Abstract
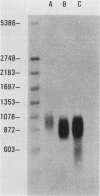
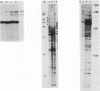
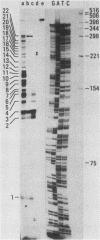
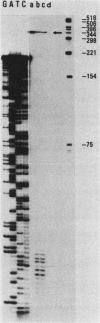
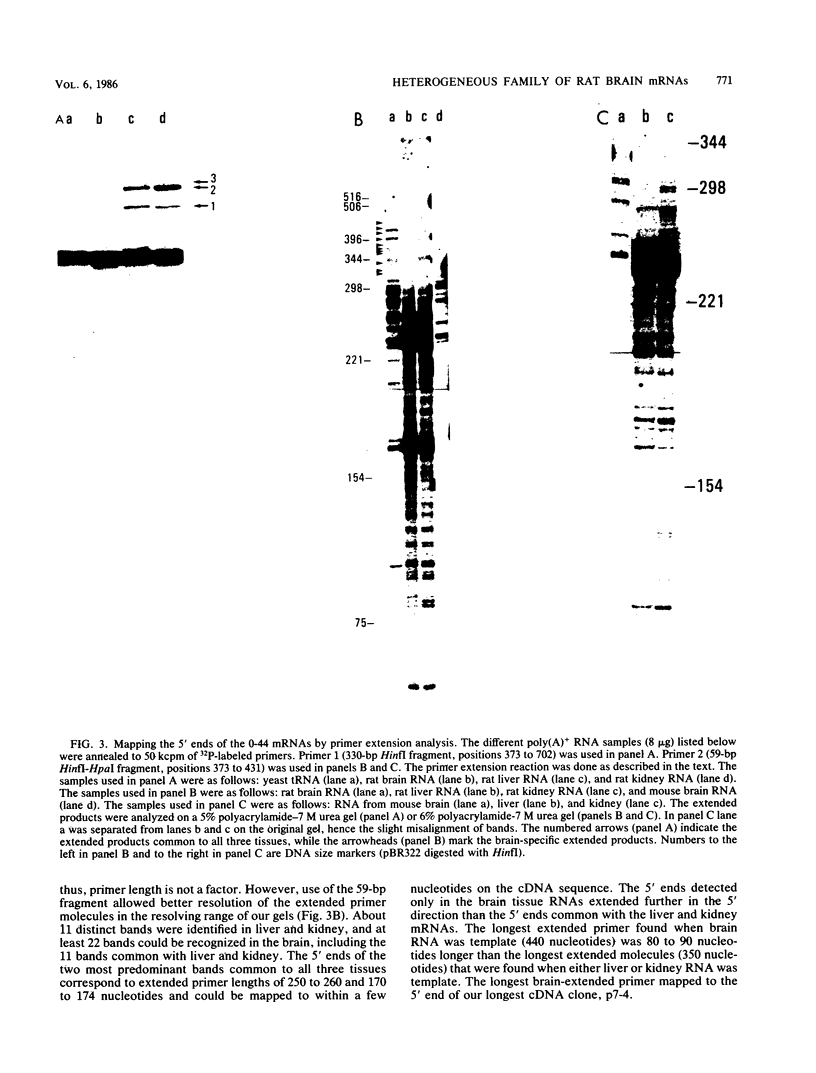
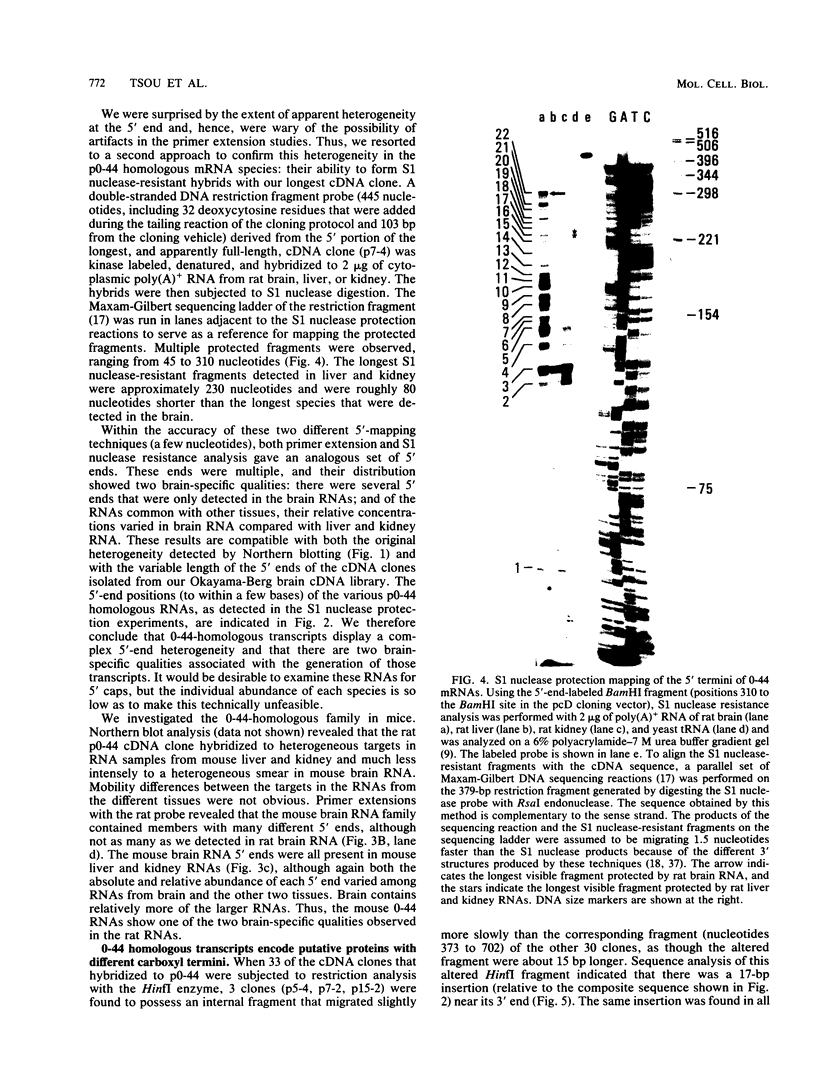
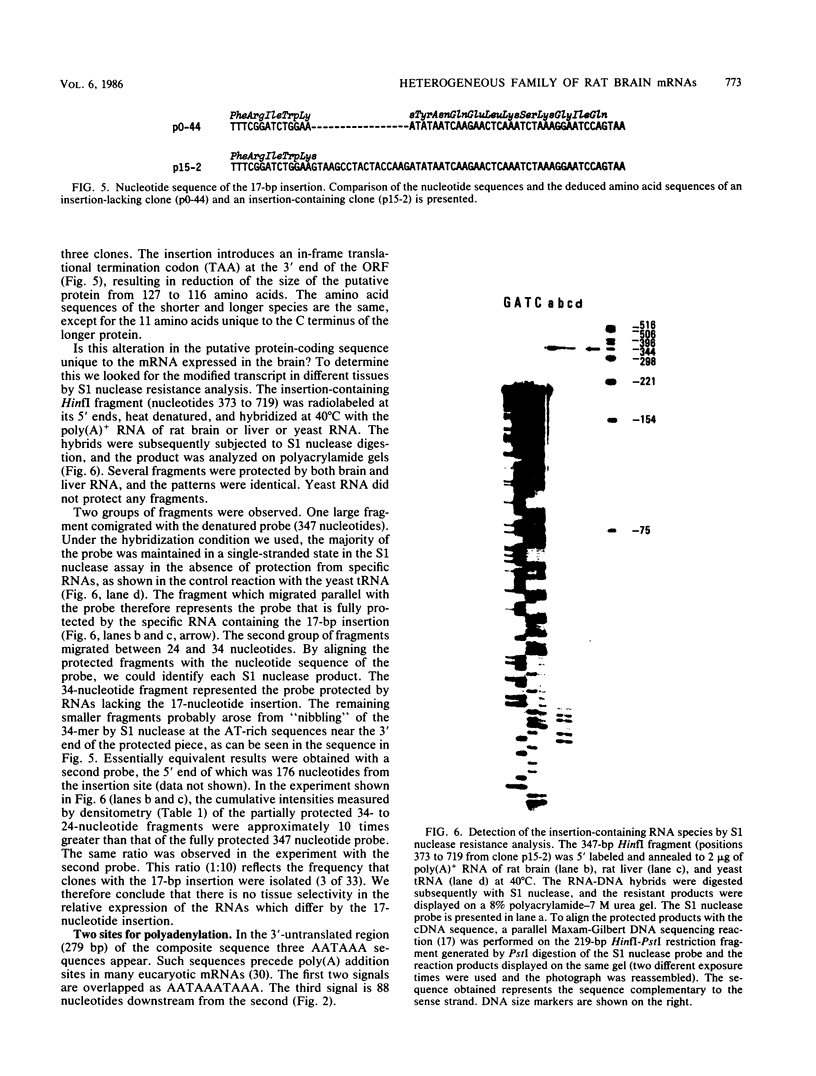
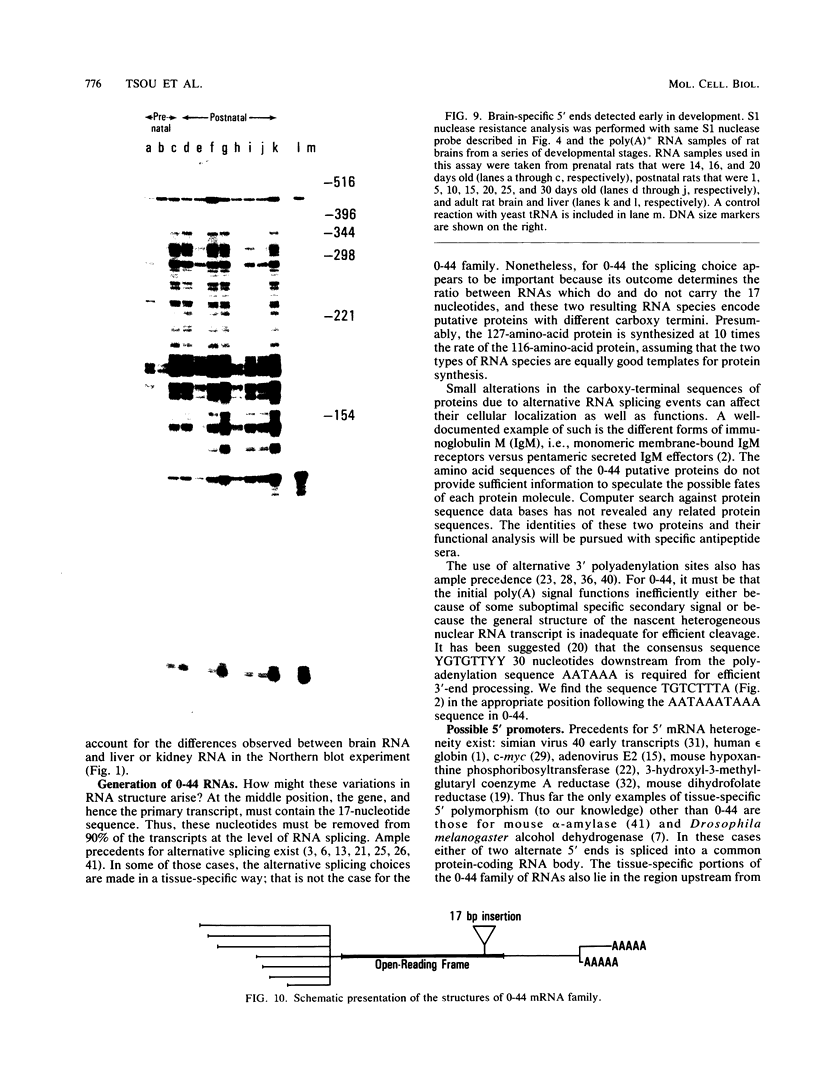
A large heterogeneous family of RNAs derived from a single rat gene contains members that differ from each other at one or more of three positions. Their 5' ends are nested and transcription can begin at 22 or more sites covering 265 nucleotides. Many of the 5' ends are detectable only in brain RNAs, and even 5' ends common with other tissues appear with different absolute and relative abundances in brain RNA. The central portions of the RNAs are of two forms, differing only by the presence or absence of 17 nucleotides; these forms are probably produced by alternative splicing. Polyadenylation occurs at either of two sites. This complicated family of 88 RNAs encodes two novel putative proteins that differ at their C termini.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allan M., Lanyon W. G., Paul J. Multiple origins of transcription in the 4.5 Kb upstream of the epsilon-globin gene. Cell. 1983 Nov;35(1):187–197. doi: 10.1016/0092-8674(83)90221-0. [DOI] [PubMed] [Google Scholar]
- Alt F. W., Bothwell A. L., Knapp M., Siden E., Mather E., Koshland M., Baltimore D. Synthesis of secreted and membrane-bound immunoglobulin mu heavy chains is directed by mRNAs that differ at their 3' ends. Cell. 1980 Jun;20(2):293–301. doi: 10.1016/0092-8674(80)90615-7. [DOI] [PubMed] [Google Scholar]
- Amara S. G., Jonas V., Rosenfeld M. G., Ong E. S., Evans R. M. Alternative RNA processing in calcitonin gene expression generates mRNAs encoding different polypeptide products. Nature. 1982 Jul 15;298(5871):240–244. doi: 10.1038/298240a0. [DOI] [PubMed] [Google Scholar]
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantle J. A., Hahn W. E. Complexity and characterization of polyadenylated RNA in the mouse brain. Cell. 1976 May;8(1):139–150. doi: 10.1016/0092-8674(76)90195-1. [DOI] [PubMed] [Google Scholar]
- Basi G. S., Boardman M., Storti R. V. Alternative splicing of a Drosophila tropomyosin gene generates muscle tropomyosin isoforms with different carboxy-terminal ends. Mol Cell Biol. 1984 Dec;4(12):2828–2836. doi: 10.1128/mcb.4.12.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benyajati C., Spoerel N., Haymerle H., Ashburner M. The messenger RNA for alcohol dehydrogenase in Drosophila melanogaster differs in its 5' end in different developmental stages. Cell. 1983 May;33(1):125–133. doi: 10.1016/0092-8674(83)90341-0. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Brow M. A., Pesin R., Sutcliffe J. G. The tetracycline repressor of pSC101. Mol Biol Evol. 1985 Jan;2(1):1–12. doi: 10.1093/oxfordjournals.molbev.a040334. [DOI] [PubMed] [Google Scholar]
- Casey J., Davidson N. Rates of formation and thermal stabilities of RNA:DNA and DNA:DNA duplexes at high concentrations of formamide. Nucleic Acids Res. 1977;4(5):1539–1552. doi: 10.1093/nar/4.5.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Early P., Rogers J., Davis M., Calame K., Bond M., Wall R., Hood L. Two mRNAs can be produced from a single immunoglobulin mu gene by alternative RNA processing pathways. Cell. 1980 Jun;20(2):313–319. doi: 10.1016/0092-8674(80)90617-0. [DOI] [PubMed] [Google Scholar]
- Giorgi C., Blumberg B. M., Kolakofsky D. Sendai virus contains overlapping genes expressed from a single mRNA. Cell. 1983 Dec;35(3 Pt 2):829–836. doi: 10.1016/0092-8674(83)90115-0. [DOI] [PubMed] [Google Scholar]
- Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984 Jan 25;12(2):857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell D. J., Searcy D. G., Sutcliffe J. G. A restriction enzyme Tha I from the thermophilic mycoplasma Thermoplasma acidophilum. Nucleic Acids Res. 1978 Jun;5(6):1729–1739. doi: 10.1093/nar/5.6.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrogan M., Simonsen C. C., Smouse D. T., Farnham P. J., Schimke R. T. Heterogeneity at the 5' termini of mouse dihydrofolate reductase mRNAs. Evidence for multiple promoter regions. J Biol Chem. 1985 Feb 25;260(4):2307–2314. [PubMed] [Google Scholar]
- McLauchlan J., Gaffney D., Whitton J. L., Clements J. B. The consensus sequence YGTGTTYY located downstream from the AATAAA signal is required for efficient formation of mRNA 3' termini. Nucleic Acids Res. 1985 Feb 25;13(4):1347–1368. doi: 10.1093/nar/13.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medford R. M., Nguyen H. T., Destree A. T., Summers E., Nadal-Ginard B. A novel mechanism of alternative RNA splicing for the developmentally regulated generation of troponin T isoforms from a single gene. Cell. 1984 Sep;38(2):409–421. doi: 10.1016/0092-8674(84)90496-3. [DOI] [PubMed] [Google Scholar]
- Melton D. W., Konecki D. S., Brennand J., Caskey C. T. Structure, expression, and mutation of the hypoxanthine phosphoribosyltransferase gene. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2147–2151. doi: 10.1073/pnas.81.7.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner R. J., Lai C., Nave K. A., Lenoir D., Ogata J., Sutcliffe J. G. Nucleotide sequences of two mRNAs for rat brain myelin proteolipid protein. Cell. 1985 Oct;42(3):931–939. doi: 10.1016/0092-8674(85)90289-2. [DOI] [PubMed] [Google Scholar]
- Milner R. J., Sutcliffe J. G. Gene expression in rat brain. Nucleic Acids Res. 1983 Aug 25;11(16):5497–5520. doi: 10.1093/nar/11.16.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima Y., Fujii-Kuriyama Y., Muramatsu M., Ogata K. Alternative transcription and two modes of splicing results in two myosin light chains from one gene. Nature. 1984 Mar 22;308(5957):333–338. doi: 10.1038/308333a0. [DOI] [PubMed] [Google Scholar]
- Nawa H., Kotani H., Nakanishi S. Tissue-specific generation of two preprotachykinin mRNAs from one gene by alternative RNA splicing. Nature. 1984 Dec 20;312(5996):729–734. doi: 10.1038/312729a0. [DOI] [PubMed] [Google Scholar]
- Okayama H., Berg P. A cDNA cloning vector that permits expression of cDNA inserts in mammalian cells. Mol Cell Biol. 1983 Feb;3(2):280–289. doi: 10.1128/mcb.3.2.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnes J. R., Robinson R. R., Seidman J. G. Multiple mRNA species with distinct 3' termini are transcribed from the beta 2-microglobulin gene. 1983 Mar 31-Apr 6Nature. 302(5907):449–452. doi: 10.1038/302449a0. [DOI] [PubMed] [Google Scholar]
- Prehn J., Mercola M., Calame K. Translocation affects normal c-myc promoter usage and activates fifteen cryptic c-myc transcription starts in plasmacytoma M603. Nucleic Acids Res. 1984 Dec 11;12(23):8987–9007. doi: 10.1093/nar/12.23.8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Reddy V. B., Ghosh P. K., Lebowitz P., Piatak M., Weissman S. M. Simian virus 40 early mRNA's. I. Genomic localization of 3' and 5' termini and two major splices in mRNA from transformed and lytically infected cells. J Virol. 1979 Apr;30(1):279–296. doi: 10.1128/jvi.30.1.279-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds G. A., Basu S. K., Osborne T. F., Chin D. J., Gil G., Brown M. S., Goldstein J. L., Luskey K. L. HMG CoA reductase: a negatively regulated gene with unusual promoter and 5' untranslated regions. Cell. 1984 Aug;38(1):275–285. doi: 10.1016/0092-8674(84)90549-x. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Sargent T. D., Wu J. R., Sala-Trepat J. M., Wallace R. B., Reyes A. A., Bonner J. The rat serum albumin gene: analysis of cloned sequences. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3256–3260. doi: 10.1073/pnas.76.7.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler U., Tosi M., Pittet A. C., Fabiani L., Wellauer P. K. Tissue-specific expression of mouse alpha-amylase genes. J Mol Biol. 1980 Sep 5;142(1):93–116. doi: 10.1016/0022-2836(80)90208-9. [DOI] [PubMed] [Google Scholar]
- Setzer D. R., McGrogan M., Schimke R. T. Nucleotide sequence surrounding multiple polyadenylation sites in the mouse dihydrofolate reductase gene. J Biol Chem. 1982 May 10;257(9):5143–5147. [PubMed] [Google Scholar]
- Sollner-Webb B., Reeder R. H. The nucleotide sequence of the initiation and termination sites for ribosomal RNA transcription in X. laevis. Cell. 1979 Oct;18(2):485–499. doi: 10.1016/0092-8674(79)90066-7. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosi M., Young R. A., Hagenbüchle O., Schibler U. Multiple polyadenylation sites in a mouse alpha-amylase gene. Nucleic Acids Res. 1981 May 25;9(10):2313–2323. doi: 10.1093/nar/9.10.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. A., Hagenbüchle O., Schibler U. A single mouse alpha-amylase gene specifies two different tissue-specific mRNAs. Cell. 1981 Feb;23(2):451–458. doi: 10.1016/0092-8674(81)90140-9. [DOI] [PubMed] [Google Scholar]