Summary
Pore-forming proteins containing the structurally conserved membrane attack complex/perforin fold play an important role in immunity and host-pathogen interactions. Intermedilysin (ILY) is an archetypal member of a cholesterol-dependent cytolysin subclass that hijacks the complement receptor CD59 to make cytotoxic pores in human cells. ILY directly competes for the membrane attack complex binding site on CD59, rendering cells susceptible to complement lysis. To understand how these bacterial pores form in lipid bilayers and the role CD59 plays in complement regulation, we determined the crystal structure of human CD59 bound to ILY. Here, we show the ILY-CD59 complex at 3.5 Å resolution and identify two interfaces mediating this host-pathogen interaction. An ILY-derived peptide based on the binding site inhibits pore formation in a CD59-containing liposome model system. These data provide insight into how CD59 coordinates ILY monomers, nucleating an early prepore state, and suggest a potential mechanism of inhibition for the complement terminal pathway.
Graphical Abstract
Highlights
-
•
Crystal structure of the ILY-CD59 complex defines two interfaces
-
•
Our two binding interfaces are supported by previous mutagenesis studies
-
•
An ILY-derived peptide competes for binding in a liposome model system
-
•
Our model provides a structural basis for CD59 nucleation of an ILY early prepore
The crystal structure of human CD59 in complex with the bacterial toxin intermedilysin (ILY) is now presented by Bubeck and colleagues. These results suggest that CD59 positions the cholesterol-binding motif of ILY in proximity to the membrane and nucleates oligomerization of ILY monomers through two binding faces. This model for CD59 function gives insight into the mechanism of cholesterol-dependent cytolysin pore formation and provides a framework for future investigation probing regulation of the complement terminal pathway.
Introduction
Bacterial toxins of the cholesterol-dependent cytolsysin (CDC) superfamily form pores in lipid bilayers through the oligomerization of their membrane attack complex/perforin fold (MACPF) domain. CDCs are produced by over five different genera of Gram-positive bacteria, resulting in a wide range of animal and human diseases (Tweten et al., 2001). Intermedilysin (ILY) is a CDC secreted by Streptococcus intermedius that causes brain and liver abscesses in its host (Nagamune et al., 1996) and is thought to be the major virulence factor for the bacterium (Nagamune et al., 1996). Cholesterol is essential for ILY cytolytic activity, and crystal structures of ILY and other CDCs show strong structural homology (Bourdeau et al., 2009; Polekhina et al., 2005; Rossjohn et al., 1997; Rossjohn et al., 2007; Xu et al., 2010). Most CDCs bind to cholesterol-rich membranes via a cholesterol recognition motif (CRM) and neighboring undecapeptide. This binding triggers the oligomerization and allosteric regulation of the transition to the pore state (Dowd and Tweten, 2012). ILY, on the other hand, has evolved to recruit the human complement regulator CD59 as a receptor providing the basis for its species specificity (Giddings et al., 2004). It is CD59-binding that promotes both oligomerization of ILY monomers on the cell surface (LaChapelle et al., 2009) and the structural rearrangements required for pore formation (Dowd and Tweten, 2012). In contrast, the interaction of cholesterol with the CRM and undecapeptide is required for the maintenance of the ILY prepore at the membrane as CD59 is released (LaChapelle et al., 2009). Mutational analysis of ILY has determined residues involved in both lipid and receptor binding (Wickham et al., 2011) as well as those crucial for mediating structural transitions (Dowd and Tweten, 2012; Hotze et al., 2012; LaChapelle et al., 2009; Soltani et al., 2007a, b). Our structure of the ILY-CD59 complex now provides a framework with which to integrate these biochemical and biophysical data.
CD59 is the only membrane-bound inhibitor of the complement terminal pathway (Meri et al., 1990). This small (77 amino acid) GPI-anchored cell-surface receptor binds complement components C8 and C9 (Ninomiya and Sims, 1992) and directly inhibits the formation of the membrane attack complex (MAC), an immune pore that kills microbes by the disruption of lipid bilayers. CD59 is expressed on a variety of cell types, including leukocytes, epithelial, and endothelial cells (Meri et al., 1991) and plays an important role in preventing complement-mediated tissue damage in hosts. CD59 has been implicated in a number of human diseases, including tumor cell evasion during monoclonal antibody treatments (Fishelson et al., 2003; Gorter and Meri, 1999) and paroxysmal nocturnal hemoglobinuria (Hill et al., 2006). Structural studies of soluble CD59 (Fletcher et al., 1994; Kieffer et al., 1994; Leath et al., 2007) and direct binding assays of CD59 with complement components (Huang et al., 2006) have provided some insight into the role of this receptor in regulating MAC assembly. However, a lack of structural information characterizing CD59 with its binding partners has limited our ability to control complement susceptibility.
Here, we report the crystal structure of CD59 bound to ILY, a bacterial toxin that directly competes for MAC binding. We define two interfaces on CD59 that coordinate ILY monomers and validate this interaction through the development of a liposome-based model system. Cholesterol-containing liposomes decorated with cytotopic CD59 capture ILY monomers and support pore formation in a fluorescence-based assay. We also find that an ILY-derived peptide based on our structure disrupts the interface and competes for pore formation. In addition, cryo-electron microscopy (cryo-EM) images of the crystallized ILY mutant bound to CD59-decorated liposomes suggest that the linear arrays observed in our crystal could also occur in the context of a membrane. Altogether, our structural and functional data provide mechanistic insight into how CD59 engages ILY for the formation of a pore in lipid bilayers, providing a basis for the development of new antibiotics as well as potential therapeutics that target CD59.
Results
Crystal Structure of the ILY-CD59 Complex Reveals Two Interfaces
An early prepore intermediate comprising the initial attachment of ILY to CD59 was captured with a disulfide-locked ILY mutant (ILYEPP) and a CD59 construct lacking the GPI anchor. Both proteins were expressed and purified independently. The complex was assembled in vitro, subjected to further purification, and crystallized. The crystals diffracted to a resolution of 3.5 Å, and the structure was solved by molecular replacement. The final model, consisting of all four domains of ILY and the ectodomain of CD59, was refined to a final Rwork/Rfree of 21.2%/26.6% and displayed good geometry (Figures 1 and S1A; Table 1).
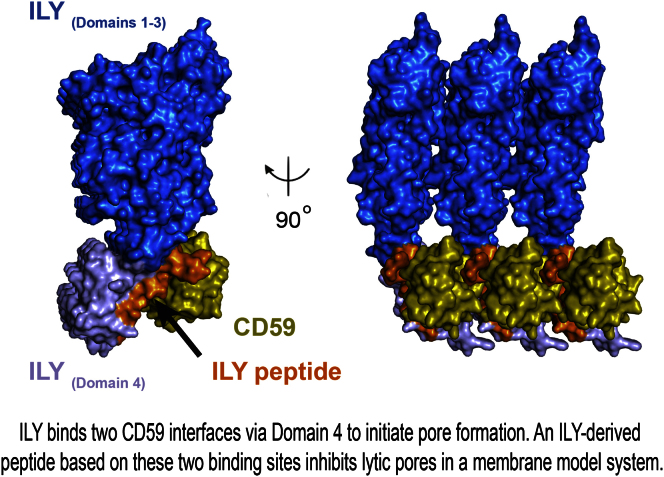
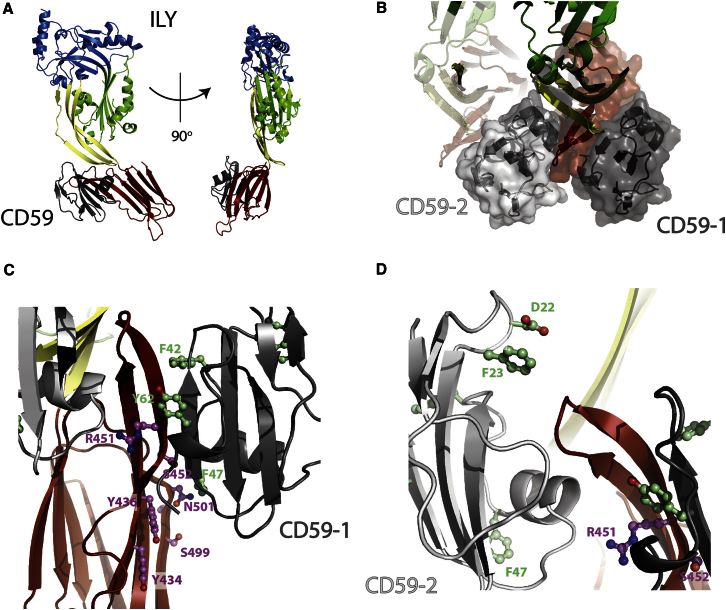
Figure 1.
Structure of the ILY-CD59 Complex
(A) Two views of the complex related by 90°. The protein is shown in cartoon representation and colored by domain. ILY-D1, blue; ILY-D2, yellow; ILY-D3, green; ILY-D4, red; and CD59, gray.
(B) A view of the crystal packing at the ILY-CD59 interface. ILY D1–D3 are shown as drawings and colored as in (A). ILY-D4 is colored red and shown as a drawing with a transparent surface. CD59 is colored gray and shown as a drawing with a transparent surface. A neighboring copy of the complex from the next unit cell is also shown with ILY rendered as a transparent drawing, and CD59 is shown in a light gray drawing with a transparent surface.
(C–D) Zoom on the two ILY-CD59 interfaces in the crystal colored as in (B). Residues on CD59 and ILY that impact binding and function when mutated are shown in ball and stick and colored green and purple, respectively.
Figures were rendered with PyMOL. See also Figure S1.
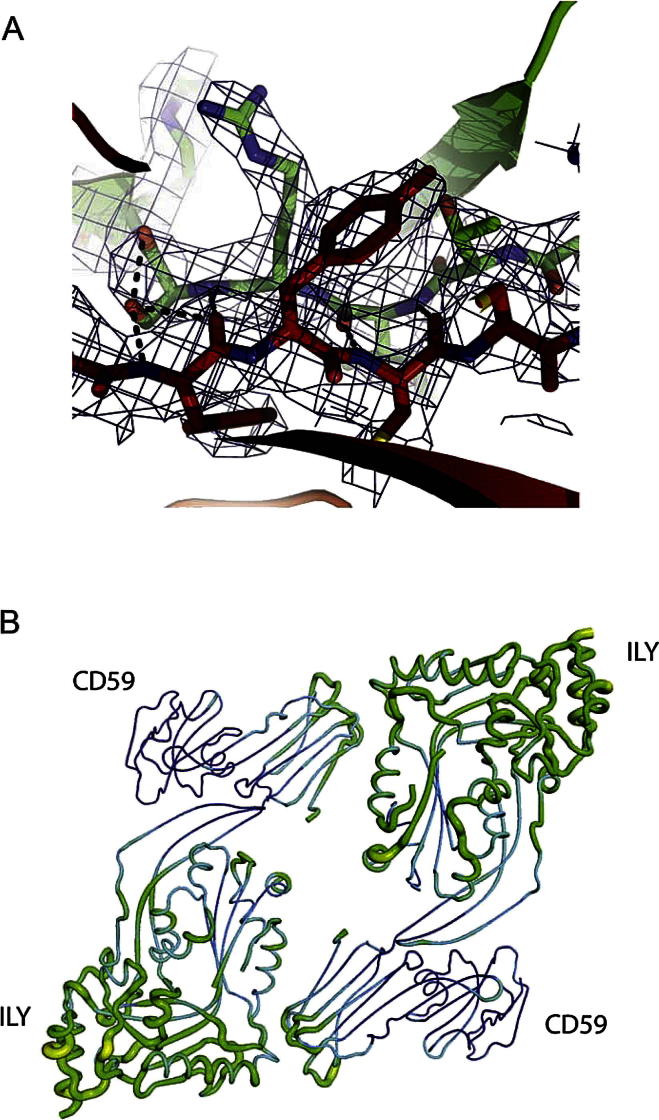
Figure S1.
Crystal Structure of the ILY-CD59 Complex, Related to Figure 1
(A) Electron density (2Fo-Fc, blue mesh) overlaid with atomic models for ILY (green) and CD59 (red) at the complex interface. Black dashed lines show hydrogen bonds between β strands.
(B) The two complexes in the asymmetric unit colored by atomic B factors (blue to red) and shown as Putty representation where the tube radius corresponds to the B factor.
Table 1.
Data Collection and Refinement Statistics
| ILYEPP/CD59 | |
|---|---|
| Space group | P1 |
| Unit cell parameters (Å) | a = 27.4, b = 101.45, c = 112.88, α = 63.75°, β = 89.95°, γ = 90.02° |
| Resolution (Å) | 45.45-3.49 (3.62-3.49) |
| No. unique reflections | 13247 (1303) |
| Rmerge | 0.111 (0.304) |
| I/σ (I) mean | 7.3 (2.0) |
| Completeness (%) | 96.0 (96.9) |
| Multiplicity | 2.0 (2.0) |
| Rwork /Rfree | 0.212/0.266 (0.239/0.267) |
| No. atoms | |
| Protein | 8387 |
| Ligand/ion | 0 |
| Water | 0 |
| Mean B factors (Å2) | 103.00 |
| RMSDs | |
| Bond lengths (Å) | 0.004 |
| Bond angles (°) | 0.96 |
| Ramachandran plot analysis | |
| Most favored regions (%) | 95.0 |
| Disallowed regions (%) | 0.2 |
| Molprobity score | 2.28 |
| PDB code | 4BIK |
Numbers in brackets refer to the highest resolution shell.
As described previously, ILY is comprised of four structural domains, the first three of which are formed from discontinuous stretches of sequence. Domain 1 (D1) (residues 56–76, 117–201, 258–299, and 379–399) forms a cap at the top of the molecule and is the least well-ordered region of our crystal (Figure S1B). Domain 2 (D2) (residues 77–116 and 400–417) consists of three kinked β strands that span 50 Å and bridge domains 1 and 4. Domain 3 (D3) (residues 202–257 and 300–378) packs against D2 and contains the helical bundles that are predicted to refold into transmembrane β-hairpins. This domain, along with D1, makes up the MACPF domain and also contains the site of the engineered disulfide bond (C346-C361) that prevents the structural rearrangements required for full pore formation. Finally, domain 4 (D4) (residues 418–528) is a discrete β sandwich structure extending from the end of D2. CD59 is a compact, heavily disulfide-bonded structure with a central β sheet bookended by a short α helix and a two-strand β hairpin.
The asymmetric unit of the crystal contains two copies of the ILY-CD59 complex, which overlay with an RMSD of 1.0 Å over 530 residues. CD59 predominantly interacts with D4 of ILY (Figure 1A), which is in agreement with previous studies (Nagamune et al., 2004; Polekhina et al., 2005). Strikingly, there are two major ILY-CD59 interfaces within the crystal: one within the asymmetric unit and one between neighboring unit cells (Figure 1B).
The primary ILY-CD59 binding site (665 Å2 buried surface area) is composed of a side-to-side contact between the β hairpin extension of ILY D4 (the β2-β3 loop) and the core β sheet of CD59 (Figure 1C), effectively creating an extension of the CD59 sheet. The interface is further stabilized by the stacking of hydrophobic side-chains (ILY:Y436, ILY:I450, CD59:F47, CD59:Y61, and CD59:F42) as well as an extensive network of hydrogen bonds (both sidechain-sidechain and sidechain-backbone) and salt bridges (ILY:R480, CD59:E58; ILY:448, CD59:K65; ILY:D445, and CD59:K66). This interface is supported by two independent mutagenesis studies (Hughes et al., 2009; Wickham et al., 2011). Specifically, ILY double (Y434A/Y436A) and triple (S499N/K500R/N501T) mutants located within our primary CD59 interface (Figure 1C) dramatically inhibited pore formation and decreased the affinity of ILY for CD59. In addition, double- and single-point mutations in CD59 have implicated aromatic residues that map to this interface (F42, F47, and Y62) as being involved in ILY binding (Wickham et al., 2011).
The secondary binding site is formed by the same region of ILY (the β2-β3 loop in D4) interacting with CD59 from a neighboring unit cell. This interface buries 500 Å2 and is stabilized by a series of hydrogen bonds and three salt bridges (ILY:D443, CD59:R55; ILY:D445, CD59:K38; ILY:E93, and CD59:K41). The second interface is again validated by previous mutagenesis studies of both ILY and CD59 (Wickham et al., 2011). In particular, a double mutation in ILY (R451A/S452A) that reduces CD59-binding and hemolysis maps to this region (Figure 1D). CD59 mutations D22 and F23, also located in this interface (Figure 1D), play a role in ILY binding and function. Therefore, CD59 displays two distinct binding sites for ILY that can be simultaneously occupied. This suggests that, in addition to localizing ILY to the membrane surface, CD59 helps nucleate an early prepore complex by bridging neighboring ILY molecules, thereby creating the linear array observed in our crystals.
Probing the ILY-CD59 Interfaces within the Context of a Cholesterol-Containing Membrane
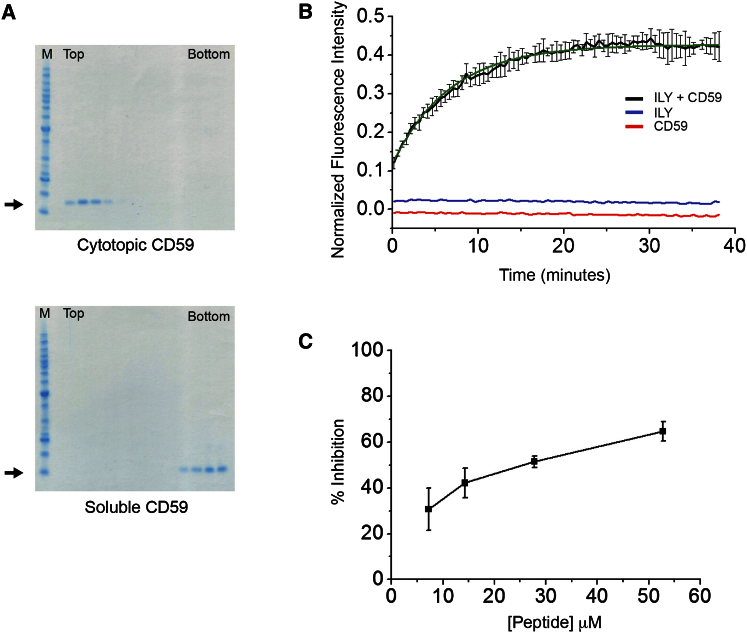
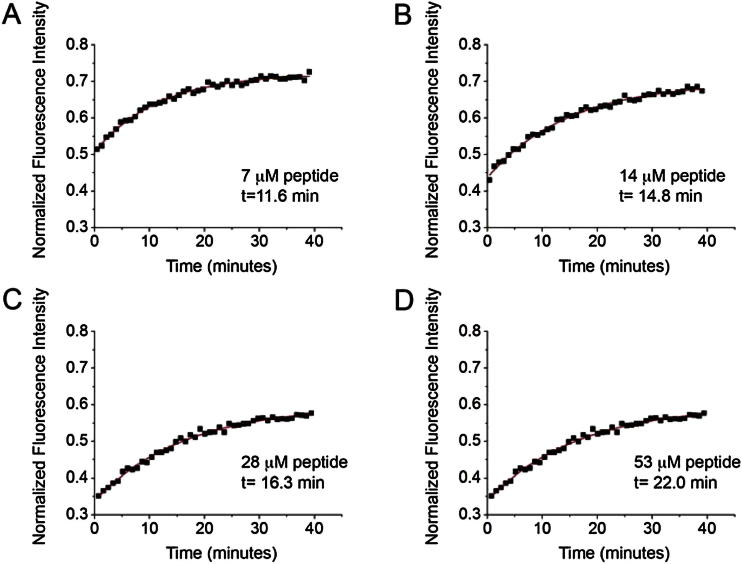
To address the functional significance of the ILY-CD59 interfaces observed in our crystal structure, we developed a simple model membrane system to probe ILY pore formation in a fluorescence-based assay (Koçer et al., 2007). Calcein-containing liposomes were decorated with soluble CD59 and modified to possess a myristoylated lysine-rich “cytotopic” peptide (Fraser et al., 2003). Lipid attachment of cytotopic CD59 was confirmed by the colocalization of protein in lipid-containing fractions across a Ficoll gradient (Figure 2A). The fluorescence intensity of calcein released from liposomes upon lysis was used to determine the rate of pore formation. Exposure of wild-type ILY (ILYWT) with cholesterol-containing CD59-decorated liposomes resulted in functional transmembrane pores (Figure 2B), whereas undecorated liposomes did not support lysis (Figure 2B). Furthermore, incubation of liposomes with cytoptic CD59 alone did not leak calcein (Figure 2B). To confirm binding of the ILY β hairpin extension to CD59, a peptide comprised of ILY residues 438–452 was synthesized with bridging N- and C-terminal cysteines. CD59-containing liposomes were incubated with increasing concentrations of peptide and subjected to ILY lysis. The peptide successfully competed for CD59 binding and inhibited ILY pore formation with an IC50 of approximately 25 μM (Figures 2C and S2).
Figure 2.
ILY Pore Formation in a Model Membrane System
(A) Cytotopic (top) and soluble CD59 (bottom) were incubated with liposomes and subjected to flotation through Ficoll (0%–20%). Gradients were fractionated from the top and aliquots run on Coomassie-stained SDS-PAGE gels. Lissamine rhodamine B-labeled liposomes banded at fractions 3 and 4. M, molecular weight markers. Arrows indicate the position of CD59 on the gel.
(B) Calcein-containing liposomes were incubated with either cytotopic CD59 (red), ILYWT (blue), or both proteins (black). The fluorescence due to calcein released from liposomes was measured as a function of time. The background was subtracted from the fluorescence intensity, and this was normalized according to the maximum intensity obtained after treatment with detergent. Error bars indicate the SD in normalized fluorescence intensity over three independent experiments. The average curve was fitted (green) with a nonlinear fitting algorithm (OriginPro 8.6), and a rate constant of 6.74 ± 0.13 min was determined.
(C) Lytic activity of ILY in the presence of increasing concentrations of peptide expressed as a percentage relative to wild-type ILY. Each sample was tested in three different experiments. SEs are indicated.
See also Figure S2.
Figure S2.
Normalized Fluorescence Intensity of Calcein-Containing Liposomes, Related to Figure 2
Calcein-containing liposomes were incubated with cytotopic CD59, wild-type ILY and increasing concentrations of an ILY-derived peptide (residues 438-452) shown in (A–D). The fluorescence intensity of calcein released from liposomes was measured as a function of time and normalized to the background and detergent solubilized samples. A nonlinear fitting algorithm (OriginPro 8.6) was used to fit the data to an exponential decay and determine the time constants (t) for each concentration of peptide. Panels show representative normalized fluorescence curves for one of three independent experiments whose average was reported in Figure 2C.
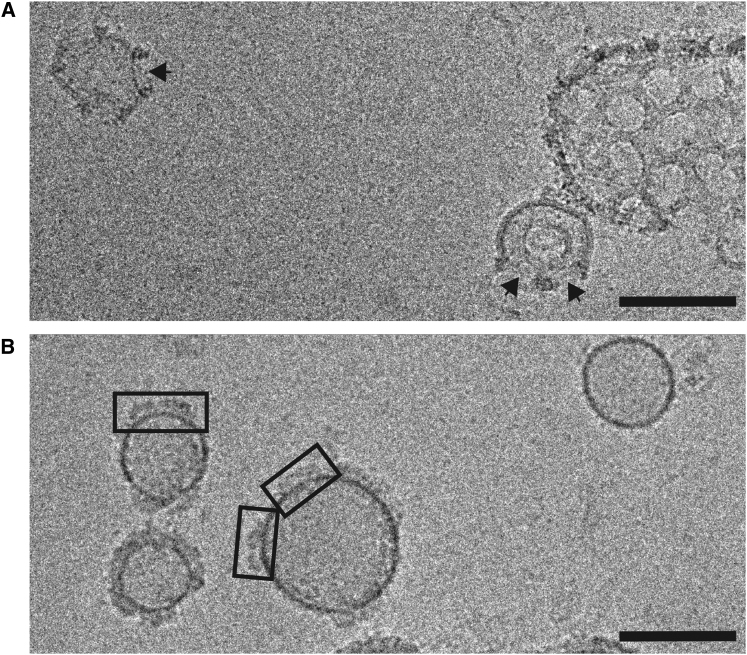
To understand how CD59 coordinates ILY monomers to form the pore, we characterized the complex in a lipid environment using electron microscopy. Because negatively stained specimens could lead to distortions of liposome curvature and possible liposome collapse, we used cryo-EM to visualize membrane-bound complexes. Examination of cryo-EM fields of ILYWT-CD59-liposome complexes revealed a population of liposomes with multiple instances of embedded circular pores (Figure 3A). Density is clearly visible for the ILYWT protein rings with obvious disruption to the lipid bilayer. The diameter of the ILYWT pore is approximately 300 Å, consistent with the dimensions previously reported for other CDC and perforin pores (Law et al., 2010; Tilley et al., 2005). To test the ability of the crystallized ILY mutant (ILYEPP) to form linear arrays in the context of a membrane, we examined two-dimensional cryo-EM images of the mutant form bound to CD59-decorated liposomes (Figure 3B). As expected, no perforation of the lipid bilayer was observed with this prepore intermediate. Protein density on the periphery of liposomes appears to be organized in linear clusters; however, due to the fragility of the system, we were unable to collect in-plane images to confirm this. Reversible linear arrays of a prepore-trapped mutant form of perfringolysin have demonstrated that CDCs undergo transient, yet specific, intermolecular interactions that precede ring closure (Hotze et al., 2012). These data suggest that the linear organization of ILY-CD59 complexes observed in our crystal structure is relevant for understanding how ILY engages with CD59 in the context of a membrane.
Figure 3.
CD59 Coordinates ILY Monomers to Form the Pore
(A) A cryo-EM image of ILYWT pores formed on CD59-decorated liposomes. Black arrows indicate sites of bilayer disruption. The scale bar represents 100 nm.
(B) A cryo-EM image field of the crystallized mutant form of ILY (ILYEPP) bound to CD59-decorated liposomes. ILYEPP clusters on the surface of liposomes are highlighted within boxes. The scale bar represents 100 nm.
Model for the ILY Early Prepore
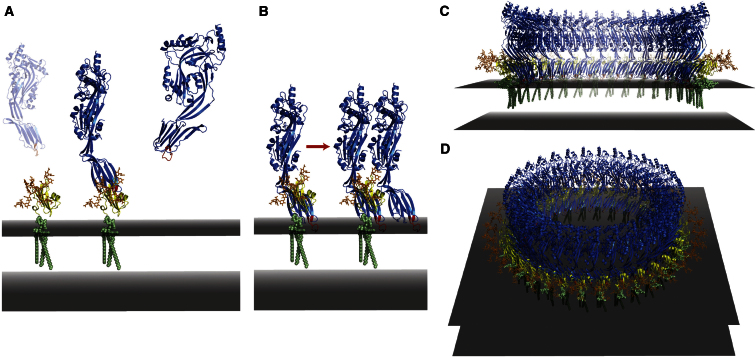
Previously reported cryo-EM reconstructions of the pneumolysin prepore and pore structures have provided a model for how CDCs oligomerize on membranes (Tilley et al., 2005). Preserving the two ILY-CD59 interfaces observed in our crystal structure, we used this as a starting point to turn the linear array of our lattice into a model for the ILY early prepore state (see Experimental Procedures for details). This arrangement places CD59 at the periphery of the ring (Figure 4), and we find that the presence of CD59 would not obstruct ILY oligomerization, in concordance with data showing that CD59 remains bound during prepore assembly (LaChapelle et al., 2009). These data, along with the high concentration of CD59 on the cell surface (Anstee, 1990), have led us to include CD59 in our model of an ILY prepore oligomeric ring. The subsequent release of CD59 during the pore transition, observed previously by biophysical studies (LaChapelle et al., 2009), could be due in part to conformational changes that are incompatible with maintaining CD59 in the complex. In our model, the C-terminal residue of CD59 preceding the GPI anchor is positioned approximately 15 Å from the plane of the membrane (Figure 4). This observation is consistent with distances measured for other GPI-anchored proteins (Lehto and Sharom, 2002) and with a model previously proposed for CD59 glycosylation (Rudd et al., 1997). Therefore, our structural and functional data together suggest that the role of CD59 in ILY pore formation is to orient the ILY CRM and undecapeptide proximal to cholesterol-containing lipid bilayers and to propagate ILY monomer-monomer interactions through two binding interfaces.
Figure 4.
Model for ILY Pore Formation
(A) Soluble ILY (blue ribbons) targets human cell membranes by binding GPI-anchored CD59 (yellow ribbons). Modeling of CD59 glycosylation reveals that binding is not obstructed by sugars (orange sticks) and that the ILY undecapeptide loop (red) is positioned proximal to the plane of the membrane (gray slab). CD59 GPI anchor is shown in light green.
(B) CD59 coordinates ILY monomers on the membrane through two distinct interfaces, nucleating an early prepore state.
(C–D) Circularization of the linear array observed in our crystal structure maintains these two interfaces and suggests that CD59 binds on the periphery of the oligomeric ring.
(C) Cross-section through the prepore oligomer.
(D) Full oligomeric ring.
Figures were rendered with PyMOL. See also Movie S1.
Discussion
Structural studies of CDCs (Bourdeau et al., 2009; Polekhina et al., 2005; Rossjohn et al., 1997; Rossjohn et al., 2007; Xu et al., 2010) and components of the complement MAC (Aleshin et al., 2012; Hadders et al., 2007; Hadders et al., 2012; Lovelace et al., 2011) reveal that the fundamental pore-forming mechanism of these MACPF-containing proteins are similar. Both involve the conversion of water-soluble proteins to transmembrane oligomeric pores. However, within this conserved mechanism, one of the most important differences remains in its regulation. For many CDCs, membrane binding by the CRM and undecapeptide initiates structural transitions (Dowd and Tweten, 2012), which are propagated between membrane-bound monomers (Hotze et al., 2012). Circularization of the CDC oligomeric ring then triggers a dramatic conformational change in which helical segments from D3 convert to amphipathic hairpins and arrange into a transmembrane β-barrel pore (Tilley et al., 2005). For a subclass of CDCs, of which ILY is an archetypal member, control of regulating prepore transitions is transferred from the undecapeptide to CD59 binding (Dowd and Tweten, 2012).
Our structural and functional analysis of the ILY-CD59 complex supports a model for these pores in which membrane-anchored CD59 recruits soluble ILY monomers to the plasma membrane of human cells, orienting the CRM and undecapeptide for insertion into cholesterol-containing lipid bilayers. Through two distinct ILY-binding interfaces on opposite sides of the molecule, CD59 brings together ILY monomers in order to facilitate intermolecular interactions that drive the formation of an oligomeric prepore ring. Relatively small alterations to the packing of our crystal lattice produces a 31-fold symmetric model for this early prepore state that agrees with a pneumolysin prepore model (Tilley et al., 2005). Even smaller manipulations of our lattice produce alternate models for ILY pores in line with the proposed stoichiometries observed for other CDCs by different biophysical techniques (31- to 44-fold) (Czajkowsky et al., 2004; Tilley et al., 2005).
Monomer-monomer interactions of CDCs have been shown to propagate early structural transitions required for pore formation (Hotze et al., 2012). Specifically, the positioning of neighboring molecules has been suggested to allow the intermolecular pairing of β1 from D3 of one molecule with β4 from the equivalent domain of the next. Analysis of the impact of applying curvature to our linear array shows that these contacts are indeed partially formed in our model (Movie S1). In order to complete the zippering of the sheet-formation, β5 would need to swing out of the way. This transition is prevented in our crystal by the engineered disulfide bond in ILYEPP locking β5 to β4, thus trapping the earliest prepore state.
Although ILY and MAC components (C8 and C9) all bind CD59, the result of this interaction has profoundly different outcomes on pore formation. Although CD59 promotes the oligomerization of ILY monomers (LaChapelle et al., 2009), the receptor inhibits MAC formation by preventing membrane perforation and incorporation of multiple C9 molecules (Farkas et al., 2002; Meri et al., 1990; Rollins and Sims, 1990). ILY directly competes for the MAC binding site on CD59 (LaChapelle et al., 2009; Wickham et al., 2011), and a mutational analysis has confirmed that the ILY and MAC binding sites on the surface of CD59 overlap, at least in part (Bodian et al., 1997; Huang et al., 2005; Wickham et al., 2011). CD59-binding MAC components lack the structural equivalent of ILY D4, therefore precluding further insight into how specific residues of C8 and C9 interact with the complement receptor. However, it is possible that CD59 engages the transmembrane segments of complement components after their transition to β hairpins but prior to membrane insertion, similar to the interaction of CD59 with the β hairpin extension of ILY D4. It is interesting to note that the residues on CD59 involved in MAC binding map to the same two opposing faces observed in our ILY interaction. CD59 inhibits MAC assembly most efficiently when incorporated prior to the addition of C9 (Lehto and Meri, 1993). Subsequently, it appears to lock a single C9 molecule into the complex in a conformation that prevents membrane perforation and further C9 recruitment (Lehto and Meri, 1993; Meri et al., 1990). These observations, along with the ILY-CD59 crystal structure, suggest a mechanism for MAC inhibition whereby CD59 intercalates between C8 and C9 in an analogous manner to the ILY nucleation, locking the MAC in a nonfunctional state.
In summary, our findings provide mechanistic insights into how ILY pores form, revealing a dual role of CD59 in facilitating the assembly of the oligomeric prepore. Our structural and functional data have defined two CD59-ILY interaction interfaces, providing a framework for the development of new antibiotics and CD59-targeted therapeutics that could improve human health.
Experimental Procedures
Expression and Purification of the ILY-CD59 Complex
Recombinant soluble human CD59 with an additional C-terminal cysteine residue was expressed in E.coli, refolded, and purified from solubilized inclusion bodies as described previously (Leath et al., 2007). Purified CD59 (approximately 0.5 mM in phosphate-buffered saline [pH7.3] PBS) was treated for 16 hr at ambient temperature with two molar equivalents of tris-2-carboxyethyl phosphine (TCEP, Sigma-Aldrich) for the generation of a free thiol at the carboxyl terminal cysteine of the protein. This was incubated for 2 hr at ambient temperature with a 3-fold molar excess of the cytotopic modification reagent bis-myristoyl lysyl SSKKSPSKKDDKKPGD (S-2-thiopyridyl)-cysteine acid (APT3146, Cambridge Research Biochemicals) for the generation of cytotopic CD59 (Hill et al., 2006). Cytotopic CD59 was further purified by hydrophobic interaction chromatography and ammonium sulfate precipitation. The main protein peak fractions were pooled, buffer exchanged into PBS, and stored at –70°C until use.
An ily gene lacking cysteine residues (ILYWT) and a disulfide-locked ily mutant containing the substitutions T346C and I361C (ILYEPP) were cloned into a pTrcHisA vector and were generous gifts from R. Tweten. This early prepore form of ILY was characterized previously (LaChapelle et al., 2009) and shown to be converted to functional pores by the addition of reducing agent. Recombinant ILY proteins were expressed in Rosetta-2 cells (Novagen). Cells were grown to an optical density of 0.6 at 37°C and induced with 0.5 mM IPTG for 4 hr. Cultures were pelleted and lysed in a buffer containing 1% Triton, 200 mM NaCl, 20 mM Tris (pH 6.8), Protease Inhibitor Mixture (Sigma-Aldrich, 50 μl per gram of cell pellet), 100 μg of DNase I, and 100 μg of RNase A (Sigma-Aldrich). Cleared lysates were loaded onto cobalt-chelated TALON beads (Clontech) and eluted with 500 mM imidazole. ILY was further purified by size exclusion chromatography in 150 mM NaCl and 20 mM Tris (pH 6.8). ILY and CD59 were incubated at room temperature in a 1:2 molar ratio, and the complex was separated on a Superdex 200 HR 10/30 column (Pharmacia Biotech).
Crystallization and Data Collection
Crystals were grown by vapor diffusion at room temperature in 96-well plates with 0.4 μl droplets at 21°C. Equal volumes of preformed ILYEPP-CD59 complex (9.9 mg/ml in 150 mm NaCl, and 20 mM Tris [pH 6.8]) and precipitant (20% PEG6000, 0.1 M HEPES [pH 7], and 0.2 M LiCl) were mixed. Crystals were cryo-protected with reservoir solution plus 20% glycerol before cryo-cooling in liquid nitrogen. Data were collected at Diamond Light Source (Harwell Science and Innovation) at beamline I04.
Structure Determination and Refinement
Diffraction data were processed with xia2 (Winter, 2010) in 3diir mode and initially assigned as P21 with a single copy of the complex per asymmetric unit (indexing initially suggested C222 (1), but this failed upon scaling, see the twinning discussion below). The structure of the ILY-CD59 complex was solved by molecular replacement with the use of Phaser (McCoy et al., 2007). A solution was reached with the use of the known structures of ILY (PDB 1S3R) (Polekhina et al., 2005) and CD59 (PDB 2UWR) (Leath et al., 2007), with the ILY split into two search models encompassing D1–D3 and D4. Analysis of the data with Xtriage (Adams et al., 2010) suggested that the data were twinned, and the structure was refined with Phenix (Adams et al., 2010) with the pseudomerohedral twinning operator (h,-k,-h-l; twin fraction = 0.08) modeled. Analysis of the resultant maps showed poor density for large stretches of D1 with no extra density or residuals, and the Rfree did not drop below 0.30. Therefore, we reprocessed the data in P1 (xia2, 3da mode) and resolved the structure with two copies of the complex in the asymmetric unit. Analysis of the P1 data set with Xtriage suggested twinning with only one of the three possible pseudomerohedral twinning operators (h,-k-l) significantly occupied (twin fraction = 0.46). Although the data merge well to 3.5 Å in both P21 (Rpim = 0.077, Rmerge = 0.105) and P1 (Rpim = 0.098, Rmerge = 0.111), refinement carried out in the P1 data set in Phenix with the h,-k,-l operator modeled produced lower R factors than the P21 refinement. After rebuilding in Coot (Emsley et al., 2010), the Rwork/Rfree converged to 0.212/0.266 (compared to 0.248/0.300 for the same model in the P21 data). Furthermore, the P1 maps showed clear features that differed between the two independent copies of the complex—in particular, the engineered disulfide (Cys346-Cys361) was clearer in one copy—and the pattern of thermal motion factors over the two molecules reproducibly refined to different distributions. Some stretches of D1 still had weak or no electron density associated with the model, although this varied between the two copies. However, refinement statistics worsen when these regions are removed, and molecular replacement of this domain placed it with Z score > 15 in Phaser. Therefore, we decided to refine D1 in place and not rebuild in the density. The final refined P1 model gave good MolProbity statistics for the resolution (MolProbity score = 2.28, 99th percentile). Running Xtriage with the final refined model in P1 gave R versus R statistics (Lebedev et al., 2006) for the h,-k,-l operator of 0.132/0.373 (Robs/Rcalc), consistent with twinning with rotational pseudosymmetry.
Building a Model of the Early Prepore
A model for an early prepore was generated from the crystal lattice by bending a linear array containing 31 copies of the 1:1 ILY-CD59 complex into a circle. In brief, the linear array was placed onto the deposited pneumolysin prepore model (PDB 2BK2) by the superposition of D1–D3 with secondary structure mapping (lsqkab). Then, a 15.3° rotation was applied to make the linear array tangential to the circular prepore density (EMDB 1106). The linear array was then bent into the circular form by the application of a 0°, 0°, 11.6° rotation (polar angles) to a monomer. The CD59 N-linked glycan on Asn18 and the GPI anchor were modeled with the coordinates described in Rudd et al., 1997, and kindly supplied by M. Wormald.
Preparation of Liposomes
1,2-dioleoyl-sn-glycero-3-phospho-L-serine (DOPS), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), and cholesterol (supplied in chloroform from Avanti Polar Lipids) were mixed in molar ratios of 3:3:2:2, respectively. Chloroform was evaporated with a stream of nitrogen gas, leaving a thin lipid film. Dried lipid was rehydrated with four freeze, thaw, and vortex cycles followed by extrusion through a 0.1 μm pore-size membrane (Avanti) for the formation of a homogeneous population of unilamellar liposomes, freshly prepared for each experiment. Fluorescently-labeled liposomes were generated by in the inclusion of 50 mM calcein (Sigma-Aldrich) in the rehydration buffer. This initial calcein concentration of 50 mM is sufficient to cause almost complete self-quenching of its fluorescence; if calcein is subsequently released from the liposomes, its concentration is reduced and an increase in fluorescence is observed. Unencapsulated dye was removed by passage through a G-50 Sephadex column (Sigma-Aldrich) run in a buffer containing 500 mM sucrose.
Lipid Flotation Assay
Lipids were tracked by the inclusion of phosphatidylethanolamine with lissamine rhodamine B-labeled head groups (Avanti) in the lipid mixture described above at a final concentration of 2% (w/w) prior to the evaporation of chloroform. Extruded liposomes at a concentration of 1 mg/ml were incubated with either soluble or cytotopic CD59 at a final concentration of 18 μM. The liposome sample was adjusted to contain Ficoll at a final concentration of 20% (w/v) and was overlaid with 10% (w/v) Ficoll followed by buffer alone. After centrifugation at 100,000 × g at 20°C for 45 min, gradients were fractionated from the top. Fractions were analyzed for the presence of liposomes by the visualization of rhodamine and for the presence of CD59 by SDS-PAGE and Coomassie staining.
Fluorescence-Based Liposome Lysis Assay
Pore-formation assays were performed at 20°C in a Varian Cary Eclipse Fluorescence Spectrophotometer (Agilent Technologies) with excitation and emission wavelengths of 490 nm and 520 nm, respectively, and a slit width of 5 nm. The kinetics mode of the spectrophotometer was used with an average read time of 0.15 s, and measurements were made every minute. A peptide corresponding to the CD59-binding interface of ILY (residues 438–452) was synthesized with bridging N- and C-terminal cysteines (CEELGHDADGYETIRSC) (Severn Biotech). Cytotopic CD59 at a final concentration of 0.7 μM was incubated with 200 μl of fluorescently-labeled liposomes in the presence or absence of peptide, and the average background fluorescence intensity was measured for 10 min. Subsequently, ILYWT at a final concentration of 0.7 μM was added to the reaction and the fluorescence was measured for 45 min. Liposomes were burst at the end of the experiment by the addition of 1 μl 0.2M C12E8 detergent (Sigma-Aldrich), and the maximum fluorescence was found by monitoring the sample for an additional 10 min. The fluorescence measurements for each reaction were normalized according to the background and detergent readings. Rates of calcein release (which corresponds to pore formation) were calculated by fitting the normalized fluorescence intensity data with a nonlinear fitting algorithm in OriginPro 8.6.
Cryo-Electron Microscopy
We incubated 0.75 mg ml−1 liposomes with cytotopic CD59 and either ILYWT or ILYEPP in equimolar ratios so that the final protein concentration was 1 mg/ml. We applied 2.5 μl of the mixture to glow discharged holey carbon grids (QUANTIFOIL R 2/2) and vitrified in liquid ethane with an FEI Vitrobot. Images were acquired on a CM200 FEG electron microscope (FEI) operating at 200 kV. Images were recorded with a defocus range of −2.5 to −3.5 microns underfocus at a magnification of 38,000× and an electron dose of 20 e−/Å2 with a 4k × 4k TemCam-F415MP CCD camera (Tietz Video and Image Processing Systems).
Acknowledgments
We thank T. Pape for technical support, R. Tweten for ILY constructs, and M. Wormald for CD59 coordinates containing modeled glycosylation and GPI anchor. We thank Diamond Light Source for synchrotron access and the staff at beamline I04. Cryo-EM images were collected at the Imperial College Electron Microscopy Centre. This work was supported by a Medical Research Council (MRC) grant (G0900888) to S.M.L and an Engineering and Physical Sciences Research Council Platform Grant (EP/G00465X) to the membrane biophysics group at Imperial College London. R.A.G.S. is supported by the National Institute for Health Research (NIHR) Biomedical Research Centre based at Guy’s and St Thomas’s National Health Service Foundation Trust and the MRC Centre for Transplantation, King’s College London. Work in S.M.L.’s lab is supported by the Oxford Martin School.
Published: May 9, 2013
Footnotes
Supplemental Information includes two figures and one movie and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2013.04.029.
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Accession Numbers
The Protein Databank accession number for the ILY-CD59 complex reported in this paper is 4BIK.
Supplemental Information
A morph between the linear array observed in our crystal structure and the curved prepore intermediate. The structural transition between these two states was calculated with PyMOL. β1 and β4 strands of ILY are colored blue and red, respectively.
References
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleshin A.E., Schraufstatter I.U., Stec B., Bankston L.A., Liddington R.C., DiScipio R.G. Structure of complement C6 suggests a mechanism for initiation and unidirectional, sequential assembly of membrane attack complex (MAC) J. Biol. Chem. 2012;287:10210–10222. doi: 10.1074/jbc.M111.327809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anstee D.J. The nature and abundance of human red cell surface glycoproteins. J. Immunogenet. 1990;17:219–225. doi: 10.1111/j.1744-313x.1990.tb00875.x. [DOI] [PubMed] [Google Scholar]
- Bodian D.L., Davis S.J., Morgan B.P., Rushmere N.K. Mutational analysis of the active site and antibody epitopes of the complement-inhibitory glycoprotein, CD59. J. Exp. Med. 1997;185:507–516. doi: 10.1084/jem.185.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdeau R.W., Malito E., Chenal A., Bishop B.L., Musch M.W., Villereal M.L., Chang E.B., Mosser E.M., Rest R.F., Tang W.J. Cellular functions and X-ray structure of anthrolysin O, a cholesterol-dependent cytolysin secreted by Bacillus anthracis. J. Biol. Chem. 2009;284:14645–14656. doi: 10.1074/jbc.M807631200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czajkowsky D.M., Hotze E.M., Shao Z., Tweten R.K. Vertical collapse of a cytolysin prepore moves its transmembrane beta-hairpins to the membrane. EMBO J. 2004;23:3206–3215. doi: 10.1038/sj.emboj.7600350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd K.J., Tweten R.K. The cholesterol-dependent cytolysin signature motif: a critical element in the allosteric pathway that couples membrane binding to pore assembly. PLoS Pathog. 2012;8:e1002787. doi: 10.1371/journal.ppat.1002787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas I., Baranyi L., Ishikawa Y., Okada N., Bohata C., Budai D., Fukuda A., Imai M., Okada H. CD59 blocks not only the insertion of C9 into MAC but inhibits ion channel formation by homologous C5b-8 as well as C5b-9. J. Physiol. 2002;539:537–545. doi: 10.1113/jphysiol.2001.013381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishelson Z., Donin N., Zell S., Schultz S., Kirschfink M. Obstacles to cancer immunotherapy: expression of membrane complement regulatory proteins (mCRPs) in tumors. Mol. Immunol. 2003;40:109–123. doi: 10.1016/s0161-5890(03)00112-3. [DOI] [PubMed] [Google Scholar]
- Fletcher C.M., Harrison R.A., Lachmann P.J., Neuhaus D. Structure of a soluble, glycosylated form of the human complement regulatory protein CD59. Structure. 1994;2:185–199. doi: 10.1016/s0969-2126(00)00020-4. [DOI] [PubMed] [Google Scholar]
- Fraser D.A., Harris C.L., Williams A.S., Mizuno M., Gallagher S., Smith R.A., Morgan B.P. Generation of a recombinant, membrane-targeted form of the complement regulator CD59: activity in vitro and in vivo. J. Biol. Chem. 2003;278:48921–48927. doi: 10.1074/jbc.M302598200. [DOI] [PubMed] [Google Scholar]
- Giddings K.S., Zhao J., Sims P.J., Tweten R.K. Human CD59 is a receptor for the cholesterol-dependent cytolysin intermedilysin. Nat. Struct. Mol. Biol. 2004;11:1173–1178. doi: 10.1038/nsmb862. [DOI] [PubMed] [Google Scholar]
- Gorter A., Meri S. Immune evasion of tumor cells using membrane-bound complement regulatory proteins. Immunol. Today. 1999;20:576–582. doi: 10.1016/s0167-5699(99)01537-6. [DOI] [PubMed] [Google Scholar]
- Hadders M.A., Beringer D.X., Gros P. Structure of C8alpha-MACPF reveals mechanism of membrane attack in complement immune defense. Science. 2007;317:1552–1554. doi: 10.1126/science.1147103. [DOI] [PubMed] [Google Scholar]
- Hadders M.A., Bubeck D., Roversi P., Hakobyan S., Forneris F., Morgan B.P., Pangburn M.K., Llorca O., Lea S.M., Gros P. Assembly and regulation of the membrane attack complex based on structures of C5b6 and sC5b9. Cell Rep. 2012;1:200–207. doi: 10.1016/j.celrep.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill A., Ridley S.H., Esser D., Oldroyd R.G., Cullen M.J., Kareclas P., Gallagher S., Smith G.P., Richards S.J., White J. Protection of erythrocytes from human complement-mediated lysis by membrane-targeted recombinant soluble CD59: a new approach to PNH therapy. Blood. 2006;107:2131–2137. doi: 10.1182/blood-2005-02-0782. [DOI] [PubMed] [Google Scholar]
- Hotze E.M., Wilson-Kubalek E., Farrand A.J., Bentsen L., Parker M.W., Johnson A.E., Tweten R.K. Monomer-monomer interactions propagate structural transitions necessary for pore formation by the cholesterol-dependent cytolysins. J. Biol. Chem. 2012;287:24534–24543. doi: 10.1074/jbc.M112.380139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Smith C.A., Song H., Morgan B.P., Abagyan R., Tomlinson S. Insights into the human CD59 complement binding interface toward engineering new therapeutics. J. Biol. Chem. 2005;280:34073–34079. doi: 10.1074/jbc.M504922200. [DOI] [PubMed] [Google Scholar]
- Huang Y., Qiao F., Abagyan R., Hazard S., Tomlinson S. Defining the CD59-C9 binding interaction. J. Biol. Chem. 2006;281:27398–27404. doi: 10.1074/jbc.M603690200. [DOI] [PubMed] [Google Scholar]
- Hughes T.R., Ross K.S., Cowan G.J., Sivasankar B., Harris C.L., Mitchell T.J., Morgan B.P. Identification of the high affinity binding site in the Streptococcus intermedius toxin intermedilysin for its membrane receptor, the human complement regulator CD59. Mol. Immunol. 2009;46:1561–1567. doi: 10.1016/j.molimm.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer B., Driscoll P.C., Campbell I.D., Willis A.C., van der Merwe P.A., Davis S.J. Three-dimensional solution structure of the extracellular region of the complement regulatory protein CD59, a new cell-surface protein domain related to snake venom neurotoxins. Biochemistry. 1994;33:4471–4482. [PubMed] [Google Scholar]
- Koçer A., Walko M., Feringa B.L. Synthesis and utilization of reversible and irreversible light-activated nanovalves derived from the channel protein MscL. Nat. Protoc. 2007;2:1426–1437. doi: 10.1038/nprot.2007.196. [DOI] [PubMed] [Google Scholar]
- LaChapelle S., Tweten R.K., Hotze E.M. Intermedilysin-receptor interactions during assembly of the pore complex: assembly intermediates increase host cell susceptibility to complement-mediated lysis. J. Biol. Chem. 2009;284:12719–12726. doi: 10.1074/jbc.M900772200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law R.H., Lukoyanova N., Voskoboinik I., Caradoc-Davies T.T., Baran K., Dunstone M.A., D’Angelo M.E., Orlova E.V., Coulibaly F., Verschoor S. The structural basis for membrane binding and pore formation by lymphocyte perforin. Nature. 2010;468:447–451. doi: 10.1038/nature09518. [DOI] [PubMed] [Google Scholar]
- Leath K.J., Johnson S., Roversi P., Hughes T.R., Smith R.A., Mackenzie L., Morgan B.P., Lea S.M. High-resolution structures of bacterially expressed soluble human CD59. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2007;63:648–652. doi: 10.1107/S1744309107033477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebedev A.A., Vagin A.A., Murshudov G.N. Intensity statistics in twinned crystals with examples from the PDB. Acta Crystallogr. D Biol. Crystallogr. 2006;62:83–95. doi: 10.1107/S0907444905036759. [DOI] [PubMed] [Google Scholar]
- Lehto T., Meri S. Interactions of soluble CD59 with the terminal complement complexes. CD59 and C9 compete for a nascent epitope on C8. J. Immunol. 1993;151:4941–4949. [PubMed] [Google Scholar]
- Lehto M.T., Sharom F.J. Proximity of the protein moiety of a GPI-anchored protein to the membrane surface: a FRET study. Biochemistry. 2002;41:8368–8376. doi: 10.1021/bi012038+. [DOI] [PubMed] [Google Scholar]
- Lovelace L.L., Cooper C.L., Sodetz J.M., Lebioda L. Structure of human C8 protein provides mechanistic insight into membrane pore formation by complement. J. Biol. Chem. 2011;286:17585–17592. doi: 10.1074/jbc.M111.219766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meri S., Morgan B.P., Davies A., Daniels R.H., Olavesen M.G., Waldmann H., Lachmann P.J. Human protectin (CD59), an 18,000-20,000 MW complement lysis restricting factor, inhibits C5b-8 catalysed insertion of C9 into lipid bilayers. Immunology. 1990;71:1–9. [PMC free article] [PubMed] [Google Scholar]
- Meri S., Waldmann H., Lachmann P.J. Distribution of protectin (CD59), a complement membrane attack inhibitor, in normal human tissues. Lab. Invest. 1991;65:532–537. [PubMed] [Google Scholar]
- Nagamune H., Ohnishi C., Katsuura A., Fushitani K., Whiley R.A., Tsuji A., Matsuda Y. Intermedilysin, a novel cytotoxin specific for human cells secreted by Streptococcus intermedius UNS46 isolated from a human liver abscess. Infect. Immun. 1996;64:3093–3100. doi: 10.1128/iai.64.8.3093-3100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamune H., Ohkura K., Sukeno A., Cowan G., Mitchell T.J., Ito W., Ohnishi O., Hattori K., Yamato M., Hirota K. The human-specific action of intermedilysin, a homolog of streptolysin O, is dictated by domain 4 of the protein. Microbiol. Immunol. 2004;48:677–692. doi: 10.1111/j.1348-0421.2004.tb03479.x. [DOI] [PubMed] [Google Scholar]
- Ninomiya H., Sims P.J. The human complement regulatory protein CD59 binds to the alpha-chain of C8 and to the “b”domain of C9. J. Biol. Chem. 1992;267:13675–13680. [PubMed] [Google Scholar]
- Polekhina G., Giddings K.S., Tweten R.K., Parker M.W. Insights into the action of the superfamily of cholesterol-dependent cytolysins from studies of intermedilysin. Proc. Natl. Acad. Sci. USA. 2005;102:600–605. doi: 10.1073/pnas.0403229101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins S.A., Sims P.J. The complement-inhibitory activity of CD59 resides in its capacity to block incorporation of C9 into membrane C5b-9. J. Immunol. 1990;144:3478–3483. [PubMed] [Google Scholar]
- Rossjohn J., Feil S.C., McKinstry W.J., Tweten R.K., Parker M.W. Structure of a cholesterol-binding, thiol-activated cytolysin and a model of its membrane form. Cell. 1997;89:685–692. doi: 10.1016/s0092-8674(00)80251-2. [DOI] [PubMed] [Google Scholar]
- Rossjohn J., Polekhina G., Feil S.C., Morton C.J., Tweten R.K., Parker M.W. Structures of perfringolysin O suggest a pathway for activation of cholesterol-dependent cytolysins. J. Mol. Biol. 2007;367:1227–1236. doi: 10.1016/j.jmb.2007.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd P.M., Morgan B.P., Wormald M.R., Harvey D.J., van den Berg C.W., Davis S.J., Ferguson M.A., Dwek R.A. The glycosylation of the complement regulatory protein, human erythrocyte CD59. J. Biol. Chem. 1997;272:7229–7244. doi: 10.1074/jbc.272.11.7229. [DOI] [PubMed] [Google Scholar]
- Soltani C.E., Hotze E.M., Johnson A.E., Tweten R.K. Specific protein-membrane contacts are required for prepore and pore assembly by a cholesterol-dependent cytolysin. J. Biol. Chem. 2007;282:15709–15716. doi: 10.1074/jbc.M701173200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltani C.E., Hotze E.M., Johnson A.E., Tweten R.K. Structural elements of the cholesterol-dependent cytolysins that are responsible for their cholesterol-sensitive membrane interactions. Proc. Natl. Acad. Sci. USA. 2007;104:20226–20231. doi: 10.1073/pnas.0708104105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilley S.J., Orlova E.V., Gilbert R.J., Andrew P.W., Saibil H.R. Structural basis of pore formation by the bacterial toxin pneumolysin. Cell. 2005;121:247–256. doi: 10.1016/j.cell.2005.02.033. [DOI] [PubMed] [Google Scholar]
- Tweten R.K., Parker M.W., Johnson A.E. The cholesterol-dependent cytolysins. Curr. Top. Microbiol. Immunol. 2001;257:15–33. doi: 10.1007/978-3-642-56508-3_2. [DOI] [PubMed] [Google Scholar]
- Wickham S.E., Hotze E.M., Farrand A.J., Polekhina G., Nero T.L., Tomlinson S., Parker M.W., Tweten R.K. Mapping the intermedilysin-human CD59 receptor interface reveals a deep correspondence with the binding site on CD59 for complement binding proteins C8alpha and C9. J. Biol. Chem. 2011;286:20952–20962. doi: 10.1074/jbc.M111.237446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter G. xia2: an expert system for macromolecular crystallography data reduction. J. Appl. Cryst. 2010;43:186–190. [Google Scholar]
- Xu L., Huang B., Du H., Zhang X.C., Xu J., Li X., Rao Z. Crystal structure of cytotoxin protein suilysin from Streptococcus suis. Protein Cell. 2010;1:96–105. doi: 10.1007/s13238-010-0012-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A morph between the linear array observed in our crystal structure and the curved prepore intermediate. The structural transition between these two states was calculated with PyMOL. β1 and β4 strands of ILY are colored blue and red, respectively.