Abstract
This study investigated the effects of genetic polymorphisms in organic cation transporter (OCT) genes, such as OCT1-3, OCTN1, MATE1, and MATE2-K, on metformin pharmacokinetics. Of particular interest was the influence of genetic polymorphisms as covariates on the variability in the population pharmacokinetics (PPK) of metformin using nonlinear mixed effects modeling (NONMEM). In a retrospective data analysis, data on subjects from five independent metformin bioequivalence studies that used the same protocol were assembled and compared with 96 healthy control subjects who were administered a single oral 500 mg dose of metformin. Genetic polymorphisms of OCT2-808 G > T and OCTN1-917C > T had a significant (P < 0.05) effect on metformin pharmacokinetics, yielding a higher peak concentration with a larger area under the serum time–concentration curve. The values obtained were 102 ± 34.5 L/h for apparent oral clearance (CL/F), 447 ± 214 L for volume of distribution (Vd/F), and 3.1 ± 0.9 h for terminal half-life (mean ± SD) by non-compartmental analysis. The NONMEM method gives similar results. The metformin serum levels were obtained by setting the one-compartment model to a first-order absorption and lag time. In the PPK model, the effects of OCT2-808 G > T and OCTN1-917C > T variants on the CL/F were significant (P < 0.001 and P < 0.05, respectively). Thus, genetic variants of OCTN1-917C > T, along with OCT2-808 G > T genetic polymorphisms, could be useful in titrating the optimal metformin dose.
Electronic supplementary material
The online version of this article (doi:10.1208/s12248-013-9460-z) contains supplementary material, which is available to authorized users.
KEY WORDS: genetic polymorphism, metformin, OCTs, population pharmacokinetics
INTRODUCTION
Metformin is in broad clinical use to treat type 2 diabetes, acting by improving sensitivity to insulin and overcoming insulin resistance (1). In addition, metformin has received increasing interest because of its effects in decreasing cancer risk and cancer-related mortality (2,3).
Drug transporters play a key role in the distribution of metformin over tissues and the elimination of metformin through renal excretion. Metformin is eliminated primarily by the kidney without significant metabolism by hepatic enzymes. Previous studies suggest that there is considerable variation in metformin renal clearance, and genetic factors were found to contribute highly, more than 90%, to the inter-subject variation (4,5).
Organic cation transporters (OCTs) of the solute carrier (SLC) family 22 and multidrug and toxin extrusion (MATE) transporters of the SLC47 family have been identified as uptake and efflux transporters of metformin, respectively (6–10). Previous studies have shown that genetic polymorphisms in OCTs are associated with variations in the pharmacokinetics (PKs)/pharmacodynamics profiles of metformin (7,11–13).
Shu et al. (11) showed that OCT1 genotypes had a significant (P < 0.05) effect on metformin pharmacokinetics in subjects of European ancestry with reduced OCT1 allele function (148C > T, 1,201 G > A, 1258delATG, or 1,393 G > A). However, those variants are quite rare in the Korean population (14). Recently, Chen et al. (6) reported that genetic variants of OCT1 (289C > A, 350C > T, and 616C > T) also have reduced function in vitro. Song et al. (12) found that the variants of OCT2 (596C > T, 602C > T, and 808 G > T) resulted in markedly higher values for area under the serum concentration–time curve (AUC), peak concentration (Cmax) values, but lower volume of distribution (Vd/F), apparent oral clearance (CL/F) values, compared to the wild types in the 26 Korean volunteer subjects. For confirmation, a larger study in a Korean population was needed.
To date, there have been few studies on the effects of genetic polymorphisms of OCT3 on metformin disposition or pharmacologic action (8,13). The cation/carnitine transporter 1 (OCTN1) is a multispecific, bidirectional, and pH-dependent organic cation transporter with low carnitine transport activity (15) that is markedly expressed in the kidney, trachea, bone marrow, and fetal liver (16). The high expression of OCTN1 in renal proximal tubules implies the importance of OCTN1 as a transporter for renal disposition of substrate drugs (17). One study reported that the OCTN1-1507C > T variant showed no association with the PKs of metformin in Caucasians (13). Metformin has been identified as a substrate for MATE1 and MATE2-K (9,18). MATE1 and MATE2-K may be involved in the tubular secretion of metformin across the brush-border membranes of the kidneys (19), and the genetic polymorphisms of MATEs are reported to be associated with metformin efficacy (20,21). However, their effect on the PKs of metformin has not been well evaluated in the Korean population; most of these studies were performed in vitro or in Caucasians.
Therefore, to clarify the role of OCTs in the disposition of metformin, the inter-subject variations in the pharmacokinetics of orally administered metformin in the environment of genetic variants of OCT1-3, OCTN1, MATE1, and MATE2-K in the Korean population were analyzed. In addition, we characterized the population pharmacokinetics of metformin utilizing a nonlinear mixed effects modeling (NONMEM) method and assessed the effect of genetic polymorphisms in OCTs on the population pharmacokinetics of metformin.
METHODS
Subjects and Study Design
Ninety-six healthy Korean male volunteers participated in this study. The subjects’ ages ranged from 19 to 31 years (mean ± SD, 22.41 ± 2.43 years) and weights from 53.1 to 95.6 kg (mean ± SD, 67.74 ± 8.24 kg). Written informed consent was obtained from all subjects to undergo genotyping and pharmacokinetic studies. The study protocol was approved by the Institutional Review Board of the Institute of Bioequivalence and Bridging Study, Chonnam National University, Gwangju, Korea. This study was conducted in compliance with the revised Declaration of Helsinki for biomedical research involving human subjects and the rules of Good Clinical Practice.
In this retrospective data analysis, data on subjects from five independent metformin bioequivalence studies which used the same protocol were assembled; only data from the reference formulation were used for the current analysis. The BE studies were performed as single-dose, randomized, two-way, open-label, and crossover studies at the Institute of Bioequivalence and Bridging Study, College of Pharmacy, Chonnam National University. After fasting overnight, the subjects in each study were administered a single oral dose of 500 mg metformin with 240 mL of water (Glucophage tablet in each study, 500 mg; lot no. 07MK003B, Boehringer Ingelheim Korea Ltd, Seoul, Korea). Blood samples were collected in Vacutainer® tubes before dosing and at 0.5, 1, 1.5, 2, 2.5, 3, 4, 6, 8, 10, 12, and 24 h after drug administration. After centrifugation (3,000×g, 20 min, 4°C), serum samples were transferred to polyethylene tubes and immediately stored at −70°C until analysis.
Analytical Methods
Metformin concentrations in serum were determined using a validated high-performance liquid chromatography method as in the previous report (22). Briefly, to 200 μL of serum, a 50-μL aliquot of internal standard (IS, phenformin hydrochloride 2 μg/mL) and 200 μL of solution (0.5 g ZnSO4 dissolved in 100 mL methanol, plus l mL ethylene glycol) were added. The samples were extracted and centrifuged. The supernatant (300 μL) was transferred and diluted with an equal amount of distilled water, and 50 μL of the mixture was then injected onto the HPLC system. The separation was performed on a Nucleosil 100–5 SA column (5 μm particle size, 125 × 4.6 mm I.D.; MACHEREY-NAGEL GmbH & Co. KG, Postfach, Düren, Germany) at ambient temperature. The mobile phase was a mixture of acetonitrile–water–methanol (50:45:5, v/v/v) in 12 mM potassium dihydrogen phosphate at a flow rate of 1.5 mL/min. Detection was carried out at 236 nm with a UV detector. The HPLC system consisted of a model LC-10ADvp isocratic pump (Shimadzu, Kyoto, Japan) equipped with a Rheodyne 7,725 injection valve (Rheodyne, Cotati, CA, USA), a model SPD-10Avp UV detector (Shimadzu, Kyoto, Japan), and a degasser (model DGU-12A). Detector output was quantitated on a model Class LC-10 integrator (Shimadzu, Kyoto, Japan).
In this HPLC method, no interference from any endogenous substances was observed in human serum. The retention times for metformin and IS were approximately 9.2 and 11.5 min, respectively. The calibration curve was linear from 10 to 2,000 ng/mL (r2 = 0.9999, n = 9). The mean intra- and inter-assay coefficients of variation for human serum were lower than 15% at relevant concentrations (n = 9). The lower limit of quantification was 10 ng/mL. The mean relative recovery of metformin was 99.4 ± 2.43% (mean ± SD).
Genotype Analysis
DNA Direct Sequencing
The polymerase chain reaction (PCR) method was used to amplify the seven fragments (OCT1-289C > T, OCT1-350C > T, OCT1-616C > T, OCT2-596C > T, OCT2-602C > T, OCTN1-1507C > T, MATE2-K-632_633GC > TT). The final volume of the PCR was 10 μL, consisting of 10 ng of DNA, 0.5 μM of each primer pair, 0.25 mM dNTPs, 3 mM MgCl2, 1 μL 1× reaction buffer, and 0.25 U Taq DNA polymerase (Intron Biotechnology, Seongnam-Si, Gyeonggi-do, Korea). The following PCR conditions were used: initial denaturation at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 60–65°C for 30 s, initial extension at 72°C for 30–60 s, and final extension at 72°C for 10 min. The PCR products were purified using a MultiScreen384-PCR Filter Plate (Millipore, Billerica, MA, USA). The purified products were then sequenced using a BigDye Terminator Cycle Sequencing Kit and an ABI 3,730 × l automated sequencer (Applied Biosystems, Foster City, CA, USA). The sequencing primers were the same as those used for the PCR amplification (Supplementary Table S1). Mutation analyses were performed using Phred, Phrap, Consed, Polyphred 5.04 software (http://droog.mbt.washington.edu/PolyPhred.html).
SNaPshot Assay
The genotyping was screened with single base primer extension assay using ABI PRISM SNaPShot Multiplex kit (ABI, Foster City, CA, USA) according to the manufacturer’s recommendation. DNA was extracted from blood and amplified using PCR. Primer sequences and annealing temperatures are described in Supplementary Table S1. The PCR was performed on an ABI 9,700 ThermalCycler (ABI, Foster City, CA). The PCR product was purified after amplification using shrimp alkaline phosphatase (USB Corporation, Cleveland, OH, USA) and exonuclease I (USB Corporation, Cleveland, OH, USA). One microliter of the purified amplification products was added to a SNaPshot Multiplex Ready reaction mixture containing 0.15 pmol of genotyping primer for primer extension reaction. The sequences were then analyzed on an ABI Prism 3730xl DNA analyzer (Applied Biosystems, USA). Analysis was performed using GeneMapper software (version 4.0; Applied Biosystems). To control genotyping validity, genotype analyses were also conducted by PCR-restriction fragment length polymorphism method or Pyrosequencing, and only the genotyping results that were in 100% concordance were used in the actual genotype run.
Statistical Analysis for the Influence of Genetic Polymorphisms
Allele prevalence for the various SNPs was assessed for deviation from the Hardy–Weinberg equilibrium using the Pearson χ2 test. The pharmacokinetic parameters from three or more different genotype groups were compared using the Kruskal–Wallis test, a nonparametric one-way ANOVA. Differences between the pharmacokinetic parameters of the two genotypic groups were determined with the Mann–Whitney test. Two-tailed null hypotheses of no difference were rejected if p values were less than 0.05. All statistical analyses were conducted with SPSS 12.0 K for Windows (version 12.0.1; SPSS Inc, Chicago, IL, USA).
Population Analysis
Non-compartmental Analysis
Pharmacokinetic parameters were calculated by non-compartmental analysis of serum concentration–time curve data with WinNonlin software (Pharsight Corporation, Mountain View, CA, USA). The area under the serum concentration–time curve from zero to time infinity (AUCinf) and the Cmax for metformin were determined from individual serum concentration–time profiles. The AUCinf was calculated as AUC0−t + Ct/λZ, where Ct is the last measurable concentration and λZ is the terminal rate constant. AUC0−t was calculated by the linear trapezoidal rule from zero to the last measurable time point. The CL/F was calculated as the dose of metformin divided by AUCinf, where F is the oral bioavailability.
Population Pharmacokinetic Analysis
The population parameters, inter-subject (η), and residual (ε) random effects were estimated using nonlinear mixed-effects modeling as implemented in NONMEM [version VI, level 2.0; GloboMax, Hanover, MD (23)]. The basic population model was implemented within the PREDPP library subroutine ADVAN2 and TRANS2 in NONMEM and estimated using the first-order conditional estimation method with η–ε interaction. The inter-subject variability of each structural parameter of the basic model was modeled with an exponential error model:
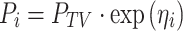 |
where Pi signifies the parameter value in an ith subject, ηi represents a random variable and the difference between Pi and PTV, which is the value of the parameter in a typical subject. It is assumed that the values of ηi are normally distributed with a mean of zero and a variance of ω2. The residual variability was evaluated (i.e., additive error, proportional error, and combined additive and proportional error) to describe the intra-subject variability. The residual variability was described best by a proportional error model:
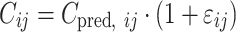 |
where Cij represents the jth observed value in the ith subject, Cpred, ij signifies the jth predicted value in the ith subject, and εij is the residual intra-subject variability with a mean of zero and a variance of σ2. The influence of subject covariate on the pharmacokinetic parameters was analyzed. The covariates included in this analysis were weight, age, body surface area (BSA), creatinine clearance (CrCL), liver function [alanine aminotransferase (ALT), aspartate aminotransferase (AST), and alkaline phosphatase (ALP)], five independent metformin BE studies, and genetic polymorphisms of OCT1-1022C > T, OCT2-808 G > T, OCTN1-917C > T, and MATE2-K-130 G > A which were selected on the basis of the genotyping results. The most appropriate pharmacostatistical model was selected based on goodness-of-fit plots, precision of estimates, and the likelihood ratio test within NONMEM. With respect to non-nested models, the Akaike Information Criteria value was used (24). The significance level was P < 0.05 during forward inclusion and P < 0.01 during backward deletion. The goodness-of-fit plots included plots of the observed and predicted subject profiles, and the population predicted estimates and conditional weighted residuals (25), which were obtained using Xpose (26) software. The precision of the population estimates was evaluated on the basis of relative standard errors (RSE%). The inter-subject variability was estimated in terms of the coefficient of variance.
The accuracy and robustness of the final population model were evaluated using visual predictive checks, implemented in Xpose with the Pearl-speaks-NONMEM. The predicted estimates-time profiles for 1,000 data sets at each point were generated from the parameters and variances of the final model selected. The 90% prediction intervals (5th to 95th percentile) of simulated metformin concentrations corresponding to the observed values were calculated and plotted for comparison with observed values.
RESULTS
Population Characteristics
In the 96 subjects analyzed, the allele frequencies of OCT1-1022C > T was 13.5%, OCT2-602C > T was 0.5%, OCT2-808 G > T was 7.8%, OCTN1-917C > T was 62.5%, MATE1-191 G > A was 0.5%, MATE1-373C > T was 0.5%, and MATE2-K-130 G > A was 45.8% (Table I). The other allele frequencies were 0%. These were comparable to previously observed results (6,8,9,21,27–34). The observed frequencies were in Hardy–Weinberg equilibrium. We selected four SNPs (OCT1-1022C > T, OCT2-808 G > T, OCTN1-917C > T, and MATE2-K-130 G > A) with frequencies >0.5% because the allele frequencies of other variants analyzed in this study were quite low. In our data analysis, subjects were categorized into two groups based upon their genotypes: variants (homozygous and heterozygous variants) and wild types. Dropouts did not occur in the five metformin BE studies and SNP analyses.
Table I.
Allele Frequencies of OCT1, OCT2, OCT3, OCTN1, MATE1, and MATE2-K Variants in Korean Population
| Gene | Nucleotide change (A > Ba) | Protein variation | n | Frequency (%) | ||||
|---|---|---|---|---|---|---|---|---|
| AA | AB | BB | Korean | Asian (6,8,9,21,27,30–32) | European (8,21,27,32–34) | |||
| OCT1 | 289C > A | Q97K | 96 | 0 | 0 | 0 | 2 (JA) | |
| 350C > T | P117L | 96 | 0 | 0 | 0 | 2 (JA) | ||
| 616C > T | R206C | 96 | 0 | 0 | 0 | 1 (JA) | ||
| 1022C > T | P341L | 72 | 22 | 2 | 13.5 | 16.8 (JA) | 0 (EA) | |
| OCT2 | 596C > T | T199I | 96 | 0 | 0 | 0 | 0.9 (JA) | 0 (EC) |
| 602C > T | A201M | 95 | 1 | 0 | 0.5 | 1.3 (JA) | 0 (EC) | |
| 808 G > T | A270S | 81 | 15 | 0 | 7.8 | 16.8 (JA) | 16 (EA) | |
| OCT3 | 1199C > T | T400I | 96 | 0 | 0 | 0 | 0 (AS) | 0.5 (EA) |
| 1,267 G > T | V423F | 96 | 0 | 0 | 0 | 6.8 (JA) | 0 (CEU) | |
| OCTN1 | 917C > T | T306I | 10 | 52 | 34 | 62.5 | 64.2 (AS) | 33.8 (EA) |
| 1507C > T | L503F | 96 | 0 | 0 | 0 | 0 (AS) | 41.2 (EA) | |
| MATE1 | 191 G > A | G64D | 95 | 1 | 0 | 0.5 | 0.6 (JA) | |
| 373C > T | L125F | 95 | 1 | 0 | 0.5 | 0.7 (AS) | 0 (EA) | |
| 929C > T | A310V | 96 | 0 | 0 | 0 | 2.2 (JA) | ||
| 983A > C | D328A | 96 | 0 | 0 | 0 | 0.6 (JA) | ||
| 1421A > G | N474S | 96 | 0 | 0 | 0 | 0.6 (JA) | ||
| MATE2-K | 130 G > A | 30 | 44 | 22 | 45.8 | 48.5 (AS) | 26.2 (EA) | |
| 192 G > T | K64N | 96 | 0 | 0 | 0 | 0.6 (JA) | ||
| 632_633GC > TT | G211V | 96 | 0 | 0 | 0 | 1.7 (JA) | ||
AS Asian American; CEU Caucasians in Utah; EA European American; EC European Caucasian; JA Japanese Asian
aThe major (i.e., more frequent or active) alleles were designated A and the minor alleles B. A/A, A/B, and B/B represent individuals homozygous for the major allele, heterozygous for the minor allele, and homozygous for the minor allele, respectively. A/A denotes carriers of no inactive allele and B/B denotes carriers of two inactive alleles
Effects of Genetic Polymorphisms on Metformin Pharmacokinetics
The AUCinf and the Cmax of metformin in relation to the different OCT1-1022C > T, OCT2-808 G > T, OCTN1-917C > T, and MATE2-K-130 G > A genotypes are summarized in Table II. Significant differences were observed between wild types and variants in OCT1-1022C > T, OCT2-808 G > T, and OCTN1-917C > T, although not for MATE2-K-130 G > A.
Table II.
Pharmacokinetic Parameters of Metformin in Relation to OCT1-1022C > T, OCT2-808 G > T, OCTN1-917C > T, and MATE2-K-130 G > A Genotypes After a Single Oral Administration of 500 mg Metformin
| Gene/genotype | AUCinf (mg h L−1) | P | C max (mg/L) | P | |
|---|---|---|---|---|---|
| OCT1 | 1022CC (n = 72) | 5.14 ± 1.48 | 0.047 | 0.84 ± 0.24 | 0.124 |
| 1022CT/TT (n = 24) | 6.23 ± 2.06 | 0.97 ± 0.31 | |||
| OCT2 | 808GG (n = 81) | 5.14 ± 1.46 | 0.007 | 0.84 ± 0.25 | 0.012 |
| 808GT (n = 15) | 6.24 ± 1.07 | 0.98 ± 0.21 | |||
| OCTN1 | 917CC (n = 10) | 3.98 ± 1.21 | 0.021 | 0.63 ± 0.16 | 0.018 |
| 917CT/TT (n = 86) | 5.49 ± 1.64 | 0.88 ± 0.28 | |||
| MATE2-K | 130GG (n = 30) | 5.34 ± 1.80 | 0.565 | 0.91 ± 0.29 | 0.761 |
| 130GA/AA (n = 66) | 5.84 ± 1.41 | 0.93 ± 0.25 | |||
Data shown as mean value ± SD. P values by Mann–Whitney test between two genotypes
AUC inf area under the serum concentration–time curve from zero to time infinity, C max peak concentration
These results indicated that genetic polymorphisms of OCT1-1022C > T, OCT2-808 G > T, and OCTN1-917C > T can potentially influence the pharmacokinetics of orally administered metformin.
However, since the disposition of metformin may involve several transporters, we also examined the association between AUCinf and Cmax of metformin and different combinations of the OCT1-1022C > T, OCT2-808 G > T, and OCTN1-917C > T genotypes. Five different combinations (out of eight possible) of the OCT1-1022C > T, OCT2-808 G > T, and OCTN1-917C > T genotypes were observed (Fig. 1a, b). Significant associations were observed between the combined genotypes of OCT1-1022C > T, OCT2-808 G > T, and OCTN1-917C > T and the pharmacokinetic parameters (AUCinf and Cmax) of metformin (P = 0.010 and P = 0.028, respectively). The combined genotype of OCT1-1022CC, OCT2-808GG, and OCTN1-917CT/TT differed significantly (P = 0.007 and P = 0.008) in both of the metformin pharmacokinetic parameters (AUCinf and Cmax) relative to individuals lacking mutations in each gene. The combined genotype of OCT1-1022CC, OCT2-808GT, and OCTN1-917CT/TT showed a statistically significant difference (P = 0.029) in the AUCinf compared with that of OCT1-1022CC, OCT2-808GG, and OCTN1-917CT/TT. The combined genotype of OCT1-1022CC, OCT2-808GT, and OCTN1-917CT/TT showed statistically significant differences (P = 0.001 and P = 0.008) in the AUCinf and Cmax, respectively, compared with those in individuals lacking mutations in each gene.
Fig. 1.
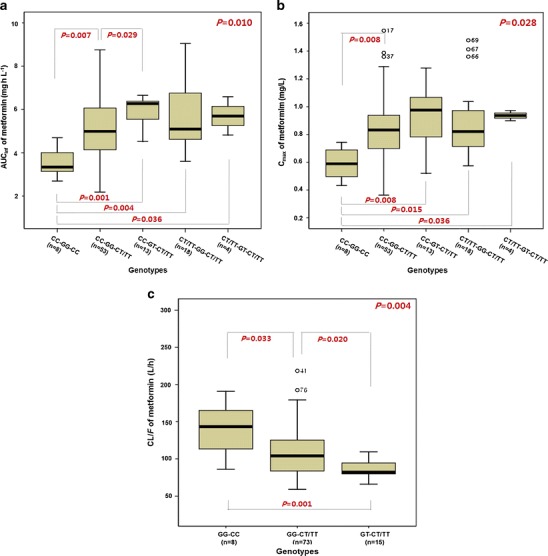
Effects of OCT1-1022C > T, OCT2-808 G > T, and OCTN1-917C > T variants on the pharmacokinetics of metformin. The AUCinf (a) and C max (b) in the combination of OCT1-1022C > T, OCT2-808 G > T, and OCTN1-917C > T genotypes after a single oral administration of 500 mg of metformin. Comparison of population pharmacokinetic parameter estimates (CL/F) in the different OCT2-808 G > T and OCTN1-917C > T genotypes (c). Significance was calculated using the Kruskal–Wallis test for simultaneous comparison of three or more genotypes, and the Mann–Whitney test for comparing two genotypes
The combined genotype of OCT1-1022CT/TT, OCT2-808GG, and OCTN1-917CT/TT differed significantly (P = 0.004 and P = 0.015) in the AUCinf and Cmax relative to individuals lacking mutations in each gene. However, these results may come from the effect of the OCTN1-917C > T genetic polymorphism. If the genetic polymorphism of OCT1-1022C > T affects the pharmacokinetics of metformin, a significant difference would be observed between the combined genotypes of OCT1-1022CC, OCT2-808GG, and OCTN1-917CT/TT, and OCT1-1022CT/TT, OCT2-808GG, and OCTN1-917CT/TT. However, when OCT1-1022CC and OCT1-1022CT/TT subjects who were OCT2-808GG with OCTN1-917CT/TT were compared, no differences were observed in the pharmacokinetic parameters of metformin (Fig. 1a, b). As a result, we observed that the genetic polymorphisms of OCT2-808 G > T and OCTN1-917C > T had a significant effect on the pharmacokinetics of metformin, while OCT1-1022C > T did not.
Population Pharmacokinetic Model
The serum concentration profile of metformin vs. time was well described by a one-compartment model with first-order absorption to the central compartment and a lag time. In the initial analyses, the one-compartment model decreased the OFV significantly (P < 0.001) when compared to the two-compartment model. When a random effect on CL/F, V/F, and ka (P < 0.001) was fit to the model, the OFV of the one-compartment model decreased significantly. Residual variability was modeled using the proportional error. A significant decrease in the OFV (ΔOFV = −129.33, P < 0.001) resulted from the inclusion of a lag time in the model. This was selected as a base model.
The model development is summarized in Supplementary Table S2 with covariates and the steps that resulted in statistically significant changes in the OFV while fitting the pharmacostatistical model. Screening the effects of covariates on PK parameters during the GAM analysis suggested that the inclusion of body weight and certain genotypes had an effect on V/F and CL/F (Fig. 1c). Inclusion of the OCT2-808 G > T genotype had a significant effect (ΔOFV = −14.08, P < 0.001) on the CL/F. In addition, the effect of the OCTN1-917C > T genotype (dominant model) on the CL/F was significant (ΔOFV = −5.88, P < 0.05). Furthermore, a combination of OCT2-808 G > T and OCTN1-917C > T genotypes was associated with a statistically significant difference (ΔOFV = −4.69, P < 0.05) in the CL/F (Supplementary Table S2). Inclusion of body weight had a significant effect (ΔOFV = −5.07, P < 0.05) on the V/F. Other covariates including age, BSA, CrCL, blood urea nitrogen, liver function (ALT, AST, and ALP), five separate metformin bioequivalence studies, OCT1-1022C > T, and MATE2K-130 G > A genotypes did not affect any PK parameters. As a result, Model 7 (Supplementary Table S2) was arrived at as the final population PK model:
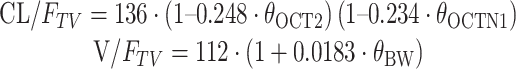 |
where 136 L/h is the estimated oral clearance for subjects with the combined OCT2-808 G > T and OCTN1-917C > T genotypes and the fractional change in the oral clearance of OCT2-808 G > T and OCTN1-917C > T genotypes is −0.248 and −0.234, respectively. θcovariates is the index variable for covariates.
The effect of OCT2-808 G > T variants on the CL/F of metformin was significant (P = 0.005), and the estimated CL/F in subjects with the OCT2-808GT allele (84.1 L/h) was lower than in those with the OCT2-808GG allele (109.5 L/h). In addition, statistically significant differences (P = 0.019) in CL/F were observed between OCTN1-917C > T genotypes, and the estimated CL/F in subjects with the OCTN1-917CT/TT alleles (103.6 L/h) was lower than in those with the OCTN1-917CC allele (140.4 L/h). Furthermore, the combined genotype of OCT2-808GT and OCTN1-917CT/TT (87.0 L/h) showed statistically significant differences in CL/F compared with OCT2-808GG and OCTN1-917CT/TT (107.2 L/h, P = 0.020) or those from individuals lacking mutation in both genes (140.4 L/h, P = 0.001; Fig. 1c). The final model’s equation gave metformin’s estimated CL/F at about 40% lower in subjects with the combined genotype of OCT2-808GT and OCTN1-917CT/TT than in those of OCT2-808GG and OCTN1-917CC. The Cmax of metformin ranged from 363.2 to 1546.9 mg/L (about 4.5-fold). Mean Cmax of OCT2-808GG with OCTN1-919CC and OCT2-808GT with OCTN1-917CT/TT subjects was 589.8 and 936.5 mg/L, respectively. The values obtained were 102 ± 34.5 L/h for CL/F, 447 ± 214 L for Vd/F, and 3.1 ± 0.9 h for terminal half-life (t1/2, mean ± SD) by non-compartmental analysis. The NONMEM method gives similar results.
Table III showed the population parameter values and inter- and intra-subject variability, which were estimated by the final model. Generally, the PK parameters were well estimated, with the relative standard error (%RSE) for estimation being approximately 3.00–18.4% of the estimated population parameter mean values. The relative standard error (%RSE) of the inter-subject variability of parameter estimates was low (9.63–24.6%). In the final model, the η shrinkage values were relatively small (all < 24.8%). Residual error was low at 17% of the proportional error, meaning low intra-subject variability, measurement error, and model mis-specification. Supplementary Figure S1 shows the basic goodness-of-fit plots. The diagnostic plots did not reveal any systematic bias, while predicted and observed values were in good agreement. The weighted predictions for the final population PK model were generally distributed around zero and were relatively symmetric. The population and individual post hoc predictions were distributed around the line of identity.
Table III.
Population Pharmacokinetic Parameters for Metformin in 96 Subjects and the Results of Bootstrap Validation (Final Model)
| Bootstrap replicates | |||||
|---|---|---|---|---|---|
| Population parameter | Estimate (%RSE) | Shrinkage (%) | Median | CI (95%)a | |
| TV (CL/F) (L/h) | 136 (18.4) | 135 | 104–172 | ||
| CL/F, θ OCT2 | −0.248 (15.8) | −0.222 | −0.329 to −0.0814 | ||
| CL/F, θ OCTN1 | −0.234 (61.1) | −0.234 | −0.399 to −0.0150 | ||
| TV (V/F) (L) | 112 (6.88) | 112 | 97.4–129 | ||
| V/F, θ BW | 0.0183 (42.8) | 0.0180 | 0.00330–0.0343 | ||
| TV (k a) (h−1) | 0.248 (3.00) | 0.248 | 0.234–0.263 | ||
| TV (T lag) | 0.182 (9.78) | 0.182 | 0.147–0.217 | ||
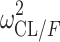
|
0.0800 (13.4) | 1.20 | 0.0800 | 0.0600–0.106 | |
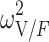
|
0.295 (15.4) | 11.7 | 0.287 | 0.207–0.387 | |
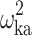
|
0.0572 (9.63) | 12.0 | 0.0561 | 0.0461–0.0680 | |
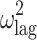
|
0.281 (24.6) | 24.8 | 0.278 | 0.167–0.441 | |
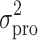
|
0.0291 (8.25) | 0.0292 | 0.0250–0.0340 | ||
A total of 1,000 replicates of bootstrap analysis
CI confidence interval, θ COVARIATE influential factor for covariate, ω interindividual variability, σ residual variability
aThe 2.5th and 97.5th percentile of the bootstrap parameter estimates
Model 7 was used to predict concentrations of a single oral 500-mg dose of metformin over 1,000 simulated datasets, with results shown in Fig. 2. The majority of observed metformin serum concentrations fell within the range of the lower (5%) and upper (95%) percentiles of the simulated concentrations.
Fig. 2.
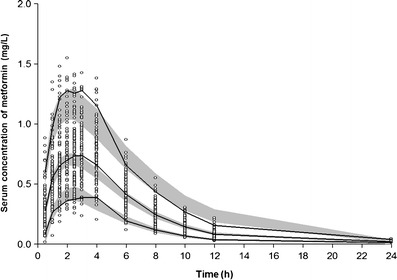
Visual predictive check of the final model between 0 and 24 h after a single oral administration of 500 mg metformin. A total of 1,000 datasets were simulated using the final pharmacokinetic parameter estimates. Open circle, observed metformin serum concentrations; the 90% confidence interval of the simulated concentrations (gray area) and observed concentration (solid line) of the 5th, median, and 95th percentile
DISCUSSION
Previous studies reported that metformin could be a substrate of MATE1 and MATE2-K, as well as OCTs (10,35). However, few investigations (12) have focused upon the relationship between polymorphisms of OCTs and the full pharmacokinetic patterns of metformin as a phenotype in the Korean population. Therefore, the objective of the current study was to evaluate whether genetic polymorphisms of OCTs have significant effects on the pharmacokinetics of metformin in healthy Korean subjects. Except for OCT1-1022C > T, OCT2-808 G > T, OCTN1-917C > T, and MATE2-K-130 G > A variants, genetic polymorphisms in OCTs are quite rare among Koreans (Table I), and our data analysis was conducted with those four variants.
In the present study, except for MATE2-K-130 G > A variant, significant differences were observed in OCT1-1022C > T, OCT2-808 G > T, and OCTN1-917C > T genotypes in the pharmacokinetic parameters of metformin. However, the combined analysis revealed that the genetic polymorphism of OCT1-1022C > T does not influence the metformin pharmacokinetics (Fig. 1a, b), even though a trend towards higher AUCinf and Cmax appeared in subjects with the variant allele (Table II). This was consistent with the result of the NONMEM method. We showed only the results of non-parametric data analyses in which we couldn’t conduct any adjustments. If any adjustment or multiple testing were available in a statistical package for a non-parametric method, the results would be more reliable.
The current study provides a population PK analysis of a single oral dose of 500 mg of metformin in healthy volunteers. A one-compartment model with a first-order absorption and lag time yielded good estimates of the metformin serum concentrations. The one-compartment model has been employed to describe the PPK of metformin (36,37). Metformin exhibited apparent flip-flop kinetics, consistent with the previous report (38), which could blunt the effects of polymorphism-associated elimination. Therefore, the values of t1/2 did not differ significantly between the variants and wild types. The values of t1/2 in the combined genotype of OCT2-808GG and OCTN1-1022CC was 3.00 ± 0.880, OCT2-808GG and OCTN1-1022CT/TT was 2.96 ± 0.875, and OCT2-808GT and OCTN1-1022CT/TT was 3.15 ± 0.972 h (mean ± SD).
The model identified key factors influencing the pharmacokinetics of metformin, such as body weight and genetic polymorphisms of OCT2-808 G > T and OCTN1-917C > T. The results showed that the inter-subject variability decreased by including body weight as a covariate for V/F, and OCT2-808 G > T and OCTN1-917C > T genotypes as a covariate for CL/F (Supplementary Table S2). Variability in CL/F of metformin among different genotypes, which is in inverse proportion to drug concentration, could influence significantly the likelihood of adverse effects such as lactic acidosis. Therefore, it might be useful to analyze an individual’s genotypes of OCT2 and OCTN1 to individualize the use of metformin. Such genetic differences may explain the inter-subject variability in the disposition of metformin in humans. Serum concentrations vs. time profiles were simulated 1,000 times for all subjects, with incorporation of the parameters, inter-subject and residual error of the final model.
The genetic polymorphisms of OCT2-808 G > T and OCTN1-917C > T both appeared to affect the CL/F, which indicates that metformin is a strong substrate of both OCT2 and OCTN1. The high expressions of OCT2 (basolateral) and OCTN1 (luminal) in renal tubules (16,39) suggest that defects in OCTs transport function may cause a decrease in the renal excretion and an increase in the serum concentration of metformin. Variant alleles led to significantly higher AUCinf and Cmax values, possibly related to increased efficacy and toxicity. The deficient OCTs proteins might delay renal excretion of metformin, prolonging the glucose-lowering effect and increasing the possibility of adverse reactions. The CL/F and CLr have been observed to be 74–78% lower in subjects with moderate and severe chronic renal impairment than in healthy young subjects (40). As the CL/F and CLr of metformin decrease approximately proportionately to CrCL, metformin dose should be reduced in patients with renal impairment in proportion to the reduced CrCL (41). Furthermore, the metformin dosage should be titrated when administered to renal impaired patients with OCT2-808GT and OCTN1-1022CT/TT variants. This recommendation, however, awaits more extensive research for confirmation.
Previous studies suggest that metformin is a substrate of OCT2 (12,13), and that OCT2 plays an important role in the disposition of metformin because renal secretion is the major elimination route of metformin. In the case of OCTN1, an in vitro study is needed to clarify whether metformin is a substrate for OCTN1 and whether the genetic polymorphism of OCTN1-917C > T has a significant effect on the transport of metformin. Renal secretion of metformin includes an entry step at the basolateral membrane and an exit step at the apical membrane of renal tubules. While OCT2 is involved with the entry of metformin into the renal tubular cells (42), OCTN1 may contribute to the flux of metformin from the tubule cells to the tubule lumen.
The frequency of OCT2-808 G > T in our study population was close to that in other Asians and Caucasians (30,33). However, the prevalence of the OCTN1-917TT allele has been reported to be about 60% in Asians and 33% in Caucasians (27,29). Therefore, the impact of OCTN1 genetic polymorphism on metformin pharmacokinetics seems to be greater in Asians than it does in Caucasians in the context of clinical therapy.
Shu et al. (43) previously demonstrated that OCT1-1022C > T variants are not related to decreased metformin uptake in HEK-293 cells and our in vivo study showed similar results. Chen et al. (6) reported that the uptake of metformin in cells expressing OCT1-289C > A, -350C > T, and -616C > T was significantly reduced relative to the OCT1 wild types. However, in the current study, the genetic polymorphisms of OCT1-289C > T, -350C > T, -616C > T display no significant impact on the pharmacokinetics of metformin because there was no subject carrying the variant in the Korean population (Table I). The genetic polymorphism of OCT2-808 G > T had a significant effect on metformin pharmacokinetics, in agreement with the previous reports conducted in Asian populations (12,44). The OCT2-808GT variant is associated with greater renal clearance of metformin in European and African-American populations (7), and while clearly involved with renal excretion of metformin, is also present in other tissues. This may explain the apparent contrasting effects on CL/F and CLr of metformin. As a side point, CLr of metformin is much smaller than its CL/F. The values of population CLr and CL/F were 510 ± 130 and 1,140 ± 330 mL/min (mean ± SD), respectively (41). Ethnic differences or genetic polymorphisms in other transporters may explain the remaining differences since renal secretion of metformin might involve several transporters (10).
We could not confirm the effects of OCT2-596C > T and -602C > T variants on metformin pharmacokinetics because of low allele frequencies. Tzetkove et al. (13) proposed that there was no relationship between OCTN1-1507C > T variant and metformin pharmacokinetics in Caucasians, and this finding seems to be in agreement with our study. However, the present study identified that the genetic polymorphism of OCTN1-917C > T influences the pharmacokinetics of metformin in the Korean population. Therefore, in addition to OCT2 genetic polymorphisms, genetic variants in OCTN1 may contribute to the variation in response to metformin.
Genotyping on 96 Korean subjects of this study showed that the allele frequencies of MATE variants (except for MATE2-K-130 G > A) were quite low (Table I), which is consistent with previous studies (9,32). The MATE2-K-130 G > A variant has recently been found to be associated with pharmacodynamic response to metformin (21). However, the investigators could not determine whether the effect of this variant for pharmacokinetics was responsible for metformin’s response, and were unable to assess the contribution of this variant to metformin pharmacokinetics. In this study, the genetic variant of MATE2-K-130 G > A apparently is not significantly associated with metformin pharmacokinetics. Therefore, we could conclude that MATE variants do not seem to affect the variability of metformin disposition in the Korean population.
The limitations of this study include the single dose evaluation and the use of only healthy subjects in well-controlled experimental conditions. A prospective, large-scale study of diabetic patients is required to determine the clinical relevance of OCTs genetic polymorphisms and their effects on clinical pharmacokinetics, therapeutic effects, and side effects in metformin therapy. If renal metformin clearance data were available, it would be helpful to further clarify the impact of OCTs genetic polymorphisms on metformin disposition. However, the present investigation using serum samples could provide valuable information.
In conclusion, the present study finds that the pharmacokinetics of metformin is significantly affected by OCT2-808 G > T and OCTN1-917C > T polymorphisms, and this may influence the clinical response to metformin therapy. In conclusion, it is recommended to consider both genetic variants of OCTN1-917C > T and OCT2-808 G > T genetic polymorphisms when titrating metformin dose.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
(DOC 173 kb)
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MEST) [No. 2011–0029209].
Competing Interests
The authors declare no conflict of interest.
Footnotes
The first two authors contributed equally to this work.
References
- 1.Bailey CJ, Turner RC. Metformin. N Engl J Med. 1996;334(9):574–9. doi: 10.1056/NEJM199602293340906. [DOI] [PubMed] [Google Scholar]
- 2.Bowker SL, Majumdar SR, Veugelers P, Johnson JA. Increased cancer-related mortality for patients with type 2 diabetes who use sulfonylureas or insulin. Diabetes Care. 2006;29(2):254–8. doi: 10.2337/diacare.29.02.06.dc05-1558. [DOI] [PubMed] [Google Scholar]
- 3.Evans JM, Donnelly LA, Emslie-Smith AM, Alessi DR, Morris AD. Metformin and reduced risk of cancer in diabetic patients. BMJ. 2005;330(7503):1304–5. doi: 10.1136/bmj.38415.708634.F7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yin OQ, Tomlinson B, Chow MS. Variability in renal clearance of substrates for renal transporters in Chinese subjects. J Clin Pharmacol. 2006;46(2):157–63. doi: 10.1177/0091270005283838. [DOI] [PubMed] [Google Scholar]
- 5.Leabman MK, Giacomini KM. Estimating the contribution of genes and environment to variation in renal drug clearance. Pharmacogenetics. 2003;13(9):581–4. doi: 10.1097/00008571-200309000-00007. [DOI] [PubMed] [Google Scholar]
- 6.Chen L, Takizawa M, Chen E, Schlessinger A, Segenthelar J, Choi JH, et al. Genetic polymorphisms in organic cation transporter 1 (OCT1) in Chinese and Japanese populations exhibit altered function. J Pharmacol Exp Ther. 2010;335(1):42–50. doi: 10.1124/jpet.110.170159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y, Li S, Brown C, Cheatham S, Castro RA, Leabman MK, et al. Effect of genetic variation in the organic cation transporter 2 on the renal elimination of metformin. Pharmacogenetics Genom. 2009;19(7):497–504. doi: 10.1097/FPC.0b013e32832cc7e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen L, Pawlikowski B, Schlessinger A, More SS, Stryke D, Johns SJ, et al. Role of organic cation transporter 3 (SLC22A3) and its missense variants in the pharmacologic action of metformin. Pharmacogenetics Genom. 2010;20(11):687–99. doi: 10.1097/FPC.0b013e32833fe789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kajiwara M, Terada T, Ogasawara K, Iwano J, Katsura T, Fukatsu A, et al. Identification of multidrug and toxin extrusion (MATE1 and MATE2-K) variants with complete loss of transport activity. J Hum Genet. 2009;54(1):40–6. doi: 10.1038/jhg.2008.1. [DOI] [PubMed] [Google Scholar]
- 10.Takane H, Shikata E, Otsubo K, Higuchi S, Ieiri I. Polymorphism in human organic cation transporters and metformin action. Pharmacogenomics. 2008;9(4):415–22. doi: 10.2217/14622416.9.4.415. [DOI] [PubMed] [Google Scholar]
- 11.Shu Y, Brown C, Castro RA, Shi RJ, Lin ET, Owen RP, et al. Effect of genetic variation in the organic cation transporter 1, OCT1, on metformin pharmacokinetics. Clin Pharmacol Ther. 2008;83(2):273–80. doi: 10.1038/sj.clpt.6100275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song IS, Shin HJ, Shim EJ, Jung IS, Kim WY, Shon JH, et al. Genetic variants of the organic cation transporter 2 influence the disposition of metformin. Clin Pharmacol Ther. 2008;84(5):559–62. doi: 10.1038/clpt.2008.61. [DOI] [PubMed] [Google Scholar]
- 13.Tzvetkov MV, Vormfelde SV, Balen D, Meineke I, Schmidt T, Sehrt D, et al. The effects of genetic polymorphisms in the organic cation transporters OCT1, OCT2, and OCT3 on the renal clearance of metformin. Clin Pharmacol Ther. 2009;86(3):299–306. doi: 10.1038/clpt.2009.92. [DOI] [PubMed] [Google Scholar]
- 14.Choi MK, Song IS. Organic cation transporters and their pharmacokinetic and pharmacodynamic consequences. Drug Metabol Pharmacokinet. 2008;23(4):243–53. doi: 10.2133/dmpk.23.243. [DOI] [PubMed] [Google Scholar]
- 15.Yabuuchi H, Tamai I, Nezu J, Sakamoto K, Oku A, Shimane M, et al. Novel membrane transporter OCTN1 mediates multispecific, bidirectional, and pH-dependent transport of organic cations. J Pharmacol Exp Ther. 1999;289(2):768–73. [PubMed] [Google Scholar]
- 16.Tamai I, Yabuuchi H, Nezu J, Sai Y, Oku A, Shimane M, et al. Cloning and characterization of a novel human pH-dependent organic cation transporter, OCTN1. FEBS Lett. 1997;419(1):107–11. doi: 10.1016/S0014-5793(97)01441-5. [DOI] [PubMed] [Google Scholar]
- 17.Tamai I, Nakanishi T, Kobayashi D, China K, Kosugi Y, Nezu J, et al. Involvement of OCTN1 (SLC22A4) in pH-dependent transport of organic cations. Mol Pharm. 2004;1(1):57–66. doi: 10.1021/mp0340082. [DOI] [PubMed] [Google Scholar]
- 18.Meyer zu Schwabedissen HE, Verstuyft C, Kroemer HK, Becquemont L, Kim RB. Human multidrug and toxin extrusion 1 (MATE1/SLC47A1) transporter: functional characterization, interaction with OCT2 (SLC22A2), and single nucleotide polymorphisms. Am J Physiol Renal Physiol. 2010;298(4):F997–1005. doi: 10.1152/ajprenal.00431.2009. [DOI] [PubMed] [Google Scholar]
- 19.Tanihara Y, Masuda S, Sato T, Katsura T, Ogawa O, Inui K. Substrate specificity of MATE1 and MATE2-K, human multidrug and toxin extrusions/H(+)-organic cation antiporters. Biochem Pharmacol. 2007;74(2):359–71. doi: 10.1016/j.bcp.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 20.Becker ML, Visser LE, van Schaik RH, Hofman A, Uitterlinden AG, Stricker BH. Interaction between polymorphisms in the OCT1 and MATE1 transporter and metformin response. Pharmacogenetics Genom. 2010;20(1):38–44. doi: 10.1097/FPC.0b013e328333bb11. [DOI] [PubMed] [Google Scholar]
- 21.Choi JH, Yee SW, Ramirez AH, Morrissey KM, Jang GH, Joski PJ, et al. A common 5′-UTR variant in MATE2-K is associated with poor response to metformin. Clin Pharmacol Ther. 2011;90(5):674–84. doi: 10.1038/clpt.2011.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho HY, Moon JD, Lee YB. Bioequivalence of Glycomin tablet to Glucophage tablet (Metformin HCl 500 mg) J Kor Pharmaceut Sci. 2002;32:223–9. [Google Scholar]
- 23.Beal SL, Sheiner LB. NONMEM user’s guide, part I. San Francisco: University of California at San Francisco; 1992. [Google Scholar]
- 24.Ludden TM, Beal SL, Sheiner LB. Comparison of the Akaike Information Criterion, the Schwarz criterion and the F test as guides to model selection. J Pharmacokinet Biopharm. 1994;22(5):431–45. doi: 10.1007/BF02353864. [DOI] [PubMed] [Google Scholar]
- 25.Hooker AC, Staatz CE, Karlsson MO. Conditional weighted residuals (CWRES): a model diagnostic for the FOCE method. Pharm Res. 2007;24(12):2187–97. doi: 10.1007/s11095-007-9361-x. [DOI] [PubMed] [Google Scholar]
- 26.Jonsson EN, Karlsson MO. Xpose—an S-PLUS based population pharmacokinetic/pharmacodynamic model building aid for NONMEM. Comput Meth Programs Biomed. 1999;58(1):51–64. doi: 10.1016/S0169-2607(98)00067-4. [DOI] [PubMed] [Google Scholar]
- 27.Urban TJ, Yang C, Lagpacan LL, Brown C, Castro RA, Taylor TR, et al. Functional effects of protein sequence polymorphisms in the organic cation/ergothioneine transporter OCTN1 (SLC22A4) Pharmacogenetics Genom. 2007;17(9):773–82. doi: 10.1097/FPC.0b013e3281c6d08e.. [DOI] [PubMed] [Google Scholar]
- 28.Kang HJ, Song IS, Shin HJ, Kim WY, Lee CH, Shim JC, et al. Identification and functional characterization of genetic variants of human organic cation transporters in a Korean population. Drug Metabol Dispos. 2007;35(4):667–75. doi: 10.1124/dmd.106.013581. [DOI] [PubMed] [Google Scholar]
- 29.Toh DS, Koo SH, Limenta LM, Yee JY, Murray M, Lee EJ. Genetic variations of the SLC22A4 gene in Chinese and Indian populations of Singapore. Drug Metabol Pharmacokinet. 2009;24(5):475–81. doi: 10.2133/dmpk.24.475. [DOI] [PubMed] [Google Scholar]
- 30.Fukushima-Uesaka H, Maekawa K, Ozawa S, Komamura K, Ueno K, Shibakawa M, et al. Fourteen novel single nucleotide polymorphisms in the SLC22A2 gene encoding human organic cation transporter (OCT2) Drug Metabol Pharmacokinet. 2004;19(3):239–44. doi: 10.2133/dmpk.19.239. [DOI] [PubMed] [Google Scholar]
- 31.Itoda M, Saito Y, Maekawa K, Hichiya H, Komamura K, Kamakura S, et al. Seven novel single nucleotide polymorphisms in the human SLC22A1 gene encoding organic cation transporter 1 (OCT1) Drug Metabol Pharmacokinet. 2004;19(4):308–12. doi: 10.2133/dmpk.19.308. [DOI] [PubMed] [Google Scholar]
- 32.Chen Y, Teranishi K, Li S, Yee SW, Hesselson S, Stryke D, et al. Genetic variants in multidrug and toxic compound extrusion-1, hMATE1, alter transport function. Pharmacogenomics J. 2009;9(2):127–36. doi: 10.1038/tpj.2008.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leabman MK, Huang CC, Kawamoto M, Johns SJ, Stryke D, Ferrin TE, et al. Polymorphisms in a human kidney xenobiotic transporter, OCT2, exhibit altered function. Pharmacogenetics. 2002;12(5):395–405. doi: 10.1097/00008571-200207000-00007. [DOI] [PubMed] [Google Scholar]
- 34.Shu Y, Leabman MK, Feng B, Mangravite LM, Huang CC, Stryke D, et al. Evolutionary conservation predicts function of variants of the human organic cation transporter, OCT1. Proc Natl Acad Sci U S A. 2003;100(10):5902–7. doi: 10.1073/pnas.0730858100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Terada T, Inui K. Physiological and pharmacokinetic roles of H+/organic cation antiporters (MATE/SLC47A) Biochem Pharmacol. 2008;75(9):1689–96. doi: 10.1016/j.bcp.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 36.Hong Y, Rohatagi S, Habtemariam B, Walker JR, Schwartz SL, Mager DE. Population exposure-response modeling of metformin in patients with type 2 diabetes mellitus. J Clin Pharmacol. 2008;48(6):696–707. doi: 10.1177/0091270008316884. [DOI] [PubMed] [Google Scholar]
- 37.Bardin C, Nobecourt E, Larger E, Chast F, Treluyer JM, Urien S. Population pharmacokinetics of metformin in obese and non-obese patients with type 2 diabetes mellitus. Eur J Clin Pharmacol. 2012;68(6):961–8. doi: 10.1007/s00228-011-1207-0. [DOI] [PubMed] [Google Scholar]
- 38.Scheen AJ. Clinical pharmacokinetics of metformin. Clin Pharmacokinet. 1996;30(5):359–71. doi: 10.2165/00003088-199630050-00003. [DOI] [PubMed] [Google Scholar]
- 39.Motohashi H, Sakurai Y, Saito H, Masuda S, Urakami Y, Goto M, et al. Gene expression levels and immunolocalization of organic ion transporters in the human kidney. J Am Soc Nephrol. 2002;13(4):866–74. doi: 10.1681/ASN.V134866. [DOI] [PubMed] [Google Scholar]
- 40.Sambol NC, Chiang J, Lin ET, Goodman AM, Liu CY, Benet LZ, et al. Kidney function and age are both predictors of pharmacokinetics of metformin. J Clin Pharmacol. 1995;35(11):1094–102. doi: 10.1002/j.1552-4604.1995.tb04033.x. [DOI] [PubMed] [Google Scholar]
- 41.Graham GG, Punt J, Arora M, Day RO, Doogue MP, Duong JK, et al. Clinical pharmacokinetics of metformin. Clin Pharmacokinet. 2011;50(2):81–98. doi: 10.2165/11534750-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 42.Wright SH. Role of organic cation transporters in the renal handling of therapeutic agents and xenobiotics. Toxicol Appl Pharmacol. 2005;204(3):309–19. doi: 10.1016/j.taap.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 43.Shu Y, Sheardown SA, Brown C, Owen RP, Zhang S, Castro RA, et al. Effect of genetic variation in the organic cation transporter 1 (OCT1) on metformin action. J Clin Invest. 2007;117(5):1422–31. doi: 10.1172/JCI30558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang ZJ, Yin OQ, Tomlinson B, Chow MS. OCT2 polymorphisms and in-vivo renal functional consequence: studies with metformin and cimetidine. Pharmacogenetics Genom. 2008;18(7):637–45. doi: 10.1097/FPC.0b013e328302cd41. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 173 kb)


