Background: USP7 is a ubiquitin-specific protease that regulates the turnover of target proteins in the cell.
Results: UbE2E1 was identified to interact with USP7 through its ASTS motif.
Conclusion: USP7 interacts with and deubiquitinates UbE2E1.
Significance: USP7 regulates the stability of UbE2E1, the first example of a deubiquitinating enzyme, regulating an E2 conjugation enzyme.
Keywords: Deubiquitination, E3 Ubiquitin Ligase, Protein Structure, Ubiquitin-conjugating Enzyme (Ubc), Ubiquitination
Abstract
Ubiquitin-specific protease 7 (USP7) is a deubiquitinating enzyme found in all eukaryotes that catalyzes the removal of ubiquitin from specific target proteins. Here, we report that UbE2E1, an E2 ubiquitin conjugation enzyme with a unique N-terminal extension, is a novel USP7-interacting protein. USP7 forms a complex with UbE2E1 in vitro and in vivo through the ASTS USP7 binding motif within its N-terminal extension in an identical manner with other known USP7 binding proteins. We show that USP7 attenuates UbE2E1-mediated ubiquitination, an effect that requires the N-terminal ASTS sequence of UbE2E1 as well as the catalytic activity of USP7. Additionally, USP7 is critical in maintaining the steady state levels of UbE2E1 in cells. This study reveals a new cellular mechanism that couples the opposing activities of the ubiquitination machinery and a deubiquitinating enzyme to maintain and modulate the dynamic balance of the ubiquitin-proteasome system.
Introduction
Ubiquitination is a post-translational modification that regulates protein turnover, function, and localization. The attachment of ubiquitin takes place in several steps and involves at least three distinct types of enzymes. First, ubiquitin is activated through adenylation at its C terminus by the E1 activating enzyme. The activated ubiquitin is then transferred to an E2 conjugating enzyme. Finally, an E3 ligase transfers the ubiquitin from the E2 to the ϵ-amino group of a lysine residue on a target protein in a substrate-specific manner (1, 2). There are two ubiquitin E1 activating enzymes, approximately 40 E2 conjugating enzymes, and over 600 E3 ligases in the human genome, allowing precise spatial and temporal regulation of ubiquitination (3–5).
All E2s have a conserved ubiquitin-conjugating (UBC)5 domain, consisting of about 150 residues with a central cysteine essential in formation of the ubiquitin thioester bond. E2s can be further divided into four classes: Class I E2s consist of only the UBC domain; Class II E2s have C-terminal extensions to the UBC core; Class III E2s have N-terminal extensions in addition to the UBC core; and Class IV contains both N-terminal and C-terminal extensions to the core domain. Emerging evidence suggests that the additional N- or C-terminal extensions are essential for the functions of the E2s, contributing to substrate specificity and/or regulation (6, 7). UbE2E (UbE2E1, UbE2E2, and UbE2E3) is a subfamily of class III E2 enzymes, homologs of yeast UBC4 and -5, that mediate degradation of misfolded and damaged proteins (8). They interact with a broad range of ubiquitin E3 ligases and are involved in ubiquitination of a large number of substrates (9). UbE2E1 was shown to play a role in histone ubiquitination, and UbE2E3 was implicated in the redox homeostasis following oxidative stress, required for retinal pigment epithelial cell proliferation (10, 11). All three UbE2E proteins contain the core UBC domain with ∼94% sequence similarity and a less conserved N-terminal extension of about 50 amino acids. The function of the N-terminal extensions of the UbE2E proteins is not clear. However, these extensions are naturally disordered and rich in serine and lysine residues, suggesting that they are subject to protein-protein interaction and post-translational modifications, which may regulate UbE2E family members.
Ubiquitination, like other post-translational modifications, is reversible. Reversal of ubiquitination, or deubiquitination, is carried out by deubiquitinating enzymes (DUBs), which belong to the metallo and cysteine families of proteases (12). By reversing the actions of ubiquitin ligases, DUBs offer a way to fine-tune the effects of ubiquitination as a post-translational modification. This is perhaps best illustrated by the regulation of the p53 tumor suppressor by the DUB, USP7. USP7 not only deubiquitinates and stabilizes p53, but also Hdm2, the primary E3 ubiquitin ligase of p53 (13, 14). The effect of USP7 on Hdm2 is through direct interaction and independently of p53. Coexistence of both the DUB and the E3 ligase activity in the same protein complex offers an elegant way to tightly regulate p53 levels, which are key in determining the fate of the cell by governing cell growth arrest and apoptosis. Interestingly, the coupling of opposing activities of ubiquitination and deubiquitination is not unique to the USP7-Hdm2 complex. Indeed, USP7 was first discovered as a protein in complex with the herpes simplex virus protein ICP0, a viral E3 ligase (15), and was termed HAUSP for herpes-associated ubiquitin-specific protease. Several other E3 ligases have been identified that are also coupled to USP7 including CHFR, HLTF, ARF-BP1, and MARCH7 (16–19) USP7 has emerged as a key regulator of a range of cellular processes as exemplified by the large number of USP7 substrate proteins, which include p53, Hdm2, HdmX, claspin, histone H2B, PTEN, and FOXO4 (13, 16, 20–22). Although USP7 generally regulates the levels, function, and localization of its interacting proteins through its deubiquitinase activity, it has also been shown to negatively regulate the PML (promyelocytic leukemia protein) proteins and PML nuclear bodies in a manner independent of its catalytic activity (23).
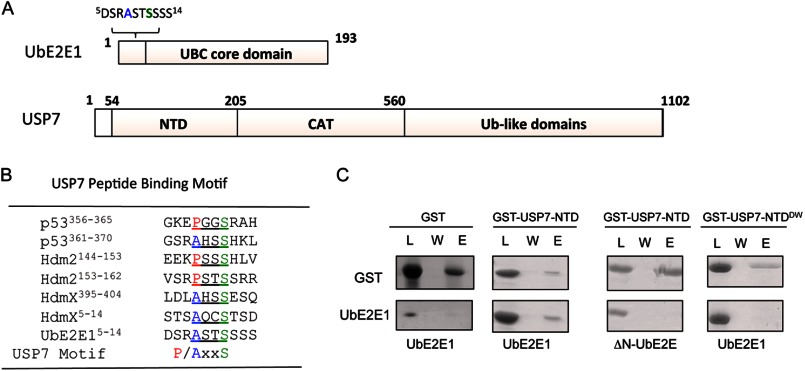
Our previous studies have revealed that the N-terminal domain of USP7 (USP7-NTD) recognizes a sequence motif ((P/A)XXS) found in its interaction partners such as p53, Hdm2, and HdmX (24, 25). Interestingly, the class III E2 conjugating enzymes, UbE2Es, contain a conserved (P/A)XXS motif within their N-terminal extensions. In this study, UbE2E1 was identified as novel USP7 interaction partner. Further structural and functional analysis revealed that USP7 deubiquitinates UbE2E1 and is important for UbE2E1 stability in vivo. This study provides a new facet of USP7 in modulating and sustaining the basic ubiquitination machinery through the regulation of yet another component of the ubiquitination machinery: the E2 conjugation enzymes.
EXPERIMENTAL PROCEDURES
Cell Culture and Antibodies
HCT116 wild type and USP7−/− cells were kindly provided by Bert Vogelstein (Johns Hopkins Medical Institutions). Human U2OS cells were grown in McCoy's medium. HCT116, 293T, and HeLa cells were grown in DMEM supplemented with 10% FBS. The antibodies used for immunoblotting and immunostaining experiments include the mouse antibody to ubiquitin (Covance MMS-258R), actin (Calbiochem, CP01), the Myc epitope tag (Millipore 05-724) and the FLAG epitope tag (Sigma F3165), the mouse, rabbit, and goat antibodies to UbE2E1 (BD Biosciences, 611218; Boston Biochem, A-630; and Santa Cruz Biotechnology, sc-47547), and the rabbit antibody to USP7 (Bethyl Laboratories A300-033A).
Expression and Purification of USP7-NTD and UbE2E1 Proteins
USP7-NTD, UbE2E1, and ΔN-UbE2E1 were expressed from pET15b plasmids in Escherichia coli BL21(DE3) cells and purified using affinity chromatography. Briefly, the cells were harvested and lysed using sonication in 50 mm Tris, pH 7.5, 500 mm NaCl, 10 mm imidazole, 1 mm benzamidine, and 0.5 mm PMSF. The lysates were cleared and poured onto nickel-nitrilotriacetic acid beads (Qiagen). After extensive washing with 50 mm Tris, pH 7.5, 500 mm NaCl, and 30 mm imidazole, the proteins were eluted with the addition of 50 mm Tris, pH 7.5, 500 mm NaCl, and 250 mm imidazole. USP7-NTD was incubated with thrombin to remove the His tag and further purified using size-exclusion chromatography (Sephacryl S200 16/60) on an ÄKTApurifier 10 UPC (GE Healthcare) prior to crystallization trials.
Peptide Synthesis
The UbE2E1 peptides were synthesized by CanPeptide Inc. (Montreal, Canada) with both N-terminal acetylation and C-terminal amidation to mimic the native peptides. All of the peptides were greater than 95% pure and were dissolved in either the crystallization or the fluorescence binding assay buffers prior to use.
Crystallization, Data Collection, and Structure Determination of the USP7·UbE2E1 Complex
USP7-NTD (100 mg/ml) was co-crystallized with UbE2E1 (5DSRASTSSSS14) peptide at ∼5-fold molar excess of peptide. Several rounds of micro-seeding using USP7-NTD·HdmXAHSS crystals as seeds were performed in crystallization tools (Qiagen). The seeded crystals grew after 1 week at 4 °C in the dark in conditions containing 30% PEG 4000, 0.1 m Tris, pH 8.5, and 0.2 m lithium sulfate. X-ray data from a frozen crystal of the USP7-NTD·UbE2E1ASTS complex was tested at 100 K on a Rigaku MicroMax007 rotating anode diffractometer with Saturn 944+ CCD detector and collected at 100 K at the 19ID Structural Biology Center beamline (Advanced Photon Source, Argonne National Laboratory, Argonne, IL). The crystal belongs to space group P41 with approximate unit cell dimensions a = b = 70.0 Å and c = 45.8 Å3. Data were integrated and scaled using HKL2000. A summary of data collection statistics is presented in Table 1. The structure was determined by using the molecular replacement component of CNS (version 1.2) with USP7-NTD (Protein Data Bank (PDB) ID 1YY6) as the search model without any peptide (26). Electron density visualization and model building were done with O (27). Rigid body and simulated annealing torsion angle refinement were normally followed by individual B-factor refinement and performed using CNS 1.2. Several rounds of refinement were combined with model rebuilding in O after inspection of both 2Fo − Fc and Fo − Fc maps. A summary of refinement statistics is presented in Table 1. PyMOL was used for the preparation of the figure.
TABLE 1.
X-ray data collection and refinement statistics
Numbers in brackets refer to the highest resolution shell, 1.98–1.95 Å.
| USP7·UbE2E1ASTS | |
|---|---|
| X-ray data | |
| Space group | P41 |
| Resolution (Å) | 50.0-1.95 |
| Unit cell axes (Å3) | 69.9 × 69.9 × 45.7 |
| Molecules/asymmetric unit | 1 |
| Total observations (No.) | 189292 |
| Unique reflections (No.) | 16086 |
| Intensity (I/σ〈I〉) | 29.0 (5.2) |
| Completeness (%) | 98.9 (95.9) |
| Rsyma | 0.133 (0.789) |
| Refinement | |
| Rwork | 0.197 |
| Rfree | 0.217 |
| Protein atoms (No.) | 1133 |
| Water molecules (No.) | |
| r.m.s.d. bonds (Å) | 0.006 |
| r.m.s.d. angles (°) | 1.4 |
| r.m.s.d. dihedrals (°) | 25.4 |
| r.m.s.d. improper (°) | 0.80 |
| Thermal factors (Å2) | 20.9 |
| Ramachandran plot | |
| Most Favored | 0.977 |
| Additionally Allowed | 0.015 |
a Rsym = Σ|I − 〈I〉|/ΣI where I is the observed intensity and 〈I〉 is the average intensity from multiple observations of symmetry-related reflections.
GST Pulldown Assays
The GST-USP7-NTD and GST-USP7-NTDDW fusion proteins were generated by PCR amplification and insertion between the BamHI and Xho1 sites of pGEX-4T-2 plasmid (GE Healthcare) as described previously (24). Proteins were expressed in E. coli BL21 pLysS cells with 3 h of 1 mm isopropyl-1-thio-β-d-galactopyranoside induction at 37 °C, purified on glutathione-Sepharose resin (GE Healthcare) using standard methods, and dialyzed against assay buffer (10 mm Na2HPO4, 1.5 mm KH2PO4, 137 mm NaCl, 2.7 mm KCl, 5% glycerol, 1 mm benzamidine, and 0.5 mm PMSF). Purified UbE2E1 and ΔN-UbE2E1 were incubated with glutathione S-transferase (GST) alone and GST-USP7-NTD and GST-USP7-NTDDW fusion proteins in a 1:1 molar ratio at 4 °C for 1 h in the presence of 50 μl of glutathione-Sepharose beads (GE Healthcare). The mixture was then transferred to a micro column, and after extensive washing with assay buffer, bound proteins were eluted with 20 mm reduced glutathione and detected by Coomassie Blue staining following SDS-PAGE.
Intrinsic Tryptophan Fluorescence Assays
UbE2E1 peptides were titrated (0–150 μm) with wild type USP7-NTD (1 μm) in 50 mm Tris, pH 7.5, 150 mm NaCl, 1 mm EDTA, and 1 mm DTT. The change in tryptophan fluorescence was monitored using the Kinetics application on a Cary Eclipse fluorescence spectrophotometer (Varian Inc.), and Kd values for USP7-NTD binding were calculated using GraphPad Prism as described previously (25). The Kd values were calculated based on three individual experiments.
In Vitro Ubiquitination and Deubiquitination Assays
The in vitro ubiquitination and deubiquitination assays were performed in a volume of 20 μl in 50 mm Tris, pH 7.6, 5 mm MgCl2, 2 mm ATP, and 2 mm DTT. The reaction mixture typically contained E1 (100 ng), E2 (200 ng), ubiquitin (5 μg), and E3 (the catalytic domains of NEDD-4L or ITCH, 0.5 μg) with increasing amounts of USP7 (0–2 μg) where indicated. The conditions for E1, E2, Ub, and ATP were optimized through titration experiments. After incubation at 30 °C for 90 min, the reactions were stopped by the addition of SDS-PAGE sample buffer and resolved on 7.5–10% SDS-polyacrylamide gels. Ubiquitinated proteins were visualized and evaluated by Western blotting using a ubiquitin-specific antibody (Covance) or other antibodies as indicated. Increasing amounts (5–20 mm) of N-ethylmaleimide (NEM, Sigma) or ubiquitin aldehyde (Boston Biochem) were added to inhibit USP7 catalytic activity. Excess NEM was removed through extensive dialysis. All assays were done independently more than three times, and consistent results were obtained.
In Vitro Ubiquitin Loading Assays
The in vitro ubiquitin loading assays were performed as described previously (28). The reactions were carried out in a volume of 10 μl in 10 mm HEPES, pH 7.5, 100 mm NaCl, 40 μm ATP, and 2 mm MgCl2 containing E1 (1 μg), E2 (1 μg), ubiquitin (5 μg), and increasing amounts of USP7 (0–2 μg). After incubation at 30 °C for 10 min, the reactions were stopped by the addition of SDS-PAGE sample buffer without DTT and resolved on 15% SDS-polyacrylamide gels. Loaded E2 protein proteins were visualized and evaluated by Western blotting using antibodies against ubiquitin (Covance) and UbE2E1 (Boston Biochem).
Immunoprecipitations
HeLa cells transfected with FLAG-tagged UbE2E1 and Myc-tagged USP7 were used for immunoprecipitations. Cells were harvested 48 h after transfection and lysed in radioimmunoprecipitation assay buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1% Nonidet P-40, 0.1% sodium deoxycholate and protease inhibitor mixture (Roche Applied Science)). Protein A-Sepharose bead precleared lysates were incubated with antibodies against the FLAG tag for FLAG-tagged UbE2E1 (Sigma) or against the Myc tag for Myc-tagged USP7 overnight at 4 °C followed by the addition of protein A beads for another 60 min. Immunoprecipitates were washed three times using radioimmunoprecipitation assay buffer. The immunoprecipitated complexes were released by boiling the beads for 5 min in SDS sample buffer and resolved on 12% SDS-polyacrylamide gels followed by immunoblotting with antibodies against UbE2E1 (Boston Biochem) or USP7 (Bethyl Laboratories). The immunoprecipitation experiments for endogenous UbE2E1 and USP7 were performed in a similar way. Lysates were immunoprecipitated with an antibody against UbE2E1 (BD Biosciences) and immunoblotted with an antibody against USP7 (Bethyl Laboratories).
Immunofluorescence
Immunostaining of endogenous UbE2E1 and USP7 was done using U2OS and HeLa cells. Cells were grown on coverslips and fixed in 2% paraformaldehyde in PBS and permeabilized with 0.5% Triton X-100 in PBS. Cells were washed once with PBST (PBS with 0.1% Tween 20) and blocked with 1% BSA in PBST for 1 h at 37 °C. Endogenous USP7 was stained with rabbit anti-USP7 (Bethyl Laboratories) and CY3-labeled anti-rabbit IgG. Endogenous UbE2E1 was stained with UbcH6 goat polyclonal IgG (Santa Cruz Biotechnology) and FITC-labeled anti-goat IgG (Sigma). Staining of the nucleus of U2OS cells was done with DRAQ5 (Enzo Life Sciences International, Inc.). To test antibody specificity for USP7 and UbE2E1, 10 μg of recombinant GST-USP7 and His6-tagged UbE2E1 were incubated with each appropriate antibody respectively for 60 min at 37 °C before the staining procedures. Negative staining controls with 1 μl of normal rabbit IgG (Santa Cruz Biotechnology; control IgG) and CY3-labeled anti-rabbit IgG and 1 μl of normal mouse IgG (Santa Cruz Biotechnology; control IgG) and FITC-labeled anti-goat IgG were prepared to validate the specific USP7 and UbE2E1 staining, respectively. Images were obtained with Zeiss LSM 700 and Olympus FluoView 300 confocal laser-scanning microscopes. Images were analyzed and superimposed using ImageJ software.
In Vivo UbE2E1 Ubiquitination and Deubiquitination
293T cells were transfected with FLAG-tagged UbE2E1 and treated with 10 μm MG-132 for 6 h. Immunoprecipitation was performed on both MG-132-treated and untreated cells using anti-FLAG. Ubiquitinated UbE2E1 was visualized by immunoblotting using both antibodies against ubiquitin and UbE2E1. Myc-tagged wild type USP7 and the catalytic inactive mutant USP7-CS were immunoprecipitated from 293T cells and incubated with ubiquitinated UbE2E1 isolated from the MG-132-treated cell lysate for 30 min at 30 °C. The reactions were stopped by the addition of SDS-PAGE sample buffer and immunoblotted with a ubiquitin antibody.
UbE2E1 Turnover Assays
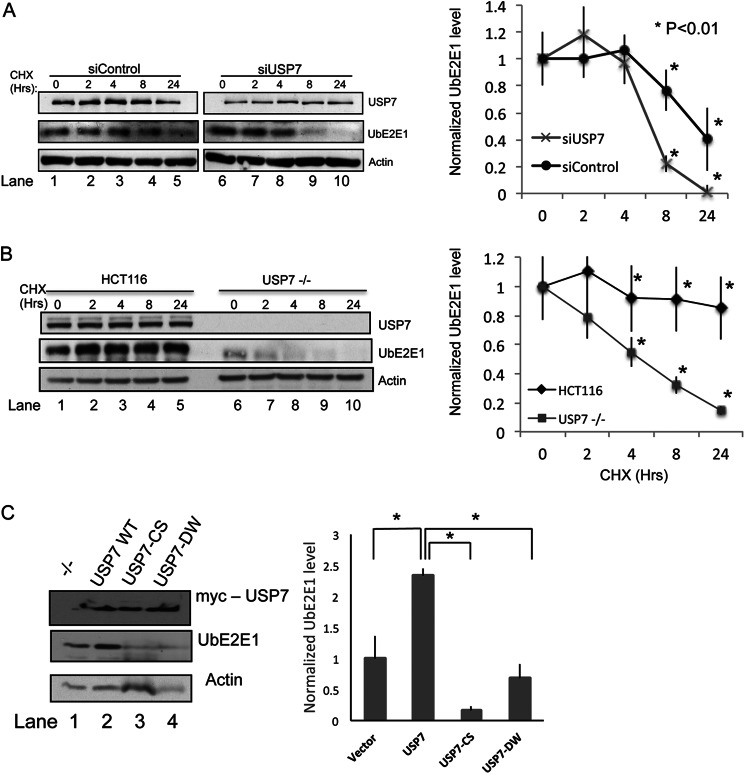
U2OS, HCT116 WT, or HCT116 USP7−/− cells were seeded in 6-cm dishes at 80% confluence. U2OS cells were transfected three times with siRNA for USP7 (5′-CCCAAAUUAUUCCGCGGCAAA as described in Tang et al. (29) or a negative control siRNA (provided by GenePharma) as outlined previously (30). siRNA-transfected U2OS cells and untransfected HCT116 cells (WT and USP7−/−) were then treated with 10 μg/ml cycloheximide (Sigma) and harvested at various times after treatment. HCT116 USP7−/− cells were transfected with empty vector, Myc-USP7, and Myc-USP7-C233S and harvested after 48 h. Cells were then lysed in radioimmunoprecipitation assay buffer. 30 μg of total protein was subjected to SDS-PAGE and Western blotting using antibodies against USP7 (Bethyl Laboratories), actin and UbE2E1 (Boston Biochem). These experiments were repeated independently, and similar results were obtained. The levels of UbE2E1 were determined by normalizing the intensity of the UbE2E1 bands to those of the actin bands. Student's t test was used to analyze the differences of the levels of UbE2E1 at each time point.
RESULTS
USP7 Interacts with UbE2E1 in Vitro
Previous studies have shown that the N-terminal domain of USP7 (USP7-NTD) interacts with substrates p53, Hdm2, and HdmX through their (P/A)XXS motifs (Fig. 1A) (24, 25). Alignment of the (P/A)XXS sequence motif among USP7 substrate proteins along with the UbE2E1 ASTS sequence suggests that UbE2E1 will interact with USP7 through this conserved motif (Fig. 1B). We therefore hypothesized that USP7 interacts with UbE2E1 and that this interaction is occurring through its N-terminal extension. Recombinant full-length UbE2E1 and a mutant lacking the N-terminal extension, ΔN-UbE2E1, were used to assess interaction with GST-USP7-NTD using GST pulldown assays (Fig. 1C). UbE2E1 was shown to form a complex with GST-USP7-NTD, but not GST alone (negative control), indicating that USP7-NTD interacts with UbE2E1. The interaction between GST-USP7-NTD and ΔN-UbE2E1 was disrupted, suggesting that the USP7 binding sequence is localized within the N-terminal extension of UbE2E1. A 164DWGF167 motif in USP7-NTD was previously shown to be essential for its interaction with substrate proteins containing the (P/A)XXS motif, and a double point mutant of USP7-NTD (USP7-NTDDW), in which Asp164 and Trp165 were changed to alanine residues, eliminated binding to previously reported (P/A)XXS-containing substrates (31). As shown in Fig. 1C, USP7-NTDDW failed to retain UbE2E1, suggesting that UbE2E1 shares the same USP7 binding surface as the other (P/A)XXS-containing substrates and that the Asp164 and Trp165 residues in USP7-NTD are critical in mediating interactions with UbE2E1. Collectively, USP7 and UbE2E1 proteins are able to directly interact in vitro through the (P/A)XXS motif in the UbE2E1 N-terminal extension and the 164DWGF167 motif of USP7-NTD.
FIGURE 1.
Analysis of the in vitro interaction between USP7-NTD and UbE2E1. A, schematics of UbE2E1 and USP7 indicating domain organization of the two proteins and the putative USP7 binding motif within the N terminus of UbE2E1. CAT, catalytic domain. B, comparison of the substrate peptide sequences recognized by USP7-NTD, featuring the (P/A)XXS motif, which is also found in the N terminus of UbE2E1. C, GST pulldowns were performed using GST-USP7-NTD and UbE2E1. GST-USP7-NTD fusion protein was incubated with UbE2E1, loaded onto glutathione resin (load (L)), washed with wash buffer (wash (W)), and eluted with Laemmli SDS-PAGE loading dye (eluate (E)). A GST pulldown of UbE2E1 protein with GST alone served as a negative control. GST pulldown experiments also tested the interaction of GST-USP7-NTD and the N-terminal UbE2E1 deletion mutant (ΔN-UbE2E1) and between the GST-USP7-NTD double mutant USP7-NTDDWand UbE2E1.
Molecular Analysis of the USP7-UbE2E1 Interaction
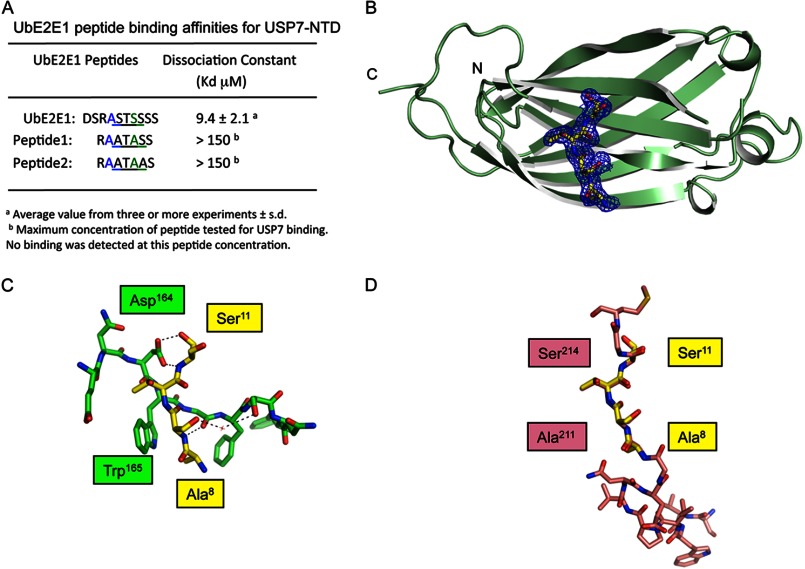
To determine the binding affinity as well as to reveal the molecular basis of the interaction between USP7-NTD and UbE2E1, a peptide corresponding to the interaction motif (5DSRASTSSSS14) in the UbE2E1 N-terminal extension and peptides with mutations in the binding motif were synthesized. The change in the intrinsic tryptophan fluorescence of USP7-NTD was monitored with increasing amounts of the UbE2E1 peptide to derive the dissociation constant between USP7-NTD and the peptide. The dissociation constant was calculated to be 9.4 ± 2.1 μm for 5DSRASTSSSS14, whereas no USP7 binding was observed with the two mutant peptides (7RAATASS13, 7RAATAAS13) at concentrations up to 150 μm. The dissociation constant between USP7 and UbE2E1 is comparable with those previously reported for Hdm2 and p53 peptides (Fig. 2A) (24).
FIGURE 2.
Molecular analysis of the USP7-NTD-UbE2E1 interaction A, dissociation constants between USP7-NTD and UbE2E1 peptides were measured by intrinsic tryptophan fluorescence assays. B, the crystal structure of the USP7-NTD·UbE2E1ASTS complex. A ribbon representation of the structure of USP7-NTD (green) with UbE2E1ASTS in stick form (yellow) is shown. The 2Fo − Fc electron density map showing the UbE2E1ASTS peptide is contoured at 1σ. C, the molecular details of the interaction between USP7-NTD and UbE2E1ASTS are shown in stick format with the same color scheme as in B. Hydrogen bonds are indicated by black dashed lines. D, superimposition of UbE2E1ASTS (yellow) with the peptide from viral interferon regulatory factor protein 4, vIRF4ASTS (salmon), showing the similarity in mode of binding within the USP7 peptide binding region.
To gain further insight into the mode of interaction between USP7 and UbE2E1, USP7-NTD was co-crystallized with the UbE2E1 5DSRASTSSSS14 peptide. The crystal structure of this USP7-NTD·UbE2E1ASTS complex was determined using molecular replacement. The overall structure of USP7-NTD has previously been described (31). Briefly, USP7-NTD forms an eight-stranded antiparallel β-sandwich identical to the TRAF domain found in receptor-associated factors of tumor necrosis factor (32). The UbE2E1 peptide binds within a groove on the USP7-NTD surface adjacent to the β-sheet formed by strands β2, β3, β4, and β7 as previously seen with other USP7 binding peptides (Fig. 2B). The final model of USP7-NTD·UbE2E1ASTS was refined to an Rwork of 0.197 and an Rfree of 0.217 at 1.75 Å resolution with 75 water molecules. Residues 54–62 and 106–111 are disordered and were not built into the final model of the protein·peptide complex. The UbE2E1ASTS peptide forms several interactions with USP7-NTD β-strand 7 (Fig. 2C). The side chain hydroxyl of UbE2E1 Ser11 makes the most contacts with USP7-NTD by forming H-bonds with the side and main chain of Asp164. Ala8 of UbE2E1 participates in van der Waals interactions with the side chains of USP7-NTD Trp165 and Phe167. The side chain hydroxyl of UbE2E1 Ser9 makes a water-mediated hydrogen bond to the main chain amide of USP7-NTD Ser168. The main chain amide and carbonyl groups of UbE2E1 Ser9 interact with main chain amide and carbonyl groups of USP7-NTD Gly166. UbE2E1 Thr10 does not make any interactions with USP7-NTD. The electron density for the remainder of the peptide is disordered, suggesting that it is not making any contacts with USP7-NTD. This is the second instance of a peptide containing an ASTS motif interacting with USP7-NTD. Viral interferon regulatory factor 4 protein (vIRF4) also interacts with USP7-NTD through an ASTS motif; however, the nature of the interaction between the vIRF4 peptide and USP7-NTD involved more residues than that seen between USP7-NTD and UbE2E1 (33). Comparison of the UbE2E1 and vIRF4 peptides indicated that the ASTS motif was completely superimposable and that the contacts made by ASTS from each peptide to USP7 were identical (Fig. 2D).
Interaction of USP7 and UbE2E1 in Vivo
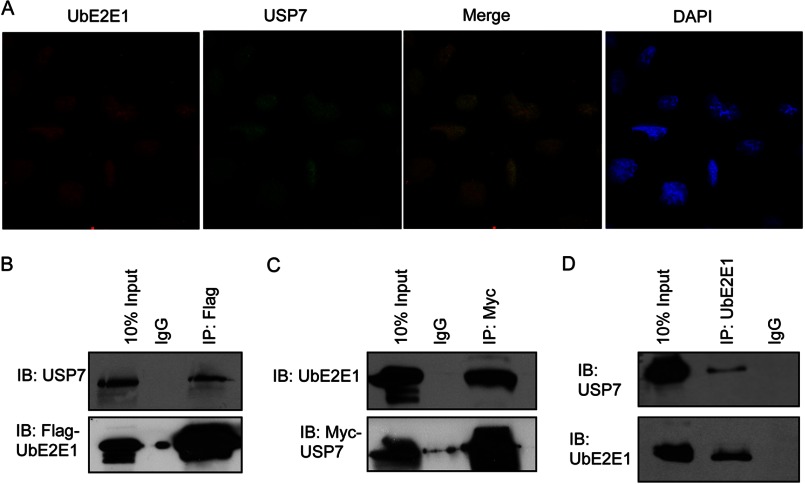
We examined the subcellular localization of USP7 and UbE2E1 in U2OS cells. Endogenous USP7 and UbE2E1 were visualized by immunofluorescence using antibodies specific for USP7 and UbE2E1. Both USP7 and UbE2E1 exhibited predominantly nuclear staining (Fig. 3A). Similar results were obtained when studying USP7 and UbE2E1 subcellular localization in HeLa cells. The immunostaining was specific for both USP7 and UbE2E1; staining for both proteins was negligible in IgG control experiments and in experiments in which USP7 and UbE2E1 antibodies were incubated with purified USP7 or UbE2E1, respectively (data not shown). Immunoprecipitation experiments were conducted in HeLa cells to study the in vivo interaction between USP7 and UbE2E1. HeLa lysate transfected with FLAG-tagged UbE2E1 was immunoprecipitated with an anti-FLAG antibody and blotted for endogenous USP7. USP7 was readily detected in the complex of FLAG-tagged UbE2E1 (Fig. 3B). A reciprocal experiment was conducted using lysate of HeLa cells ectopically expressing Myc-tagged USP7. Endogenous UbE2E1 was immunoprecipitated with Myc-USP7 (Fig. 3C). This interaction was further validated using immunoprecipitation of endogenous USP7 and UbE2E1 from untransfected HeLa cells. Endogenous USP7 was readily detected after immunoprecipitation with anti-UbE2E1 but not with IgG alone (Fig. 3D). All of the above results suggest that USP7 and UbE2E1 interact in vivo.
FIGURE 3.
USP7 and UbE2E1 localization and interaction in vivo. A, immunofluorescence images of U2OS cells are shown after staining for endogenous UbE2E1 (Cy-3) and USP7 (Alexa Fluor 488) proteins. The merged image indicates a partially overlapping localization of USP7 and UbE2E1 in the nucleus. The nuclei were counterstained with DAPI. B, 293T cells transfected with FLAG-UbE2E1 were subject to immunoprecipitation (IP) using mouse IgG (negative control) and anti-FLAG followed by immunoblotting (IB) using antibodies against USP7 and UbE2E1. C, 293T cells transfected with Myc-USP7 were subject to immunoprecipitation using rabbit IgG and anti-Myc followed by immunoblotting against USP7 and UbE2E1 D, endogenous UbE2E1 from 293T cells was immunoprecipitated with a monoclonal antibody against UbE2E1 or mouse IgG followed by immunoblotting with polyclonal rabbit antibodies against USP7 or UbE2E1.
USP7 Attenuates UbE2E1-mediated Ubiquitination
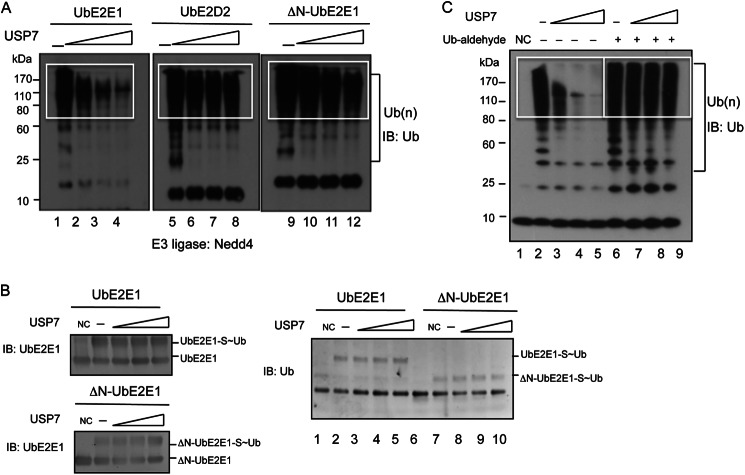
The effect of USP7 on UbE2E1-mediated ubiquitination was tested using in vitro ubiquitination assays. NEDD-4L is a UbE2E1-interacting E3 ligase and known to catalyze ubiquitination in its presence (34–36). The level of total ubiquitination is greatly reduced in the presence of increasing amounts of full-length USP7 protein (Fig. 4A, lanes 1–4). However, when these assays were repeated using UbE2D2/UbcH5b, an E2 enzyme lacking the N-terminal extension seen in UbE2E1, no difference in the total level of ubiquitination in the presence of increasing amounts of USP7 was observed (Fig. 4A, lanes 5–8). To confirm that the decreased levels of total ubiquitination were due to the interaction between USP7 and UbE2E1, the in vitro ubiquitination assays were repeated using ΔN-UbE2E1, which lacks the N-terminal extension sequence and is unable to interact with USP7. Although ΔN-UbE2E1 was active in mediating ubiquitination, USP7 did not show any effect on the total level of ubiquitination as was seen with full-length UbE2E1 (Fig. 4A, lanes 9–12), suggesting that USP7 modulates UbE2E1-mediated ubiquitination through the interaction between the N-terminal domain of USP7 and the UbE2E1 N-terminal extension. In contrast to the effect of full-length USP7, increasing amounts of USP7-NTD did not have any effects on the level of ubiquitination in the presence of full-length UbE2E1 or ΔN-UbE2E1 (data not shown). The effect of USP7 on UbE2E1-mediated ubiquitination was further investigated using ITCH as an E3 ligase. Similar results were obtained, suggesting that the effect of USP7 on UbE2E1 is E3-independent (data not shown). Collectively, these results indicate that the interaction between USP7 and UbE2E1 attenuates the total levels of UbE2E1-mediated ubiquitination and that this effect requires an intact USP7 protein rather than USP7-NTD alone.
FIGURE 4.
USP7 attenuates UbE2E1-mediated ubiquitination. A, in vitro ubiquitination assays were performed using UbE2E1, UbE2D2, or ΔN-UbE2E1 (as the E2) in the presence of ATP, E1, Ub, the catalytic domain of NEDD4 (as the E3 ligase), and increasing amounts of USP7 (0–2 μg). Total ubiquitinated products (Ub(n))were detected by immunoblotting (IB) using a specific antibody against ubiquitin. Polyubiquitinated products were highlighted in boxes. B, ubiquitin loading assays were carried out in a reaction containing E1, His6-Ub, and ATP with increasing amounts of USP7 (0–2 μg) in the presence of 1 μg of full-length UbE2E1 or ΔN-UbE2E1. The UbE2E2-S∼Ub and ΔN-UbE2E2-S∼Ub intermediates were visualized by immunoblotting, using anti-UbE2E1 (left panel) or anti-His6 epitope tag (right panel). C, in vitro ubiquitination assays were performed as in A with the addition of active USP7 (lanes 3–5) or USP7 pretreated with ubiquitin-aldehyde (lanes 7–9), which blocks the catalytic activity of USP7. NC (negative control) represents a standard ubiquitination reaction in the absence of ATP.
To examine whether the decreased levels of total ubiquitination were due to compromised ubiquitin loading of UbE2E1 by USP7, the ability of full-length UbE2E1 and ΔN-UbE2E1 to load ubiquitin was tested in the presence of E1 and ATP with increasing amounts of USP7. As shown in Fig. 4B, both full-length UbE2E1 and ΔN-UbE2E1 were able to load ubiquitin and the amount of loaded ubiquitin was not affected by the addition of USP7. Therefore, USP7 does not affect UbE2E1 ubiquitin loading.
To test whether the deubiquitination activity of USP7 was required for the observed reduction of UbE2E1-mediated ubiquitination, ubiquitin aldehyde was used to inhibit USP7 catalytic activity in an in vitro ubiquitination assay. As shown in Fig. 4C, untreated USP7 attenuated UbE2E1-mediated ubiquitination in a dose-dependent manner, whereas ubiquitin aldehyde-treated USP7 had no effect on ubiquitination. Similar results were obtained when USP7 was inactivated by preincubation with NEM, an alkylating reagent that irreversibly blocks cysteine thiol groups. NEM effectively abolishes the effect of USP7 on UbE2E1-mediated ubiquitination (data not shown). Collectively, these results indicate that the catalytic activity of USP7 is essential for the attenuation of UbE2E1 ubiquitination by USP7.
USP7 Deubiquitinates UbE2E1
Because UbE2E1 has been shown to undergo polyubiquitination in in vitro ubiquitination assays (37), we tested whether USP7 affects UbE2E1-specific polyubiquitination in vitro. Consistent with the previous study, polyubiquitinated UbE2E1 was detected in the in vitro ubiquitination assays using a UbE2E1-specific antibody. The addition of USP7 reduced the levels of polyubiquitinated UbE2E1, even at the lowest tested concentration of USP7 (Fig. 5A). This suggests that UbE2E1 is a substrate of USP7 and that USP7 can deubiquitinate UbE2E1 in vitro. It has previously been shown that E2s are subject to regulation by ubiquitination, which leads to their degradation by the 26 S proteasome (38–40). Therefore, we wanted to establish whether UbE2E1 was polyubiquitinated and degraded in vivo. High molecular weight species indicative of UbE2E1 polyubiquitination were observed in the presence of the proteasomal inhibitor, MG-132, but not in its absence (Fig. 5B). The ability of USP7 to deubiquitinate UbE2E1 was further confirmed using Myc-tagged USP7 and a catalytically inactive mutant, Myc-tagged USP7-C233S (USP7-CS (13)), both expressed and purified from 293T cells. Polyubiquitinated UbE2E1 was enriched in the cells using the proteasome inhibitor MG-132, purified using a UbE2E1-specific antibody, and incubated with USP7 or USP7-CS. USP7 but not USP7-CS catalyzed the deubiquitination of UbE2E1 (Fig. 5C). These data strongly support that UbE2E1 is a bona fide deubiquitination substrate of USP7.
FIGURE 5.
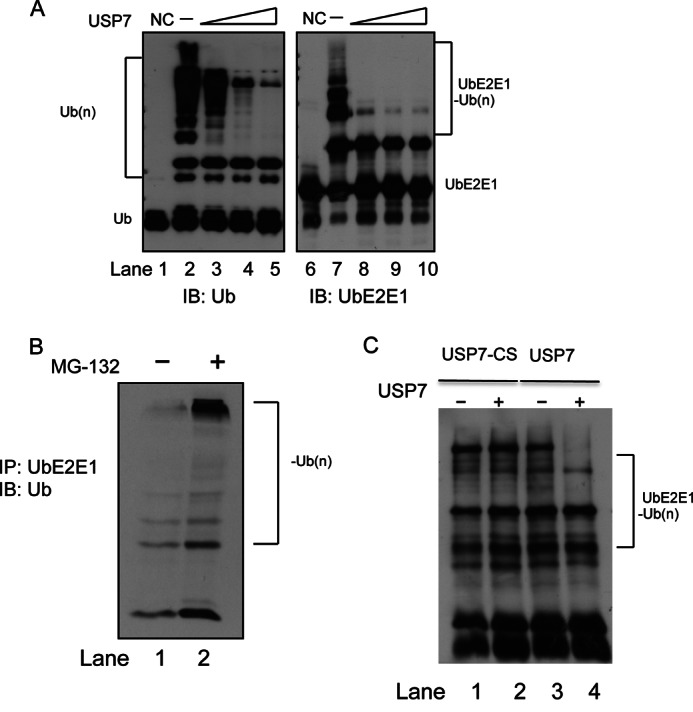
USP7 deubiquitinates UbE2E1. A, in vitro ubiquitination assays were performed as in Fig. 4A with increasing amounts of USP7. Total ubiquitination (Ub(n)) was detected using anti-Ub (left panel) and ubiquitinated UbE2E1 was detected using anti-UbE2E1 (right panel). NC (negative control) represents a standard ubiquitination reaction in the absence of ATP. IB, immunoblot. B, UbE2E1 is subject to ubiquitination and accumulated in the presence of MG132 in U2OS cells. Ubiquitinated UbE2E1 was immunoprecipitated (IP) and visualized by immunoblot using anti-ubiquitin. C, ubiquitinated UbE2E1 was subject to USP7 deubiquitination using USP7 or USP7-CS and visualized by immunoblot using anti-ubiquitin.
USP7 Is Important for UbE2E1 Stabilization in Cells
USP7 regulates the stability of many of its substrates by removing polyubiquitin chains and rescuing these substrates from proteasomal degradation. The fact that UbE2E1 is a substrate of USP7 prompted us to investigate whether USP7 regulates UbE2E1 stability in cells. We monitored UbE2E1 protein turnover in U2OS cells transfected with either control siRNA (siControl) or siRNA for USP7 (siUSP7). The cells were treated with cycloheximide to block de novo protein synthesis or left untreated (Fig. 6A). When compared with control siRNA transfection, transfection with USP7 siRNA noticeably decreased the level of endogenous USP7 (Fig. 6A, top panel). USP7 knockdown resulted in much faster UbE2E1 turnover when compared with control cells (Fig. 6A, middle panel), suggesting that USP7 is important for UbE2E1 stability. We also examined the turnover of UbE2E1 in the wild type colorectal carcinoma cell line HCT116 and a derivative cell line HCT116 USP7−/−, in which the USP7 gene is deleted (Fig. 6B, top panel). Consistent with our observations in U2OS cells, we found UbE2E1 to be stable for over 24 h in the WT HCT116 cells. However, in the HCT116 USP7−/− cells, UbE2E1 turned over much faster and was undetectable within 24 h (Fig. 6B, middle panel). It was also noted that the starting level of UbE2E1 was much lower in the absence of USP7. These observations show that USP7 is critical for the stability of UbE2E1 protein in vivo.
FIGURE 6.
USP7 stabilizes UbE2E1 in vivo. A, U2OS cells were transfected with nonspecific control siRNA (siControl) or siRNA targeting USP7 (siUSP7) and treated with 10 μg/ml cycloheximide (CHX) for the indicated number of hours or left untreated (lane 0). Cell lysates were subjected to SDS-PAGE and Western blot analysis using antibodies indicated. B, WT HCT116 cells or HCT116 USP7−/− were treated with cycloheximide and harvested for Western blotting as in A. The levels of UbE2E1 were normalized using actin as a loading control and presented as line graphs in the cycloheximide chase experiments. The asterisk represents the statistical analysis comparing levels of UbE2E1 after siRNA treatment with p < 0.01. Error bars indicate S.D. C, WT USP7 and the catalytically inactive mutant C223S, USP7-CS, were transfected into HCT116 USP7−/− cells. Cells were lysed and blotted for UbE2E1 48 h after transfection. The levels of UbE2E1 were normalized using actin as a loading control and presented in a bar graph (bottom). The asterisk represents the statistical analysis comparing levels of UbE2E1 after USP7 rescue with p < 0.01. Error bars indicate S.D.
To further confirm that the observed decrease in UbE2E1 stability in the HCT116 USP7−/− cells was due to the lack of USP7 expression, we introduced USP7, USP7-CS, and USP7-DW into HCT116 USP7−/− cells. Expression of USP7, USP7-CS, and USP7-DW was readily detectable by Western blotting using an antibody against USP7, 48 h after transfection (Fig. 6C, top panel). When compared with untransfected cells, cells with ectopic USP7 expression showed a recovery in UbE2E1 levels (Fig. 6C, middle panel, compare lanes 1 and 2). This suggested that the lack of USP7 expression is in large part responsible for the lower stability of UbE2E1 in the HCT116 USP7−/− cells. Interestingly, expression of USP7-CS led to a further decrease in detectable UbE2E1 protein below the background levels observed in untransfected HCT116 USP7−/− cells (Fig. 6C, middle panel, compare lanes 1 and 3). Expression of USP7-DW showed similar results to USP7-CS, whereas levels of UbE2E1 were below background levels (Fig. 6C, middle panel, compare lanes 1 and 4). It should be noted that transfection efficiency was observed to be about 50%; thus the effect of USP7 on UbE2E1 levels would likely be even greater if the sample did not include untransfected cells.
DISCUSSION
The N-terminal domain of USP7 binds p53, Hdm2, and HdmX through distinct regions bearing the sequence motif (P/A)XXS, which make contact with a shallow groove lined by the β7 strand residues 164DWGF167 on the surface of USP7-NTD. Through biochemical, structural, and mutational analysis, USP7 was found to interact with the 8ASTS11 motif harbored in the N-terminal extension of UbE2E1. GST pulldown assays indicated that USP7-NTD but not USP7-NTDDW was able to interact with UbE2E1. Conversely, UbE2E1 lacking its N-terminal extension was unable to interact with GST-USP7-NTD. The interactions formed between USP7 and UBE2E1 are similar to the previously described USP7·p53, USP7·Hdm2, and USP7·HdmX complex structures (24, 25, 31). Specifically, residue Ala8 of the (P/A)XXS motif within UbE2E1 makes contact with Trp165 of USP7, and Ser11 forms two H-bonds with Asp164 of USP7. Indeed, mutations at the Ser11 residue of 8ASTS11 in UbE2E1 or at the Asp and Trp residues of 164DWGF167 in USP7 abolish the interaction, suggesting that USP7-NTD 164DWGF167 is a general substrate binding site, responsible for recognizing a broad range of substrates including UbE2E1. This binding site is also targeted by the viral proteins, EBNA1 (31) and vIRF4 (33), to suppress USP7 function.
The effect of the USP7 UbE2E1 interaction on UbE2E1 was investigated by in vitro ubiquitination assays. UbE2E1-mediated polyubiquitination was found to be susceptible to the deubiquitination activity of USP7. This effect of USP7 was contingent upon the ability of USP7 to physically interact with UbE2E1 as USP7 was unable to affect the activity of ΔN-UbE2E1 and UbE2D2, both shown to be lacking USP7 binding. This observation suggests that USP7 has to be in complex with UbE2E1 to confer its regulatory effect rather than nonspecifically breaking down polyubiquitin chains formed in the ubiquitination reaction. USP7 did not negatively affect the ubiquitin loading activity of UbE2E1. Thus USP7 was conjectured to be attenuating UbE2E1 activity through noncanonical steric hindrance or through its deubiquitination activity. To explore these possibilities, we conducted ubiquitination experiments in the presence or absence of USP7 incubated with two different inhibitors, NEM and ubiquitin aldehyde, to render USP7 inactive. These experiments showed that USP7 catalytic activity is critical and required to negatively regulate UbE2E1 activity. The outcome of the USP7 UbE2E1 interaction offers an interesting contrast to the interaction between OTUB-1 and UbE2N, another DUB and E2, respectively. OTUB-1 binds to UBE2N and inhibits UBE2N-mediated ubiquitination (41). However, this inhibition does not require OTUB-1 catalytic activity because OTUB-1 prevents polyubiquitination through physical interaction and sequestering of Ub-charged UbE2N.
Because USP7 cleaves polyubiquitin from UbE2E1 and polyubiquitination of proteins in cells is generally associated with proteasomal degradation, this observation implies that USP7 plays a role in regulating UbE2E1 stability in cells. This role of USP7 was validated using both siRNA and gene knock-out approaches. Partial silencing of USP7 expression in U2OS cells led to a measurable decrease in the protein levels of UbE2E1. Lack of USP7 expression in HCT116 USP7−/− cells was accompanied by a substantial reduction in both levels and stability of the UbE2E1 protein. When USP7 was reintroduced using ectopic expression into HCT116 USP7−/− cells, UbE2E1 levels increased. Taken together, these observations are consistent with the idea that USP7 stabilizes UbE2E1. Note that neither partial silencing nor complete ablation of USP7 expression led to a complete absence of UbE2E1 from cells, suggesting that factors other than USP7 may also contribute to the stability of UbE2E1. This possibility is supported by our data that the expression of USP7-CS or USP7-DW in HCT116 USP7−/− cells led to a further decrease in UbE2E1 levels when compared with mock-transfected control cells. This suggests that other factors may contribute to the stability of UbE2E1; however, our observations clearly show USP7 to be a key regulator of UbE2E1 stability.
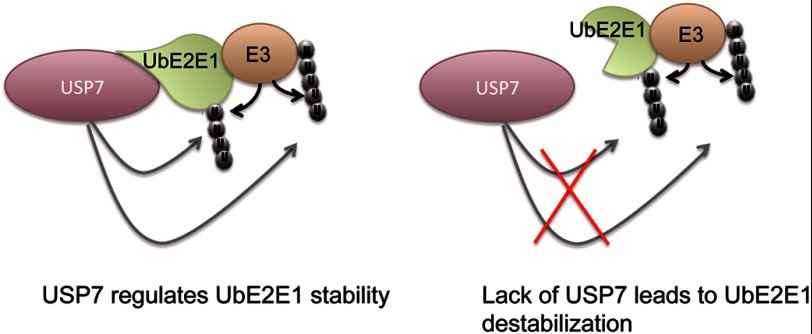
In this study, we describe a novel interaction between the class III ubiquitin conjugating enzyme UbE2E1 and the deubiquitinase USP7. Our study revealed that USP7-NTD interacts with the N-terminal extension of UbE2E1 and that the co-crystal structure of USP7-NTD with a UbE2E1 peptide resembles the complex structures of USP7 with its other three substrates, p53, Hdm2, and HdmX. The biological significance of the interaction between USP7 and UbE2E1 was also investigated. We showed that USP7 specifically attenuates UbE2E1-mediated ubiquitination and plays an important role in regulating the stability of UbE2E1 in cells (Fig. 7). To the best of our knowledge, this represents the first example of a DUB regulating the stability of an E2 enzyme and provides new insights into the complex network of the ubiquitin-proteasome pathway.
FIGURE 7.
USP7 regulates the stability of UbE2E1. USP7 attenuates UbE2E1-mediated total ubiquitination and stabilizes UbE2E1 through an interaction between USP7-NTD and the N-terminal extension of UbE2E1. Inactivation or disruption of the interaction between USP7 and UbE2E1 leads to UbE2E1 destabilization.
Although our studies were limited to the interaction between USP7 and UbE2E1, it is noteworthy that UbE2E2 and UbE2E3, members of the UbE2E subfamily, also contain a (P/A)XXS motif in their N-terminal extensions. We predict that USP7 will also interact with and regulate the levels of UbE2E2 and UbE2E3 as it does UbE2E1. Our studies with UbE2E1 as a representative of this subfamily clearly establish USP7 as an important regulator of the UbE2Es. E2 ubiquitination is often observed in vitro, but not in vivo. For an efficient ubiquitination machinery to target substrates, instead of targeting itself for degradation, cells need to develop a self-protecting and self-rescuing mechanism built into the ubiquitination machinery. In this regard, USP7 serves as a guardian of UbE2E1, ensuring its stability and availability for its biological function(s).
Acknowledgments
We thank Dr. Bert Vogelstein for generously providing the HCT116 USP7−/− and the HCT116 parental cell lines. We thank Dr. Sirano Dhe-Paganon from Structural Genomics Consortium (SGC) Toronto for the HECT domain E3 ligase constructs (NEDD-4L and ITCH). Infrastructure was funded by the Canada Foundation for Innovation (CFI) Leaders Opportunity Fund Program and the Ontario Research Fund Research Infrastructure Program (ORF-RI). Results shown in this study are derived from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. The Argonne National Laboratory is operated by the University of Chicago, Argonne, LLC, for the United States Department of Energy, Office of Biological and Environmental Research under Contract DE-AC02-06CH11357.
This work was supported by Operating Grant 106583 from the Canadian Institutes of Health Research (CIHR) (to V. S. and Y. S.) as well as York University startup funds (to V. S. and Y. S.).
The atomic coordinates and structure factors (code 4JJQ) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- UBC
- ubiquitin-conjugating
- Ub
- ubiquitin
- DUB
- deubiquitinating enzyme
- NTD
- N-terminal domain
- NEM
- N-ethylmaleimide
- vIRF4
- viral interferon regulatory factor 4 protein
- r.m.s.d.
- root mean square deviation.
REFERENCES
- 1. Pickart C. M. (2001) Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70, 503–533 [DOI] [PubMed] [Google Scholar]
- 2. Schwartz A. L., Ciechanover A. (2009) Targeting proteins for destruction by the ubiquitin system: implications for human pathobiology. Annu. Rev. Pharmacol. Toxicol. 49, 73–96 [DOI] [PubMed] [Google Scholar]
- 3. Jin J., Li X., Gygi S. P., Harper J. W. (2007) Dual E1 activation systems for ubiquitin differentially regulate E2 enzyme charging. Nature 447, 1135–1138 [DOI] [PubMed] [Google Scholar]
- 4. van Wijk S. J., Timmers H. T. (2010) The family of ubiquitin-conjugating enzymes (E2s): deciding between life and death of proteins. FASEB J. 24, 981–993 [DOI] [PubMed] [Google Scholar]
- 5. Deshaies R. J., Joazeiro C. A. (2009) RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 [DOI] [PubMed] [Google Scholar]
- 6. Block K., Appikonda S., Lin H. R., Bloom J., Pagano M., Yew P. R. (2005) The acidic tail domain of human Cdc34 is required for p27Kip1 ubiquitination and complementation of a cdc34 temperature sensitive yeast strain. Cell Cycle 4, 1421–1427 [DOI] [PubMed] [Google Scholar]
- 7. Summers M. K., Pan B., Mukhyala K., Jackson P. K. (2008) The unique N terminus of the UbcH10 E2 enzyme controls the threshold for APC activation and enhances checkpoint regulation of the APC. Mol. Cell 31, 544–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Seufert W., Jentsch S. (1990) Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO J. 9, 543–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. van Wijk S. J., de Vries S. J., Kemmeren P., Huang A., Boelens R., Bonvin A. M., Timmers H. T. (2009) A comprehensive framework of E2-RING E3 interactions of the human ubiquitin-proteasome system. Mol. Syst. Biol. 5, 295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Plafker K. S., Singer J. D., Plafker S. M. (2009) The ubiquitin conjugating enzyme, UbcM2, engages in novel interactions with components of cullin-3 based E3 ligases. Biochemistry 48, 3527–3537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhu B., Zheng Y., Pham A. D., Mandal S. S., Erdjument-Bromage H., Tempst P., Reinberg D. (2005) Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol. Cell 20, 601–611 [DOI] [PubMed] [Google Scholar]
- 12. Nijman S. M., Luna-Vargas M. P., Velds A., Brummelkamp T. R., Dirac A. M., Sixma T. K., Bernards R. (2005) A genomic and functional inventory of deubiquitinating enzymes. Cell 123, 773–786 [DOI] [PubMed] [Google Scholar]
- 13. Li M., Chen D., Shiloh A., Luo J., Nikolaev A. Y., Qin J., Gu W. (2002) Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature 416, 648–653 [DOI] [PubMed] [Google Scholar]
- 14. Cummins J. M., Vogelstein B. (2004) HAUSP is required for p53 destabilization. Cell Cycle 3, 689–692 [PubMed] [Google Scholar]
- 15. Meredith M., Orr A., Everett R. (1994) Herpes simplex virus type 1 immediate-early protein Vmw110 binds strongly and specifically to a 135-kDa cellular protein. Virology 200, 457–469 [DOI] [PubMed] [Google Scholar]
- 16. Oh Y. M., Yoo S. J., Seol J. H. (2007) Deubiquitination of Chfr, a checkpoint protein, by USP7/HAUSP regulates its stability and activity. Biochem. Biophys. Res. Commun. 357, 615–619 [DOI] [PubMed] [Google Scholar]
- 17. Nathan J. A., Sengupta S., Wood S. A., Admon A., Markson G., Sanderson C., Lehner P. J. (2008) The ubiquitin E3 ligase MARCH7 is differentially regulated by the deubiquitylating enzymes USP7 and USP9X. Traffic 9, 1130–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Qing P., Han L., Bin L., Yan L., Ping W. X. (2011) USP7 regulates the stability and function of HLTF through deubiquitination. J. Cell. Biochem. 112, 3856–3862 [DOI] [PubMed] [Google Scholar]
- 19. Khoronenkova S. V., Dianov G. L. (2013) USP7S-dependent inactivation of Mule regulates DNA damage signalling and repair. Nucleic Acids Res. 41, 1750–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Faustrup H., Bekker-Jensen S., Bartek J., Lukas J., Mailand N. (2009) USP7 counteracts SCFβTrCP- but not APCCdh1-mediated proteolysis of Claspin. J. Cell Biol. 184, 13–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. van der Knaap J. A., Kumar B. R., Moshkin Y. M., Langenberg K., Krijgsveld J., Heck A. J., Karch F., Verrijzer C. P. (2005) GMP synthetase stimulates histone H2B deubiquitylation by the epigenetic silencer USP7. Mol. Cell 17, 695–707 [DOI] [PubMed] [Google Scholar]
- 22. Song M. S., Salmena L., Carracedo A., Egia A., Lo-Coco F., Teruya-Feldstein J., Pandolfi P. P. (2008) The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature 455, 813–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sarkari F., Wang X., Nguyen T., Frappier L. (2011) The herpesvirus associated ubiquitin specific protease, USP7, is a negative regulator of PML proteins and PML nuclear bodies. PLoS One 6, e16598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sheng Y., Saridakis V., Sarkari F., Duan S., Wu T., Arrowsmith C. H., Frappier L. (2006) Molecular recognition of p53 and MDM2 by USP7/HAUSP. Nat. Struct. Mol. Biol. 13, 285–291 [DOI] [PubMed] [Google Scholar]
- 25. Sarkari F., La Delfa A., Arrowsmith C. H., Frappier L., Sheng Y., Saridakis V. (2010) Further insight into substrate recognition by USP7: structural and biochemical analysis of the HdmX and Hdm2 interactions with USP7. J. Mol. Biol. 402, 825–837 [DOI] [PubMed] [Google Scholar]
- 26. Brunger A. T. (2007) Version 1.2 of the Crystallography and NMR system. Nat. Protoc. 2, 2728–2733 [DOI] [PubMed] [Google Scholar]
- 27. Jones T. A., Kjeldgaard M. (1997) Electron-density map interpretation. Methods Enzymol. 277, 173–208 [DOI] [PubMed] [Google Scholar]
- 28. Sheng Y., Hong J. H., Doherty R., Srikumar T., Shloush J., Avvakumov G. V., Walker J. R., Xue S., Neculai D., Wan J. W., Kim S. K., Arrowsmith C. H., Raught B., Dhe-Paganon S. (2012) A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen. Mol. Cell Proteomics 11, 329–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tang J., Qu L. K., Zhang J., Wang W., Michaelson J. S., Degenhardt Y. Y., El-Deiry W. S., Yang X. (2006) Critical role for Daxx in regulating Mdm2. Nat. Cell Biol. 8, 855–862 [DOI] [PubMed] [Google Scholar]
- 30. Sarkari F., Sanchez-Alcaraz T., Wang S., Holowaty M. N., Sheng Y., Frappier L. (2009) EBNA1-mediated recruitment of a histone H2B deubiquitylating complex to the Epstein-Barr virus latent origin of DNA replication. PLoS pathogens 5, e1000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Saridakis V., Sheng Y., Sarkari F., Holowaty M. N., Shire K., Nguyen T., Zhang R. G., Liao J., Lee W., Edwards A. M., Arrowsmith C. H., Frappier L. (2005) Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1 implications for EBV-mediated immortalization. Mol. Cell 18, 25–36 [DOI] [PubMed] [Google Scholar]
- 32. Park Y. C., Ye H., Hsia C., Segal D., Rich R. L., Liou H. C., Myszka D. G., Wu H. (2000) A novel mechanism of TRAF signaling revealed by structural and functional analyses of the TRADD-TRAF2 interaction. Cell 101, 777–787 [DOI] [PubMed] [Google Scholar]
- 33. Lee H. R., Choi W. C., Lee S., Hwang J., Hwang E., Guchhait K., Haas J., Toth Z., Jeon Y. H., Oh T. K., Kim M. H., Jung J. U. (2011) Bilateral inhibition of HAUSP deubiquitinase by a viral interferon regulatory factor protein. Nat. Struct. Mol. Biol. 18, 1336–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fotia A. B., Cook D. I., Kumar S. (2006) The ubiquitin-protein ligases Nedd4 and Nedd4–2 show similar ubiquitin-conjugating enzyme specificities. Int. J. Biochem. Cell Biol. 38, 472–479 [DOI] [PubMed] [Google Scholar]
- 35. Anan T., Nagata Y., Koga H., Honda Y., Yabuki N., Miyamoto C., Kuwano A., Matsuda I., Endo F., Saya H., Nakao M. (1998) Human ubiquitin-protein ligase Nedd4: expression, subcellular localization and selective interaction with ubiquitin-conjugating enzymes. Genes Cells 3, 751–763 [DOI] [PubMed] [Google Scholar]
- 36. Debonneville C., Staub O. (2004) Participation of the ubiquitin-conjugating enzyme UBE2E3 in Nedd4–2-dependent regulation of the epithelial Na+ channel. Mol. Cell. Biol. 24, 2397–2409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. David Y., Ziv T., Admon A., Navon A. (2010) The E2 ubiquitin-conjugating enzymes direct polyubiquitination to preferred lysines. J. Biol. Chem. 285, 8595–8604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yamanaka A., Hatakeyama S., Kominami K., Kitagawa M., Matsumoto M., Nakayama K. (2000) Cell cycle-dependent expression of mammalian E2-C regulated by the anaphase-promoting complex/cyclosome. Mol. Biol. Cell 11, 2821–2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Banerjee A., Gregori L., Xu Y., Chau V. (1993) The bacterially expressed yeast CDC34 gene product can undergo autoubiquitination to form a multiubiquitin chain-linked protein. J. Biol. Chem. 268, 5668–5675 [PubMed] [Google Scholar]
- 40. Seol J. H., Feldman R. M., Zachariae W., Shevchenko A., Correll C. C., Lyapina S., Chi Y., Galova M., Claypool J., Sandmeyer S., Nasmyth K., Deshaies R. J. (1999) Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 13, 1614–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nakada S., Tai I., Panier S., Al-Hakim A., Iemura S., Juang Y. C., O'Donnell L., Kumakubo A., Munro M., Sicheri F., Gingras A. C., Natsume T., Suda T., Durocher D. (2010) Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature 466, 941–946 [DOI] [PubMed] [Google Scholar]