Introduction
Discovery of new adverse drug events (ADEs) in the post-approval period is an important goal of the health system. Data mining methods that can transform data into meaningful knowledge to inform patient safety have proven to be essential. New opportunities have emerged to harness data sources that have not been used within the traditional framework. This article provides an overview of recent methodological innovations and data sources used in support of ADE discovery and analysis.
Keywords: Pharmacovigilance, Adverse Drug Events, Data Mining
1. Background
Pharmacovigilance (PhV), also referred to as drug safety surveillance, is defined as: “the science and activities relating to the detection, assessment, understanding and prevention of adverse effects or any other drug problem”1. PhV starts at the pre-approval stage, where information about adverse drug events (ADEs) is collected during phase I-III clinical trials without causal relationship to the investigational product or concomitant therapies, and continues in post-approval stage throughout a drug’s life on the market. During the initial post-approval stage PhV may continue through phase IV clinical trials, often mandated by regulatory agencies to obtain additional safety data on a product during routine use. While clinical trials are used to evaluate safety issues, they are limited in the number, duration, characteristics of patients exposed, and the type of data collected. As a result, the complete safety profile associated with a new drug cannot be fully established through clinical trials.
Post-approval ADEs are a major global health concern accounting for more than 2 million injuries, hospitalizations, and deaths each year in the US alone2, 3, and associated costs estimated at $75 billion annually4. Hence, the timely and accurate detection of ADEs in the post-approval period is now an urgent goal of the public health system. Computational methods at the intersection of statistics, computer science, medicine, epidemiology, chemoinformatics, and biology that can translate data into meaningful knowledge to benefit patient safety have proven to be a critical component in PhV. These methods have commonly been referred to as data mining algorithms (DMAs).
Historically PhV relied on a clinical review process of case reports collected at designated organizations. Strained by the vast quantities and complexity of data that needed to be examined, DMAs were originally designed to aid this process and allow evaluators to peruse large volumes of data and focus their attention on issues that may be more important to public health. Since then however, the role, quality, and capabilities of DMAs have dramatically expanded in order to address new challenges, leverage new information sources, and overall improve drug safety surveillance. In what follows, DMAs will be used to describe automated high-throughput methods that are used to uncover hidden relationships of potential clinical significance to drug safety.
DMAs can be classified along several axes depending on the data source to which they are applied, and the scientific function they are designed to perform. The main PhV data sources in current use are spontaneous reporting systems. Research focus is now being shifted towards the use of large healthcare databases such as electronic health records and administrative claims. Other sources that have recently been considered include: the biomedical literature, chemical and biological information sources, and patient-generated data in health related web forums. The main class of DMAs represent methods designed to generate measures of statistical association for large sets of drug-outcome pairs, which can be used to prioritize and identify risk signals that warrant further attention5, 6. Newer approaches have been designed to facilitate identification of higher-order or multivariate associations that represent more complex safety phenomena such as drug-drug interactions, syndromic events, or class effects. A large class of methods has been designed to address the issues of confounding. Other approaches have been designed to abstract the data in meaningful ways to uncover interesting patterns, such as clusters or networks of ADEs that may convey clinically important information, while a new wave of methods have been designed to leverage non-traditional data sources or link information from multiple data sources.
In recognition of their importance, research into the application of DMAs to PhV has steadily grown in the past decade (Figure 1-DMA related publication trends), and much progress have been made. The aim of this article is to provide an overview of recent DMA methodological innovations and data sources used in support of PhV. Extending the theme of several related reviews6-8, we aim to cover a broader range of methods and data sources. In addition, our discussion is restricted to works published in the past 5 years. We do not exhaustively list all relevant work. Nor is it our goal to critically examine the works, but rather present an informational synopsis of basic concepts, contributions, and major findings. We begin our discussion with a description of the data and information sources covered in this article, highlighting their strengths and limitations. Methods discussion is organized according to the data source axis: spontaneous reports, healthcare data, and other data sources. We also provide a brief overview of traditional approaches (disproportionality analysis), as a foundation for discussion of other approaches.
Figure 1.
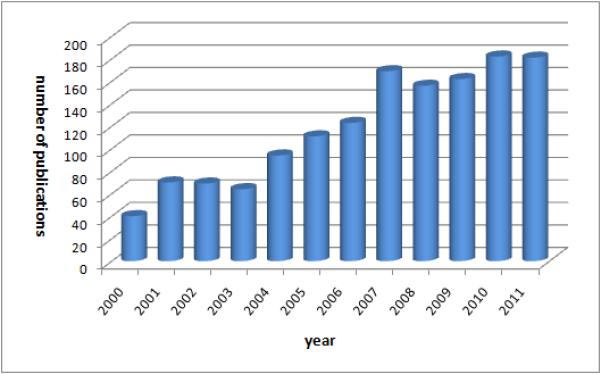
PhV DMA research evolution described by volume of publications per year indexed in PubMed. 2011 volume is effectively larger due to delayed indexing.
2. Data and information sources used in support of pharmacovigilance
Drug safety surveillance has predominantly relied on spontaneous reporting systems (SRS), which are passive systems comprised of reports of suspected ADEs collected from healthcare professionals, consumers, and pharmaceutical companies, and maintained largely by regulatory and health agencies. Among the prominent SRS are the US Food and Drug Administration (FDA) Adverse Event reporting System (AERS) and the VigiBase maintained by World Health Organization (WHO). Although the structure and content of each SRS may differ, most are based on voluntary reporting (except for pharmaceutical companies that are required to report to regulators suspected ADEs once they come to their attention), and typically capture suspected and concomitant drugs, indications, suspected events, and limited demographic information in a structured format directly amenable to data mining. The FDA uses a data mining engine to compute signal scores (statistical reporting associations) for all of the millions of drug-event combinations in AERS, which offers a “hypothesis-free” view of the safety characteristics in the underlying data. It should be stressed however, that these signals by themselves do not establish a causal ADE relationship, but are rather considered initial warnings that require further assessment using other sources of support. Typically, after this initial signal generation step, an intertwined process of signal strengthening and signal confirmation follows, where drug safety evaluators look for signs such as a temporal relationship, coherence with published case reports, biological and clinical plausibility, similarity with other drugs, supporting data from clinical trials, or by conducting epidemiological studies in several large health care databases to establish causality9, 10.
SRS are pre-focused on drug-adverse event relationships, the collection and processing is centralized, they communicate genuine health concerns, cover large populations, are accessible for analysis, and since their inception have supported regulatory decisions for a long list of marketed drugs11. Notwithstanding, SRS suffer from a range of limitations including: over-reporting where drugs with known and publicized ADEs are more likely to be reported than other drugs, misattributed drug-event combinations, missing and incomplete data, duplicated reporting, and unspecified causal links5, 12.
Recent drug safety events, such as the Rofecoxib (Vioxx) case – a widely used anti-inflammatory drug estimated to have caused 88,000 episodes of myocardial infarction (MI)13, have highlighted the need to identify new data sources and improved analytic methods to create a more effective PhV system9, 14-16. US Congress has recently mandated the FDA to establish an active surveillance system17. Subsequently, several large scale research initiatives, such as the Sentinel Initiative 9, 18 and the Observational Medical Outcomes Partnership (OMOP)15, 19, were established in the US. A similar research called the EU-ADR project was initiated in Europe by the European commission20. These new developments rely on the expanded secondary use of electronic healthcare data such as electronic health records and administrative claims that typically contain: time-stamped interventions, procedures, diagnoses, medications, medical narratives, and billing codes. Unlike spontaneous reports, electronic healthcare data are representative of routine clinical care recorded over long periods of time. As such, they contain a more complete record of the patient’s medical history, treatments, conditions, and potential risk factors. They are also not restricted to patients experiencing ADEs. Consequently, electronic healthcare data offer several advantages that may be used to complement SRS, especially confirmatory analysis and the potential for active surveillance. Several retrospective studies have demonstrated that the Vioxx case could have been signaled earlier using this type of data21-24. However, the secondary use of healthcare data presents other challenges. The data often require complex preprocessing to support analysis. The data are not oriented to capture adverse events, which are typically not identified per se, but as diagnoses (usually based on billing codes). There are logistical issues in storing, accessing, and sharing data across healthcare providers, that are compounded by legal and privacy issues concerning access to patient data9, 15. There are varying data capture and documentation styles, and varying standards for data encoding9, 15. There is also a need for automated methods that can extract relevant information from free-text clinical narratives25, and methods that can address the pervasiveness of confounding inherent in observational studies10, 26, 27.
In the recent past, researchers have begun to focus on data and information sources that have not traditionally been used for PhV. Each source offers unique prospects that may be leveraged to complement or augment existing approaches and we discuss several of these in turn.
The public availability of chemical and biological knowledge bases such as DrugBank28, which contains information on both chemical structure and drug targets, is opening new opportunities to bridge the gap between the molecular and clinical domains and further the study of ADEs29, 30. By leveraging this type of knowledge, e.g., protein binding sites, biological pathways of drug action and metabolism, linking chemical substructures to specific toxicities, and chemical similarity, the molecular determinants of ADE can better be understood. Moreover, predictive models can be created, thereby allowing a more proactive approach to PhV. A central premise in this domain is that ADEs are largely predictable consequences of certain molecular actors30. Several aspects of this premise have been validated by long experience, and exploited for high-throughput screening of active compounds in computer aided drug design and development. It has also been used by pharmaceutical companies in the preclinical drug design stage to predict toxicological effects, with the main goal of decreasing late stage attrition of new drugs due to toxic effects31. In contrast to the preclinical stage, recent successful studies have linked this knowledge source with knowledge on post-marketed ADEs to create better predictive models and as a tool to augment existing ADE discovery methodlogies30, 32, 33.
Mining the biomedical literature holds the promise of consolidating large amounts of biomedical knowledge for new discoveries. It has been successfully used to discover new relationships between biomedical entities such as genes, biological pathways, diseases, as well as for of drug repurposing (discovering new indications)34. Among others, the biomedical literature contains ADE related information based on clinical studies and anecdotal observations. Current use of biomedical literature by drug safety researchers (to evaluate or confirm new ADEs) suggests that automated or data mining approaches can supplement existing ADE discovery techniques. However, extracting information from the biomedical literature is non-trivial and requires elaborate Natural Language Processing (NLP) tools. Recent work has demonstrated its potential as a strategy for prioritizing ADE associations under consideration35.
Patient social networks and forums such as Ask a Patient, DailyStrength, Yahoo Health and Wellness, and PatientsLikeMe collect patient self-reports of drug side-effects and provide a platform for patients to discuss and share their experiences with medications. Although the information provided by patients may be inaccurate or even questionable, such forums can provide valuable supplementary information on drug effectiveness and side-effects as they cover large and diverse populations and offer unsolicited, uncensored data directly from patients. However, extracting such information is very challenging and requires deep statistical and linguistic methods to interpret colloquial language, correct grammatical and spelling errors, and distinguish real experiences from hearsay. Nonetheless, recent work has shown that the information contained in these forums is extractable and relevant to PhV36.
3. Methods applied to spontaneous reporting systems
3.1 Disproportionality analysis & basic concepts
Disproportionality analysis (DPA) is the main driving force behind most computerized PhV methods for SRS. DPA methodologies use frequency analysis of 2×2 contingency tables to estimate surrogate measures of statistical association between specific drug-event combinations mentioned in spontaneous reports. Their name stems from the idea that they intend to quantify the degree to which a drug-event combination co-occurs “disproportionally” compared to what would be expected if there were no association5.
DPA methodologies differ by the exact measures that are used, the statistical adjustments made to account for low counts, and can generally be classified into two categories: frequentist and Bayesian. Both approaches use the entries of Table 1a (or stratified versions thereof) to derive a statistical association/disproportionality measure. This table is usually computed for each drug-event pair in the SRS. The most widely discussed measure is the Relative Reporting Ratio (RRR)6 defined as the ratio of the observed incidence rate of a drug-event combination to its “baseline” expected rate under the assumption that the drug and event occur independently. Both the FDA and WHO use a Bayesian version of RRR as a basis for monitoring safety signals in their SRS37, 38. Other widely used measures including their mathematical definitions are displayed in Table 1b. A true value of close to 1 for any of these measures supports the hypothesis that there is no association between the drug and event. A value of 3 in the case of RRR for example indicates that there are 3 times as many drug-event reports in the database than would be expected, and might support the hypothesis of an ADE association.
Table 1 (a).
Contingency table used in SRS based DPA. Reports are classified according to the presence/absence of specific drug-adverse event (AE) combinations. Each cell contains report counts.
| with target AE | without target AE | Total | |
|---|---|---|---|
| with target drug | a | b | n=a+b |
| without target drug | c | d | c+d |
| Total | m=a+c | b+d | t=a+b+c+d |
Table 1 (b).
Mathematical definitions of measures of association.
| Measure of association | Mathematical Definition |
|---|---|
| Relative Reporting Ratio (RRR) | (t·a)/(m·n) |
| Proportional Reporting Ratio (PRR) | [a·(t-n)]/(c·n) |
| Reporting Odds Ratio (ROR) | (a·d)/(c·b) |
| Information Component (IC) | log2(RRR) |
Frequentist approaches use one of the measures listed in Table 1b to estimate associations, and are typically accompanied by hypothesis tests of independence (chi-squared test or Fisher’s exact test), which are used as an extra precautionary measure to also account for the sample size used to compute an association. Bayesian approaches attempt to account for the uncertainty in the disproportionality measure associated with small observed and expected counts, by “shrinking” the measure towards the baseline case of no association, by an amount that is proportional to the variability of the disproportionality statistic. The result of this shrinkage is a reduction of spurious associations when there are not enough data to support them.
Among the Bayesian approaches is the Multi-item Gamma Poisson Shrinker (MGPS)8, 39. MGPS is the predominant DMA used in the US and the UK, and is currently used by the FDA40 as well as several pharmaceutical companies to detect ADE signals in their databases. MGPS is based on a modeling framework called empirical Bayes, and computes a measure called EBGM (empirical Bayes geometric mean), which is a Bayesian interpretation of the RRR measure (posterior expectation of the RRR distribution). Typically, the EB05 measure, which corresponds to the lower 5th percentile of the posterior RRR distribution is used instead for extra conservatism. The WHO uses a Bayesian approach similar to MGPS, called Bayesian Confidence Propagation Neural Network (BCPNN)38, which estimates a Bayesian version of the Information Component.
Ad-hoc thresholds are typically applied to the association measures (regardless of the approach or measure) in order to highlight strong associations worthy of further investigation. The thresholds selected usually do not have theoretical or empirical justification. They are rather used as a preliminary means of filtering or sorting. Deshpande et al. provide a review of published threshold criteria for qualifying signals of disproportionate reporting in SRS41. A graphical illustration of DMA output is provided in Figure 2.
Figure 2.
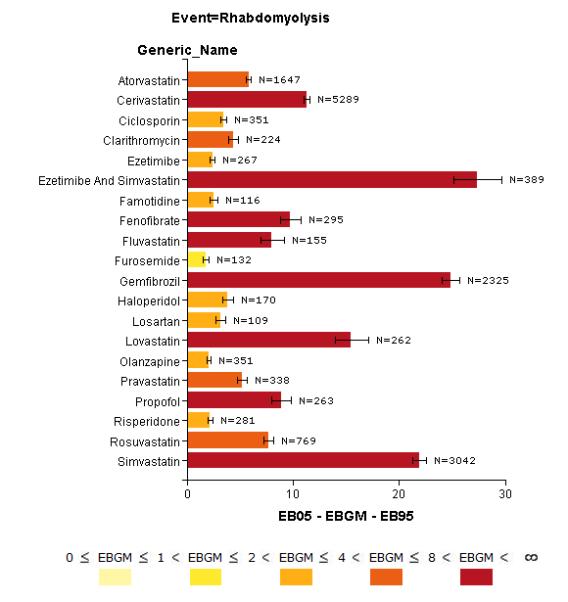
Bar-plot of drugs statistically associated with rhabdomyolysis in AERS as an example of DPA output. Bar colors and length reflect statistical association strength based on the EBGM score. Each bar also includes the 90% confidence interval (EB05-EB95) and report count (N) for the corresponding drug. The plot and underlying DPA were performed using Oracle’s Empirica Signal 7.3 based on AERS data up to and including the year 2011 Q2. Only the top 20 associations consisting of drugs reported as “suspected” with N ≥ 100 were selected for display. Expectedly, the majority of drugs come from the class of statins known to cause rhabdomyolysis as a rare ADE.
As of yet there is no consensus on which DPA approach is best, and no gold standard has been established to evaluate their performance. It is widely accepted that none of the approaches is universally better than any other 5, 6. As the number of reports of a specific drug-event combination increases, the different methods tend to give similar results. Some have argued that for small counts frequentist approaches are more prone to extreme values and therefore generate more false positives. Others have argued that the Bayesian approaches are too conservative delaying the detection of novel ADEs. Frequentist approaches are computationally more efficient than Bayesian approaches, but the latter offer the convenience of being able to sort associations along a single dimension as they incorporate information about both disproportionality and sample size. That said, none of the approaches can effectively address reporting biases or confounding in SRS.
3.2 Multivariate methods
Although cumulative experience with DPA has shown it to be a promising adjunct in safety analysis, the reduction of ADE analysis to two dimensions may result in loss of clinically crucial information. 2-D DPA approaches do not support the discovery and/or analysis of more complex or higher-dimensional drug safety phenomena that involve more than just one drug and one event. The importance and difficulty associated with the detection of these more complex drug safety phenomena was noted in several prominent PhV reports5, 7, 8, suggesting that more elaborate methods, henceforth collectively referred to as “multivariate” methods, are required.
More complex drug safety patterns may correspond to drug-drug interaction adverse events, such as the pharmacodynamic drug interaction between Tramadol and Fluoxetine, where Tramadol (a pain reliever) can enhance the effect of Fluoxetine (Prozac) increasing serotonin levels, which may lead to seizures. Recent studies showed that many ADEs (close to 50% in hospital patients42) are due to drug interactions, suggesting that many of the ADEs reported to SRS are plausibly due to drug interactions and not due to the single suspected drug that was reported. Other more complex drug safety patterns of clinical interest are class effects and syndromic events (drug induced syndromes). For example, the class of statins (cholesterol lowering drugs) is known to cause rhabdomyolysis. The drug varenicline (indicated for smoking cessation) may cause a syndrome of sleeping disorders and other neuro-psychiatric disorders. Additionally, these patterns are important in highlighting the etiology of other ADEs, the further probing of simpler associations, and overall contribute to greater understanding of drug safety risk factors7.
Another limitation of the 2-D DPA approaches is that they are not properly equipped to deal with confounding, which is key to association analysis. A confounder is an extraneous variable, either observed or unobserved, that mediates an association between two other variables. If not properly accounted for, confounding may lead to the discovery of spurious associations and therefore erroneous study conclusions. Confounding can be addressed either through experimental design prior to data collection (e.g., selection of appropriate controls), or in the analysis stage when the data has already been collected (as in the case of SRS). Simpler types of confounding such as confounding by age, gender, and year, have been effectively dealt with within DPA approaches through stratification and Mantel-Haenszel type adjustments6, 7. Although there are many types of confounding, e.g., confounding by indication where the reported event is associated with the indication for treatment, most SRS related publications have focused on confounding by drug co-administration, where a drug is associated with an event just because it is frequently co-prescribed or reported with another drug, which is the real cause of the adverse event.
In recent years several SRS multivariate approaches have been proposed to address these issues. They can generally be classified as DPA extensions, multivariate logistic regression based approaches, and unsupervised machine learning approaches such as associations rule mining, clustering, and network analysis. DPA extensions to larger dimensions have been applied to mostly 3-D associations corresponding to drug-drug interactions43, where observed to expected ratios are calculated in a similar manner but based on 3 elements (drug1-drug2-event). Logistic regression based approaches have been applied mostly to eliminate confounding by co-medication (due to lack of other confounding information in SRS), whereas unsupervised machine learning approaches have been used for the identification of more complex or higher-dimensional drug safety phenomena, as well as for data abstraction and pattern discovery.
3.2.1 Logistic regression based approaches
The traditional approach to handle confounding during the analysis stage, i.e., stratification, is not effective in situations where a large number of potential confounders need to be examined44. A more appropriate approach to handle confounding is by the use of multiple logistic regression, which allows the estimation of a drug-event association by controlling or adjusting for the presence of other covariates (potential confounders)44. Confounding by co-medication can theoretically be addressed by using all drugs in a SRS as regression predictors for an event. However, the regression of a specific event against all the thousands (>10,000) of drugs included in a SRS represented a significant computational as well a theoretical barrier until recently. New extensions of logistic regression to very large dimensional data called Regularized or Bayesian Logistic Regression (BLR) can now carry out regressions with millions of covariates45. Caster et al.46 describe an application of BLR to the WHO SRS, in an attempt to address confounding by co-medication and a “masking” effect. The latter corresponding to cases where an increase of background reporting for a specific event (e.g., due to media influences) can attenuate disproportionally measures of true associations towards lower values of no association, thereby masking the true association. The authors describe several real examples of false positive associations due to confounding by co-medication that were corrected by their method, and true ADEs masked by media influences surrounding the withdrawal of a drug causing rhabdomyolysis. In earlier work, Solomon and DuMouchel47 applied standard DPA along with BLR to AERS as part of a study to estimate associations between several contrast media (CM) agents and events related to contrast-induced nephropathy. All methods were adjusted for demographic variables. BLR was also adjusted for 200 drugs co-reported with CM as potential confounders and as proxies for unobserved confounders. The authors found that the results were consistent among the different methods (including the rank order of associations), but that BLR odds ratio estimates were generally 50% larger. The authors explain that this difference stems from the different comparators used by each method and a masking effect related to the agents investigated.
3.2.2 Unsupervised machine learning approaches
Multi-item ADE associations are associations relating multiple drugs to possibly multiple adverse events. Association rule mining (ARM)48 is a well established data mining method for discovering interesting relationships between variables in large databases. ARM can be applied to discover multi-item ADE associations - a special case of association rules. For example,
chantix, darvocet → memory impairment, abnormal dreams, fatigue, insomnia
(Chantix may interact with darvocet - a pain reliever, and cause various sleeping or mental disorders).
Computing association rules is inherently a very hard combinatorial problem that can easily become computationally intractable. The Apriori algorithm48 can alleviate the problem, but does not completely resolve the computational challenge. Rouane et al.49 applied ARM to the SRS of the French Medicines Agency to identify rules related to anti-HIV drugs. The authors proposed the use of formal concept analysis as a means for reducing the computational complexity, but their approach was restricted to only 3-item associations. In a recent study Harpaz et al.50 applied ARM with rules of up to 6 items to AERS. Noting the inappropriateness of standard ARM scores to ADE applications, the authors used the RRR score instead, with the additional constraint that each rule must have an RRR larger than any of its subsets. The latter was used to exclude rules that can better be explained by smaller sets of drugs or events43. The authors showed that roughly 66% of the rules corresponded to known associations, thereby demonstrating the potential value of SRS for the discovery of multi-item clinically relevant ADE associations. A promising Bayesian approach to ARM has recently been proposed by McCormick et al.51, which has direct application to ADE discovery and can address the sparseness of SRS data when computing association rules.
Currently the main bottleneck in the widespread application of ARM to SRS is its computationally intensive requirements. It is likely that its adaptation will grow as computing power increases. As an alternative, a recent pilot study by Fan et al.52 have demonstrated the potential applicability of the highly parallelized and distributed computing paradigm - MapReduce - to the same problem.
Clustering is routinely used in many biomedical areas, but until recently its potential was not investigated in the context of ADE analysis. Harpaz et al.53 proposed a non-standard clustering approach suited to deal with the high-dimensional nature and sparseness of SRS data. The method, called biclustering was applied to AERS and defines an ADE cluster as a group of drugs that are all statistically associated with the same group of adverse events. The authors demonstrated how biclustering can be used as an exploratory tool in PhV with which the underlying large and complex structure of SRS can be summarized and described in a macroscopic manner (e.g., 40% of ADEs in AERS are cancer related). They demonstrated how biclustering can be used to highlight class effects (e.g., bisphosphonates), and syndromic events (e.g., sleeping disorders), and how it could be used to support the discovery of potentially new ADEs. They found that a large proportion (41%) of the clustered relationships contained associations that are currently unrecognized, signaling potentially new ADEs by allowing these unrecognized associations to borrow support from confirmed ADE associations within the same cluster. Examples include the associations: chlorpromazine–hepatotoxicity, methotrexate–pancytopenia, and bosentan–hepatic steatosis, which are supported by published case reports.
Ball et al.54 proposed the use of Network Analysis (NA) to facilitate the identification of clinically interesting multi-dimensional patterns of adverse events. The authors applied NA to FDA’s Vaccine Adverse Event Reporting System, where nodes in the network correspond to vaccines and events. They focused on identifying “hubs”, which are tightly clustered elements within the network that reveal strong informative structures. The authors found patterns linking the vaccine HPV4 with syncope and syncope with seizures in adolescents. They also found patterns of serious gastrointestinal adverse events with the vaccine rotavirus. Last, they demonstrated that VAERS has the characteristics of a “scale free” (non-random) network, where certain vaccines and events act as hubs.
4. Methods applied to electronic healthcare data
Methods applied to electronic healthcare data (HCD) can generally be classified as those based on modified DPA ported from spontaneous reporting, and those based on epidemiological study designs such as the cohort, case-control, and self-controlled study designs. One of the major challenges in the use of HCD is the pervasiveness of confounding. While DPA approaches are simpler, methods based on epidemiological study designs may be better equipped to deal with confounding, but present challenges in scaling to high-throughput settings and require many design decisions to be made. Another major challenge in the use of HCD, are the definition and ascertainment of exposures and outcomes. Because HCD are not collected for PhV purposes it must be ensured that the data contain sufficient clinical information to correctly capture and validate the exposures and outcomes of interest. Outcomes can be defined in various different ways, each of which may have different operating characteristics55. Often the exposures and outcomes of interest may not be captured or do not reflect actual experience, e.g., over-the-counter or dietary supplements may not be captured because they are not prescribed or associated with reimbursement. Mild symptoms, which are not treated, or extreme conditions such as death without medical care may also not be captured. Actual ingestion, dosage, or prescription fulfillment of a drug is hard to ascertain. Once defined, actual identification of exposures and outcomes may be challenging when portions of the data are in unstructured uncoded format, such as with health record medical narratives, which may require NLP. A key distinguishing feature of HCD based methods is the use of temporal information to identify time frames (known as surveillance windows, drug/condition eras, hazard periods) in which drug-outcome pairs are identified and analyzed, e.g., outcomes recorded 30 days from drug exposure. Essentially all HCD based methods define and use some form of time frames to detect ADEs. Using this temporal information , drug safety analysis with HCD can be visualized and analyzed using patient timeline graphs – Figure 3.
Figure 3.
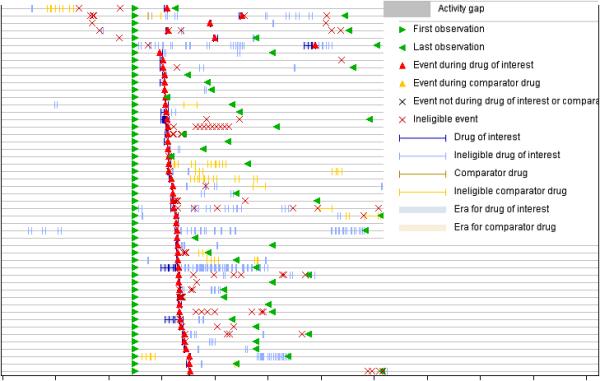
Patient timelines used to visualize and analyze healthcare data in drug safety. Each patient is represented by a horizontal line capturing time, and symbols on the line represent clinical events, e.g., diagnoses, test results, treatments, and drug eras. The figure shows timelines of patients experiencing various events such as headache (red triangle) within eligible periods of acetaminophen administration. The patients are sorted according to the first occurrence of an event (red triangle).
4.1 Disproportionality analysis
There are several ways in which drug-outcome pairs can be counted and mapped into 2×2 contingency tables. Zorych et al.56 from OMOP discuss three approaches called ‘distinct patients’, ‘SRS’ and ‘modified SRS’. The first counts the number of distinct patients that experience an outcome within a drug era (even though the same patient may experience multiple outcomes in several drug eras). The second approach attempts to mimic SRS and treats each drug-outcome occurrence as a spontaneous report. The last approach attempts to take advantage of other information and to augment SRS like reporting with denominator information by also counting exposures without outcomes and outcomes without exposure (SRS only counts exposures reported with an event). Another distinction is made between incident conditions where only the first occurrence of an event is counted, and prevalent conditions where all occurrences are counted, giving a total of six ways to map the data into 2×2 tables. Based on a large scale systematic evaluation of these counting approaches using the DPA metrics described in Section 3.1, the authors conclude that the SRS and modified SRS approaches using Bayesian metrics provide the best performance.
Some have hypothesized that metrics based on person-time rather than person-counts could produce more accurate association estimates, because length of exposure is a more granular and information carrying quantity than number of persons. Exploiting the temporal information available in HCD, Schuemie57 proposed an approach called Longitudinal GPS (LGPS), which is a modification of the original MGPS approach, that uses person-time rather than person-counts to estimate the expected number of events. In LGPS the expected number of occurrences of an event is calculated as the total time patients are exposed to a specific drug multiplied by the number of occurrences of the event per unit time when the patients are not exposed. Schuemie also proposed a heuristic to apply in conjunction with LGPS in order to remove spurious associations caused by protopathic bias. The heuristic is based on the assumption that an increase of the number of prescriptions after an event compared to before the event is an indication of protopathic bias. LGPS has been shown to outperform related methods, including MGPS, and was the winner of the 2010 OMOP cup competition based on simulated data19. A similar approach to LGPS was proposed by Noren et al., who argue that their approach has several important advantages over LGPS that better protect against confounding58. Using their method applied to longitudinal HCD from the UK the authors were able to demonstrate the timely identification of the association between terbinafine and angioedema.
4.2 Cohort designs
Although there are many variants, the basic concept underlying cohort designs is to partition the subject population into two groups: those that are “exposed” (taking a specific drug) and those that are “unexposed” (taking a comparator drug/s). The relationship between the exposure and the outcome is then examined by comparing the prevalence of the outcome in both groups. An association is identified when the outcome occurs more often in the exposed than the unexposed group. Comparators can be those not taking the drug or those taking a drug/s from the same therapeutic drug class. Yet, due to the non-random assignment of groups, increased attention must be given to the selection of appropriate comparators. Inappropriate selection may lead to confounding and biases such as channeling (commonly observed when comparing drugs with similar indications), where imbalances of risk or prognostic factors between groups results in biased effect estimates and thus unreliable conclusions. To address and minimize these issues - matching, where the two groups are matched based on a set of covariates (e.g., gender, age, length of exposure, and co-morbidities), or propensity scores are often employed.
Propensity Score (PS) methods have become a common analytic approach to control confounding in cohort designs by impersonating the role of randomization in clinical trials10, 27. A PS is the conditional probability that a subject receives treatment given a set of measured preselected covariates (potential confounders). Among subjects with the same propensity to receive treatment, the treatment is conditionally independent of the confounders, suggesting that within groups of subjects with the same PS any difference in outcome between the treated and untreated cannot be attributed to the confounders. A PS can also be viewed as 1-D (scalar) value that summarizes a large number of covariates. Treatment-outcome effects can then estimated by using the PS for matching, stratification, or as an adjustment factor in regression models59. A central challenge in the use of PS is the selection of covariates to be included in the model. Incorrect selection may introduce bias into the analysis. There are differing views as to the type of covariates that should be included, i.e., whether the covariates should be related to: exposure only, both outcome and exposure, or outcome regardless of exposure60.
Schneeweiss et al.61 proposed an algorithm for PS covariate selection that has received much attention lately called High-Dimensional Propensity Score (HDPS). The method automatically identifies and selects empirical confounders, estimates propensity scores, and integrates them into an exposure-outcome PS-based confounder adjustment model. The authors claim that adjusting for large numbers of covariates ascertained from patients’ healthcare claims data may improve control of confounding, as these variables may collectively be proxies for unobserved factors. Empirical confounders are automatically identified based on a function that incorporates both the prevalence of a covariate and its association with the outcome. Covariates are then ranked based on this function and the top k covariates are selected as the final set of empirical confounders (in addition to the usual demographic covariates). Based on the empirical confounders, a logistic regression is used to estimate a PS for each subject. These PSs are converted into indictor variables based on PS deciles, and then used in the final logistic regression model of exposure-outcome to estimate confounder adjusted associations. In one experiment conducted by OMOP, HDPS achieved a sensitivity of 56%, specificity of 82%, and positive predictive value of 38% in the detection of 53 associations corresponding to true ADEs and negative controls19.
4.3 Case-control designs
In a case-control study the subject population is divided into those experiencing the outcome under investigation called “cases” and a comparator group called “controls”. The relationship between the exposure and the outcome is then examined by comparing the prevalence of the exposure in both groups. An association is identified when the exposure occurs more often in the cases than the comparator group. In case-control designs the controls will typically consist of subjects that did not experience the outcome being studied, but are otherwise similar. Other options for selecting controls include subjects experiencing another set of conditions of interest, or subjects with conditions indicated for the same types of drugs. The main advantage of case-control studies over alternative study designs such as cohort designs is in their data efficiency, which permits the study of rare events44. Matching is often employed to control for potential confounding factors. In a matched case-control study each case is matched to one or more controls based on a set of predetermined covariates. Over-matching may introduce bias and should be discouraged62. As part of OMOP’s experiment in which HDPS was evaluated, an implementation of a case-control design achieved close to 100% sensitivity but at the expense of extremely low 15% specificity19.
4.3 Self-controlled designs
A self-controlled design can be viewed as a special variant of the case-control design, where subjects are used as their own controls, and outcome rates are compared between periods when a subject is exposed to periods when the subject is unexposed. Because exposure data is provided by the same person, these designs implicitly control for all time-invariant confounders that do not vary within a subject (e.g., comorbidities, smoking status, and chronic use of drugs) without the need for confounders to be measured. They also eliminate selection bias. Another advantage of this design is that only “cases” need to be included in the analysis. Self-controlled designs can be used when subject data include multiple exposure risk periods.
The self-controlled case series (SCCS)63 is a type of a self-controlled design. SCCS assumes that adverse events arise according to a non-homogeneous Poisson process, where each subject has an individual baseline (non-exposure) event rate constant over time, and periods of exposure result in a multiplicative effect on the baseline rate. The goal is to estimate the multiplicative effect, which corresponds to the relative risk of an adverse event during exposure. Simpson et al.21 from OMOP were able to demonstrate that applying SCCS to one of OMOP’s observational data sources (i3 claims data-approximately 50 million subjects) would have led to detection of the Vioxx-MI association 3 years prior to the drug withdrawal (2004), when AERS based DPA failed to detect the association. It was also demonstrated that SCCS outperformed DPA methods on most performance metrics.
Although much progress has been made, methodological research into the use of HCD is currently in its early stages64. Ultimately, it is unlikely that an optimal solution will apply a one-size-fits-all methodological solution; instead, a process may be developed to refine the analysis to the characteristics of the medical product, outcome, and databases in question. Gagne et al.65 provided a taxonomy of study design considerations based on anticipated drug safety questions but substantial research is required to validate such recommendations. Establishing best practices requires further empirical evaluation to measure performance of alternative methods across the continuum of expected scenarios. Absent such information, heuristics are applied using expert subjective assessment without supporting empirical evidence.
5. Methods using non-standard data sources or linking multiple data sources
Chemical and Biological information
In a series of articles Matthews et al. 32, 33 from FDA’s Center for Drug Evaluation and Research discuss an implementation of a system based on Quantitative Structure Activity Relationship (QSAR) models to predict ADEs and possible mechanisms of action (MOA) responsible for adverse events. QSARs are mathematical models that are used to predict measures of toxicity from physical characteristics of the structure of chemicals. Drug candidates for QSAR modeling were indentified from AERS using standard DPA and were supplemented with literature findings. Commercial QSAR software was then applied to the drug candidates to identify chemical properties of molecules that correlate with adverse events. The authors built separate QSAR models for several adverse events including: cardiac, liver, and urinary related ADEs. They report an average of 78% specificity and 56% sensitivity noting that when data based on the literature was added there was usually a substantial improvement in performance. They remark that roughly half of drugs related to hepatobiliary and urinary tract ADEs that were missed in pre-market clinical trials could have been predicted using QSAR models. They also found that cardiac ADEs correlate with MOAs affecting cardiovascular and cardioneurological functions, such as the alpha/beta adrenoceptors, the dopamine and hydroxytryptomine receptors, and that screening new drugs based on these MOAs could predict the majority of cardiac ADEs. The QSAR models are now being used internally by the FDA to provide decision support information for a variety of regulatory activities.
Villar et al.66 proposed a SAR modeling technique to prioritize ADE associations generated from AERS. The authors compiled a reference set of drugs related to rhabdomyolysis from the literature, and mapped them to 2-D molecular fingerprints (bit vectors that represent the presence/absence of specific structural features) using information available in DrugBank and commercial software. The initial drug candidates, generated from AERS by the MGPS algorithm, were then screened by comparing their structural fingerprints with the reference set of fingerprints, and highly similar candidates were then retained as the final set of drug candidates. Using this approach the authors achieved 70% sensitivity, 45% positive predictive value, and an over 2-fold enrichments of AERS signals.
Publicly available preclinical molecular screening assays such as those available in PubChem can be mined to correlate a drug’s bioactivity with postmarketing ADEs. Pouliot et al.30 created models to correlate a drug’s propensity to cause specific system organ class (SOC) ADEs. They used data from over 487,000 drug activity screens from the National Center for Biotechnology Information’s PubChem BioAssay database and SOC-specific ADE information from the Canadian Adverse Drug Reaction database to create logistic regression models for 9 SOCs. They validated these models by performing retroprediction for eight individual drugs and report that 75% of the predicted adverse reactions in humans could be substantiated by the literature or drug labeling information. Using these validated models, they predicted yet unrecognized ADEs for 3 drugs that were recently approved or not yet approved in the US. The authors note that making such predictions can generate testable hypotheses for the identification of ADEs in the clinical setting and thus shorten the duration for which new ADEs go unrecognized.
Biomedical literature
Shetty et al.35 describe an approach for collecting, filtering, and analyzing biomedical literature as a complementary strategy for prioritizing ADE associations generated from SRS. First, all articles mentioning drug-outcome pairs from a predefined set of drugs and events were retrieved from PubMed. Then, NLP was used to identify and exclude retrieved articles mentioning irrelevant pairs (e.g., pairs that capture a treatment relationship). Last, DPA was applied to the pairs mentioned in the remaining articles to highlight statistically significant drug-event associations. The authors show that the method discovered true associations with over 70% sensitivity and 40% positive predictive value, using a reference set of true ADE associations obtained from the ‘Warnings’ section of drug labels. They also demonstrate that using their approach 54% of the associations analyzed could have been detected prior to FDA warning, where the Vioxx-MI association could have been identified using literature published before the year 2002.
User generated content in health forums
In a recent study, Leaman et al.36 demonstrated that user posts on health related websites contain extractable information relevant to PhV, and describe a prototype system to mine this type information source. Raw data was automatically collected using a webcrawler from the web site DailyStrength. NLP techniques were used to process the raw data and extract clinical concepts related to ADEs. Special procedures were used to deal with colloquial phrases, e.g., “zoned out” meaning somnolence, and user spelling errors. The system was evaluated using an expert annotated set of 3,600 user generated posts corresponding to 6 drugs. The system achieved 78% precision and 70% recall in correctly labeling the user generated data. Importantly, the authors found that the incidence of ADEs reported by users is highly correlated with documented incidence rates listed by the FDA, noting that the most frequent ADEs identified corresponded to well known ADEs. PatientsLikeMe recently published67 a study based on their data, where the user community essentially self-assigned themselves to assess lithum efficacy in patients with amyotrophic lateral sclerosis. While not safety surveillance, it is a study that illustrates the promise of patient-initiated observational studies.
Linking multiple knowledge sources
By linking information from multiple sources Cami et al.68 proposed a network-based model to predict ADEs. The authors first constructed a network representation of drug-event associations that were known as of 2005. Then, using network topological indices (e.g., node degree), supplemented with ontological features (e.g., distance between two events in the MedDRA hierarchy) and molecular descriptors (e.g., drug’s molecular weight and melting point) the authors trained a logistic regression model to predict the probability of an unknown ADE association (edge in the network graph). The predictive performance of the model was prospectively validated by predicting ADEs reported in the years 2006-2010. The model achieved an Area Under the Receiver Operating Characteristic Curve of 0.87, sensitivity of 42%, and specificity of 95%. The authors were also able to predict seven of eight ADEs that emerged after 2005 including: the seizure drug zonisamide causing suicidal thoughts, the antibiotic norfloxacin linked to ruptured tendons, and the controversial diabetes drug rosiglitazone (Avandia) linked to heart attacks. The authors claim that unlike related work the prospective characteristics of their model make it a realistic method for predicting future ADEs.
By integrating information from AERS and several HCD sources, Tatonetti et al.69 discovered a potentially new drug interaction between two widely used drugs - the antidepressant paroxetine and the cholesterol-lowering medication pravastatin-that can lead to unexpected increases in blood glucose levels. The motivating idea behind their data mining approach is the observation that side effects are not independent of each other and latent evidence for an (unreported) adverse event can be found by examining other (reported) side effects. By scanning AERS for pairs of drugs having matching side effect profiles when taken together but not when taking individually, the authors created a candidate set of drug-drug interactions. The list of candidates was then narrowed down to the paroxetine-pravastatin interaction by conducting retrospective studies using electronic health records from Stanford University Hospital, Vanderbilt University Hospital, and Partners Healthcare. Last, the interaction was confirmed by a prospective study in an insulin resistant mouse model.
6. Concluding remarks, future perspectives, and challenges
We have shown that a rich and diverse portfolio of data mining approaches aligned to different strategies and objectives is now available for the analysis and detection of post-approval ADEs. Each approach may offer unique prospects that collectively can advance the science of drug safety surveillance.
New opportunities and interest have emerged to harness data that has not been traditionally used in PhV, allowing for new active and proactive paradigms of surveillance. Although methodological research is now shifting away from SRS, this will not diminish the important role or value of spontaneous reports. That said, the use of spontaneous report narratives to enhance SRS based discovery is yet to be explored. It is also evident that a new trend is emerging by which data mining is used to link human safety information with experimental platforms that have been traditionally used in the preclinical drug discovery phase. The diversity of approaches highlights the value of systems that can span data and expertise across multiple domains. Nonetheless, to fully realize this potential, new and creative methods will be required to integrate these disparate sources in a more synergistic manner.
It has been suggested that a revisit of randomized clinical trials data, by synthesis or pooling of several related trials, should be employed to augment findings from other sources. The main benefit to this approach is that it enjoys the scientific and statistical benefits of randomization70. Although beyond the scope of this article, it is important emphasize the role of pharmacogenomics research29, 71 and the utility of knowledge bases such as PharmGKB72 that would further enhance our understanding of ADEs by correlating human genetic variation with drug toxicity.
While much progress has been made in utilizing healthcare data, a substantial amount of further empirical assessment is required using both real and simulated data sets. A key OMOP finding is that the heterogeneity of data sources and methods strongly affects results. Thus, consistent methods that can be applied to multiple data sources will be required. There are also existing opportunities to improve data quality, coding standards, sharing, and access. The relative value of information contained in the electronic health records compared to administrative claims needs to be further explored. The continued development of simulated data for which ground-truth is available is critically important to further the understanding of method efficacy.
A central challenge in PhV research is the lack of established standards to evaluate DMAs. One of the main contributors to this problem is the lack of a gold standard, as the set and nature of all possible drugs safety issues is unknown. Although suggestions have been made, there is an ongoing debate as to testing strategies and what should constitute reference standards for DMA evaluation7, 73. With that said, there is currently little empirical evidence to support or prefer the use of one method or data source over the other, and efforts such as those made by OMOP and EU-ADR are of paramount importance so that method and data source value can be assessed on solid scientific footing.
DMA activity is typically conducted at certain granularity levels of medical terminologies, which are not optimally designed to support PhV. Often, similar medical concepts are fragmented across distinct terms, weakening the potential for statistical discovery. Therefore, methods that make better use and integrate knowledge from lexical resources74 may prove beneficial. Relatedly, Bayesian approaches allowing for information borrowing75 across similar terms and drugs, should be further developed and evaluated. Additionally, improved NLP methods to process unstructured textual data, whether from clinical narratives, literature, or health forums, will continue to play an important role25.
Last but not least, it is important to recognize that at its core, data mining is a tool to formulate or refine new hypothesis and thus will not eliminate the important role of medical review that will always be required for final adjudication of causality.
Acknowledgments
This work was supported in part by grants 1R01LM010016, 3R01LM010016-01S1, 3R01LM010016-02S1, and 5T15-LM007079-19(HS) from the National Library of Medicine, U54-HG004028 from the National Center for Biomedical Ontology, and 2U54LM008748 from the i2b2 National Center for Biomedical Computing. We thank Nicholas Tatonetti from Stanford University and Roee Sa’adon from First Life Research for their valuable comments, and extend our gratitude to Oracle’s Health Sciences Division for supplying us with data and figures.
Reference List
- (1).World Health Organization . The Importance of Pharmacovigilance - Safety Monitoring of Medicinal Products. World Health Organization; Geneva: 2002. [Google Scholar]
- (2).Lazarou J, Pomeranz BH, Corey PN. Incidence of adverse drug reactions in hospitalized patients: a meta-analysis of prospective studies. JAMA. 1998 Apr 15;279(15):1200–5. doi: 10.1001/jama.279.15.1200. [DOI] [PubMed] [Google Scholar]
- (3).Classen DC, Pestotnik SL, Evans RS, Lloyd JF, Burke JP. Adverse drug events in hospitalized patients. Excess length of stay, extra costs, and attributable mortality. JAMA. 1997 Jan 22;277(4):301–6. [PubMed] [Google Scholar]
- (4).Ahmad SR. Adverse drug event monitoring at the Food and Drug Administration - Your report can make a difference. Journal of General Internal Medicine. 2003 Jan;18(1):57–60. doi: 10.1046/j.1525-1497.2003.20130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Bate A, Evans SJ. Quantitative signal detection using spontaneous ADR reporting. Pharmacoepidemiol Drug Saf. 2009 Jun;18(6):427–36. doi: 10.1002/pds.1742. [DOI] [PubMed] [Google Scholar]
- (6).Hauben M, Madigan D, Gerrits CM, Walsh L, van Puijenbroek EP. The role of data mining in pharmacovigilance. Expert Opin Drug Saf. 2005 Sep;4(5):929–48. doi: 10.1517/14740338.4.5.929. [DOI] [PubMed] [Google Scholar]
- (7).Hauben M, Bate A. Decision support methods for the detection of adverse events in post-marketing data. Drug Discov Today. 2009 Apr;14(7-8):343–57. doi: 10.1016/j.drudis.2008.12.012. [DOI] [PubMed] [Google Scholar]
- (8).Almenoff JS, Pattishall EN, Gibbs TG, DuMouchel W, Evans SJ, Yuen N. Novel statistical tools for monitoring the safety of marketed drugs. Clin Pharmacol Ther. 2007 Aug;82(2):157–66. doi: 10.1038/sj.clpt.6100258. [DOI] [PubMed] [Google Scholar]
- (9).Platt R, Wilson M, Chan KA, Benner JS, Marchibroda J, McClellan M. The New Sentinel Network - Improving the Evidence of Medical-Product Safety. New England Journal of Medicine. 2009 Aug 13;361(7):645–7. doi: 10.1056/NEJMp0905338. [DOI] [PubMed] [Google Scholar]
- (10).Schneeweiss S. A basic study design for expedited safety signal evaluation based on electronic healthcare data. Pharmacoepidemiology and Drug Safety. 2010 Aug;19(8):858–68. doi: 10.1002/pds.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Wysowski DK, Swartz L. Adverse drug event surveillance and drug withdrawals in the United States, 1969-2002 - The importance of reporting suspected reactions. Archives of Internal Medicine. 2005 Jun 27;165(12):1363–9. doi: 10.1001/archinte.165.12.1363. [DOI] [PubMed] [Google Scholar]
- (12).Stephenson W, Hauben M. Data mining for signals in spontaneous reporting databases: proceed with caution. Pharmacoepidemiol Drug Saf. 2007;16(4):359–65. doi: 10.1002/pds.1323. [DOI] [PubMed] [Google Scholar]
- (13).Graham DJ, Campen D, Hui R, et al. Risk of acute myocardial infarction and sudden cardiac death in patients treated with cyclo-oxygenase 2 selective and non-selective non-steroidal anti-inflammatory drugs: nested case-control study. Lancet. 2005;365(9458):475–81. doi: 10.1016/S0140-6736(05)17864-7. [DOI] [PubMed] [Google Scholar]
- (14).Avorn J, Schneeweiss S. Managing Drug-Risk Information - What to Do with All Those New Numbers. New England Journal of Medicine. 2009 Aug 13;361(7):647–9. doi: 10.1056/NEJMp0905466. [DOI] [PubMed] [Google Scholar]
- (15).Stang PE, Ryan PB, Racoosin JA, et al. Advancing the Science for Active Surveillance: Rationale and Design for the Observational Medical Outcomes Partnership. Annals of Internal Medicine. 2010 Nov 2;153(9):600–W206. doi: 10.7326/0003-4819-153-9-201011020-00010. [DOI] [PubMed] [Google Scholar]
- (16).McClellan M. Drug Safety Reform at the FDA - Pendulum Swing or Systematic Improvement. N Engl J Med. 2007;356:1700–2. doi: 10.1056/NEJMp078057. [DOI] [PubMed] [Google Scholar]
- (17). [accessed Feb 2012];Food and Drug Administration Amendments Act (FDAAA) of 2007. http://www.fda.gov/
- (18). [accessed Feb 2012];The Sentinel Initiative: a national strategy for monitoring medical product safety. http://www.fda.gov/downloads/Safety/FDAsSentinelInitiative/UCM124701.pdf.
- (19). [accessed Feb 2012];Observational Medical Outcomes Partnership (OMOP) http://omop.fnih.org/
- (20).Coloma PM, Schuemie MJ, Trifiro G, et al. Combining electronic healthcare databases in Europe to allow for large-scale drug safety monitoring: the EU-ADR Project. Pharmacoepidemiol Drug Saf. 2011 Jan;20(1):1–11. doi: 10.1002/pds.2053. [DOI] [PubMed] [Google Scholar]
- (21).Simpson SE. Ph.D. dissertation. Columbia University; 2011. Self-controlled methods for postmarketing drug safety surveillance in large-scale longitudinal data. [Google Scholar]
- (22).LePendu P, Iyer SV, Fairon C, Shah NH. Annotation Analysis for Testing Drug Safety Signals using Unstructured Clinical Notes. Journal of Biomedical Semantics. 2012 doi: 10.1186/2041-1480-3-S1-S5. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Brownstein JS, Sordo M, Kohane IS, Mandl KD. The tell-tale heart: population-based surveillance reveals an association of rofecoxib and celecoxib with myocardial infarction. PLoS One. 2007;2(9):e840. doi: 10.1371/journal.pone.0000840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Brown JS, Kulldorff M, Chan KA, et al. Early detection of adverse drug events within population-based health networks: application of sequential testing methods. Pharmacoepidemiol Drug Saf. 2007 Dec;16(12):1275–84. doi: 10.1002/pds.1509. [DOI] [PubMed] [Google Scholar]
- (25).Chapman WW, Nadkarni PM, Hirschman L, D’Avolio LW, Savova GK, Uzuner O. Overcoming barriers to NLP for clinical text: the role of shared tasks and the need for additional creative solutions. J Am Med Inform Assoc. 2011 Sep;18(5):540–3. doi: 10.1136/amiajnl-2011-000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Brookhart MA, Stürmer T, Robert JG, Rassen J, Schneeweiss S. Confounding Control in Healthcare Database Research: Challenges and Potential Approaches. Med Care. 2010;48(6 Sippl):114–S120. doi: 10.1097/MLR.0b013e3181dbebe3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Schneeweiss S, Avorn J. A review of uses of health care utilization databases for epidemiologic research on therapeutics. J Clin Epidemiol. 2005;58(4):323–37. doi: 10.1016/j.jclinepi.2004.10.012. [DOI] [PubMed] [Google Scholar]
- (28).Wishart DS, Knox C, Guo AC, et al. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008 Jan;36(Database issue):D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Chiang AP, Butte AJ. Data-driven methods to discover molecular determinants of serious adverse drug events. Clin Pharmacol Ther. 2009 Mar;85(3):259–68. doi: 10.1038/clpt.2008.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Pouliot Y, Chiang AP, Butte AJ. Predicting adverse drug reactions using publicly available PubChem BioAssay data. Clin Pharmacol Ther. 2011 Jul;90(1):90–9. doi: 10.1038/clpt.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Bender A, Glen RC. Molecular similarity: a key technique in molecular informatics. Organic & Biomolecular Chemistry. 2004;2(22):3204–18. doi: 10.1039/B409813G. [DOI] [PubMed] [Google Scholar]
- (32).Matthews EJ, Ursem CJ, Kruhlak NL, et al. Identification of structure-activity relationships for adverse effects of pharmaceuticals in humans: Part B. Use of (Q)SAR systems for early detection of drug-induced hepatobiliary and urinary tract toxicities. Regulatory Toxicology and Pharmacology. 2009 Jun;54(1):23–42. doi: 10.1016/j.yrtph.2009.01.009. [DOI] [PubMed] [Google Scholar]
- (33).Frid AA, Matthews EJ. Prediction of drug-related cardiac adverse effects in humans--B: use of QSAR programs for early detection of drug-induced cardiac toxicities. Regul Toxicol Pharmacol. 2010 Apr;56(3):276–89. doi: 10.1016/j.yrtph.2009.11.005. [DOI] [PubMed] [Google Scholar]
- (34).Deftereos SN, Andronis C, Friedla EJ, Persidis A, Persidis A. Drug repurposing and adverse event prediction using high-throughput literature analysis. Wiley Interdiscip Rev Syst Biol Med. 2011 May;3(3):323–34. doi: 10.1002/wsbm.147. [DOI] [PubMed] [Google Scholar]
- (35).Shetty KD, Dalal SR. Using information mining of the medical literature to improve drug safety. J Am Med Inform Assoc. 2011 Sep;18(5):668–74. doi: 10.1136/amiajnl-2011-000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Leaman R, Wojtulewicz L, Sullivan R, Skariah A, Yang J, Gonzalez G. Towards Internet-Age Pharmacovigilance: Extracting Adverse Drug Reactions from User Posts in Health-Related Social Networks. Proceedings of the 2010 Workshop on Biomedical Natural Language Processing.2010. pp. 117–25. [Google Scholar]
- (37).Szarfman A, Machado SG, O’Neill RT. Use of screening algorithms and computer systems to efficiently signal higher-than-expected combinations of drugs and events in the US FDA’s spontaneous reports database. Drug Saf. 2002;25(6):381–92. doi: 10.2165/00002018-200225060-00001. [DOI] [PubMed] [Google Scholar]
- (38).Bate A, Lindquist M, Edwards IR, et al. A Bayesian neural network method for adverse drug reaction signal generation. Eur J Clin Pharmacol. 1998 Jun;54(4):315–21. doi: 10.1007/s002280050466. [DOI] [PubMed] [Google Scholar]
- (39).DuMouchel W. Bayesian data mining in large frequency tables, with an application to the FDA Spontaneous Reporting System. Am Stat. 1999;53(3):177–90. [Google Scholar]
- (40).Szarfman A. [accessed Dec 2011];Safety Data Mining. FDA Advisory Committee Meeting Briefing Document. 2006 http://www.fda.gov/ohrms/dockets/as/06/briefing/2006-4266b1-02-06-FDA-appendic-f.pdf.
- (41).Deshpande G, Gogolak V, Weiss Smith S. Data Mining in Drug Safety: Review of Published Threshold Criteria for Defining Signals of Disproportionate Reporting. Pharmaceutical Medicine. 2010;24(1) [Google Scholar]
- (42).Ramirez E, Carcas AJ, Borobia AM, et al. A pharmacovigilance program from laboratory signals for the detection and reporting of serious adverse drug reactions in hospitalized patients. Clin Pharmacol Ther. 2010 Jan;87(1):74–86. doi: 10.1038/clpt.2009.185. [DOI] [PubMed] [Google Scholar]
- (43).Almenoff JS, DuMouchel W, Kindman LA, Yang X, Fram D. Disproportionality analysis using empirical Bayes data mining: a tool for the evaluation of drug interactions in the post-marketing setting. Pharmacoepidemiol Drug Saf. 2003 Sep;12(6):517–21. doi: 10.1002/pds.885. [DOI] [PubMed] [Google Scholar]
- (44).Jewell NP. Statistics for Epidemiology. 1 ed Chapman and Hall; 2003. [Google Scholar]
- (45).Genkin A, Lewis DD, Madigan D. Large-Scale Bayesian Logistic Regression for Text Categorization. Technometrics. 2007;49(3):291–304. [Google Scholar]
- (46).Caster O, Noren GN, Madigan D, Bate A. Large-scale regression-based pattern discovery: The example of screening the WHO global drug safety database. Statistical Analy Data Mining. 2010;3(4):197–208. [Google Scholar]
- (47).Solomon R, DuMouchel W. Contrast media and nephropathy: findings from systematic analysis and Food and Drug Administration reports of adverse effects. Invest Radiol. 2006 Aug;41(8):651–60. doi: 10.1097/01.rli.0000229742.54589.7b. [DOI] [PubMed] [Google Scholar]
- (48).Agrawal R, Imielinski T, Swami A. Mining association rules between sets of items in large databases. 1993. pp. 207–216. SIGMOD.
- (49).Rouane H, Toussaint Y, Valtchev P. AIME 2009. Springer; Berlin/Heidelberg: 2009. Mining signals in spontaneous reports database using concept analysis. [Google Scholar]
- (50).Harpaz R, Chase HS, Friedman C. Mining multi-item drug adverse effect associations in spontaneous reporting systems. BMC Bioinformatics. 2010;11(Suppl 9):S7. doi: 10.1186/1471-2105-11-S9-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).McCormick TH, Rudin C, Madigan D. Bayesian Hierarchical Rule Modeling for Predicting Medical Conditions. Annals of Applied Statistics. 2012 (in press) [Google Scholar]
- (52).Fan K, Sun X, Tao Y, et al. High-Performance Signal Detection for Adverse Drug Events using MapReduce Paradigm. AMIA Annu Symp Proc. 2010;2010:902–6. [PMC free article] [PubMed] [Google Scholar]
- (53).Harpaz R, Perez H, Chase HS, Rabadan R, Hripcsak G, Friedman C. Biclustering of adverse drug events in the FDA’s spontaneous reporting system. Clin Pharmacol Ther. 2011 Feb;89(2):243–50. doi: 10.1038/clpt.2010.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Ball R, Botsis T. Can network analysis improve pattern recognition among adverse events following immunization reported to VAERS? Clin Pharmacol Ther. 2011 Aug;90(2):271–8. doi: 10.1038/clpt.2011.119. [DOI] [PubMed] [Google Scholar]
- (55).Stang PE, Ryan PB, Dusetzina SB, et al. Health Outcomes of Interest in Observational Data: Issues in Identifying Definitions in the Literature. Health Outcomes Research in Medicine. 2012 Feb;3(1):e37–e44. [Google Scholar]
- (56).Zorych I, Madigan D, Ryan P, Bate A. Disproportionality methods for pharmacovigilance in longitudinal observational databases. Stat Methods Med Res. 2011 Aug 30; doi: 10.1177/0962280211403602. [DOI] [PubMed] [Google Scholar]
- (57).Schuemie MJ. Methods for drug safety signal detection in longitudinal observational databases: LGPS and LEOPARD. Pharmacoepidemiol Drug Saf. 2011 Mar;20(3):292–9. doi: 10.1002/pds.2051. [DOI] [PubMed] [Google Scholar]
- (58).Noren GN, Hopstadius J, Bate A, Edwards IR. Safety surveillance of longitudinal databases: methodological considerations. Pharmacoepidemiol Drug Saf. 2011 Jul;20(7):714–7. doi: 10.1002/pds.2151. [DOI] [PubMed] [Google Scholar]
- (59).D’Agostino RB, Jr., D’Agostino RB., Sr. Estimating treatment effects using observational data. JAMA. 2007 Jan 17;297(3):314–6. doi: 10.1001/jama.297.3.314. [DOI] [PubMed] [Google Scholar]
- (60).Patrick AR, Schneeweiss S, Brookhart MA, et al. The implications of propensity score variable selection strategies in pharmacoepidemiology: an empirical illustration. Pharmacoepidemiol Drug Saf. 2011 Jun;20(6):551–9. doi: 10.1002/pds.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Schneeweiss S, Rassen JA, Glynn RJ, Avorn J, Mogun H, Brookhart MA. High-dimensional propensity score adjustment in studies of treatment effects using health care claims data. Epidemiology. 2009;20(4):512–22. doi: 10.1097/EDE.0b013e3181a663cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Bloom MS, Schisterman EF, Hediger ML. The use and misuse of matching in case-control studies: the example of polycystic ovary syndrome. Fertil Steril. 2007 Sep;88(3):707–10. doi: 10.1016/j.fertnstert.2006.11.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Madigan D, Ryan P, Simpson SE, Zorych I. Bayesian methods in pharmacovigilance (with discussion) In: Bernardo JM, Bayarri MJ, Berger JO, Dawid AP, Heckerman D, Smith AFM, West M, editors. Bayesian Statistics 9. Oxford University Press; 2010. pp. 421–438. [Google Scholar]
- (64).Madigan D, Ryan P. What can we really learn from observational studies?: the need for empirical assessment of methodology for active drug safety surveillance and comparative effectiveness research. Epidemiology. 2011 Sep;22(5):629–31. doi: 10.1097/EDE.0b013e318228ca1d. [DOI] [PubMed] [Google Scholar]
- (65).Gagne JJ, Fireman B, Ryan PB, et al. Design considerations in an active medical product safety monitoring system. Pharmacoepidemiol Drug Saf. 2012 Jan;21(Suppl 1):32–40. doi: 10.1002/pds.2316. [DOI] [PubMed] [Google Scholar]
- (66).Vilar S, Harpaz R, Chase HS, Costanzi S, Rabadan R, Friedman C. Facilitating adverse drug event detection in pharmacovigilance databases using molecular structure similarity: application to rhabdomyolysis. J Am Med Inform Assoc. 2011 Dec;18(Suppl 1):i73–i80. doi: 10.1136/amiajnl-2011-000417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Wicks P, Vaughan TE, Massagli MP, Heywood J. Accelerated clinical discovery using self-reported patient data collected online and a patient-matching algorithm. Nat Biotech. 2011 May;29(5):411–4. doi: 10.1038/nbt.1837. [DOI] [PubMed] [Google Scholar]
- (68).Cami A, Arnold A, Manzi S, Reis B. Predicting adverse drug events using pharmacological network models. Sci Transl Med. 2011 Dec 21;3(114):114ra127. doi: 10.1126/scitranslmed.3002774. [DOI] [PubMed] [Google Scholar]
- (69).Tatonetti NP, Denny JC, Murphy SN, et al. Detecting drug interactions from adverse-event reports: interaction between paroxetine and pravastatin increases blood glucose levels. Clin Pharmacol Ther. 2011 Jul;90(1):133–42. doi: 10.1038/clpt.2011.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Gibbons RD, Amatya AK, Brown CH, et al. Post-approval drug safety surveillance. Annu Rev Public Health. 2010 Apr 21;31:419–37. doi: 10.1146/annurev.publhealth.012809.103649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Becquemont L. Pharmacogenomics of adverse drug reactions: practical applications and perspectives. Pharmacogenomics. 2009 Jun;10(6):961–9. doi: 10.2217/pgs.09.37. [DOI] [PubMed] [Google Scholar]
- (72). [accessed Feb 2012];The Pharmacogenomics Knowledge Base (PharmGKB) http://www.pharmgkb.org/
- (73).Hochberg AM, Hauben M, Pearson RK, et al. An evaluation of three signal-detection algorithms using a highly inclusive reference event database. Drug Saf. 2009;32(6):509–25. doi: 10.2165/00002018-200932060-00007. [DOI] [PubMed] [Google Scholar]
- (74).Musen MA, Noy NF, Shah NH, et al. The National Center for Biomedical Ontology. J Am Med Inform Assoc. 2011 Nov 10; doi: 10.1136/amiajnl-2011-000523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Berry SM, Berry DA. Accounting for multiplicities in assessing drug safety: a three-level hierarchical mixture model. Biometrics. 2004 Jun;60(2):418–26. doi: 10.1111/j.0006-341X.2004.00186.x. [DOI] [PubMed] [Google Scholar]


