Abstract
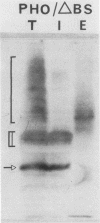
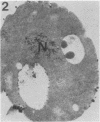
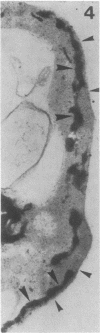
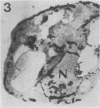
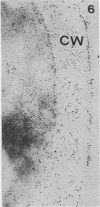
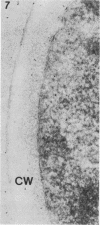
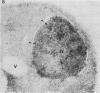
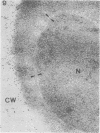
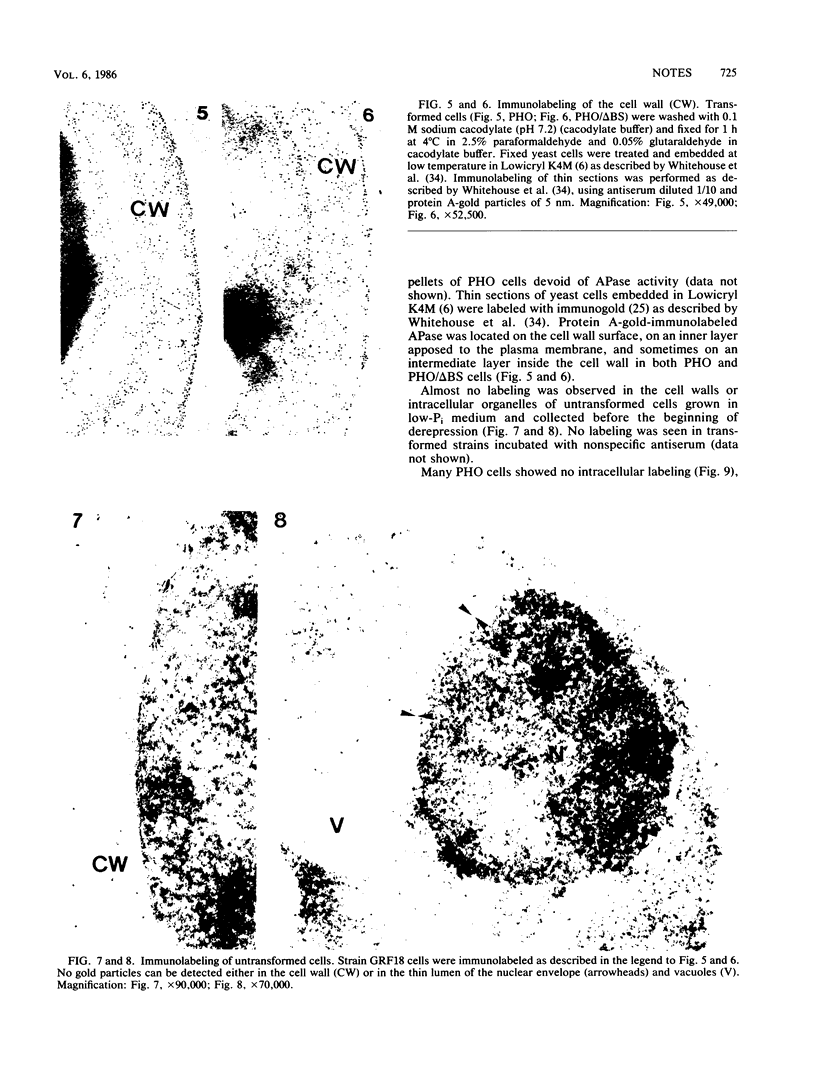
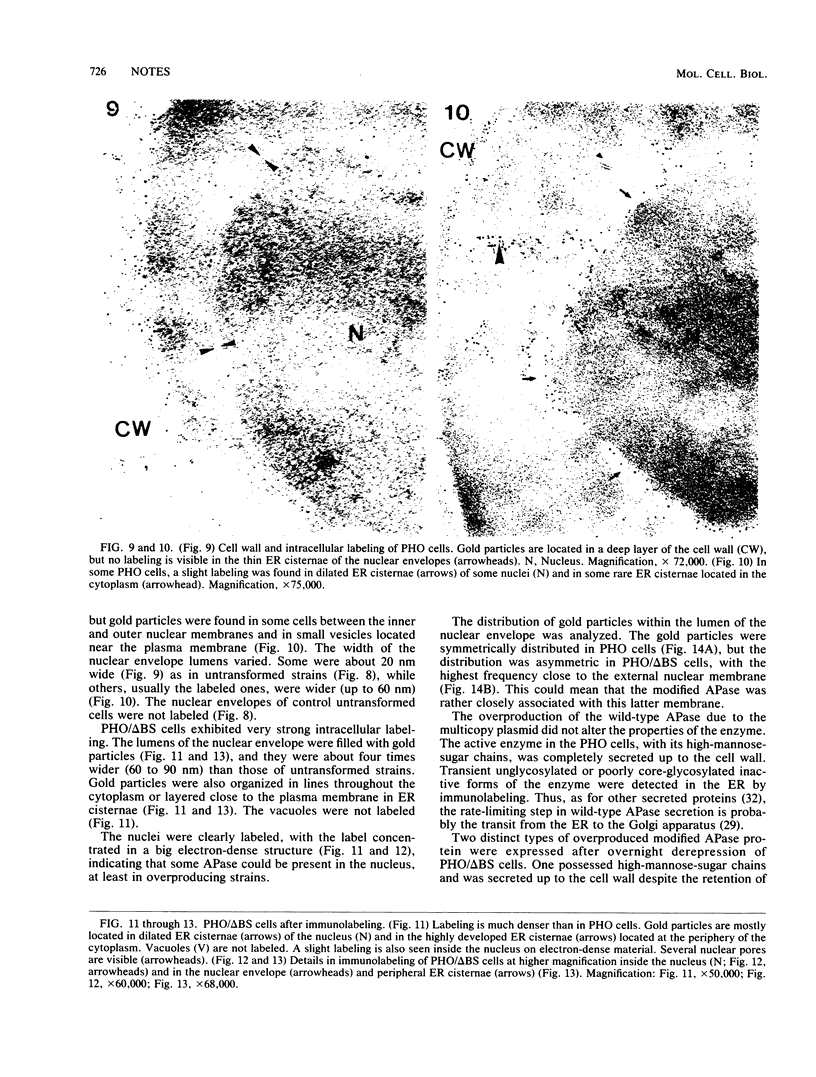
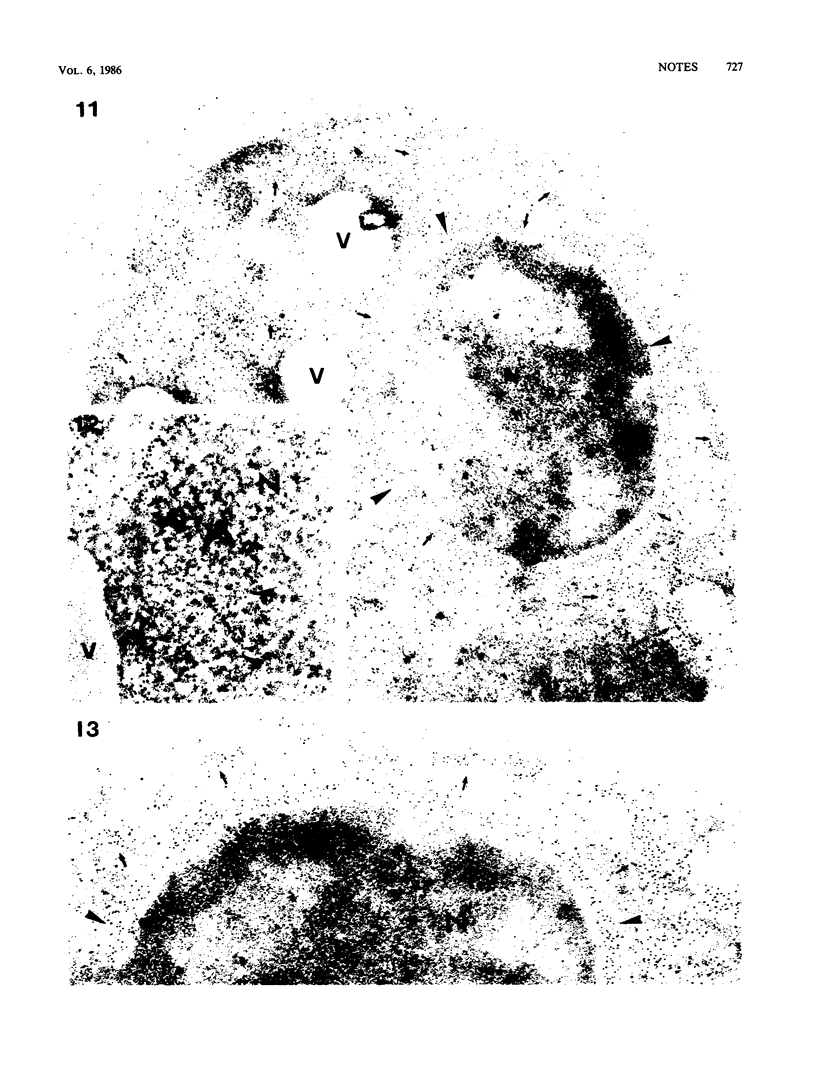
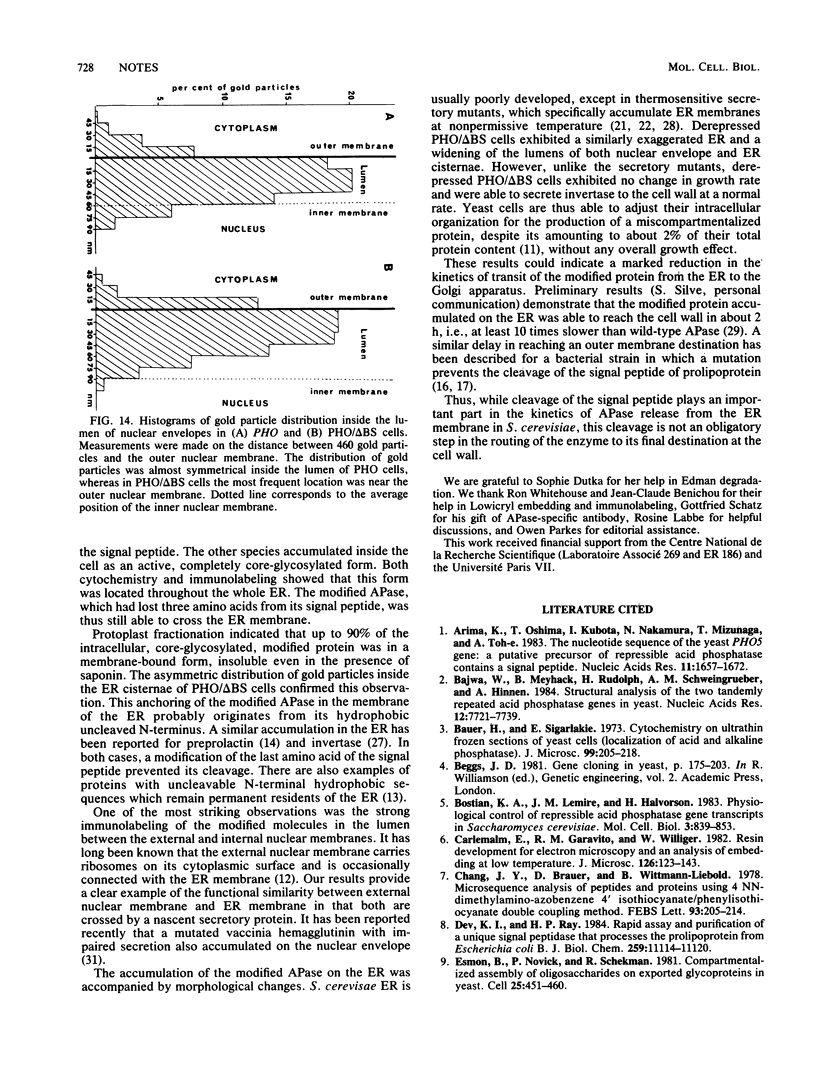
We studied ultrastructural localization of acid phosphatase in derepressed Saccharomyces cerevisiae cells transformed with a multicopy plasmid carrying either the wild-type PHO5 gene or a PHO5 gene deleted in the region overlapping the signal peptidase cleavage site. Wild-type enzyme was located in the cell wall, as was 50% of the modified protein, which carried high-mannose-sugar chains. The remaining 50% of the protein was active and core glycosylated, and it accumulated in the endoplasmic reticulum cisternae. The signal peptide remained uncleaved in both forms. Cells expressing the modified protein exhibited an exaggerated endoplasmic reticulum with dilated lumen.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arima K., Oshima T., Kubota I., Nakamura N., Mizunaga T., Toh-e A. The nucleotide sequence of the yeast PHO5 gene: a putative precursor of repressible acid phosphatase contains a signal peptide. Nucleic Acids Res. 1983 Mar 25;11(6):1657–1672. doi: 10.1093/nar/11.6.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajwa W., Meyhack B., Rudolph H., Schweingruber A. M., Hinnen A. Structural analysis of the two tandemly repeated acid phosphatase genes in yeast. Nucleic Acids Res. 1984 Oct 25;12(20):7721–7739. doi: 10.1093/nar/12.20.7721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostian K. A., Lemire J. M., Halvorson H. O. Physiological control of repressible acid phosphatase gene transcripts in Saccharomyces cerevisiae. Mol Cell Biol. 1983 May;3(5):839–853. doi: 10.1128/mcb.3.5.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dev I. K., Ray P. H. Rapid assay and purification of a unique signal peptidase that processes the prolipoprotein from Escherichia coli B. J Biol Chem. 1984 Sep 10;259(17):11114–11120. [PubMed] [Google Scholar]
- Esmon B., Novick P., Schekman R. Compartmentalized assembly of oligosaccharides on exported glycoproteins in yeast. Cell. 1981 Aug;25(2):451–460. doi: 10.1016/0092-8674(81)90063-5. [DOI] [PubMed] [Google Scholar]
- Goldstein A., Lampen J. O. Beta-D-fructofuranoside fructohydrolase from yeast. Methods Enzymol. 1975;42:504–511. doi: 10.1016/0076-6879(75)42159-0. [DOI] [PubMed] [Google Scholar]
- Haguenauer-Tsapis R., Hinnen A. A deletion that includes the signal peptidase cleavage site impairs processing, glycosylation, and secretion of cell surface yeast acid phosphatase. Mol Cell Biol. 1984 Dec;4(12):2668–2675. doi: 10.1128/mcb.4.12.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris J. R. The biochemistry and ultrastructure of the nuclear envelope. Biochim Biophys Acta. 1978 Apr 10;515(1):55–104. doi: 10.1016/0304-4157(78)90008-4. [DOI] [PubMed] [Google Scholar]
- Hortin G., Boime I. Transport of an uncleaved preprotein into the endoplasmic reticulum of rat pituitary cells. J Biol Chem. 1981 Feb 25;256(4):1491–1494. [PubMed] [Google Scholar]
- Hortsch M., Meyer D. I. Pushing the signal hypothesis: what are the limits? Biol Cell. 1984;52(1 Pt A):1–8. doi: 10.1111/j.1768-322x.1985.tb00319.x. [DOI] [PubMed] [Google Scholar]
- Kreil G., Mollay C., Kaschnitz R., Haiml L., Vilas U. Prepromelittin: specific cleavage of the pre- and the propeptide in vitro. Ann N Y Acad Sci. 1980;343:338–346. doi: 10.1111/j.1749-6632.1980.tb47262.x. [DOI] [PubMed] [Google Scholar]
- Lin J. J., Kanazawa H., Ozols J., Wu H. C. An Escherichia coli mutant with an amino acid alteration within the signal sequence of outer membrane prolipoprotein. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4891–4895. doi: 10.1073/pnas.75.10.4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. J., Kanazawa H., Wu H. C. Assembly of outer membrane lipoprotein in an Escherichia coli mutant with a single amino acid replacement within the signal sequence of prolipoprotein. J Bacteriol. 1980 Feb;141(2):550–557. doi: 10.1128/jb.141.2.550-557.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnemans W. A., Boer P., Elbers P. F. Localization of acid phosphatase in Saccharomyces cerevisiae: a clue to cell wall formation. J Bacteriol. 1977 Aug;131(2):638–644. doi: 10.1128/jb.131.2.638-644.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyhack B., Bajwa W., Rudolph H., Hinnen A. Two yeast acid phosphatase structural genes are the result of a tandem duplication and show different degrees of homology in their promoter and coding sequences. EMBO J. 1982;1(6):675–680. doi: 10.1002/j.1460-2075.1982.tb01229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy M., Laporte J., Penverne B., Hervé G. Nuclear localization of aspartate transcabamoylase in Saccharomyces cerevisiae. J Cell Biol. 1982 Mar;92(3):790–794. doi: 10.1083/jcb.92.3.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P., Ferro S., Schekman R. Order of events in the yeast secretory pathway. Cell. 1981 Aug;25(2):461–469. doi: 10.1016/0092-8674(81)90064-7. [DOI] [PubMed] [Google Scholar]
- Novick P., Field C., Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980 Aug;21(1):205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- Rogers D. T., Lemire J. M., Bostian K. A. Acid phosphatase polypeptides in Saccharomyces cerevisiae are encoded by a differentially regulated multigene family. Proc Natl Acad Sci U S A. 1982 Apr;79(7):2157–2161. doi: 10.1073/pnas.79.7.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth J., Bendayan M., Orci L. Ultrastructural localization of intracellular antigens by the use of protein A-gold complex. J Histochem Cytochem. 1978 Dec;26(12):1074–1081. doi: 10.1177/26.12.366014. [DOI] [PubMed] [Google Scholar]
- Rott R., Nelson N. Purification and immunological properties of proton-ATPase complexes from yeast and rat liver mitochondria. J Biol Chem. 1981 Sep 10;256(17):9224–9228. [PubMed] [Google Scholar]
- Schauer I., Emr S., Gross C., Schekman R. Invertase signal and mature sequence substitutions that delay intercompartmental transport of active enzyme. J Cell Biol. 1985 May;100(5):1664–1675. doi: 10.1083/jcb.100.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwencke J., Canut H., Flores A. Simultaneous isolation of the yeast cytosol and well-preserved mitochondria with negligible contamination by vacuolar proteinases. FEBS Lett. 1983 Jun 13;156(2):274–280. doi: 10.1016/0014-5793(83)80512-2. [DOI] [PubMed] [Google Scholar]
- Schönholzer F., Schweingruber A. M., Trachsel H., Schweingruber M. E. Intracellular maturation and secretion of acid phosphatase of Saccharomyces cerevisiae. Eur J Biochem. 1985 Mar 1;147(2):273–279. doi: 10.1111/j.1432-1033.1985.tb08747.x. [DOI] [PubMed] [Google Scholar]
- Shida H., Matsumoto S. Analysis of the hemagglutinin glycoprotein from mutants of vaccinia virus that accumulates on the nuclear envelope. Cell. 1983 Jun;33(2):423–434. doi: 10.1016/0092-8674(83)90424-5. [DOI] [PubMed] [Google Scholar]
- Strous G. J., Lodish H. F. Intracellular transport of secretory and membrane proteins in hepatoma cells infected by vesicular stomatitis virus. Cell. 1980 Dec;22(3):709–717. doi: 10.1016/0092-8674(80)90547-4. [DOI] [PubMed] [Google Scholar]
- Whitehouse R. L., Benichou J. C., Couture-Tosi E., Schenkman S., Ryter A. Immunolabelling of bacteriophage lambda receptor protein (LamB) on thin sections of E. coli embedded in Lowicryl. Biol Cell. 1984;51(3):389–394. doi: 10.1111/j.1768-322x.1984.tb00314.x. [DOI] [PubMed] [Google Scholar]
- van Rijn H. J., Linnemans W. A., Boer P. Localization of acid phosphatase in protoplasts from Saccharomyces cerevisiae. J Bacteriol. 1975 Sep;123(3):1144–1149. doi: 10.1128/jb.123.3.1144-1149.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]