. Data collection and refinement statistics are shown in Table 1.
TABLE 1.
Data collection and refinement statistics
| hSNX11-142C | hSNX11-170C | |
|---|---|---|
| Data collection | ||
| Space group | P21 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 53.48, 38.74, 73.46 | 40.12, 62.40, 66.06 |
| α, β, γ | 90.00°, 101.13°, 90.00° | 90.00°, 90.00°, 90.00° |
| Resolution (Å) | 34.12-1.78 (1.87-1.78)a | 34.29-1.60 (1.69-1.60) |
| Total observations | 145,434 (21,005) | 94,444 (13,827) |
| Unique reflections | 27,836 (3973) | 22,474 (3254) |
| Rsym or Rmergeb | 0.064 (0.132) | 0.123 (0.521) |
| I/σ | 17.6 (9.8) | 7.2 (2.8) |
| Completeness (%) | 97.4 (96.3) | 99.7 (99.8) |
| Redundancy | 5.2 (5.3) | 4.2 (4.2) |
| Wilson B-factor | 16.8 | 20.5 |
| Refinement | ||
| Resolution (Å) | 34.12-1.78 | 34.29-1.60 |
| No. of reflections | 26,427 | 21,180 |
| Rworkc/Rfreed (%) | 18.67/22.13 | 18.60/23.74 |
| No. of atoms | ||
| Protein | 2146 | 1266 |
| Ligand/ion | 78 | 7 |
| Water | 247 | 158 |
| B-factors | ||
| Protein | 17.83 | 22.88 |
| Ligand/ion | 37.61 | 48.26 |
| Water | 27.99 | 35.28 |
| r.m.s.d.e | ||
| Bond lengths (Å) | 0.013 | 0.015 |
| Bond angles | 1.73° | 1.75° |
| Ramachandran plotf | ||
| Most favored (%) | 100 | 100 |
a The values in parentheses refer to statistics in the highest bin.
b Rmerge = ΣhklΣi|Ii(hkl) − 〈I(hkl)〉|/ΣhklΣiIi(hkl), where Ii(hkl) is the intensity of an observation and 〈I(hkl)〉 is the mean value for its unique reflection. Summations are over all reflections.
c Rwork = Σh|Fo(h) − Fc(h)|/ΣhFo(h), where Fo and Fc are the observed and calculated structure factor amplitudes, respectively.
d Rfree was calculated with 5% of the data excluded from the refinement.
e Root mean square deviation from ideal values.
f Categories were defined by MolProbity.
Protein-Lipid Overlay Assay
Protein-lipid overlay assays were performed as described previously (26). Nitrocellulose membranes spotted with 100 pmol of the indicated lipids were obtained from Echelon Research. Membranes were blocked with 5% BSA in 10 mm Tris-HCl (pH 8.0), 150 mm NaCl, and 0.1% (v/v) Tween 20 for 1 h at room temperature. Membranes were incubated for 1 h at room temperature with 1 μg/ml His tag fusion protein in the blocking solution. The membranes were then washed six times for 5 min each with Tris-buffered saline. To detect bound protein, the membranes were incubated with mouse anti-His polyclonal antibody for 1 h, washed six times for 5 min each with Tris-buffered saline, incubated with horseradish peroxidase-conjugated rabbit anti-mouse antibody, washed again, and finally visualized using chemiluminescence substrate.
Cell Culture and Transfection
MCF-7 and HeLa cells were cultured in RPMI 1640 medium and 10% FBS at 37 °C with 5% CO2. Plasmids were transfected with Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's protocol. Vacuoles induced by SNX10 were analyzed 24 h after transfection as described (17).
Western Blotting
Western blotting was performed as described (16). Briefly, cells were lysed 50 mm Tris-HCl (pH 8.0), 200 mm NaCl, 2 mm EDTA, 1% Nonidet P-40, and protease inhibitor mixture (Roche Applied Science) and clarified by centrifugation at 14,000 rpm for 5 min. The supernatants were then mixed with SDS loading buffer and separated by SDS-PAGE. Western blotting was performed following the standard protocol, and mouse anti-HA (1:1000; Beyotime) and mouse anti-FLAG (1:5000; Sigma) antibodies were used.
Immunoblotting and Immunofluorescence Staining
Immunoblotting and immunofluorescence staining were performed as described (16). The antibodies used included mouse anti-HA (1:100) and Cy5-conjugated goat anti-mouse IgG (1:400; Beyotime). Confocal images were acquired using a Leica TCS SP2 spectral confocal system.
RESULTS
Crystal Structure of hSNX11
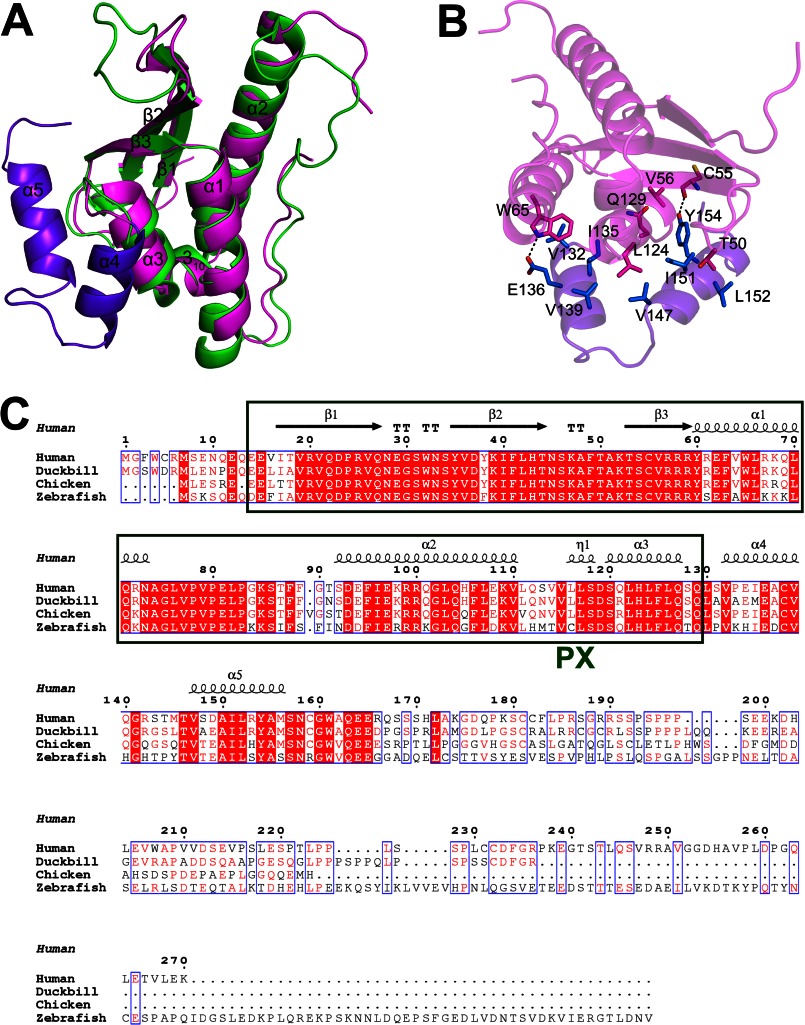
Two constructs of hSNX11 were generated for the structural study, one with residues 7–142 (hSNX11-142C) and the other with residues 7–170 (hSNX11-170C). hSNX11-142C crystallized in space group P21 and contains two molecules in the asymmetric unit. hSNX11-170C crystallized in space group P212121 and contains one molecule in the asymmetric unit. The crystal structures of hSNX11-142C and hSNX11-170C were determined at resolutions of 1.78 and 1.60 Å, respectively. Both structures contain a conventional PX domain fold composed of a β-sheet with three antiparallel β-strands, a helical subdomain consisting of three α-helices, and a polyproline II (PPII) loop. In addition to the PX domain, the hSNX11-142C structure contains an additional α-helix at the C terminus from residues 132 to 140 (α4) (Fig. 1A), and the hSNX11-170C structure contains two α-helices at the C terminus from residues 132 to 140 (α4) and from residues 147 to 156 (α5) (Fig. 1B). In the hSNX11-170C structure, no electron density was observed after Gly-160, indicating that the last 10 residues (positions 161–170) are very flexible.
FIGURE 1.
Overall structures of hSNX11-142C and hSNX11-170C. A, overall structure of hSNX11-142C. B, overall structure of hSNX11-170C. The conventional PX region is shown in magenta, and the additional C-terminal helices are purple. C, superposition of hSNX11-142C molecule A (magenta), hSNX11-142C molecule B (cyan), and hSNX11-170C (purple).
The two molecules in the asymmetric unit of hSNX11-142C can be superimposed with a root mean square deviation of 1.03 Å. hSNX11-170C and molecule A from hSNX11-142C can be superimposed with a root mean square deviation of 0.99 Å. Of the three molecules from these two structures, significant differences are observed in the β2/β3, α1/PPII, and PPII/α2 loop and the 310 and α4 helices (Fig. 1C). A notable difference between these two forms of hSNX11 is that the hSNX11-142C structure contains sulfate ions, whereas the hSNX11-170C structure does not. This is due to the existence of ammonium sulfate in the crystallization buffer used for hSNX11-142C but not for hSNX11-170C.
Interestingly, hSNX11-142C forms a dimer through crystallographic symmetry operation (supplemental Fig. 1). The interactions involved in the dimer formation consist mainly of hydrophobic contacts. Key residues that form hydrophobic contacts include Trp-32, Trp-65, and Val-132. However, gel filtration of the purified hSNX11-142C showed that it eluted as a monomer. This indicates that the dimerization of hSNX11-142C was probably induced by the crystallization conditions, especially the addition of sulfate to the crystallization buffer.
PI-binding Pocket and Lipid Interaction Specificity of SNX11
Both molecules A and B of hSNX11-142C in the asymmetric unit contain bound sulfate ions. Analogous to the PI 3-phosphate (PI3P)-binding pocket of p40phox-PX (27), one sulfate ion lies in a positively charged pocket formed by the N terminus of β2, the β3/α1 loop, the PPII/α2 loop, and the N terminus of α2 (supplemental Fig. 2A). The sulfate forms hydrogen bonds with the guanidino group of Arg-59 in the β3/α1 loop and the amide group of Arg-61 and interacts with Lys-85 and Arg-99 through water molecules. When superimposed with the PI3P-binding pocket in the p40phox-PX structure, the bound sulfate is orientated exactly at the 3-phosphate group of the PI3P in p40phox-PX. The four residues involved in the sulfate binding in SNX11 match closely Arg-58, Arg-60, Lys-92, and Arg-105 in the p40phox-PX structure (Fig. 2A). In addition, the side chains of Tyr-60 from hSNX11-142C and Tyr-59 from p40phox-PX assume the same conformation, such that a stacking interaction with the inositol ring of PI3P is formed in the p40phox-PX structure (27).
FIGURE 2.
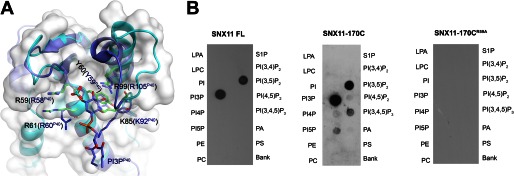
SNX11 interaction with PI3P and PI(3,5)P2. A, superposition of critical residues in the PI-binding pocket of hSNX11-142C and p40phox-PX. hSNX11-142C is shown as a white surface and cyan schematic. The side chains of residues that are critical for PI binding are shown for hSNX11-142C (green) and p40phox-PX (blue). The sulfate in the PI-binding pocket of hSNX11-142C (show as a stick model: orange for sulfur and red for oxygen) coincides with the location of the 3-phosphate of PI3P from p40phox-PX. B, the ability of His-tagged full-length (FL) SNX11, hSNX11-170C, and hSNX11-170C(R59A) to interact with various PIs was determined by lipid overlay assay. LPA, lysophosphatidic acid; LPC, lysophosphatidylcholine; PE, phosphatidylethanolamine; PC, phosphatidylcholine; S1P, sphingosine 1-phosphate; PI(3,4,5)P3, PI 3,4,5-trisphosphate; PA, phosphatidic acid; PS, phosphatidylserine.
Unlike in the hSNX11-142C structure, in the hSNX11-170C structure, there is no bound sulfate ion in the PI-binding pocket, and the electron density for Lys-85 and Ser-86 in the PPII/α2 loop is unclear. Phe-88 and Phe-89 also adopt different conformations between the two structures: in hSNX11-142C, the side chain of Phe-88 is exposed to the solvent, whereas in hSNX11-170C, Phe-89 is exposed to the solvent, and Phe-88 is facing the α2 helix (supplemental Fig. 2B). The structural variation in this region infers that the PPII/α2 loop is flexible when the PI-binding pocket is not occupied. However, sulfate ions or membrane binding may stabilize the conformation of the PPII/α2 loop.
It has been reported that p40phox-PX specifically interacts with PI3P (27). Overall, the PI-binding pocket in SNX11 is superimposable with that in p40phox-PX. However, this pocket in SNX11 has a wider opening between the PPII/α2 loop and the N terminus of β2 (Fig. 2A). The maximal movement of backbone atoms in the PPII/α2 loop between the two structures is ∼9Å. This indicates that SNX11 may interact with PIs containing a highly phosphorylated headgroup. We next test the interactions between SNX11 and various PIs using the lipid overlay assay. As shown in Fig. 2B, both hSNX11-170C and full-length SNX11 interacted with PI3P and PI 3,5-bisphosphate (PI(3,5)P2), with slightly weaker binding to PI(3,5)P2. No significant binding was observed for other lipids. Meanwhile, the hSNX11-170C(R59A) mutant could not interact with any lipids, suggesting that the conserved Arg-59 in the PI-binding pocket of SNX11 is essential for lipid interaction. In a related report, we also showed that SNX11 localized at both early and late endosomes.3 This corroborates well with our lipid overlay results showing that SNX11 can interact with PI3P and PI(3,5)P2.
Structure of hSNX11-170C Represents a Novel PX Domain
Currently, 17 different PX domain structures have been reported. Their structures are very similar, with an N terminus of three β-strands, followed by three α-helices, although their sequences are not well conserved. Six SNXs (SNX3, SNX10, SNX11, SNX12, SNX22, and SNX24) have been proposed to contain only the PX domain (5). Of these proteins, the structures of Grd19p (yeast homologue of human SNX3) (28), SNX12 (Protein Data Bank code 2CSK),4 and SNX22 (29) have been reported, and they contain only a conventional PX domain without any additional structure motif.
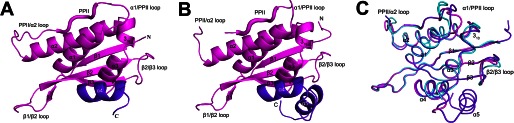
Structure superposition showed that the crystal structure of hSNX11-170C superposes well with Grd19p when excluding the two α-helices (positions 132–156) at the C terminus of hSNX11-170C (Fig. 3A and supplemental Fig. 3). As shown in Fig. 3B, the α4 and α5 helices in hSNX11-170C interact with the preceding conventional PX domain through hydrophilic and hydrophobic interactions. Key residues that form hydrogen bonds with the conventional PX domain of SNX11 include Glu-136 (from α4) and Tyr-154 (from α5). Key residues involved in hydrophobic interactions include Gln-129 (from the α3/α4 loop); Val-132, Ile-135, and V-139 (from α4); and Val-147, Ile-151, and Leu-152 (from α5).
FIGURE 3.
Novel PXe domain of SNX11. A, superposition of hSNX11-170C with Grd19p (Protein Data Bank code 1OCS). Grd19p is colored green. SNX11 is colored as in Fig. 1A. B, interactions between the conventional PX domain (magenta) and the two C-terminal α-helices (purple). Key residues involved in the interaction are shown as sticks. C, multiple sequence alignment of SNX11 from different species. The conventional PX domain is boxed with green lines.
Multiple sequence alignment showed that the conventional PX domain, α4, and α5 are highly conserved among SNX11 sequences from human, chicken, zebrafish, and duckbill, whereas the sequence downstream of these two α-helices is not conserved (Fig. 3C). The structure and sequence analysis suggested that the conventional PX region, α4, and α5 in SNX11 represent a novel PX domain, hereby named the PXe domain.
The Intact PXe Domain Is Critical for SNX11 Function
In a related report, we showed that SNX11 inhibited the enlargement of the late endosome induced by the overexpression of SNX10.3 To investigate whether the α4 and α5 helices are essential for the function of SNX11, we made serial deletions in SNX11 (Fig. 4A) and determined the activity of these deletion mutants of SNX11. They were constructed in the pCR3.1 vector with a GFP-FLAG tag at the C terminus. When cotransfected into cells with HA-tagged SNX10, SNX11(1–156) showed full activity in inhibiting SNX10-induced vacuole formation. This suggests that the C-terminal region after Met-156 is not required for SNX11 to inhibit SNX10-induced vacuolation. SNX11(1–140) was not able to inhibit SNX10-induced vacuolation (Fig. 4, B and C). However, this was not due to any change in the expression level of SNX11(1–140), as it was not significantly reduced (Fig. 4D). Therefore, the α5 helix is required for the intact function of SNX11 in inhibiting SNX10-induced vacuolation.
FIGURE 4.
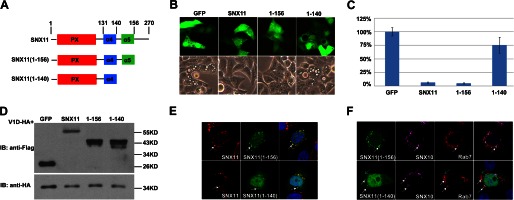
The α5 helix is required for SNX11 activity. A, SNX11 deletion constructs used in this study. B, full-length SNX11 or SNX11(1–156), but not SNX11(1–140), inhibited SNX10-induced vacuolation. p < 0.001 for SNX11 and SNX11(1–156). C, statistical analysis of B. D, Western blot analysis of the protein expression levels of wild-type and mutant SNX11 proteins. IB, immunoblot. V1D (D subunit of V-ATPase) was used as the expression control. E, co-localization of various GFP-tagged SNX11 mutants with mCherry-tagged full-length SNX11. They localized to the same vesicles. White arrows point to SNX11- and SNX11(1–156)/SNX11(1–140)-containing endosomes. F, both SNX11(1–156) and SNX11(1–140) co-localized with SNX10 to Rab7-positive vesicles. White arrows point to SNX11(1–156)/SNX11(1–140)-, SNX10-, and Rab7-containing endosomes. Cyan arrows point to large vesicles caused by SNX10 overexpression.
As shown in Fig. 3B, the α5 helix exhibits strong interaction with the conventional PX region. The conventional PX domain of hSNX11-170C superposed well with that of hSNX11-142C (Fig. 1C), suggesting that the structural stability of the conventional PX domain does not rely on interaction with the α5 helix. We then investigated whether the α5 helix is required for the subcellular distribution of SNX11. As shown in Fig. 4E, the subcellular distribution of SNX11(1–156) was almost identical to that of full-length SNX11. SNX11(1–140) had a more diffused distribution, and its enhanced staining was detected at the nucleus. SNX11(1–140)-positive vesicles were detected around the nucleus, and they co-localized well with full-length SNX11 (Fig. 4E). In a separate report, we showed that SNX10 and SNX11 co-localized at small vesicles with Rab7.3 Here, we observed that SNX11(1–140) or SNX11(1–156) also co-localized with SNX10 in the Rab7-positive vesicles (Fig. 4F). Thus, it is not likely that the loss of inhibitory activity of SNX11(1–140) in SNX10-induced vacuolation was caused by the misdistribution or the instability of the protein.
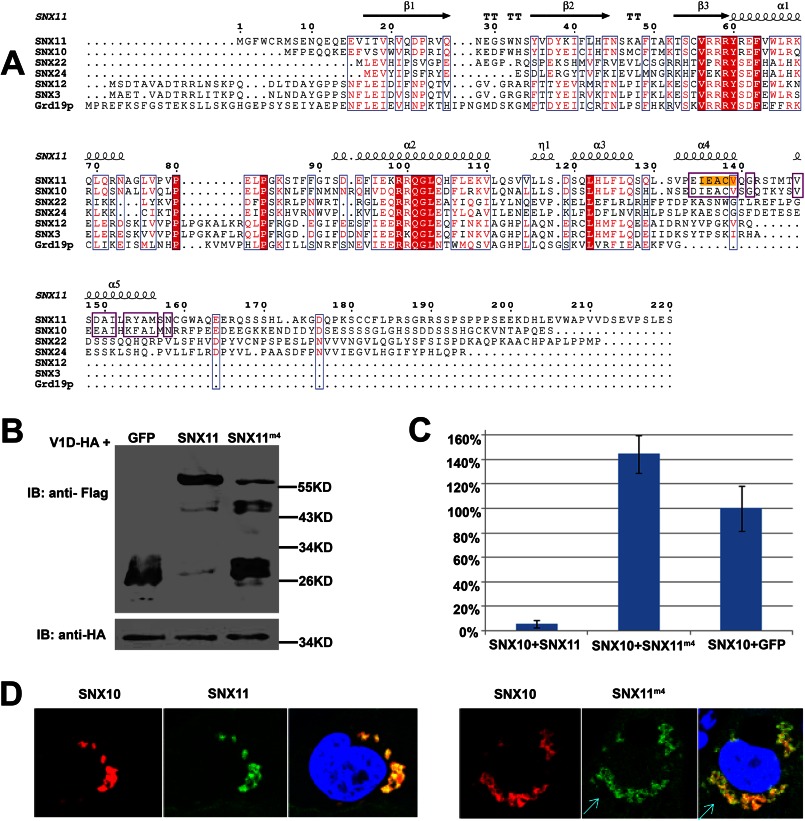
To further verify that the α4 helix is important for the function of SNX11, we generated a mutation on the α4 helix of full-length SNX11, termed SNX11m4 (135IEACV139 to AAAAA) (Fig. 5A). As shown in Fig. 5 (B and C), SNX11m4 was not able to inhibit SNX10-induced vacuolation. As Ile-135, Glu-136, and Val-139 are involved in the interaction with the conversional PX domain of SNX11 (Fig. 3B), it was necessary to determine whether or not SNX11m4 had any change in protein stability and subcellular distribution. As shown in Fig. 5D, SNX11m4 also co-localized well with SNX10. This confirmed that the loss of activity in SNX11m4 was not due to a failure in correct subcellular localization.
FIGURE 5.
The α4 helix is required for SNX11 activity. A, multiple sequence alignment of SNXPX proteins. The secondary structure elements and residue positions of hSNX11-170C are shown on top. The regions containing the highly conserved residues downstream of the PX domain between SNX10 and SNX11 are boxed with purple lines. Residues highlighted in yellow were mutated to Ala for full-length SNX11, and the SNX11 mutant was named SNX11m4. B, expression levels of SNX11m4 constructs as determined by Western blotting. IB, immunoblot. V1D (D subunit of V-ATPase) was used as the expression control. C, SNX11m4 could not inhibit SNX10-induced vacuolation. D, SNX11m4 and wild-type SNX11 co-localized well with SNX10. Cyan arrows point to large vesicles caused by SNX10 overexpression.
Taken together, these results demonstrate that the additional two α-helices are essential for SNX11 to inhibit the vacuolation induced by SNX10. The α5 and α4 helices are required for the unique function of the PXe domain.
SNX10 May Also Contain the Functional PXe Domain
Of the SNXPX proteins, SNX3, SNX10, and SNX12 share high sequence homology with SNX11 (4). We next investigated, by sequence analysis, if this novel PXe domain exists in other SNXPX proteins. Sequence alignment showed that SNX3, SNX10, SNX11, and SNX12 share high homology in the conventional PX domain. However, of the SNXPX proteins, only the downstream region (residues 123–153) of the putative SNX10 PX domain shares high homology with SNX11 in the corresponding region (α4 and α5) (Fig. 5A). This strongly indicates that SNX10 is the only other SNXPX protein that may also contain the same novel PXe domain with the additional two α-helices downstream of the conventional PX domain.
Our previous results showed that SNX10(1–153) is the minimal functional domain required for the vacuolation activity exhibited in full-length SNX10, whereas SNX10(1–122) and SNX10(1–143) are inactive (30). This corroborates with our current hypothesis that the PXe domain is a functional domain in SNX10 and is essential for the activity of SNX10.
DISCUSSION
The PX domain (containing three β-strands, followed by three α-helices) is well characterized as a PI-binding motif. It mediates the targeting of PX domain-containing proteinss to the membrane. SNXs are proteins that contain the PX domain and are involved in protein trafficking. Most SNXs contain other domains in addition to the PX domain, whereas some SNXs contain only the PX domain (SNXPX, including SNX3, SNX10, SNX11, SNX12, SNX22, and SNX24). Besides membrane binding, the PX domain in SNXPX may play additional functional roles. Of the SNXPX proteins, SNX11 and SNX10 share the highest homology and are proposed to regulate endosome morphology,3 but the structural basis for their function is largely unknown.
To better understand the PX domain in SNXPX proteins and to investigate the structure-function relationship of the two SNXPX proteins SNX10 and SNX11, we carried out a structural study of SNX11. We successfully obtained the structures of two forms of hSNX11 (hSNX11-142C and hSNX11-170C). The hSNX11-170C structure reveals a novel PXe domain with two additional α-helices downstream of the conventional PX domain. We systematically analyzed this motif in all SNXPX family proteins and found that it is present in SNX10 and SNX11, but not in any other SNXPX family members.
In this study, we further verified that this intact PXe domain is critical for the activity of SNX11. It is interesting to note that SNX10 also requires this PXe domain to be active, whereas partial removal of this motif reduces the vacuolation activity (30). These results suggest that the PXe domain is both structurally and functionally significant.
With SNX10 and SNX11 sharing this unique PXe domain, it is intriguing how SNX11 acts as an antagonist of the SNX10 function in endosome vacuolation. We speculate that they share a common partner using the PXe domain, and through competitive binding to the partner, SNX11 will then inhibit the function of SNX10. Further study to determine the protein partner and the complex structure is needed for clarification.
Acknowledgments
We thank the staff members at beamline BL17U1 of the Shanghai Synchrotron Radiation Facility of China for support in the diffraction data collection.
The work was supported in part by National Basic Research Program of China 973 Program Grants 2012CB917200 and 2009CB941102, National Natural Science Foundation of China Grants 30870496 and 30871404, National Science Fund for Distinguished Young Scholars Grant 30725012, and the One Hundred Person Project of the Chinese Academy of Sciences (to X. S. and J. L.).

This article contains supplemental Figs. 1–3.
The atomic coordinates and structure factors (codes 4IKB and 4IKD) have been deposited in the Protein Data Bank (http://wwpdb.org/).
B. Wu, B. Qin, Y. Chen, Y. Ye, X. Shu, and D. Pei, submitted for publication.
T. Suetake, F. Hayashi, and S. Yokoyama, unpublished data.
- PI
- phosphatidylinositol
- PI3P
- PI 3-phosphate
- PI(3,5)P2
- PI 3,5-bisphosphate
- PX
- phox homology
- PXe
- extended PX
- SNX
- sorting nexin
- hSNX11
- human SNX11
- PPII
- polyproline II.
REFERENCES
- 1. Mellman I. (1996) Endocytosis and molecular sorting. Annu. Rev. Cell Dev. Biol. 12, 575–625 [DOI] [PubMed] [Google Scholar]
- 2. Simonsen A., Wurmser A. E., Emr S. D., Stenmark H. (2001) The role of phosphoinositides in membrane transport. Curr. Opin. Cell Biol. 13, 485–492 [DOI] [PubMed] [Google Scholar]
- 3. Seet L. F., Hong W. (2006) The Phox (PX) domain proteins and membrane traffic. Biochim. Biophys. Acta 1761, 878–896 [DOI] [PubMed] [Google Scholar]
- 4. Cullen P. J. (2008) Endosomal sorting and signalling: an emerging role for sorting nexins. Nat. Rev. Mol. Cell Biol. 9, 574–582 [DOI] [PubMed] [Google Scholar]
- 5. Teasdale R. D., Collins B. M. (2012) Insights into the PX (phox-homology) domain and SNX (sorting nexin) protein families: structures, functions and roles in disease. Biochem. J. 441, 39–59 [DOI] [PubMed] [Google Scholar]
- 6. Carlton J., Bujny M., Rutherford A., Cullen P. (2005) Sorting nexins–unifying trends and new perspectives. Traffic 6, 75–82 [DOI] [PubMed] [Google Scholar]
- 7. Kurten R. C., Cadena D. L., Gill G. N. (1996) Enhanced degradation of EGF receptors by a sorting nexin, SNX1. Science 272, 1008–1010 [DOI] [PubMed] [Google Scholar]
- 8. Cozier G. E., Carlton J., McGregor A. H., Gleeson P. A., Teasdale R. D., Mellor H., Cullen P. J. (2002) The phox homology (PX) domain-dependent, 3-phosphoinositide-mediated association of sorting nexin-1 with an early sorting endosomal compartment is required for its ability to regulate epidermal growth factor receptor degradation. J. Biol. Chem. 277, 48730–48736 [DOI] [PubMed] [Google Scholar]
- 9. Rojas R., Kametaka S., Haft C. R., Bonifacino J. S. (2007) Interchangeable but essential functions of SNX1 and SNX2 in the association of retromer with endosomes and the trafficking of mannose 6-phosphate receptors. Mol. Cell. Biol. 27, 1112–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mari M., Bujny M. V., Zeuschner D., Geerts W. J., Griffith J., Petersen C. M., Cullen P. J., Klumperman J., Geuze H. J. (2008) SNX1 defines an early endosomal recycling exit for sortilin and mannose 6-phosphate receptors. Traffic 9, 380–393 [DOI] [PubMed] [Google Scholar]
- 11. Wassmer T., Attar N., Harterink M., van Weering J. R., Traer C. J., Oakley J., Goud B., Stephens D. J., Verkade P., Korswagen H. C., Cullen P. J. (2009) The retromer coat complex coordinates endosomal sorting and dynein-mediated transport, with carrier recognition by the trans-Golgi network. Dev. Cell 17, 110–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hong Z., Yang Y., Zhang C., Niu Y., Li K., Zhao X., Liu J. J. (2009) The retromer component SNX6 interacts with dynactin p150Glued and mediates endosome-to-TGN transport. Cell Res. 19, 1334–1349 [DOI] [PubMed] [Google Scholar]
- 13. van Weering J. R., Verkade P., Cullen P. J. (2012) SNX-BAR-mediated endosome tubulation is coordinated with endosome maturation. Traffic 13, 94–107 [DOI] [PubMed] [Google Scholar]
- 14. Zhang P., Wu Y., Belenkaya T. Y., Lin X. (2011) SNX3 controls Wingless/Wnt secretion through regulating retromer-dependent recycling of Wntless. Cell Res. 21, 1677–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Harterink M., Port F., Lorenowicz M. J., McGough I. J., Silhankova M., Betist M. C., van Weering J. R., van Heesbeen R. G., Middelkoop T. C., Basler K., Cullen P. J., Korswagen H. C. (2011) A SNX3-dependent retromer pathway mediates retrograde transport of the Wnt sorting receptor Wntless and is required for Wnt secretion. Nat. Cell Biol. 13, 914–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen Y., Wu B., Xu L., Li H., Xia J., Yin W., Li Z., Shi D., Li S., Lin S., Shu X., Pei D. (2012) A SNX10/V-ATPase pathway regulates ciliogenesis in vitro and in vivo. Cell Res. 22, 333–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Qin B., He M., Chen X., Pei D. (2006) Sorting nexin 10 induces giant vacuoles in mammalian cells. J. Biol. Chem. 281, 36891–36896 [DOI] [PubMed] [Google Scholar]
- 18. Zhu C. H., Morse L. R., Battaglino R. A. (2012) SNX10 is required for osteoclast formation and resorption activity. J. Cell. Biochem. 113, 1608–1615 [DOI] [PubMed] [Google Scholar]
- 19. Aker M., Rouvinski A., Hashavia S., Ta-Shma A., Shaag A., Zenvirt S., Israel S., Weintraub M., Taraboulos A., Bar-Shavit Z., Elpeleg O. (2012) An SNX10 mutation causes malignant osteopetrosis of infancy. J. Med. Genet. 49, 221–226 [DOI] [PubMed] [Google Scholar]
- 20. Rayment I. (1997) Reductive alkylation of lysine residues to alter crystallization properties of proteins. Methods Enzymol. 276, 171–179 [PubMed] [Google Scholar]
- 21. Battye T. G., Kontogiannis L., Johnson O., Powell H. R., Leslie A. G. (2011) iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 23. Langer G., Cohen S. X., Lamzin V. S., Perrakis A. (2008) Automated macromolecular model building for x-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cowtan K. (2006) The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 62, 1002–1011 [DOI] [PubMed] [Google Scholar]
- 25. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dowler S., Currie R. A., Campbell D. G., Deak M., Kular G., Downes C. P., Alessi D. R. (2000) Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities. Biochem. J. 351, 19–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bravo J., Karathanassis D., Pacold C. M., Pacold M. E., Ellson C. D., Anderson K. E., Butler P. J., Lavenir I., Perisic O., Hawkins P. T., Stephens L., Williams R. L. (2001) The crystal structure of the PX domain from p40phox bound to phosphatidylinositol 3-phosphate. Mol. Cell 8, 829–839 [DOI] [PubMed] [Google Scholar]
- 28. Zhou C. Z., Li de La Sierra-Gallay I., Quevillon-Cheruel S., Collinet B., Minard P., Blondeau K., Henckes G., Aufrère R., Leulliot N., Graille M., Sorel I., Savarin P., de la Torre F., Poupon A., Janin J., van Tilbeurgh H. (2003) Crystal structure of the yeast Phox homology (PX) domain protein Grd19p complexed to phosphatidylinositol-3-phosphate. J. Biol. Chem. 278, 50371–50376 [DOI] [PubMed] [Google Scholar]
- 29. Song J., Zhao K. Q., Newman C. L., Vinarov D. A., Markley J. L. (2007) Solution structure of human sorting nexin 22. Protein Sci. 16, 807–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yao D., Wu B., Qin B., Pei D. (2009) PX domain and CD domain play different roles in localization and vacuolation of Sorting Nexin 10. Chinese Sci. Bull. 54, 3965–3971 [Google Scholar]