Abstract
The Zinc Finger Homeodomain Enhancer-binding Protein (Zfhep) is involved in skeletal patterning, immune cell, muscle, and brain development, and is necessary for life. Zfhep contains a single central homeodomain (HD) adjacent to an isolated zinc finger, the function of which is unknown. The placement of a zinc finger so close to a homeodomain is novel in nature. The aim of this work was to characterize the Zfhep homeodomain (HD) or the zinc finger homeodomain (ZHD), with respect to DNA-binding and protein-protein interactions. Glutathione-S-transferase (GST) fusion proteins containing either just the HD or both the zinc finger and HD (ZHD) were expressed in E. coli. The GST fusion protein affinity-binding assay demonstrated that Zfhep ZHD interacts specifically with the POU domain of the Oct-1 transcription factor. The adjacent zinc finger is required since Zfhep HD alone does not interact with Oct-1 POU domain. Furthermore, ZHD does not bind to the POU homeodomain lacking the POU specific region. These results demonstrate that the Zfhep zinc finger homeodomain motif functions as a protein-binding domain in vitro, and suggests that Zfhep may modulate the activity of POU domain transcription factors. However, neither the Zfhep ZHD nor the HD bound DNA in EMSA or selected a DNA-binding site from a pool of random oligonucleotides. This is the first demonstration of a function for the HD region of Zfhep, which is the first case of a bi-partite domain requiring both a zinc finger and a HD for binding to protein.
Keywords: AREB6, Oct-1, POU domain, Zfhep, ZEB, deltaEF1
Introduction
Zinc Finger Homeodomain Enhancer-binding Protein (Zfhep) is a member of the zinc finger homeodomain (zfh) family of proteins. These unusual proteins contain both homeodomains and multiple zinc fingers. Two subfamilies are present, having either a single homeodomain (HD) in the Zfh-1 subfamily, or 3-4 HDs in the Zfh-2 subfamily. Zfhep (deltaEF-1, ZEB, AREB6, Nil-2-a) is implicated in regulation of skeletal patterning [1], lymphocyte [2], muscle [3], and brain development [4]. Zfhep has an N-terminal domain of four zinc fingers, a separate C-terminal domain with three zinc fingers, a single central homeodomain, and an isolated zinc finger adjacent to the homeodomain [5-10]. The single HD is similar to the POU family, but diverges from the canonical HD sequence. Typically, HDs contain the amino acid sequence WFxNxR which is conserved due to important DNA contacts [11, 12]. Members of the Zfh-2 subfamily, and invertebrate members of the Zfh-1 subfamily contain the canonical HD sequence. However, in vertebrate homologues of Zfhep this sequence is altered to WFEKMQ, which would likely either decrease or destroy the ability to bind DNA.
The adjacent zinc finger may contribute to the activity of the HD. Tandem arrays of 2 or more zinc fingers are commonly used for binding to specific DNA sequences [13]. In addition, zinc fingers, especially of the CCHC subtype, can have important roles in protein-protein interactions [14]. For example, binding of the terminal zinc finger in GATA4 to the bHLH domain of dHAND is essential for synergistic activation of cardiac-specific target genes [15].
While HDs are not usually closely aligned with zinc fingers (except in the zfh family) they are often closely associated with other domains, including paired, LIM, and POUspecific domains. The POU domain is bipartite, comprising the POUspecific and POUHD, with a spacer of approximately 23 amino acids. Both parts are necessary for high affinity binding to DNA. This structure is similar to the ZHD region of rat Zfhep in which the isolated zinc finger is juxtaposed to the HD with a spacer of 25 aa. An artificial domain similar to the Zfhep ZHD has been previously described. Pomerantz et al. [16] demonstrated that an artificial chimera containing two Zif268 zinc fingers and an Oct-1 POUHD selected a hybrid homeodomain-zinc finger binding site from a pool of random oligonucleotides, and bound with high affinity. This suggests that the ZHD region of Zfhep might target a novel DNA sequence, or that the zinc finger might compensate for the non-canonical sequence of the HD in Zfhep. However, no function has been ascribed to this region of Zfhep.
As with zinc fingers, homeodomains also can have a role in protein-protein interactions. For example, Oct-1 and Pit-1 both bind protein through the POU domain [17, 18]. Such interactions can create homodimers, or heterodimers with other POU proteins, or allow binding of unrelated transcription factors. These interactions can result in synergistic activation of a target gene. Similarly, HOP is a divergent homeodomain protein that cannot bind DNA, but regulates cardiac development by interaction with the transcription factor SRF [19].
Since the HD is a prominent feature of this family of transcription factors, and the bipartite ZHD is unique to this family, we investigated possible functions of this domain. The ZHD has the potential to function in DNA-binding and/or, protein-protein interactions, and both were tested. We report that the ZHD functions as a protein-binding domain and interacts specifically with the Oct-1 POU domain. Binding requires both the zinc finger and the HD indicating that this is truly a bipartite structure.
Materials and methods
Construction of plasmids expressing fusion proteins
A cDNA fragment encoding the rat Zfhep homeodomain alone (HD; amino acids 550 to 646) and a larger fragment encoding the zinc finger and adjacent homeodomain (ZHD; amino acids 484 to 646) were subcloned by PCR to create glutathione-S-transferase (GST) fusion proteins. The following oligonucleotide primers (Genosys, The Woodlands,TX) were used for PCR and cloning: GS1, (5′-AAAGGGATCCAAAAGTGAGAAGTCACCA); GS2 (5′-GCCTGGATCCTCTGCTTCATCAGCCG), GS3 (5′-GGCTGAATTCCATCCACAGGTTGAGG). The template for PCR was the full-length rat Zfhep cDNA clone. ZHD was amplified using upper primer GS1 and lower primer GS3. HD was amplified with GS2 (upper) and GS3 (lower). Upper strand primers incorporated a Bam HI site to facilitate cloning and maintain the reading frame with pGEX-2TK, while the lower primer incorporated an Eco RI site. PCR amplified fragments were incubated with Bam HI and Eco RI, gel purified and cloned into the vector pGEX-2TK (Amersham Biosciences, Arlington Heights, USA). All clones were confirmed by DNA sequence analysis. pGEX-POU and pGEX-POUHD were kindly provided by Dr. Peter C. van der Vliet [18]. pGEX-POU encodes the POUspecific and POUHD of the Oct-1 transcription factor. pGEX-POUHD encodes only the HD portion of Oct-1. pMALc2-ZFC encodes a maltose binding protein (MBP) fusion with 3 zinc fingers from Zfhep located between amino acids 867 - 1001 (MBP-ZFC), and has been previously described [7].
Expression of fusion proteins
BL-21 or TB1 bacteria containing an expression plasmid were grown to an OD600 of 0.6-0.8, and protein expression was induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Bacteria harboring pGEX-2TK, pGEX-HD, pGEX-POU, pGEX-POUHD or pMALc2-ZFC plasmids were induced 3.5 hours at 37 °C. The pGEX-ZHD plasmid was induced 18 hours at 25 °C. After centrifugation, the bacterial pellet was resuspended in 12.5 ml ice cold PBS, 1 mM PMSF and sonicated ten times 10 seconds on ice. The mixture was brought to 1 % Triton X-100, 1 mM PMSF, 10 mM CaCl2, 6 mM MgCl2 and 6.7 U/ml DNase I (Sigma, St. Louis, MO) and mixed for 30 min at 25 °C to aid cell lysis and protein solubilization, and centrifuged at 12,000 × g for 20 minutes at 4 °C. The supernatant was mixed with 250 μl of a 50 % slurry of glutathione-sepharose (Amersham Biosciences) or amylose resin (New England Biolabs, Beverly, MA). Binding was performed on a rotator for 30 minutes at 25 °C. The resin was washed extensively with PBS, and eluted with a total of 420 μl elution buffer (10 mM glutathione for GST fusions and 10 mM maltose for MBP fusions).
Radiolabeling GST fusion proteins
Fusion proteins expressed from the pGEX-2TK plasmid contain a consensus protein kinase site that was used for radiolabeling the expressed protein. The kinase site is located between GST and the cloned protein. Radiolabeling was performed according to the manufacturer’s instructions (Amersham Biosciences). Briefly, 125 μl of glutathione sepharose beads loaded with GST fusion protein were incubated with Protein Kinase buffer (20 mM Tris pH 7.5, 100 mM NaCl, 12 mM MgCl2) with 12.5 U Bovine Heart Muscle Kinase (Sigma, St Louis, MO) and 5 μCi α-32P-ATP (6000 Ci/mmol, Amersham Biosciences) for 30 minutes at 4 °C. The reaction was terminated with 1.25 ml stop solution (10 mM sodium phosphate pH 8.0, 10 mM sodium pyrophosphate, 10 mM EDTA, and 1 mg/ml BSA). The beads were washed 5 times with 1.25 ml PBS. The radioactive GST-fusion protein was digested with thrombin to free the radioactive protein from GST. To cleave with thrombin, the beads were incubated with 7 U thrombin (Amersham Biosciences) for 16 hours at 25 °C with gentle shaking. The mixture was centrifuged to collect the supernatant containing the 32P-protein cleaved from GST.
Electrophoretic Mobility Shift Assay (EMSA)
Double-stranded oligonucleotides were designed to contain homeodomain-binding sites. The H2B oligo (5′-GATCGTGTATGCAAATAAGGTC) contains sequence from positions −35 to −55 of the human Histone 2B promoter, and is bound by the Oct-1 POU domain protein [20]. The AFP1 oligo (5′-GATCCCTGATTAATAATTACACTAAGT) contains sequence from the alpha feto-protein gene enhancer that is bound by ATBF-1, a member of the zfh family [21]. A single-stranded oligonucleotide was end-labeled with polynucleotide kinase and α-32P-ATP prior to annealing to its complement. The labeled oligonucleotide and affinity purified fusion proteins (3 - 10 μl) were incubated at 25 °C for 30 minutes in 30 μl Binding Buffer (25 mM Hepes pH 7.4, 5 mM MgCl2, 2 mM DTT, 4 mM EDTA, 10 μg/ml BSA and 20 μg/ml sheared salmon sperm DNA). The reaction mixture was electrophoresed on a 5% non-denaturing polyacrylamide gel, and exposed to film, as described [7].
Binding Site Selection
The ability of the ZHD to bind DNA was tested by binding site selection from a pool of random oligonucleotides, by the selected and amplified binding method (SAAB) [22]. A pool of oligonucleotides (49 bp) was synthesized, containing the sequence for Primer A (5′-ACCGAATTCCTACAG) at the 5′ end, an internal random sequence of 18 nucleotides and the reverse complement of Primer B (5′-AGACGGATCCATAGCA) at the 3′ end. The random oligonucleotides were annealed with Primer B, and Klenow polymerase was used to synthesize the second strand. For the first round of selection 260 ng of gel purified random oligonucleotides were reacted with a GST fusion protein for 30 minutes at 25 °C in Binding Buffer. 50 μl of a 1:5 slurry of glutathione-sepharose was added to the reaction and incubated for 10 minutes at 25 °C. The sepharose bead-GST fusion protein-DNA complexes were washed four times in Binding Buffer with 50 mM KCl and separated from unbound DNA fragments by centrifugation. The bound DNA was amplified by PCR for 15 cycles consisting of 94 °C for 15 seconds, 60 °C for 15 seconds, and 72 °C for 15 seconds. Amplified DNA was ethanol precipitated and used for additional rounds of binding site selection.
GST fusion protein affinity binding assay
Protein-protein interactions were identified by affinity chromatography. To block non-specific binding, the reactions were performed in the presence of 10 mg/ml E. coli cell extract as a competitor [23]. Approximately 20 μg of GST-fusion protein bound to glutathione sepharose beads was mixed with 5 μl of 32P-HD or 32P-ZHD protein (5 × 105 cpm) in 400 μl E. coli extract in Affinity Buffer (50 mM potassium phosphate pH 7.5, 150 mM KCl, 1 mM MgCl2) with 1 % Triton X-100and 10 % glycerol, and incubated 2 hours at 4 °C. The beads were washed 3 times with 600 μl cold Affinity Buffer. Bound proteins were eluted by boiling in 20 μl 2 × SDS sample buffer and analyzed by SDS-PAGE followed by autoradiography.
Results
Expression of fusion proteins
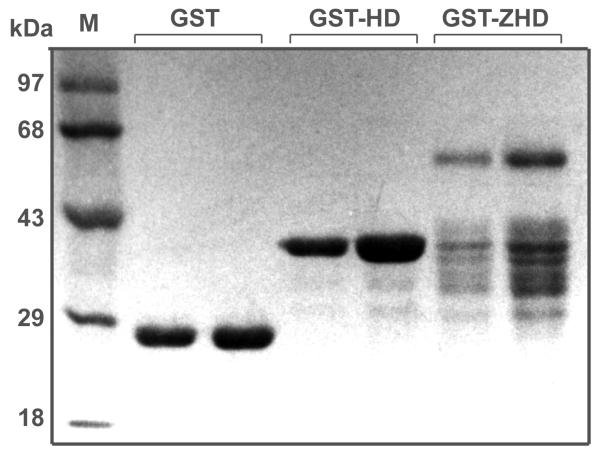
Members of the Zfh-1 subfamily typically contain a single zinc finger adjacent to the central HD (Figure 1). Bacterially expressed Zfhep homeodomain (GST-HD) or the Zfhep zinc finger homeodomain (GST-ZHD) was purified by glutathione-sepharose affinity chromatography as described above. SDS-PAGE analysis of the purified proteins shows a prominent band of the expected size for both fusion proteins (Figure 2). Proteolytic fragments were present in the GST-ZHD due to cleavage of the C-terminal end during bacterial expression. This process was minimized by growing the bacteria at a lower temperature (25 °C), such that the full-length GST-ZHD comprised about 35% of the preparation. GST-HD was expressed as essentially a single species.
Fig 1.
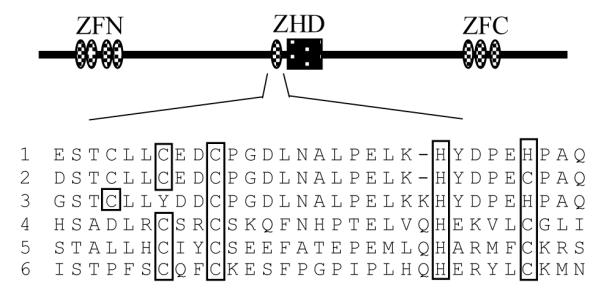
Domain structure of Zfhep. The Zfhep structure contains tandem repeats of zinc fingers near the N- and C-terminal ends (ZFN and ZFC), and a central ZHD. The amino acid sequence of the central zinc finger region in the Zfh-1 subfamily typically is similar to the zinc finger consensus (C-X2,4-C-X3-F/L-X8-H-X2-5-C/H) [13]. Sequences are from; 1, rat Zfhep (Q62947); 2, mouse Zfhx1a (NP 035676); 3, hamster BZP (Q60542); 4, Drosophila Zfh-1 (P28166); 5, mosquito (EAA06926); 6, mouse SIP-1 (NP 056568).
Fig 2.
SDS-PAGE of GST-fusion proteins. pGEX-2TK vector, or subclones of Zfhep were expressed in bacteria and purified by affinity chromatography. GST; GST-HD; or GST-ZHD proteins were analyzed by SDS-PAGE. High Range protein molecular size standards (Invitrogen, Carlsbad, USA) are shown (M).
Zfhep ZHD binds the POU domain of Oct-1
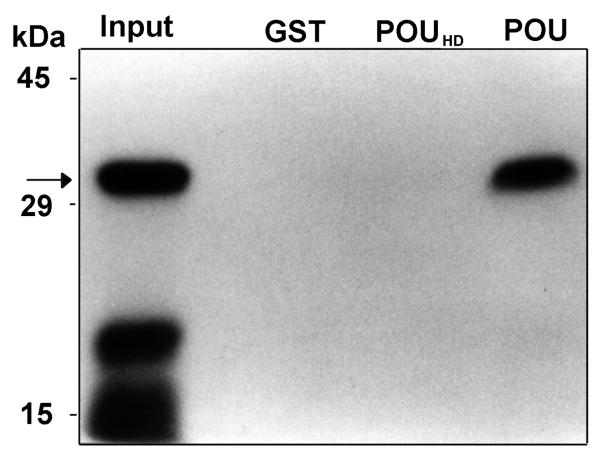
We investigated the ability of 32P-ZHD to interact with either the entire Oct-1 POU domain (i.e., the POUspecific plus the POUHD) or just the POUHD using GST fusion protein affinity binding assays. Oct-1 was used because it is capable of interacting with other specific proteins [18], and because it’s ubiquitous distribution makes it likely to be co-expressed with Zfhep in several tissues. GST-ZHD was phosphorylated at the N-terminal of the ZHD, and then the GST portion was cleaved to isolate 32P-ZHD. This labeled protein was then incubated with proteins bound to sepharose beads. GST alone served as a negative control and showed no binding. However, 32P-ZHD interacted strongly with GST-POU (Figure 3). The input 32P-ZHD appears as three proteins, the full-length 32 kDa protein and two fragments of 19 and 15 kDa. Interestingly, only full length 32P-ZHD, and not the smaller proteolytic fragments, bound to GST-POU. These smaller fragments are necessarily C-terminal truncations of ZHD since the protein was purified by an N-terminal GST tag, and the kinase site is at the N-terminal end. Therefore, the Zfhep HD is absolutely required for binding to Oct-1. In contrast to the interaction with GST-POU, 32P-ZHD did not interact with GST-POUHD (Figure 3). This indicates that amino acids in the Oct-1 POUspecific domain are necessary for interaction with 32P-ZHD. Furthermore, 32P-ZHD was unable to form dimers by binding to GST-ZHD bound to beads (not shown).
Fig 3.
Zfhep ZHD binds the Oct-1 POU domain. 32P-ZHD (5 × 105 cpm) was incubated with 20 μl GST, GST-POUHD or GST-POU bound to glutathione-sepharose in the presence of 4 mg E. coli extract as competitor. Pellets were washed as described. 32P-ZHD bound by the fusion protein was eluted in SDS sample buffer, separated by 12.5 % SDS-PAGE and analyzed by autoradiography. Lane 1 received 5 % 32P-ZHD input in the reaction (Input). Full-length 32P-ZHD is indicated by the arrow.
Saturable binding is an important characteristic of a specific interaction between proteins. We tested whether the interaction between 32P-ZHD and GST-POU is saturable by performing the reaction in the presence of 100-fold excess non-radioactive ZHD as competitor. If the binding of 32P-ZHD to GST-POU is specific, then including non-radioactive ZHD in the reaction would reduce or eliminate binding by 32P-ZHD. Binding was reduced to background levels in the presence of 100-fold competitor (Figure 4). Similar results were obtained with 50-fold competitor. We conclude from these results that the interaction of 32P-ZHD with GST-POU is saturable.
Fig 4.
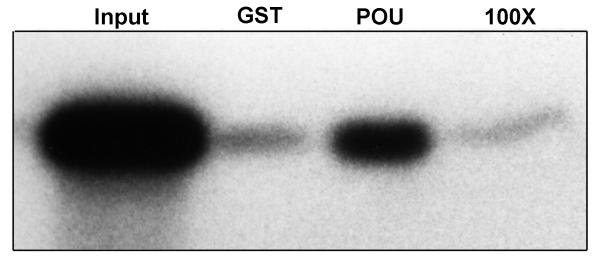
ZHD interaction with Oct-1 POU domain is specific. 32P-ZHD (2.5 μl, approximately 2.3 × 105 cpm) was incubated with 7 μl glutathione-sepharose beads with GST-POU (or GST alone) with or without a 100-fold excess non-radioactive ZHD (250 μl). Beads were washed and eluted as described. The SDS-PAGE gel lanes received either 5 % 32P-ZHD input (Input), or 32P-ZHD eluted from the GST beads (GST), GST-POU beads (POU), or GST-POU beads with 100-fold excess non-radioactive ZHD (100x).
To determine which region of the Zfhep ZHD is necessary for binding GST-POU, radiolabeled Zfhep homeodomain (32P-HD) was incubated with GST-POU or GST-POUHD. Zfhep 32P-HD does not bind either GST-POU or GST-POUHD (Figure 5). This result suggests that the region containing the solitary zinc finger is necessary for binding to Oct-1 POU domain. However, the zinc finger is not sufficient for binding since C-terminal deletions in 32P-ZHD prevent interaction with GST-POU. Therefore, both the zinc finger and homeodomain are necessary for binding to GST-POU (Figure 6).
Fig 5.

The Zfhep HD alone does not bind GST-POU or GST-POU. 32HD P-HD (5 × 105 cpm) was incubated with 20 μl of either GST, GST-HD, GST-POUHD or GST-POU bound to glutathione sepharose, and washed as described. 32P-HD bound by the fusion protein was eluted in SDS sample buffer, separated by 15 % SDS-PAGE and analyzed by autoradiography. Five percent of the 32P-HD input was loaded in lane 1 for comparison. Zfhep HD did not form homodimers under these conditions, as indicated by a lack of binding to GST-HD, nor did 32P-HD bind to Oct-1 proteins.
Fig 6.
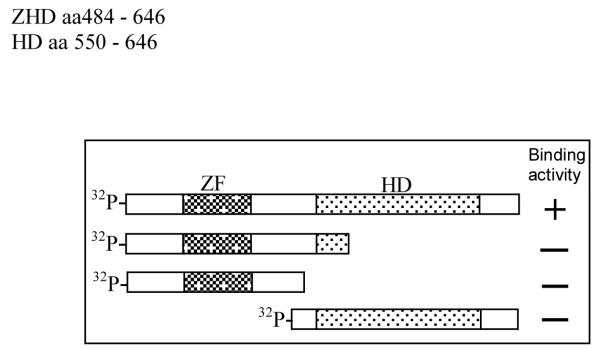
Domains in Zfhep ZHD required for interaction with Oct-1 POU domain. A schematic of the domains that bind GST-POU are shown. Neither the HD alone, nor the zinc finger alone, are able to bind the Oct-1 POU domain.
Zfhep ZHD does not bind DNA
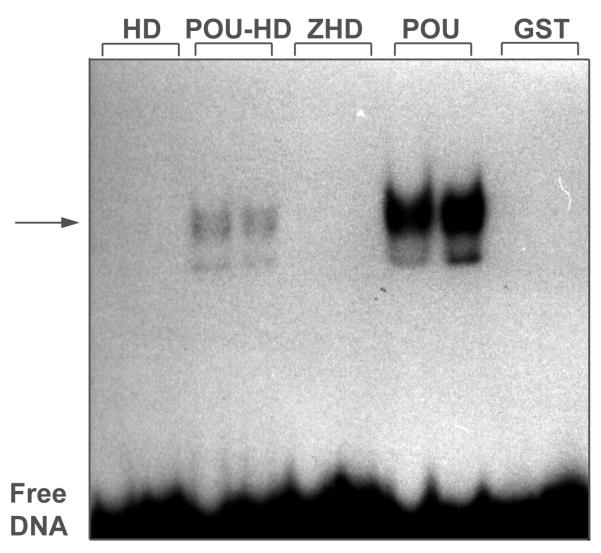
We examined the DNA-binding activity of Zfhep GST-HD or GST-ZHD with two different homeodomain-binding sites using the EMSA method. For positive control proteins, we used GST-POU and GST-POUHD. GST-POU contains the entire Oct-1 POU domain (i.e., the POUspecific plus the POUHD). GST-POUHD has only a low affinity for DNA due the lack of the POUspecific region [17]. GST alone was used as a negative control. The homeodomain-binding sites used in EMSA were double stranded oligonucleotides termed H2B and AFP1. Figure 7A shows that the H2B oligonucleotide was bound strongly by GST-POU and very weakly by GST-POUHD. However, GST-HD and GST-ZHD did not bind the H2B DNA, even as compared to the crippled POUHD. Identical results were obtained in three experiments. Increasing the amount of GST-ZHD or GST-HD in the DNA-binding reaction had no effect. No GST-ZHD, or GST-HD, complexes with H2B were detected even after long (three day) exposures of the gel. As expected, GST exhibited no binding to the H2B oligonucleotide. Similarly, 32P-AFP1 was bound effectively by GST-POU and somewhat weakly by GST-POUHD as expected (Figure 7B). In contrast, neither GST-ZHD nor GST-HD bound the 32P-AFP1 DNA in three experiments. Increasing the amount of fusion protein in the DNA-binding reaction had no effect.
Fig 7.
Zfhep HD and ZHD do not bind DNA in EMSA. (A) The Zfhep HD and ZHD do not bind the 32P-H2B probe. Each GST-fusion protein (5 μl in the first lane, and 10 μl in the second lane) was incubated with the 32P-H2B oligonucleotide. GST-POUHD and GST-POU were used as positive controls. The reaction mixture was separated on a 5 % non-denaturing gel, and exposed to film. (B) The Zfhep HD and ZHD do not bind the 32P-AFP1 probe. 5 or 10 μl of the indicated GST-fusion protein was incubated with the 32P-AFP1 oligonucleotide. The reaction mixture was separated on a 5 % non-denaturing gel, and exposed to film.
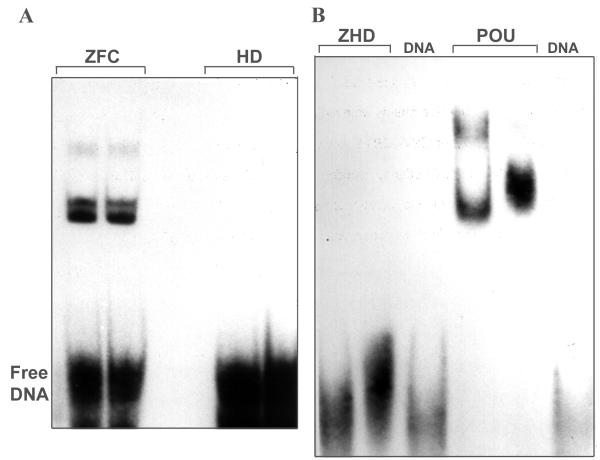
The DNA-binding activity of GST-HD and GST-ZHD was also investigated by selection of binding sites from a pool of random oligonucleotides. Either GST-POU or MBP-ZFC, a fusion protein expressing the C-terminal zinc finger domain from Zfhep, served as positive control proteins for DNA-binding. GST served as a negative control. Figure 8A shows strong binding to the SAAB oligonucleotides selected by MBP-ZFC after eight rounds of binding. In contrast, GST-HD did not shift the radiolabeled oligonucleotides, indicating that the Zfhep HD did not select a binding site. GST alone had no DNA-binding activity, as expected (not shown). Similarly, after five rounds of selection, GST-POU selected a binding site as indicated by the formation of a protein-DNA complex (Figure 8B). In contrast, GST-ZHD did not bind the SAAB oligonucleotides after five rounds of selection. These results clearly indicate that the SAAB procedure was successful, but GST-ZHD showed no DNA-binding activity.
Fig 8.
Zfhep HD and ZHD cannot select a DNA binding site. (A) Zfhep HD did not select a DNA binding site from a pool of random oligonucleotides. Oligonucleotides obtained after eight rounds of binding were radiolabeled with 32P and used in EMSA with either 5 μl (first lane) or 10 μl (second lane) of GST-HD or MBP-ZFC (positive control) fusion proteins. (B) The Zfhep ZHD did not select a DNA-binding site after five rounds of binding. GST-ZHD or GST-POU (the positive control) were used for five rounds of oligonucleotide selection. The resulting oligonucleotides were labeled and incubated with either 5 μl (first lane) or 10 μl (second lane) of the indicated GST-fusion protein. The binding reactions were separated on a 5% non-denaturing gel, and exposed to film. Labeled oligonucleotides without protein are run on adjacent lanes (DNA). Unbound oligonucleotides migrate to the bottom of the gel (Free DNA).
We concluded from these binding experiments that bacterially expressed Zfhep ZHD does not bind the AT rich enhancer bound by the zfh family protein ATBF1, or the octamer sequence bound by the POU family of proteins. The Zfhep ZHD also did not bind DNA under the SAAB experimental conditions.
Discussion
We have demonstrated that the Zfhep ZHD can function as a protein interaction domain. 32P-ZHD interacts specifically with Oct-1 POU as demonstrated by two criteria; protein specificity, and saturable binding. Competition with non-radioactive ZHD demonstrates that the binding is saturable (Figure 4). Specificity is also indicated by the fact that Zfhep ZHD does not bind to GST alone, or the Oct-1 POUHD, and does not bind to itself to form homodimers. Examples of protein interactions with either zinc fingers or homeodomains have been described, and are important for regulation of transcription [14, 17]. However, the ZHD within Zfhep is unique in that neither the HD alone, nor the zinc finger alone are sufficient for binding to Oct-1 POU (Figure 6). Therefore, we have identified a novel structure requiring the combination of a homeodomain and zinc finger for protein-binding activity.
Members of the Zfh-2 subfamily also contain single zinc fingers adjacent to HDs. ATBF1 and ZFH-4 proteins each have 4 HDs, three of which are closely followed by a zinc finger [24]. Kawaguchi et al. [25] demonstrate that both the HD and zinc finger are required for DNA/RNA-dependent ATPase activity of one of these bipartite domains, demonstrating that cooperative activity is also observed in this zfh sub-family.
The functional significance of the interaction between ZHD and the Oct-1 POU domain is unknown. The Oct-1 POU domain mediates binding to POU proteins, as well as several other proteins, including VP16, TFIIB, OBF-1, and the nuclear receptors GR, AR, RXR, VDR, and TR [17, 18, 26, 27]. This interaction can be mediated by either the POUspecific or POUHD portions of Oct-1. The formation of complexes of Oct-1 and AR, or GR requires binding of both to DNA for synergistic activation of the target promoters [26]. Our results suggest a possible interaction when Zfhep and Oct-1 regulate a target gene, for example, Zfhep represses the interleukin-2 promoter through an element that is adjacent to an octamer site [9], and both Zfhep and Oct-1 regulate the immunoglobulin heavy chain promoter [6]. Given the nearly ubiquitous distribution of Oct-1, interaction between Oct-1 and Zfhep is possible in many different tissues. Future investigation should define the functional results of this interaction, and also determine whether closely related proteins, such as Brn1/3, can bind to Zfhep.
We have also shown that neither the HD nor ZHD of rat Zfhep bind DNA by either EMSA or SAAB. The oligonucleotides used for EMSA were chosen as likely HD binding sites. H2B contains the octamer consensus sequence that is bound by many POU domain proteins. This sequence is found in the human Histone 2b, interleukin-2 and immunoglobulin heavy chain gene promoters [20]. HD proteins in the POU domain family would be expected to bind the H2B oligomer [17]. This oligomer was a good choice for testing the rat Zfhep homeodomain, since it is most similar to POU-type HDs, and since Zfhep is known to regulate both the interleukin-2 and immunoglobulin heavy chain promoters [6, 9, 28]. In addition, AFP1 is an AT rich enhancer from the α-fetoprotein gene promoter that is bound tightly by a homeodomain in ATBF1, a member of the zfh family which includes Zfhep [21]. Hence, H2B and AFP1 oligonucleotides contain potential binding sites for the Zfhep HD, but neither GST-HD nor GST-ZHD exhibited any evidence of complex formation with the H2B or AFP1 oligonucleotides.
We also examined the ability of rat Zfhep GST-HD or GST-ZHD to select a binding site, using the SAAB procedure. While MBP-ZFC and GST-POU selected and bound DNA well (Figure 8), neither Zfhep HD protein could form a DNA complex. Hence, the zinc finger in ZHD does not function to enable the variant HD to bind DNA under these condidions. The observation that the rat Zfhep ZHD is not active in DNA-binding, but is a protein-binding site, is analogous to the HOP homeodomain, which also functions only through protein-interaction [18].
Acknowledgements
We thank Dr. Peter C. van der Vliet for providing clones, and Dr. Ana Cabanillas for helpful discussions. This work was supported by NIH DE 13614 and the University of Louisville School of Medicine.
Abbreviations
- EMSA
electrophoretic mobility shift assay
- GST
glutathione-S-transferase
- HD
homeodomain
- IPTG
isopropyl-β-d-thiogalactopyranoside
- MBP
maltose binding protein
- PMSF
phenylmethyl-sulfonyl flouride
- ZHD
zinc finger homeodomain
References
- 1.Takagi T, Moribe H, Kondoh H, Higashi Y. Development. 1998;125:21–31. doi: 10.1242/dev.125.1.21. [DOI] [PubMed] [Google Scholar]
- 2.Higashi Y, Moribe H, Takagi T, Sekido R, Kawakami K, Kikutani H, Kondoh H. Journal of Experimental Medicine. 1997;185:1467–79. doi: 10.1084/jem.185.8.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Postigo AA, Dean DC. EMBO Journal. 1997;16:3935–43. doi: 10.1093/emboj/16.13.3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yen G, Croci A, Dowling A, Zhang S, Zoeller RT, Darling DS. Brain Res Mol Brain Res. 2001;96:59–67. doi: 10.1016/s0169-328x(01)00267-4. [DOI] [PubMed] [Google Scholar]
- 5.Cabanillas AM, Darling DS. DNA & Cell Biology. 1996;15:643–51. doi: 10.1089/dna.1996.15.643. [DOI] [PubMed] [Google Scholar]
- 6.Genetta T, Ruezinsky D, Kadesch T. Molecular & Cellular Biology. 1994;14:6153–63. doi: 10.1128/mcb.14.9.6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darling DS, Gaur NK, Zhu B. Molecular & Cellular Endocrinology. 1998;139:25–35. doi: 10.1016/s0303-7207(98)00076-8. [DOI] [PubMed] [Google Scholar]
- 8.Funahashi J, Sekido R, Murai K, Kamachi Y, Kondoh H. Development. 1993;119:433–446. doi: 10.1242/dev.119.2.433. [DOI] [PubMed] [Google Scholar]
- 9.Williams TM, Moolten D, Burlein J, Romano J, Bhaerman R, Godillot A, Mellon M, Rauscher FJ, III, Kant JA. Science. 1991;254:1791–1794. doi: 10.1126/science.1840704. [DOI] [PubMed] [Google Scholar]
- 10.Watanabe Y, Kawakami K, Hirayama Y, Nagano K. Journal of Biochemistry. 1993;114:849–855. doi: 10.1093/oxfordjournals.jbchem.a124267. [DOI] [PubMed] [Google Scholar]
- 11.Hovde S, Abate-Shen C, Geiger J. Biochemistry. 2001;40:12013–12021. doi: 10.1021/bi0108148. [DOI] [PubMed] [Google Scholar]
- 12.Wilson DS, Sheng G, Jun S, Desplan C. PNAS. 1996;93:6886–6891. doi: 10.1073/pnas.93.14.6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pabo CO, Peisach E, Grant RA. Annu. Rev. Biochem. 2001;70:313–340. doi: 10.1146/annurev.biochem.70.1.313. [DOI] [PubMed] [Google Scholar]
- 14.Matthews JM, Kowalski K, Liew CK, Sharpe BK, Fox AH, Crossley M, Mackay JP. Eur. J. Biochem. 2000;267:1030–1038. doi: 10.1046/j.1432-1327.2000.01095.x. [DOI] [PubMed] [Google Scholar]
- 15.Dai Y-S, Cserjesi P, Markham BE, Molkentin JD. J. Biol. Chem. 2002;277:24390–24398. doi: 10.1074/jbc.M202490200. [DOI] [PubMed] [Google Scholar]
- 16.Pomerantz J, Sharp P, Pabo C. Science. 1995;267:93–96. doi: 10.1126/science.7809612. [DOI] [PubMed] [Google Scholar]
- 17.Andersen B, Rosenfeld MG. Endocr Rev. 2001;22:2–35. doi: 10.1210/edrv.22.1.0421. [DOI] [PubMed] [Google Scholar]
- 18.Verrijzer CP, van Oosterhout JA, van der Vliet PC. Mol. Cell. Biol. 1992;12:542–551. doi: 10.1128/mcb.12.2.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen F, Kook H, Milewski R, Gitler AD, Lu MM, Li J, Nazarian R, Schnepp R, Jen K, Biben C, Runke G, Mackay JP, Novotny J, Schwartz RJ, Harvey RP, Mullins MC, Epstein JA. Cell. 2002;110:713–723. doi: 10.1016/s0092-8674(02)00932-7. [DOI] [PubMed] [Google Scholar]
- 20.Sive H, Roeder R. 1986;83:6382–6386. doi: 10.1073/pnas.83.17.6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yasuda H, Mizuno A, Tamaoki T, Morinaga T. Mol. Cell. Biol. 1994;14:1395–1401. doi: 10.1128/mcb.14.2.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pollock R. In: Current Protocols in Molecular Biology. Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. John Wiley and Sons, Inc.; New York: 1996. Chapter 12. [Google Scholar]
- 23.Swaffield JC, Johnston SA. In: Current Protocols in Molecular Biology. Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. John Wiley and Sons, Inc.; New York: 1996. Chapter 20. [Google Scholar]
- 24.Sakata N, Hemmi K, Kawaguchi M, Miura Y, Noguchi S, Ma D, Sasahara M, Kato T, Hori M, Tamaoki T. Biochem Biophys Res Commun. 2000;273:686–693. doi: 10.1006/bbrc.2000.2990. [DOI] [PubMed] [Google Scholar]
- 25.Kawaguchi M, Miura Y, Ido A, Morinaga T, Sakata N, Oya T, Hashimoto-Tamaoki T, Sasahara M, Koizumi F, Tamaoki T. Biochim Biophys Acta. 2001;1550:164–174. doi: 10.1016/s0167-4838(01)00284-9. [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez MI, Robins DM. J. Biol. Chem. 2001;276:6420–6428. doi: 10.1074/jbc.M008689200. [DOI] [PubMed] [Google Scholar]
- 27.Kakizawa T, Miyamoto T, Ichikawa K, Kaneko A, Suzuki S, Hara M, Nagasawa T, Takeda T, Mori J-i, Kumagai M, Hashizume K. J. Biol. Chem. 1999;274:19103–19108. doi: 10.1074/jbc.274.27.19103. [DOI] [PubMed] [Google Scholar]
- 28.Yasui DH, Genetta T, Kadesch T, Williams TM, Swain SL, Tsui LV, Huber BT. Journal of Immunology. 1998;160:4433–4440. [PubMed] [Google Scholar]