Background: The complex formed by the terminase and the connector provides the structural framework and the chemical energy for DNA packaging during bacteriophage morphogenesis.
Results: The terminase builds a pentamer that presents two conformational topologies.
Conclusion: The conformational change of the terminase might be coupled to DNA packaging.
Significance: Terminase function might be based on concerted intersubunit movement within the connector-terminase complex.
Keywords: ATPases, Bacteriophage, Electron Microscopy (EM), Protein Conformation, Virus Assembly, DNA Packaging, DNA Translocation, Single Particle Reconstruction, Terminase
Abstract
During bacteriophage morphogenesis DNA is translocated into a preformed prohead by the complex formed by the portal protein, or connector, plus the terminase, which are located at an especial prohead vertex. The terminase is a powerful motor that converts ATP hydrolysis into mechanical movement of the DNA. Here, we have determined the structure of the T7 large terminase by electron microscopy. The five terminase subunits assemble in a toroid that encloses a channel wide enough to accommodate dsDNA. The structure of the complete connector-terminase complex is also reported, revealing the coupling between the terminase and the connector forming a continuous channel. The structure of the terminase assembled into the complex showed a different conformation when compared with the isolated terminase pentamer. To understand in molecular terms the terminase morphological change, we generated the terminase atomic model based on the crystallographic structure of its phage T4 counterpart. The docking of the threaded model in both terminase conformations showed that the transition between the two states can be achieved by rigid body subunit rotation in the pentameric assembly. The existence of two terminase conformations and its possible relation to the sequential DNA translocation may shed light into the molecular bases of the packaging mechanism of bacteriophage T7.
Introduction
The assembly pathway is highly conserved in DNA bacteriophages (such as tailed phages) and in animal viruses such as herpes and adenovirus (1–3). These viruses a first assemble a protein prohead where the DNA is later packaged through a portal located at a unique icosahedral vertex. The high forces required to push the DNA against the internal pressure of the capsid are up to 110 piconewtons (4–7), and the packaging process requires the hydrolysis of one ATP molecule for the translocation of each 2.5 bp (8–10). The powerful motor involved is the complex formed by the connector (or portal protein) plus the terminase. The connector is a dodecameric assembly that sits in the prohead portal vertex and builds a channel for the passage of the DNA (11, 12). The terminase is composed by two proteins; the small one recognizes the DNA, whereas the large one provides the chemical energy for translocation. This large terminase is also involved in the processing of the concatemeric DNA to ensure the encapsidation of one single genome (in those cases where the replication process yields multimeric DNA). Once the DNA is packaged, the terminase detaches from the connector-terminase complex, and the tail assembles to the connector building the infective final virion (for review, see Refs. 13 and 14).
The connector shows an overall common architecture that has been extensively characterized in different viral systems (11). The toroidal dodecameric connectors present distinct domains to accommodate the connection between the prohead and the outer viral components (terminase, tail), with a conspicuous central channel that fits the dsDNA molecule. The symmetry mismatch between the connector (12-folded), the capsid vertex (5-fold), and the DNA (pseudo 10-fold) has been suggested as instrumental to the translocation mechanism (15), although a rotation inside the capsid has been discarded (16, 17). On the other hand, small terminases are quite variable in stoichiometry and dimensions (18–23), and the large terminases show conformational heterogeneity (14, 24–26), thus hindering in both cases their structural characterization. A common feature in large terminases is the presence of sites for ATP binding and DNA cleavage. Based on their structural similarity to other ATPases associated with diverse activities (27), it has been proposed that the large subunit undergoes a conformational change in response to ATP hydrolysis to physically move the DNA (28–31). A more detailed knowledge of the connector-terminase complex is required to understand the precise role of its components in the translocation mechanism.
The genus T7-like bacteriophages include T7 coliphage and the closely related phage T3 (32). They present an icosahedral head filled with a dsDNA of 40 kb and a short non-contractile tail. The prohead is formed by the capsid proteins (gp10, A and B), the scaffolding (gp9), the core (gp14, gp15, and gp16) and the connector (gp8). The prohead interacts with the large subunit of the terminase complex (gp19), whereas the DNA binds to the small subunit (gp18). When the DNA interacts with the prohead the packaging starts, and the capsid undergoes a maturation process (for review, see Ref. 33). The transition between the prohead and the mature capsid involves structural modifications in the shell and the core components (34–36). Although the structure of the connector of T7 has been determined at 8 Å resolution (37, 38), there is far less information on the terminase. Using a T3 defined in vitro packaging system (24, 39–41) it has been possible to quantify the energy consumption of gp19 during DNA packaging (42) as well as to determine its functional domains (25, 43).
The location of the ATPase and nuclease regions in the amino and carboxyl domains, respectively, is well conserved in many large terminases (42, 44–48). Furthermore, certain folding motifs such as the Rossmann fold in the amino domain (30) and the RNase H-like-fold are also present in the terminase carboxyl domain of T4, human citomegalovirus, SPP1, and P22 (26, 30, 49, 50). In addition, although most large terminases exist as monomers in solution (26, 47, 51, 52), its oligomerization state may change during the packaging machinery assembly, as shown in Φ29 and T4 (30, 53). However, due to its aggregation tendency, the structure or even the stoichiometry of the T7 gp19 terminase have not been defined so far (25, 43).
Here, we report the structures of the T7 pentameric large terminase both isolated and bound to the connector as part of the connector-terminase complex, obtained by electron microscopy (EM). These structures reveal two conformational states for the terminase, achieved by reordering the topology of its monomers. The coupling of symmetries between the 12-fold connector and the 5-fold terminase and the features of the gp19 morphological change provide clues for the understanding of the T7 translocation mechanism.
EXPERIMENTAL PROCEDURES
Samples Preparation
Gp19 was purified from Escherichia coli strain BL21 pLys DE3 using a two-step purification protocol as described elsewhere (54, 55). Briefly, the soluble gp19 was concentrated at 4 °C by 3–30% ammonium sulfate precipitation, purified by nickel affinity chromatography, and centrifuged either on a glycerol gradient (for ATPase activity assays) or GraFix gradient (for EM analysis). Before the gradient, the sample was dialyzed and concentrated by using Amicon ultra centrifugal tubes 0.5 ml, 10 K (Millipore). ATPase activity assays were performed using thin layer chromatography (52, 56) and radioactivity-labeled [γ-32P]ATP (Taper) as described previously (55).
For GraFix experiments (57), gp19-enriched fractions were dialyzed in buffer C: 50 mm sodium phosphate buffer, pH 7, 300 mm NaCl, 10 mm MgCl2, 1 mm ADP, 5 mm DTT, and 20% (v/v) glycerol. A density gradient primer (Gradient Master, Biocomp) was used to prepare the GraFix tubes: 20–50% glycerol and 0.09–0.15% glutaraldehyde in buffer C. Gradients were centrifuged at 35,000 rpm for 16 h at 4 °C using SW55Ti rotor. The cross-linking was finished by adding glycine 80 mm (final concentration) to the collected fractions. Aliquots of the fractions were stored at −20 °C. Bovine Serum Albumin, aldolase, and ferritin (Amersham Biosciences) were used as molecular weight markers in a control gradient (without glutaraldehyde) under identical conditions.
Gene 8 was cloned in the plasmid pET28Aa (Novagen) with 5 histidines on its carboxyl-terminal domain (kindly provided by R. Perez-Luque and M. Coll). A 1-liter culture of BL21 DE3 pLys E. coli was grown to stationary phase in LB medium containing 35 μg/ml chloramphenicol and 50 μg/ml kanamycin at 37 °C and then induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside for 2–3 h. The cells were centrifuged for 15 min at 4 °C (6000 rpm). The pellet was resuspended in 20 ml of lysis buffer: 20 mm Tris-HCl, pH 7.7, 500 mm NaCl, 50 mm MgCl2, 3 mm 2-mercaptoethanol, 10 mm imidazole, 20 μg/ml DNase, 10 μg/ml RNase, and protease inhibitors EDTA free (Roche Applied Science). The solution was incubated for 30 min on ice and centrifuged (15 min, 7000 rpm at 4 °C), and the pellet was discarded. The supernatant was processed on Cobalt Talon resin (Clontech) (2 ml resin/1l culture) using wash buffer (20 mm Tris-HCl, pH 7.7, 500 mm NaCl, 3 mm 2-mercaptoethanol, and 10 mm imidazole) and up to 350 mm imidazole in wash buffer for elution. Finally, the sample was dialyzed on 50 mm sodium phosphate buffer, pH 7, 300 mm NaCl, 10 mm MgCl2, and 20% (v/v) glycerol for the connector-terminase interaction assay.
The production of the connector-terminase complex was carried out as follows. Purified gp19 and gp8 were concentrated and dialyzed in buffer D (50 mm sodium phosphate buffer, pH 7, 300 mm NaCl, 20 mm MgCl2, 5 mm DTT, and 20% (v/v) glycerol) using Amicon ultracentrifugal tubes. Equimolar quantities (2.5:1) of both oligomeric proteins were added to the reaction volume (250 μl) containing 1 mm ATP in buffer D. The reaction mixture was incubated for 90 min at 30 or 37 °C to induce complex formation. Ultracentrifugation conditions were the same as for gp19 GraFix.
Electron Microscopy and Image Processing
For negative staining microscopy, 10-μl aliquots of the fractions containing the purified oligomeric terminase were applied to glow discharged freshly evaporated carbon copper-collodion grids for 1 h at 4 °C and stained with a 2% uranyl acetate solution (w/v). The connector and connector-terminase complex were prepared for negative staining microscopy by applying the carbon sandwich technique as described in Ohi et al. (58).
Images were collected on a Philips TecnaiG2 FEG 200 (FEI) electron microscope at 100 kV (terminase) and 200 kV (connector and connector-terminase complex) by using an Eagle 4k CCD camera to a final magnification of 67,000× under minimum dose conditions. The defocus of the selected images ranged from 1.5 to 3.2 μm. The pixel size corresponded to 4.4 Å/pixel in both the terminase and the connector and 4.2 Å/pixel in the case of the complex (59, 60).
The contrast transfer function correction was performed using the Wiener filter approach. All particle images were initially classified using reference-free methods in XMIPP: Maximum Likelihood (ML2D) (61) for the terminase and clustering approach classification methods (CL2D) (62) for the connector and the complex to select the best ones for the three-dimensional model building. The initial three-dimensional references for the three reconstructions were obtained by common line procedures using the EMAN software (63). These initial models were subjected to three-dimensional maximum likelihood classification XMIPP (ML3D) (64) or multi-reference alignment methods in EMAN software until the resolution of the preliminary models did not significantly increase. The final refinement of the three-dimensional reconstructions was performed using the projection matching protocol implemented in XMIPP software (64). Finally, on the connector-terminase complex, rotational analysis of the harmonic components of the different planes of the unsymmetrized reconstruction was used to calculate the actual symmetry of each region. The values obtained show an excellent correlation corresponding with the terminase (5-fold) and the connector (12-fold); based on that analysis, symmetry was applied to improve the quality of the reconstruction. The final resolutions using the 0.3 Fourier shell correlation criterion were 16 Å for the terminase reconstruction, 20 Å for the connector, and 29 Å for the complex. The cryo-EM model of the connector (37) was filtered to 20 Å for comparison using Spider software (65).
Structural Analysis and Segmentation
The difference volume between the connector and the connector-terminase complex was calculated using Chimera Software (66). Segmentation of the terminase from the complex reconstructed density was performed manually using Chimera software (66) by visual inspection of the density displayed at different σ levels. The electrostatic surface potential was calculated with the Coulomb Surface Coloring option in Chimera.
Generation of the gp19 Atomic Model in the Terminase EM Volumes
In this work we used HHpred (67), which is based on the comparison of the profile HMM (Hidden Markov Model), generated for the target sequence, and those in a library, generated for proteins of known structure. In this way we obtained several template candidates, the best being the gp17 protein of bacteriophage T4 (PDB code 3CPE). Based on curated alignments of the different domains of the terminases (14, 28), we refined the alignment between gp19 and gp17 and generated a final three-dimensional model with Swiss-PdbViewer and Swiss-Model (68). We also used these programs for evaluating the quality of the model.
The gp19 monomer model was manually fitted as a rigid body into the EM electron density using Chimera and refined computationally using CoLoRes program from the SITUS package (69). The calculation of the correlation coefficients at different thresholds, by applying either standard cross-correlation or Laplacian methods, leads to an unambiguous assignation of the hand in the terminase volume. The correlation coefficient between the complete pentameric model and the terminase EM volume was found to be 0.79.
RESULTS AND DISCUSSION
Purification and Characterization of Bacteriophage T7 Large Terminase
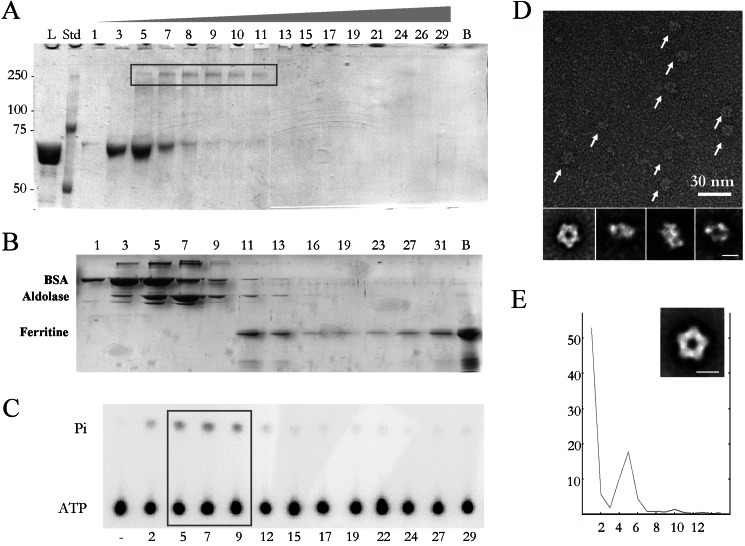
Large terminases from dsDNA bacteriophages present intrinsic instability during purification (25, 43), something that has hindered their analysis in different viral systems (14, 26). The large terminase of bacteriophage T7 is coded by gene 19, and the complete gp19 protein was expressed and purified (see “Experimental Procedures”). In parallel to the standard glycerol gradient ultracentrifugation (see above), we applied the GraFix technique (57), which combines a light fixation with ultracentrifugation to ensure the stabilization of the sample. The Grafix gradient (Fig. 1A) showed a peak of gp19 near the top of the gradient in fractions 3–6 that may correspond to its monomer form. A cross-linked, higher molar mass band was also present centered in fractions 7–11 (box, Fig. 1A). The analysis of these later fractions by EM showed the presence of ring-shaped structures consistent with oligomeric assemblies of gp19. Higher order assemblies were also found toward the bottom of the gradient, suggesting an aggregation tendency that has been characterized in previous work (55). To elucidate the approximate size of the gp19 assemblies, a control gradient was performed with several proteins acting as molecular weight markers (Fig. 1B). The bovine serum albumin (66 kDa) profile mimics the monomeric terminase (67 kDa) in fractions 3–5. The mobility of the cross-linked gp19 oligomers (fractions 7–11) falls between the aldolase (158 kDa, fractions 5–9) and ferritin (440 kDa, fractions 11–13). Considering the molecular mass of the gp19 monomer, such a profile is consistent with the pentameric assembly (335 kDa) previously reported for the T4 large terminase ring (30). To assess the functional relevance of the gp19 oligomeric states, we checked the ATPase activity, an intrinsic property of large viral terminases (42, 44–48), in the equivalent non-fixed gp19 gradient fractions. Fig. 1C shows that ATPase activity was found among fractions 5–9 (box), which are those containing the gp19 oligomers. The analysis of these fractions by electron microscopy revealed the presence of ring-like oligomers of around 160 Å in diameter (Fig. 1D, arrows). These results show that the full-length gp19 terminase oligomerizes in solution forming functionally active ring complexes, in accordance with the previous finding of ATPase activity in the gp19 oligomer provided by a completely different approach (54). The gp19 ring oligomers showed dimensions and an overall shape consistent with other previously described oligomeric terminases (29, 30). The two-dimensional averaged images (Fig. 1D, lower row) could be interpreted as end-on, tilted, and side views of the gp19 ring. Rotational analysis of end-on views showed a main peak on the fifth harmonic (Fig. 1E). The two-dimensional 5-fold averaged symmetrization of the gp19 oligomer front views (Fig. 1E, inset) supports the interpretation that the gp19 ring is, indeed, a pentamer. This is a novel result, as, although pentameric symmetry is present when terminases are assembled into the viral prohead (30, 53), most of the terminases described so far exist as monomers in solution (26, 47, 51, 52).
FIGURE 1.
Purification of the oligomeric active terminase and EM analysis. A, SDS-polyacrylamide gel electrophoresis of the fractions from a GraFix centrifugation show the presence of the gp19 monomer (fractions 3–8) and oligomer (box, fractions 5–11). Bands were detected by staining with Coomassie Brilliant Blue. L, load; Std, protein standards; lane B, bottom of the centrifuge tube. B, shown is SDS-polyacrylamide gel electrophoresis of the fractions from a gradient centrifugation of a mixture of molecular mass markers: bovine serum albumin (66 kDa, peak in fraction 3–5), aldolase (158 kDa, peak in fraction 5–9) and ferritin (440 kDa, peak in fraction 11–13). Lane B, bottom of the centrifuge tube. C, shown is measurement of the gp19 ATPase activity. ATPase assays were tested in duplicate; − represents buffer control. The concentration of gp19 tested in each assay was 0.45 μm. The box highlights the active oligomeric fractions from 5 to 9. Pi represents inorganic phosphate. D shows a negatively stained sample of a field of purified large terminases (arrows). Lower row, two-dimensional averaged images of the oligomeric terminase. E, rotational analysis of the harmonic components (x axis) of the end-on averaged image of the terminase shows the existence of 5-fold symmetry (percentage of total rotational power in the y axis). Inset, averaged image with imposed pentameric symmetry. The scale bar represents 100 Å.
Structure of the Large Terminase Pentamer and Modeling of gp19 into the EM Volume
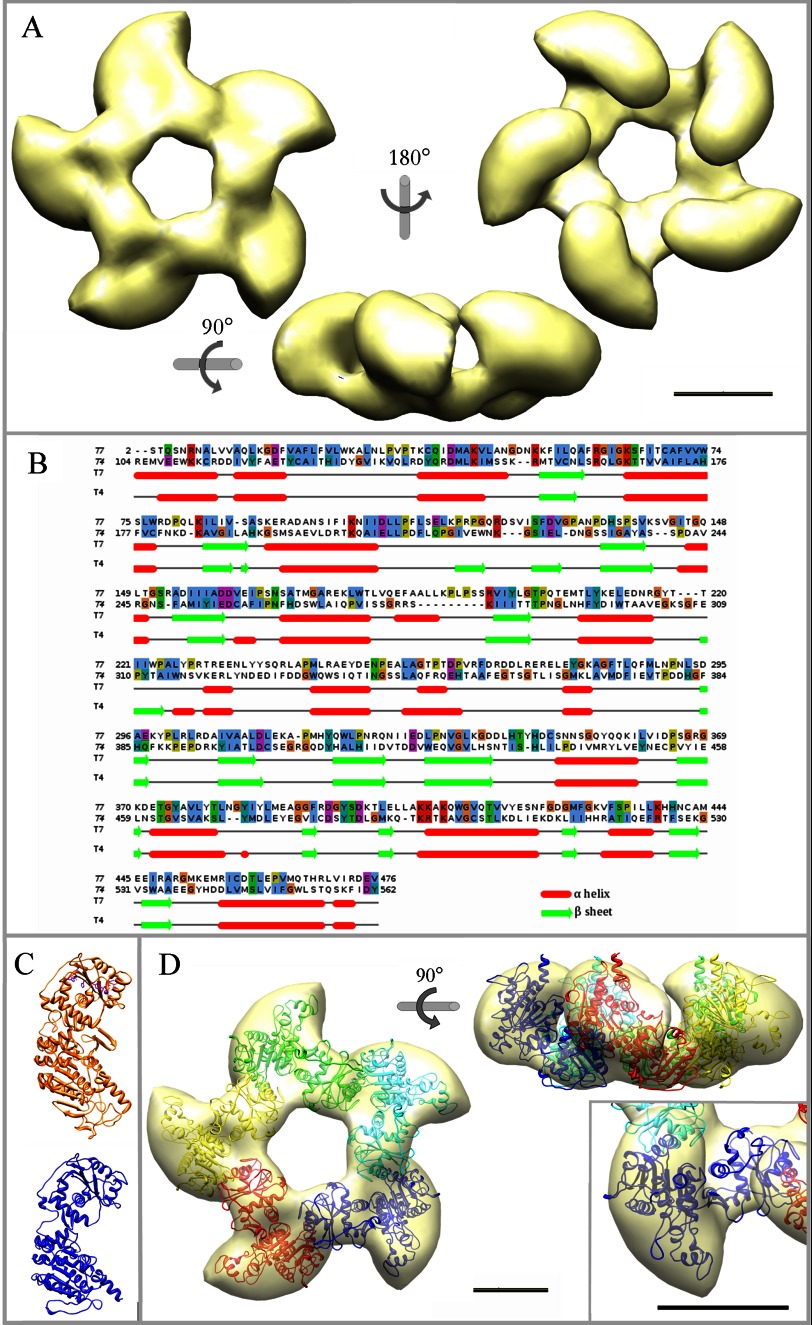
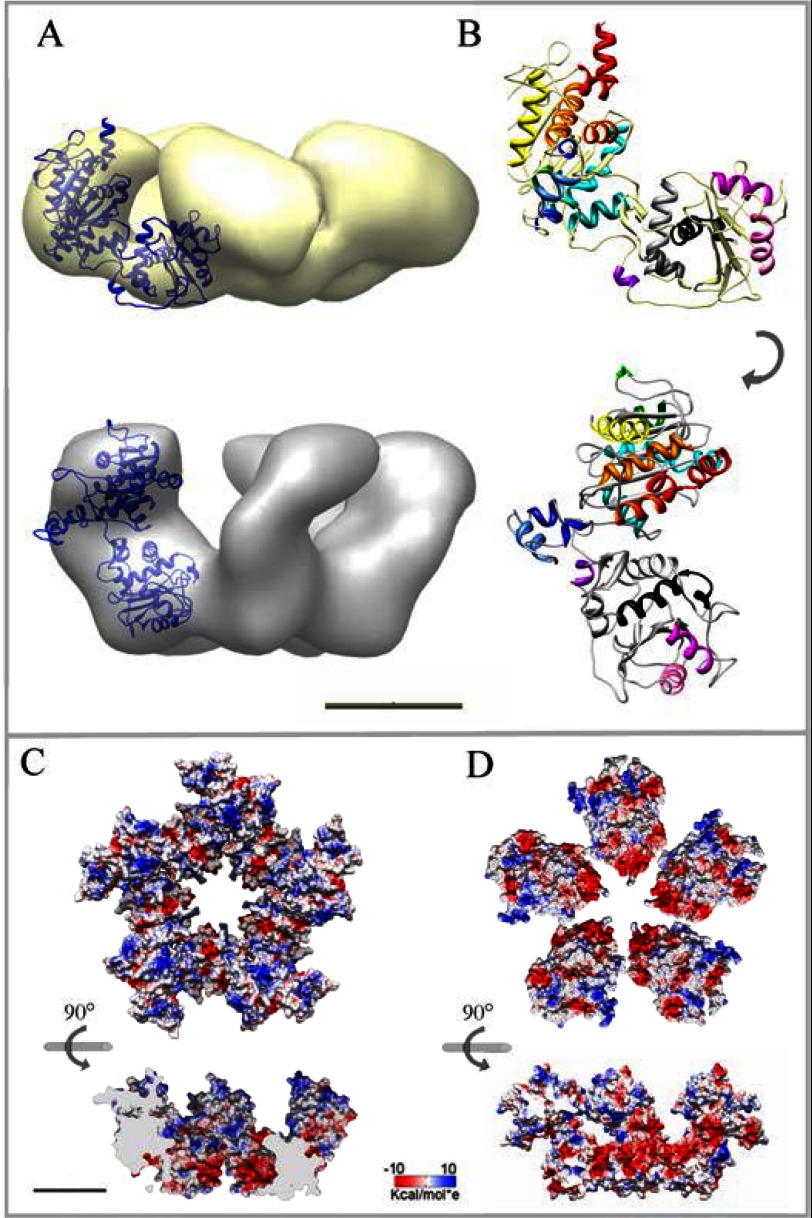
14,097 images were used to generate the three-dimensional reconstruction of the pentameric gp19 large terminase (see “Experimental Procedures”). The imposition of 5-fold symmetry to the reconstructed volume rendered a model with an estimated resolution of 16 Å (Fig. 2A). The T7 large terminase oligomer shows a 5-lobe star-shaped morphology with an external diameter of ∼160 Å and 65 Å in height. Each lobe is divided in two domains with different size (Fig. 2A, side view). Adjacent lobes are linked through their small domains, and the complete structure forms an open channel of ∼35 Å in diameter, wide enough to accommodate dsDNA (70). This general topology resembles the gp17 terminase structure derived from cryo EM of whole T4 proheads (30). The comparison of the sequences of gp19 from phage T7 and gp17 from T4 is shown in the Fig. 2B. Despite a modest homology in sequence alignments (12%), there is a consistent similarity in secondary structure prediction. This fact together with the presence in all viral terminases of functional ATPase motifs, also conserved in helicases, translocases, and restriction endonucleases (28), supported the generation of an atomic model of the gp19 monomer by-fold recognition using the T4 gp17 structure as a template (Fig. 2C, orange). The gp19 atomic model (Fig. 2C, blue) presents a major amino domain (residues 1–229) linked by a hinge region (residues 230–305) to the smaller carboxyl domain (residues 306–476). The overall shape and size of this model fit well with the two subdomains of each lobe of the EM gp19 ring (Fig. 2D, inset). The model for the pentameric form of gp19 was generated by 5-fold symmetrization of the best fit of the gp19 atomic model to the EM volume followed by a global structure refinement. The final molecular model (achieved using rigid modeling without major bending or partitioning of the gp19 molecule) closely resembles the features of the EM volume with an overall correlation coefficient of 0.79 (Fig. 2D).
FIGURE 2.
Three-dimensional reconstruction of the pentameric large terminase and fitting of the gp19 atomic model. A, shown is an EM reconstruction of the pentameric terminase. The structure is shown in three different orientations, as indicated. B, sequence alignment used for the modeling of the T7 large terminase (gp19, target protein) based on the template structure of the T4 large terminase (gp17, 3CPE) shows conserved motifs at the amino acid level and the secondary structures correspondence. C, shown is a comparison of the gp17 atomic structure (in orange) and the gp19 final model (in blue). D, shown is a translucent model of the terminase structure together with the fitted atomic model of the gp19 pentamer in end-on and side views. Each gp19 monomer is presented in a different color. Inset, two detailed views of the fitted monomer in different orientations. The scale bar represents 50 Å.
Although the gp19 pentameric terminase showed ATPase activity by itself (see Fig. 1C), its assembly with the rest of the packaging machinery must be instrumental to define its physiological structure (25). In fact, the topology and orientation of the terminase during its transient interaction with the prohead is mainly directed by its binding to the connector to yield the complex responsible for the DNA translocation inside the viral prohead (71). For that reason we decided to purify the connector-terminase complex to understand the specific arrangement of the packaging motor that constitutes the viral DNA translocation mechanism.
Structural Characterization of the Connector-Terminase Complex
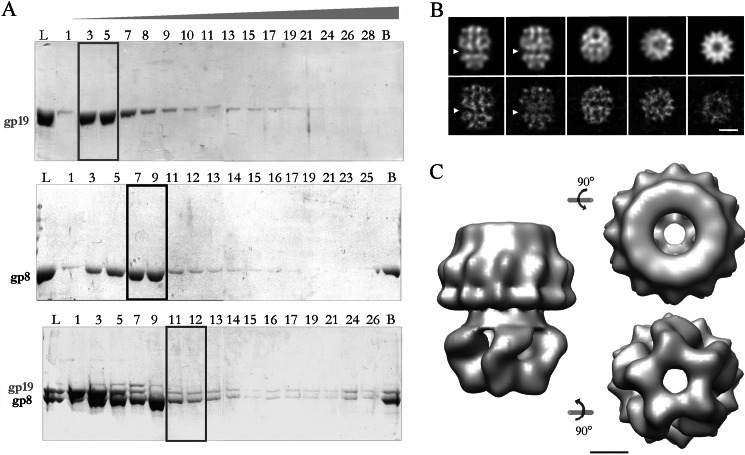
Equimolar amounts of gp19 (terminase) and purified gp8 (connector protein assembled as dodecamers) were incubated in the presence of different amounts of magnesium ions and ATP to eventually favor their interaction (43). This reaction mixture was analyzed by glycerol gradient ultracentrifugation to isolate the putative gp8-gp19 complexes (Fig. 3A). Although gp19 moved mainly in fractions 3–5 (box) and the dodecameric connectors moved mainly in fractions 7–9 (box), the gradient containing the gp8-gp19 mixture showed an additional peak of gp19 in fractions 11–12. This second peak together with the stoichiometry of gp19 and gp8 are consistent with the existence of a putative complex within these fractions (box).
FIGURE 3.
Purification and characterization of the connector-terminase complex. A, shown is an electrophoretic analysis of the fractions from glycerol gradient centrifugation of purified terminase (upper panel), connector (middle panel), and connector-terminase complex (lower panel). L, load; B, bottom of the gradient. The gp19 monomer was concentrated in fractions 3–5 (box), the peak of oligomeric connectors corresponded to fractions 7–9 (box), and an estequiometric proportion of both oligomeric proteins was observed in fractions 11–12 of the lower panel gradient (box), suggesting the existence of a putative complex. B, shown are projection images from the three-dimensional reconstructed model (upper row) and averaged views from the experimental images (lower row). The arrowhead points the proposed interface between the terminase and the connector assemblies. The scale bar corresponds to 100 Å. C, side, end-on, and bottom views of the three-dimensional reconstruction of the complex show the two morphologically different domains. The longitudinal axis of the complex is 220 Å, and the maximum diameter is 190 Å. The scale bar represents 50 Å.
The low proportion of complexes is in accordance with the transient interaction reported between gp19 and gp8. Thus we reproduced the complex purification by applying the GraFix technique, enhancing the stability and contrast of the complex for subsequent structural analysis.
The electron microscopy analysis of the gp8-gp19 complex showed ring-shaped projections, very similar in dimensions (190 Å in diameter) to the isolated connector end-on views described before (37). Other views exhibiting an elongated shape that could be interpreted as side views of the complex and their size (210–240 Å) fit with the sum of the connector and terminase heights (144 Å and 65 Å, respectively). The three-dimensional reconstruction of the complex revealed a barrel-shaped structure with two characteristics views, one circular end-on view and another elongated side view. The correlation between the projection images of the reconstructed complex model (Fig. 3B, upper row) and the averaged projection images (Fig. 3B, lower row) showed 12-fold symmetry in one side of the volume that would correspond to the connector protein. The side view revealed the presence of two different domains (one bigger than the other) separated by a constriction (Fig. 3B, arrowheads). Considering that the molar mass of the connector is almost two times bigger than the pentameric terminase (708 and 335 kDa, respectively), it is tempting to correlate the bigger domain with the connector and the smaller one with the terminase. Fig. 3C presents the three-dimensional reconstruction of the connector-terminase complex obtained from 11,650 particles to a final resolution of 30 Å. The model shows two distinguishable domains with different rotational symmetries, building a central channel. The entire complex is 220 Å high, and it has a maximum diameter of 190 Å in the 12-fold domain and 165 Å in the 5-fold domain. The width of the channel varies from 55 Å in the 12-fold part to around 30–35 Å in the 5-fold part. Although the overall morphologies, sizes, and symmetries of the two domains strongly suggest the correlation of each complex domain to either the terminase or the connector, we decided to complement the reported structure of the terminase with a three-dimensional reconstruction of the isolated connector to validate the topology of the complex.
Topology of the Packaging Complex and Modeling of the Pentameric Terminase within the Complex
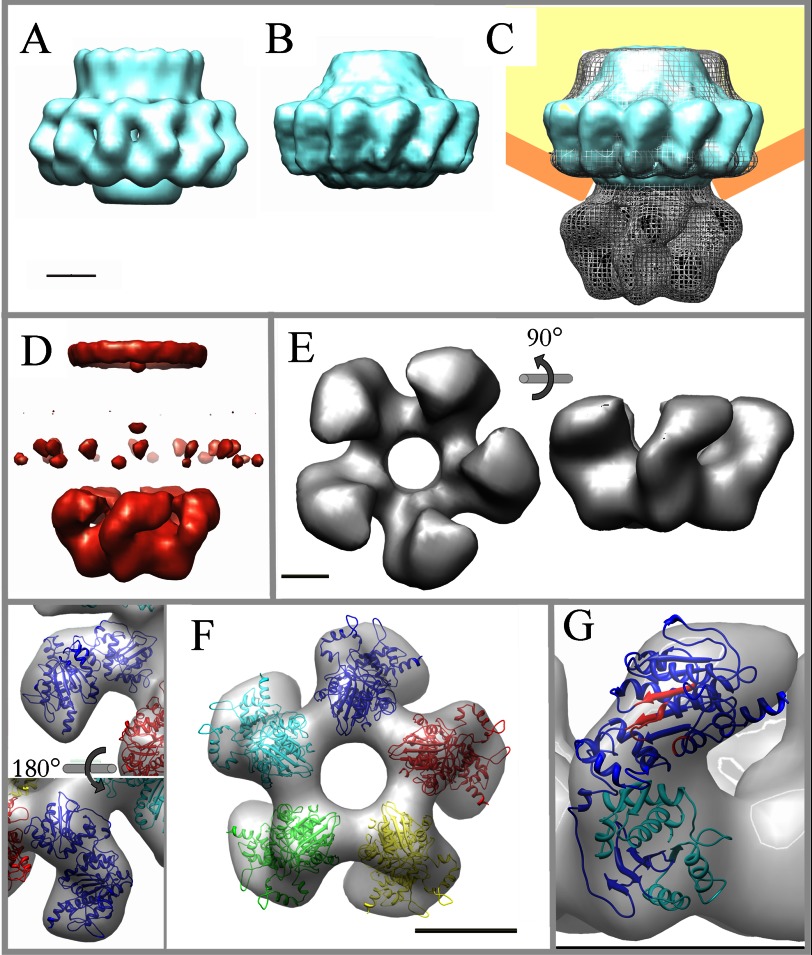
To elucidate the position of the connector within the connector-terminase complex, purified connectors were subjected to the same preparation protocol as the complex. The three-dimensional reconstruction was carried out using 17,382 images to a final resolution around 20 Å. The previously reported cryo-EM reconstruction of the connector (37, 38) was filtered to the same resolution as that obtained for the negatively stained connector (Fig. 4A) to get a direct comparison between both reconstructions. The negatively stained reconstruction (Fig. 4B) certainly resembles the cryo-EM connector and other viral connectors (72–74) considering their dimensions and overall morphology (the crown, the conspicuous wings, and the stem). The main differences involve part of the crown region (as was also reported for the SPP1 connector (72, 75)) and minor differences in the wings). Based on the good correlation between the cryo-EM and the negative staining connector structures, we compared the latter with the connector-terminase complex, both solved by negative staining (Fig. 4C, gp8 in cyan; complex in gray mesh; procapsid in orange and yellow). From the overall morphology and taking into account the internal structure of the channels, the overlay of the connector and the complex strongly supports the genuine location of gp8 within the complex (Fig. 4C). The connector builds the upper part of the complex that connects through a continuous channel to the lower region that corresponds to the large terminase. This was confirmed by the generation of a difference volume between the complex and the connector (Fig. 4D, red) that rendered, apart from minor differences in the crown and wings of the connector, the density corresponding to the terminase.
FIGURE 4.
Topology of the connector-terminase complex and docking of the gp19 atomic model. A, shown is filtered reconstruction of the cryo-EM model of the T7 connector. B, shown is three-dimensional reconstruction of the connector by negative staining. C, the three-dimensional volume of the connector (in cyan) fits well in the upper domain of the connector-terminase complex (in gray mesh). A schematic of the position of the complex in the procapsid is also included (in orange and yellow). D, shown is difference volume (in red) between the complex and the connector; besides some minor differences in the crown and wings of the connector, the main difference volume corresponds to the lower domain of the complex, which has been assigned to the terminase. E, shown is a surface representation of the pentameric terminase segmented from the connector-terminase complex in two different orientations, as indicated. F, shown is fit of the gp19 atomic model into the segmented volume of the terminase complexed with the connector. Two detailed views of monomer docking are shown in distinct orientations as indicated (left) and the docking of the pentameric atomic model in end-on view (right). G, location of the catalytic regions of the terminase structure in the connector-bound conformation is shown. ATPase motifs are shown in red, and the nuclease region is in cyan. The scale bars represent 50 Å.
Besides the general similarities among the isolated connector and the terminase to the corresponding domains of the gp8-gp19 complex, there are important differences. The interface between the connector and terminase domains revealed some structural differences possibly related to their interaction. Furthermore, there are clear differences in the arrangement of the terminase subunits comparing the isolated terminase and the structure of the terminase bound to the connector (compare Figs. 2 and 4). This observation could indicate the existence of different conformational states of gp19, as has been proposed for other packaging ATPases (29, 30). To have a deeper insight in this possibility, we segmented the terminase domain from the complex (Fig. 4E). The segmented terminase showed a star-shaped morphology with a maximum diameter of ∼160 Å and ∼85 Å height and enclosing a 35 Å diameter channel. Adjacent lobes were linked through the region opposite to the contact area with the connector, defining a vorticity in the particle. The comparison between the isolated and the connector-bound terminase pointed to a significant change in the lobes conformation, producing a global narrowing of the channel as well as a height increase of the complex-segmented terminase. To determine the reorganization of the gp19 molecule upon the interaction with the connector, we fitted the atomic model of the gp19 into the terminase volume segmented from the complex (Fig. 4F). The docking in the isolated terminase volume and the compelling similarity between the end-on views in both models (Fig. 4E, left, and Fig. 2A, right) strongly suggests the orientation of the gp19 atomic model in the complex-segmented terminase. The fitting was quite accurate (Fig. 4F, left panel), except for the domains connecting the lobes between adjacent monomers (Fig. 4F, right panel). These areas will possibly account for the carboxyl-terminal 110 amino acids of gp19 not present in our atomic model (due to the absence of corresponding residues in the threading template; see “Experimental Procedures”). Despite the considerable change among the overall shape in both terminase conformations, the fitting was obtained without modifying the structure of the monomer.
The amino acid sequence identity among the terminases from T7 and the related phage T3 is 86% (92% if we consider the similarity between the residues). The extensive biochemical characterization of gp19 in T3 (24, 25, 39–41, 43, 71, 76, 77) combined with the sequence comparison with other viral terminases (28) allowed the location of the T7 gp19 catalytic regions in the atomic model of the complex conformation (Fig. 4G). As in the rest of large terminases (44–48), the ATPase activity center is placed on its amino domain (42), which spans residue 1 to 229 and conserves the αβ folding (Fig. 4G in red), characteristic of proteins that bind and cleave ATP (revised in Draper and Rao (28)). The ATPase region comprises the characteristic nucleotide binding domains Walker A (58AFRGIGKS65), Walker B (156IIIADD161), and the adenine binding motif (38CQ39), as suggested by the absence of ATPase activity in the G61D gp19 mutant (25, 78). The gp19 carboxyl domain comprises the nuclease region, presenting a conserved RNase H/integrase folding built mostly by anti-parallel β sheets sandwiched between α helices. This conserved folding is also found, with some length variability, in the nuclease domain of terminases from virus T4, SPP1, human citomegalovirus, and P22 (26, 30, 49, 50). The T7 gp19 nuclease region, which is located in the surface of the channel entrance, encloses residues 344–429 (Fig. 4F, in cyan), and its location is further validated by the previous gp19 mutational analysis (25), which pointed to several critical residues: the H344D mutant (active in DNA packaging but defective in DNA cleavage), the H347R (defective in packaging initiation), and the G369D and G424E (defective in DNA processing).
Changes in the gp19 Oligomerization Topology Induced upon Interaction with the Connector
The comparison of the two terminase conformations revealed differences that might play a role in the terminase function during DNA translocation. The terminase bound to the connector is ∼20 Å taller than the isolated terminase (Fig. 5A), and the channel shape changes drastically, becoming narrower in the connector-bound terminase. The fitting of the monomer in both terminase structures showed that it is possible to shift from the isolated terminase to the structure of the terminase in the complex (Fig. 5B) by an outward rotation and a stand-up rigid body movement of the gp19 monomer (see supplemental Movie S1). The topology of the monomers in the complex shows the two domains almost parallel to the longitudinal axis of the complex, specifically restricting the connector interaction area to the amino domain of gp19, as proposed for the large terminase of phage T4 (79). The different topology of the gp19 monomers in the two structures generates different electrostatic environments in the channel surface (Fig. 5, C and D) as well as differences in the accessibility of the catalytic regions of gp19. The coulomb potential surface of the isolated terminase is predominantly electropositive (Fig. 5C), with stripes of negative charges nearby the channel entrance. The electrostatic surface potential of the terminase in the gp8-gp19 complex showed a conspicuous cluster of negative charges in the entrance of the channel (gp19 carboxyl domain) followed by stripes of negative and positive charges covering the rest of the channel walls (Fig. 5D). The presence of channels with a mainly electronegative surface is a common structural feature of viral connectors and other proteins involved in DNA translocation (11). The existence of a diagonal stripe of positive charges toward the amino domain and another positive cluster on top of both conformations (Fig. 5, C and D, side sections) resembles the lysine rings reported in channel walls of the Φ29 connector (80) and might be interpreted as a common feature involved someway in nucleic acids interaction.
FIGURE 5.
Two distinct conformations of the terminase pentamer; isolated terminase and complexed after interaction with the connector. A, shown is fit of the gp19 monomer atomic model into the EM envelopes of the isolated (yellow) and complex-segmented terminase (gray). B, shown is rigid body transition of the gp19 monomer atomic model from the isolated (upper site) to the complex conformation (lower site). Each helix is depicted in a different color to highlight the global movement and the absence of intramolecular bending. C, shown is electrostatic potential of the isolated terminase surface in end-on view (amino domain up) and side section of the internal channel (lower panel). D, surface potential of the complex-segmented terminase in an equivalent end-on view (upper panel) and cross-section of the internal channel (lower panel) are shown. The scale bar represents 50 Å.
The features exhibited by the T7 large terminase assembled in the complex with the connector are consistent with its role during DNA packaging. The fact that the isolated terminase is able to build also a pentameric assembly with a distinct but related structure poses the question of the possible role of different terminase conformations involved in the DNA translocation process. The existence of different conformations has been proposed previously for the T4 large terminase, where tense and relaxed states were suggested to be derived by the bending of the linker between the amino and carboxyl domains (30). These transitions involving a “power stroke” have been also described in many other hexameric ATPases (29, 81–85). Although in these cases different states are mostly related to intrasubunit changes, a more collective type of rearrangement for the terminase oligomeric assembly has been also suggested both for T4 and Φ29 terminases. The existence of large changes in translocation velocity, measured in optical tweezers experiments, was correlated to different conformational states in the T4 terminase (4). Also, the relative motion between terminase subunits has been implicated in the DNA packaging of phage Φ29 (10). In this case optical tweezers experiments have shown that the interaction between the terminase and the DNA changes during the mechano-chemical translocation cycle (8). Two defined states, the dwell and the burst, characterized by differential contacts between the terminase and the DNA, are involved in the sequential nature of DNA movement (10). In addition, the relative motion between adjacent subunits has been also observed in other DNA-translocating oligomers, as the gene 4 hexameric helicase of T7 (86).
Our results show that the large terminase of T7 can assemble in two related conformations. They configure two different electrostatic environments for the potential interaction of DNA with the terminase channel; although the isolated terminase channel has a large electropositive surface, the channel in the terminase assembled with the connector is more electronegative, thus being consistent with a smoother coaxial DNA translocation. An interesting aspect is that in both conformations, the terminase subunits maintain the accessibility of the ATPase active site in the outer surface of the oligomer. It is thus tempting to suggest that these two related topologies could be the first structural evidences of the different conformations involved in the DNA translocation suggested from the functional analysis of the T4 and Φ29 terminases. The fact that ATP hydrolysis by the T7 terminase leads to a large change in the oligomeric topology, as measured in microcantilever assays (54), also points to the relative motion of terminase subunits coupled to ATP hydrolysis. Further experiments directly correlating these structures with functional translocation states will be required to provide additional clues to understand the translocation mechanism of T7 DNA by the viral packaging machinery.
Acknowledgments
We are in debt to Sonia Moreno for the connector cloning and purification. We greatly acknowledge Melisa Lazaro and Mikel Valle for help with the GraFix experiments. We are grateful to Rocio Arranz for the EM support. We also thank Florencio Pazos for the structure prediction.
Footnotes
This work was supported by Grants BFU 2011-29038 of Ministerio de Ciencia e Innovacion (MICINN) and S2009/Mat 1507 from Comunidad de Madrid (to J. L. C.) and BFU 2011-25090/BMC of MICINN (to J. M.-B.).

This article contains supplemental Movie S1.
The atomic coordinates and structure factors (codes 4BIJ and 4BIL) have been deposited in the Protein Data Bank (http://wwpdb.org/).
The three-dimensional EM volumes have been deposited in the EM Data Base (http://www.ebi.ac.uk/pdbe/emdb/) with accession numbers EMD-2355 and EMD-2356.
REFERENCES
- 1. Bamford D. H., Grimes J. M., Stuart D. I. (2005) What does structure tell us about virus evolution? Curr. Opin. Struct. Biol. 15, 655–663 [DOI] [PubMed] [Google Scholar]
- 2. Cuervo A., Carrascosa J. L. (2012) Bacteriophages. Structure. eLS, DOI 10.1002/9780470015902.a0024053 [DOI] [Google Scholar]
- 3. Mettenleiter T. C., Klupp B. G., Granzow H. (2006) Herpesvirus assembly. A tale of two membranes. Curr. Opin. Microbiol. 9, 423–429 [DOI] [PubMed] [Google Scholar]
- 4. Fuller D. N., Raymer D. M., Kottadiel V. I., Rao V. B., Smith D. E. (2007) Single phage T4 DNA packaging motors exhibit large force generation, high velocity, and dynamic variability. Proc. Natl. Acad. Sci. U.S.A. 104, 16868–16873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fuller D. N., Raymer D. M., Rickgauer J. P., Robertson R. M., Catalano C. E., Anderson D. L., Grimes S., Smith D. E. (2007) Measurements of single DNA molecule packaging dynamics in bacteriophage λ reveal high forces, high motor processivity, and capsid transformations. J. Mol. Biol. 373, 1113–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rickgauer J. P., Fuller D. N., Grimes S., Jardine P. J., Anderson D. L., Smith D. E. (2008) Portal motor velocity and internal force resisting viral DNA packaging in bacteriophage φ29. Biophys. J. 94, 159–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Smith D. E., Tans S. J., Smith S. B., Grimes S., Anderson D. L., Bustamante C. (2001) The bacteriophage straight φ29 portal motor can package DNA against a large internal force. Nature 413, 748–752 [DOI] [PubMed] [Google Scholar]
- 8. Aathavan K., Politzer A. T., Kaplan A., Moffitt J. R., Chemla Y. R., Grimes S., Jardine P. J., Anderson D. L., Bustamante C. (2009) Substrate interactions and promiscuity in a viral DNA packaging motor. Nature 461, 669–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chemla Y. R., Aathavan K., Michaelis J., Grimes S., Jardine P. J., Anderson D. L., Bustamante C. (2005) Mechanism of force generation of a viral DNA packaging motor. Cell 122, 683–692 [DOI] [PubMed] [Google Scholar]
- 10. Moffitt J. R., Chemla Y. R., Aathavan K., Grimes S., Jardine P. J., Anderson D. L., Bustamante C. (2009) Intersubunit coordination in a homomeric ring ATPase. Nature 457, 446–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cuervo A., Carrascosa J. L. (2012) Viral connectors for DNA encapsulation. Curr. Opin. Biotechnol. 23, 529–536 [DOI] [PubMed] [Google Scholar]
- 12. Valpuesta J. M., Carrascosa J. L. (1994) Structure of viral connectors and their function in bacteriophage assembly and DNA packaging. Q. Rev. Biophys. 27, 107–155 [DOI] [PubMed] [Google Scholar]
- 13. Casjens S. R. (2011) The DNA-packaging nanomotor of tailed bacteriophages. Nat. Rev. Microbiol. 9, 647–657 [DOI] [PubMed] [Google Scholar]
- 14. Rao V. B., Feiss M. (2008) The bacteriophage DNA packaging motor. Annu. Rev. Genet. 42, 647–681 [DOI] [PubMed] [Google Scholar]
- 15. Hendrix R. W. (1978) Symmetry mismatch and DNA packaging in large bacteriophages. Proc. Natl. Acad. Sci. U.S.A. 75, 4779–4783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Baumann R. G., Mullaney J., Black L. W. (2006) Portal fusion protein constraints on function in DNA packaging of bacteriophage T4. Mol. Microbiol. 61, 16–32 [DOI] [PubMed] [Google Scholar]
- 17. Hugel T., Michaelis J., Hetherington C. L., Jardine P. J., Grimes S., Walter J. M., Falk W., Anderson D. L., Bustamante C. (2007) Experimental test of connector rotation during DNA packaging into bacteriophage varφ29 capsids. PLoS Biol. 5, e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sun S., Gao S., Kondabagil K., Xiang Y., Rossmann M. G., Rao V. B. (2012) Structure and function of the small terminase component of the DNA packaging machine in T4-like bacteriophages. Proc. Natl. Acad. Sci. U.S.A. 109, 817–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhao H., Kamau Y. N., Christensen T. E., Tang L. (2012) Structural and functional studies of the phage Sf6 terminase small subunit reveal a DNA-spooling device facilitated by structural plasticity. J. Mol. Biol. 423, 413–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Büttner C. R., Chechik M., Ortiz-Lombardía M., Smits C., Ebong I. O., Chechik V., Jeschke G., Dykeman E., Benini S., Robinson C. V., Alonso J. C., Antson A. A. (2012) Structural basis for DNA recognition and loading into a viral packaging motor. Proc. Natl. Acad. Sci. U.S.A. 109, 811–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. de Beer T., Fang J., Ortega M., Yang Q., Maes L., Duffy C., Berton N., Sippy J., Overduin M., Feiss M., Catalano C. E. (2002) Insights into specific DNA recognition during the assembly of a viral genome packaging machine. Mol. Cell 9, 981–991 [DOI] [PubMed] [Google Scholar]
- 22. Nemecek D., Lander G. C., Johnson J. E., Casjens S. R., Thomas G. J., Jr. (2008) Assembly architecture and DNA binding of the bacteriophage P22 terminase small subunit. J. Mol. Biol. 383, 494–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Roy A., Bhardwaj A., Datta P., Lander G. C., Cingolani G. (2012) Small terminase couples viral DNA binding to genome-packaging ATPase activity. Structure 20, 1403–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hamada K., Fujisawa H., Minagawa T. (1986) A defined in vitro system for packaging of bacteriophage T3 DNA. Virology 151, 119–123 [DOI] [PubMed] [Google Scholar]
- 25. Morita M., Tasaka M., Fujisawa H. (1994) Analysis of functional domains of the packaging proteins of bacteriophage T3 by site-directed mutagenesis. J. Mol. Biol. 235, 248–259 [DOI] [PubMed] [Google Scholar]
- 26. Nadal M., Mas P. J., Mas P. J., Blanco A. G., Arnan C., Solà M., Hart D. J., Coll M. (2010) Structure and inhibition of herpesvirus DNA packaging terminase nuclease domain. Proc. Natl. Acad. Sci. U.S.A. 107, 16078–16083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hanson P. I., Whiteheart S. W. (2005) AAA+ proteins. Have engine, will work. Nat. Rev. Mol. Cell Biol. 6, 519–529 [DOI] [PubMed] [Google Scholar]
- 28. Draper B., Rao V. B. (2007) An ATP hydrolysis sensor in the DNA packaging motor from bacteriophage T4 suggests an inchworm-type translocation mechanism. J. Mol. Biol. 369, 79–94 [DOI] [PubMed] [Google Scholar]
- 29. Mancini E. J., Kainov D. E., Grimes J. M., Tuma R., Bamford D. H., Stuart D. I. (2004) Atomic snapshots of an RNA packaging motor reveal conformational changes linking ATP hydrolysis to RNA translocation. Cell 118, 743–755 [DOI] [PubMed] [Google Scholar]
- 30. Sun S., Kondabagil K., Draper B., Alam T. I., Bowman V. D., Zhang Z., Hegde S., Fokine A., Rossmann M. G., Rao V. B. (2008) The structure of the phage t4 DNA packaging motor suggests a mechanism dependent on electrostatic forces. Cell 135, 1251–1262 [DOI] [PubMed] [Google Scholar]
- 31. Sun S., Kondabagil K., Gentz P. M., Rossmann M. G., Rao V. B. (2007) The structure of the ATPase that powers DNA packaging into bacteriophage T4 procapsids. Mol. Cell 25, 943–949 [DOI] [PubMed] [Google Scholar]
- 32. Davis R. W., Hyman R. W. (1971) A study in evolution. The DNA base sequence homology between coliphages T7 and T3. J. Mol. Biol. 62, 287–301 [DOI] [PubMed] [Google Scholar]
- 33. Cerritelli M. E., Conway J. F., Cheng N., Trus B. L., Steven A. C. (2003) Molecular mechanisms in bacteriophage T7 procapsid assembly, maturation, and DNA containment. Adv. Protein Chem. 64, 301–323 [DOI] [PubMed] [Google Scholar]
- 34. Agirrezabala X., Martín-Benito J., Castón J. R., Miranda R., Valpuesta J. M., Carrascosa J. L. (2005) Maturation of phage T7 involves structural modification of both shell and inner core components. EMBO J. 24, 3820–3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Agirrezabala X., Velázquez-Muriel J. A., Gómez-Puertas P., Scheres S. H., Carazo J. M., Carrascosa J. L. (2007) Quasi-atomic model of bacteriophage t7 procapsid shell. Insights into the structure and evolution of a basic fold. Structure 15, 461–472 [DOI] [PubMed] [Google Scholar]
- 36. Ionel A., Velázquez-Muriel J. A., Luque D., Cuervo A., Castón J. R., Valpuesta J. M., Martín-Benito J., Carrascosa J. L. (2011) Molecular rearrangements involved in the capsid shell maturation of bacteriophage T7. J. Biol. Chem. 286, 234–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Agirrezabala X., Martín-Benito J., Valle M., González J. M., Valencia A., Valpuesta J. M., Carrascosa J. L. (2005) Structure of the connector of bacteriophage T7 at 8A resolution. Structural homologies of a basic component of a DNA translocating machinery. J. Mol. Biol. 347, 895–902 [DOI] [PubMed] [Google Scholar]
- 38. Fernández J. J., Luque D., Castón J. R., Carrascosa J. L. (2008) Sharpening high resolution information in single particle electron cryomicroscopy. J. Struct. Biol. 164, 170–175 [DOI] [PubMed] [Google Scholar]
- 39. Hamada K., Fujisawa H., Minagawa T. (1987) Characterization of ATPase activity of a defined in vitro system for packaging of bacteriophage T3 DNA. Virology 159, 244–249 [DOI] [PubMed] [Google Scholar]
- 40. Shibata H., Fujisawa H., Minagawa T. (1987) Characterization of the bacteriophage T3 DNA packaging reaction in vitro in a defined system. J. Mol. Biol. 196, 845–851 [DOI] [PubMed] [Google Scholar]
- 41. Shibata H., Fujisawa H., Minagawa T. (1987) Early events in DNA packaging in a defined in vitro system of bacteriophage T3. Virology 159, 250–258 [DOI] [PubMed] [Google Scholar]
- 42. Morita M., Tasaka M., Fujisawa H. (1993) DNA packaging ATPase of bacteriophage T3. Virology 193, 748–752 [DOI] [PubMed] [Google Scholar]
- 43. Hamada K., Fujisawa H., Minagawa T. (1986) Overproduction and purification of the products of bacteriophage T3 genes 18 and 19, two genes involved in DNA packaging. Virology 151, 110–118 [DOI] [PubMed] [Google Scholar]
- 44. Goetzinger K. R., Rao V. B. (2003) Defining the ATPase center of bacteriophage T4 DNA packaging machine. Requirement for a catalytic glutamate residue in the large terminase protein gp17. J. Mol. Biol. 331, 139–154 [DOI] [PubMed] [Google Scholar]
- 45. Guo P., Peterson C., Anderson D. (1987) Prohead and DNA-gp3-dependent ATPase activity of the DNA packaging protein gp16 of bacteriophage φ29. J. Mol. Biol. 197, 229–236 [DOI] [PubMed] [Google Scholar]
- 46. Hwang Y., Feiss M. (1996) Mutations affecting the high affinity ATPase center of gpA, the large subunit of bacteriophage λ terminase, inactivate the endonuclease activity of terminase. J. Mol. Biol. 261, 524–535 [DOI] [PubMed] [Google Scholar]
- 47. Nemecek D., Gilcrease E. B., Kang S., Prevelige P. E., Jr., Casjens S., Thomas G. J., Jr. (2007) Subunit conformations and assembly states of a DNA-translocating motor. The terminase of bacteriophage P22. J. Mol. Biol. 374, 817–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Oliveira L., Henriques A. O., Tavares P. (2006) Modulation of the viral ATPase activity by the portal protein correlates with DNA packaging efficiency. J. Biol. Chem. 281, 21914–21923 [DOI] [PubMed] [Google Scholar]
- 49. Roy A., Cingolani G. (2012) Structure of p22 headful packaging nuclease. J. Biol. Chem. 287, 28196–28205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Smits C., Chechik M., Kovalevskiy O. V., Shevtsov M. B., Foster A. W., Alonso J. C., Antson A. A. (2009) Structural basis for the nuclease activity of a bacteriophage large terminase. EMBO Rep. 10, 592–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gual A., Camacho A. G., Alonso J. C. (2000) Functional analysis of the terminase large subunit, G2P, of Bacillus subtilis bacteriophage SPP1. J. Biol. Chem. 275, 35311–35319 [DOI] [PubMed] [Google Scholar]
- 52. Leffers G., Rao V. B. (2000) Biochemical characterization of an ATPase activity associated with the large packaging subunit gp17 from bacteriophage T4. J. Biol. Chem. 275, 37127–37136 [DOI] [PubMed] [Google Scholar]
- 53. Simpson A. A., Tao Y., Leiman P. G., Badasso M. O., He Y., Jardine P. J., Olson N. H., Morais M. C., Grimes S., Anderson D. L., Baker T. S., Rossmann M. G. (2000) Structure of the bacteriophage φ29 DNA packaging motor. Nature 408, 745–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mertens J., Daudén M. I., Carrascosa J. L., Tamayo J. (2012) Stepwise motion of a microcantilever driven by the hydrolysis of viral ATPases. Nanotechnology 23, 015501 [DOI] [PubMed] [Google Scholar]
- 55. Mertens J., Dauden M. I., Carrascosa J. L., Tamayo J. (2012) Interaction of viral ATPases with nucleotides measured with a microcantilever. Sensors and Actuators B: Chemical 171–172, 263–270 [Google Scholar]
- 56. Manne V., Rao V. B., Black L. W. (1982) A bacteriophage T4 DNA packaging related DNA-dependent ATPase-endonuclease. J. Biol. Chem. 257, 13223–13232 [PubMed] [Google Scholar]
- 57. Kastner B., Fischer N., Golas M. M., Sander B., Dube P., Boehringer D., Hartmuth K., Deckert J., Hauer F., Wolf E., Uchtenhagen H., Urlaub H., Herzog F., Peters J. M., Poerschke D., Lührmann R., Stark H. (2008) GraFix. Sample preparation for single-particle electron cryomicroscopy. Nat. Methods 5, 53–55 [DOI] [PubMed] [Google Scholar]
- 58. Ohi M., Li Y., Cheng Y., Walz T. (2004) Negative staining and image classification. Powerful tools in modern electron microscopy. Biol. Proced. Online 6, 23–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Marabini R., Masegosa I. M., San Martin M. C., Marco S., Fernandez J. J., de la Fraga L. G., Vaquerizo C., Carazo J. M. (1996) Xmipp. An image processing package for electron microscopy. J. Struct. Biol. 116, 237–240 [DOI] [PubMed] [Google Scholar]
- 60. Sorzano C. O., Marabini R., Velázquez-Muriel J., Bilbao-Castro J. R., Scheres S. H., Carazo J. M., Pascual-Montano A. (2004) XMIPP. A new generation of an open-source image processing package for electron microscopy. J. Struct. Biol. 148, 194–204 [DOI] [PubMed] [Google Scholar]
- 61. Scheres S. H., Valle M., Carazo J. M. (2005) Fast maximum-likelihood refinement of electron microscopy images. Bioinformatics 21, ii243–244 [DOI] [PubMed] [Google Scholar]
- 62. Sorzano C. O., Bilbao-Castro J. R., Shkolnisky Y., Alcorlo M., Melero R., Caffarena-Fernández G., Li M., Xu G., Marabini R., Carazo J. M. (2010) A clustering approach to multireference alignment of single-particle projections in electron microscopy. J. Struct. Biol. 171, 197–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ludtke S. J., Baldwin P. R., Chiu W. (1999) EMAN. Semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 [DOI] [PubMed] [Google Scholar]
- 64. Scheres S. H., Gao H., Valle M., Herman G. T., Eggermont P. P., Frank J., Carazo J. M. (2007) Disentangling conformational states of macromolecules in 3D-EM through likelihood optimization. Nat. Methods 4, 27–29 [DOI] [PubMed] [Google Scholar]
- 65. Frank J., Radermacher M., Penczek P., Zhu J., Li Y., Ladjadj M., Leith A. (1996) SPIDER and WEB. Processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 [DOI] [PubMed] [Google Scholar]
- 66. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera. A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 67. Söding J., Biegert A., Lupas A. N. (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Guex N., Peitsch M. C. (1997) SWISS-MODEL and the Swiss-PdbViewer. An environment for comparative protein modeling. Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 69. Wriggers W., Milligan R. A., McCammon J. A. (1999) Situs. A package for docking crystal structures into low-resolution maps from electron microscopy. J. Struct. Biol. 125, 185–195 [DOI] [PubMed] [Google Scholar]
- 70. Vlieghe D., Turkenburg J. P., Van Meervelt L. (1999) B-DNA at atomic resolution reveals extended hydration patterns. Acta Crystallogr. D. Biol. Crystallogr. 55, 1495–1502 [DOI] [PubMed] [Google Scholar]
- 71. Morita M., Tasaka M., Fujisawa H. (1995) Analysis of the fine structure of the prohead binding domain of the packaging protein of bacteriophage T3 using a hexapeptide, an analog of a prohead binding site. Virology 211, 516–524 [DOI] [PubMed] [Google Scholar]
- 72. Orlova E. V., Dube P., Beckmann E., Zemlin F., Lurz R., Trautner T. A., Tavares P., van Heel M. (1999) Structure of the 13-fold symmetric portal protein of bacteriophage SPP1. Nat. Struct. Biol. 6, 842–846 [DOI] [PubMed] [Google Scholar]
- 73. Trus B. L., Cheng N., Newcomb W. W., Homa F. L., Brown J. C., Steven A. C. (2004) Structure and polymorphism of the UL6 portal protein of herpes simplex virus type 1. J. Virol. 78, 12668–12671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Valpuesta J. M., Sousa N., Barthelemy I., Fernández J. J., Fujisawa H., Ibarra B., Carrascosa J. L. (2000) Structural analysis of the bacteriophage T3 head-to-tail connector. J. Struct. Biol. 131, 146–155 [DOI] [PubMed] [Google Scholar]
- 75. Orlova E. V., Gowen B., Dröge A., Stiege A., Weise F., Lurz R., van Heel M., Tavares P. (2003) Structure of a viral DNA gatekeeper at 10 Å resolution by cryo-electron microscopy. EMBO J. 22, 1255–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Fujisawa H., Shibata H., Kato H. (1991) Analysis of interactions among factors involved in the bacteriophage T3 DNA packaging reaction in a defined in vitro system. Virology 185, 788–794 [DOI] [PubMed] [Google Scholar]
- 77. Morita M., Tasaka M., Fujisawa H. (1995) Structural and functional domains of the large subunit of the bacteriophage T3 DNA packaging enzyme. Importance of the C-terminal region in prohead binding. J. Mol. Biol. 245, 635–644 [DOI] [PubMed] [Google Scholar]
- 78. Kimura M., Fujisawa H. (1991) Dissection of functional domains of the packaging protein of bacteriophage T3 by site-directed mutagenesis. Virology 180, 709–715 [DOI] [PubMed] [Google Scholar]
- 79. Hegde S., Padilla-Sanchez V., Draper B., Rao V. B. (2012) Portal-large terminase interactions of the bacteriophage T4 DNA packaging machine implicate a molecular lever mechanism for coupling ATPase to DNA translocation. J. Virol. 86, 4046–4057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Guasch A., Pous J., Ibarra B., Gomis-Rüth F. X., Valpuesta J. M., Sousa N., Carrascosa J. L., Coll M. (2002) Detailed architecture of a DNA translocating machine. The high-resolution structure of the bacteriophage φ29 connector particle. J. Mol. Biol. 315, 663–676 [DOI] [PubMed] [Google Scholar]
- 81. Lee J. Y., Yang W. (2006) UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell 127, 1349–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Massey T. H., Mercogliano C. P., Yates J., Sherratt D. J., Löwe J. (2006) Double-stranded DNA translocation. Structure and mechanism of hexameric FtsK. Mol. Cell 23, 457–469 [DOI] [PubMed] [Google Scholar]
- 83. Nishizaka T., Oiwa K., Noji H., Kimura S., Muneyuki E., Yoshida M., Kinosita K., Jr. (2004) Chemomechanical coupling in F1-ATPase revealed by simultaneous observation of nucleotide kinetics and rotation. Nat. Struct. Mol. Biol. 11, 142–148 [DOI] [PubMed] [Google Scholar]
- 84. Oster G., Wang H. (2000) Reverse engineering a protein. The mechanochemistry of ATP synthase. Biochim. Biophys. Acta 1458, 482–510 [DOI] [PubMed] [Google Scholar]
- 85. Wang H., Oster G. (1998) Energy transduction in the F1 motor of ATP synthase. Nature 396, 279–282 [DOI] [PubMed] [Google Scholar]
- 86. Singleton M. R., Sawaya M. R., Ellenberger T., Wigley D. B. (2000) Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell 101, 589–600 [DOI] [PubMed] [Google Scholar]