Abstract
Classical opioids have been historically used for the treatment of pain and are among the most widely used drugs for both acute severe pain and long-term pain. Morphine and endogenous mu-opioid peptides exert their pharmacological actions mainly through the mu-opioid receptor (MOR). However, the expression of opioid receptor (OR) proteins is controlled by extensive transcriptional and post-transcriptional processing. Previously, the 5′-untranslated region (UTR) of the mouse MOR was found to be important for post-transcriptional regulation of the MOR gene in neuronal cells. To identify proteins binding to the 5′-UTR as potential regulators of the mouse MOR gene, affinity column chromatography using 5′-UTR-specific RNA oligonucleotides was performed using neuroblastoma NS20Y cells. Chromatography was followed by two-dimensional gel electrophoresis and MALDI-TOF mass spectrometry. We identified two heterogeneous ribonucleoproteins (hnRNPs) that bound to RNA sequences of interest: hnRNP H1 and hnRNP F. Binding of these proteins to the RNA region was M4-region sequence-specific as confirmed by Western-blot analysis and RNA supershift assay. Furthermore, a cotransfection study showed that the presence of hnRNP H1 and F resulted in repressed expression of the mouse MOR. Our data suggest that hnRNP H1 and F can function as repressors of MOR translation dependent on the M4 (−75 to −71 bp upstream of ATG) sequences. We demonstrate for the first time a role of hnRNPs as post-transcriptional repressors in MOR gene regulation.
Keywords: Mu-opioid receptor (MOR), Post-transcriptional regulation, RNA binding protein
Introduction
Opioids remain the most widely prescribed analgesics and among the most widely abused categories of recreational drugs. The specific biological targets of opioids, opioid receptors (ORs), were first detected by use of radioactive ligand binding assays in the early 1970s [1–3]. The opioid receptors, classified into three major types (μ, δ, and κ) have been characterized by molecular cloning and in numerous pharmacological reports [4, 5]. The use of opioids has been associated with many side-effects, such as nausea, vomiting, constipation, respiratory depression, sedation, pruritus, tolerance, and dependence, although brief, short-term prescription of an opioid rarely leads to addiction or abuse of the drug [6]. Among the three major types of opioid receptors, several studies have suggested that the mu-opioid receptor (MOR) plays a key role in mediating the major clinical effects of morphine, as well as the development of tolerance and physical dependence with chronic administration [7].
Gene expression starts with transcription and is followed by multiple post-transcriptional processes that carry out the splicing, capping, polyadenylation, and export of each mRNA. In the mouse MOR gene, over 28 splice variants have been isolated [8, 9]. The diversity and complexity created by alternative splicing of the MOR gene may provide important insights of understanding the diverse responses to the various mu opioids. However, the MOR protein is expressed mainly in the central nervous system, with receptors varying in densities in different regions [10]. To achieve its unique spatial expression pattern, the expression of MOR must be tightly regulated by multiple mechanisms, including transcriptional and post-transcriptional events. Our laboratory and others have demonstrated that MOR promoter activity is regulated by many enhancer elements and their related transcriptional factors and post-transcriptional control [5, 7, 11–21]. While interest in post-transcriptional regulation has increased recently with explosive discoveries of large numbers of non-coding RNAs such as microRNAs, post-transcriptional processes depend largely on the functions of RNA-binding proteins as well [22]. Post-transcriptional control of gene expression essentially relies on specific interactions between cis-acting elements mainly localized in the untranslated region (UTR) of the transcript and the trans-acting factors [RNA-binding proteins (RBPs) and non-coding regulatory RNAs] that bind to these sequences [23, 24].
The heterogeneous nuclear ribonucleoproteins (hnRNPs) constitute a family of more than 20 proteins designated with letter from A to U. Among them, the hnRNPs F/H include hnRNPs F, H1 (H), H2 (H′), and H3 (2H9). This is unlike most hnRNPs, which can bind both RNA and SSDNA independently of sequence [25]. Therefore, determination of RBP specificities is a critical step in the elucidation and analysis of mechanisms involved in co- and post-transcriptional gene regulation [26]. Several methods have been developed to identify and characterize RBPs and the RNAs with which they interact, including ultraviolet (UV) cross-linking of RNA–protein complexes in vivo [27, 28], cross-linking and immunoprecipitation (CLIP) [29], systemic evolution of ligands by exponential enrichment (SELEX) and electrophoretic mobility shift assay (EMSA) [30], RIP assay [31], and RNA-based affinity purification [32].
To identify proteins potentially associated with the MOR 5′-UTR RNA (M4 RNA), we used affinity column chromatography containing a specific competitor, two-dimensional gel electrophoresis, and mass spectrometry to purify and identify the factors that interact specifically with the M4 RNA from the mouse neuronal cells. The results showed that the original M4 sequence from the mouse MOR 5′-UTR region specifically interacts with two RNA-binding proteins, heterogeneous ribonucleoprotein (hnRNP) H1 and F, to regulate the mouse MOR gene expression. Thus, these proteins serve as post-transcriptional regulators of the mouse MOR.
Materials and methods
Plasmid construction and toeprinting assay
The luciferase fusion constructs [uAUG (+) and uAUG (−)] and toeprinting assay used here have been described previously [15, 19]. The multi-G sequence-mutated constructs (M1, M2, M3, and M4) were prepared via site-directed mutagenesis using the uAUG (+) and uAUG (−) constructs as the template. The following oligonucleotides were used for each C-sequence mutation: M1: 5′-TTCTAAGGTGGcAGGGGGCTA-3′ (forward) and 5′-TAGCCCCCTgCCACCTTAGAA-3′ (reverse); M2: 5′-TTCTAAGGTGGGAGcaGGCTA-3′ (forward) and 5′-TAGCCtgCTCCCACCTTAGAA-3′ (reverse); M3: 5′-GTTCCACTAGcGCTTGTCCTT-3′ (forward) and 5′-AAGGACAAGCgCTAGTGGAAC-3′ (reverse); M4, 3: 5′-AAGAGGCTGcGGCGCCTGGAA-3′ (forward) and 5′-TTCCAGGCGCCgCAGCCTCTT-3′ (reverse). The hnRNP H1 and F plasmid DNAs (pCMV-Sports6-H1 and pCMV-Sports6-F, respectively) were purchased from Thermo Scientific. DNA sequences of all constructs were confirmed by sequencing.
Cell culture, DNA transfection, and reporter gene assay
Mouse neuroblastoma NS20Y cells were grown in Dulbecco’s minimum essential medium supplemented with 10% heat-inactivated fetal bovine serum at 37°C in a humidified atmosphere of 5% CO2. Transfection and reporter gene assays were carried out as described previously [14, 15].
Quantification of LUC and LacZ transcripts by real-time PCR and reverse transcription (RT)-PCR
The above assays were performed as described previously [15].
Cytosolic extract preparation
Extract preparation was carried out as described previously [33]. Briefly, cells were lysed in cytoplasmic extract buffer (CEB) (10 mM HEPES–KOH, pH 7.6, 100 mM KCl, 2.5 mM MgCl2, 1 mM dithiothreitol [DTT], 0.25% NP-40, EDTA-free Complete Mini protease inhibitor,1 mM NaF, and 1 mM Na3VO4). The lysates were centrifuged at 16,200 × g for 15 min at 4°C, and the supernatants were used for RNA affinity pulldown.
RNA-affinity purification
RNA-affinity purification was performed as previously described in [18], with slight modifications (Fig. 3b). The following procedure is based on the interaction between biotin and streptavidin. RNA oligonucleotides were synthesized and purified using HPLC. In a sterile tube, 500 μl of 0.5× SSC solution was added to 500 pmol of each 3′-terminal-biotinylated RNA. Meanwhile, 500 pmol of streptavidin-paramagnetic particles (Promega) were resuspended by gently flicking the bottom of the tube until they were completely dispersed, which were then captured by placing the tube in a magnetic stand. The supernatant was carefully removed. The magnetic particles were washed three times with 0.5× SSC and resuspended in 100 μl of 0.5× SSC.
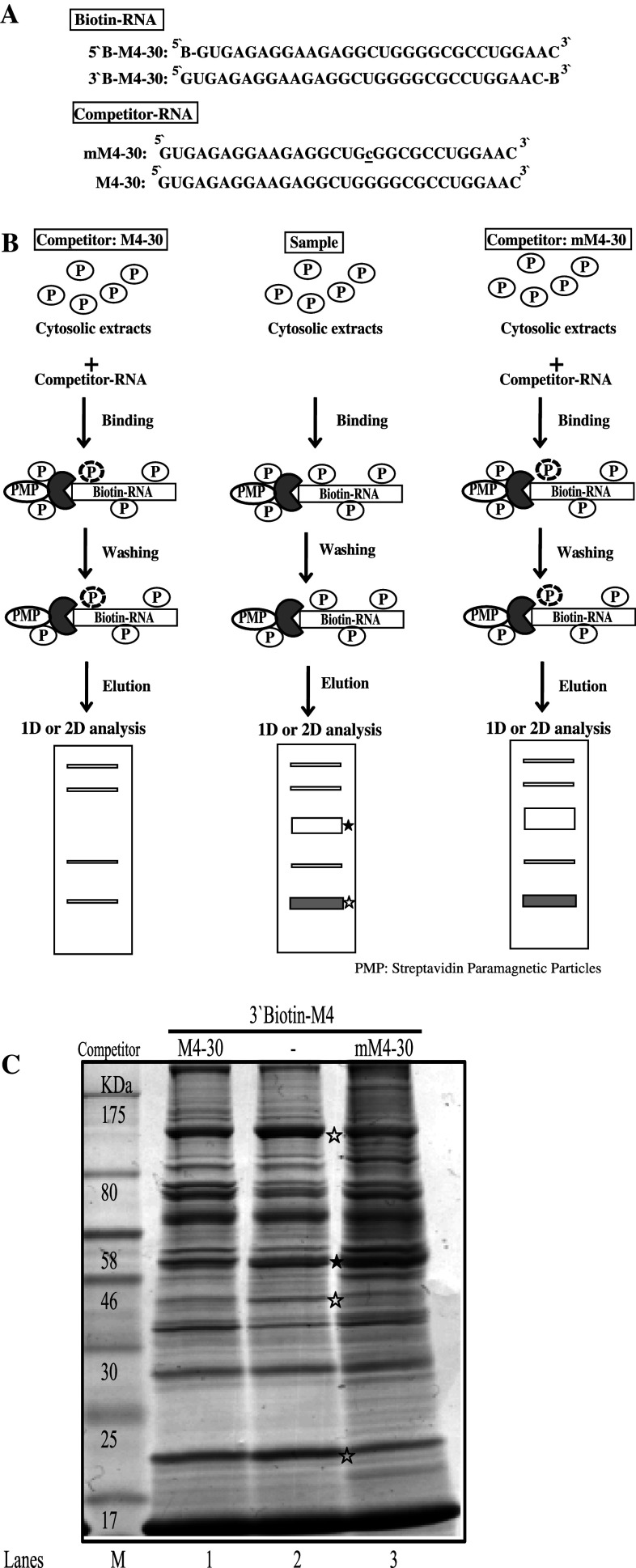
Fig. 3.
Schematic representation of the procedure for one-step purification of RNA binding proteins. a RNA oligonucleotides containing wild M4 and mutated M4 (mM4) nucleotide sequences with or without biotin label. b Outline of the modified one-step purification of RBPs using an affinity column. RNA oligonucleotides biotinylated on the 5′- or 3′-terminus were used as affinity particles. Cytosolic proteins were added to the affinity particles, incubated, and washed. Proteins bound to the particles were released by heating in SDS sample buffer. Competitor experiments to eliminate nonspecific binding (empty stars) and to identify specific binding (filled star) were performed by preincubating the cytosolic proteins with a two-fold excess of nonbiotinylated RNAs (M4-30 or mM4-30) as competitors prior to affinity binding. c Coomassie-stained gel of RNA binding proteins purified from NS20Y cytosolic extracts with competitor. The competitor RNAs M4-30 and mM4-30 are shown in panel a
Then, 500 pmol of biotinylated RNA and 500 pmol of the streptavidin-paramagnetic particles were combined and incubated for 15 min at room temperature. Samples were mixed by gentle inversion every 2 min. The magnetic beads were captured using a magnetic stand. The particles were washed three times with 300 μl of CEB buffer 1 (10 mM HEPES–KOH, 2.5 mM MgCl2, 100 mM KCl, 1 mM DTT, 0.25% NP-40, 1 mM NaF, 1 mM Na3VO4, and 1× protease inhibitors), pH 7.6. One mg of cytosolic proteins was added to the affinity particles and incubated for 1 h at 4°C. The particles were washed three times with CEB buffer 1 and CEB buffer 2 (10 mM HEPES–KOH, 2.5 mM MgCl2, 200 mM KCl, 1 mM DTT, 0.25% NP-40, 1 mM NaF, 1 mM Na3VO4, and 1× protease inhibitors), pH 7.6. Proteins bound to the particles were released by incubation in 50 μl of 1× SDS sample buffer for 10 min at 95°C in a heating block.
In order to eliminate cytosolic proteins that might bind non-specifically, control experiments were performed as follows: 1,000 pmol of non-biotinylated RNA (2× competitor) were mixed with 1 mg of cytosolic proteins for 15 min on ice. The cytosolic extracts containing the 2× competitor were added to the affinity particles and incubated for 1 h at 4°C. The remainder of the procedure was performed as above. The resultant protein solutions with and without competitor were electrophoresed on a 4–20% gradient gel (Invitrogen) and stained with Coomassie blue (Simply Blue safe-Stain, Invitrogen).
Immunoblot analysis
Purified proteins were resolved by SDS-PAGE using a 4–20% gradient polyacrylamide gel (Invitrogen). Gels were electroblotted onto polyvinylidene difluoride membranes (Amersham Bioscience) in transfer buffer (48 mM Tris–HCl, 39 mM glycine, and 20% methanol). Membranes were blocked in a blocking solution of 5% dry milk and 0.1% Tween 20 in Tris-buffered saline overnight at 4°C. Immunoblotting with anti-hnRNP H1 (Novus Biologicals) and anti-hnRNP F antibodies (AVIVA System Biology) was performed according to the manufacturer’s instructions (Amersham Biosciences). Signals were detected using a Storm 860 PhosphorImager system (Amersham Biosciences).
Two-dimensional gel electrophoresis (2-DE) in gel tryptic digestion and MALDI-TOF mass spectrometric analysis of RNA binding proteins
Purified proteins were resolved by 2-DE. 2-DE was performed as described by Görg et al., with minor modifications [34]. IPG strips were used according to the manufacturer’s instruction. Isoelectric focusing (IEF) as the first dimension was carried out on Protean IEF cell (Bio-Rad). Briefly, purified samples were mixed with an aliquot (185 μl) of rehydration solution (7 M urea, 2 M thiourea, 4% CHAPS [w/v], 60 mM DTT, a trace of bromophenol blue, and 0.5% IPG buffer [v/v]; Amersham Pharmacia Biotech) and then applied to IPG strips. After rehydration (12 h), IEF was carried out with the following voltage–time program: 500 V for 1 h; 1,000 V for 1 h: 8,000 V over a gradient up to 50,000 V h. Prior to SDS-PAGE, the IPG strips were incubated for 15 min with a solution of Tris–HCl buffer (50 mM, pH 8.8), urea (6 M), glycerol (30%, v/v), SDS (2%, w/v), and DTT (2%, w/v). The strips were then equilibrated for another 15 min in the same buffer that contained iodoacetamide (2.5%, w/v) instead of DTT. SDS-PAGE as the second dimension was carried out at 90 V constant for 3 h. Molecular masses were determined by running standard protein markers (DualColor PrecisionPlus Protein™ standard; Bio-Rad). Triplicate 2-DE gels (controls and sample) were run under the same conditions. Three gels were subjected to colloidal Coomassie staining (Simply Blue, Invitrogen) to visualize the protein spots, and gel slices of interest (differential bands) were subjected to in-gel tryptic digestion as described previously [35]. Tryptic peptides were extracted with 5% acetic acid followed by 5% acetic acid and 50% acetonitrile. Samples were dissolved in 5% acetic acid and desalted using ZipTip™ C18 reverse-phase desalting Eppendorf tips (Millipore). The peptides were eluted with 2% acetonitrile containing 0.1% TFA in a volume of 20 μl. Samples were analyzed using a MALDI-TOF mass spectrometer (Applied Biosystems).
Tandem mass spectra were extracted and charge state deconvoluted by BioWorks version 2.0. Deisotoping was not performed. All MS/MS samples were analyzed using Sequest (Thermo Fisher Scientific; version 27, rev. 13) and X! Tandem (The GPM, thegpm.org; version 2007.01.01.1). X! Tandem was set up to search the rs_mus_v200810_cRAP database (35180 entries) assuming the digestion enzyme trypsin. Sequest was set up to search the rs_mus_v200810_cRAP database (35180 entries) also assuming trypsin. Sequest and X! Tandem were searched with a fragment ion mass tolerance of 1.00 Da and a parent ion tolerance of 0.80 Da. Iodoacetamide derivative of cysteine was specified in Sequest and X! Tandem as a fixed modification. Oxidations of methionine and iodoacetic acid derivative of cysteine were specified in Sequest and X! Tandem as variable modifications.
Criteria for protein identification
Scaffold (version Scaffold_2_06_01, Proteome Software Inc., Portland, OR) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability as specified by the Peptide Prophet algorithm [36]. Protein identifications were accepted if they could be established at greater than 99.0% probability and contained at least two identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm [37]. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
In vitro translation and autoradiography
In vitro translation was performed as described [17]. Reactions were carried out with pCMV-Sports6-hnRNPH1 and -F in a mixture containing [35S]-methionine (Amersham Biosciences) or non-labeled methionine using the TnT Quick Coupled Transcription/Translation System (Promega). The labeled proteins were then analyzed via SDS-PAGE on a 10% gel, and their sizes were compared with the predicted sizes.
RNA electromobility shift analysis (REMSA)
REMSAs were performed as described [38], with modifications. We used oligonucleotides corresponding to the wild-type M4 RNA sequence [M4-30: 5′-GUGAGAGGAAGAGGCUGGGGCGCCUGGAAC-3′] and mutant sequences [mM4-30: 5′-GUGAGAGGAAGAGGCUGcGGCGCCUGGAAC-3′]. Briefly, the [35S]-methionine-labeled protein probes were incubated with non-labeled RNA oligonucleotides (Fig. 6c) or the non-labeled in vitro translated proteins were incubated with [γ–32P]-labeled RNA oligonucleotide (Fig. 6d) in a final volume of 20 μl of REMSA buffer [10% glycerol, 1 mM MgCl2, 0.5 mM EDTA, 10 mM Tris–HCl (pH 7.8), 0.1 mg/ml BSA, and 0.5 mg/ml yeast tRNA], at room temperatures for 30 min. Following the formation of ribonucleoprotein (RNP) complexes, the REMSA reactions were electrophoresed on a 4% polyacrylamide gel in 0.5× TBE (45 mM Tris–borate and 1 mM EDTA) at 4°C and visualized by autoradiography.
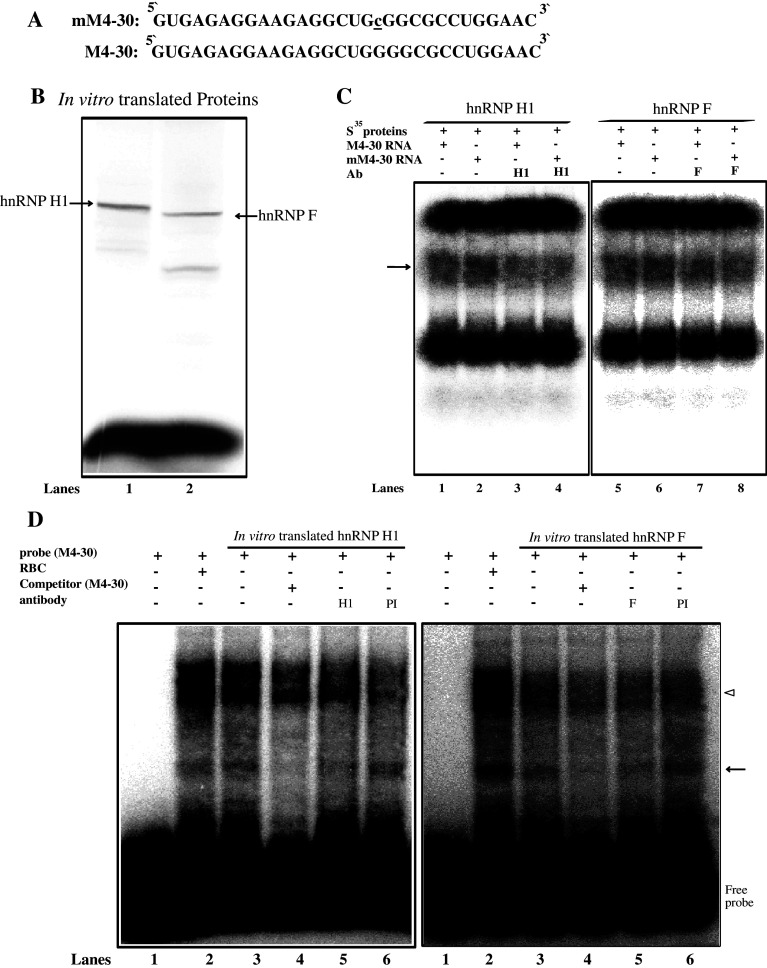
Fig. 6.
Interaction of hnRNP H1 and F with M4 RNA derived from the mouse MOR 5′-UTR. a The mouse MOR M4-30 and mutated M4-30 RNA sequences. b HnRNP H1 and F proteins radiolabeled in vitro with [S35]-methionine (lanes 1 and 2, respectively). c REMSAs performed with both RNA sequences and in vitro-labeled proteins. Lanes 1 and 5 M4-30 RNA nucleotide without antibody; lanes 2 and 6 mM4-30 RNA nucleotide without antibody; lanes 3 and 4 anti-hnRNP H1; lanes 7 and 8 anti-hnRNP F. The protein–RNA complexes are indicated by the arrow. d REMSAs were performed using 32P-labeled M4-30 as a probe with non-labeled in vitro translated proteins. Lane 1 probe alone, lane 2 reticulocyte (RBC) without antibody, lane 3 no added antibody, lane 4 self-competitor without antibody, lane 5 anti-hnRNP H1 or F, lane 6 preimmune serum (PI). The protein–RNA complexes are indicated by the arrow; the empty arrow head indicates non-specific binding between RRL and RNA
For competition assays, the cell mixture was preincubated with 200 pmol (100X) of an unlabeled RNA for 5 min prior to the addition of the radiolabeled RNA. For antibody supershift assays, the reaction mixture was preincubated with 1 μg of the indicated antibody on ice for 10 min prior to the addition of the radiolabeled RNA.
Heterogeneous expression of hnRNP H1 and F
For heterogeneous expression, the hnRNP H1 (pCMV-Sports6-H1) and F (pCMV-Sports6-F) plasmids were transfected into NS20Y cells using Effectene transfection reagent (Qiagen). To examine the regulation of MOR by these proteins, proteins were isolated from NS20Y cells transfected with hnRNP H1 or hnRNP F. To correct for the differences in transfection efficiency, a one-fifth molar ratio of a pCH110 plasmid (Amersham) containing the β-galactosidase gene under the SV40 promoter was included in each transfection for normalization. The MOR (luciferase) and galactosidase (internal control) activities of each lysate were determined according to the manufacturer’s instructions (Promega and Tropix, respectively). The relative MOR mRNA (LUC) level was reported as the ratio of LUC mRNA/LacZ mRNA by real-time PCR.
Results
Characterization of trans (cis)-acting element sites
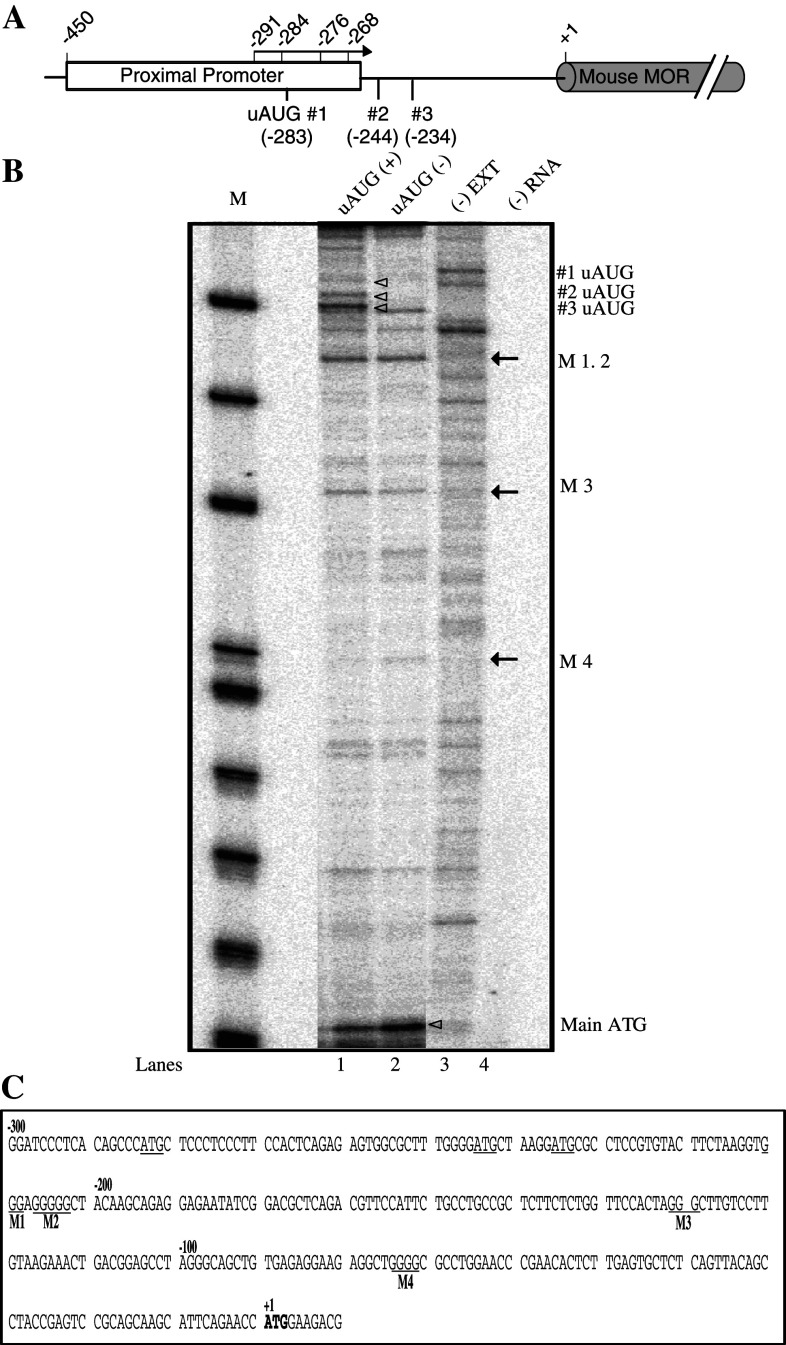
Toeprinting can be used to map the interactions of translational components such as ribosomes with mRNA, and should prove useful for analyzing the association of other factors with mRNA and for analyzing mRNA structures [39–41]. Previous work has shown that mouse MOR expression is inhibited at the translational level by the presence of upstream open reading frames (uORFs). The binding region of trans (cis)-acting elements to the 5′-untranslated region (UTR) of MOR (Fig. 1a) was further analyzed here via toeprinting, and these uORF initiation sites were confirmed by our analysis [15]. However, our toeprinting data (Fig. 1b) also revealed unexpected bands (arrows in lanes 1 and 2) in addition to the uAUG and main initiation sites (empty arrowheads). If present in the negative control lanes, the bands may have represented either authentic truncated mRNAs or false priming bands [39], however, they were not detected in the negative control lanes (lanes 3 and 4). After confirming their presence, we calculated the band sites (Fig. 1c) by size markers and sequencing markers (not shown). The unexpected bands (M1–M4) consisted of only three to five G sequences, but were not known uAUG initiation sites or uORF termination sites.
Fig. 1.
Schematic representation of the mouse MOR 5′-UTR and toeprinting analysis. a The mouse MOR 5′-UTR contains the proximal promoter and three uAUGs. The mouse MOR translation initiation site is indicated by +1. Four transcription initiation sites (−291, −284, −276, and −268 bp from translational initiation site) are indicated by the arrow. b Toeprint analyses of initiation of uAUGs and unidentified bands. Synthetic RNA transcripts (100 ng) were used to program translation mixtures derived from RRL (Promega). Mouse MOR-LUC transcripts containing all three uORFs (i.e., wild-type; uAUG (+); or mutated at all three uAUGs [uAUG (−)]) were incubated at 30°C for 15 min in micrococcal nuclease-treated RRL. Radiolabeled mToe primer was used for primer extension analyses and for sequencing of the uAUG (+) templates. The sequence of the template can be directly deduced by 5′–3′ sequence reads from top to bottom. M Dephosphorylated ΦX174 HinfI Markers (Promega); arrow position of unidentified bands; open arrowhead toeprint of each uAUG and main AUG. c Unidentified bands were calculated by molecular weight marker and sequencing ladder. The unknown bands consisted of several G sequences (M1–M4)
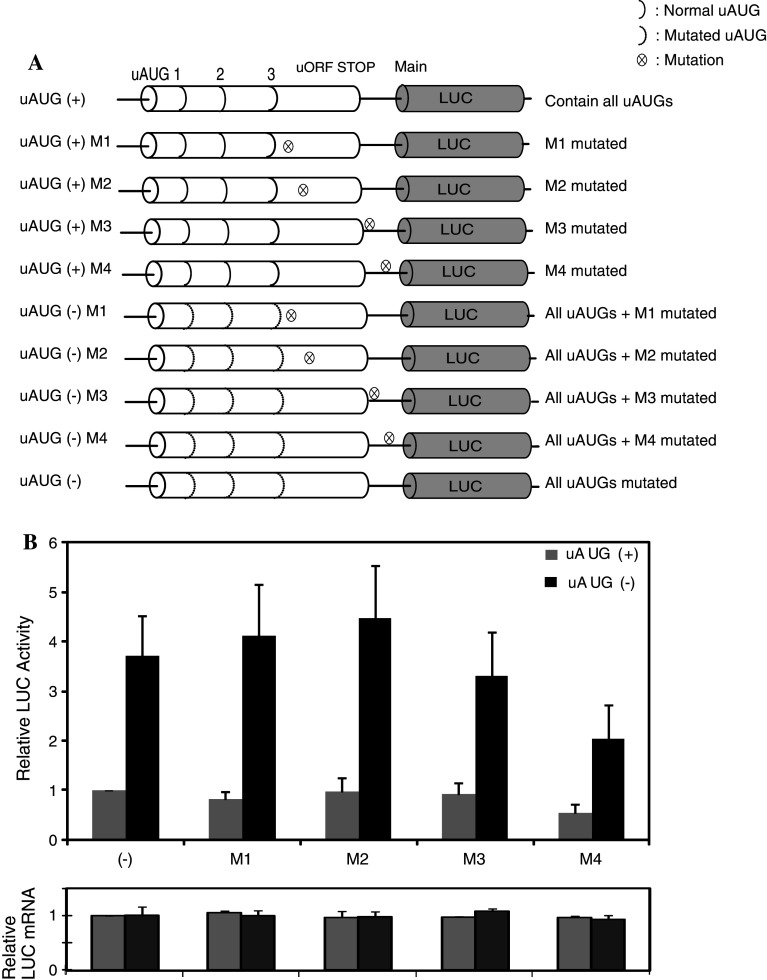
Next, we elucidated the role of the sequences in MOR gene regulation. First, we mutated the G sequences to C (see Materials and methods section and Fig. 2a). Second, we confirmed the sites’ role in both transcription and translation by real-time PCR and luciferase assay (Fig. 2b). The levels of the transcripts were very similar among all constructs, indicating that the point mutations did not alter transcription levels. In contrast, LUC activity was differentially affected by the mutations of the M sequences. Particularly, mutation within the M4 [with both uAUG (+) and uAUG (−) constructs] caused a two-fold decrease in LUC activity. Mutations of the M2 had no significant effect, but M1 and M3 mutations resulted in a slight decrease [uAUG (+) M1 and uAUG (−) M3, respectively]. However, both uAUG (+) and uAUG (−) related data displayed the same negative effect on the 5′-UTR of MOR mRNA fused reporter (MOR/LUC) gene translational level (Fig. 2b, upper panel), not transcriptional level (Fig. 2b, lower panel). Therefore, the M4 sequences were not related to the uORF mechanisms, but had a negative effect on MOR/LUC gene regulation.
Fig. 2.
Translation of the mouse MOR gene is controlled by M4 mutation. a Schematic representation of reporter constructs with wild-type and mutated mouse MOR 5′-UTRs with M mutations (M1–M4). Vertical dotted lines represent ATGs converted to ACGs by point mutations; circled X’s indicate each M sequence mutation as described in Materials and methods. b Transient transfection of each mutant construct in NS20Y cells. After transfection, cells were trypsinized and half were used for luciferase and β-galactosidase activity assays, while half were used for RNA extraction and transcript quantification. Relative LUC activity and mRNA levels were determined as the ratio of LUC/β-gal and LUC/LacZ as described in Materials and methods. The error bars indicate the standard errors of triplicate LUC assays
Isolation and identification of RNA binding factors that interact with M4 sequences in the mouse MOR 5′-UTR region using a new affinity column-containing competitor
In general, many RBPs remain yet to be characterized. Several methods have been developed to identify and characterize the RBPs and the RNAs with which they interact. The messenger RNP (mRNP) complexes were initially isolated by several methods [42]. Our previous data showed that the M4 region, mutated itself, could regulate MOR/LUC gene expression activity at the post-transcriptional level. We therefore speculated that regulation of MOR expression by M4 could involve the interaction between the 5′-UTR of MOR mRNA and binding factors. To confirm this inference, we prepared cytosolic extracts from mouse NS20Y cells and tested the ability of protein constituents of the extracts to bind the M4 region in the 5′-UTR of MOR mRNA by affinity column assay (Fig. 3).
For these studies, we used biotin-labeled-RNA oligonucleotides (Fig. 3a). We used two kinds of competitors (non-biotinylated M4-30 and mM4-30; Fig. 3b) to identify the specific RBPs to M4 sequences (Fig. 3c). Using this protocol, we were able to purify and identify proteins from NS20Y cytosolic extracts that bound to the entire 30 bp-region of RNA (M4-30) (empty star) or specifically bound to the GGGG sequences of the M4 RNA sequence (filled star). The bands indicating binding to the 30-bp region (empty star) were decreased after treatment with the M4-30 competitor, whereas treatment with the mM4-30 competitor increased the intensity of only the GGGG sequence-specific bands.
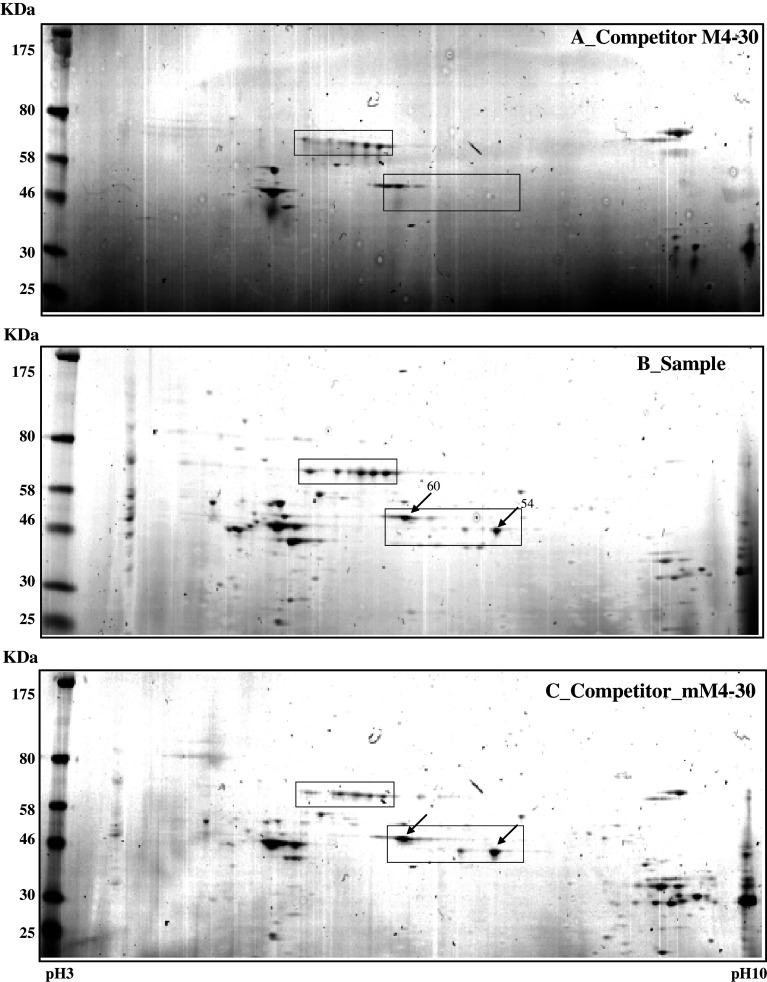
After we confirmed the M4-specific binding proteins by competitor assay, we used two-dimensional electrophoresis (2-DE) to further characterize the major proteins binding to the M4 RNA sequence (Fig. 4). On average, over 50 protein spots were detected in pH 3–pH 10 2-DE images of competitor-treated controls and samples. Most spots were distributed within a pH range of 4–8 with molecular masses of 30–80 kDa. Comparisons of the 2-DE images identified two protein spots (54 and 60 kDa) present in sample gels. These spots were analyzed using MALDI-TOF mass spectrometry and bioinformatics. The two spots were identified as heterogeneous nuclear ribonucleoprotein (hnRNP) H1 and hnRNP F (Table 1).
Fig. 4.
Simply Blue safe-stained 2-DE images of RBPs purified using an affinity column. Purified samples were separated on pH 3–10 IPG strips followed by separation by 12% SDS-PAGE. Competitor assay is shown in a and c; sample assay shown in b. Molecular weight markers are indicated on the left, and PI values are indicated the bottom. Indicated arrow spots were subjected to analysis by MALDI-TOF mass spectrometry and bioinformatics. Detailed information on each spot is listed in Table 1
Table 1.
Analysis by MALDI-TOF MS of the tryptic peptide profiles of the protein spots
| Spot numbera | Accession number | Protein nameb | MW (kDa) (obs)c | % coverage (amino acids) | Unique peptides |
|---|---|---|---|---|---|
| 54 | gi19527048 | Heterogeneous nuclear ribonucleoprotein F | 46 | 9 (39/415) | 3 |
| 60 | gi10946928 | Heterogeneous nuclear ribonucleoprotein H1 | 49 | 13 (58/449) | 5 |
hnRNP H1 and F bind specifically to the M4 RNA sequence derived from mouse MOR 5′-UTR region
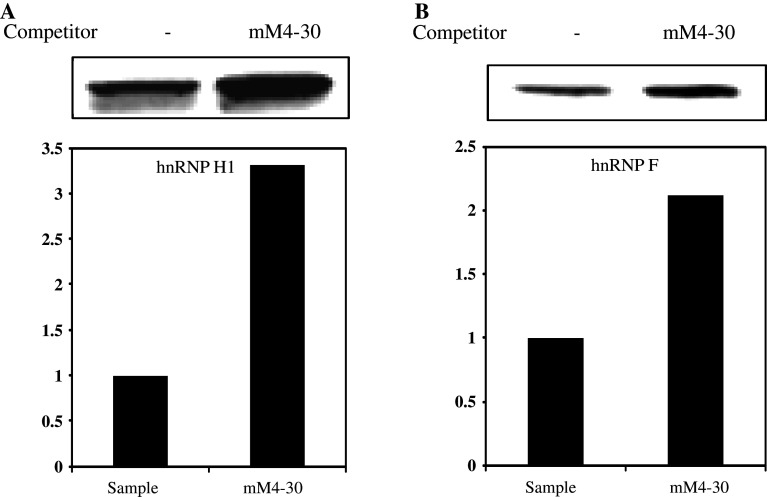
We next used independent methods (Western blotting and REMSA) to validate the interactions of hnRNP H1 and F with the mouse MOR M4 sequences. Western-blot analyses were carried out using purified proteins from NS20Y cells. The results showed that the purified proteins contained higher amounts of hnRNP H1 (Fig. 5a) and hnRNP F (Fig. 5b) compared to the non-competitive sample, indicating that the mutated M4 (mM4-30) competitor could increase the binding activity of both hnRNP H1 and F. Furthermore, REMSAs were performed using two alternative labeling methods and are shown in Fig. 6. One method utilized the in vitro translated [35S]-methionine-labeled proteins (Fig. 6b) with non-labeled RNA oligonucleotides (Fig. 6a) derived from the mouse MOR 5′-UTR region. The RNA–protein complex was formed using both M4-30 and mM4-30 RNA oligonucleotides (Fig. 6c, arrow), while significant formation of a major complex was not observed in control lanes with the antibodies (Fig. 6c, lanes 3, 4 and 7, 8). The alternative EMSA experiments were carried out using 32P-labeled M4-30 RNA probe and unlabeled in vitro translated proteins (Fig. 6d). Two proteins were able to bind the target M4-30 probe (Fig. 6d, compare lane 3 and lane 1 in each panel). The M4-30 probe also bound to reticulocytes, which are known express hnRNP H1 and F (Fig. 6d, lane 2 of each panel). A 100-fold molar excess of unlabeled M4-30 RNA oligonucleotide (Fig. 6d, lane 4 of each panel) completely inhibited the complex formation. The specificity of the RNA–protein interaction was verified using anti-hnRNP H1 and F antibodies. The hnRNP H1- and F-RNA complexes were supershifted by addition of their own antibodies (Fig. 6d, lane 5 of each panel), but the pre-immune serum (PI) was ineffective in the super-shift assay (Fig. 6d, lane 6 of each panel). Our REMSA data could not show the super-shift band, because the broad ranges of non-specific band areas (empty arrowhead) were the same as in the control reaction (Fig. 6d, lane 2 of each panel). However, both the Western blotting and REMSA assays confirmed that the M4 mutated competitor (mM4-30) could increase the activity of protein binding compared to the normal circumstances.
Fig. 5.
Identification of hnRNP H1 and F proteins as M4 binding proteins using mM4-30 competitor. Western-blot analysis of purified RBPs performed with anti-hnRNP H1 (a) and anti-hnRNP F (b) antibodies. HnRNP H1 (a) and hnRNP F (b) proteins level were measured in NS20Y cells by Western blotting after purification. Intensities of each signal were analyzed by ImageQuant 5.2 software
The hnRNP H1 and F negatively regulated the mouse MOR gene expression by M4 sequence in the 5′-UTR of MOR
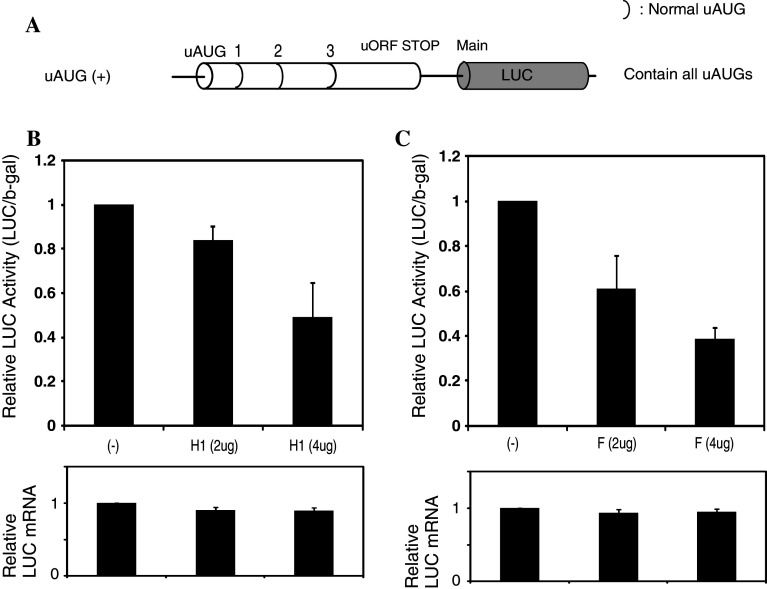
Our western and REMSA data showed that the hnRNP H1 and F proteins could bind to the normal M4 RNA oligonucleotides derived from the 5′-UTR of mouse MOR. To examine the functional role of hnRNP H1 and F in normal mouse MOR regulation, we used an uAUG (+) reporter construct containing the mouse 5′-UTR region (minus promoter) fused in-frame to a luciferase reporter construct (Fig. 7a). Luciferase assay and real-time PCR were performed with both proteins and RNAs from NS20Y cells transfected with varying amounts (0–4 μg) of hnRNP H1 and F protein expression vectors with normal mouse uAUG (+) reporter construct. As shown in Fig. 7b and c, the protein levels of MOR were decreased by co-transfection with hnRNP H1 and F compared to uAUG (+) construct alone. Furthermore, the hnRNP H1 and F constructs all down-regulated MOR/LUC gene expression in a dose-dependent manner, but these effects are not shown at the transcriptional level. These differences may differentially regulate their cellular location and their ability to express the protein, but there is no evidence to date to support this possibility [9].
Fig. 7.
Mouse MOR expression levels in hnRNP H1- and F-transfected NS20Y cells. a Schematic representation of the mouse MOR 5′-UTR containing the reporter construct shown previously [15]. b Graphic representation of relative luciferase activity determined by luciferase assay (Promega) of the constructs shown in a with 2–4 μg of RBP expression constructs. Relative LUC activity and mRNA levels were determined as the ratio of LUC/β-gal and LUC/LacZ as described in the Materials and methods section. The error bars indicate the standard errors of triplicate LUC assays
Discussion
Protein levels in cells are regulated not only by the rate of transcription but also by rates of subsequent events, including RNA processing, nuclear RNA export, translation and RNA decay [24]. These post-transcriptional events are controlled by RNA-binding proteins. Many studies have suggested that RBPs indirectly couple transcriptional and subsequent post-transcriptional steps by interacting with their target transcript [43]. Although progressive coupling of these processes is widely accepted, both coupling and coordination are important in determining how, when, and where to translate functionally related subpopulations of mRNAs [44]. RBPs are universal in living cells, and, in eukaryotes, are estimated to number approximately 650 in yeast and over 2,500 in mammals [45, 46]. Although some RBPs are thought to bind RNA with little or no sequence specificity, many and possibly most RBPs are specific for binding to distinct subpopulations of RNA [22]. The heterogeneous nuclear ribonucleoproteins (hnRNPs) comprise a family of primarily nuclear RBPs that bind to nascent transcripts produced by RNA polymerase II, and which do not stably associate with other RNA–protein complexes [25]. The complexity and diversity associated with the hnRNPs render them multifunctional, involved not only in processing heterogeneous nuclear RNAs (hnRNAs) into mature mRNAs, but also as trans-factors in regulating gene expression [47].
Moreover, toeprinting method offers an easy way to study in vitro how mRNA conformational changes alter ribosome binding at the initiation site. These changes can be induced either by environmental cues (temperature and ion concentration), or by the binding of metabolites, regulatory proteins, or trans-acting RNAs [41, 48]. Our study analyzed the binding region of trans (cis)-acting elements to mouse MOR 5′-UTR by toeprinting assay, and investigated the interaction of trans (cis)-acting elements with the mouse MOR 5′-UTR RNA and their importance on MOR post-transcriptional regulation.
To date, existing RNA tags can be classified into two categories: tags developed based on RNA–protein interactions found in nature, and tags developed from apatamers evolved in vitro to possess affinity for specific ligands such as streptavidin, streptomycin, or tobramycin [32]. In the present study, we identified new candidate RBPs (hnRNP H1 and F) associated with M4 RNA sequences from the mouse MOR 5′-UTR regions using streptavidin paramagnetic particles (PMP). The hnRNPs-mediated negative regulation is dependent on the presence of a G-rich region in the 5′-UTR of mouse MOR mRNA, specifically, M4 sequences. We used typical methods to isolate mRNA bound to RBPs (RNA affinity purification assays with or without competition). Western and REMSAs further revealed the characteristics of the sequence-specific interaction between these hnRNPs and the M4 RNA sequence of the mouse MOR 5′-UTR. Previous reports showed that hnRNP H and F are two hnRNPs with high structural homology, although they are expressed from distinct genes [49]. Both hnRNP H and F have three RNA recognition motifs with binding preferences for GGGA [50] or DGGGD (where D represents a U, G, or A nucleotide) motifs [51]. However, our results showed that both hnRNPs could bind to GGGG (M4) sequences in the mouse MOR 5′-UTR.
Our functional analyses suggested that hnRNP H1 and F regulated the mouse MOR containing mutated and wild-type M4 RNA sequences. These results were confirmed by studies in NS20Y cells, a mouse neuroblastoma cell line endogeneously expressing MOR. Increasing the exogenous expression of hnRNP H1 or hnRNP F in these cells down-regulated MOR/LUC expression in a dose-dependent manner. Taken together, the results indicate that hnRNP H1 and F act as repressors of mouse MOR expression in neuronal cells via a mechanism dependent on the M4 RNA sequence of the mouse MOR. Studies such as ours offer new perspectives on the role of the hnRNP family in MOR gene regulation and a better understanding of the molecular mechanisms underlying MOR expression. Future experiments would be extended to the well-known region- and cell-specific expression and functions of the MOR in MOR-expressing cell lines or primary cells isolated from different regions of the mouse brain.
Acknowledgements
This work was supported by the National Institutes of Health (Grants DA000564, DA001583, DA011806, K05-DA070554, DA011190, DA013926), and by the A&F Stark Fund of the Minnesota Medical Foundation.
Conflict of interest
The authors declare no conflicts of interest.
Footnotes
K. Y. Song and H. S. Choi contributed equally to this work.
References
- 1.Pert CB, Snyder SH. Properties of opiate-receptor binding in rat brain. Proc Natl Acad Sci USA. 1973;70:2243–2247. doi: 10.1073/pnas.70.8.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simon EJ, Hiller JM, Edelman I. Stereospecific binding of the potent narcotic analgesic [3H]Etorphine to rat-brain homogenate. Proc Natl Acad Sci USA. 1973;70:1947–1949. doi: 10.1073/pnas.70.7.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Terenius L. Characteristics of the “receptor” for narcotic analgesics in synaptic plasma membrane fraction from rat brain. Acta Pharmacol Toxicol. 1973;33:377–384. doi: 10.1111/j.1600-0773.1973.tb01539.x. [DOI] [PubMed] [Google Scholar]
- 4.Min BH, Augustin LB, Felsheim RF, Fuchs JA, Loh HH. Genomic structure analysis of promoter sequence of a mouse μ opioid receptor gene. Proc Natl Acad Sci USA. 1994;91:9081–9085. doi: 10.1073/pnas.91.19.9081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei LN, Loh HH. Regulation of opioid receptor expression. Curr Opin Pharmacol. 2002;2:69–75. doi: 10.1016/S1471-4892(01)00123-0. [DOI] [PubMed] [Google Scholar]
- 6.Porter J, Jick H. Addiction rare in patients treated with narcotics. N Engl J Med. 1980;302:123. doi: 10.1056/nejm198001103020221. [DOI] [PubMed] [Google Scholar]
- 7.Law PY, Loh HH, Wei LN. Insights into the receptor transcription and signaling: implications in opioid tolerance and dependence. Neuropharmacology. 2004;47:300–311. doi: 10.1016/j.neuropharm.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 8.Pan YX. Diversity and complexity of the mu opioid receptor gene: alternative pre-mRNA splicing and promoters. DNA Cell Biol. 2005;24(11):736–750. doi: 10.1089/dna.2005.24.736. [DOI] [PubMed] [Google Scholar]
- 9.Xu J, Xu M, Rossi GC, Pasternak GW, Pan YX. Identification and characterization of seven new exon 11-associated splice variants of the rat μ opioid receptor gene, OPRM1. Mol Pain. 2011;7(1):9–22. doi: 10.1186/1744-8069-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mansour A, Fox CA, Akil H, Watson SJ. Opioid receptor mRNA expression in the rat CNS: anatomical and functional implications. Trends Neurosci. 1995;18:22–29. doi: 10.1016/0166-2236(95)93946-U. [DOI] [PubMed] [Google Scholar]
- 11.Wei LN, Law PY, Loh HH. Post-transcriptional regulation of opioid receptors in the nervous system. Front Biosci. 2004;9:1665–1679. doi: 10.2741/1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim CS, Hwang CK, Choi HS, Song KY, Law PY, Wei LN, Loh HH. Neuron-restrictive silencer factor (NRSF) functions as a repressor in neuronal cells to regulate the mu opioid receptor gene. J Biol Chem. 2004;279:46464–46473. doi: 10.1074/jbc.M403633200. [DOI] [PubMed] [Google Scholar]
- 13.Kim CS, Choi HS, Hwang CK, Song KY, Lee BK, Law PY, Wei LN, Loh HH. Evidence of the neuron-restrictive silencer factor (NRSF) interaction with Sp3 and its synergic repression to the mu opioid receptor (MOR) gene. Nucleic Acid Res. 2006;34:6392–6403. doi: 10.1093/nar/gkl724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi HS, Kim CS, Hwang CK, Song KY, Law PY, Wei LN, Loh HH. Novel function of the poly(C)-binding protein alpha CP3 as a transcriptional repressor of the mu opioid receptor gene. FASEB J. 2007;21:3673–3963. doi: 10.1096/fj.07-8561com. [DOI] [PubMed] [Google Scholar]
- 15.Song KY, Hwang CK, Kim CS, Choi HS, Law PY, Wei LN, Loh HH. Translational repression of mouse mu opioid receptor expression via leaky scanning. Nucleic Acid Res. 2007;35:1501–1513. doi: 10.1093/nar/gkm034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hwang CK, Song KY, Kim CS, Choi HS, Guo XH, Law PY, Wei LN, Loh HH. Evidence of endogenous mu opioid receptor regulation by epigenetic control of the promoters. Mol Cell Biol. 2007;27:4720–4736. doi: 10.1128/MCB.00073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi HS, Hwang CK, Kim CS, Song KY, Law PY, Loh HH, Wei LN. Transcriptional regulation of mouse mu opioid receptor gene in neuronal cells by poly(ADP-ribose) polymerase-1. J Cell Mol Med. 2008;12:2319–2333. doi: 10.1111/j.1582-4934.2008.00259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi HS, Song KY, Hwang CK, Kim CS, Law PY, Wei LN, Loh HH. A proteomics approach for identification of single strand DNA-binding proteins involved in transcriptional regulation of mouse mu opioid receptor gene. Mol Cell Proteomics. 2008;7:1517–1529. doi: 10.1074/mcp.M800052-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song KY, Kim CS, Hwang CK, Choi HS, Law PY, Wei LN, Loh HH. uAUG-mediated translational initiations are responsible for human mu opioid receptor gene expression. J Cell Mol Med. 2010;14(5):1113–1124. doi: 10.1111/j.1582-4934.2009.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hwang CK, Song SY, Kim CS, Choi HS, Guo XH, Law PY, Wei LN, Loh HH. Epigenetic programming of mu-opioid receptor gene in mouse brain is regulated by MeCP2 and Brg1 chromatin remodelling factor. J Cell Mol Med. 2009;13(9):3591–3615. doi: 10.1111/j.1582-4934.2008.00535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song KY, Choi HS, Hwang CK, Kim CS, Law PY, Wei LN, Loh HH. Differential use of an in-frame translation initiation codon regulates human mu opioid receptor (OPRM1) Cell Mol Life Sci. 2009;66(17):2933–2942. doi: 10.1007/s00018-009-0082-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keene JD. Minireview: global regulation and dynamics of ribonucleic Acid. Endocrinology. 2010;154(4):1391–1397. doi: 10.1210/en.2009-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delestiennem N, Wauquier C, Soin R, Dierick JF, Gueydan C, Kruys V. The splicing factor ASF/SF2 is associated with TIA-1-related/TIA-1-containing ribonucleoproteic complexes and contributes to post-transcriptional repression of gene expression. FEBS J. 2010;277(11):2496–2514. doi: 10.1111/j.1742-4658.2010.07664.x. [DOI] [PubMed] [Google Scholar]
- 24.Mittal N, Roy N, Babu MM, Janga SC. Dissecting the expression dynamics of RNA-binding proteins in posttranscriptional regulatory networks. Proc Natl Acad Sci U S A. 2009;106(48):20300–20305. doi: 10.1073/pnas.0906940106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dreyfuss G, Matunis MJ, Piñol-Roma S, Burd CG. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 26.Ray D, Kazan H, Chan ET, Peña Castillo L, Chaudhry S, Talukder S, Blencowe BJ, Morris Q, Hughes TR. Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nat Biotechnol. 2009;27(7):667–670. doi: 10.1038/nbt.1550. [DOI] [PubMed] [Google Scholar]
- 27.Piñol-Roma S, Adam SA, Choi YD, Dreyfuss G. Ultraviolet-induced cross-linking of RNA to proteins in vivo. Methods Enzymol. 1989;180:418. doi: 10.1016/0076-6879(89)80115-6. [DOI] [PubMed] [Google Scholar]
- 28.Dreyfuss G, Adam SA, Choi YD. Physical change in cytoplasmic messenger ribonucleoproteins in cells treated with inhibitors of mRNA transcription. Mol Cell Biol. 1984;4(3):415–423. doi: 10.1128/mcb.4.3.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ule J, Jensen K, Mele A, Darnell RB. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods. 2005;37(4):376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 30.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249(4968):505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 31.Niranjanakumari S, Lasda E, Brazas R, Garcia-Blanco MA. Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods. 2002;26(2):182–190. doi: 10.1016/S1046-2023(02)00021-X. [DOI] [PubMed] [Google Scholar]
- 32.Hogg J, Collins K. RNA-based affinity purification reveals 7SK RNPs with distinct composition and regulation. RNA. 2007;13(6):868–880. doi: 10.1261/rna.565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Majumder M, Yaman I, Gaccioli F, Zeenko VV, Wang C, Caprara MG, Venema RC, Komar AA, Snider MD, Hatzoglou M. The hnRNA-binding proteins hnRNP L and PTB are required for efficient translation of the Cat-1 arginine/lysine transporter mRNA during amino acid starvation. Mol Cell Biol. 2009;29(10):2899–2912. doi: 10.1128/MCB.01774-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gorg A, Obermaier C, Boguth G, Harder A, Scheibe B, Wildgruber R, Weiss W. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis. 2000;21(6):1037–1053. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1037::AID-ELPS1037>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 35.Patterson SD, Aebersold R. Mass spectrometric approaches for the identification of gel-separated proteins. Electrophoresis. 1995;16(10):1791–1814. doi: 10.1002/elps.11501601299. [DOI] [PubMed] [Google Scholar]
- 36.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74(20):5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 37.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75(17):4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 38.Gerbasi VR, Link AJ. The myotonic dystrophy type 2 protein ZNF9 is part of an ITAF complex that promotes cap-independent translation. Mol Cell Proteomics. 2007;6(6):1049–1058. doi: 10.1074/mcp.M600384-MCP200. [DOI] [PubMed] [Google Scholar]
- 39.Sachs M, Wang Z, Gaba A, Fang P, Belk J, Ganesan R, Amrani N, Jacobson A. Toeprint analysis of the positioning of translation apparatus components at initiation and termination codons of fungal mRNAs. Methods. 2002;26:105–114. doi: 10.1016/S1046-2023(02)00013-0. [DOI] [PubMed] [Google Scholar]
- 40.Dhar D, Venkataramana M, Ponnuswamy A, Das S. Role of polypyrimidine tract binding protein in mediating internal initiation of translation of interferon regulatory factor 2 RNA. PLoS One. 2009;4(9):e7049. doi: 10.1371/journal.pone.0007049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fechter P, Chevalier C, Yusupova G, Yusupov M, Romby P, Marzi S. Ribosomal initiation complexes probed by toeprinting and effect of trans-acting translational regulators in bacteria. Methods Mol Biol. 2009;540:247–263. doi: 10.1007/978-1-59745-558-9_18. [DOI] [PubMed] [Google Scholar]
- 42.Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582(14):1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416(6880):499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 44.Keene J, Tenenbaum SA. Eukaryotic mRNPs may represent posttranscriptional operons. Mol cell. 2002;9(6):1161–1167. doi: 10.1016/S1097-2765(02)00559-2. [DOI] [PubMed] [Google Scholar]
- 45.Keene JD. Ribonucleoprotein infrastructure regulating the flow of genetic information between the genome and the proteome. Proc Natl Acad Sci USA. 2001;98(13):7018–7024. doi: 10.1073/pnas.111145598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309(5740):1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 47.Chaudhury A, Chander P, Howe PH. Heterogeneous nuclear ribonucleoproteins (hnRNPs) in cellular processes: focus on hnRNP E1’s multifunctional regulatory roles. RNA. 2010;16(8):1449–1462. doi: 10.1261/rna.2254110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simonetti A, Marzi S, Myasnikov AG, Fabbretti A, Yusupov M, Gualerzi CO, Klaholz BP. Structure of the 30S translation initiation complex. Nature. 2008;455:416–420. doi: 10.1038/nature07192. [DOI] [PubMed] [Google Scholar]
- 49.Matunis M, Xing J, Dreyfuss G. The hnRNP F protein: unique primary structure, nucleic acid-binding properties, and subcellular localization. Nucleic Acids Res. 1994;22(6):1059–1067. doi: 10.1093/nar/22.6.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Caputi M, Zahler AM. Determination of the RNA binding specificity of the heterogeneous nuclear ribonucleoprotein (hnRNP) H/H’/F/2H9 family. J Biol Chem. 2001;276(47):43850–43859. doi: 10.1074/jbc.M102861200. [DOI] [PubMed] [Google Scholar]
- 51.Schaub MCLS, Caputi M. Members of the heterogeneous nuclear ribonucleoprotein H family activate splicing of an HIV-1 splicing substrate by promoting formation of ATP-dependent spliceosomal complexes. J Biol Chem 2007. 2007;282(18):13617–13626. doi: 10.1074/jbc.M700774200. [DOI] [PubMed] [Google Scholar]