Background: The Far3-7-8-9-10-11 complex, part of the yeast striatin-interacting phosphatase and kinase (STRIPAK) complex, mediates target of rapamycin complex 2 (TORC2) signaling.
Results: The Far3-7-8-9-10-11 complex follows tiered assembly at the endoplasmic reticulum (ER).
Conclusion: ER localization of Far9 is required for optimal function in TORC2 signaling.
Significance: Our study provides insights into the organization of the yeast STRIPAK complex.
Keywords: Endoplasmic Reticulum (ER), PP2A, Protein Complexes, TOR Complex (TORC), Yeast, Far3-7-8-9-10-11, STRIPAK Complex, Saccharomyces cerevisiae, TORC2 Signaling, Tail-anchored Proteins
Abstract
Target of rapamycin signaling is a conserved, essential pathway integrating nutritional cues with cell growth and proliferation. The target of rapamycin kinase exists in two distinct complexes, TORC1 and TORC2. It has been reported that protein phosphatase 2A (PP2A) and the Far3-7-8-9-10-11 complex (Far complex) negatively regulate TORC2 signaling in yeast. The Far complex, originally identified as factors required for pheromone-induced cell cycle arrest, and PP2A form the yeast counterpart of the STRIPAK complex, which was first isolated in mammals. The cellular localization of the Far complex has yet to be fully characterized. Here, we show that the Far complex localizes to the endoplasmic reticulum (ER) by analyzing functional GFP-tagged Far proteins in vivo. We found that Far9 and Far10, two homologous proteins each with a tail-anchor domain, localize to the ER in mutant cells lacking the other Far complex components. Far3, Far7, and Far8 form a subcomplex, which is recruited to the ER by Far9/10. The Far3-7-8- complex in turn recruits Far11 to the ER. Finally, we show that the tail-anchor domain of Far9 is required for its optimal function in TORC2 signaling. Our study reveals tiered assembly of the yeast Far complex at the ER and a function for Far complex's ER localization in TORC2 signaling.
Introduction
Protein phosphorylation plays important roles in many cellular processes. Protein phosphorylation is catalyzed by specific protein kinases, and protein dephosphorylation is carried out by protein phosphatases. Thus, the phosphorylation state of proteins is finely controlled by the opposing activities of protein kinases and phosphatases. Multiple mechanisms exist to fine-tune the activity of these protein kinases and phosphatases through the regulation of their expression levels, their activity, cellular localization, and availability to substrates, among others. Recently, a large multiprotein complex known as the striatin-interacting phosphatase and kinase (STRIPAK)2 complex was found in mammals (Table 1). STRIPAK contains PP2A catalytic and scaffolding subunits, striatins, the striatin-associated protein Mob3, two homologous novel proteins STRIP1 and STRIP2, members of the germinal center kinase III family of Ste20 kinases, and the cerebral cavernous malformation 3 protein (1, 2). The STRIPAK assembly maintains mutually exclusive interactions with either the CTTNBP2 (cortactin-binding protein 2) proteins or a second subcomplex consisting of sarcolemmal membrane-associated protein (SLMAP) and two related coiled-coil proteins SIKE and FGFR1OP2. The N-terminal region of CCM3 mediates heterodimerization with Ste20 kinases, and its C-terminal domain interacts with striatin (3–7), which also interacts with the regulatory and catalytic subunits of PP2A, thus bridging a kinase to a phosphatase. This arrangement likely facilitates the regulation of the activity of protein kinases by PP2A (3, 8). Both Striatin 3 and STRIP1 localize to the Golgi, and depletion of either results in similar defects suggesting they perform similar functions in regulation of Golgi morphology and mitosis (3, 9). The SLMAP gene has several splice variants, encoding tail-anchored membrane proteins that associate with the sarcolemmal membrane in muscle cells, the endoplasmic reticulum and the mitochondrial membrane in nonmuscle cells, and the centrosome and the outer nuclear envelope (9–13). SLMAP is required for myoblast fusion, centrosome function, and structural arrangement of the excitation-contraction coupling apparatus in cardiomyocytes (9, 12, 14, 15). Much remains to be determined about the role of the STRIPAK components, the mechanism of their regulation, and the identity of their substrates.
TABLE 1.
Components of the STRIPAK complex in different species
Only components that are known to form a complex or perform similar cellular functions are listed. Hs, Homo sapiens; Dm, Drosophila melanogaster; Sc, S. cerevisiae, Sp, S. pombe; Sm, S. macrospora; Nc, N. crassa.
| Description | Hs (1, 3) | Dm (2) | Sc (22, 24, 25) | Sp (9, 20, 47) | Sm (18) | Nc (16, 48) |
|---|---|---|---|---|---|---|
| PP2A A subunit | PP2A Aα/Aβ | Pp2A-29B | Tpd3 | Paa1p | SmPP2AA | |
| PP2A B-type subunit | Striatins | Cka | Far8 | Csc3p | PRO11 | ham-3 |
| PP2A C subunit | PP2Acα/cβ | Mts | Pph21/22 | Ppa3p | SmPP2AC | pp2A |
| Striatin-interacting protein | STRIP1/2 | CG11526 | Far11 | Csc2p | PRO22 | ham-2 |
| Tail-anchor domain protein | SLMAP | CG17494 | Far9/10 | Csc1p | ham-4 | |
| Coiled-coil domain protein | SIKE/ FGFR1OP2 | FGOP2 | Far3/7 | Csc4p | ||
| Striatin-associated protein | Mob3 | Mob4 | Mob1p | SmMOB3 | mob3 | |
| Ste20 family kinase | STK24/STK25/Mst4 | GckIII | ||||
| Cerebral cavernous malformation 3 | CCM3 | CG5073 | ||||
| Cortactin-binding protein 2 | CTTNBP2 | CG10915 |
Orthologs of mammalian STRIPAK components have been reported to exist in many eukaryotes (Table 1). The Drosophila STRIPAK complex has been reported to be involved in Hippo signaling by mediating phosphorylation of the Hippo kinase and the transcriptional activator Yorkie (2). In Neurospora crassa, orthologs of STRIPAK complex components are required for hyphal fusion (16, 17). In the ascomycete Sordaria macrospora, the STRIPAK complex is required for sexual development and vegetative hyphal fusion (18, 19). In the fission yeast Schizosaccharomyces pombe, components of the STRIPAK complex localize to the mitotic spindle pole body in early mitosis and are required for the establishment of asymmetry of the septation initiation network, a conserved signaling pathway that is required for cytokinesis and mitotic transitions (9, 20).
In the budding yeast Saccharomyces cerevisiae, the STRIPAK complex has been reported to mediate pheromone signaling, the TORC2 signaling pathway, and the toxicity due to expression of human caspase-10 in yeast (21–24). The yeast STRIPAK complex contains Far3, Far7, Far8, Far9, Far10, Far11, Tpd3 (the A scaffolding subunit of PP2A), and Pph21/22 (the two redundant catalytic subunits of PP2A) (22–26). Far11 is an ortholog of human STRIP1/2; Far8 shares limited sequence similarity with human striatins; Far9 and Far10 are homologous tail-anchored proteins similar to human SLMAP (Table 1) (1, 27). Yeast cells secrete pheromones to induce cell cycle arrest to prepare for mating as part of the fungal life cycle (28). Mutations in FAR genes lead to increased resistance to pheromone-induced cell cycle arrest (22, 29), but the underlying mechanism is still unclear. The targets of rapamycin kinases are conserved in eukaryotes and exist in two distinct multiprotein complexes, TORC1 and TORC2 (30, 31), and mutations in the yeast STRIPAK complex components lead to suppression of cell lethality specifically due to TORC2 deficiency possibly by restoring phosphorylation of TORC2 substrates Slm1, Slm2, Ypk1, and Ypk2 (21, 24, 32, 33). The role of STRIPAK in human caspase-10-induced toxicity in yeast likely results from promoting Atg13 dephosphorylation and subsequent activation of autophagy (23, 34).
In yeast, Far3, Far7, Far8, Far9, Far10, and Far11 have been reported to form a complex (22, 24, 25). However, it is unclear how these proteins assemble together to form the final complex, and identification of the cellular component of this complex could potentially provide insights into the mechanism of its function. Cellular localization of subsets of the Far complex components has been reported in three different studies; however, the results were not consistent (23, 27, 35). A genome-wide study on the localization of yeast proteins C-terminally tagged with green fluorescent protein (GFP) found that Far3, Far7, and Far8 localize to the ER (35). In that study, Far9 was shown to be localized in the cytoplasm; Far10 localization was ambiguous, and there were no data on Far11. In another study, Beilharz et al. (27) showed that N-terminal GFP-tagged Far9 and Far10 localize to the ER and clusters within the bounds of the ER, respectively. In the third reported study on the localization of Far proteins with a C-terminal fluorescent tag, Far11 was reported to co-localize with Chc1, a late-Golgi protein, Far3 with Cop1, an early Golgi protein, and Far9 with Sec13, an ER-to-Golgi protein that is located on ER-derived transport vesicles (23). To gain insights into how the Far proteins assemble into a complex and address the inconsistency in their cellular localization, we constructed functional GFP-tagged Far proteins and analyzed their localization in various far mutants. Our data show that all of the Far proteins localize in a tiered fashion at the endoplasmic reticulum and ER localization of Far9 is required for its optimal function in TORC2 signaling.
EXPERIMENTAL PROCEDURES
Strains, Plasmids, Growth Media, and Growth Conditions
Yeast strains and plasmids used in this study are listed in Tables 2 and 3, respectively. Yeast cells were grown in SD (0.67% yeast nitrogen base plus 2% dextrose), YNBcasD (SD medium plus 1% casamino acids), or YPD (1% yeast extract, 2% peptone, 2% dextrose) medium at temperatures as indicated in the text and in the figure legends. When necessary, amino acids, adenine, and/or uracil were added to the growth medium at standard concentrations to cover auxotrophic requirements (36).
TABLE 2.
S. cerevisiae strains used in this study
| Strain | Genotype | Source | Application |
|---|---|---|---|
| SY2227 (WT) | MATa ade1-1 leu2-2,113 trp1 ura3-52 bar1 HIS3::pFUS1::HIS3 mfa2-1::FUS1-lacZ rad16::pGAL1::STE4 | 22 | |
| TPY1010 (far3) | SY2227 far3::kanMX4 | This study | Figs. 1A, 3A, and 4 |
| TPY1013 (far7) | SY2227 far7::kanMX4 | This study | Figs. 1A and 3B |
| TPY1015 (far8) | SY2227 far8::kanMX4 | This study | Figs. 1A and 3C |
| TPY1048 (far9) | SY2227 far9::kanMX4 | This study | Figs. 1A and 7B |
| TPY1072 (far10) | SY2227 far10::kanMX4 | This study | Figs. 1A and 4 |
| SY4064 (far11) | SY2227 far11::kanMX4 | 22 | Figs. 1A, 3D, and 6 |
| SH100 (WT) | MATa leu2 - 3,112 ura3 - 52 rme1 trp1 his4 HMLa ade2 tor2::ADE2 [YCplac111::TOR2] | 43 | Fig. 7C |
| SH121 (tor2-21) | MATa leu2 - 3,112 ura3 - 52 rme1 trp1 his4 HMLa ade2 tor2::ADE2 [YCplac111::tor2-21] | 43 | Fig. 7C |
| TPY157 (tor2-21 far3) | SH121 far3::kanMX4 | 24 | Fig. 1B |
| TPY147 (tor2-21 far7) | SH121 far7::kanMX4 | 24 | Fig. 1B |
| TPY213 (tor2-21 far8) | SH121 far8::kanMX4 | 24 | Fig. 1B |
| TPY357 (tor2-21 far9) | SH121 far9::kanMX4 | 24 | Figs. 1B and 7, C and D |
| TPY151 (tor2-21 far10) | SH121 far10::kanMX4 | 24 | Fig. 1B |
| TPY116 (tor2-21 far11) | SH121 far11::kanMX4 | 24 | Fig. 1B |
| SY4075 (far sext) | SY2227 far3::LEU2 far7::CgTRP1 far8::URA3 far9::HYGB far10::KAN far11::NAT | 22 | |
| TPY845 (far sext ura3) | SY4075 ura3::kanMX4 | This study | Figs. 2 and 5, A and C |
| TPY1358 (far3 far7) | SY2227 far3::kanMX4 far7::TRP1 [pRS416-FAR3-GFP] | This study | Fig. 3A |
| TPY1361 (far3 far8) | SY2227 far3::kanMX4 far8::TRP1 [pRS416-FAR3-GFP] | This study | Fig. 3A |
| TPY1348 (far3 far9) | SY2227 far3::kanMX4 far9::TRP1 [pRS416-FAR3-GFP] | This study | Fig. 3A |
| TPY1363 (far3 far10) | SY2227 far3::kanMX4 far10::TRP1 [pRS416-FAR3-GFP] | This study | Fig. 3A |
| TPY1402 (far3 far11) | SY2227 far3::kanMX4 far11::TRP1 [pRS416-FAR3-GFP] | This study | Fig. 3A |
| TPY1408 (far7 far3) | SY2227 far7::kanMX4 far3::TRP1 [pRS416-FAR7-GFP] | This study | Fig. 3B |
| TPY1366 (far7 far8) | SY2227 far7::kanMX4 far8::TRP1 [pRS416-FAR7-GFP] | This study | Fig. 3B |
| TPY1350 (far7 far9) | SY2227 far7::kanMX4 far9::TRP1 [pRS416-FAR7-GFP] | This study | Fig. 3B |
| TPY1368 (far7 far10) | SY2227 far7::kanMX4 far10::TRP1 [pRS416-FAR7-GFP] | This study | Fig. 3B |
| TPY1352 (far7 far11) | SY2227 far7::kanMX4 far11::TRP1 [pRS416-FAR7-GFP] | This study | Fig. 3B |
| TPY1369 (far8 far3) | SY2227 far8::kanMX4 far3::TRP1 [pRS416-FAR8-GFP] | This study | Figs. 3C and 5B |
| TPY1370 (far8 far7) | SY2227 far8::kanMX4 far7::TRP1 [pRS416-FAR8-GFP] | This study | Figs. 3C and 5B |
| TPY1351 (far8 far9) | SY2227 far8::kanMX4 far9::TRP1 [pRS416-FAR8-GFP] | This study | Fig. 3C |
| TPY1373 (far8 far10) | SY2227 far8::kanMX4 far10::TRP1 [pRS416-FAR8-GFP] | This study | Fig. 3C |
| TPY1405 (far8 far11) | SY2227 far8::kanMX4 far11::TRP1 [pRS416-FAR8-GFP] | This study | Fig. 3C |
| TPY1374 (far11 far3) | SY2227 far11::kanMX4 far3::TRP1 [pRS416-MKS1-FAR11-GFP] | This study | Fig. 3D |
| TPY1377 (far11 far7) | SY2227 far11::kanMX4 far7::TRP1 [pRS416-MKS1-FAR11-GFP] | This study | Fig. 3D |
| TPY1406 (far11 far8) | SY2227 far11::kanMX4 far8::TRP1 [pRS416-MKS1-FAR11-GFP] | This study | Fig. 3D |
| TPY1410 (far11 far9) | SY2227 far11::kanMX4 far9::TRP1 [pRS416-MKS1-FAR11-GFP] | This study | Fig. 3D |
| TPY1379 (far11 far10) | SY2227 far11::kanMX4 far10::TRP1 [pRS416-MKS1-FAR11-GFP] | This study | Fig. 3D |
| SY4078 (FAR7-myc) | SY2227 FAR7-MYC13-KAN [pSL2771, CEN LEU2] | 22 | Fig. 4 |
| SY4079 (FAR8-myc) | SY2227 FAR8-MYC13-KAN [pSL2771, CEN LEU2] | 22 | Fig. 4 |
| SY4082 (FAR11-myc) | SY2227 FAR11-MYC13-KAN [pSL2771, CEN LEU2] | 22 | Fig. 4 |
| SY4080 (FAR9-myc) | SY2227 FAR9-MYC13-KAN [pSL2771, CEN LEU2] | 22 | |
| TPY1411 (FAR9-myc) | SY4080 without the pSL2771 plasmid | This study | Fig. 6 |
| SY4070 (FAR9-myc far3) | SY2227 far3::LEU2 FAR9-MYC13-KAN [pSL2784, 2μ URA3 FAR3-HA] | 22 | |
| TPY1412 (far3 FAR9-myc) | SY4070 without the pSL2784 plasmid | This study | Fig. 6 |
| TPY1413 (far7 FAR9-myc) | TPY1411 far7::TRP1 | This study | Fig. 6 |
| TPY1416 (far8 FAR9-myc) | TPY1411 far8::TRP1 | This study | Fig. 6 |
| TPY1341 (far9ΔC) | SH121 far9ΔC | This study | Fig. 7C |
TABLE 3.
Plasmids used in this study
| Plasmid | Description | Source | Application |
|---|---|---|---|
| pTP201 | pRS414-SHR3-RFP expressing Shr3 from its own promoter with an RFP tag at the C terminus | This study | Fig. 1B |
| pTP143 | pRS416-FAR3-GFP, expressing Far3 from its own promoter with a GFP tag at the C terminus | This study | Figs. 1, A and B, 2, 3A, 5A |
| pTP164 | pRS416-FAR7-GFP, expressing Far7 from its own promoter with a GFP tag at the C terminus | This study | Figs. 1, A and B, 2, 3B, and 5A |
| pTP131 | pRS416-FAR8-GFP, expressing Far8 from its own promoter with a GFP tag at the C terminus | This study | Figs. 1, A and B, 2, 3C, and 5B |
| pTP179 | pRS416-MKS1-GFP-FAR9, expressing Far9 from the MKS1 promoter with a GFP tag at the N terminus | This study | Figs. 1, A and B, 2, and 7, B and D |
| pTP203 | pRS416-MKS1-GFP-FAR10, expressing Far10 from the MKS1 promoter with a GFP tag at the N terminus | This study | Figs. 1, A and B, and 2 |
| pZL2564 | pRS416-FAR11-GFP, expressing Far11 from its own promoter with a GFP tag at the C terminus | This study | Figs. 1, A and B, 2, and 3D |
| pTP543 | pRS418; ADE1 was cloned into pRS416 to replace the URA3 | This study | |
| pTP203 | pRS416-MKS1-GFP-FAR10, expressing Far10 from the MKS1 promoter with a GFP tag at the N terminus | This study | Fig. 4 |
| pTP646 | pRS418-FAR7-HA, expressing Far7 from its own promoter with a 3×HA tag at the C terminus | This study | Fig. 5, A–C |
| pTP658 | pRS418-FAR8-HA, expressing Far8 from its own promoter with a 3×HA tag at the C terminus | This study | Fig. 5A |
| pTP655 | pRS418-FAR3-HA, expressing Far3 from its own promoter with a 3×HA tag at the C terminus | This study | Figs. 4 and 5, A–C |
| pTP664 | pRS418-FAR7-FAR3-HA, expressing nontagged Far7 from its own promoter and Far3 from its own promoter with a 3×HA tag at the C terminus | This study | Fig. 5C |
| pTP673 | pRS418-FAR3-FAR7-HA, expressing nontagged Far3 from its own promoter and Far7 from its own promoter with a 3×HA tag at the C terminus | This study | Fig. 5C |
| pZL2762 | pRS416-FAR11-HA, expressing Far11 from its own promoter with a 3×HA tag at the C terminus | 24 | Fig. 6 |
| pTP554 | pRS416-MKS1p-GFP-FAR9ΔC, expressing Far9ΔC from the MKS1 promoter with a GFP tag at the N terminus | This study | Fig. 7, B and D |
Cellular Extract Preparation and Co-immunoprecipitation
Total cellular protein extracts were prepared by disrupting yeast cells in extraction buffer (1.85 n NaOH, 7.5% β-mercaptoethanol) followed by precipitation with trichloroacetic acid as described (37). For co-immunoprecipitation experiments, cellular lysates were prepared in IP buffer (50 mm Tris-HCl, pH 7.6, 150 mm NaCl, 0.5% Triton X-100, and protease inhibitors). Cell extracts (∼3 mg of proteins) were incubated at 4 °C for 1 h with anti-Myc antibody (9E10, Roche Applied Science) or anti-HA antibody (12CA5, Roche Applied Science) as indicated, after which 30 μl of a 50% slurry of protein G-Sepharose (Roche Applied Science) was added to each sample, and the samples were further incubated at 4 °C for 2 h. Washed immunoprecipitates bound to the Sepharose beads were released by boiling in 1× SDS-PAGE loading buffer. The released immune complexes were analyzed by SDS-PAGE and immunoblotting. Myc-, HA-, and GFP-tagged proteins were probed with anti-Myc antibody 9E10, high affinity anti-HA antibody 3F10 (Roche Applied Science), and anti-GFP antibody B-2 (Santa Cruz Biotechnology, Inc), respectively. Chemiluminescence images of Western blots were captured using the Bio-Rad Chemi-Doc photo documentation system (Bio-Rad) and processed using Bio-Rad Quantity One software.
Fluorescence Microscopy
Fluorescence of GFP- and RFP-tagged proteins was analyzed in live cells grown in SD medium to the mid-logarithmic growth phase by fluorescence microscopy. Cells were concentrated by centrifugation at 5000 × g for 2 min, and fluorescence images were immediately captured using a Nikon Eclipse E800 microscope equipped with an HBO 100 W/2 mercury arc lamp, a Nikon Plan Fluor ×100 objective lens, a Photometrics Coolsnap fx CCD camera, and a Nikon B-2E/C filter set (excitation light wavelengths 465–495 nm, emission light wavelengths 515–555 nm, dichromatic mirror cut-on wavelength 505 nm) for GFP images and a Y-2E/C filter set (excitation light wavelengths 540–580 nm, emission light wavelengths 600–660 nm, dichromatic mirror cut-on wavelength 595 nm) for RFP images. Digital images were acquired using the Metamorph Imaging Software and processed using ImageJ (National Institutes of Health) and Adobe Photoshop software.
Pheromone Response Halo Assay
Sensitivity to the mating pheromone α-factor was assayed by standard plate halo assays as described previously (29, 38). Briefly, 2 μg of α-factor was applied to a sterile 6-mm filter paper disc placed onto a lawn of 1 × 106 cells spread on a YNBcasD plate. Halo formation was documented after 3 days of cell growth.
Generation of Spheroplasts, Membrane Isolation, and Extraction
Cells expressing epitope-tagged Far proteins were grown at 30 °C overnight to a concentration of A600 1.5–2.0. Cells were converted into spheroplasts as described by Jazwinski (39). Briefly, cell pellets were prepared and resuspended to a concentration of A600 5 in reduction buffer (1 m sorbitol, 50 mm Tris-HCl, pH 7.6, 10 mm dithiothreitol (DTT)) and incubated for 10 min at room temperature. Cells were again harvested, and pellets were resuspended to a concentration of A600 25 in spheroplasting buffer (1 m sorbitol, 50 mm Tris-HCl, pH 7.6, 1 mm DTT). Spheroplasts were prepared by treatment with Zymolyase 20T, rinsed in 1 m sorbitol solution, suspended in lysis buffer (50 mm Tris-HCl, pH 7.6, 200 mm sorbitol, 1 mm EDTA, 1 mm DTT, and protease inhibitors), and homogenized using a glass homogenizer. Cell debris was sedimented two times by centrifugation at 2000 × g for 5 min, and the remaining supernatant was centrifuged at 100,000 × g for 15 min to generate a crude membrane fraction. The resulting membrane pellet was resuspended in lysis buffer. Aliquots of the membrane resuspension were treated with an equal volume of either H2O, 200 mm Na2CO3, or 2% Triton X-100 for 60 min at 4 °C with occasional gentle agitation. Membrane pellets were re-isolated by ultracentrifugation at 200,000 × g for 30 min. 10% trichloroacetic acid and 0.015% deoxycholic acid were used to precipitate proteins from supernatant fractions. Equivalent amounts of proteins from the supernatant and pellet fractions were analyzed using Western blotting and enhanced chemiluminescence.
RESULTS
GFP-tagged Far3, Far7, Far8, Far9, Far10, and Far11 Localize on the Endoplasmic Reticulum
Elucidating the cellular localization of the Far complex thus far has not been straightforward. Several studies report inconsistent data with the components of the Far complex localizing to different cellular compartments from one study to the next (23, 27, 35). Furthermore, none of the studies comprehensively analyzed the intracellular localization of all components of the complex. In all cases, fluorescent tags were fused to each Far protein to localize each component. One explanation for the differential localization of the Far complex components is that some of the fusion proteins may not have been functional as the functionality of the fusions of the previous studies was not indicated. Another factor that could affect the localization of a fusion protein is the placement of the fluorescent tag. For instance, Far9 and Far10 contain a hydrophobic tail-anchor domain; therefore, the addition of a fluorescent protein at the C-terminal end of Far9 and Far10 could affect their cellular localization. Indeed, in the genome-wide cellular localization study on yeast proteins and in the study by Lisa-Santamaria et al. (23), Far9 and Far10 were tagged with a C-terminal fluorescent protein, and in the study by Beilharz et al. (27), Far9 and Far10 carried an N-terminal GFP tag (35). Therefore, we generated N-terminal GFP-tagged Far9 and Far10 fusion constructs and C-terminal GFP-tagged Far3, Far7, Far8, and Far11 constructs on centromeric plasmids and determined their functionality and cellular localization. Expression of GFP-tagged Far3, Far7, Far8, and Far11 fusion proteins was under the control of their respective endogenous promoters. Expression of GFP-tagged Far9 and Far10 was under the control of a stronger but still relatively weak promoter of MKS1 (40) due to our initial observations that N-terminal GFP-tagged Far9 and Far10 under the control of their endogenous promoters did not yield enough signal to determine their cellular localization. To minimize potential interference of GFP with the functionality and thus localization of Far proteins, we also introduced a 10-alanine linker between the Far proteins and the GFP tag.
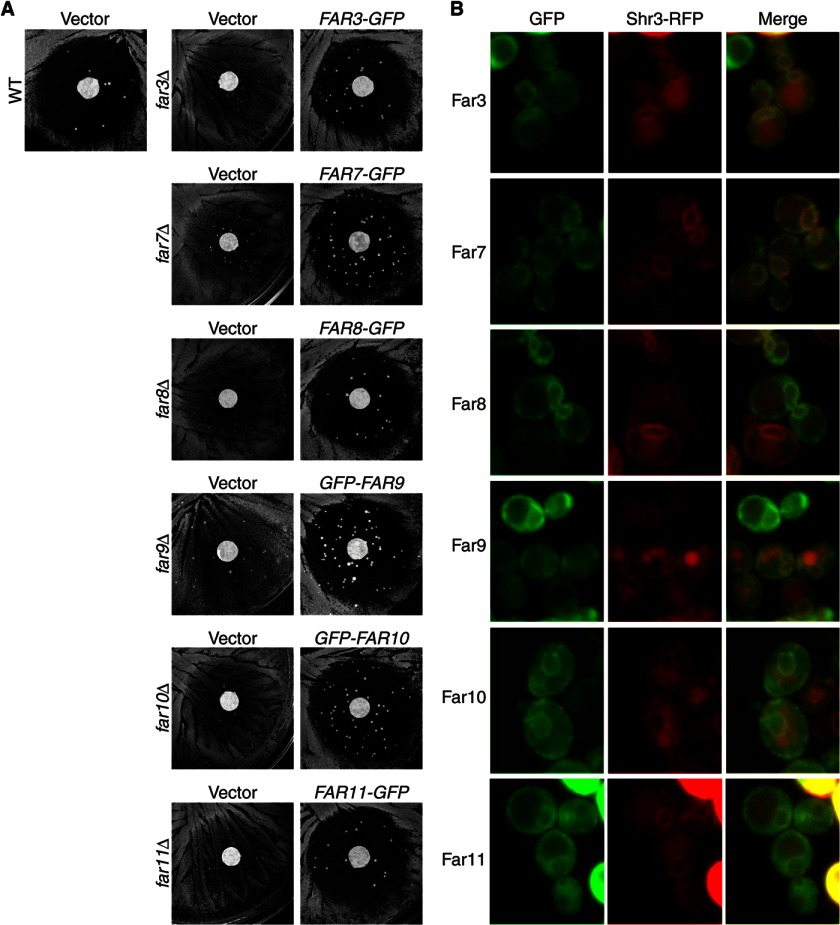
We first sought to determine the functionality of our GFP-tagged Far proteins. A quantitative assay is best suited for determining the function of epitope-tagged proteins. However, expression of FUS1-lacZ, a pheromone-responsive reporter gene, is not affected by the far mutations in our study despite the role of these Far proteins in mediating cell cycle arrest in response to the mating pheromone and thus is not useful in the determination of the functionality of GFP-tagged Far fusion proteins (22). Furthermore, we conducted a quantitative mating assay to assess whether far mutations affect mating efficiency, and we found that they reduced the mating efficiency marginally by ∼10% (data not shown), which is not sensitive enough to provide quantitative information on the functionality of GFP-tagged Far proteins. Therefore, to assess the functionality of GFP-tagged Far proteins, we used a pheromone response halo assay (Fig. 1A). Wild-type mating type a (MATa) cells normally arrest cell growth around a paper disc infused with α-factor and create a cell-free zone in the shape of a halo. We tested far3, far7, far8, far9, far10, and far11 single deletion mutants and found that they became resistant to α-factor-induced cell cycle arrest, which was manifested by increased cell growth around the disc containing α-factor, consistent with previous results (22). After these farΔ mutants were transformed with centromeric plasmids encoding respective wild-type FAR genes tagged with GFP, the resultant transformants were as sensitive to α-factor as wild-type cells, indicating that GFP-tagged Far3, Far7, Far8, Far9, Far10, and Far11 proteins were all functional.
FIGURE 1.
Far3-7-8-9-10-11 complex localizes to the ER. A, GFP-tagged FAR constructs complement respective far deletion mutations by halo assay. Wild-type (WT, SY2227) and isogenic farΔ mutant cells (far3Δ, TPY1010; far7Δ, TPY1013; far8Δ, TPY1015; far9Δ, TPY1048; far10Δ, TPY1072; far11Δ, SY4064) carrying the empty vector pRS416 (Vector) or plasmids encoding respective FAR-GFP fusions (FAR3-GFP, pTP143; FAR7-GFP, pTP164; FAR8-GFP, pTP131; GFP-FAR9, pTP179; GFP-FAR10, pTP203; FAR11-GFP, pZL2564) were grown on YNBcasD medium in the presence of a paper filter disc containing α-factor as described under “Experimental Procedures.” B, co-localization of GFP-tagged Far proteins with ER-localized Shr3-RFP. farΔ mutant cells (far3Δ, TPY157; far7Δ, TPY147; far8Δ, TPY213; far9Δ, TPY357; far10Δ, TPY151; far11Δ, TPY116) co-expressing respective GFP-tagged Far proteins as described for A, and RFP-tagged Shr3 (pTP201) were grown in SD medium and observed by fluorescence microscopy. GFP and RFP fluorescence images were captured and processed using the same parameters for each channel. Vacuolar autofluorescence in the RFP channel was sometimes observed.
We next examined the cellular localization of the GFP-tagged Far proteins in their respective far deletion mutant strains. Fig. 1B, left column, shows that all exhibited localization patterns suggestive of ER localization. To support this assumption, in cells expressing GFP-tagged Far proteins, we co-expressed the C-terminal RFP-tagged Shr3, an ER-localized chaperone for packaging amino acid permeases into COPII-coated transport vesicles (41). Fig. 1B shows that Shr3-RFP and each of the six Far-GFP fusion proteins co-localize, indicating that the Far complex localizes to the ER. We expressed GFP-tagged Far proteins in respective far deletion mutant strains of two other strain backgrounds, BY4741, which is derived from S288c, and SY2227 (22), and we found that in these strains, GFP-tagged Far proteins also localized to the ER (data not shown), indicating that ER localization of the Far complex is not strain-dependent.
Far9 and Far10 Localize to the ER Independently of the Other Far Proteins
To better understand how the Far complex is organized on the ER, we sought to determine which Far protein(s) establishes a foothold on the ER. To that end, we analyzed the cellular localization of individual GFP-tagged Far proteins in a far3/7/8/9/10/11Δ sextuple mutant strain. Fig. 2 shows that Far3-GFP, Far7-GFP, Far8-GFP, and Far11-GFP, when expressed individually in the sextuple mutant strain, did not exhibit ER localization, indicating that they were unable to localize to the ER in the absence of other Far complex components. In contrast, GFP-Far9 and GFP-Far10 fusion proteins could still localize to the ER in the sextuple mutant, indicating that Far9 and Far10 establish ER localization for the complex. Far9 and Far10 contain a tail-anchor domain at their C termini. Therefore, it is not surprising that they may function as the ER anchor for the Far complex. Furthermore, the localization of Far9 and Far10 on the ER when expressed individually in the sextuple mutant suggests that these two homologous proteins do not require hetero-oligomerization for ER recruitment.
FIGURE 2.
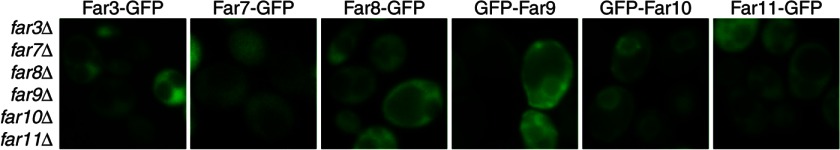
Far9 and Far10, but not Far3, Far7, Far8, or Far11, are able to localize to the ER in the absence of the other Far complex components. Sextuple farΔ mutant cells (TPY845) expressing GFP-tagged Far proteins as indicated were grown in SD medium and observed by fluorescence microscopy.
Tiered Assembly of the Components of the Far Complex at the ER
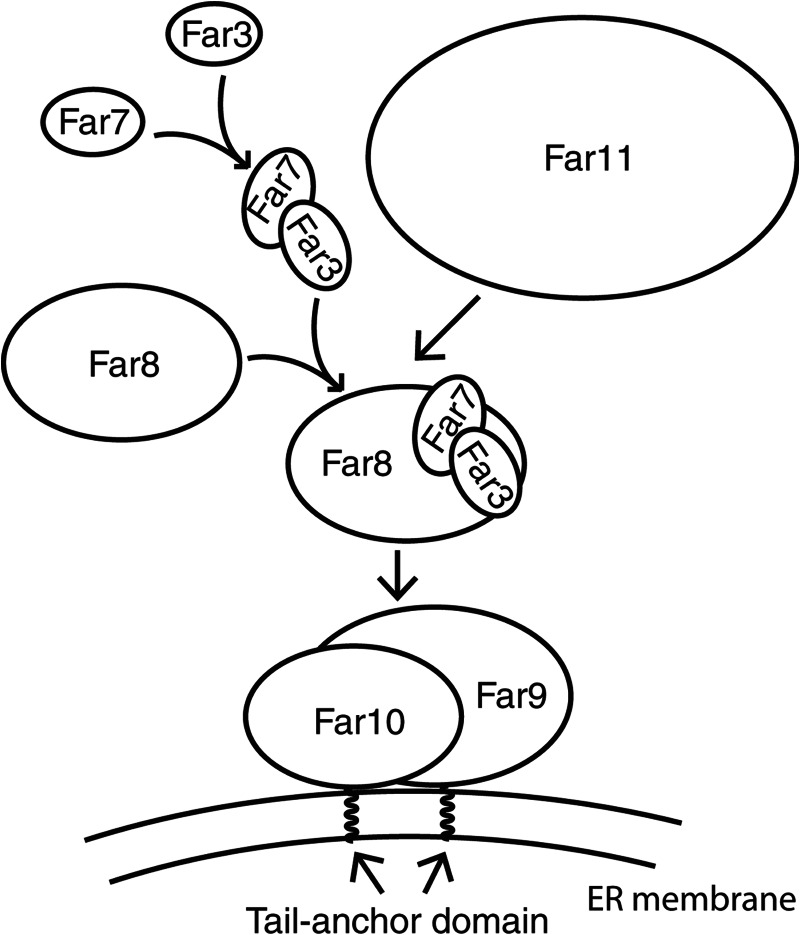
To further characterize the organization of the Far complex on the ER, we sought to determine the order in which the rest of the complex localizes to the ER. To achieve this, we determined whether ER localization of GFP-tagged Far3, Far7, Far8, or Far11 could be altered by the absence of just one of the other five Far complex components. Accordingly, we characterized the cellular localization of Far3-GFP in far3Δ far7Δ, far3Δ far8Δ, far3Δ far9Δ, far3Δ far10Δ, and far3Δ far11Δ double deletion mutant cells. Far3-GFP localization at the ER was abolished by far7Δ, far8Δ, far9Δ, and far10Δ mutations but still showed normal ER localization in a far11Δ mutant, indicating that ER localization of Far3-GFP requires Far7, Far8, Far9, and Far10 but not Far11 (Fig. 3A). Using the same strategy, we determined the cellular localization of Far7-GFP, Far8-GFP, and Far11-GFP in respective double deletion mutant cells. Likewise, ER localization of Far7-GFP and Far8-GFP was abolished by all respective farΔ mutations except a far11Δ mutation (Fig. 3, B and C). Interestingly, ER localization of Far11-GFP was disrupted by the deletion of any of the other five Far complex components (Fig. 3D), suggesting that Far11 is the most peripheral component of this complex at the ER. These data also suggest that Far3, Far7, Far8, and Far11 are peripheral membrane proteins because their ER localization requires Far9 and Far10. This possibility was supported by our initial observation early in our studies that the localization of GFP-tagged Far3, Far7, Far8, and Far11 became more cytoplasmic when they were expressed in wild-type cells in comparison with the respective deletion mutant cells, suggesting that these four GFP-tagged Far proteins compete with their nontagged counterparts for Far9/10-dependent ER localization (data not shown). The interdependence of Far3, Far7, and Far8 for ER localization also suggests that they might form a subcomplex before their ER recruitment.
FIGURE 3.
Cellular localization of GFP-tagged Far3, Far7, Far8, or Far11 in the absence of individual components of the Far complex. A, localization of Far3-GFP in the mutant strains as indicated (far3Δ, TPY1010; far3Δ far7Δ, TPY1358; far3Δ far8Δ, TPY1361; far3Δ far9Δ, TPY1348; far3Δ far10Δ, TPY1363; far3Δ far11Δ, TPY1402). B, localization of Far7-GFP in the mutant strains as indicated (far7Δ, TPY1013; far7Δ far3Δ, TPY1408; far7Δ far8Δ, TPY1366; far7Δ far9Δ, TPY1350; far7Δ far10Δ, TPY1368; far7Δ far11Δ, TPY1352). C, localization of Far8-GFP in the mutant strains as indicated (far8Δ, TPY1015; far8Δ far3Δ, TPY1369; far8Δ far7Δ, TPY1370; far8Δ far9Δ, TPY1351; far8Δ far10Δ, TPY1373; far8Δ far11Δ, TPY1405). D, localization of Far11-GFP in the mutant strains as indicated (far11Δ, SY4064; far11Δ far3Δ, TPY1374; far11Δ far7Δ, TPY1377; far11Δ far8Δ, TPY1406; far11Δ far9Δ, TPY1410; far11Δ far10Δ, TPY1379). All cells were grown in SD medium and observed by fluorescence microscopy.
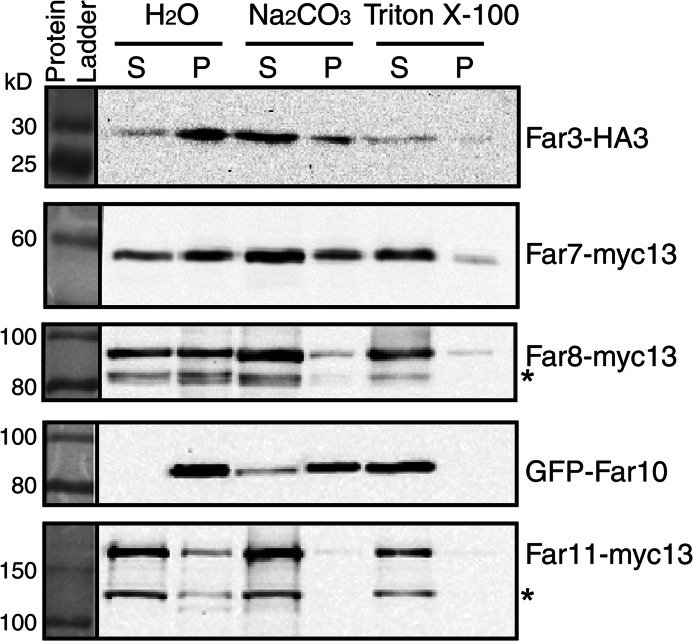
To confirm that Far3, Far7, Far8, and Far11 are peripherally associated with the ER membrane, we conducted membrane extraction experiments using carbonate or Triton X-100. Alkaline carbonate extraction removes peripheral membrane proteins but not integral membrane proteins from membrane fractions. In contrast, Triton X-100 treatment disrupts membranes and releases both peripheral and integral membrane proteins. A crude membrane fraction was isolated and treated with 100 mm carbonate, 1% Triton X-100, or water. Fig. 4 shows that after water treatment, Far3, Far7, Far8, and Far11 fractionated in both the supernatant and the pellet, whereas Far10 fractionated only in the pellet. Furthermore, 100 mm carbonate treatment released most of Far3, Far7, and Far8, all of Far11, yet very little of Far10 into the supernatant fraction. However, 1% Triton X-100 treatment efficiently released Far10 into the supernatant fraction. Surprisingly, for unknown reasons a small portion of Far3, Far7, and Far8 was resistant to Triton X-100 extraction. Together, these data indicate that Far3, Far7, Far8, and Far11 are peripheral membrane proteins, whereas Far10 is an integral membrane protein. Based on the efficiency of carbonate extraction of Far proteins from membranes and the fluorescence microscopy data, the Far complex appears to assemble at the ER in the spatial order of Far9/10, Far3/7/8, and then Far11.
FIGURE 4.
Analysis of membrane association of epitope-tagged Far3, Far7, Far8, Far10, and Far11 by membrane extraction with alkaline carbonate or Triton X-100. A membrane pellet fraction was prepared from cells expressing epitope-tagged Far proteins as indicated and subjected to extraction with mock treatment (H2O), 0.1 m Na2CO3, or 1% Triton X-100 as described under “Experimental Procedures.” Epitope-tagged Far proteins in the supernatant (S) and pellet (P) fractions were separated by SDS-PAGE and visualized by immunoblotting. The result is representative of two independent experiments. * denotes degradation products of Far8 and Far11.
Far3, Far7, and Far8 Form a Subcomplex
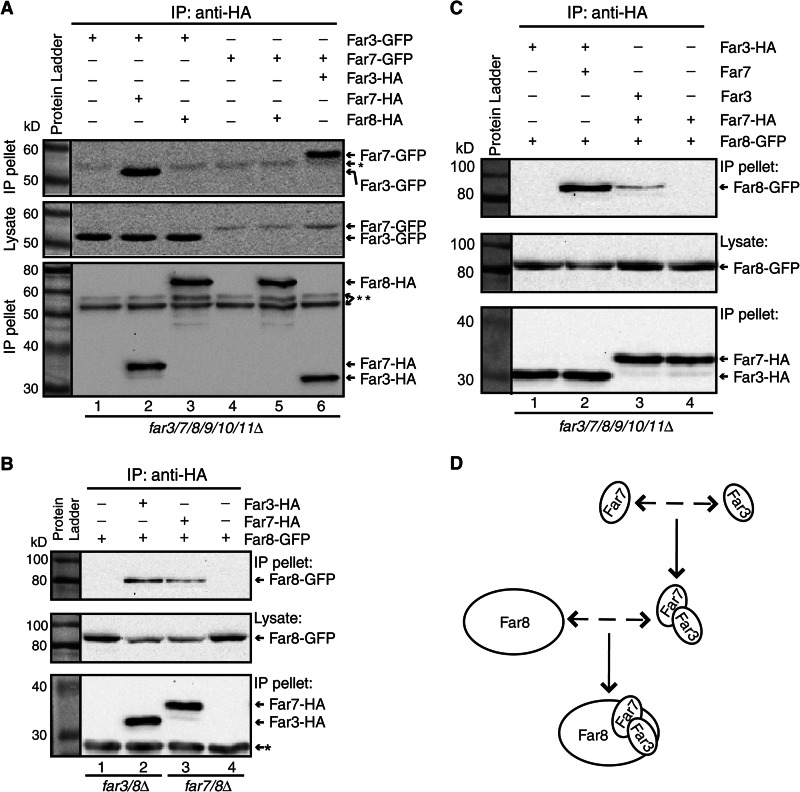
To test whether Far3, Far7, and Far8 are able to form a subcomplex independent of Far9, Far10, and Far11, we determined whether Far3, Far7, and Far8 could form pairwise interactions in far3/7/8/9/10/11Δ sextuple mutant cells by co-immunoprecipitation. Accordingly, Far3-GFP was co-expressed with either 3×HA epitope-tagged Far7 or Far8, and Far7-GFP was co-expressed with either 3×HA epitope-tagged Far3 or Far8 in far3/7/8/9/10/11Δ sextuple mutant cells. HA-tagged proteins from cell lysates were immunoprecipitated with anti-HA antibody, and immunoprecipitates were probed with anti-GFP antibody to detect GFP-tagged proteins via Western blotting. We found that Far7-HA, but not Far8-HA, was able to pull down Far3-GFP (Fig. 5A, lanes 1–3). Similarly, Far3-HA, but not Far8-HA, was able to pull down Far7-GFP (Fig. 5A, lanes 4–6). Together, these data indicate that Far3 and Far7 are able to interact with each other in the absence of the other Far complex components.
FIGURE 5.
Far3, Far7, and Far8 form a subcomplex. A, Far3 and Far7 are able to interact in the absence of Far8, Far9, Far10, and Far11. Cell lysates of sextuple farΔ mutant cells (TPY845) co-expressing Far3-GFP (pTP143) and Far7-HA (pTP646) or Far8-HA (pTP658), Far7-GFP (pTP164), and Far3-HA (pTP655) or Far8-HA (pTP658) as indicated were subjected to immunoprecipitation (IP) with anti-HA antibody, and epitope-tagged proteins were detected by immunoblotting. * and ** indicate the heavy chain of the anti-HA antibody used for immunoprecipitation that was detected by goat anti-mouse IgG light chain specific and standard secondary antibody, respectively. B, Far8 interacts with Far3 or Far7 in the presence of the other Far complex components. far3/8Δ mutant cells expressing Far8-GFP (TPY1369) without or with Far3-HA and far7/8Δ mutant cells expressing Far8-GFP (TPY1370) without or with Far7-HA were analyzed for interactions between Far8-GFP and Far3-HA or Far7-HA by immunoprecipitation and immunoblotting. C, Far8 interacts with the Far3-7 complex. Cell lysates of sextuple farΔ mutant cells expressing epitope-tagged and nontagged proteins as indicated were subjected to immunoprecipitation with anti-HA antibody. GFP- and HA-tagged proteins were detected by immunoblotting. D, model of Far3-Far7-Far8 subcomplex formation.
Far8 has been reported to interact with Far3 and Far7 by yeast two-hybrid and co-immunoprecipitation analyses in wild-type strains (22, 25). Although we carried out the interaction analysis between Far8 and Far3 or Far7 in sextuple mutant cells, which were not used in previous studies, the failure to detect their interactions in such cells was still surprising because these three proteins appear to require each other for ER localization as shown in Fig. 3. One possibility is that Far8 may only bind to the Far3-Far7 complex. To test this hypothesis, we first confirmed whether Far8 interacts with Far3 or Far7 in respective double deletion mutant cells. We generated a far3Δ far8Δ double mutant carrying plasmids encoding FAR3-HA and FAR8-GFP. Fig. 5B shows that Far8-GFP was co-immunoprecipitated specifically with Far3-HA (compare lanes 1 and 2). Similarly, we found that Far8-GFP specifically interacts with Far7-HA in a far7Δ far8Δ double mutant (Fig. 5B, lanes 3 and 4). We next examined whether Far8 interacts with the Far3-Far7 complex in the sextuple mutant. We co-expressed Far3-HA, Far8-GFP, and nontagged Far7 in far3/7/8/9/10/11Δ sextuple mutant cells and found that expression of Far7 in the sextuple mutant was sufficient for Far3-HA to interact with Far8-GFP (Fig. 5C, compare lanes 1 and 2). Similarly, reintroduction of Far3 into sextuple mutant cells co-expressing Far7-HA and Far8-GFP also enabled an interaction between Far7 and Far8 (Fig. 5C, compare lanes 3 and 4). These data together indicate that Far3, Far7, and Far8 form a subcomplex in the absence of the other components of the Far complex.
Interaction between Far9 and Far11 Requires the Far3-7-8- Subcomplex
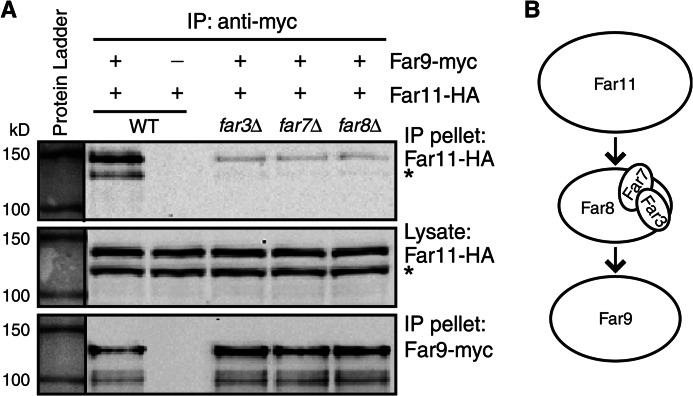
ER localization of Far11-GFP was disrupted by deletion of any of the other Far complex components as shown in Fig. 3D. These findings along with the findings that Far3, Far7, and Far8 form a subcomplex suggest that the Far3-7-8- subcomplex may bridge the interaction of Far11 and Far9/10 at the ER. To test this possibility, interaction between Myc-tagged Far9 and HA-tagged Far11 was analyzed in far9Δ far11Δ double mutant cells without (WT) or with an additional mutation of far3Δ, far7Δ, or far8Δ. Far9-myc was immunoprecipitated from cell lysates, and immunoprecipitates were probed with anti-HA antibody to detect Far11-HA via Western blotting. Fig. 6 shows that Far11-HA was co-immunoprecipitated with Far9-myc, and deletion of FAR3, FAR7, or FAR8 greatly reduced their interaction. These data indicate that ER recruitment of Far11 by Far9 requires the Far3-7-8- subcomplex.
FIGURE 6.
Interaction between Far9 and Far11 is greatly reduced in the absence of Far3, Far7, or Far8. A, cell lysates of strains TPY1411 (WT), TPY1412 (far3Δ), TPY1413 (far7Δ), and TPY1416 (far8Δ) co-expressing Far9-myc and Far11-HA (pZL2762) and the strain SY4064 (WT) expressing Far11-HA alone were subjected to immunoprecipitation (IP) with anti-Myc antibody. HA- and Myc-tagged proteins were detected by immunoblotting. An asterisk denotes a proteolytic product of Far11-HA (24). B, model of the Far3-Far7-Far8 subcomplex bridging interaction between Far11 and Far9.
ER Localization of Far9 Is Required for Its Optimal Function in TORC2 Signaling
Yeast Far9 and Far10 and their human and fly orthologs all contain a tail-anchor domain and a forkhead-associated domain (Fig. 7A). Tail-anchored proteins utilize the tail-anchor domain for membrane association (42). The finding that Far9 and Far10 are able to localize to the ER in the absence of the other Far complex components prompted us to determine the role of Far9's tail-anchor in the ER localization of Far9. Accordingly, we constructed a GFP-tagged C-terminal truncation mutant of Far9, GFP-Far9ΔC, and examined its location in far9Δ mutant cells. Unlike GFP-tagged wild-type Far9, GFP-Far9ΔC localized diffusely in the cytoplasm (Fig. 7B), indicating that the tail-anchor domain of Far9 is required for its ER localization.
FIGURE 7.
Tail-anchor domain of Far9 is required for its ER localization. A, diagrammatic representations of Far9 and its orthologs in flies and humans. A conserved forkhead-associated (FHA) domain and the tail-anchor domain are indicated by gray and white rectangles, respectively. B, ER localization of Far9 requires the tail-anchor domain. far9Δ mutant cells (TPY1048) expressing GFP-tagged Far9 (pTP179) or Far9ΔC (pTP554) were grown in SD medium and observed by fluorescence microscopy. C, tail-anchor domain of Far9 is required for the optimal function of Far9 in TORC2 signaling. Serial dilutions of indicated cells (WT, SH100; tor2-21, SH121; tor2-21 far9Δ, TPY357; tor2-21 far9ΔC, TPY1341) were grown on YPD medium at 30, 36, and 37 °C for 3–4 days. D, loss of the tail-anchor domain of Far9 does not reduce its steady-state level. Total cellular proteins of far9Δ mutant cells (TPY357) expressing GFP-Far9 or GFP-Far9ΔC were prepared and separated by SDS-PAGE and GFP-tagged proteins were detected by immunoblotting. 3-Phosphoglycerate kinase (Pgk1) was included as a loading control. DIC, differential interference contrast.
Recently, we proposed that the Far complex antagonizes TORC2 signaling by showing that a far9Δ mutation or loss of other Far complex components are able to bypass a tor2 temperature-sensitive (tor2-21) mutation (24). We sought to test whether ER localization of the Far complex is required for its function. To that end, we introduced a far9ΔC mutation, which results in the synthesis of Far9 without the tail-anchor domain, at the genomic FAR9 locus in tor2-21 mutant cells. We then tested whether the far9ΔC mutation could bypass the tor2-21 mutation and found that far9ΔC was able to partially mimic far9Δ in suppressing the growth defect of tor2-21 mutant cells grown at 36 and 37 °C (Fig. 7C). To exclude the possibility that the removal of the tail-anchor domain of Far9 may reduce the steady-state level of Far9 by reducing its stability, which could explain the partial suppression of the tor2-21 growth defect at high temperatures, we examined the levels of GFP-tagged Far9 and Far9ΔC in tor2-21 far9Δ cells and found that Far9ΔC was expressed to similar levels as full-length Far9 (Fig. 7D). Together, these data suggest that the suppression of the tor2-21 mutation by far9ΔC results from the loss of ER localization of Far9 and that ER localization of Far9 is required for its optimal function.
DISCUSSION
In this report, we systematically examined the cellular localization of functional GFP-tagged Far3, Far7, Far8, Far9, Far10, and Far11 and found that they all localized to the ER. Our findings are partly consistent with two previous studies (27, 35) yet disagree with another (23). Inconsistencies in the localization of the Far proteins may be explained by several possibilities. Cytoplasmic localization of Far9 in the genome-wide study on yeast protein localization is most likely to be due to the tagging with GFP at its C terminus, which is expected to interfere with tail-anchor domain-dependent ER membrane insertion of Far9. The clustering effect of Far10 in the study by Beilharz et al. (27) may result from a higher level of overexpression of Far10 from the relatively strong MET25 promoter than in our current study (44, 45). Our data disagree with the study by Lisa-Santamaria et al. (23) on the localization of Far3, Far9, and Far11. In that study, the authors added a C-terminal CFP tag to Far9. Furthermore, the functionality of the fusion proteins in the aforementioned study was not reported. If the fusion proteins were not functional, this could present another possibility that accounts for the localization differences.
ER/nuclear envelope localization of proteins associated with the STRIPAK complex is likely to be a conserved feature, which is expected to be mediated by the tail-anchor domain of Far9/Far10 orthologs in different species (Table 1). Apart from the ER localization of human and S. cerevisiae STRIPAK components, two components of the fission yeast S. pombe STRIPAK complex, Csc2 and Csc3 (S. cerevisiae Far11 and Far8 orthologs, respectively), also associate with the nuclear envelope/ER. Conversely, Csc1, the S. cerevisiae Far9/10 ortholog, was not found to localize to the nuclear envelope (20). However, this could be due to the addition of GFP at its C terminus, which interferes with its tail-anchor domain-dependent ER localization. ER localization of the STRIPAK components appears to be important for their function in human cells (11, 14, 15). Likewise, we show that, in yeast, ER localization of the Far complex is required for its optimal function in TORC2 signaling. Why is ER localization of the yeast Far complex required for its function in TORC2 signaling? We and others have shown that the yeast Far complex and PP2A negatively regulate TORC2 signaling. Both TORC2 and its substrates localize to the plasma membrane (30, 32, 33). The association with the Far complex could bring PP2A to the periplasmic region to facilitate dephosphorylation of Slm1 and other TORC2 substrates located at the plasma membrane.
Human SLMAP and the S. pombe STRIPAK complex also localize to the centrosome and the spindle pole body, respectively (9, 12, 20). Consistent with their localization, the STRIPAK complex from these two species has been reported to play roles in mitosis (9, 12, 20). Our failure to detect localization of the yeast Far complex components and even the Far9 truncation mutant without its tail-anchor domain to the spindle pole body is consistent with the hypothesis of Frost et al. (9) that the function of the STRIPAK complex may have been “repurposed” and the S. cerevisiae STRIPAK complex has lost its function in mitosis.
Apart from identifying the cellular localization of the Far complex, we also determined the spatial order by which the Far complex organizes itself on the ER; Far9/10 establish a foothold on the ER utilizing a tail-anchor domain, and Far3/7/8 form a subcomplex that bridges Far11 to Far9/10 at the ER (Fig. 8). Previously, we reported that mutations in genes encoding the Far complex components suppress TORC2 deficiency in the following order: far11Δ > far8/9Δ > far3/7Δ > far10Δ (24). It is interesting to note that Far11, which is the most peripheral component of the complex, is also the most important among the six Far proteins in TORC2 signaling. Although this result is surprising, it may help understand why ER localization of the Far complex is not absolutely required for its function in TORC2 signaling.
FIGURE 8.
Model of spatial assembly of the Far complex at the endoplasmic reticulum. See text for details.
We found that Far3 and Far7 are able to form a complex in the absence of the other four Far proteins. Interestingly, the mutant phenotypes of far3Δ and far7Δ on TORC2 signaling are most similar (24). Orthologs of Far3 and Far7 are only found in a restricted set of fungal species and lack apparent orthologs in animals (9). However, yeast Far3/Far7 and human SIKE/FGFR1OP2 are all relatively small proteins predicted to have coiled-coil domains (22, 25, 46). Therefore, it is possible that Far3/Far7 may be the functional or structural counterparts of human SIKE/FGFR1OP2 (Table 1). Interestingly, the STRIPAK complex in S. pombe contains a novel protein of 166 residues, Csc4, which is also predicted to have a coiled-coil domain (20). It is possible that these small coiled-coil domain proteins may play the same structural role in the STRIPAK complex in different species as a result of divergent evolution.
Human Striatin 3 and its Sordaria macrospora ortholog PRO11 have been reported to contain an N-terminal coiled-coil region, and both proteins are critical components in the organization of their respective STRIPAK complexes (3, 18). Like human striatins, yeast Far8 was also predicted to contain an N-terminal coiled-coil domain (22, 25). Interestingly, almost all components of the Far complex are predicted to contain coiled-coil domains (22, 25). These coiled-coil domains may mediate protein-protein interactions and provide a structural framework for the organization of the Far complex. Further research will be conducted to uncover the role of Far8 in the yeast STRIPAK complex and how the Far complex interfaces with PP2A.
Acknowledgments
We thank Michael Hall and George Sprague, Jr. for yeast strains, Muhammad Farooq for technical support, the W. M. Keck Foundation for the Keck Facility, and Robin Rowe for sequencing.
This work was supported, in whole or in part, by National Institutes of Health Grant 1R15GM094772-01A1. This work was also supported by State of Louisiana Board of Regents Grant LEQSF(2008-11)-RD-A-31.
- STRIPAK
- striatin-interacting phosphatase and kinase
- ER
- endoplasmic reticulum
- SLMAP
- sarcolemmal membrane-associated protein
- RFP
- red fluorescent protein.
REFERENCES
- 1. Goudreault M., D'Ambrosio L. M., Kean M. J., Mullin M. J., Larsen B. G., Sanchez A., Chaudhry S., Chen G. I., Sicheri F., Nesvizhskii A. I., Aebersold R., Raught B., Gingras A. C. (2009) A PP2A phosphatase high density interaction network identifies a novel striatin-interacting phosphatase and kinase complex linked to the cerebral cavernous malformation 3 (CCM3) protein. Mol. Cell. Proteomics 8, 157–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ribeiro P. S., Josué F., Wepf A., Wehr M. C., Rinner O., Kelly G., Tapon N., Gstaiger M. (2010) Combined functional genomic and proteomic approaches identify a PP2A complex as a negative regulator of Hippo signaling. Mol. Cell 39, 521–534 [DOI] [PubMed] [Google Scholar]
- 3. Kean M. J., Ceccarelli D. F., Goudreault M., Sanches M., Tate S., Larsen B., Gibson L. C., Derry W. B., Scott I. C., Pelletier L., Baillie G. S., Sicheri F., Gingras A. C. (2011) Structure-function analysis of core STRIPAK proteins: a signaling complex implicated in Golgi polarization. J. Biol. Chem. 286, 25065–25075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ceccarelli D. F., Laister R. C., Mulligan V. K., Kean M. J., Goudreault M., Scott I. C., Derry W. B., Chakrabartty A., Gingras A. C., Sicheri F. (2011) CCM3/PDCD10 heterodimerizes with germinal center kinase III (GCKIII) proteins using a mechanism analogous to CCM3 homodimerization. J. Biol. Chem. 286, 25056–25064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zheng X., Xu C., Di Lorenzo A., Kleaveland B., Zou Z., Seiler C., Chen M., Cheng L., Xiao J., He J., Pack M. A., Sessa W. C., Kahn M. L. (2010) CCM3 signaling through sterile 20-like kinases plays an essential role during zebrafish cardiovascular development and cerebral cavernous malformations. J. Clin. Invest. 120, 2795–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li X., Zhang R., Zhang H., He Y., Ji W., Min W., Boggon T. J. (2010) Crystal structure of CCM3, a cerebral cavernous malformation protein critical for vascular integrity. J. Biol. Chem. 285, 24099–24107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fidalgo M., Fraile M., Pires A., Force T., Pombo C., Zalvide J. (2010) CCM3/PDCD10 stabilizes GCKIII proteins to promote Golgi assembly and cell orientation. J. Cell Sci. 123, 1274–1284 [DOI] [PubMed] [Google Scholar]
- 8. Gordon J., Hwang J., Carrier K. J., Jones C. A., Kern Q. L., Moreno C. S., Karas R. H., Pallas D. C. (2011) Protein phosphatase 2a (PP2A) binds within the oligomerization domain of striatin and regulates the phosphorylation and activation of the mammalian Ste20-like kinase Mst3. BMC Biochem. 12, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Frost A., Elgort M. G., Brandman O., Ives C., Collins S. R., Miller-Vedam L., Weibezahn J., Hein M. Y., Poser I., Mann M., Hyman A. A., Weissman J. S. (2012) Functional repurposing revealed by comparing S. pombe and S. cerevisiae genetic interactions. Cell 149, 1339–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Byers J. T., Guzzo R. M., Salih M., Tuana B. S. (2009) Hydrophobic profiles of the tail anchors in SLMAP dictate subcellular targeting. BMC Cell Biol. 10, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nader M., Westendorp B., Hawari O., Salih M., Stewart A. F., Leenen F. H., Tuana B. S. (2012) Tail-anchored membrane protein SLMAP is a novel regulator of cardiac function at the sarcoplasmic reticulum. Am. J. Physiol. Heart Circ. Physiol. 302, H1138–H1145 [DOI] [PubMed] [Google Scholar]
- 12. Guzzo R. M., Sevinc S., Salih M., Tuana B. S. (2004) A novel isoform of sarcolemmal membrane-associated protein (SLMAP) is a component of the microtubule organizing centre. J. Cell Sci. 117, 2271–2281 [DOI] [PubMed] [Google Scholar]
- 13. Wielowieyski P. A., Sevinc S., Guzzo R., Salih M., Wigle J. T., Tuana B. S. (2000) Alternative splicing, expression, and genomic structure of the 3′ region of the gene encoding the sarcolemmal-associated proteins (SLAPs) defines a novel class of coiled-coil tail-anchored membrane proteins. J. Biol. Chem. 275, 38474–38481 [DOI] [PubMed] [Google Scholar]
- 14. Guzzo R. M., Salih M., Moore E. D., Tuana B. S. (2005) Molecular properties of cardiac tail-anchored membrane protein SLMAP are consistent with structural role in arrangement of excitation-contraction coupling apparatus. Am. J. Physiol. Heart Circ. Physiol. 288, H1810–H1819 [DOI] [PubMed] [Google Scholar]
- 15. Guzzo R. M., Wigle J., Salih M., Moore E. D., Tuana B. S. (2004) Regulated expression and temporal induction of the tail-anchored sarcolemmal-membrane-associated protein is critical for myoblast fusion. Biochem. J. 381, 599–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Simonin A. R., Rasmussen C. G., Yang M., Glass N. L. (2010) Genes encoding a striatin-like protein (ham-3) and a forkhead associated protein (ham-4) are required for hyphal fusion in Neurospora crassa. Fungal Genet. Biol. 47, 855–868 [DOI] [PubMed] [Google Scholar]
- 17. Xiang Q., Rasmussen C., Glass N. L. (2002) The ham-2 locus, encoding a putative transmembrane protein, is required for hyphal fusion in Neurospora crassa. Genetics 160, 169–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bloemendal S., Bernhards Y., Bartho K., Dettmann A., Voigt O., Teichert I., Seiler S., Wolters D. A., Pöggeler S., Kück U. (2012) A homologue of the human STRIPAK complex controls sexual development in fungi. Mol. Microbiol. 84, 310–323 [DOI] [PubMed] [Google Scholar]
- 19. Bloemendal S., Lord K. M., Rech C., Hoff B., Engh I., Read N. D., Kück U. (2010) A mutant defective in sexual development produces aseptate ascogonia. Eukaryot. Cell 9, 1856–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Singh N. S., Shao N., McLean J. R., Sevugan M., Ren L., Chew T. G., Bimbo A., Sharma R., Tang X., Gould K. L., Balasubramanian M. K. (2011) SIN-inhibitory phosphatase complex promotes Cdc11p dephosphorylation and propagates SIN asymmetry in fission yeast. Curr. Biol. 21, 1968–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Baryshnikova A., Costanzo M., Kim Y., Ding H., Koh J., Toufighi K., Youn J. Y., Ou J., San Luis B. J., Bandyopadhyay S., Hibbs M., Hess D., Gingras A. C., Bader G. D., Troyanskaya O. G., Brown G. W., Andrews B., Boone C., Myers C. L. (2010) Quantitative analysis of fitness and genetic interactions in yeast on a genome scale. Nat. Methods 7, 1017–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kemp H. A., Sprague G. F., Jr. (2003) Far3 and five interacting proteins prevent premature recovery from pheromone arrest in the budding yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 23, 1750–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lisa-Santamaría P., Jiménez A., Revuelta J. L. (2012) The protein factor-arrest 11 (Far11) is essential for the toxicity of human caspase-10 in yeast and participates in the regulation of autophagy and the DNA damage signaling. J. Biol. Chem. 287, 29636–29647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pracheil T., Thornton J., Liu Z. (2012) TORC2 signaling is antagonized by protein phosphatase 2A and the Far complex in Saccharomyces cerevisiae. Genetics 190, 1325–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lai F., Wu R., Wang J., Li C., Zou L., Lu Y., Liang C. (2011) Far3p domains involved in the interactions of Far proteins and pheromone-induced cell cycle arrest in budding yeast. FEMS Yeast Res. 11, 72–79 [DOI] [PubMed] [Google Scholar]
- 26. Uetz P., Giot L., Cagney G., Mansfield T. A., Judson R. S., Knight J. R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Qureshi-Emili A., Li Y., Godwin B., Conover D., Kalbfleisch T., Vijayadamodar G., Yang M., Johnston M., Fields S., Rothberg J. M. (2000) A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403, 623–627 [DOI] [PubMed] [Google Scholar]
- 27. Beilharz T., Egan B., Silver P. A., Hofmann K., Lithgow T. (2003) Bipartite signals mediate subcellular targeting of tail-anchored membrane proteins in Saccharomyces cerevisiae. J. Biol. Chem. 278, 8219–8223 [DOI] [PubMed] [Google Scholar]
- 28. Ferguson B., Horecka J., Printen J., Schultz J., Stevenson B. J., Sprague G. F., Jr. (1994) The yeast pheromone response pathway: new insights into signal transmission. Cell. Mol. Biol. Res. 40, 223–228 [PubMed] [Google Scholar]
- 29. Horecka J., Sprague G. F., Jr. (1996) Identification and characterization of FAR3, a gene required for pheromone-mediated G1 arrest in Saccharomyces cerevisiae. Genetics 144, 905–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Loewith R., Hall M. N. (2011) Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 189, 1177–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Laplante M., Sabatini D. M. (2012) mTOR signaling in growth control and disease. Cell 149, 274–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fadri M., Daquinag A., Wang S., Xue T., Kunz J. (2005) The pleckstrin homology domain proteins Slm1 and Slm2 are required for actin cytoskeleton organization in yeast and bind phosphatidylinositol-4,5-bisphosphate and TORC2. Mol. Biol. Cell 16, 1883–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Niles B. J., Mogri H., Hill A., Vlahakis A., Powers T. (2012) Plasma membrane recruitment and activation of the AGC kinase Ypk1 is mediated by target of rapamycin complex 2 (TORC2) and its effector proteins Slm1 and Slm2. Proc. Natl. Acad. Sci. U.S.A. 109, 1536–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kamada Y. (2010) Prime-numbered Atg proteins act at the primary step in autophagy: unphosphorylatable Atg13 can induce autophagy without TOR inactivation. Autophagy 6, 415–416 [DOI] [PubMed] [Google Scholar]
- 35. Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. (2003) Global analysis of protein localization in budding yeast. Nature 425, 686–691 [DOI] [PubMed] [Google Scholar]
- 36. Amberg D. C., Burke D. J., Strathern J. N. (2005) Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 37. Yaffe M. P., Schatz G. (1984) Two nuclear mutations that block mitochondrial protein import in yeast. Proc. Natl. Acad. Sci. U.S.A. 81, 4819–4823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sprague G. F., Jr. (1991) Assay of yeast mating reaction. Methods Enzymol. 194, 77–93 [DOI] [PubMed] [Google Scholar]
- 39. Jazwinski S. M. (1990) Preparation of extracts from yeast. Methods Enzymol. 182, 154–174 [DOI] [PubMed] [Google Scholar]
- 40. Sekito T., Liu Z., Thornton J., Butow R. A. (2002) RTG-dependent mitochondria-to-nucleus signaling is regulated by MKS1 and is linked to formation of yeast prion [URE3]. Mol. Biol. Cell 13, 795–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kuehn M. J., Schekman R., Ljungdahl P. O. (1996) Amino acid permeases require COPII components and the ER resident membrane protein Shr3p for packaging into transport vesicles in vitro. J. Cell Biol. 135, 585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Borgese N., Fasana E. (2011) Targeting pathways of C-tail-anchored proteins. Biochim. Biophys. Acta 1808, 937–946 [DOI] [PubMed] [Google Scholar]
- 43. Helliwell S. B., Howald I., Barbet N., Hall M. N. (1998) TOR2 is part of two related signaling pathways coordinating cell growth in Saccharomyces cerevisiae. Genetics 148, 99–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mumberg D., Müller R., Funk M. (1994) Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 22, 5767–5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu Z., Sekito T., Spírek M., Thornton J., Butow R. A. (2003) Retrograde signaling is regulated by the dynamic interaction between Rtg2p and Mks1p. Mol. Cell 12, 401–411 [DOI] [PubMed] [Google Scholar]
- 46. Grand E. K., Grand F. H., Chase A. J., Ross F. M., Corcoran M. M., Oscier D. G., Cross N. C. (2004) Identification of a novel gene, FGFR1OP2, fused to FGFR1 in 8p11 myeloproliferative syndrome. Genes Chromosomes Cancer 40, 78–83 [DOI] [PubMed] [Google Scholar]
- 47. Hou M. C., Salek J., McCollum D. (2000) Mob1p interacts with the Sid2p kinase and is required for cytokinesis in fission yeast. Curr. Biol. 10, 619–622 [DOI] [PubMed] [Google Scholar]
- 48. Fu C., Iyer P., Herkal A., Abdullah J., Stout A., Free S. J. (2011) Identification and characterization of genes required for cell-to-cell fusion in Neurospora crassa. Eukaryot. Cell 10, 1100–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]