Background: The regulation of NOTCH signaling under inflammatory conditions in the nucleus pulposus is unknown.
Results: Expression of select NOTCH pathway genes, including NOTCH2 and NOTCH signaling, is regulated by IL-1β and TNF-α.
Conclusion: Inflammatory cytokines promote NOTCH signaling in disc.
Significance: NOTCH signaling may play a role in pathogenesis of disc disease.
Keywords: Chondrocytes, Cytokine, NF-kappa B (NF-KB), Notch Pathway, Osteoarthritis, Intervertebral Disc, Nucleus Pulposus
Abstract
The objective of the study was to investigate how inflammatory cytokines, IL-1β, and TNF-α control NOTCH signaling activity in nucleus pulposus (NP) cells. An increase in expression of selective NOTCH receptors (NOTCH1 and -2), ligand (JAGGED2), and target genes (HES1, HEY1, and HEY2) was observed in NP cells following cytokine treatment. A concomitant increase in NOTCH signaling as evidenced by induction in activity of target gene HES1 and HEY1 promoters and reporter 12xCSL was seen. Moreover, treatment increased activity of a 2-kb NOTCH2 promoter. Treatment of cells with NF-κB and MAPK inhibitors abolished the inductive effect of cytokines on NOTCH2 promoter and its expression. Gain and loss-of-function studies confirmed the inductive effect of p65 on NOTCH2 promoter activity. In contrast, p50 blocked the cytokine induction of promoter activity. Supporting promoter studies, lentiviral delivery of sh-p65, and sh-IKKβ significantly decreased cytokine dependent change in NOTCH2 expression. Interestingly, MAPK signaling showed an isoform-specific control of NOTCH2 promoter; p38α/β2/δ, ERK1, and ERK2 contributed to cytokine dependent induction, whereas p38γ played no role. Analysis of human NP tissues showed that NOTCH1 and -2 and HEY2 expression correlated with each other. Moreover, expression of NOTCH2 and IL-1β as well as the number of cells immunopositive for NOTCH2 significantly increased in histologically degenerate discs compared with non-degenerate discs. Taken together, these results explain the observed dysregulated expression of NOTCH genes in degenerative disc disease. Thus, controlling IL-1β and TNF-α activities during disc disease may restore NOTCH signaling and nucleus pulposus cell function.
Introduction
Intervertebral disc (IVD)2 degeneration and related disorders have been recognized as the major cause of low back pain (1) affecting ∼80% of the population during their life time with high economic cost (2). One of the hallmarks of the disease state is increased production of degradative enzymes coupled with decreased synthesis of the extracellular matrix (3, 4). These catabolic processes are thought to be mediated by a number of cytokines, including IL-1β and TNF-α (5, 6). Although IL-1β has been shown to be directly involved in the decrease in matrix synthesis and increased matrix degradation (7), TNF-α has been linked to disc cell apoptosis, herniation, and nerve irritation (8). Our previous studies demonstrated that there was an increase in expression of both IL-1β and TNF-α mRNA in human degenerate nucleus pulposus (9, 10).
It has been postulated that with aging and degeneration, NP cells undergo phenotypic transformation and are replaced by fibroblastic cells (11) that secrete atypical fibrous extracellular matrix. This suggests that changes in cellular microenvironment and presence of catabolic mediators likely promote abnormal proliferation and differentiation. That cytokines mediate shift in nucleus pulposus cell function is supported by other studies (7, 9, 12, 13). Moreover, similar to chondrocytes, our own investigations have shown that disc cell proliferation and differentiation is dependent on the NOTCH signaling pathway (14–16). This is a highly conserved pathway (17) regulated by interactions between neighboring cells: four cell surface NOTCH receptors (in mammals, NOTCH1–4) are activated by ligands, the Delta-like (Dll1,-3, and -4), and JAGGED (JAG1 and -2) on neighboring cells. Upon activation, the intracellular domain of NOTCH is cleaved by the presenilin proteases (PSEN1/2) of the γ-secretase complex. It translocates to the nucleus to associate with recombination signal-binding protein for immunoglobulin κ J region (RBP-Jκ) (also known as CSL (CBF-1, Su(H), Lag-1)) and Mastermind-like (Maml) proteins to activate the transcription of target genes, including HES and HEY (18). Our previous work has shown that all components of the NOTCH signaling pathway, namely receptors, ligands, and target genes, are expressed in intervertebral disc cells (16). Moreover, our studies suggest that NOTCH signaling is critical for the maintenance of NP cell proliferation in the hypoxic niche of the disc (16).
The purpose of this study was to evaluate whether inflammatory cytokines IL-1β and TNF-α that are associated with disc degeneration promote the activity of NOTCH signaling proteins in NP cells. It was found that cytokine activation of NOTCH signaling was dependent on both NF-κB and MAPK pathways.
EXPERIMENTAL PROCEDURES
Reagents and Plasmids
Plasmids were provided as follows: NOTCH responsive reporters 12xCSL-Luc and HES1-Luc (19) from Dr. Urban Lendahl (Karolinska Institute, Stockholm, Sweden); NOTCH responsive 4xwtCBF1-Luc and 4xmtCBF1-Luc reporters containing four copies of the wild-type or mutant CBF1 binding elements (20) from Dr. Diane Hayward (The Johns Hopkins University); human NOTCH2 promoter N2PR −2327/−99 and N2PR −246/−99, in pGL3 (21) from Dr. Jonas Ungerbäck (Linköpings University); p38aAF, p38b2AF, p38dAF, and p38gAF from Dr. Jiahui Han (Scripps Research Institute, La Jolla, CA); and ERK-1K71R and ERK-2K52R from Dr. Melanie Cobb (University of Texas Southwestern Medical Center, Dallas, TX). Expression plasmids for sh-p65, and sh-IKKb in lentiviral FSVsi vector that co-expresses YFP was from Dr. Andree Yeremian (University of Lleida), and Hey1 reporter was from Manfred Gessler. Plasmids pCMX-IkBM (catalogue no. 12330), RelA/p65 (no. 20012), p50 (no. 20018), RelB (no. 20017) and cRel (no. 20013) developed by Dr. Inder Verma and psPAX2 (no. 12260) and pMD2G (no. 12259) developed by Dr. Didier Trono were obtained from Addgene. pRL-TK (Promega) containing the Renilla reniformis luciferase gene was used as an internal transfection control (22). Fibroblasts derived from p65 null and wild type mice were a kind gift from Dr. Denis Guttridge (University of Ohio, Columbus, OH). NOTCH1, NOTCH2, NOTCH3, p65, and IKKb antibodies were from Cell Signaling; Hey2 was from Peprotech; β-tubulin was from Developmental Studies Hybridoma Bank; and GAPDH was from Novus Biologicals. TNF-α and IL-1β were purchased from Peprotech. Real time PCR predesigned primer/probe mixes were purchased from Applied Biosystems (Warrington, UK). NOTCH2 antibody for immunohistochemical localization was purchased from Abcam (Cambridge, UK).
Isolation of Nucleus Pulposus Cells and Treatments
Rat and human nucleus pulposus cells from seven patients (Pfirrmann grades 2–5; mean age, 46.71 years (range of 36–64 years), 4 males:3 females) were isolated using a method described previously by Risbud et al. (22). Nucleus pulposus cells were maintained in DMEM and 10% FBS supplemented with antibiotics. To investigate effect of cytokines, cells were treated with IL-1β (10 ng/ml) and TNF-α (25–50 ng/ml) for 24 h.
Human Tissue Collection and Grading
Human lumbar IVD tissue was obtained either at surgery or postmortem examination with informed consent of the patient or relatives (Sheffield Research Ethics Committee no. 09/H1308/70). Six postmortem IVDs were recovered from two donors. They consisted of intact IVDs within the complete motion segment from which the IVDs were removed. Sixty-two surgical IVD tissue samples were obtained from patients undergoing microdiscectomy procedures for the treatment of low back pain and root pain as caused by prolapse of the IVD. NP tissue was divided into two, and one-half was fixed in 10% neutral buffered formalin and processed for histological and immunohistochemical examination. The remaining tissue was used for RNA isolation. H&E-stained sections were used to score the degree of morphological degeneration; sections were scored numerically between 0 and 3 for four features of degeneration: 1) the presence of cell clusters; 2) the presence and extent of fissures; 3) the loss of demarcation between NP and AF regions; and 4) the loss of hematoxophilia (indicating reduced proteoglycan content) from the NP. The scores from each category were then summed to provide histological grades between 0–12, where scores of 0 to 3 are considered as representing histologically non-degenerate tissue samples and scores ≥4 represent degenerate tissue samples (7). Gene expression study samples were classified as non-degenerate (≤3.9), midgrade degenerate (4–6.9) and moderate/severe degenerate (≥7) based on histological examination. Grading was performed independently by two researchers, and grades were averaged.
Quantitative Real-time RT-PCR
Human tissues were processed as described previously. Extracted RNA was subjected to treatment with DNase (Qiagen, Crawley, UK) and purified using Qiagen MinElute Cleanup kit prior to cDNA synthesis using MMLV reverse transcriptase (Bioline, London, UK) and random hexamers. Real-time PCR analysis was performed using predesigned, FAM-labeled Taqman® gene expression assays (Applied Biosystems). A total of 49 IVDs were used for this component of the study. Twelve histologically non-degenerate discs (mean histological grade, 2.66 (range, 1–3.5); mean age, 36.7 years (range, 20–49); 11 males:1 female). Nineteen histologically midgrade degenerate discs (mean histological grade, 5.53 (range, 4–6.9); mean age, 44.7 years (range, 25–73); 11 males:8 females). Eighteen moderate/serve degenerate discs (mean histological grade, 7.87 (range, 7–9); mean age, 37.8 years (range, 28–52); 5 males:14 females). For cultured rat and human NP cells, 1–2 μg of total DNA free-RNA was used to synthesize cDNA using SuperScript III cDNA synthesis kit (Invitrogen). Reactions were set up in triplicate in 96 well plates using cDNA and appropriate PCR Master Mix (Applied Biosystems). PCR reactions were performed in a StepOnePlus real-time PCR system (Applied Biosystems) according to the manufacturer's instructions. Hypoxanthine phosphoribosyltransferase (HPRT) and β-actin were used to normalize the mRNA expression for in vitro experiments, whereas GAPDH and 18 S were used to normalize mRNA in human samples. For human tissue analysis, an average of housekeeping gene Ct value was used to normalize data using 2−ΔCt. Gene primers used in analysis of cultured cells were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA).
Protein Extraction and Western Blotting
Cells were placed on ice immediately following treatment. They were lifted, washed with ice-cold PBS, and harvested in mammalian protein extraction reagent buffer (Pierce). All of the wash buffers and mammalian protein extraction reagent buffer included 1× protease inhibitor mixture (Roche Applied Science), NaF (5 mm), and Na3VO4 (200 μm). Total cell proteins were resolved on 8–12% SDS-polyacrylamide gels and transferred by electroblotting to PVDF membranes (Bio-Rad). The membranes were blocked with 5% nonfat dry milk in TBST (50 mm Tris, pH 7.6, 150 mm NaCl, 0.1% Tween 20) and incubated overnight at 4 °C in 3% nonfat dry milk in TBST with the antibodies against NOTCH1 (1:2000), NOTCH2 (1:5000), NOTCH3 (1:1000), HEY2 (1:1000), p65 (1:1000), IKKβ (1:1000), or β-tubulin (1:1000). Immunolabeling was detected using the ECL reagent (Amersham Biosciences).
Transfections and Dual-Luciferase Assay
Rat NP cells were transferred to 48-well plates (2 × 104 cells/well) 1 day before transfection. To measure the effect of cytokine treatments on NOTCH signaling, cells were transfected with 250 ng of reporter plasmids with 250 ng of pRL-TK plasmid. To investigate the effect of NF-κB and MAPK on NOTCH2, cells were cotransfected with 50–150 ng of p65 or p50 or p65 with p50 or dominant negative (DN)-p38α/β2/γ/δ, or DN-ERK1/-ERK2 with or without appropriate backbone vector(s) and 175 ng N2PR-W1 reporter and 175 ng pRL-TK plasmid. In some experiments, cells were treated with the inhibitors SM7368 (10 μm), SB203580 (10 μm), PD98059 (50 μm), or SP60025 (10 μm) (Calbiochem) 1 h prior to treatment with cytokines. Lipofectamine 2000 (Invitrogen) was used as a transfection reagent. For each transfection, plasmids were premixed with the transfection reagent. 24 h after transfection, some cells were treated with IL-1β or TNF-α. 48 h after transfection, the cells were harvested, and a Dual-LuciferaseTM reporter assay system (Promega) was used for sequential measurements of firefly and Renilla luciferase activities. Quantification of luciferase activities and calculation of relative ratios were carried out using a luminometer (TD-20/20, Turner Designs). At least three independent transfections were performed, and all analyses were carried out in triplicate.
Lentiviral Particle Production and Viral Transduction
HEK 293T cells were seeded in 10-cm plates (1.3 × 106 cells/plate) in DMEM with 10% heat-inactivated FBS 2 days before transfection. Cells were transfected with 2.5 μg of sh-control, or sh-p65 or sh-IKKβ plasmids along with 1.875 μg of psPAX2 and 0.625 μg of pMD2.G. After 16 h, transfection medium was removed and replaced with DMEM with 5% heat-inactivated FBS and penicillin/streptomycin. Lentiviral particles were harvested at 48 and 60 h post-transfection. Human NP cells (grade 2) were plated in DMEM with 5% heat-inactivated FBS 1 day before transduction. Cells in 10-cm plates were transduced with 5 ml of conditioned medium containing viral particles along with 6 μg/ml polybrene. After 24 h, conditioned medium was removed and replaced with DMEM with 5% heat-inactivated FBS. Cells were harvested for mRNA and protein extraction 5 days after viral transduction.
Site-directed Mutagenesis
N2PR-W1 reporter plasmid was used to mutate either the NF-κB site a (AGGATTTCCC to ATGATCTCTC) or the NF-κB site b (GGGAAACCCC to GTGATACCAC) or both. Mutants were generated using QuikChange II XL site-directed mutagenesis kit (Stratagene, Wilmington, DE), using forward and reverse primer pairs containing the desired mutation following the manufacturer's instructions. The mutations were verified by sequencing.
Immunohistochemical Analysis
Immunohistochemistry was used to confirm and localize production of NOTCH2 in 31 IVDs; 4 postmortem and 27 surgical samples. Five histologically non-degenerate discs (mean histological grade, 2.86 (range of 2–3.5); mean age, 37.2 years (range of 26–45); 4 males:1 female). Thirteen histologically midgrade degenerate discs (mean histological grade, 5.31 (range of 4–6); mean age, 48.66 years (range of 25–82); 7 males:6 females). Ten moderate/severe degenerate discs (mean histological grade, 7.86 (range of 7–9); mean age, 47.5 years (range of 35–82); 2 males:8 females). 4-μm tissue sections were dewaxed and rehydrated, and endogenous peroxidases were quenched and, following heat antigen retrieval, blocked in goat serum. Sections were incubated overnight at 4 °C with rabbit polyclonal antibodies against human NOTCH2 (1:200). Preimmune rabbit IgG (Abcam) was used as a negative control. After washing, sections were incubated with biotinylated goat anti-rabbit antiserum (1:400 dilution; Abcam) and binding detected by the formation of streptavadin-biotin complex (Vector Laboratories, Peterborough, UK) with 3,3′-diaminobenzidine tetrahydrochloride solution (Sigma-Aldrich). Sections were counterstained with Mayers Hematoxylin (Leica Microsystems, Milton Keynes, UK), dehydrated, cleared, and mounted in Pertex (Leica Microsystems). Sections were visualized images captured using an Olympus BX60 microscope and QCapture Pro software (version 8.0, MediaCybernetics, Marlow, UK). Random fields of view were assessed for immunopositive and immunonegative cells until a total of 200 NP cells were counted in each section, and the number of immunopositive cells were expressed as a percentage of the total count.
Statistical Analysis
All measurements were performed in triplicate. Results are presented as the mean ± S.E. Differences between groups were assessed by analysis of variance. p values < 0.05 were considered statistically significant. RT- PCR expression data of human tissues was found to be non-parametric in distribution, and so the Kruskal-Wallis test combined with post hoc analysis by Conover-Inman test was used to determine significance between 2−ΔCt values and percentage immunopositivity between study groups. In addition, Spearman's rank correlation was deployed to determine correlations between gene expressions for different targets.
RESULTS
IL-1β and TNF-α Regulation of NOTCH Signaling in Rat Nucleus Pulpous Cells
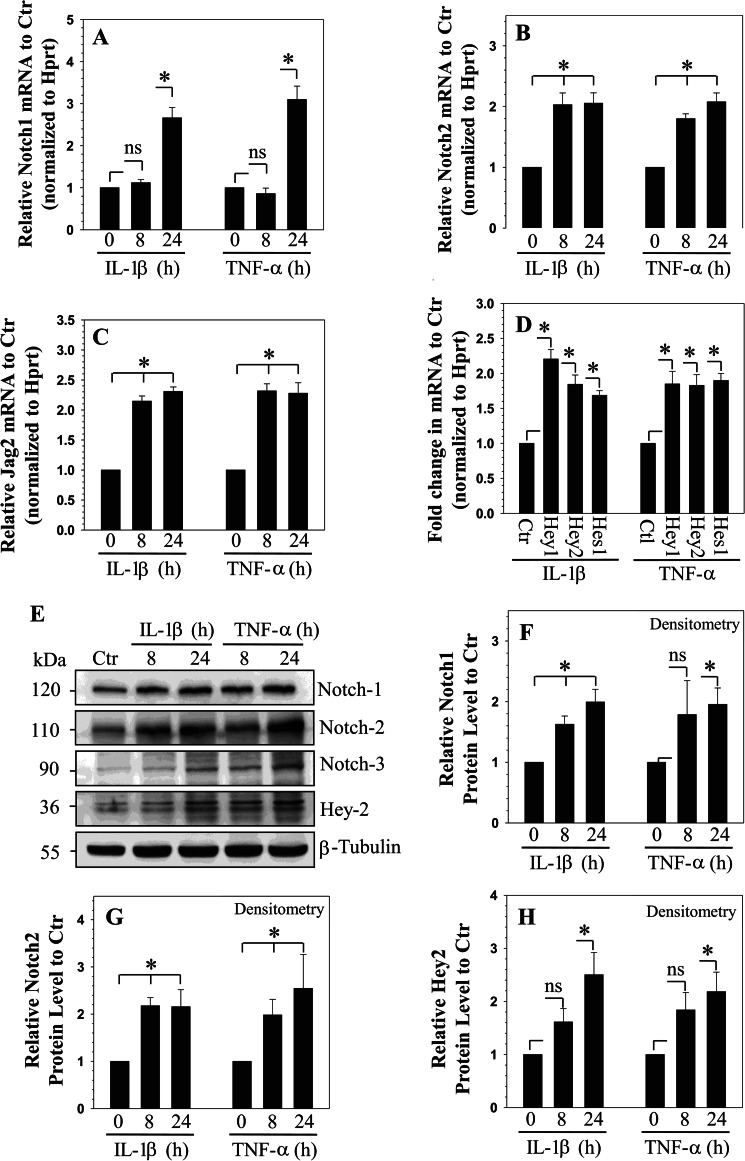
To test the premise that cytokines concerned with disc degeneration control NOTCH signaling, rat NP cells were treated with IL-1β or TNF-α, and expression of NOTCH signaling components was analyzed. Cytokine treatment induces expression of NOTCH1 (Fig. 1A) and NOTCH2 (Fig. 1B) but not NOTCH3 mRNA (data not shown). NOTCH1 mRNA expression exhibits a delayed increase compared with NOTCH2, which is induced as early as 8 h following treatment (Fig. 1B). Similarly, mRNA expression of NOTCH ligand, JAGGED2 showed an induction (Fig. 1C), whereas no change in expression is seen for JAGGED1 and Dll4 (data not shown). In contrast, expression of Dll1 is suppressed by the cytokines (data not shown). Moreover, there is increased expression of NOTCH target genes HEY1, HEY2, and HES1 following treatment with both cytokines (Fig. 1D).
FIGURE 1.
Cytokine-dependent expression of NOTCH receptors, ligands, and target genes in rat NP cells. Real-time RT-PCR analysis showed selective induction in expression of NOTCH1 (A), NOTCH2 (B), and JAGGED2 (C) by NP cells treated with IL-1β and TNF-α. ctr: control, Hprt: hypoxanthine phosphoribosyltransferase. D, expression of NOTCH target genes HEY1, HEY2, and HES1 was induced by IL-1β and TNF-α treatment of NP cells and represented as fold change relative to corresponding untreated control (considered as 1). E, Western blot analysis shows that when treated with IL-1β or TNF-α, NP cells increase levels of cleaved NOTCH1, cleaved NOTCH2, and HEY2 but not of cleaved NOTCH3. Densitometric analysis of multiple blots from experiment (E) above showed that levels of cleaved NOTCH1 (F), cleaved NOTCH2 (G), and HEY2 (H). Values shown are mean ± S.E. of three independent experiments; *, p < 0.05.
To further examine the activation of NOTCH signaling after cytokine treatment, NOTCH1, NOTCH2, NOTCH3, and HEY2 protein expression were evaluated by Western blot analysis. Fig. 1E shows that cytokine treatment increases the levels of cleaved/active NOTCH1 and NOTCH2, but not of NOTCH3 in rat NP cells. Increase in HEY2 level can also be seen (Fig. 1E). Densitometric analysis confirmed that formation of cleaved NOTCH1 (Fig. 1F) and NOTCH2 (Fig. 1G) but not that of cleaved NOTCH3 (data not shown) is induced by the cytokines. Similarly, a significant increase in HEY2 protein levels by rat NP cells is seen in presence of the cytokines (Fig. 1, E and H).
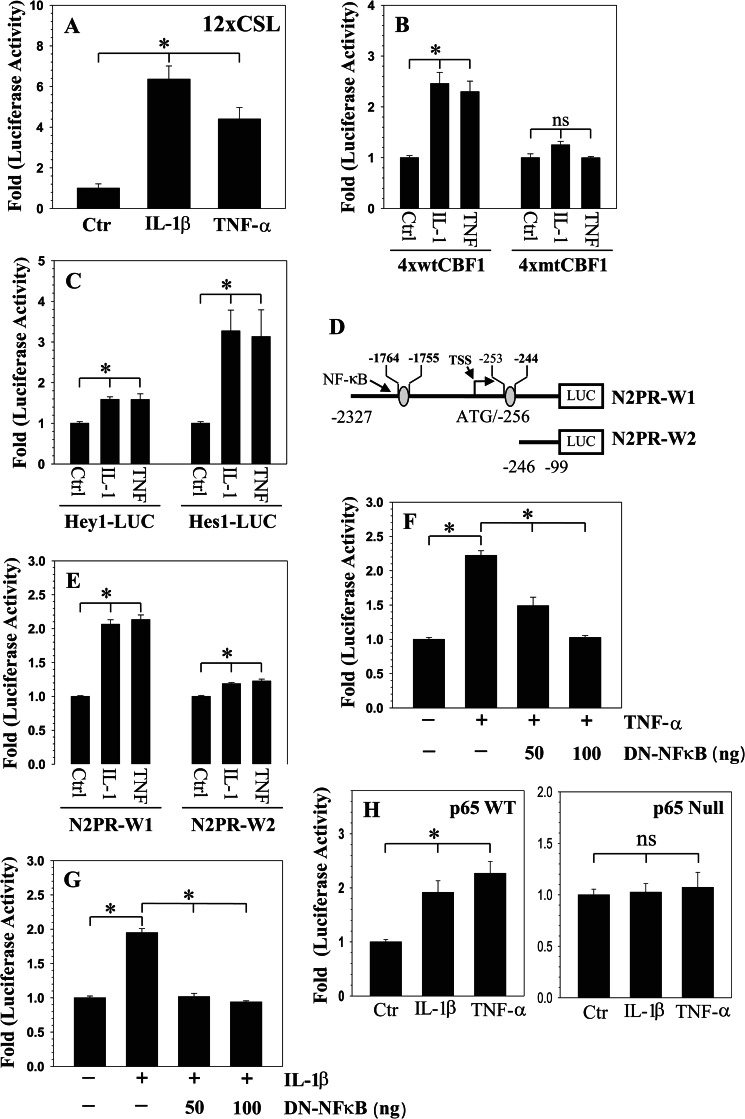
To further investigate whether cytokines enhance NOTCH signaling activity in rat NP cells, we transfected cells with NOTCH-responsive reporters 12xCSL-Luc and 4xwtCBF1-Luc and measured their activity following IL-1β and TNF-α treatment. Results of these experiments show that both of the cytokines increased the activity of 12xCSL-Luc (Fig. 2A) and 4xwtCBF1 (Fig. 2B) reporters. As expected, RBPjκ site mutation in CBF1 promoter (4xmtCBF1) made it unresponsive to the treatment (Fig. 2B). Similarly, the activity of NOTCH target gene promoters HEY1 and HES1 is enhanced by cytokine treatment (Fig. 2C). We then investigated the mechanism of cytokine action on NOTCH2 expression by measuring the activity of two promoter fragments N2PR-W1 (−2327/−99) and N2PR-W2 (−246/−99) (Fig. 2D). The activity of N2PR-W1 reporter is significantly induced by the cytokines (∼2.2-fold), whereas N2PR-W2 exhibits a much smaller increase in activity (∼1.2-fold) (Fig. 2E). Due to robust activation of the N2PR-W1 reporter, it was chosen for subsequent mechanistic studies to investigate cytokine regulation of NOTCH2 expression.
FIGURE 2.
Inflammatory cytokines promote NOTCH signaling in rat NP cells. Activity of NOTCH responsive reporters 12xCSL (A), 4xwtCBF1Luc (B), and target gene promoters HEY1 and HES1 (C) was induced by IL-1β and TNF-α. In contrast, activity of reporter 4xmtCBF1Luc that contained mutation in RBPjκ site was unaffected by cytokines. ctr: control. D, schematic of different length human NOTCH2 promoter constructs used in the study, putative NF-κB elements are shown; numbers are relative to the translational start site (ATG). E, NP cells were transfected with different length NOTCH2 promoter fragments (N2PR-W1 and N2PR-W2), and the induction in luciferase activity was measured following IL-1β and TNF-α treatment. In contrast to a small increase in N2PR-W2 activity, a significant induction in N2PR-W1 reporter activity was seen. F and G, co-transfection of DN-NF-κB completely blocked IL-1β (F) and TNF-α-dependent (G) induction in Nocth2 (N2PR-W1) promoter activity. H, p65 WT and p65 null MEFs were transfected with NOTCH2 reporter and treated with IL-1β or TNF-α. Only wild type cells evidenced an increase in NOTCH2 reporter activity. Values shown are mean ± S.E. of three independent experiments; *, p < 0.05.
NF-κB Signaling Controls NOTCH2 Promoter Activity in NP Cells
To explore the role of NF-κB in the transcriptional regulation of NOTCH2 expression, we used the JASPAR core database to identify putative NF-κB binding motifs in the NOTCH2 promoter (23). Sequence analysis reveals two putative binding sites; location in the promoter, sequence, and relative score were as follows: −253/−244 (GGGGTTTCCC, 0.94) and −1764/−1755 (AGGATTTCCC, 0.87) (Fig. 2D). To confirm that NOTCH2 promoter activity is responsive to NF-κB signaling, we first performed a loss of function study. When rat NP cells were co-transfected with DN-NF-κB (IκBαM) plasmid, cytokine-dependent induction of promoter activity is completely inhibited (Fig. 2, F and G). To determine whether p65/RelA subunit of NF-κB controlled NOTCH2 promoter activity and whether this was cell type-specific, we measured promoter activity in p65 wild type and null mouse fibroblasts. Only in the wild type cells was the promoter activity cytokine inducible (Fig. 2H).
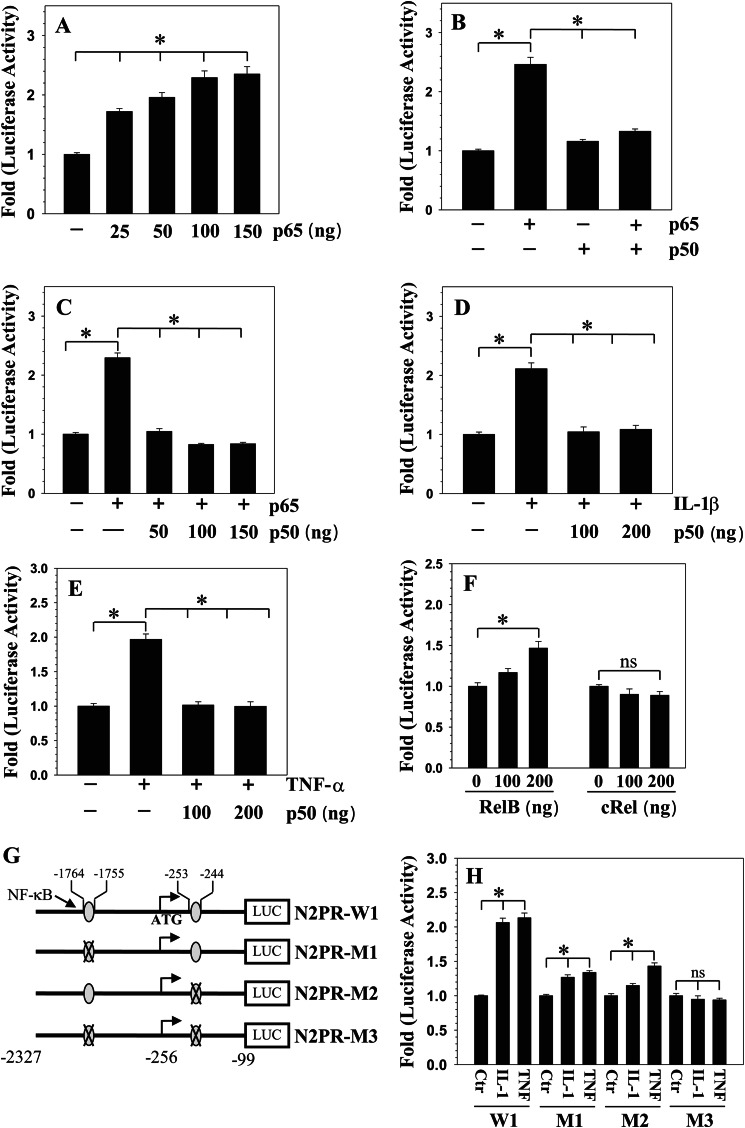
To elucidate the role of NF-κB in controlling promoter activity in rat NP cells, we measured N2PR-W1 activity following overexpression of NF-κB subunits p65/RelA or p50 or RelB or c-Rel. Co-transfection with p65 results in a dose-dependent increase in N2PR-W1 activity (Fig. 3A). Interestingly, co-transfection of 100 ng each of p50 with p65 blocked the inductive effect of p65 on NOTCH2 promoter (Fig. 3B), this suppression was evident even when p50 dose was reduced to 50 ng in presence of 100 ng of p65 (Fig. 3C). Noteworthy, p50 alone has no effect on the promoter activity. Furthermore, p50 suppresses the inductive effect of IL-1β (Fig. 3D) as well as TNF-α (Fig. 3E) on N2PR-W1 promoter activity. Co-transfection with RelB elicits a small but significant up-regulation in NOTCH2 promoter activity, only at a dose of 100 ng, whereas addition of c-Rel has no effect at any given concentrations (Fig. 3F).
FIGURE 3.
NF-κB regulation of NOTCH2 expression. A, rat NP cells were transfected with 25–150 ng of p65 plasmid and NOTCH2 promoter activity measured. There was a dose-dependent increase in NOTCH2 promoter activity. B and C, NP cells were co-transfected with p65 and/or p50, and promoter activity was measured. Transfection with p65 but not p50 resulted in increased promoter activity. Together, p50 significantly blocked p65-mediated induction in NOTCH2 promoter activity with suppression evident even at a 50-ng dose. D and E, co-transfection with p50 completely abolished both IL-1β (D) and TNF-α (E) mediated induction in NOTCH2 promoter activity. F, in contrast to p65, RelB and cRel had small or no effect on NOTCH2 promoter activity. G, schematic of constructs N2PR-M1, N2PR-M2, and N2PR-M3 containing mutation in either one or both NF-κB sites generated by site-directed mutagenesis. H, NP cells were transfected with either wild type (N2PR-W1) or NF-κB mutant reporter plasmids, and the induction of luciferase activity was determined following IL-1β and TNF-α treatment. Compared with the wild type reporter, treatment caused a much smaller induction in activity of single site mutants. Activity of double mutant reporter was unresponsive to the treatment. Values shown are mean ± S.E. of three independent experiments; *, p < 0.05.
NF-κB Interaction with the NOTCH2 Promoter Controls Its Activity in NP Cells
Next, we evaluated the role of conserved NF-κB binding sites in controlling NOTCH2 promoter activity (Fig. 3G). For this purpose, using N2PR-W1 fragment as a template, we generated three mutant reporters that contained mutation in either one (M1, M2) or both (M3) NF-κB binding sites (Fig. 3G) and examined the effect of IL-1β and TNF-α on their activity in rat NP cells. In contrast to the wild-type promoter, the two single site mutants are less responsive to cytokine treatment (Fig. 3H). Noteworthy, there is a complete lack of induction in activity of double mutant by TNF-α or IL-1β (Fig. 3H).
NF-κB Signaling Controls IL-1β- and TNF-α-dependent NOTCH2 Expression in NP Cells
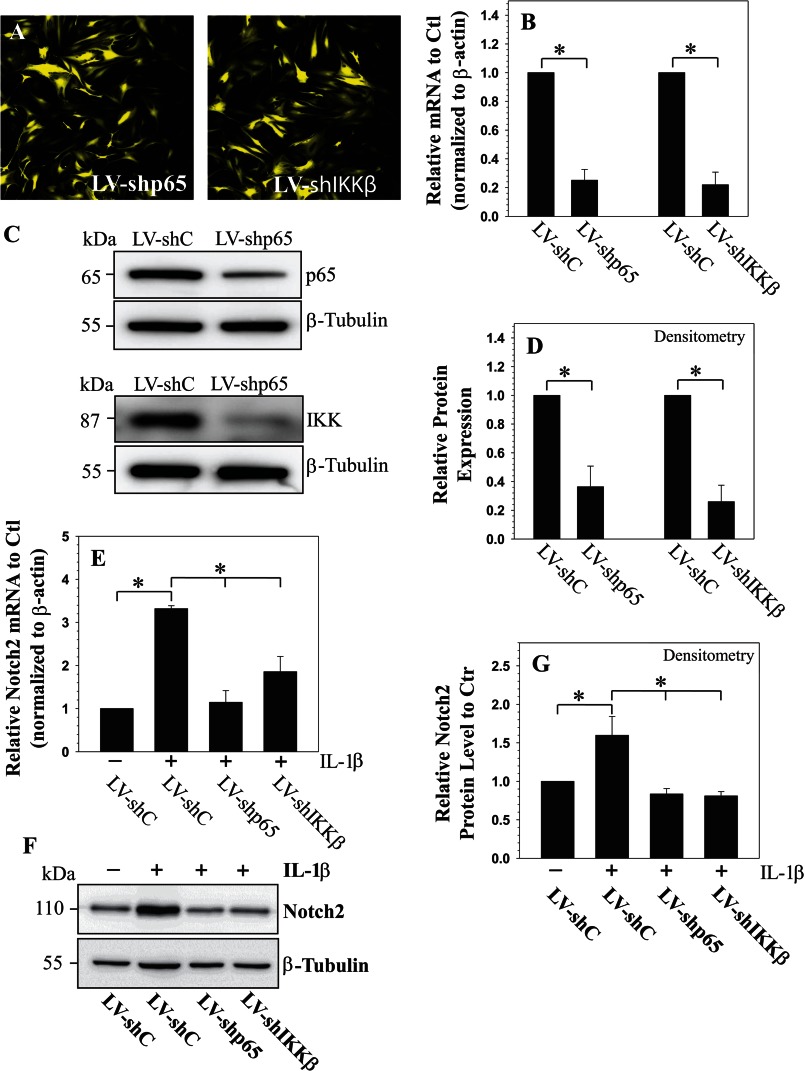
To further elucidate the role of the NF-κB signaling pathway on the regulation of NOTCH2 expression, we silenced the expression of individual NF-κB signaling components and transduced human NP cells with lentivirus co-expressing YFP and p65-shRNA or IKKβ-shRNA. Fig. 4A shows that there is a robust YFP expression in the virally transduced cells, indicating a high level of transduction efficiency and transgene expression. There is a significant decrease in the expression of p65 and IKKβ mRNA (Fig. 4B) and protein (Fig. 4, C and D) in cells transduced with sh-p65 and sh-IKKβ, respectively, when compared with cells transduced with control shRNA. Importantly, suppression of NF-κB signaling components significantly decreases the inductive effect of IL-1β on the expression of NOTCH2 mRNA (Fig. 4E) and the level of cleaved NOTCH2 protein (Fig. 4, F and G).
FIGURE 4.
A, immunofluorescence detection of YFP in human NP cells (grade 2) transduced with lentivirus co-expressing YFP and either shp65 or shIkkβ show high transduction efficiency (magnification of ×20). LV: lentiviral. Shown is real-time RT-PCR (B) and Western blot (C and D) and corresponding densitometric analysis of cells transduced with LV-shC, LV-shp65, and LV-shIkkβ. Expression of p65 and Ikkβ was significantly suppressed by the respective shRNAs compared with cells transduced with a lentivirus expressing control-shRNA. Shown is a real-time RT-PCR (E) and Western blot (F and G) and densitometric analysis of NOTCH2 expression in cells infected with LV-shC and LV-shp65 and LV-shIkkβ following IL-1β treatment. Note that the IL-1β-dependent induction in NOTCH2 mRNA (E) and protein (F and G) is significantly blocked by suppression of components of the NF-κB pathway. Values shown are mean ± S.E. of three independent experiments; *, p < 0.05.
TNF-α and IL-1β Promote NOTCH2 Expression and Signaling through MAPK and NF-κB Pathways
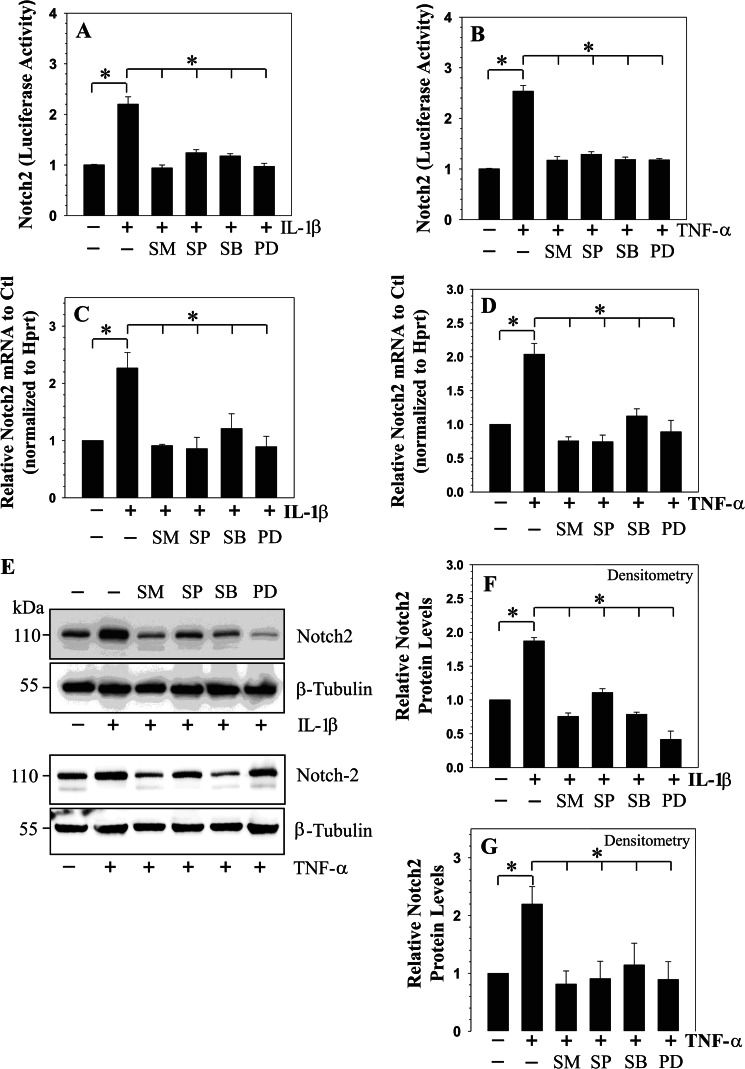
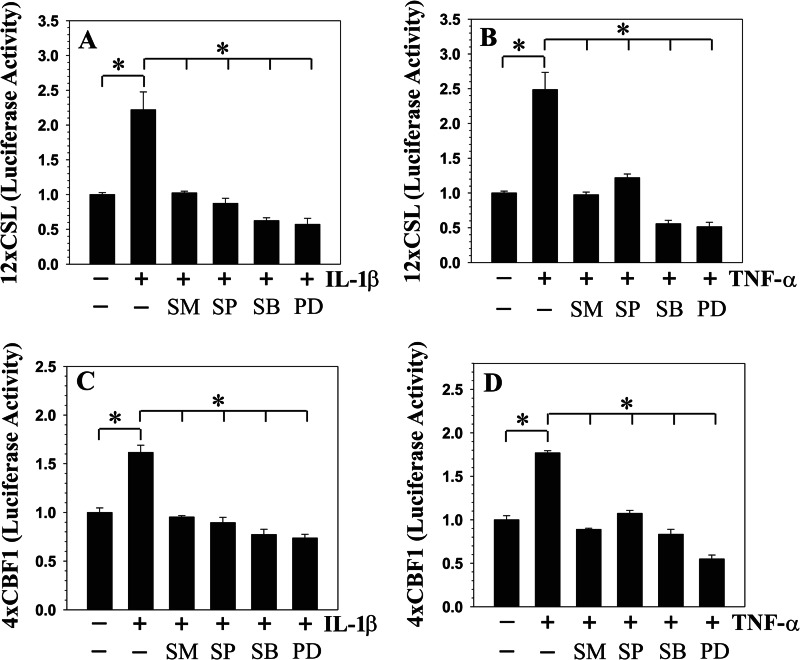
We determined whether, in addition to the NF-κB pathway, MAPK signaling controls cytokine-dependent induction of NOTCH2 expression and activity. For these studies, rat NP cells were treated with pathway specific inhibitors prior to the exposure of IL-1β or TNF-α. When induced with the cytokines, pretreatment with inhibitors results in a significant suppression in NOTCH2 promoter activity (Fig. 5, A and B) and mRNA expression (Fig. 5, C and D). Similarly, Western blot and densitometric analysis showed a pronounced suppression in cytokine-dependent increase in cleaved NOTCH2 protein in the presence of all of the pathway specific inhibitors (Fig. 5, E–G). In addition to NOTCH2, we evaluated activity of NOTCH responsive reporters: 12xCSL-Luc and 4xwtCBF1 to NF-κB and MAPK inhibitors in rat NP cells. Results show that cytokine-mediated induction in the activity of 12xCSL-Luc (Fig. 6, A and B) and 4xwtCBF1-Luc (Fig. 6, C and D) was completely blocked by NF-κB and MAPK pathway inhibitors.
FIGURE 5.
Cytokines modulate NOTCH2 expression through NF-κB/MAPK signaling in rat NP cells. A and B, activity of NOTCH2 promoter following IL-1β (A) and TNF-α (B) treatment for 24 h with or without NF-κB inhibitor (SM7368 (SM)) or JNK inhibitor (SP60025 (SP)) or p38 inhibitor (SB203580 (SB)) or ERK inhibitor (PD98059 (PD)). A significant blocking of cytokine-dependent induction in NOTCH2 promoter activity is seen by all inhibitors. C and D, real-time RT-PCR analysis of NOTCH2 expression shows significant blocking of cytokine dependent induction in NOTCH2 mRNA expression by the inhibitors. E and F, Western blot (E) and corresponding densitometric analysis shows that IL-1β-dependent (F) and TNF-α-dependent (G) increase in cleaved NOTCH2 level in NP cells was blocked by signaling inhibitors. Values shown are mean ± S.E. of three independent experiments; *, p < 0.05.
FIGURE 6.
Cytokines modulate NOTCH signaling activity through NF-κB and MAPK pathways in rat NP cells. The activity of NOTCH responsive reporters 12xCSL (A and B) and 4xCBF1 (C and D) following IL-1β and TNF-α treatment for 24 h with or without inhibitors is shown: SM7368 (SM), SP60025 (SP), SB203580 (SB), or PD98059 (PD). Increased activation of both the reporters by IL-1β (A and C) or TNF-α (B and D) was suppressed by NF-κB and MAPK inhibitors. Values shown are mean ± S.E. of three independent experiments; *, p < 0.05.
MAPKs Control NOTCH2 Promoter Activity in Isoform-specific Manner in NP Cells
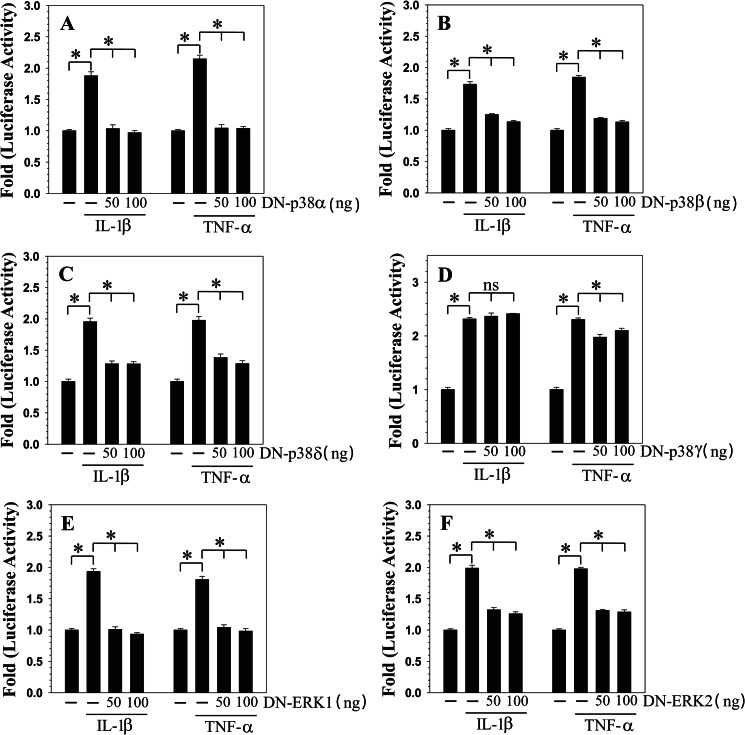
We investigated whether MAPK regulates NOTCH2 expression in an isoform-specific manner. Rat NP cells were transfected with DN-p38α/β2/γ/δ, DN-ERK1, or DN-ERK2 expression plasmids, and NOTCH2 promoter activity was measured in the presence of IL-1β or TNF-α. Cytokine-dependent induction in NOTCH2 promoter activity was significantly inhibited by co-transfection with DN-P38α (Fig. 7A) or DN-p38β2 (Fig. 7B) or DN-p38δ (Fig. 7C). In contrast, another p38 isoform, p38γ (Fig. 7D), had little or no effect on the induction of promoter activity by IL-1β and TNF-α, respectively. Co-transfection with DN-ERK1 (Fig. 7E) or DN-ERK2 (Fig. 7F) also inhibited cytokine induction of NOTCH2 promoter activity.
FIGURE 7.
MAPK regulation of cytokine-mediated induction of NOTCH2 promoter activity in rat NP cells. NP cells were cotransfected with NOTCH2 promoter construct along with either DN-p38α (A) or DN-p38β2 (B) or DN-p38δ (C) or DN-p38γ (D) or DN-ERK1 (E) or DN-ERK2 (F) or empty backbone vector pcDNA3. Transfected cells were treated with IL-1β or TNF-α, and reporter activity was measured 24 h after the treatment. Co-transfection with DN-p38α, DN-p38β2, DN-p38δ, DN-ERK1, and DN-ERK2 resulted in nearly complete suppression of cytokine-dependent induction of the NOTCH2 promoter activity. However, cotransfection of DN-p38γ did not block IL-1β-mediated induction of the reporter activity, and there was a small inhibition in TNF-α treatment group. Values shown are mean ± S.E. of three independent experiments; *, p < 0.05.
NOTCH Expression Correlates to Elevated Cytokine Levels in Degenerate Human Intervertebral Discs
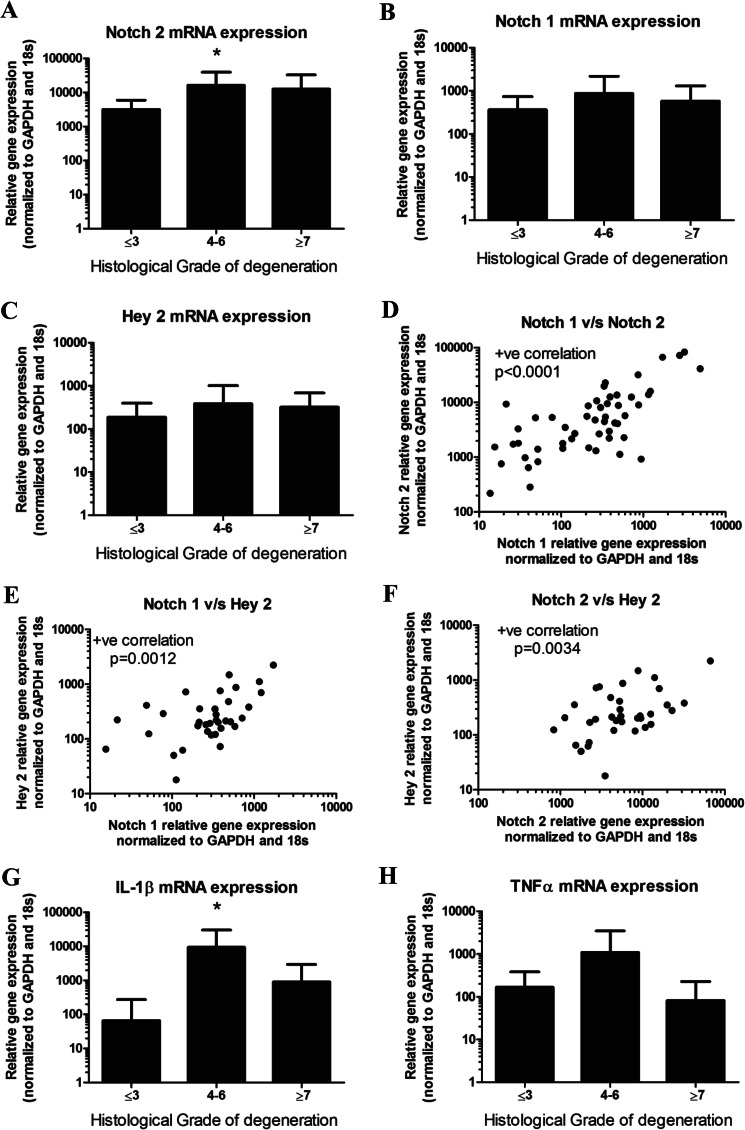
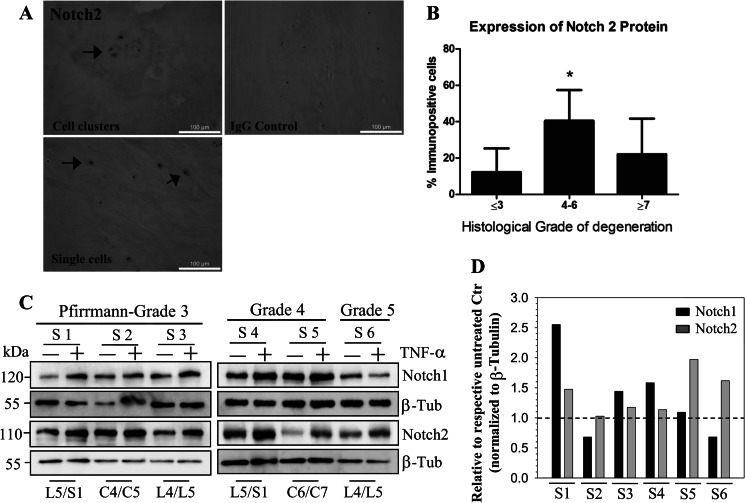
An increase in NOTCH2 mRNA level is seen in mid-grade degenerate discs (graded 4–6) (Fig. 8A; p = 0.04). Increased expression of NOTCH1 (Fig. 8B) and HEY2 (Fig. 8C) within the mid-grade degenerate discs (graded 4–6) were also seen, although they failed to reach statistical significance (NOTCH1, p = 0.19; Hey 2, p = 0.35). Importantly, expression of NOTCH1 and NOTCH2 (Fig. 8D; p < 0.0001), NOTCH1 and HEY2 (Fig. 8E; p = 0.0012), and NOTCH2 and HEY2 (Fig. 8F; p = 0.0034) significantly correlated with each other. Interestingly, this disease group also displayed highest levels of IL-1β (Fig. 8G; p = 0.018); this trend failed to reach significance for TNF-α (Fig. 8H; p = 0.38). IL-1β and TNF-α showed a positive correlation with each other (p = 0.0001; data not shown). NOTCH2 expression was further investigated at the protein level. NOTCH2 was localized to the membrane and cytosol of NP cells (Fig. 9A), and a significant increase in number of cells displaying NOTCH2 immunopositivity was seen in the degenerate discs (graded 4–6) compared with non-degenerate (p = 0.002) or high grade degenerate discs (Fig. 9B; p = 0.018). In addition to direct expression analysis from human tissues, we treated NP cells that were isolated from six degenerate human tissue samples with TNF-α for 24 h. Fig. 9C shows that cells exhibit an TNF-α-dependent increase in levels of both cleaved NOTCH1 and NOTCH2. NOTCH2 shows a more consistent response to the treatment, although patient to patient variation was clearly evident (Fig. 9D).
FIGURE 8.
mRNA expression of NOTCH2 and inflammatory cytokines IL-1β and TNF-α in 49 human disc samples. A, NOTCH2 mRNA expression was highest in mid-grade degenerate discs (grades 4–6), which was significant compared with non-degenerate discs (p = 0.04). B, NOTCH1 mRNA expression was highest in mid-grade degenerate discs (grades 4–6), although this failed to reach significance (p = 0.19). C, HEY2 mRNA expression was highest in mid-grade degenerate discs (grades 4–6), although this failed to reach significance (p = 0.35). D, a positive (+ve) correlation in mRNA levels between NOTCH1 and NOTCH2 was observed (p < 0.0001). v/s: versus. E, a positive correlation in mRNA levels between NOTCH 1 and HEY2 was observed (p = 0.0012). F, a positive correlation in mRNA levels between NOTCH2 and HEY2 was noted (p = 0.034). G, IL-1β mRNA expression was significantly increased in mid-grade degenerate discs compared with non-degenerate discs (p = 0.018). H, TNF-α mRNA expression was increased in mid-grade degenerate discs, although this failed to reach significance (p = 0.38).
FIGURE 9.
NOTCH2 protein level is elevated in degenerated human tissues and is responsive to TNF-α stimulation. A, immunohistochemical analysis demonstrated that NOTCH2 was localized to NP cells within human discs, whereas IgG controls were negative for staining. B, the number of cells that displayed immunopositivity for NOTCH2 was significantly higher in discs histologically graded 4–6, compared with non-degenerate (≤3; p = 0.002) and high-grade degenerate discs (≥7; p = 0.018). C, NP cells isolated from six patient tissue samples (S) with different Pfirrmann grades were treated with TNF-α, and NOTCH1 and NOTCH2 expression was analyzed by Western blot. Spinal level is indicated at the bottom of the blot. D, densitometric analysis of Western blot. NOTCH expression levels are normalized to β-tubulin and represented as fold change relative to corresponding untreated control (considered as 1). As expected, sample-to-sample variation was seen, although NOTCH2 levels were higher in 5/6 treated samples, whereas NOTCH1 expression in 4/6 samples was increased by TNF-α.
DISCUSSION
The experiments described in this work demonstrated for the first time that expression of specific NOTCH pathway genes, concerned with regulation of cell proliferation, differentiation, and fate determination was up-regulated by the inflammatory cytokines IL-1β and TNF-α in NP cells. Another observation was that the cytokine-mediated expression and activation of the NOTCH pathway was dependent on MAPK and NF-κB signaling. Importantly, in terms of clinical relevance, analysis of human tissues indicated that the NOTCH1/2 and HEY2 were expressed by degenerate discs, and expression levels of NOTCH2 were higher in mid-grade degenerate discs, which also displayed highest expression levels for the inflammatory cytokines. These results, for the first time, provide a mechanistic link between the high levels of inflammatory cytokines and the dysregulated expression of NOTCH signaling components observed during disc disease (16).
To test the premise that NOTCH signaling is modulated by the inflammatory conditions, we first investigated the impact of cytokine exposure on expression of NOTCH signaling proteins. The marked increase in mRNA and protein expression of receptors NOTCH1 and -2, ligand JAGGED2, and target genes HES1, HEY1, and HEY2 in the NP cells indicated that the cytokines implicated in degeneration affect NOTCH signaling. Cytokine-dependent induction at both the mRNA and protein level suggested that the response was primarily transcriptional. Importantly, this induction was specific only to a select group of NOTCH receptors and ligands; in contrast to JAGGED2, Dll1 showed a decrease in expression. The physiological significance of the selectivity of this response is currently under investigation.
In addition to the effects of cytokines on NOTCH receptors and ligands, we noted that there was a cytokine-dependent increase in NOTCH target genes HES1, HEY1, and HEY2 and a robust increase in HEY2 protein was also apparent. The cytokines increased the activity of synthetic reporters 12xCSL, 4xwtCBF1, and also NOTCH target gene promoters HES1 and HEY1. Moreover, when a CBF1 reporter with a mutation in CSL binding site was used (4xmutCBF1), cytokine-dependent inducibility was abolished. Taken together, these studies indicated that cytokines increase expression of NOTCH pathway genes, resulting in activation of NOTCH signaling. These results are consistent with previous observations that showed increased NOTCH signaling in stem cells and other cell types in response to cytokines (24–27). In osteoarthritic cartilage, a condition linked to production of proinflammatory cytokines, the role of NOTCH1, JAGGED1, and HES5 has been associated to the expression of a reparative phenotype (28), although increase in NOTCH signaling promotes dedifferentiated phenotype of articular chondrocytes (29). Likewise, in the NP, the activity of this signaling system, in the presence of TNF-α and IL-1β possibly indicates an attempt by the cells to promote tissue repair. The increased expression of NOTCH2 in midgrade degenerate discs but not high grade degenerate discs supports this hypothesis.
Based on results of recent studies (9, 16), we hypothesized that cytokine-dependent NOTCH expression and activation in NP cells is regulated by NF-κB and MAPK signaling (30, 31). For this purpose, we focused on NOTCH2 as a prototype gene and tested two promoter fragments, N2PR-W1 (−2327/−99) and N2PR-W2 (−246/−99). Because the N2PR-W1 fragment was most sensitive to IL-1β and/or TNF-α treatment, the regulatory elements contained within −2347/−247 was chosen for further study. Analysis of this promoter sequence revealed two putative IκB binding motifs in N2PR-W1, suggesting that this pathway is likely involved in regulation. Gain of function studies and use of p65 null cells clearly suggested that the inductive effect is restricted to p65, whereas p50 suppressed p65 as well as cytokine-dependent induction of the NOTCH2 promoter.
The observation that the cytokine induction of NOTCH2 expression is suppressed by p50 is in line with previous studies that demonstrate repressive function of p50 homodimers in controlling expression of a number of genes, including CCL2, CxCL10, GM-CSF, and MMP-13 (32–34). Moreover, our own recent findings have clearly identified p50 as a negative regulator of cytokine dependent transcription of CCL3 as well as ADAMTS-4, genes concerned with inflammatory and catabolic responses (35, 36). On the other hand, our studies have shown that p65 and p50 act synergistically to induce the expression of syndecan-4, one of the target genes of TNF-α and IL-1β in the NP cells (9). These results thus highlight the importance of both context and target gene specificity in the regulatory machinery of NP cells. Moreover, it is likely that formation of p50 homodimers and their binding to the κB motifs could promote recruitment of a transcriptional repressor such as HDAC1 to the NOTCH2 promoter, thereby suppressing RelA response (32). That NF-κB signaling modulated NOTCH2 promoter function was further supported by the loss of function studies. Transfection studies using IκBαM that functions as DN-NF-κB, and site-directed mutagenesis studies suggested that cytokine-dependent induction of NOTCH2 required direct interaction between NF-κB/p65 with the promoter. Importantly, lentivral shRNA silencing studies showed that there was inhibition of IL-1β dependent NOTCH2 expression following suppression of p65 and IKKβ signaling, highlighting the importance of this pathway in controlling NOTCH2 gene expression.
It is important to acknowledge that silenced NP cells or cells treated with NF-κB inhibitor maintain basal levels of activated NOTCH2, suggesting that there are NF-κB-independent mechanism(s) that control base-line transcription. To explore this regulatory system further, because the activity of the MAPK pathway is responsive to both NF-κB and C/EBP-β, we investigated whether it controlled NOTCH2 expression. Our recent work has shown that both TNF-α and IL-1β promoted MAPK activation (35) and that the signaling pathways controlled NOTCH2 expression. We noted that there was differential sensitivity of NOTCH2 expression to MAPK isoforms: thus, although p38α, β2, δ, and ERK1 and ERK2 positively controlled cytokine-dependent induction in NOTCH2, the p38γ was not involved in the response. Based on these findings, it is not unreasonable to assume that by controlling the activity of specific MAPK isoforms, cytokines regulate the expression of NOTCH2 through NF-κB in NP cells. It is important to note that NOTCH signaling has been shown to modulate in both positive and negative fashion, NF-κB-dependent signaling and inflammatory cytokine production in many cell types and in disease contexts (37–41). However, as function of NOTCH signaling is context- and cell type-specific, whether such a feedback between NOTCH and NF-κB or cytokines exists in nucleus pulposus cells remains to be seen.
Relevant to the discussion of increased expression and activity of NOTCH proteins by cytokines, in human and animal models of disc degeneration, increased levels of both IL-1β and TNF-α have been reported (7, 42); likewise, elevated levels of several NOTCH pathway genes in degenerate discal tissues has been shown (16). In concert with these previous studies, (7, 16, 42), our studies confirmed that NOTCH2 expression is elevated in the degenerate human disc tissue and that staining is localized to NP cells. Importantly, expression of NOTCH2 mRNA and protein expression was highest in the mid-grade degenerate discs, which also displayed the highest levels of cytokines IL-1β and TNF-α. Results of the present study clearly demonstrate it is likely that increased level of cytokines alter NOTCH expression and activity during degeneration. Further support to this notion is provided by in vitro studies using human NP cells isolated from degenerate tissues; cells evidenced preservation of sensitivity of NOTCH/2 expression to cytokine exposure. Likewise, TNF-α and IL-1β treatment did not suppress NP cell proliferation, inhibition is seen when NOTCH activity is suppressed (not shown). One possible explanation for the increase in NOTCH activation is that it serves as a protective mechanism and is a part of a compensatory response that maintains resident NP cell proliferation necessary to replace the lost or nonfunctional cells. However, NOTCH signaling has been shown to form a positive feedback with NF-κB and cytokines under inflammatory conditions (37–39). Thus, the therapeutic implications of targeting the NOTCH signaling pathway depend on the role it plays during disease pathogenesis.
This work was supported by National Institutes of Health Grants AR050087 and AR055655, a DISCs grant, and a biomedical research PhD studentship from Sheffield Hallam University.
- IVD
- intervertebral disc
- NP
- nucleus pulposus
- DN
- dominant negative
- Luc
- luciferase.
REFERENCES
- 1. Luoma K., Riihimäki H., Luukkonen R., Raininko R., Viikari-Juntura E., Lamminen A. (2000) Low back pain in relation to lumbar disc degeneration. Spine 25, 487–492 [DOI] [PubMed] [Google Scholar]
- 2. Maniadakis N., Gray A. (2000) The economic burden of back pain in the UK. Pain 84, 95–103 [DOI] [PubMed] [Google Scholar]
- 3. Weiler C., Nerlich A. G., Zipperer J., Bachmeier B. E., Boos N. (2002) 2002 SSE Award Competition in Basic Science: expression of major matrix metalloproteinases is associated with intervertebral disc degradation and resorption. Eur. Spine J. 11, 308–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Le Maitre C. L., Freemont A. J., Hoyland J. A. (2004) Localization of degradative enzymes and their inhibitors in the degenerate human intervertebral disc. J. Pathol. 204, 47–54 [DOI] [PubMed] [Google Scholar]
- 5. Ahn S. H., Cho Y. W., Ahn M. W., Jang S. H., Sohn Y. K., Kim H. S. (2002) mRNA expression of cytokines and chemokines in herniated lumbar intervertebral discs. Spine 27, 911–917 [DOI] [PubMed] [Google Scholar]
- 6. Kang J. D., Georgescu H. I., McIntyre-Larkin L., Stefanovic-Racic M., Donaldson W. F., 3rd, Evans C. H. (1996) Herniated lumbar intervertebral discs spontaneously produce matrix metalloproteinases, nitric oxide, interleukin-6, and prostaglandin E2. Spine 21, 271–277 [DOI] [PubMed] [Google Scholar]
- 7. Le Maitre C. L., Freemont A. J., Hoyland J. A. (2005) The role of interleukin-1 in the pathogenesis of human intervertebral disc degeneration. Arthritis Res. Ther. 7, R732–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Igarashi T., Kikuchi S., Shubayev V., Myers R. R. (2000) 2000 Volvo Award winner in basic science studies: Exogenous tumor necrosis factor-α mimics nucleus pulposus-induced neuropathology. Molecular, histologic, and behavioral comparisons in rats. Spine 25, 2975–2980 [DOI] [PubMed] [Google Scholar]
- 9. Wang J., Markova D., Anderson D. G., Zheng Z., Shapiro I. M., Risbud M. V. (2011) TNF-α and IL-1β promote a disintegrin-like and metalloprotease with thrombospondin type I motif-5-mediated aggrecan degradation through syndecan-4 in intervertebral disc. J. Biol. Chem. 286, 39738–39749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Le Maitre C. L., Hoyland J. A., Freemont A. J. (2007) Catabolic cytokine expression in human IVD degeneration: IL-1β and TNFα expression profile. Arthritis Res. Ther. 9, R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Peng B., Chen J., Kuang Z., Li D., Pang X., Zhang X. (2009) Expression and role of connective tissue growth factor in painful disc fibrosis and degeneration. Spine 34, E178–182 [DOI] [PubMed] [Google Scholar]
- 12. Pockert A. J., Richardson S. M., Le Maitre C.L., Lyon M., Deakin J. A., Buttle D. J., Freemont A. J., Hoyland J. A. (2009) Modified Expression of the ADAMTS Enzymes and TIMP-3 During Human Intervertebral Disc Degeneration. Arthritis Rheum. 60, 482–491 [DOI] [PubMed] [Google Scholar]
- 13. Hoyland J. A., Le Maitre C., Freemont A. J. (2008) Investigation of the role of IL-1 and TNF in matrix degradation of the intervertebral disc. Rheumatology 47, 809–814 [DOI] [PubMed] [Google Scholar]
- 14. Dong Y., Jesse A. M., Kohn A., Gunnell L.M., Honjo T., Zuscik M. J., O'Keefe R. J., Hilton M. J. (2010) RBPjκ-dependent Notch signaling regulates mesenchymal progenitor cell proliferation and differentiation during skeletal development. Development 137, 1461–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mead T. J., Yutzey K. E. (2009) Notch pathway regulation of chondrocyte differentiation and proliferation during appendicular and axial skeleton development. Proc. Natl. Acad. Sci. U.S.A. 106, 14420–14425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hiyama A., Skubutyte R., Markova D., Anderson D. G., Yadla S., Sakai D., Mochida J., Albert T. J., Shapiro I. M., Risbud M. V. (2011) Hypoxia activates the Notch signaling pathway in cells of the intervertebral disc: implications in degenerative disc disease. Arthritis Rheum. 63, 1355–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Richards G. S., Degnan B. M. (2009) The dawn of developmental signaling in the metazoa. Cold Spring Harb. Symp. Quant. Biol. 74, 81–90 [DOI] [PubMed] [Google Scholar]
- 18. Iso T., Kedes L., Hamamori Y. (2003) HES and HERP families: multiple effectors of the Notch signaling pathway. J. Cell. Physiol. 194, 237–255 [DOI] [PubMed] [Google Scholar]
- 19. Gustafsson M. V., Zheng X., Pereira T., Gradin K., Jin S., Lundkvist J., Ruas J. L., Poellinger L., Lendahl U., Bondesson M. (2005) Hypoxia requires Notch signaling to maintain the undifferentiated cell state. Dev. Cell 9, 617–628 [DOI] [PubMed] [Google Scholar]
- 20. Hsieh J. J., Henkel T., Salmon P., Robey E., Peterson M. G., Hayward S. D. (1996) Truncated mammalian Notch1 activates CBF1/RBPJK-repressed genes by a mechanism resembling that of Epstein Barr virus EBNA2. Mol. Cell. Biol. 16, 952–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ungerbäck J., Elander N., Grünberg J., Sigvardsson M., Söderkvist P. (2011) The Notch-2 gene is regulated by Wnt signaling in cultured colorectal cancer cells. PLoS One 6, e17957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Risbud M. V., Guttapalli A., Stokes D. G., Hawkins D., Danielson K. G., Schaer T. P., Albert T. J., Shapiro I. M. (2006) Nucleus pulposus cells express HIF-1 α under normoxic culture conditions: a metabolic adaptation to the intervertebral disc microenvironment. J. Cell. Biochem. 98, 152–159 [DOI] [PubMed] [Google Scholar]
- 23. Wasserman W. W., Sandelin A. (2004) Applied bioinformatics for the identification of regulatory elements. Nat. Rev. Genet. 5, 276–287 [DOI] [PubMed] [Google Scholar]
- 24. Ottaviani S., Tahiri K., Frazier A., Hassaine Z. N., Dumontier M. F., Baschong W., Rannou F., Corvol M. T., Savouret J. F., Richette P. (2010) Hes1, a new target for interleukin 1β in chondrocytes. Ann. Rheum. Dis. 69, 1488–1494 [DOI] [PubMed] [Google Scholar]
- 25. Boonyatecha N., Sangphech N., Wongchana W., Kueanjinda P., Palaga T. (2012) Involvement of Notch signaling pathway in regulating IL-12 expression via c-Rel in activated macrophages. Mol. Immunol. 51, 255–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Johnston D. A., Dong B., Hughes C. C. (2009) TNF induction of jagged-1 in endothelial cells is NFκB-dependent. Gene 435, 36–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ando K., Kanazawa S., Tetsuka T., Ohta S., Jiang X., Tada T., Kobayashi M., Matsui N., Okamoto T. (2003) Induction of Notch signaling by tumor necrosis factor in rheumatoid synovial fibroblasts. Oncogene 22, 7796–7803 [DOI] [PubMed] [Google Scholar]
- 28. Karlsson C., Brantsing C., Egell S., Lindahl A. (2008) Notch1, Jagged1, and HES5 are abundantly expressed in osteoarthritis. Cells Tissues Organs 188, 287–298 [DOI] [PubMed] [Google Scholar]
- 29. Blaise R., Mahjoub M., Salvat C., Barbe U., Brou C., Corvol M. T., Savouret J. F., Rannou F., Berenbaum F., Bausero P. (2009) Involvement of the Notch pathway in the regulation of matrix metalloproteinase 13 and the dedifferentiation of articular chondrocytes in murine cartilage. Arthritis Rheum. 60, 428–439 [DOI] [PubMed] [Google Scholar]
- 30. Li H., Lin X. (2008) Positive and negative signaling components involved in TNFα-induced NF-κB activation. Cytokine 41, 1–8 [DOI] [PubMed] [Google Scholar]
- 31. Glossop J. R., Cartmell S. H. (2009) Effect of fluid flow-induced shear stress on human mesenchymal stem cells: differential gene expression of IL1B and MAP3K8 in MAPK signaling. Gene Expr. Patterns 9, 381–388 [DOI] [PubMed] [Google Scholar]
- 32. Elsharkawy A. M., Oakley F., Lin F., Packham G., Mann D. A., Mann J. (2010) The NF-κB p50:p50:HDAC-1 repressor complex orchestrates transcriptional inhibition of multiple pro-inflammatory genes. J. Hepatol. 53, 519–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Panzer U., Steinmetz O. M., Turner J. E., Meyer-Schwesinger C., von Ruffer C., Meyer T. N. (2009) Resolution of renal inflammation: a new role for NF-κB1(p50) in inflammatory kidney diseases. Am. J. Physiol. Renal Physiol. 297, F429–39 [DOI] [PubMed] [Google Scholar]
- 34. Oakley F., Mann J., Nailard S., Smart D. E., Mungalsingh N., Constandinou C. (2005) Nuclear factor-κB1 (p50) limits the inflammatory and fibrogenic responses to chronic injury. Am. J. Pathol. 166, 695–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang J., Tian Y., Phillips K. L., Chiverton N., Haddock G., Bunning R. A., Cross A. K., Shapiro I. M., Le Maitre C. L., Risbud M. V. (2013) TNF-α and IL-1β dependent induction of CCL3 expression by nucleus pulposus cells promotes macrophage migration through CCR1. Arthritis Rheum. 65, 832–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tian Y., Yuan W., Fujita N., Wang J., Wang H., Shapiro I. M., Risbud M. V. (2013) Inflammatory cytokines associated with degenerative disc disease control aggrecanase1/ADAMTS-4 expression in nucleus pulposus cells through MAPK and NF-κB Am. J Pathol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wei Z., Chigurupati S., Arumugam T. V., Jo D. G., Li H., Chan S. L. (2011) Notch activation enhances the microglia-mediated inflammatory response associated with focal cerebral ischemia. Stroke 42, 2589–2594 [DOI] [PubMed] [Google Scholar]
- 38. Wongchana W., Palaga T. (2012) Direct regulation of interleukin-6 expression by Notch signaling in macrophages. Cell Mol. Immunol. 9, 155–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fukushima H., Nakao A., Okamoto F., Shin M., Kajiya H., Sakano S., Bigas A., Jimi E., Okabe K. (2008) The association of Notch2 and NF-κB accelerates RANKL-induced osteoclastogenesis. Mol. Cell. Biol. 28, 6402–6412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang Q., Wang C., Liu Z., Liu X., Han C., Cao X., Li N. (2012) Notch signal suppresses Toll-like receptor-triggered inflammatory responses in macrophages by inhibiting extracellular signal-regulated kinase 1/2-mediated nuclear factor κB activation. J. Biol. Chem. 287, 6208–6217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Curry C. L., Reed L. L., Broude E., Golde T. E., Miele L., Foreman K. E. (2007) Notch inhibition in Kaposi's sarcoma tumor cells leads to mitotic catastrophe through nuclear factor-κB signaling. Mol. Cancer Ther. 6, 1983–1992 [DOI] [PubMed] [Google Scholar]
- 42. Weiler C., Nerlich A. G., Bachmeier B. E., Boos N. (2005) Expression and distribution of tumor necrosis factor α in human lumbar intervertebral discs: a study in surgical specimen and autopsy controls. Spine 30, 44–53 [DOI] [PubMed] [Google Scholar]