Abstract
Purpose
Children diagnosed at age ≥ 18 months with metastatic MYCN-nonamplified neuroblastoma (NBL-NA) are at high risk for disease relapse, whereas those diagnosed at age < 18 months are nearly always cured. In this study, we investigated the hypothesis that expression of genes related to tumor-associated inflammatory cells correlates with the observed differences in survival by age at diagnosis and contributes to a prognostic signature.
Methods
Tumor-associated macrophages (TAMs) in localized and metastatic neuroblastomas (n = 71) were assessed by immunohistochemistry. Expression of 44 genes representing tumor and inflammatory cells was quantified in 133 metastatic NBL-NAs to assess age-dependent expression and to develop a logistic regression model to provide low- and high-risk scores for predicting progression-free survival (PFS). Tumors from high-risk patients enrolled onto two additional studies (n = 91) served as independent validation cohorts.
Results
Metastatic neuroblastomas had higher infiltration of TAMs than locoregional tumors, and metastatic tumors diagnosed in patients at age ≥ 18 months had higher expression of inflammation-related genes than those in patients diagnosed at age < 18 months. Expression of genes representing TAMs (CD33/CD16/IL6R/IL10/FCGR3) contributed to 25% of the accuracy of a novel 14-gene tumor classification score. PFS at 5 years for children diagnosed at age ≥ 18 months with NBL-NA with a low- versus high-risk score was 47% versus 12%, 57% versus 8%, and 50% versus 20% in three independent clinical trials, respectively.
Conclusion
These data suggest that interactions between tumor and inflammatory cells may contribute to the clinical metastatic neuroblastoma phenotype, improve prognostication, and reveal novel therapeutic targets.
INTRODUCTION
The concept of tumor-promoting inflammation is a recognized enabling characteristic of cancers.1 Recent studies have demonstrated the prognostic significance of tumor-associated macrophages (TAMs) in some adult cancers, including Hodgkin's lymphoma and breast cancer.2–6 However, the prognostic significance of tumor-associated inflammatory cells in metastatic disease and in childhood cancers is unknown.
Neuroblastoma, an embryonal tumor of the sympathetic nervous system, is one of the most common solid tumors in children, with approximately 40% of patients presenting with metastatic disease at diagnosis.7 Among those with metastatic disease, amplification of the MYCN proto-oncogene (30% of tumors) is associated with high risk of disease relapse, whereas those lacking MYCN amplification (NBL-NA) have clinical behaviors that are distinctly associated with age at diagnosis.8–10 Patients diagnosed with metastatic NBL-NA at age ≥ 18 months often have tumors with recurrent segmental genomic alterations and have high-risk disease with only 45% long-term disease-free survival.10–16 In contrast, children diagnosed with metastatic NBL-NA at age < 18 months of age frequently have tumors with whole chromosomal alterations and have greater than 90% overall survival (OS) after receiving only moderate-intensity chemotherapy.17–20 Biologic mechanisms responsible for the age-dependent genomic and clinical phenotypes of metastatic NBL-NA and for different responses to treatment among those age ≥18 months at diagnosis have been unclear.
Our previous gene expression profiling study of metastatic NBL-NA tumors suggested that there may be age-dependent differences in expression of genes representing tumor-associated inflammatory cells.21 In the current study, we focused on intratumor inflammatory cells, especially TAMs, and their relationship with clinical behavior of metastatic NBL-NA. We examined the infiltration of macrophages in locoregional and metastatic tumors with immunohistochemistry (IHC). We also used a TaqMan low-density array (TLDA; Life Technologies, Carlsbad, CA) assay to assess expression of inflammatory and tumor cell–related genes in metastatic NBL-NA tumors diagnosed before and after 18 months of age. Our findings provide new insights about intratumor inflammation in metastatic NBL-NA tumors and provide the basis for constructing a novel 14-gene model that predicts risk of disease progression in those diagnosed at age ≥ 18 months.
METHODS
Tissue microarray (TMA) details are provided in the Data Supplement. Macrophages were identified using IHC analysis of primary neuroblastoma tissues using antibodies directed against CD163 and allograft inflammatory factor 1 (AIF1). Tissue section scores ranged from 0 to 7 for each marker, with higher scores indicating a greater proportion of positive cells.
The details of the 48-gene TLDA assay are provided in the Data Supplement. All patients included in the gene expression study had metastatic NBL-NA tumors and were enrolled onto Children's Cancer Group (CCG), German Society for Pediatric Oncology-Hematology (GPOH), or Children's Oncology Group (COG) trials at diagnosis (Table 1 and Data Supplement). Details of treatment for the three cohorts were described previously and are provided in the Data Supplement.12,14,16,21 Informed consent was obtained in accordance with institutional review board policies.
Table 1.
Characteristics of Patients With Metastatic Neuroblastoma Lacking MYCN Gene Amplification
| Characteristic | Training Cohort (age at diagnosis, months) |
Validation Cohorts (age at diagnosis, months) |
||||||
|---|---|---|---|---|---|---|---|---|
| CCG |
GPOH |
COG |
||||||
| < 18 (n = 39) |
≥ 18 (n = 94) |
≥ 18 (n = 39) |
≥ 18 (n = 52) |
|||||
| No. | % | No. | % | No. | % | No. | % | |
| Age at diagnosis, months | ||||||||
| Mean | 9.3 | 53.6 | 56 | 55.9 | ||||
| Range | 0.1-17.3 | 18.2-151 | 18.4-182 | 19.5-186 | ||||
| COG risk stratification | Intermediate* | High | High | High | ||||
| INPC classification | ||||||||
| Favorable | 34 | 87 | 2 | 2 | 3 | 8 | 3 | 6 |
| Unfavorable | 5 | 13 | 91 | 97 | 34 | 87 | 42 | 81 |
| Unknown | 0 | 0 | 1 | 1 | 2 | 5 | 7 | 13 |
| Clinical trials | 323P, 3881 | 323P, 321-2, 321-3, 3891 | NB90, NB95, NB97, NB2004 | A3973† | ||||
| 5-year EFS rate, % | 95 | 23 | 34 | 31 | ||||
| 95% CI, % | 81 to 99 | 15 to 32 | 19 to 50 | 19 to 44 | ||||
| 5-year OS rate, % | 95 | 35 | 50 | 38 | ||||
| 95% CI, % | 81 to 99 | 25 to 44 | 32 to 66 | 23 to 54 | ||||
Abbreviations: CCG, Children's Cancer Group; COG, Children's Oncology Group; EFS, event-free survival; GPOH, German Society for Pediatric Oncology-Hematology (Gesellschaft für Paediatrische Onkologie und Haematologie); INPC, International Neuroblastoma Pathology Classification; OS, overall survival.
Patients age > 12 months were classified as high risk and treated accordingly per CCG guidelines. On the basis of current COG guidelines, four of 12 patients diagnosed at 12 to 18 months of age with unfavorable tumor histology would be considered high risk.
Three patients were treated on ANBL0532.
Statistical Analysis
The Data Supplement illustrates the flow of statistical and validation methods used in this report. Because our primary interest was to identify genes that are predictive of outcome in the cohort of patients older than 18 months of age, we first conducted a univarite logistic regression model based on TLDA gene expression data from 133 samples from the training cohort (CCG), which includes patients older and younger than 18 months of age at the time of diagnosis. Genes that were independent of age at diagnosis with a P value of ≤ .25 were included in a final multivariate logistic model to predict PFS. Our aim was to build a robust model that was predictive of disease progression in patients older than 18 months of age and that could be used as the basis for classification into signature-based low- and high-risk tumor-progression groups. Disease progression was defined a priori (Data Supplement). The effective period for risk of disease progression in the training cohort was 4 years from diagnosis. Because few patients were censored before the end of the effective period for risk of disease progression, ignoring this censoring had little practical effect on the logistic regression analysis of whether disease progression had occurred. Age was included as a continuous covariate in the final multivariate logistic regression analysis to assess for residual significance. Logit values, representing the tumor-progression scores, were computed for each patient. Measures of accuracy based on resubstitution analysis and leave-one-out cross validation (LOOCV) are presented. Classification accuracy was assessed using receiver operating characteristic curves and areas under the curve (AUC). External validation of the prediction model was performed using the independent GPOH and COG samples, with the tumor-progression score for each patient calculated using the regression coefficients from the prediction model derived from the training cohort. The relative contributions to the accuracy of the 14-gene NBL-NA prediction score of age at diagnosis, tumor cell–related genes, and inflammation-related genes were assessed using 5,000 permutations of the data set (Data Supplement).
Tumor-progression risk scores obtained from the multivariate logistic regression model were used to define signature-based risk groups. The median tumor-progression risk score from the training cohort (n = 133) was used as the cutoff point to define signature-based high-risk (tumor-progression score ≥ median score) or low-risk (tumor-progression score < median score) scores.
Statistical Methods
Details of the statistical analyses for developing the prognostic score are described here and summarized in the Data Supplement. In addition, survival analysis methods22 are used to describe outcome in low- and high-risk groups defined by the prognostic score. The primary end point for these analyses was progression-free survival (PFS), defined as the minimum interval from date of diagnosis to date of disease progression, date of death (four patients only), or date of last follow-up. Patients who did not experience progression or die were censored at the time of last follow-up. The Kaplan-Meier method was used to compute PFS probabilities and produce survival curves. CIs are based on Greenwood SEs. Unless otherwise stated, the reported probabilities are based on 5-year PFS rates. Tests of the difference in PFS between risk groups are based on the log-rank statistic. Other common statistics23 (eg, t test, Spearman rank correlation) were used where appropriate and are indicated in the text. Bonferroni adjustments to account for multiple comparisons were used where appropriate. Statistical computations were performed using STATA software (version 9.0; STATA, College Station, TX) or the R project (http://www.r-project.org).
RESULTS
Infiltration of Inflammatory Cells in Metastatic Neuroblastoma
We performed IHC analysis of 71 neuroblastoma tumors (29 patients with locoregional, 31 with metastatic disease [stage 4], and 11 with metastatic disease with special designation [stage 4S]; Data Supplement) using antibodies directed against two macrophage markers (CD163 and AIF1). There were significantly greater numbers of infiltrating macrophages observed only with CD163 staining, which identifies alternatively activated macrophage (M2), in samples of patients with metastatic (stage 4) compared with locoregional neuroblastoma (t test P = .003; Bonferroni-adjusted P < .017 considered significant; Fig 1A). There was no statistically significant difference in the number of intratumor CD163+ macrophages between metastatic tumors with special designation (stage 4S), which undergo spontaneous regression, and locoregional tumors (Fig 1B).
Fig 1.
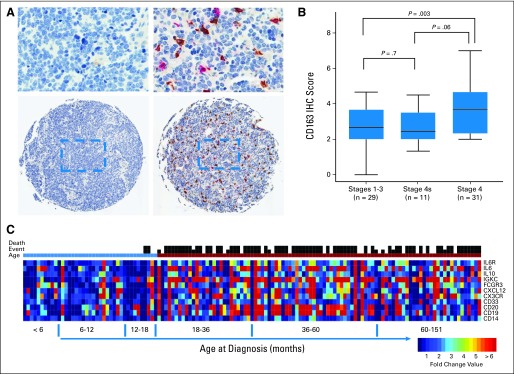
Evidence of tumor-associated macrophages and inflammation in neuroblastoma. (A) Representative immunohistochemical analyses of staining of CD163 (brown staining) and AIF1 (red staining) in primary tumor samples from a patient with stage 1 tumor (left panel) lacking any infiltrating macrophages and a patient with metastatic disease (right panel) with extensive infiltration of CD163+ macrophages. (B) Average scores for the presence of CD163+ infiltrating macrophages reveals significant infiltration in tumor samples of patients with metastatic disease compared with those with locoregional tumors. Patients with stage 4S tumors, who are known to undergo spontaneous regression, had CD163+ macrophages comparable to those with locoregional tumors (Bonferroni adjusted P < .017 considered significant). (C) Heatmap of gene expression levels of inflammation-related genes displayed along increasing age at diagnosis of 133 children with metastatic MYCN-nonamplified neuroblastoma. Heatmap colors reflect fold-change value of individual tumor relative to the average expression level of inflammation-related genes in children diagnosed at age < 18 months. Occurrences of an event and of death are indicated by black vertical bars above the color-coded bar for age at diagnosis.
We also performed gene expression analysis of 133 metastatic NBL-NA tumors (CCG cohort: 94 children diagnosed at age ≥ 18 months and 39 diagnosed at age < 18 months; Table 1; Data Supplement) with a custom-built TLDA containing 31 tumor-related and 13 inflammation-related genes (Data Supplement). We identified greater expression of inflammation-related genes associated with monocyte/macrophage, myeloid, and B cells in tumors of children diagnosed at age ≥ 18 months compared with those diagnosed at age < 18 months (Fig 1C). Although inflammation-related genes CD33, FCGR3 (CD16), and IGKC showed significant association with PFS in univariate analysis (Data Supplement), we did not identify any single gene model that could accurately predict PFS in children diagnosed at age ≥ 18 months with AUC > 0.7. These data suggest that inflammatory cells within tumors, especially TAMs, contribute to the age-associated clinical behavior of metastatic NBL-NA tumors.
Expression of Inflammation- and Tumor Cell–Related Genes Comprises a Prognostic Signature
We further examined the expression of the 31 tumor-related and 13 inflammation-related genes in the CCG cohort and identified 14 genes that contributed to a model predictive of PFS (Table 2). Among the 14 genes used in our model, nine (64%) were tumor cell related, and five (36%) were inflammation related. The accuracy of the model for predicting PFS using LOOCV AUC estimates was 0.82 for patients in all age groups and 0.74 for patients age ≥ 18 months at diagnosis (Data Supplement).
Table 2.
Characteristics and Prognostic Significance of the 14 Genes of the Neuroblastoma Signature
| Symbol | Gene Name | Gene Location | Univariate OR | 95% CI† | P | Prediction Accuracy(AUC; CCG patients age ≥ 18 months)‡ |
|---|---|---|---|---|---|---|
| Tumor-related genes | ||||||
| H2AFV | H2A histone family, member V | 7p13 | 0.42 | 0.26 to 0.68 | < .001 | 0.6275 |
| GPATC4 | G patch domain containing 4 | 1q22 | 2.08 | 1.33 to 3.24 | < .001 | 0.6060 |
| PTPN5 | Protein tyrosine phosphatase, nonreceptor type 5 | 11p15.1 | 1.28 | 1.10 to 1.48 | < .001 | 0.5636 |
| PGM2L1 | Phosphoglucomutase 2-like 1 | 11q13.4 | 0.62 | 0.45 to 0.85 | .002 | 0.5127 |
| GFRA3 | GDNF family receptor alpha 3 | 5q31.1 | 0.79 | 0.62 to 1.01 | .05 | 0.4729 |
| THAP2 | THAP domain containing, apoptosis-associated protein 2 | 12q21.1 | 0.59 | 0.34 to 1.03 | .05 | 0.4703 |
| BTBD3 | BTB (POZ) domain containing 3 | 20p12.2 | 0.76 | 0.56 to 1.02 | .06 | 0.4514 |
| CAMTA1 | Calmodulin-binding transcription activator 1 | 1p36.31 | 0.80 | 0.60 to 1.05 | .1 | 0.4494 |
| NTRK2 | Neurotrophic tyrosine kinase receptor type 2 | 9q22.1 | 1.16 | 0.91 to 1.48 | .2 | 0.4886 |
| Inflammation-related genes | ||||||
| FCGR3 (CD16) | Fc fragment of immunoglobulin G, CD16 | 1q23 | 1.36 | 1.08 to 1.72 | .006 | 0.5649 |
| IL-6R | Interleukin-6 receptor | 1q21 | 1.26 | 0.97 to 1.65 | .08 | 0.4977 |
| CD33 | CD33 antigen | 19q13.3 | 1.34 | 1.00 to 1.80 | .04 | 0.5179 |
| IL-10 | Interleukin-10 | 1q31-q32 | 1.17 | 0.91 to 1.50 | .2 | 0.4814 |
| CD14 | CD14 antigen | 5q31.1 | 1.25 | 0.92 to 1.70 | .15 | 0.5036 |
Abbreviations: AUC, area under the curve; CCG, Children's Cancer Group; OR, odds ratio; ROC, receiver operating characteristic curve.
Univariate OR for each gene after adjusting for age at diagnosis for the training CCG cohort (n = 133). Coefficients were calculated based on a two-fold increase in gene expression (ie, equivalent to a change of 1 ΔCT).
AUC values are reported for the patients diagnosed at age ≥ 18 months in the training cohort. A logistic regression model that included the individual gene expression value plus age at diagnosis as a continuous variable was fit to the training cohort (n = 133). The logit scores were used in ROC analysis to obtain AUC values.
Tumors from the CCG cohort were categorized as low or high risk based on their 14-gene tumor-progression risk score using LOOCV analysis. Figures 2A and 2B show that patients with a low-risk score had significantly better PFS (72% at 5 years; 95% CI, 60% to 82%) than those in the high-risk score group (16% at 5 years; 95% CI, 8% to 26%) using LOOCV analysis (P < .001). The overall 5-year PFS for the 94 patients who were age ≥ 18 months at diagnosis and treated on CCG high-risk protocols was 23%. Among these patients, 30 (32%) had a low-risk score with a 5-year PFS rate of 47% (95% CI, 28% to 63%), and 64 (68%) had a high-risk score with a 5-year PFS of 12% (95% CI, 5% to 22%; P = .002), demonstrating that a subset of patients at extremely high risk of disease progression can be identified by the 14-gene signature among these otherwise clinically indistinguishable patients. Five-year OS for the 94 patients whose tumors had low- or high-risk scores also was significantly different (60% v 23%; P = .003; Data Supplement). Classification by resubstitution analysis showed higher accuracy in prediction and more divergent Kaplan-Meier curves (Data Supplement), reflecting the optimistically biased approach of this analysis and the need for cross validation.24–28
Fig 2.
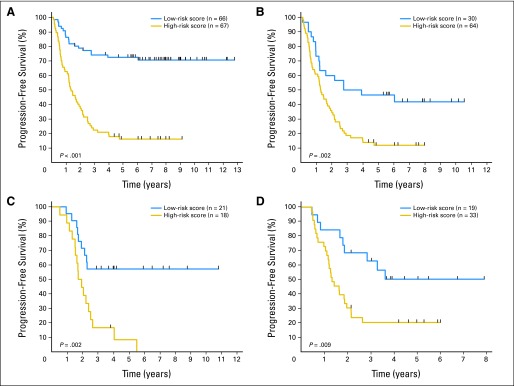
Progression-free survival (PFS) for patients in the training and validation cohorts with metastatic MYCN-nonamplified neuroblastoma (NBL-NA) 14-gene signature low- and high-risk scores. The cutoff value used to categorize patients into signature-based high- and low-risk score groups depended on the median score obtained in the Children's Cancer Group (CCG) cohort. The high-risk score group had prediction scores higher than the median score, whereas the low-risk score group had prediction scores lower than the median. The graphs show Kaplan-Meier estimates of PFS for patients with NBL-NA according to 14-gene signature risk classification. PFS estimates using the leave-one-out cross-validation signature classification for (A) the entire CCG cohort (n = 133) and (B) CCG patients diagnosed at age ≥ 18 months (clinically defined high-risk group; n = 94). The classification model developed using the CCG samples was then used to identify signature-based groups for patients age ≥ 18 months treated on (C) German Society for Pediatric Oncology-Hematology high-risk protocols (n = 39) and (D) Children's Oncology Group high-risk protocols (n = 52). All P values are based on log-rank test.
Independent validation of the 14-gene signature to predict PFS was obtained from analysis of metastatic NBL-NA tumors from two independent cohorts of patients (GPOH, n = 39; COG, n = 52) diagnosed at age ≥ 18 months (Table 1; Data Supplement). Those patients whose tumors had signature-based high-risk scores had significantly worse 5-year PFS than those with low-risk scores (Figs 2C and 2D). Among the 39 GPOH patients, 21 (54%) had low-risk scores with a 5-year PFS rate of 57% (95% CI, 34% to 75%), and 18 (46%) had high-risk scores with a 5-year PFS of 8% (95% CI, 1% to 29%; P = .002). Nineteen COG patients (36%) had low-risk scores with a 5-year PFS rate of 50% (95% CI, 26% to 70%), and 33 (64%) had high-risk scores with a 5-year PFS of 20% (95% CI, 8% to 35%; P = .009). Five-year OS for patients in these two cohorts whose tumors had low- or high-risk scores also was significantly different (GPOH: 65% v 31%; P = .012; COG: 51% v 31%; P = .039; Data Supplement). These data show the validity of the 14-gene signature and the prognostic information obtained from inclusion of inflammation-related genes to identify subsets of patients with different outcomes in a clinically indistinguishable population.
Prognostic Contribution of Genes Related to Tissue-Associated Macrophages
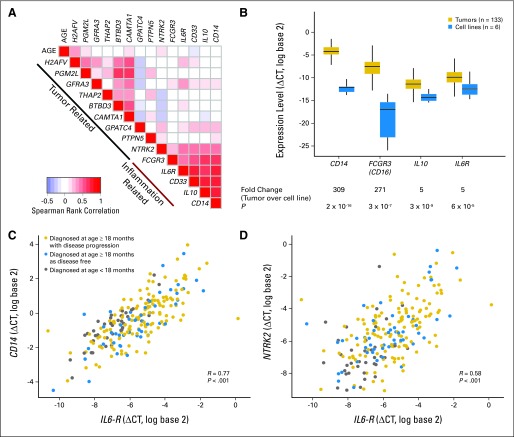
The five inflammation-related genes in our 14-gene model included CD14, CD33, FCGR3 (CD16), interleukin-6 receptor (IL6R), and interleukin-10 (IL10), which are mainly expressed by macrophages and myeloid cells and, along with CD163, signify intratumor macrophage polarization to the anti-inflammatory M2-like phenotype.28,29 Our previous research demonstrated that expression of these inflammation-related genes in neuroblastoma tumors correlates with microscopic presence of IL6-producing CD68+ cells, which are considered to be TAMs.30 Comparison of levels of expression of inflammation-related genes in neuroblastoma tumors with five neuroblastoma cell lines demonstrated that these markers are expressed 150-fold (range, five to 309; t test P < .001) higher on average in tumors than in cell lines (Fig 3B), suggesting that these genes are primarily expressed by tumor-associated inflammatory cells and consistent with IHC findings.
Fig 3.
Inflammation- and tumor-related gene-gene correlations. (A) Heatmap of the Spearman rank correlation matrix of the 14 genes in the metastatic MYCN-nonamplified neuroblastoma (NBL-NA) signature. Pairwise rank correlation analyses were performed for all 14 genes and age at diagnosis. The patterns of correlation using samples from Children's Cancer Group (CCG) patients diagnosed at age ≥ 18 months were similar to the pairwise rank correlations obtained using samples from the German Society for Pediatric Oncology-Hematology and Children's Oncology Group validation cohorts (Data Supplement). Red represents positive rank correlation level above zero (white color) for a given gene pair, and blue represents negative rank correlation level. The inflammation-related genes (FCGR3/CD16, CD33, CD14, IL6R, IL10) show high levels of correlation across all cohorts. NTRK2 has the strongest correlation of any tumor cell–related gene with inflammation-related genes. (B) Expression of inflammation-related genes is predominantly present in neuroblastoma tumors and not on cell lines. Normalized expression (ΔCT) values of four of the five inflammation-related genes (CD14, FCGR3/CD16, IL10, IL6R) in the CCG tumors (n = 133) were compared with six neuroblastoma MYCN nonamplified cell lines (CHLA-15, CHLA-20, CHLA-255, CHLA-42, CHLA-90, LAN-6). Data on the cell lines were generated on a TaqMan low-density array (Life Technologies, Carlsbad, CA) card that did not include CD33 probes. On average, there was 309-, 271-, seven-, and five-fold higher expression of CD14, FCGR3/CD16, IL10, and IL6R in tumors than in cell lines, respectively, suggesting that these genes are primarily expressed by tumor-associated inflammatory cells. (C) Scatter diagram of expression of CD14, a macrophage marker, and IL6R reveals a high correlation in patients with metastatic NBL-NA. (D) Similarly, NTRK2, a tumor cell–related gene, shows moderate correlation with expression of IL6R, an inflammation-related gene. Gray, patients diagnosed at age < 18 months with metastatic NBL-NA; blue, disease-free patients diagnosed at age ≥ 18 months; gold, disease progression in patients diagnosed at age ≥ 18 months.
We next investigated the contribution of gene categories and age at diagnosis to the predictive accuracy of the NBL-NA signature. Using a permutation strategy (Data Supplement), we discovered that on average, the inclusion of inflammation-related genes explained 25% of the accuracy of the 14-gene model in predicting PFS and added to the 63% provided by tumor cell–related genes. Age at diagnosis explained an additional 12% of the accuracy.
Inflammation-related genes were also found to be highly correlated to one another in the CCG cohort, a pattern that was also observed in the GPOH and COG cohorts (Fig 3A; Data Supplement). The strongest gene-gene correlations were observed between IL6R and CD14 (Spearman r = 0.77; P < .001; Fig 3C) and between IL6R and CD33 (Spearman r = 0.75; P < .001). IL6R is primarily expressed on cells of the monocytic lineage, but it has also been reported to be expressed on some neuroblastoma cell lines.31
The nine genes categorized as tumor cell–related included neurotrophic kinase receptor 2 (NTRK2) and calmodulin-binding transcription activator 1 (CAMTA1), genes known to be associated with neuroblastoma growth and suppression, respectively.24–27 Interestingly, IL6R was also found to be moderately correlated with the expression of NTRK2 (Spearman r = 0.58; P < .001; Fig 3D). Together, these data suggest that inflammation-related genes, especially genes related to polarization of TAMs, contribute to prognosis and are associated with a poor clinical outcome.
DISCUSSION
The diverse outcomes of children with NBL-NA have been largely unexplained. Our study suggests for the first time that infiltrating inflammatory cells, especially TAMs, may contribute to this diversity. We demonstrate that TAMs are more prevalent in tumors of children with metastatic rather than locoregional neuroblastoma. Furthermore, we show that expression of inflammation-related genes is higher in tumors of children diagnosed at age ≥ 18 months and that a subset of these genes representing TAMs is associated with an extremely poor outcome in this group. Including expression of both inflammatory and tumor cell genes in a 14-gene signature enables prediction of disease progression for the first time in the clinically indistinguishable group of patients diagnosed at age ≥ 18 months with metastatic NBL-NA. The novel finding that five inflammation-related genes contribute to 25% of the accuracy of the 14-gene model emphasizes the role of inflammation in neuroblastoma and uncovers previously unrecognized potential targets for therapy. This 14-gene expression scoring model, which was validated in two independent cohorts of patients, has clinical applicability and may be of used for managing high-risk patients.
In recent years, the concept of inflammatory cells in the tumor microenvironment as critical participants in tumor progression has gained acceptance.1,32 A tumor-infiltrating macrophage and T-cell signature (CD68high/CD4high/CD8low) has been reported to predict PFS and OS in patients with breast cancer.4 In our study, IL6R expression was found to be highly correlated with expression of CD14 (macrophage marker) and CD33 (myeloid marker), and their expression, along with that of IL10 and FCGR3/CD16 (M2 polarization), contributed to the accuracy of our model in predicting disease progression. This novel finding provides a validated clinical context for the recently established role of TAMs and bone marrow mesenchymal stem cells in promoting neuroblastoma growth via activation of the IL6/IL6R pathway.30,31 Melanomas and myelomas, similar to neuroblastomas, have also been shown to co-opt their tumor microenvironment cells to produce IL6, leading to STAT3 activation and promotion of tumor growth.30,31,33,34 Studies using immunocompetent mouse cancer models have demonstrated that antibody responses against transformed cells could lead to recruitment and polarization of macrophages, leading to production of cytokines such as IL6, IL10, and IL4 that in turn stimulate tumor growth and angiogenesis.35,36 In the current study, using a highly specific and sensitive TLDA assay, expression of genes related to the humoral immune system was observed in children diagnosed at age ≥ 18 months but did not contribute to the final predictive model. Additional studies are needed to elucidate the role of antibody response in neuroblastoma and its relation to recruitment and polarization of TAMs.
Genes related to tumor cells contributed most to the accuracy of the 14-gene NBL-NA signature and included NTRK2, which binds brain-derived neurotrophic factor and plays an important role in the survival and differentiation of neuroblastoma cells.25,26,37,38 The association of high NTRK2 expression with aggressive behavior in metastatic NBL-NA was previously reported by our group21 and is further supported in this study. Our present work also reveals a novel association between expression of IL6R and NTRK2. Although additional mechanistic studies are required to understand interactions between these two pathways, our data point to the role of a pro-tumor inflammatory microenvironment in enabling a highly aggressive neuroblastoma phenotype.
Several gene expression and genomic studies of neuroblastoma tumors obtained at diagnosis have previously reported associations with patient outcome.20,39–43 However, these studies analyzed groups of patients who were heterogeneous with respect to MYCN gene amplification status, clinical stage, and age at diagnosis. Segmental genomic alterations identified in high-risk NBL-NA tumors have not been predictive of outcome in this high-risk group but have had clinical utility in children with intermediate-risk neuroblastoma.20,41,44–48 Gene expression–based models built using neuroblastoma samples from clinically heterogeneous groups of patients lack predictive accuracy in those diagnosed at age ≥ 18 months with metastatic NBL-NA.42,43 Similarly, our previously reported 55-gene NBL-NA–specific microarray signature21 was not predictive of outcome for patients with MYCN-amplified tumors, which represent a distinct molecular subgroup of neuroblastomas (unpublished data).49 Overall, these findings suggest that prognostic studies in neuroblastomas should focus on well-defined molecular and clinical subgroups (eg, using MYCN status). Our current finding also highlights the importance of assessing the neuroblastoma tumor microenvironment in prognostic studies.
Our current study defines a clinically applicable 14-gene expression signature that identifies two subsets of patients with different PFS. Children with high-risk tumor-progression scores uniformly had a poor outcome, with 8% to 20% PFS at 5 years after diagnosis, whereas those with low-risk scores had 47% to 57% PFS. It is possible that the addition of treatment response evaluations such as imaging with 123I-metaiodobenzylguanidine and quantification of bone marrow disease with polymerase chain reaction assays will further improve risk classification, especially for patients with a low-risk tumor-progression score.50,51
In summary, our study reports the first evidence of a role for intratumor inflammation in metastatic neuroblastomas and provides a validated prognostic signature for children with metastatic NBL-NA. The increase in expression of inflammation-related genes in children age ≥ 18 months with poor outcome allows the identification of a subgroup of patients at extremely high risk who may benefit from treatments targeting the tumor microenvironment along with tumor cells. The recent success of therapies directed at tumor-associated immune system cells in adult cancers52–54 suggests opportunities for their application in children with neuroblastoma.
Supplementary Material
Acknowledgment
We acknowledge the Children's Oncology Group and the German Society for Pediatric Oncology-Hematology for providing neuroblastoma specimens. We thank Ronnie Houston, BS, for his technical assistance in immunohistochemistry and Janahan Gnanachandran, MS, for his data analysis. We would also like to thank Martine Torres, PhD, for her editorial review of the manuscript and Yves DeClerck, MD, for his comments and suggestions.
Footnotes
Supported by Grant No. 2R01 CA60104-16 from the National Cancer Institute (R.C.S.), a grant from Amgen (J.A.S.), Grant No. K12-CA60104 from the National Institute of Child Health and Human Development, grants from Alex's Lemonade Stand Foundation and St Baldrick's Foundation (S.A.), and grants from the Nautica Malibu Triathlon and Bogart Pediatric Cancer Research Program (S.A., R.C.S.).
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The author(s) indicated no potential conflicts of interest.
AUTHOR CONTRIBUTIONS
Conception and design: Shahab Asgharzadeh, Robert C. Seeger
Collection and assembly of data: Shahab Asgharzadeh, Jill A. Salo, André Oberthuer, Matthias Fischer, Frank Berthold, Michael Hadjidaniel, Cathy Wei-Yao Liu, Leonid S. Metelista, Roger Pique-Regi, Peter Wakamatsu, Judith G. Villablanca, Susan G. Kreissman, Katherine K. Matthay, Hiroyuki Shimada, Wendy B. London, Richard Sposto, Robert C. Seeger
Data analysis and interpretation: Shahab Asgharzadeh, Jill A. Salo, Lingyun Ji, Michael Hadjidaniel, Roger Pique-Regi, Hiroyuki Shimada, Wendy B. London, Richard Sposto, Robert C. Seeger
Manuscript writing: All authors
Final approval of manuscript: All authors
REFERENCES
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Steidl C, Lee T, Shah SP, et al. Tumor-associated macrophages and survival in classic Hodgkin's lymphoma. N Engl J Med. 2010;362:875–885. doi: 10.1056/NEJMoa0905680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ueno T, Toi M, Saji H, et al. Significance of macrophage chemoattractant protein-1 in macrophage recruitment, angiogenesis, and survival in human breast cancer. Clin Cancer Res. 2000;6:3282–3289. [PubMed] [Google Scholar]
- 4.DeNardo DG, Brennan DJ, Rexhepaj E, et al. Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy. Cancer Discov. 2011;1:54–67. doi: 10.1158/2159-8274.CD-10-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bronkhorst IH, Ly LV, Jordanova ES, et al. Detection of M2-macrophages in uveal melanoma and relation with survival. Invest Ophthalmol Vis Sci. 2011;52:643–650. doi: 10.1167/iovs.10-5979. [DOI] [PubMed] [Google Scholar]
- 6.Kurahara H, Shinchi H, Mataki Y, et al. Significance of M2-polarized tumor-associated macrophage in pancreatic cancer. J Surg Res. 2011;167:e211–e219. doi: 10.1016/j.jss.2009.05.026. [DOI] [PubMed] [Google Scholar]
- 7.Cohn SL, Pearson AD, London WB, et al. The International Neuroblastoma Risk Group (INRG) classification system: An INRG Task Force report. J Clin Oncol. 2009;27:289–297. doi: 10.1200/JCO.2008.16.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deyell RJ, Attiyeh EF. Advances in the understanding of constitutional and somatic genomic alterations in neuroblastoma. Cancer Genet. 2011;204:113–121. doi: 10.1016/j.cancergen.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt ML, Lukens JN, Seeger RC, et al. Biologic factors determine prognosis in infants with stage IV neuroblastoma: A prospective Children's Cancer Group study. J Clin Oncol. 2000;18:1260–1268. doi: 10.1200/JCO.2000.18.6.1260. [DOI] [PubMed] [Google Scholar]
- 10.Matthay KK, Reynolds CP, Seeger RC, et al. Long-term results for children with high-risk neuroblastoma treated on a randomized trial of myeloablative therapy followed by 13-cis-retinoic acid: A Children's Oncology Group study. J Clin Oncol. 2009;27:1007–1013. doi: 10.1200/JCO.2007.13.8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simon T, Hero B, Faldum A, et al. Long term outcome of high-risk neuroblastoma patients after immunotherapy with antibody ch14.18 or oral metronomic chemotherapy. BMC Cancer. 2011;11:21. doi: 10.1186/1471-2407-11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berthold F, Boos J, Burdach S, et al. Myeloablative megatherapy with autologous stem-cell rescue versus oral maintenance chemotherapy as consolidation treatment in patients with high-risk neuroblastoma: A randomised controlled trial. Lancet Oncol. 2005;6:649–658. doi: 10.1016/S1470-2045(05)70291-6. [DOI] [PubMed] [Google Scholar]
- 13.Cheung NK, Kushner BH, Kramer K. Monoclonal antibody-based therapy of neuroblastoma. Hematol Oncol Clin North Am. 2001;15:853–866. doi: 10.1016/s0889-8588(05)70255-0. [DOI] [PubMed] [Google Scholar]
- 14.Matthay KK, Villablanca JG, Seeger RC, et al. Treatment of high-risk neuroblastoma with intensive chemotherapy, radiotherapy, autologous bone marrow transplantation, and 13-cis-retinoic acid: Children's Cancer Group. N Engl J Med. 1999;341:1165–1173. doi: 10.1056/NEJM199910143411601. [DOI] [PubMed] [Google Scholar]
- 15.George RE, Li S, Medeiros-Nancarrow C, et al. High-risk neuroblastoma treated with tandem autologous peripheral-blood stem cell-supported transplantation: Long-term survival update. J Clin Oncol. 2006;24:2891–2896. doi: 10.1200/JCO.2006.05.6986. [DOI] [PubMed] [Google Scholar]
- 16.Yu AL, Gilman AL, Ozkaynak MF, et al. Anti-GD2 antibody with GM-CSF, interleukin-2, and isotretinoin for neuroblastoma. N Engl J Med. 2010;363:1324–1334. doi: 10.1056/NEJMoa0911123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt ML, Lal A, Seeger RC, et al. Favorable prognosis for patients 12 to 18 months of age with stage 4 nonamplified MYCN neuroblastoma: A Children's Cancer Group Study. J Clin Oncol. 2005;23:6474–6480. doi: 10.1200/JCO.2005.05.183. [DOI] [PubMed] [Google Scholar]
- 18.Baker DL, Schmidt ML, Cohn SL, et al. Outcome after reduced chemotherapy for intermediate-risk neuroblastoma. N Engl J Med. 2010;363:1313–1323. doi: 10.1056/NEJMoa1001527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.George RE, London WB, Cohn SL, et al. Hyperdiploidy plus nonamplified MYCN confers a favorable prognosis in children 12 to 18 months old with disseminated neuroblastoma: A Pediatric Oncology Group study. J Clin Oncol. 2005;23:6466–6473. doi: 10.1200/JCO.2005.05.582. [DOI] [PubMed] [Google Scholar]
- 20.Janoueix-Lerosey I, Schleiermacher G, Michels E, et al. Overall genomic pattern is a predictor of outcome in neuroblastoma. J Clin Oncol. 2009;27:1026–1033. doi: 10.1200/JCO.2008.16.0630. [DOI] [PubMed] [Google Scholar]
- 21.Asgharzadeh S, Pique-Regi R, Sposto R, et al. Prognostic significance of gene expression profiles of metastatic neuroblastomas lacking MYCN gene amplification. J Natl Cancer Inst. 2006;98:1193–1203. doi: 10.1093/jnci/djj330. [DOI] [PubMed] [Google Scholar]
- 22.Kalbfleisch J, Prentice R. The Statistical Analysis of Failure Time Data. New York, NY: John Wiley and Sons; 1980. [Google Scholar]
- 23.Mood AM, Garybill FA, Boes DC. Introduction to the Theory of Statistics. New York, NY: McGraw-Hill; 1974. [Google Scholar]
- 24.Katoh M, Katoh M. Identification and characterization of FLJ10737 and CAMTA1 genes on the commonly deleted region of neuroblastoma at human chromosome 1p36.31-p36.23. Int J Oncol. 2003;23:1219–1224. [PubMed] [Google Scholar]
- 25.Nakagawara A, Azar CG, Scavarda NJ, et al. Expression and function of TRK-B and BDNF in human neuroblastomas. Mol Cell Biol. 1994;14:759–767. doi: 10.1128/mcb.14.1.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakagawara A, Arima-Nakagawara M, Scavarda NJ, et al. Association between high levels of expression of the TRK gene and favorable outcome in human neuroblastoma. N Engl J Med. 1993;328:847–854. doi: 10.1056/NEJM199303253281205. [DOI] [PubMed] [Google Scholar]
- 27.Henrich KO, Fischer M, Mertens D, et al. Reduced expression of CAMTA1 correlates with adverse outcome in neuroblastoma patients. Clin Cancer Res. 2006;12:131–138. doi: 10.1158/1078-0432.CCR-05-1431. [DOI] [PubMed] [Google Scholar]
- 28.Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: Cancer as a paradigm. Nat Immunol. 2010;11:889–896. doi: 10.1038/ni.1937. [DOI] [PubMed] [Google Scholar]
- 29.Leidi M, Gotti E, Bologna L, et al. M2 macrophages phagocytose rituximab-opsonized leukemic targets more efficiently than m1 cells in vitro. J Immunol. 2009;182:4415–4422. doi: 10.4049/jimmunol.0713732. [DOI] [PubMed] [Google Scholar]
- 30.Song L, Asgharzadeh S, Salo J, et al. Vα24-invariant NKT cells mediate antitumor activity via killing of tumor-associated macrophages. J Clin Invest. 2009;119:1524–1536. doi: 10.1172/JCI37869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ara T, Song L, Shimada H, et al. Interleukin-6 in the bone marrow microenvironment promotes the growth and survival of neuroblastoma cells. Cancer Res. 2009;69:329–337. doi: 10.1158/0008-5472.CAN-08-0613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan TT, Coussens LM. Humoral immunity, inflammation and cancer. Curr Opin Immunol. 2007;19:209–216. doi: 10.1016/j.coi.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 33.Bromberg JF, Wrzeszczynska MH, Devgan G, et al. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 34.Klein B, Zhang X, Jourdan M, et al. Paracrine rather than autocrine regulation of myeloma-cell growth and differentiation by interleukin-6. Blood. 1989;73:517–526. [PubMed] [Google Scholar]
- 35.de Visser KE, Korets LV, Coussens LM. De novo carcinogenesis promoted by chronic inflammation is B lymphocyte dependent. Cancer Cell. 2005;7:411–423. doi: 10.1016/j.ccr.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 36.Andreu P, Johansson M, Affara NI, et al. FcRgamma activation regulates inflammation-associated squamous carcinogenesis. Cancer Cell. 2010;17:121–134. doi: 10.1016/j.ccr.2009.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki T, Bogenmann E, Shimada H, et al. Lack of high-affinity nerve growth factor receptors in aggressive neuroblastomas. J Natl Cancer Inst. 1993;85:377–384. doi: 10.1093/jnci/85.5.377. [DOI] [PubMed] [Google Scholar]
- 38.Aoyama M, Asai K, Shishikura T, et al. Human neuroblastomas with unfavorable biologies express high levels of brain-derived neurotrophic factor mRNA and a variety of its variants. Cancer Lett. 2001;164:51–60. doi: 10.1016/s0304-3835(00)00715-1. [DOI] [PubMed] [Google Scholar]
- 39.Ohira M, Nakagawara A. Global genomic and RNA profiles for novel risk stratification of neuroblastoma. Cancer Sci. 2010;101:2295–2301. doi: 10.1111/j.1349-7006.2010.01681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fischer M, Spitz R, Oberthür A, et al. Risk estimation of neuroblastoma patients using molecular markers. Klin Padiatr. 2008;220:137–146. doi: 10.1055/s-2008-1065345. [DOI] [PubMed] [Google Scholar]
- 41.Attiyeh EF, London WB, Mossé YP, et al. Chromosome 1p and 11q deletions and outcome in neuroblastoma. N Engl J Med. 2005;353:2243–2253. doi: 10.1056/NEJMoa052399. [DOI] [PubMed] [Google Scholar]
- 42.Oberthuer A, Hero B, Berthold F, et al. Prognostic impact of gene expression–based classification for neuroblastoma. J Clin Oncol. 2010;28:3506–3515. doi: 10.1200/JCO.2009.27.3367. [DOI] [PubMed] [Google Scholar]
- 43.De Preter K, Vermeulen J, Brors B, et al. Accurate outcome prediction in neuroblastoma across independent data sets using a multigene signature. Clin Cancer Res. 2010;16:1532–1541. doi: 10.1158/1078-0432.CCR-09-2607. [DOI] [PubMed] [Google Scholar]
- 44.Guo C, White PS, Hogarty MD, et al. Deletion of 11q23 is a frequent event in the evolution of MYCN single-copy high-risk neuroblastomas. Med Pediatr Oncol. 2000;35:544–546. doi: 10.1002/1096-911x(20001201)35:6<544::aid-mpo10>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 45.Plantaz D, Vandesompele J, Van Roy N, et al. Comparative genomic hybridization (CGH) analysis of stage 4 neuroblastoma reveals high frequency of 11q deletion in tumors lacking MYCN amplification. Int J Cancer. 2001;91:680–686. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1114>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 46.Bagatell R, Rumcheva P, London WB, et al. Outcomes of children with intermediate-risk neuroblastoma after treatment stratified by MYCN status and tumor cell ploidy. J Clin Oncol. 2005;23:8819–8827. doi: 10.1200/JCO.2004.00.2931. [DOI] [PubMed] [Google Scholar]
- 47.Bown N, Cotterill S, Lastowska M, et al. Gain of chromosome arm 17q and adverse outcome in patients with neuroblastoma. N Engl J Med. 1999;340:1954–1961. doi: 10.1056/NEJM199906243402504. [DOI] [PubMed] [Google Scholar]
- 48.Carén H, Kryh H, Nethander M, et al. High-risk neuroblastoma tumors with 11q-deletion display a poor prognostic, chromosome instability phenotype with later onset. Proc Natl Acad Sci. 2010;107:4323–4328. doi: 10.1073/pnas.0910684107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guo X, Chen QR, Song YK, et al. Exon array analysis reveals neuroblastoma tumors have distinct alternative splicing patterns according to stage and MYCN amplification status. BMC Med Genomics. 2011;4:35. doi: 10.1186/1755-8794-4-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beiske K, Burchill SA, Cheung IY, et al. Consensus criteria for sensitive detection of minimal neuroblastoma cells in bone marrow, blood and stem cell preparations by immunocytology and QRT-PCR: Recommendations by the International Neuroblastoma Risk Group Task Force. Br J Cancer. 2009;100:1627–1637. doi: 10.1038/sj.bjc.6605029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Naranjo A, Parisi MT, Shulkin BL, et al. Comparison of 123I-metaiodobenzylguanidine (MIBG) and 131I-MIBG semi-quantitative scores in predicting survival in patients with stage 4 neuroblastoma: A report from the Children's Oncology Group. Pediatr Blood Cancer. 2011;56:1041–1045. doi: 10.1002/pbc.22991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hodi FS, O'Day SJ, McDermott DF, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beatty GL, Chiorean EG, Fishman MP, et al. CD40 agonists alter tumor stroma and show efficacy against pancreatic carcinoma in mice and humans. Science. 2011;331:1612–1616. doi: 10.1126/science.1198443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brahmer JR, Drake CG, Wollner I, et al. Phase I study of single-agent anti–programmed death-1 (MDX-1106) in refractory solid tumors: Safety, clinical activity, pharmacodynamics, and immunologic correlates. J Clin Oncol. 2010;28:3167–3175. doi: 10.1200/JCO.2009.26.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.