Abstract
The innate immune response (IIR) is a coordinated intracellular signaling network activated by the presence of pathogen-associated molecular patterns that limits pathogen spread and induces adaptive immunity. Although the precise temporal activation of the various arms of the IIR is a critical factor in the outcome of a disease, currently there are no quantitative multiplex methods for its measurement. In this study, we investigate the temporal activation pattern of the IIR in response to intracellular double-stranded RNA stimulation using a quantitative 10-plex stable isotope dilution–selected reaction monitoring–MS assay. We were able to observe rapid activation of both NF-κB and IRF3 signaling arms, with IRF3 demonstrating a transient response, whereas NF-κB underwent a delayed secondary amplification phase. Our measurements of the NF-κB-IκBα negative feedback loop indicate that about 20% of IκBα in the unstimulated cell is located within the nucleus and represents a population that is rapidly degraded in response to double-stranded RNA. Later in the time course of stimulation, the nuclear IκBα pool is repopulated first prior to its cytoplasmic accumulation. Examination of the IRF3 pathway components shows that double-stranded RNA induces initial consumption of the RIG-I PRR and the IRF3 kinase (TBK1). Stable isotope dilution–selected reaction monitoring–MS measurements after siRNA-mediated IRF3 or RelA knockdown suggests that a low nuclear threshold of NF-κB is required for inducing target gene expression, and that there is cross-inhibition of the NF-κB and IRF3 signaling arms. Finally, we were able to measure delayed noncanonical NF-κB activation by quantifying the abundance of the processed (52 kDa) NF-κB2 subunit in the nucleus. We conclude that quantitative proteomics measurement of the individual signaling arms of the IIR in response to system perturbations is significantly enabled by stable isotope dilution–selected reaction monitoring–MS-based quantification, and that this technique will reveal novel insights into the dynamics and connectivity of the IIR.
The innate immune response (IIR)1 is the major initial host signaling pathway activated in response to pathogen infection. It is triggered when invading microorganisms are identified by pattern recognition receptors (PRRs), a class of receptors that includes the plasma membrane Toll-like receptors and the cytoplasmic RIG-I-like RNA helicases, which are activated upon binding pathogen-associated molecular patterns. This event triggers a coupled intracellular serine kinase–ubiquitin ligase cascade that produces protein complex formation, proteolysis of inhibitory proteins, and nuclear translocation of transcriptional effectors (1). The primary transcription effectors of the IIR are nuclear factor (NF)-κB, IFN regulatory factor (IRF), and signal transducer and activator of transcription. Upon post-translational modifications, these effectors in turn activate the expression of inflammatory cytokine and type I interferon genes (2). These genes encode paracrine factors that limit pathogen replication and activate the adaptive immune response.
Molecular and biochemical analyses have indicated that the transcriptional effectors of the IIR are under control of a network of inter-related pathways. The NF-κB pathway represents two physically distinct signaling arms, referred to as the “canonical” and “noncanonical” NF-κB pathways. Mammalian cells contain NF-κB family members RelA/p65, c-Rel, RelB, p105/p50, and p100/p52, all of which form homo- or hetero-dimeric inducible cytoplasmic DNA complexes tethered in the cytoplasm by association with IκB inhibitors (3). The well-known canonical NF-κB pathway is initiated by either activated TNF superfamily receptors or the cytoplasmic PRR RIG-I that converges on IκB kinase (IKK), the rate-limiting kinase controlling IκBα degradation. Importantly, IKK activation absolutely requires the IKKγ regulatory subunit (4–7), which mediates its oligomerization and recruitment to the activated TNF receptor, or the activated RIG-I·MAVS complex (8, 9). By contrast, the noncanonical NF-κB pathway is activated by RIG-I in an IKKγ-independent manner to converge on the NF-κB-inducing kinase (NIK)–IKKα complex that initiates NF-κB2/p100 processing to liberate both sequestered RelB/p52 (10, 11) and RelA complexes from cytoplasmic NF-κB2/p100 precursor. This latter arm has been termed the “cross-talk” pathway (11), because it is dependent on noncanonical NIK-IKKα kinases but liberates the canonical RelA transcriptional activator.
The IRF family of transcription factors are a distinct effector arm of the IIR that controls type I IFN expression in response to viral patterns. So far, nine human IRFs have been reported (IRF1–9); of these, IRF3, -1, and -7 are the key regulators of type 1 IFN gene expression (12, 13). IRF3 is the major early signaling protein constitutively expressed in the cytoplasm. The molecular mechanisms by which RIG-I·MAVS activates IRF3 signaling are partly understood. Here, RIG-I binds to short- or 5′-phosphorylated double-stranded (ds)-RNA, inducing its ubiquitylation and complex formation with the mitochondrial antiviral signaling protein (MAVS). RIG-I-complexed MAVS recruits TRAF3, a signal adapter that binds to specific motifs on MAVS (14). The MAVS–TRAF3 complex in turn mediates activation of tank binding kinase (TBK)-1-IKKε/ι, a complex representing the major rate-limiting kinases of IRF3 activation (15). Phosphorylation of IRF3 on COOH-terminal domain serine residues allows it to homodimerize, associate with p300/CBP, and translocate into the nucleus (16). In this manner, phospho-IRF3 activates target genes containing interferon-stimulated response elements, such as IFNβ/IFNα4, and induces downstream IRF-1 and -7. IRF-1 and -7 induce other type I IFNs, whose actions induce antiviral responses in neighboring cells (17).
Presently, we understand that the arms of the IIR may vary in response to different cues and fluctuate over time following stimulation. Understanding the IIR at the systems level will result in novel insight into the mechanisms underlying host defense, pathogen–host interaction, and cellular stress response. However, quantification of this response is extremely challenging (18, 19). Firstly, the major regulatory components of the IIR, consisting of PRRs, kinases, and transcription factors, are all low-abundance proteins. Secondly, the pathogen-inducible post-translational modifications are labile and usually present at sub-stoichiometric amounts, and few high-affinity detection reagents are available that can detect them in complex cellular samples. Finally, identifying the dynamic nature of the IIR requires precise quantification of each component and its post-translational modifications over time. Traditional immunoassays such as Western blotting are not suitable for precise quantification. Instead, high-accuracy, high-sensitivity multiplex assays are required.
Advances in mass spectrometry (MS) technologies have enhanced our understanding of signaling networks at the systems level (20–24). Classical “shotgun” proteomics combined with stable isotope labeling and/or spectral counting is limited by the fact that this approach samples only a fraction of the proteome and is usually biased toward higher abundance proteins. The poor reproducibility of target selection results in the identification of only partially overlapping sets of proteins from substantially similar samples (25). Many important low-abundance proteins usually cannot be consistently identified across samples. Such fragmentary data sets are not satisfactory for understanding the signaling pathway at the systems level, for which a complete quantification profile for each node of the pathway is required. Therefore, new approaches that can deliver consistent, precise quantification data from pre-defined sets of proteins across multiple samples are needed. Recently, selected reaction monitoring (SRM) has been developed as a new “targeted” MS approach for the detection and accurate quantification of a predetermined set of proteins in a complex background (25–27). In an SRM-MS assay, one or two signature proteotypic peptides unique to the protein of interest are selected to represent the protein. SRM-MS quantification of these signature peptides is performed on a triple quadrupole mass spectrometer, an instrument with the capability to selectively isolate precursor ions corresponding to the signature peptide m/z value and to monitor peptide-specific fragment ions that provide near-absolute structural specificity for the target protein. SRM-MS assays have multiplexing capability. Hundreds to thousands of precursor-product ion transitions can be monitored in SRM mode over one liquid chromatography run, allowing the simultaneous quantification of all of the components in a signaling network in parallel. Specifically, SRM-MS has been used for studying the dynamics of signaling pathways (28, 29).
In this study, we carried out a quantitative proteomics analysis aimed at studying the dynamics of IIR in response to ds-RNA stimulation. We developed stable isotope dilution (SID)-SRM-MS assays for 10 proteins that are major regulators of IIR. We then used these assays to reproducibly measure the concentrations of these proteins in cytoplasmic and nuclear compartments of A549 cells over a time course of ds-RNA stimulation. These results reveal, for the first time, the presence of a second NF-κB activation pathway, the existence of a negative NF-κB-IRF3 cross-talk pathway, and the consumption of RIG-I-TBK1 in IRF3 signaling.
EXPERIMENTAL PROCEDURES
Reagents and Chemicals
All reagents were American Chemical Society grade or higher. All solvents used, including water, methanol, and acetonitrile (ACN), were LC/MS grade. Sequence-grade modified trypsin was purchased from Promega (Madison, WI). Recombinant TNFα was from PeproTek (Rocky Hill, NJ). The antibodies used were rabbit anti-NF-κB/RelA (sc-372) and IRF3 (sc-9082) Abs from Santa Cruz Biotechnology (Santa Cruz, CA). The recombinant proteins full-length human GST-tagged IKKγ and GST-tagged IκBα were made by our laboratory. GST-tagged human NFκB2 (1–454 amino acid), N-terminal proprietary tagged human IKKα, His-tagged human MAVS, GST-tagged human TBK1, and recombinant human Rela (1–537 amino acid) were purchased from Creative Biomart (Shirley, NY). GST-tagged full-length human IRF3 was purchased from Abnova (Walnut, CA), and C-terminal MYC/DDK-tagged human RIG-I protein was purchased from Origene (Rockville, MD).
Cell Culture
Human A549 pulmonary epithelial cells (American Type Culture Collection) were grown in F12K medium (Invitrogen) with 10% fetal bovine serum, penicillin (100 U/ml), and streptomycin (100 g/ml) at 37 °C in a 5% CO2 incubator.
ds-RNA Treatment
Duplex RNA was synthesized using a fragment of human β-tubulin cDNA as a template flanked by T7 RNA polymerase binding sites (30). Duplex RNA was generated in vitro using T7 RNA polymerase (Megascript, Ambion, Inc., Austin, TX). The 300 nt ds-RNA was purified according to the manufacturer's recommendation; 4 μg was transfected into 2 × 106 freshly isolated A549 cells in suspension as described elsewhere (6, 30).
siRNA Knockdown Experiments
RelA and IRF3 gene-specific siRNAs were reverse-transfected at a 100-nm concentration into A549 cells using TransIT-siQUEST transfection reagent (Mirus Bio Corp., Madison, WI) as described elsewhere (30). Transfection was performed 64 h before dsRNA stimulation.
Confocal Microscopy
Dynamic live cell imaging of EGFP-RelA/monomeric Strawberry-IRF3-stably transfected A549 cells was performed using a Zeiss LSM-510 META confocal microscope with a 63 × 1.4 numerical aperture oil immersion objective with an incubation system in which cells were stably maintained at 37 °C with humidified 5% CO2. Samples were excited using the 488-nm line at 1% of transmission, and emission was collected with a low-pass 505-nm filter. The time lapse images for each sample were acquired at 6-min intervals. Focus was automatically corrected before each time point by a customized Zeiss Multitime autofocus macro. Cell images were segmented, quantified, and tracked by a custom-written pipeline program using CellTracker as described elsewhere (6).
Quantitative Real Time PCR
Quantitative real time PCR (Q-RT-PCR) assays were performed as described elsewhere (30). Briefly, 1 μg of total RNA was reverse-transcribed using random hexamers in a 20-μl reaction mixture. One microliter of cDNA product was amplified in SYBR Green Supermix (Bio-Rad) and 0.4 μm each of forward and reverse gene-specific primers. The reactions were denatured for 90 s at 95 °C and then subjected to 40 cycles of 15 s at 94 °C, 60 s at 60 °C, and 1 min at 72 °C in an iCycler (Bio-Rad). PCR products were subjected to melting curve analysis to ensure that a single amplification product was produced.
Relative changes in gene expression were calculated using the ΔΔCt method, whereby the product was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) for each sample and the fold difference between experimental and control samples was calculated using the formula 2(−ΔΔCt). Primers for the mRNA expression of RelA, IRF3, RIG-I, IKKγ, TNFAIP3/A20, NFKBIA/IκBα, IL8, IL6, IFNβ, and CCL5/RANTES were described previously (30). For hISG15 (GenBank Accession No. NM 005101), the forward primer was 5′-CGCAGATCACCCAGAAGATCG-3′, and the reverse primer was 5′- TTCGTCGCATTTGTCCACCA-3′.
Western Blot
Cytoplasmic or nuclear extracts from a constant number of cells were boiled in Laemmli buffer, fractionated on 10% SDS-PAGE, and electro-transferred to polyvinylidene difluoride membranes (Millipore, Bedford, MA). Membranes were blocked in 5% milk/Tris-buffered saline (TBS) with 0.1% Tween for 1 h and immunoblotted with the indicated primary antibody for 1 h at 4 °C (Santa Cruz Biotechnology, Santa Cruz, CA). Membranes were washed four times in TBS with 0.1% Tween. Immune complexes were detected by Infrared dye (IRD)-labeled goat-anti-rabbit antibody and imaged in an Odyssey Infrared Imager (LiCor, Lincoln, NE).
Signature Peptide Selection
The selection of high-responding peptides was performed using a workflow developed previously (31). Briefly, if a lab-based discovery experiment had been conducted on a target protein, the MS response of sequence-identified proteotypic peptides of the target proteins was calculated from the extracted ion chromatogram of the monoisotopic peak of each peptide. The peptides with the highest MS responses were selected as high-responding signature peptides. For proteins that had not been observed in previous discovery experiments, an approach combining data mining of public databases and computational prediction was used for selecting the candidate high-responding signature peptides. The protein of interest was in silico digested with trypsin. The tryptic peptides were filtered applying the following criteria: the proteotypic peptides were unique to the target protein; the length was between 6 and 25 residues; and peptides with missed cleavages, chemically active amino acids (such as cysteine or methionine), Lys (K)-Pro (P) and Arg (R)-Pro (P), or two neighboring basic amino acids (K,R) at either cleavage site of the peptide sequence were excluded (25). The candidate peptides passing these initial filters were searched within the Global Proteome Machine Database (32) to identify the number of observations in the database. The sensitivity of peptide MS responses was computationally predicted using ESPPredictor (33), and the Krokhin hydrophobicity factor was calculated using Thermo Scientific Pinpoint™ software. Peptides were eliminated that had (i) a low enhanced signature peptide score, (ii) a low number of Global Proteome Machine Database observations, and/or (iii) an extremely low (10) or high (<40) Krokhin hydrophobicity factor. The remaining peptides were considered as signature peptide candidates and chemically synthesized in stable isotope labeled, crude, and unpurified forms for selecting SRM precursor-product ion (Q1/Q3) transitions and evaluating SRM assay specificity.
The stable isotope labeled, crude, unpurified peptides were infused into a TSQ Vantage triple quadrupole mass spectrometer via a nanospray source and analyzed by means of SRM-triggered MS/MS. The three or four highest intensity y ions in MS/MS were selected to generate the SRM Q1/Q3 transition. The collision energy (CE) breakdown curve of each Q1/Q3 transition was acquired so as to select the optimal CE. The S-lens voltage breakdown curve of each precursor ion was acquired in the same fashion.
The mixture containing stable isotope labeled, crude, and unpurified peptides was then spiked into a tryptic digest of A549 cell extract. The endogenous peptides of target proteins and stable isotope labeled peptides were analyzed via LC-SRM-MS. The following four analyte-specific criteria were used for evaluating the specificity of each SRM assay: precursor ion m/z, product ion m/z, chromatographic retention time, and relative product ion intensities. The MS raw data were manually examined. The analyte peptides and their stable isotopically labeled peptide standard (SIS) analogs that had same chromatographic retention times (variance below 0.05 min) and relative product ion intensities (±20% variance in the relative ratios for each fragment) were considered as high-specificity signature peptides that were free of matrix interference from co-eluting ion. These peptides were chosen as the high-responding signature peptides of the target protein.
Synthesis of Isotopically Labeled Pepscreen® Peptides
SISs were custom-synthesized by Sigma-Aldrich (St. Louis, MO). Isotopically labeled Pepscreen® peptides were synthesized using isotopically labeled amino acids, [13C615N4]Arg or [13C615N4]Lys (98% isotopic enrichment). The purity of peptides was determined with HPLC, electrospray ionization MS, and MALDI-TOF-MS as described above. Peptides were used if their desired molecular weight was one of the three major ions determined via MALDI-TOF-MS.
Synthesis of Isotopically Labeled Peptides
SISs were custom-synthesized by Sigma (St. Louis, MO). SIS peptides were synthesized using isotopically labeled amino acid, [13C615N4] Arg (98% isotopic enrichment). The peptides were HPLC-purified and were stringently tested to ensure high purity (>98%). The accurate molecular weights of SIS peptides were measured via electrospray ionization MS and MALDI-TOF-MS, and the specific peptide content was determined by means of amino acid analysis.
Synthesis of Native Tryptic Peptides
The native tryptic peptides were synthesized using Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry. The peptides were purified via reversed-phase HPLC. The accurate molecular weights of the peptides were measured with MALDI-TOF-MS, and the specific peptide content was determined by means of amino acid analysis.
Subcellular Fractionation of A549 Cells and Protein Extraction
A549 cells were scraped and subjected to hypotonic buffer/detergent lysis (34). Briefly, A549 cells were scraped from plates and washed in phosphate-buffered saline twice via centrifugation. The cell pellet was suspended in cold hypotonic buffer A (50 mm HEPES (pH 7.9), 10 mm KCl, 1 mm EDTA, 1 mm EGTA, 1 mm dithiothreitol (DTT), and 10 μl/ml of protease inhibitor mixture (Sigma-Aldrich Chemical Co., St. Louis, MO)) and 0.1% IGEPAL CA-630 and incubated on ice for 10 min (9). The lysates were centrifuged at 6000 rpm (4000 × g) for 30 s at 4 °C. The supernatant (cytoplasmic fraction) was saved, and the pellet (containing the nuclei) was resuspended in buffer B (buffer A containing 1.0 m sucrose) and centrifuged at 12,000 rpm (15,000 × g) for 10 min at 4 °C. The supernatant was discarded, and the pellet was incubated in buffer C (50 mm HEPES (pH 7.9), 10% glycerol, 400 mm KCl, 1 mm EDTA, 1 mm EGTA, 1 mm DTT, 0.1 mm PMSF, and protease inhibitor mixture) with frequent shaking for 30 min at 4 °C. After centrifugation at 15,000 × g for 10 min at 4 °C, the supernatant (nuclear extract) was saved at −80 °C. The protein content of both cytoplasmic and nuclear extracts was measured via Coomassie Brilliant Blue staining using bovine serum albumin as a standard (Bio-Rad, Hercules, CA).
Trypsin Digestion of Subcellular Protein Extraction and Addition of Internal Standard
The protein extracts were denatured with 8 m guanidinium HCl, and 50 μg of protein of each sample was reduced with 10 mm of DTT for 30 min at room temperature. Protein cysteinyl residues were alkylated with 30 mm of iodoacetamide for 2 h at 37 °C. The samples were diluted 10 times with 100 mm ammonium bicarbonate and digested with 2 μg of trypsin at 37 °C for 24 h. The trypsin digestion was stopped by the addition of 10% TFA to reduce the pH of the solution to 2.5. The volume of each sample was adjusted with water to 400 μl. The concentration of the resulting tryptic peptides of each sample was 0.125 μg/μl. The tryptic digests were dried and resuspended in 5% formic acid–0.01% TFA prior to LC-SRM-MS analysis.
Individual SIS peptide stocks of 20 pmol/μl were made in 80% ACN. A substock solution of SIS peptides was diluted to 10 fmol/μl with 0.01% TFA. Before LC-SRM-MS analysis, 30 μl of each tryptic digest was mixed with 10 μl of the substock SIS peptide solution. The peptide mixture was desalted with a C18 cartridge (Waters Corp., Milford, MA) following the manufacturer's instructions, and the peptides were eluted from the cartridge with 80% ACN and dried.
Strong Cation Exchange Chromatography
Dried peptide samples were redissolved in 20 μl of ACN and diluted with 100 μl of 5 mm ammonium formate, pH 2.7. The peptide mixture was loaded onto a strong cation exchange column (Poly-SULFOETHYL A™, 5μ, 200 Å, 0.30 × 150 mm, Poly LC, Columbia, MD) and washed with 10 min of isocratic buffer A (5 mm ammonium formate–20% ACN, pH 2.7), and peptides were eluted with a linear gradient from 100% buffer A to 40% buffer B (1 m ammonium formate–20% ACN, pH 3.0) over 20 min at a flow rate of 0.4 ml/min, followed by 5 min of isocratic 40% buffer B. Sixty fractions were collected and dried in a Speedvac. Before LC-SRM-MS analysis, each strong cation exchange fraction was reconstituted in 30 μl of 5% formic acid–0.01% TFA.
LC-SRM-MS Analysis
The LC-SRM-MS analysis was performed with a TSQ Vantage triple quadrupole mass spectrometer equipped with a nanospray source (Thermo Scientific, San Jose, CA). The online desalting and chromatography were performed using an Eksigent NanoLC-2D HPLC system (AB SCIEX, Dublin, CA). Aliquots of 10 μl of each tryptic digest were injected onto a C18 peptide trap (Agilent, Santa Clara, CA) and desalted with 0.1% formic acid at a flow rate of 2 μl/min for 45 min. Peptides were eluted from the trap and separated on a reversed-phase nano-HPLC column (PicoFrit™, 75 μm × 10 cm; tip inner diameter = 15 μm) packed in-house using Zorbax SB-C18 (5-μm diameter particles, Agilent, Santa Clara, CA). Separations were performed using a flow rate of 500 nl/min with a 20-min linear gradient from 2% to 40% mobile phase B (0.1% formic acid–90% ACN) in mobile phase A (0.1% formic acid), followed by a 0.1-min gradient from 40% to 90% mobile phase B and 5 min at 90% mobile phase B. The TSQ Vantage was operated in high-resolution SRM mode with Q1 and Q3 set to 0.2 and 0.7 Da full width at half-maximum. All acquisition methods used the following parameters: 1800 V ion spray voltage, a 275 °C ion transferring tube temperature, a collision-activated dissociation pressure of 1.5 mTorr, and an S-lens voltage using the values in the S-lens table generated during MS calibration. Each sample was analyzed via LC-SRM-MS twice. The MS raw files will be made available on the website of the Sealy Center for Molecular Medicine.
SRM Data Analysis
All data were manually inspected to ensure peak detection and accurate integration. The chromatographic retention times and the relative product ion intensities of the analyte peptides were compared with those of the SIS peptides. The variation of the retention time between the analyte peptides and their SIS counterparts was within 0.05 min, and no significant differences in the relative product ion intensities of the analyte peptides and SIS peptides were observed. The peak area in the extract ion chromatography of the native and SIS versions of each signature peptide were integrated using Xcalibur® 2.1. The default values for noise percentage and base-line subtraction window were used. The ratios between the peak areas of native and SIS versions of each peptide were calculated. Beta-actin was used as a loading control. All of the measured native versus SIS standard peptide ratios were normalized by beta-actin. The native versus SIS peptide ratios measured in the two replicate LC-SRM-MS analyses were averaged, and the mean and standard deviation were calculated.
RESULTS
Development of SRM Assays for Major Regulatory Components of IIR
In this study, we focused on understanding the kinetics of the two NF-κB pathways and the RIG-I-IRF3 pathway, the major signaling effectors of the IIR (supplemental Fig. S1). We previously developed a workflow for selecting high-responding signature peptides of low-abundance proteins (see “Experimental Procedures” and Ref. 31). Using this workflow, we selected the candidate high-responding signature peptides for 10 major regulatory components of the IIR (supplemental Fig. S1). The sensitivity of the assays was further improved by selecting the highest intensity y ions and optimizing the CE of each transition. The selected Q1/Q3 transitions and their optimized CE for signature peptides are displayed in Table I. A key feature of the SRM-MS assay is its high specificity for the target analyte. For each analyte, assay specificity was evaluated on a real sample (e.g. cytoplasmic or nuclear A549 cell extract). For this purpose, a mixture containing unpurified stable isotope labeled signature peptide candidates was spiked into a tryptic digest of A549 cell extract. The endogenous peptides of target proteins were analyzed via LC-SRM, using the stable isotope labeled peptides as a reference. The specificity of each SRM assay was evaluated based on the chromatographic retention time, precursor m/z, product ion m/z, and relative product ion intensities (“Experimental Procedures”). Among the IIR proteins shown in supplemental Fig. S1, the SRM assays of RelA, IRF3, RIG-I, MAVS, IKKα, IKKγ, IκBα, TBK1, and p100/p52 have adequate sensitivity and specificity for accurate quantification of the target proteins directly from cell extract. Here, the baseline separation of the analyzed peptides from the matrix interference was achieved. The native peptide had a chromatographic retention time that was nearly identical to that of its heavy analog, and the relative product ion intensities of the native peptide agreed precisely with the intensities of the product ions from the heavy peptide (supplemental Figs. S2 and S3). However, the assays of IKKβ, IKKε, and NIK were not sensitive enough for quantifying these three proteins directly from cell extract and require pre-fractionation enrichment (such as immunoprecipatation) for measurement.
Table I. SRM parameters of SRM assays of proteins for sample amount normalization.
| Gene name | Swissprot number | Sequence | Q1 (m/z) | Q3 (m/z) | Ion type | CE (V) | Linear range | R2 | Lower limit of quantitation (amol) |
|---|---|---|---|---|---|---|---|---|---|
| IKKa | O15111 | IQLPIIQLR | 547.358 | 529.345 | y4 | 26 | 20,000 | 0.9996 | 50 |
| 547.358 | 642.429 | y5 | 23 | ||||||
| 547.358 | 739.482 | y6 | 19 | ||||||
| 547.358 | 852.566 | y7 | 18 | ||||||
| MAVS | Q7Z434 | VSASTVPTDGSSR | 632.312 | 719.331 | y7 | 20 | 20,000 | 0.9995 | 250 |
| 632.312 | 818.400 | y8 | 16 | ||||||
| 632.312 | 919.447 | y9 | 17 | ||||||
| 632.312 | 1006.479 | y10 | 20 | ||||||
| RelA | Q04206 | TPPYADPSLQAPVR | 756.396 | 867.504 | y8 | 27 | 20,000 | 0.9990 | 250 |
| 756.396 | 982.531 | y9 | 27 | ||||||
| 756.396 | 1053.568 | y10 | 27 | ||||||
| 756.396 | 1313.684 | y12 | 30 | ||||||
| IRF3 | Q14653 | LVGSEVGDR | 466.242 | 446.235 | y4 | 24 | 100,000 | 0.9998 | 50 |
| 466.242 | 575.278 | y5 | 26 | ||||||
| 466.242 | 719.331 | y7 | 26 | ||||||
| RIG-I | O95786 | VVFFANQIPVYEQQK | 905.480 | 891.457 | y7 | 26 | 2,000 | 0.9989 | 2500 |
| 905.480 | 1004.541 | y8 | 24 | ||||||
| 905.480 | 1132.599 | y9 | 26 | ||||||
| 905.480 | 1246.642 | y10 | 27 | ||||||
| TBK1 | Q9UHD2 | TTEENPIFVVSR | 696.362 | 817.493 | y7 | 24 | 20,000 | 0.9988 | 250 |
| 696.362 | 931.535 | y8 | 24 | ||||||
| 696.362 | 1060.578 | y9 | 23 | ||||||
| 696.362 | 1189.621 | y10 | 23 | ||||||
| IKBA | P259633 | LEPQEVPR | 484.264 | 628.341 | y5 | 20 | 20,000 | 0.9992 | 50 |
| 484.264 | 725.394 | y6 | 20 | ||||||
| 484.264 | 854.436 | y7 | 20 | ||||||
| 484.264 | 967.520 | y8 | 20 | ||||||
| IKKR | Q9Y6K9 | AQVTSLLGELQESQSR | 873.455 | 976.469 | y8 | 27 | 10,000 | 0.9984 | 500 |
| 873.455 | 1033.490 | y9 | 27 | ||||||
| 873.455 | 1146.574 | y10 | 27 | ||||||
| 873.455 | 1259.658 | y11 | 27 | ||||||
| 873.455 | 1346.690 | y12 | 27 | ||||||
| p52/p100 | Q00653 | DSGEEAAEPSAPSR | 701.808 | 743.368 | y7 | 20 | 20,000 | 0.9976 | 250 |
| 701.808 | 814.405 | y8 | 20 | ||||||
| 701.808 | 885.442 | y9 | 20 | ||||||
| 701.808 | 1014.484 | y10 | 27 | ||||||
| p100 | Q00653 | ALLDYGVTADAR | 632.83 | 689.357 | y7 | 20 | 20,000 | 0.9985 | 250 |
| 632.83 | 852.421 | y8 | 20 | ||||||
| 632.83 | 967.447 | y9 | 20 | ||||||
| 632.83 | 1080.531 | y10 | 21 |
Notes: Masses listed are for the native forms of the peptides.
CE, collision energy; Q, quadropole; R2, coefficient of determination.
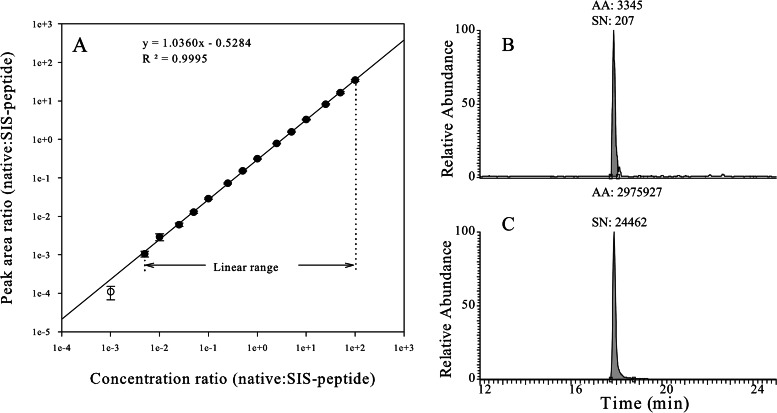
The assay dynamic range was assessed via the method of standard addition (35–40). All of the SRM assays yielded large concentration ranges with strong linear correlations (coefficient of determination R2 > 0.99). For example, the SRM assay of MAVS had a linear response over a 20,000-fold concentration range with a coefficient of determination R2 = 0.9995 (Fig. 1). We further ensured that any biological concentration encountered would fall well within the linear portion of the calibration curve. The lower limit of quantification of the SRM assay (i.e. the lowest analyte quantity that can be accurately measured) is the measurement of the assay sensitivity. The lower limit of quantification of the SRM assay can be defined as the lowest analyte concentration that can be measured with a <20% coefficient of variation (CV) (40–42). In our study, the lower limits of quantification of all [12C]-analyte peptides were within the sub-attomole to low-fentamole range. The linear dynamic ranges and lower limits of quantification of each SRM assay are listed in Table I.
Fig. 1.
Calibration curve for MAVS SID-SRM-MS assay. A stable isotope labeled [12C]-high-responding signature peptide of MAVS with its native flanking sequences (LTK-VSASTVPTDGSSR-NEE) was quantified via amino acid analysis and used as the calibrator to characterize the assay dynamic range. This peptide was trypsin digested in the same matrix as that used for the analysis of A549 cell protein extracts. We diluted the tryptic digest to generate a range of analyte concentrations spanning a 100,000-fold concentration range (from 50 amol to 5 pmol on column). These various analyte concentrations were combined with a constant amount of [13C]-analyte peptide internal standard. Four replicate LC-SRM-MS analyses of each sample dilution were performed in order from most diluted to most concentrated. Linear regression analysis was performed on the observed peak area ratios (native:heavy) versus the concentration ratios (native:heavy) to generate calibration curves. A, the linear regression analysis (1/x weighted) for MAVS illustrates the linear dynamic range of the assay. The error bars indicate the standard deviation of the measurements. B, extract ion chromatogram of the native peptide of MAVS (250 amol on column) in which the x-axis is the chromatographic retention time (min) and the y-axis is the extracted ion intensity. AA, area under the curve; SN, signal-to-noise ratio. C, extract ion chromatogram of the SIS peptide of MAVS.
Complete proteolysis of target proteins is critical for the interpretation of SID-SRM-MS assays (43). To evaluate the completeness of trypsin digestion of IIR target proteins, we digested a mix of recombinant IIR proteins with trypsin for 24 h (using the same conditions that we used for processing the cell extract). Afterward, the tryptic digest was separated via SDS-PAGE, and the proteins were visualized with SYPRO® Ruby staining. As shown in supplemental Fig. S4A, the protein bands disappeared after digestion, except for a very faint band at 20 kDa close to the dyefront. An in silico digest of RelA generated several very large tryptic peptides (>17 kDa), explaining the trypsin resistance of this low molecular weight fraction. The tryptic digest was further analyzed via LC-MS/MS, and no signature peptides with trypsin miscleavage sites were identified (not shown). We concluded from these SDS-PAGE and MS/MS analyses that the IIR proteins were completely trypsin digested.
Previous studies have shown that tryptic proteolysis of native proteins can follow distinct digestion routes, a phenomenon that results in variable rates of peptide production prior to reaching the “limit peptide” population (43). We therefore used SID-SRM-MS assays to monitor the kinetics of the release of signature peptides from the recombinant proteins during a time course of trypsin digestion. A mixture of recombinant IIR target proteins was denatured with guanidine HCl, reduced and alkylated (“Experimental Procedures”), and spiked with a mixture of SIS signature peptides of IRF3, IκBα, IKKγ, RelA, MAVS, IKKα, TBK1, RIG-I, and p100/p52. The sample was diluted and incubated with trypsin, and sequential time points were sampled for up to 30 h. With each aliquot, the proteolysis was terminated via the addition of trifluoroacetic acid and analyzed via SID-SRM-MS in duplicate. The native versus SIS peptide ratio of each signature peptide was calculated. As shown in supplemental Fig. S5, all of the signature peptides were rapidly released during the first 30 min of proteolysis and achieved plateau values within 5 h of reaction time, indicating that the signature peptides were fully released well within the reaction time. Moreover, a rapid release of signature peptides minimizes the inaccuracy of quantification caused by nonspecific degradation events (44).
Amino acid residues close to the cleavage site can affect proteolysis by changing the affinity of the endopeptidase for the substrate (43, 45). For example, acidic residues of two amino acids distal to the C-terminal cleavage site can slow trypsin hydrolysis and therefore might lead to incomplete digestion (43, 46, 47). To investigate the effect of amino acid residues close to the cleavage site on the completeness of trypsin digestion, we monitored the disappearance of the IIR signature peptides containing their native flanking sequences after trypsin digestion using SRM. A mixture of synthetic IIR signature peptides synthesized with three to four native amino acids flanking the tryptic cleavage site (e.g. MAVS signature peptide with its native flanking sequence, LTK-VSASTVPTDGSSR-NEE) was digested with trypsin using the same experimental conditions that were used for cell extract. As shown in supplemental Table S1, after 24 h of incubation, all of the IIR signature peptides with their native flanking sequences were digested to signature peptides. The completeness of trypsin digestion of each IIR signature peptide with its native flanking sequence was over 99%, indicating that the amino acid residues flanking the cleavage sites of the IIR signature peptides did not affect the tryptic digestion. Taken together, these experiments confirm the complete release of the signature peptide of IIR target proteins. Although we did not evaluate the release of p100 signature peptide from the recombinant NFκB2 protein because of a lack of the C-terminal segment of NFκB2 in the NFκB recombinant protein, we found that the release of p100 signature peptide (ALLDYGVTADAR) from the peptide with its native flanking sequence was 99.66%, indicating that p100 signature peptide can also be completely released under our experimental conditions.
SID-SRM-MS Analysis of the IIR in Response to ds-RNA Stimulation
ds-RNA is a viral pathogen pattern and represents a potent stimulus for the IIR because it engages three PRRs: TLR3, RIG-I, and MDA5 (48, 49). The engagement of PRRs initiates parallel signaling through TBK1/IKKε, IKKα/IKKβ, and NIK/IKKα kinases, leading to the activation of IRF3 and NF-κB effector pathways (50–52). Although the activation of NF-κB signaling (both canonical and noncanonical pathways) can be induced by cytokines and mitogens, IRF3 activation is selectively induced by PRRs responding to ds-RNA generated as a molecular pattern during virus infection (53). In order to investigate how the innate immune pathways fluctuate in response to ds-RNA stimulation, we transfected A549 cells with ds-RNA for varying times (0, 0.5, 1, 2, 4, and 6 h), and then the cells were fractionated into cytoplasmic and nuclear fractions. Nuclei were further purified using a sucrose step gradient centrifugation protocol (54, 55). The workflow for the analysis of IIR in response to ds-RNA stimulation is demonstrated in supplemental Fig. S6. The high quality of the nuclear preparation was demonstrated in our previous work using light microscopy and Western blot analysis of nuclear protein markers (34). In this study, we used SID-SRM-MS analysis to further assess the enrichment of the nuclear protein marker Lamin B. As shown in supplemental Fig. S7, an enrichment of over a 100-fold of Lamin B was achieved in nuclear fractions. Next, we used SID-SRM-MS to quantify the abundance of innate immune pathway components in cytoplasmic and nuclear compartments at each time point. To ensure that equivalent amounts of cytoplasmic and nuclear extract were analyzed at each time point, data were normalized on the basis of an invariant protein, beta-actin. A signature peptide of beta-actin was added to be quantified together with the target protein set in the multiplex SRM-MS assay. Normalization on the beta-actin was performed during the data analysis and yielded more reliable data.
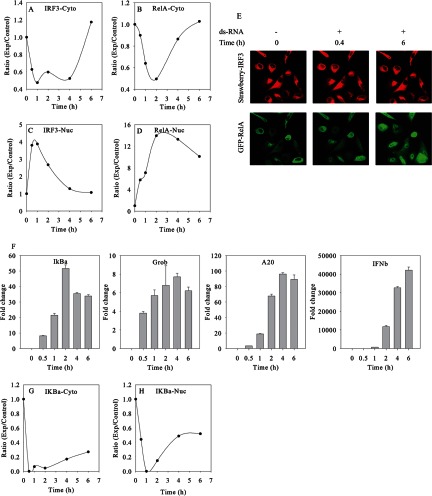
First, we focused on the IRF3 pathway and canonical NF-κB pathways. By this quantitative measure, amounts of IRF3 and RelA were increased in the nucleus after ds-RNA treatment. The accumulation of RelA and IRF3 was accompanied by the gradual depletion of cytoplasmic RelA and IRF3 (Figs. 2A–2D), suggesting that ds-RNA activated NF-κB and IRF3 pathways and induced RelA and IRF3 nuclear translocation.
Fig. 2.
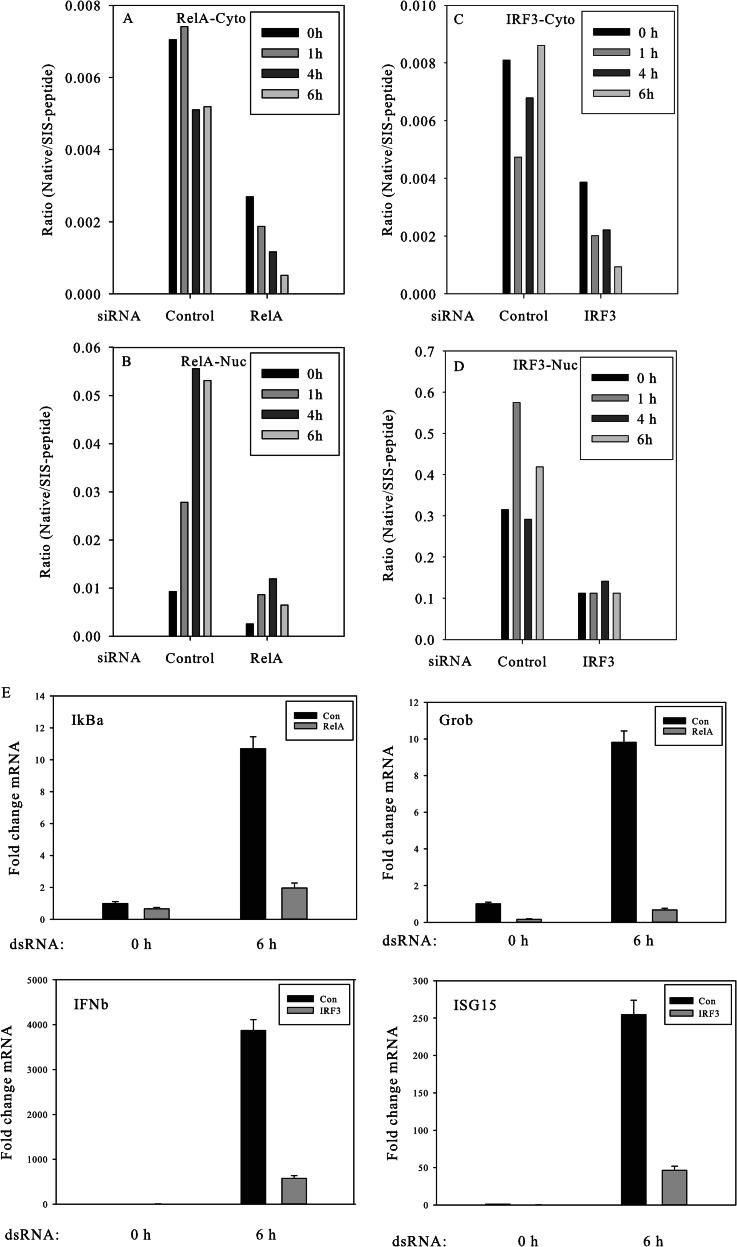
Dynamics of innate immune response to the stimulation of ds-RNA. The temporal profiles of cytoplasmic (Cyto) IRF3 (A) and nuclear (Nuc) IRF3 (C) and of cytoplasmic RelA (B) and nuclear RelA (D), derived from SID-SRM-MS analysis. E, Q-RT-PCR analysis of IRF3 and RelA downstream genes. Shown is the fold change in normalized mRNA relative to GAPDH. F, confocal microscopy of IRF3 and RelA. Shown are sequential images of the same field of double A549 transfectants expressing Strawberry-IRF3 and EGFP-RelA fusion proteins. The cells were transfected at time 0 with ds-RNA. Images of IRF3 (top) and EGFP (bottom) were taken at 0.4 and 6 h after ds-RNA exposure. G, the temporal profiles of cytoplasmic IκBα, derived from SID-SRM-MS analysis. H, the temporal profiles of nuclear IκBα derived from SID-SRM-MS analysis. Beta-actin was used to normalize between samples in SID-SRM-MS analysis.
Although both IRF3 and NF-κB pathways were rapidly activated by ds-RNA stimulation, our study revealed that IRF3 and RelA displayed different temporal nuclear translocation profiles (Figs. 2C and 2D). For example, IRF3 rapidly translocated into the nucleus and reached a peak value (3.8-fold change relative to basal level) within 0.5 h. The amount of nuclear IRF3 remained high until 1 h after stimulation and rapidly declined to the basal level thereafter. Relative to IRF3, although NF-κB could be detected as having increased at 0.5 h, the peak activation of NF-κB was delayed, stronger, and more prolonged. We observed the nuclear translocation of RelA at 0.5 h with a 5.7-fold change. The activation of NF-κB reached peak values at 2 h with a 14-fold increase and slowly declined after 4 h of stimulation. At the last measured time of 6 h, the amount of nuclear RelA was still about 10-fold higher than the basal level. We also imaged intact A549 cells expressing an enhanced green fluorescent protein (EGFP)-RelA fusion protein and Strawberry-red-tagged IRF3. The EGFP-tagged RelA and Strawberry-red-tagged IRF3 allowed us to monitor the cellular localization of RelA and IRF3 in response to ds-RNA stimulation by means of confocal microscopy. The cells were stimulated with ds-RNA and serially imaged. Shown are individual images at 0.4 h and 6 h after ds-RNA transfection. The levels of RelA and IRF3 were measured based on the fluorescent intensities of EGFP and Strawberry-red dye. As shown in upper panel of Fig. 2E, IRF3 rapidly accumulated into the nuclear fraction within 0.4 h and returned to basal levels after 6 h. By contrast, a minor degree of RelA nuclear translocation could be detected at 0.4 h and remained strong even after 6 h of stimulation (bottom panel of Fig. 2E). In all, the dynamic live-cell imaging of EGFP-RelA/monomeric Strawberry IRF3-stably transfected A549 cells was consistent with SRM-MS observations and further confirmed the differential dynamic profiles of IRF3 and NF-κB pathways in response to ds-RNA stimulation.
Once in the nucleus, RelA and IRF3 recognize the κB and IRF motifs present in the target genes, respectively, and regulate the expression of downstream genes. In order to determine whether ds-RNA activation of the canonical NF-κB pathway and IRF3 pathways results in the expression of distinct temporal waves of downstream genes, A549 cells were transfected with ds-RNA for varying times (0, 0.5, 1, 2, 4, and 6 h), and the total RNA was isolated for gene expression analysis via Q-RT-PCR. As shown in Fig. 2F, ds-RNA strongly induced the parallel expression of RelA-dependent genes NFKBIA/IκBα, CXLC2/Groβ, TNFAIP3/A20, and IFNβ with an initial increase in mRNA expression first detectable at 0.5 h. CXLC2/Groβ and TNFAIP3/A20 showed qualitatively similar profiles, with a gradual increase in mRNA expression until mRNA abundance peaked at 4 h, followed by a slight decrease. The induction of mRNA expression of NFKBIA/IκBα reached an apparent maximum after 2 h of ds-RNA stimulation and declined thereafter, despite the continued presence of nuclear RelA. These data indicate that members of the RelA-dependent gene network have a low threshold for activation of gene expression and do not exhibit an expression delay.
In contrast, the expression of IRF3-dependent IFNβ mRNA increased monotonically with an initial induction at 0.5 h, but a later, more potent induction over 6 h of over 40,000-fold. The dissociation between the IRF3 nuclear translocation profile and the expression of downstream IFNβ mRNA suggests that additional modifying factors control IRF3-dependent IFNβ expression, such as chromatin remodeling and the well-established IRF1 and IRF7 amplification loops.
The activation of the canonical NF-κB pathway by ds-RNA is well understood (56, 57). NF-κB is present in the cytoplasm as an inactive complex with its regulatory subunit IκB. Upon stimulation, IκBα undergoes phosphorylation by IκB kinases (IKKα and IKKβ), unbiquitination, and proteasome-mediated degradation, which result in the liberation of the NF-κB heterodimer, followed by its rapid nuclear translocation. As shown in Fig. 2G, the SID-SRM-MS analysis revealed that after stimulation of ds-RNA for 30 min, the amount of IκBα in cytoplasm was dramatically decreased, becoming virtually undetectable 30 min after induction with ds-RNA, which is consistent with the rapid degradation of IκBα and the initiation of RelA nuclear translocation. Once in the nucleus, RelA will bind and recruit co-activators to stimulate target gene expression. One of the RelA target genes of is IκBα. As shown in Fig. 2D, nuclear RelA increased 5.7-fold after 0.5 h, which induced an 8.1-fold increase in mRNA expression of IκBα (Fig. 2F). The de novo resynthesis of IκBα replenished the intracellular stores of IκBα. As shown in Fig. 2H, the cytoplasmic pool started to be replenished after 1 h and was maintained for up to 6 h after the stimulation of ds-RNA. However, the newly synthesized IκBα did not completely compensate for the ongoing degradation of IκBα in the presence of ds-RNA. We observed that only about one-fifth of the cytoplasmic pool of IκBα was restored. Besides its cytoplasmic expression, IκBα has also been reported to be localized in the nuclear compartment and displays nucleocytoplasmic shuttling properties in complex with NF-κB, where it exports NF-κB from the nucleus to the cytoplasm (58–60). Without stimulation, nuclear IκBα can be detected with Western blotting when the protein is overexpressed from a transfected vector or microinjected into the cytoplasm (58). The detection of basal levels of endogenous nuclear IκBα in unstimulated cells and the temporal profile of nuclear IκBα at the early stage of stimulation have not been documented. By using a more sensitive and accurate SID-SRM-MS assay, we found that at the basal level, about 20% of IκBα was located in the nucleus of A549 cells. After cell activation with ds-RNA, the amount of nuclear IκBα diminished gradually and became virtually undetectable 1 h later, perhaps because of ongoing proteolysis in the presence of ds-RNA stimulus. Unlike the cytoplasmic IκBα, the replenishment of nuclear IκBα progressed much more rapidly. After 6 h of stimulation of ds-RNA, the amount of nuclear IκBα was restored to half of its basal level. The recovery of nuclear IκBα will resume the translocation of nuclear NF-κB complexes from the nucleus to the cytoplasm, which will at least in part contribute to the decreased amount of RelA in the nucleus and the replenishment of the cytoplasmic pool of RelA.
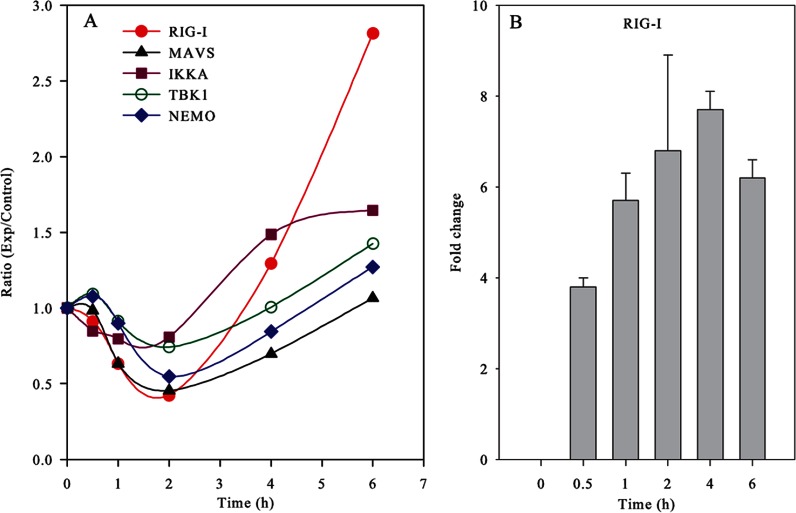
Activation of the IRF3 pathway involves the RIG-I·MAVS complex and is controlled by two IKK-related kinases, TBK1 and IKKε (15). Our recent study demonstrated that IRF3 activation also requires the participation of IKKγ as a signaling adapter (6). Using SID-SRM-MS, we quantified the temporal profiles of cytoplasmic IKKα, IKKγ, TBK1, MAVS, and RIG-I proteins (Fig. 3). All of these display similar quantitative behavior involving a decrease upon stimulation that is followed by the recovery of activity over a 6-h period. The decline of these proteins in cytoplasm during the early phase is probably caused by the inhibition of protein synthesis by ds-RNA, as ds-RNA is known as a potent inhibitor of protein synthesis in mammalian cells (61). After 2 h of ds-RNA activation, the cytoplasmic pools of these proteins were slowly replenished by protein re-synthesis. One dramatic example is RIG-I, the expression level of which decreased to less than half of the basal level at 2 h, followed by a steady increase. At the last measured time of 6 h, the expression level of RIG-I was increased 2.8-fold relative to the control sample. The up-regulation of RIG-I protein expression was consistent with the RIG-I mRNA expression profile (Fig. 3B).
Fig. 3.
Temporal profiles of cytoplasmic RIG-I, MAVS, IKKα, TBK1, and IKKγ. A, SID-SRM-MS analysis profiles of cytoplasmic RIG-I, MAVS, IKKα, TBK1, and IKKγ. Beta-actin was used to normalize between samples in SID-SRM-MS analysis. B, mRNA expression profiles of RIG-I; y-axis, fold change in mRNA relative to GAPDH.
The Effect of siRNA Knockdown of RelA and IRF3
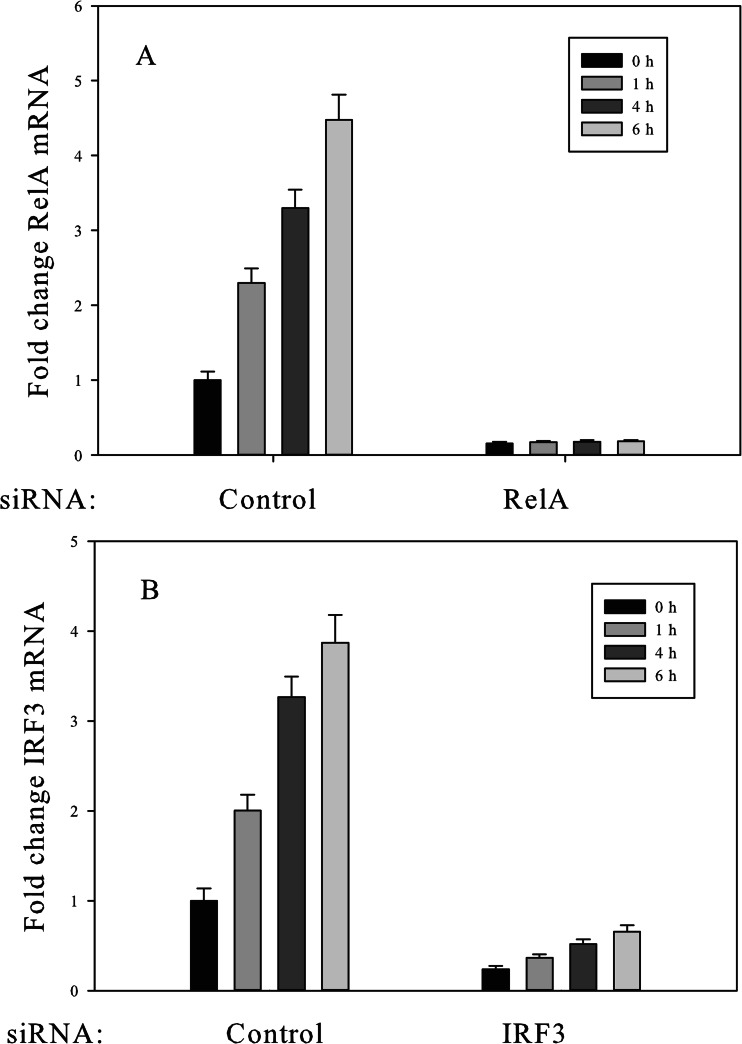
To investigate the interconnectivity of the NF-κB and IRF3 pathways, we used siRNA targeting RelA and IRF3 to inhibit the activation of NF-κB and IRF3 pathways, respectively. We transfected A549 cells with RelA siRNA and IRF3 siRNA and confirmed target gene down-regulation of RelA via Q-RT-PCR. As shown in Figs. 4A and 4B, the treatment of A549 cells with RelA siRNA resulted in a marked decrease in RelA mRNA, and, similarly, the treatment of A549 cells with IRF3 siRNA effectively knocked down IRF3 expression. We then used SID-SRM-MS to quantify the protein levels of RelA and IRF3 in A549 cells transfected with RelA siRNA, IRF3 siRNA, or control non-targeting siRNA. As shown in Figs. 5A and 5B, about 30% of RelA remained after 64 h of treatment with RelA siRNA, as measured via SID-SRM-MS, and about 40% of IRF3 remained after 64 h of treatment with IRF3 siRNA.
Fig. 4.
Suppression of RelA and IRF3 expression by siRNA knockdown. A549 cells were transfected with RelA siRNA, IRF3 siRNA, and control non-targeting siRNA for 64 h prior to ds-RNA stimulation. Cells were stimulated in the absence or presence of dsRNA for 1, 4, and 6 h. Non-targeting siRNA was used as a negative control siRNA. The total RNA was extracted and subjected to Q-RT-PCR to measure RelA mRNA abundance (A). A549 cells were transfected with IRF3 siRNA and analyzed for changes in IRF3 mRNA abundance (B).
Fig. 5.
Effect of IRF3 and RelA knockdown on the IIR. A549 cells were transfected with RelA siRNA, IRF3 siRNA, and non-targeting control siRNA for 64 h. Then the cells were treated with ds-RNA for varying times. The cytoplasmic and nuclear proteins were extracted and subjected to SID-SRM-MS analysis. The temporal profiles of cytoplasmic RelA (A) and nuclear RelA (B) in the cells transfected with RelA siRNA or non-targeting control siRNA derived from SID-SRM-MS analysis are shown. The temporal profiles of cytoplasmic IRF3 (C) and nuclear IRF3 (D) in the cells transfected with IRF3 siRNA or non-targeting control siRNA derived from SID-SRM-MS analysis also are shown. Beta-actin was used to normalize between samples for SID-SRM-MS analysis. E, inhibition of expression of downstream target genes by RelA siRNA and IRF3 siRNA. Total RNA in RelA siRNA-transfected A549 cells was assayed for NFKBIA/IκBα and Groβ expression by Q-RT-PCR in response to ds-RNA (6 h). Non-targeting siRNA was used as a negative control (top panel). Total RNA in IRF3 siRNA transfected cells was assayed for ISG15 and IFNβ expression in response to ds-RNA (bottom panel).
We next investigated whether the nuclear translocation of RelA or IRF3 and the induction of target genes were inhibited by the partial depletion of RelA or IRF3. The A549 cells were transfected with RelA siRNA, IRF3 siRNA, or control non-targeting siRNA for 64 h, and then the cells were treated with ds-RNA for varying times (0, 1, and 4 h). The levels of RelA and IRF3 in the nucleus were quantified via SID-SRM-MS. As shown in Fig. 5B, the basal abundance of RelA was significantly reduced in both the cytoplasmic and nuclear compartments in RelA siRNA-transfected cells. Similarly, ds-RNA stimulation induced only a minor amount of RelA nuclear translocation in the RelA siRNA-transfected cells. After the induction of ds-RNA for 4 h, the nuclear RelA increased about 4-fold, indicating that nuclear translocation of RelA was not completely inhibited by siRNA. However, relative to the cells transfected with control siRNA, the absolute amount of nuclear RelA was significantly lower in RelA siRNA-transfected cells. At the 4-h time point, the amount of nuclear RelA in RelA siRNA-treated cells was about the same as the basal level of nuclear RelA in the cells treated with control non-targeting siRNA. To confirm the functional effect of RelA siRNA knockdown on RelA-dependent genes, NFKBIA/IκBα and Groβ mRNA expression were measured in control or RelA siRNA-transfected cells. RelA siRNA produced significant inhibition of ds-RNA-induced IκBα and Groβ expression (Fig. 5E, top). This effect was specific because the expression of internal control β-actin and IRF3-dependent genes was not affected in the RelA silenced cells (not shown). Similar expression of IRF3-dependent genes was significantly inhibited in cells transfected with IRF3 siRNA (Fig. 5E, bottom panel). Here, the ds-RNA-induced expression of ISG15 and IFNβ was significantly inhibited. These data demonstrate that the expression level of downstream genes is controlled by the amount of RelA in the nucleus; in order to induce the expression of downstream genes, a critical threshold of nuclear RelA is required. SID-SRM-MS analysis of nuclear IRF3 in IRF3 siRNA-transfected A549 cell shows that the absolute amount of IRF3 is significantly lower in IRF3 siRNA-transfected cells than in non-targeting control siRNA-transfected cells, and also that the nuclear translocation of IRF3 is inhibited (Fig. 5D).
ds-RNA Activates the Noncanonical NF-κB Pathway
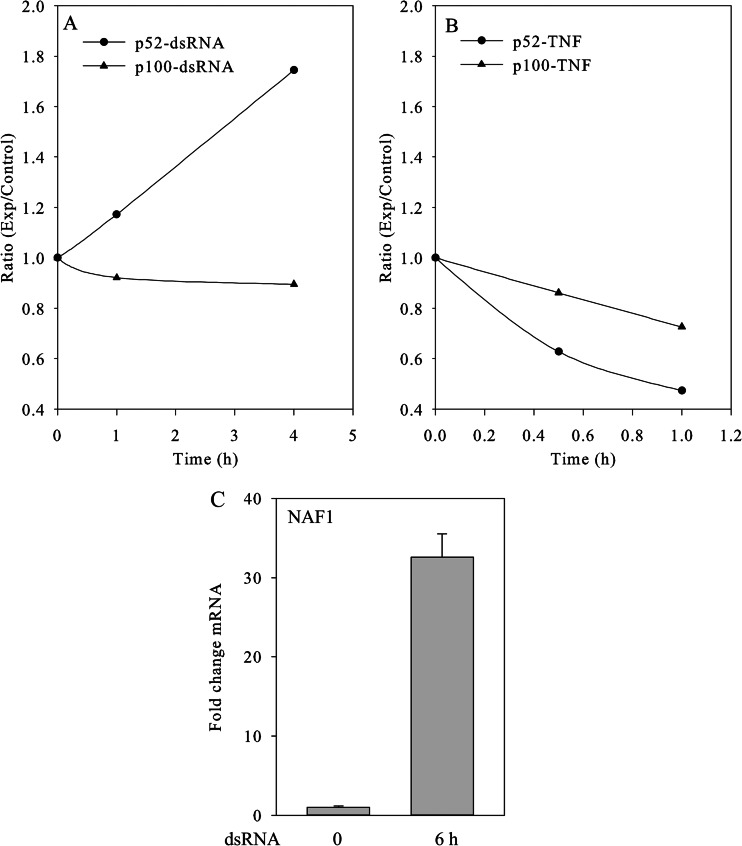
The noncanonical NF-κB pathway is induced by some stimuli distinct from those activating the canonical pathway, including lymphotoxin B, B cell activating factor, CD40 ligand, and DNA virus infection (62). Our group has shown that RNA virus infection is also a rapid inducer of the noncanonical pathway (11). This pathway utilizes an atypical IKK complex comprising two IKKα subunits, but not NF-κB essential modulator. In the noncanonical pathway, ligand-induced activation results in the activation of NIK, a serine kinase that phosphorylates and activates IKKα, which in turn phosphorylates nuclear factor NF-κB p100, leading to the processing and liberation of the RelB/p52 active heterodimer (11). The heterodimeric RelB/p52 complex is a transcriptional activator that translocates into the nucleus and binds to the κB consensus sequence 5′-GGRNNYYCC-3′, located in the enhancer region of genes involved in the immune response and acute phase reactions. Because the precise temporal relationship of the canonical and noncanonical pathway activation has not been defined in response to ds-RNA, we quantified noncanonical NF-κB pathway activation using SID-SRM-MS. A challenge in SID-SRM-MS assay design is that p52 is the proteolytically processed 454 amino acid residue of the NH2 terminus of p100. In order to distinguish these two proteins via SRM assay, we selected a signature peptide unique to p100 having the sequence ALLDYGVTADAR (amino acid position 460–471). The p52 signature peptide, belonging to both p52 and p100, was selected having the sequence DSGEEAAEPSAPSR. However, by the measure of SID-SRM-MS, the peak area ratios of (p52/p100-native) versus (p52/p100-SIS) and p100-native versus p100-SIS were compared, and we found that the amount of p52 in the nucleus of A549 cells is about 10 times greater than that of p100 (see supplemental Table S2). Therefore, nuclear p100 has little effect on the quantification of nuclear p52. In this experiment, we treated A549 cells with ds-RNA and TNF for varying times. Previous work has shown that the TNF-induced canonical pathway response is much more rapid than that of ds-RNA; for this reason, the TNF time course was shorter. We then used SID-SRM-MS to measure the temporal profiles of p52 and p100 in the nucleus after the stimulation. As shown in Fig. 6A, ds-RNA treatment led to a time-dependent increase in the nuclear accumulation of p52; at 4 h after ds-RNA stimulation, about 2-fold accumulation of p52 was seen. Meanwhile, the level of nuclear p100 was barely changed. The accumulation of p52 suggests that p52 processing and nuclear translocation are induced by ds-RNA stimulation. In a control experiment, p52 processing and nuclear translocation were not induced by TNF stimulation over the same time interval (Fig. 6B). Expression of the noncanonical pathway-dependent gene NAF1 is shown. Here, ds-RNA induced a 32-fold induction of NAF1 expression (Fig. 6C).
Fig. 6.
Activation of noncanonical NF-κB pathway by the stimulation of ds-RNA. A549 cells were treated with ds-RNA and TNF. The nuclear proteins were extracted and subjected to SID-SRM-MS analysis. A, the temporal profiles of nuclear p52 and p100 in the cells treated with dsRNA, derived from SID-SRM-MS analysis. B, the temporal profiles of nuclear p52 and p100 in the cells treated with TNF, derived from SID-SRM-MS analysis. C, the expression profiles of noncanonical NF-κB pathway-dependent NAF1 gene. Q-RT-PCR of NAF1 expression in the absence or presence of ds-RNA.
DISCUSSION
The rapid activation of the regulatory arms of the IIR is essential for host survival in response to invading pathogens. Despite significant advances in our understanding of the biochemistry and regulation of the IIR, the kinetics of the pathway response are poorly understood. In this study, we designed, characterized, and applied SID-SRM-MS assays for the quantification of 10 major IIR regulators using a versatile workflow for the quantification of low-abundance proteins (31). We were able to observe rapid activation of both NF-κB and IRF3 pathways in response to the molecular ds-RNA pattern, with the NF-κB pathway demonstrating a secondary, delayed amplification phase. Our SID-SRM-MS measurements were able to measure the nucleo-cytoplasmic shuttling of the IκBα inhibitor in the unstimulated cell, and we observed its dynamic response to ds-RNA stimulation with initial nuclear clearance followed by cytoplasmic degradation and subsequent repopulation upon its re-synthesis. We observed that in the presence of system perturbation with RelA siRNA, a low nuclear threshold of NF-κB is required for target gene expression. The quantitative measurement of the major regulatory PRR RIG-I upstream of the IRF3 pathway and of the kinases controlling IRF3 phosphorylation (TBK1 and MAVS) suggests that these proteins are initially consumed during the early phase of IRF3 activation. Finally, we were able to measure delayed noncanonical NF-κB activation by quantifying the abundance of the processed (52 kDa) NF-κB2 subunit in the nucleus. These quantitative observations enabled by SID-SRM-MS have led to new insights into the dynamics and interconnectivity of the IIR response.
Although SID-SRM-MS assays have been widely developed for the quantification of moderate- and high-abundance analytes, their application to low-abundance signaling proteins has not been widely accomplished. In this study, we optimized and extensively characterized the assay performance of 10 low-abundance regulatory kinases and transcription factors. Care in selecting SRM Q1/Q3 transitions, CE optimization, and evaluating assay specificity is critical because the detection of signature peptides of low-abundance proteins is subject to interference that can lead to false positives and quantification inaccuracy. Despite the complexity of tryptic digests of cellular extracts, most IIR-SRM assays exhibited baseline separation from other peaks on the peptide LC chromatogram (supplemental Figs. S2 and S3). Out of 13 initial target proteins, 10 (Table I) could be quantified directly from cell extracts with minimal sample fractionation. Even though the chemically synthesized SIS signature peptides of NIK, IKKε, and IKKβ had strong MS responses, we could not detect their cognate proteins directly from cellular extracts. This might have been caused by multiple factors, including low abundance of the target proteins, efficiency of release during tryptic digestion, and the presence of unrecognized post-translational modifications arising during biology or sample preparation. Moreover, our assessment of the reproducibility of peptide SRM measurements indicated that 94% of the SRM measurements had within-run CVs of 20% or less, and 80% had CVs of less than 5%. These precision values demonstrate clearly that peptide SRM measurement can be used in high-precision quantification, and that these SRM assays can be widely used in the study of inflammation, virus infection, cancer, and other IIR-related diseases.
Our observations show that ds-RNA stimulation rapidly induces both NF-κB/RelA and IRF3 arms of the IIR, producing cytoplasmic depletion and concomitant nuclear increases of both NF-κB/RelA and IRF3 transcription factors. We interpret this phenomenon as indicating ds-RNA-induced cytoplasmic-to-nuclear translocation, a well-established mechanism for activation of these transcription factors that is consistent with our confocal colocalization experiments. Our studies further indicate that in response to ds-RNA, only a fraction of the cytoplasmic transcription factor is translocated into the nucleus. Using an aptamer-SRM assay for the identification of biologically active RelA, we previously demonstrated that ∼10% of the cytoplasmic RelA is translocated into the nucleus in response to TNF (40, 63). Our studies here indicate that a greater fraction (∼50%) of cytoplasmic RelA is induced to translocate in response to ds-RNA stimulation, but a measurable fraction is still retained in the cytoplasm. Presently our SID-SRM-MS assay quantifies the presence of only the total unmodified transcription factor. In the future, the development of post-translational-modification-specific SRMs will allow us to identify the activated states (31).
The quantitative nature of the SID-SRM-MS assays further suggests asynchronous translocation of IRF3 and NF-κB effectors. Despite the common PRR-dependent activation of IRF3 and NF-κB translocation, our data suggest that IRF3 is initially translocated into the nucleus and that its nuclear residence is transient. These findings suggest the presence of a negative feedback loop controlling IRF3 translocation, a finding that will require further mechanistic investigation. Additionally, NF-κB undergoes a second inflection in nuclear abundance, suggesting the involvement of an additional positive activation pathway, perhaps related to the recruitment of other ds-RNA-activated PRRs, such as TLR3 and/or protein kinase R. More work in cells deficient in these PRRs will be required in order for us to dissect the contribution of these other pathways to the ds-RNA response.
The NF-κB pathway is actively under negative autoregulatory control, whereby the nuclear-translocation-activated NF-κB induces the expression of the IκBα inhibitor family (64, 65). Our findings suggest that a fraction of the IκBα complex is localized within the nucleus in resting cells, and that this nuclear fraction is rapidly degraded in response to ds-RNA, suggesting that inducible IκBα proteolysis might partially occur within the nucleus. In response to ds-RNA stimulation, IκBα reappears in the nucleus first, where it complexes and inactivates NF-κB (60). Within the IκBα·NF-κB complex, the IκBα nuclear export signal dominates, resulting in cytoplasmic redistribution of the complex in an attempt to restore the cell to a pre-stimulated state. Interestingly, in the presence of ds-RNA, despite partial IκBα repopulation in the cytoplasm, more NF-κB/RelA continues to translocate into the nucleus. This finding suggests that an additional NF-κB reservoir is being activated, the identity and characterization of which warrant further study.
Our SID-SRM-MS observations indicate that the ds-RNA PRR RIG-I is initially depleted in the cytosolic fraction upon ds-RNA stimulation. Previous studies have shown that RIG-I undergoes Lys (K)-63-linked polyubiquitylation, a post-translational modification mediated by the RING-finger E3 ubiquitin ligases TRIM25 (66) and RNF125 (67), and that this event is necessary for its oligomerization and binding to MAVS. Although K63-linked polyubiquitylation is not in itself linked to proteasomal degradation, RIG-I is also a target for Triad3, a K48-linked ubiquitin ligase that has been shown to mediate inducible RIG-I degradation (68). Our quantitative SID-SRM-MS observations are consistent with a model of rapid RIG-I degradation as an initial event in IRF3 activation prior to its robust re-synthesis, perhaps mediated by Triad3.
A number of studies have shown the presence of positive cross-talk between the NF-κB and IRF3 signaling pathways in the IIR. For example, activation of NF-κB up-regulates the expression of IRF isoforms, including IRF1, -5, and -7, via a pathway that enhances the downstream IFN response (69). We therefore were surprised to find that siRNA knockdown of NF-κB/RelA resulted in enhanced IRF3-dependent gene expression (supplemental Fig. s8). These intriguing findings suggest that there is negative cross-talk in the NF-κB and IRF3 signaling pathways.
The interplay between the noncanonical and canonical NF-κB pathways in the IIR has been relatively unexplored. Mechanistically, we know that activation of the noncanonical NIK-IKKα complex is dependent on RIG-I·MAVS and independent of the IKKγ/NF-κB essential modulator adapter, whereas activation of the canonical IKKα/β complex is dependent on the IKKγ/NF-κB essential modulator (5). In response to lymphotoxin b, the activation of the noncanonical pathway is delayed relative to the canonical pathway (70). Our data suggest that the noncanonical pathway is more rapidly activated in response to ds-RNA, suggesting that the timing of the two pathways is stimulus-dependent.
In summary, the precise temporal kinetics of IIR activation is a key determinate of the response to invading pathogens. Our development and characterization of a standardized, quantitative, multiplexed IIR-SRM-MS assay will significantly enhance quantitative measurements of the cellular response to PRR-induced signaling.
Supplementary Material
Acknowledgments
We thank Rovshan Sadygov for helpful discussions.
Footnotes
* This work was supported by the Institute for Translational Sciences (UL1TR000071, NCATS), HHSN272200800048C NIAID Clinical Proteomics Center, NIEHS P30(ES006676), and NIH-NHLBI-HHSN268201000037C NHLBI Proteomics Center for Airway Inflammation.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- ACN
- acetonitrile
- CE
- collision energy
- ds
- double-stranded
- EGFP
- enhanced green fluorescent protein
- HPLC
- high performance liquid chromatography
- IFN
- interferon
- IIR
- innate immune response
- IKK
- IκB kinase
- IRF
- interferon regulatory factor
- LC
- liquid chromatography
- MALDI-TOF
- matrix-assisted laser desorption/ionization time-of-flight
- MAVS
- mitochondrial antiviral signaling protein
- MS
- mass spectrometry
- NF
- nuclear factor
- NIK
- NF-κB-inducing kinase
- PRR
- pattern recognition receptor
- RT-PCR
- real time polymerase chain reaction
- SID
- stable isotopic dilution
- siRNA
- short interfering RNA
- SIS
- stable isotope-labeled standard
- SRM
- selected reaction monitoring
- TBK
- tank binding kinase.
REFERENCES
- 1. Takeda K., Akira S. (2007) Toll-like receptors. Curr. Protoc. Immunol. Chapter 14, Unit 14.12 [DOI] [PubMed] [Google Scholar]
- 2. Buonaguro L., Wang E., Tornesello M. L., Buonaguro F. M., Marincola F. M. (2011) Systems biology applied to vaccine and immunotherapy development. BMC Syst. Biol. 5, 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Siebenlist U., Franzoso G., Brown K. (1994) Structure, regulation and function of NF-kappa B. Annu. Rev. Cell Biol. 10, 405–455 [DOI] [PubMed] [Google Scholar]
- 4. Courtois G., Smahi A., Israel A. (2001) NEMO/IKK gamma: linking NF-kappa B to human disease. Trends Mol. Med. 7, 427–430 [DOI] [PubMed] [Google Scholar]
- 5. Liu P., Li K., Garofalo R. P., Brasier A. R. (2008) Respiratory syncytial virus induces RelA release from cytoplasmic 100-kDa NF-kappa B2 complexes via a novel retinoic acid-inducible gene-I{middle dot}NF- kappa B-inducing kinase signaling pathway. J. Biol. Chem. 283, 23169–23178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu P., Lu M., Tian B., Li K., Garofalo R. P., Prusak D., Wood T. G., Brasier A. R. (2009) Expression of an IKKgamma splice variant determines IRF3 and canonical NF-kappaB pathway utilization in ssRNA virus infection. PLoS One 4, e8079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yamaoka S., Courtois G., Bessia C., Whiteside S. T., Weil R., Agou F., Kirk H. E., Kay R. J., Israel A. (1998) Complementation cloning of NEMO, a component of the IkappaB kinase complex essential for NF-kappaB activation. Cell 93, 1231–1240 [DOI] [PubMed] [Google Scholar]
- 8. Poyet J. L., Srinivasula S. M., Lin J. H., Fernandes-Alnemri T., Yamaoka S., Tsichlis P. N., Alnemri E. S. (2000) Activation of the Ikappa B kinases by RIP via IKKgamma/NEMO-mediated oligomerization. J. Biol. Chem. 275, 37966–37977 [DOI] [PubMed] [Google Scholar]
- 9. Zhao T., Yang L., Sun Q., Arguello M., Ballard D. W., Hiscott J., Lin R. (2007) The NEMO adaptor bridges the nuclear factor-kappaB and interferon regulatory factor signaling pathways. Nat. Immunol. 8, 592–600 [DOI] [PubMed] [Google Scholar]
- 10. Bonizzi G., Bebien M., Otero D. C., Johnson-Vroom K. E., Cao Y., Vu D., Jegga A. G., Aronow B. J., Ghosh G., Rickert R. C., Karin M. (2004) Activation of IKKalpha target genes depends on recognition of specific kappaB binding sites by RelB:p52 dimers. EMBO J. 23, 4202–4210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Choudhary S., Boldogh S., Garofalo R., Jamaluddin M., Brasier A. R. (2005) Respiratory syncytial virus influences NF-kappaB-dependent gene expression through a novel pathway involving MAP3K14/NIK expression and nuclear complex formation with NF-kappaB2. J. Virol. 79, 8948–8959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hiscott J., Pitha P., Genin P., Nguyen H., Heylbroeck C., Mamane Y., Algarte M., Lin R. (1999) Triggering the interferon response: the role of IRF-3 transcription factor. J. Interferon Cytokine Res. 19, 1–13 [DOI] [PubMed] [Google Scholar]
- 13. Pitha P. M., Au W. C., Lowther W., Juang Y. T., Schafer S. L., Burysek L., Hiscott J., Moore P. A. (1998) Role of the interferon regulatory factors (IRFs) in virus-mediated signaling and regulation of cell growth. Biochimie (Paris) 80, 651–658 [DOI] [PubMed] [Google Scholar]
- 14. Saha S. K., Pietras E. M., He J. Q., Kang J. R., Liu S. Y., Oganesyan G., Shahangian A., Zarnegar B., Shiba T. L., Wang Y., Cheng G. (2006) Regulation of antiviral responses by a direct and specific interaction between TRAF3 and Cardif. EMBO J. 25, 3257–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fitzgerald K. A., McWhirter S. M., Faia K. L., Rowe D. C., Latz E., Golenbock D. T., Coyle A. J., Liao S. M., Maniatis T. (2003) IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4, 491–496 [DOI] [PubMed] [Google Scholar]
- 16. Imanishi D., Yamamoto K., Tsushima H., Miyazaki Y., Kuriyama K., Tomonaga M., Matsuyama T. (2000) Identification of a novel cytokine response element in the human IFN regulatory factor-1 gene promoter. J. Immunol. 165, 3907–3916 [DOI] [PubMed] [Google Scholar]
- 17. Marie I., Durbin J. E., Levy D. E. (1998) Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 17, 6660–6669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shapira S. D., Hacohen N. (2011) Systems biology approaches to dissect mammalian innate immunity. Curr. Opin. Immunol. 23, 71–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zak D. E., Aderem A. (2009) Systems biology of innate immunity. Immunol. Rev. 227, 264–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Breitkreutz A., Choi H., Sharom J. R., Boucher L., Neduva V., Larsen B., Lin Z. Y., Breitkreutz B. J., Stark C., Liu G., Ahn J., war-Darch D., Reguly T., Tang X., Almeida R., Qin Z. S., Pawson T., Gingras A. C., Nesvizhskii A. I., Tyers M. (2010) A global protein kinase and phosphatase interaction network in yeast. Science 328, 1043–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gingras A. C., Gstaiger M., Raught B., Aebersold R. (2007) Analysis of protein complexes using mass spectrometry. Nat. Rev. Mol. Cell. Biol. 8, 645–654 [DOI] [PubMed] [Google Scholar]
- 22. Kaake R. M., Wang X., Huang L. (2010) Profiling of protein interaction networks of protein complexes using affinity purification and quantitative mass spectrometry. Mol. Cell. Proteomics 9, 1650–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Choudhary C., Mann M. (2010) Decoding signalling networks by mass spectrometry-based proteomics. Nat. Rev. Mol. Cell. Biol. 11, 427–439 [DOI] [PubMed] [Google Scholar]
- 24. Rinner O., Mueller L. N., Hubalek M., Muller M., Gstaiger M., Aebersold R. (2007) An integrated mass spectrometric and computational framework for the analysis of protein interaction networks. Nat. Biotechnol. 25, 345–352 [DOI] [PubMed] [Google Scholar]
- 25. Lange V., Picotti P., Domon B., Aebersold R. (2008) Selected reaction monitoring for quantitative proteomics: a tutorial. Mol. Syst. Biol. 4, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gerber S. A., Rush J., Stemman O., Kirschner M. W., Gygi S. P. (2003) Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl. Acad. Sci. U.S.A. 100, 6940–6945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Picotti P., Bodenmiller B., Mueller L. N., Domon B., Aebersold R. (2009) Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell 138, 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bisson N., James D. A., Ivosev G., Tate S. A., Bonner R., Taylor L., Pawson T. (2011) Selected reaction monitoring mass spectrometry reveals the dynamics of signaling through the GRB2 adaptor. Nat. Biotechnol. 29, 653–658 [DOI] [PubMed] [Google Scholar]
- 29. Dong M. Q., Venable J. D., Au N., Xu T., Park S. K., Cociorva D., Johnson J. R., Dillin A., Yates J. R., III (2007) Quantitative mass spectrometry identifies insulin signaling targets in C. elegans. Science 317, 660–663 [DOI] [PubMed] [Google Scholar]
- 30. Brasier A. R., Tian B., Jamaluddin M., Kalita M. K., Garofalo R. P., Lu M. (2011) RelA Ser276 phosphorylation-coupled Lys310 acetylation controls transcriptional elongation of inflammatory cytokines in respiratory syncytial virus infection. J. Virol. 85, 11752–11769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhao Y., Brasier A. R. (2013) Applications of selected reaction monitoring (SRM)-mass spectrometry (MS) for quantitative measurement of signaling pathways (2013). Methods http://dx.doi.org/10.1016/j.ymeth.2013.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Craig R., Cortens J. P., Beavis R. C. (2004) Open source system for analyzing, validating, and storing protein identification data. J. Proteome Res. 3, 1234–1242 [DOI] [PubMed] [Google Scholar]
- 33. Fusaro V. A., Mani D. R., Mesirov J. P., Carr S. A. (2009) Prediction of high-responding peptides for targeted protein assays by mass spectrometry. Nat. Biotechnol. 27, 190–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Forbus J., Spratt H., Wiktorowicz J., Wu Z., Boldogh I., Denner L., Kurosky A., Brasier R. C., Luxon B., Brasier A. R. (2006) Functional analysis of the nuclear proteome of human A549 alveolar epithelial cells by HPLC-high resolution 2-D gel electrophoresis. Proteomics 6, 2656–2672 [DOI] [PubMed] [Google Scholar]
- 35. Addona T. A., Abbatiello S. E., Schilling B., Skates S. J., Mani D. R., Bunk D. M., Spiegelman C. H., Zimmerman L. J., Ham A. J., Keshishian H., Hall S. C., Allen S., Blackman R. K., Borchers C. H., Buck C., Cardasis H. L., Cusack M. P., Dodder N. G., Gibson B. W., Held J. M., Hiltke T., Jackson A., Johansen E. B., Kinsinger C. R., Li J., Mesri M., Neubert T. A., Niles R. K., Pulsipher T. C., Ransohoff D., Rodriguez H., Rudnick P. A., Smith D., Tabb D. L., Tegeler T. J., Variyath A. M., Vega-Montoto L. J., Wahlander A., Waldemarson S., Wang M., Whiteaker J. R., Zhao L., Anderson N. L., Fisher S. J., Liebler D. C., Paulovich A. G., Regnier F. E., Tempst P., Carr S. A. (2009) Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat. Biotechnol. 27, 633–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hoofnagle A. N., Becker J. O., Wener M. H., Heinecke J. W. (2008) Quantification of thyroglobulin, a low-abundance serum protein, by immunoaffinity peptide enrichment and tandem mass spectrometry. Clin. Chem. 54, 1796–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Keshishian H., Addona T., Burgess M., Kuhn E., Carr S. A. (2007) Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol. Cell. Proteomics 6, 2212–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Keshishian H., Addona T., Burgess M., Mani D. R., Shi X., Kuhn E., Sabatine M. S., Gerszten R. E., Carr S. A. (2009) Quantification of cardiovascular biomarkers in patient plasma by targeted mass spectrometry and stable isotope dilution. Mol. Cell. Proteomics 8, 2339–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kuhn E., Addona T., Keshishian H., Burgess M., Mani D. R., Lee R. T., Sabatine M. S., Gerszten R. E., Carr S. A. (2009) Developing multiplexed assays for troponin I and interleukin-33 in plasma by peptide immunoaffinity enrichment and targeted mass spectrometry. Clin. Chem. 55, 1108–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhao Y., Widen S. G., Jamaluddin M., Tian B., Wood T. G., Edeh C. B., Brasier A. R. (2011) Quantification of activated NF-kappaB/RelA complexes using ssDNA aptamer affinity-stable isotope dilution-selected reaction monitoring-mass spectrometry. Mol. Cell. Proteomics 10, M111.008771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Green J. M. (1996) A practical guide to analytical method validation. Anal. Chem. 68, A305–A309 [Google Scholar]
- 42. Kuzyk M. A., Smith D., Yang J., Cross T. J., Jackson A. M., Hardie D. B., Anderson N. L., Borchers C. H. (2009) Multiple reaction monitoring-based, multiplexed, absolute quantitation of 45 proteins in human plasma. Mol. Cell. Proteomics 8, 1860–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brownridge P., Beynon R. J. (2011) The importance of the digest: proteolysis and absolute quantification in proteomics. Methods 54, 351–360 [DOI] [PubMed] [Google Scholar]
- 44. Arsene C. G., Ohlendorf R., Burkitt W., Pritchard C., Henrion A., O'Connor G., Bunk D. M., Guttler B. (2008) Protein quantification by isotope dilution mass spectrometry of proteolytic fragments: cleavage rate and accuracy. Anal. Chem. 80, 4154–4160 [DOI] [PubMed] [Google Scholar]
- 45. Perona J. J., Craik C. S. (1997) Evolutionary divergence of substrate specificity within the chymotrypsin-like serine protease fold. J. Biol. Chem. 272, 29987–29990 [DOI] [PubMed] [Google Scholar]
- 46. Schechter I., Berger A. (1967) On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 27, 157–162 [DOI] [PubMed] [Google Scholar]
- 47. Siepen J. A., Keevil E. J., Knight D., Hubbard S. J. (2007) Prediction of missed cleavage sites in tryptic peptides aids protein identification in proteomics. J. Proteome Res. 6, 399–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Alexopoulou L., Holt A. C., Medzhitov R., Flavell R. A. (2001) Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413, 732–738 [DOI] [PubMed] [Google Scholar]
- 49. Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., Uematsu S., Jung A., Kawai T., Ishii K. J., Yamaguchi O., Otsu K., Tsujimura T., Koh C. S., Reis e Sousa M. Y., Fujita T., Akira S. (2006) Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441, 101–105 [DOI] [PubMed] [Google Scholar]
- 50. Iwamura T., Yoneyama M., Yamaguchi K., Suhara W., Mori W., Shiota K., Okabe Y., Namiki H., Fujita T. (2001) Induction of IRF-3/-7 kinase and NF-kappaB in response to double-stranded RNA and virus infection: common and unique pathways. Genes Cells 6, 375–388 [DOI] [PubMed] [Google Scholar]
- 51. Lenardo M. J., Fan C. M., Maniatis T., Baltimore D. (1989) The involvement of NF-kappa B in beta-interferon gene regulation reveals its role as widely inducible mediator of signal transduction. Cell 57, 287–294 [DOI] [PubMed] [Google Scholar]
- 52. Harada H., Fujita T., Miyamoto M., Kimura Y., Maruyama M., Furia A., Miyata T., Taniguchi T. (1989) Structurally similar but functionally distinct factors, IRF-1 and IRF-2, bind to the same regulatory elements of IFN and IFN-inducible genes. Cell 58, 729–739 [DOI] [PubMed] [Google Scholar]
- 53. Yoneyama M., Suhara W., Fukuhara Y., Fukuda M., Nishida E., Fujita T. (1998) Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 17, 1087–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Brasier A. R., Spratt H., Wu Z., Boldogh I., Zhang Y., Garofalo R. P., Casola A., Pashmi J., Haag A., Luxon B., Kurosky A. (2004) Nuclear heat shock response and novel nuclear domain 10 reorganization in respiratory syncytial virus-infected a549 cells identified by high-resolution two-dimensional gel electrophoresis. J. Virol. 78, 11461–11476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Han Y., Brasier A. R. (1997) Mechanism for biphasic rel A. NF-kappaB1 nuclear translocation in tumor necrosis factor alpha-stimulated hepatocytes. J. Biol. Chem. 272, 9825–9832 [DOI] [PubMed] [Google Scholar]
- 56. Ghosh S., May M. J., Kopp E. B. (1998) NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 16, 225–260 [DOI] [PubMed] [Google Scholar]
- 57. Karin M., Ben-Neriah Y. (2000) Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu. Rev. Immunol. 18, 621–663 [DOI] [PubMed] [Google Scholar]
- 58. Arenzana-Seisdedos F., Turpin P., Rodriguez M., Thomas D., Hay R. T., Virelizier J. L., Dargemont C. (1997) Nuclear localization of I kappa B alpha promotes active transport of NF-kappa B from the nucleus to the cytoplasm. J. Cell Sci. 110(Pt 3), 369–378 [DOI] [PubMed] [Google Scholar]
- 59. Arenzana-Seisdedos F., Thompson J., Rodriguez M. S., Bachelerie F., Thomas D., Hay R. T. (1995) Inducible nuclear expression of newly synthesized I kappa B alpha negatively regulates DNA-binding and transcriptional activities of NF-kappa B. Mol. Cell. Biol. 15, 2689–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Huang T. T., Kudo N., Yoshida M., Miyamoto S. (2000) A nuclear export signal in the N-terminal regulatory domain of IkappaBalpha controls cytoplasmic localization of inactive NF-kappaB/IkappaBalpha complexes. Proc. Natl. Acad. Sci. U.S.A. 97, 1014–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lenz J. R., Baglioni C. (1978) Inhibition of protein synthesis by double-stranded RNA and phosphorylation of initiation factor, eIF-2. J. Biol. Chem. 253, 4219–4223 [PubMed] [Google Scholar]
- 62. Senftleben U., Cao Y., Xiao G., Greten F. R., Krahn G., Bonizzi G., Chen Y., Hu Y., Fong A., Sun S. C., Karin M. (2001) Activation by IKKalpha of a second, evolutionary conserved, NF-kappa B signaling pathway. Science 293, 1495–1499 [DOI] [PubMed] [Google Scholar]
- 63. Kalita M. K., Sargsyan K., Tian B., Paulucci-Holthauzen A., Najm H. N., Debusschere B. J., Brasier A. R. (2011) Sources of cell-to-cell variability in canonical nuclear factor-kappaB (NF-kappaB) signaling pathway inferred from single cell dynamic images. J. Biol. Chem. 286, 37741–37757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Brasier A. R. (2006) The NF-kappaB regulatory network. Cardiovasc. Toxicol. 6, 111–130 [DOI] [PubMed] [Google Scholar]
- 65. Sun S. C., Ganchi P. A., Ballard D. W., Greene W. C. (1993) NF-kappa B controls expression of inhibitor I kappa B alpha: evidence for an inducible autoregulatory pathway. Science 259, 1912–1915 [DOI] [PubMed] [Google Scholar]
- 66. Gack M. U., Shin Y. C., Joo C. H., Urano T., Liang C., Sun L., Takeuchi O., Akira S., Chen Z., Inoue S., Jung J. U. (2007) TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446, 916–920 [DOI] [PubMed] [Google Scholar]
- 67. Arimoto K., Takahashi H., Hishiki T., Konishi H., Fujita T., Shimotohno K. (2007) Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125. Proc. Natl. Acad. Sci. U.S.A. 104, 7500–7505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Nakhaei P., Mesplede T., Solis M., Sun Q., Zhao T., Yang L., Chuang T. H., Ware C. F., Lin R., Hiscott J. (2009) The E3 ubiquitin ligase Triad3A negatively regulates the RIG-I/MAVS signaling pathway by targeting TRAF3 for degradation. PLoS Pathog. 5, e1000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Moschonas A., Ioannou M., Eliopoulos A. G. (2012) CD40 stimulates a “feed-forward” NF-kappaB-driven molecular pathway that regulates IFN-beta expression in carcinoma cells. J. Immunol. 188, 5521–5527 [DOI] [PubMed] [Google Scholar]
- 70. Basak S., Kim H., Kearns J. D., Tergaonkar V., O'Dea E., Werner S. L., Benedict C. A., Ware C. F., Ghosh G., Verma I. M., Hoffmann A. (2007) A fourth IkappaB protein within the NF-kappaB signaling module. Cell 128, 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.