Abstract
Eukaryotic RNA polymerase II (RNAPII) is a 12-subunit enzyme that is responsible for the transcription of messenger RNA. Two of the subunits of RNA polymerase II, Rpb4 and Rpb7, have been shown to dissociate from the enzyme under a number of specific laboratory conditions. However, a biological context for the dissociation of Rpb4 and Rpb7 has not been identified. We have found that Rpb4/7 dissociate from RNAPII upon interaction with specific transcriptional elongation-associated proteins that are recruited to the hyperphosphorylated form of the C-terminal domain. However, the dissociation of Rpb4/7 is likely short lived because a significant level of free Rpb4/7 was not detected by quantitative proteomic analyses. In addition, we have found that RNAPII that is isolated through Rpb7 is depleted in serine 2 C-terminal domain phosphorylation. In contrast to previous reports, these data indicate that Rpb4/7 are dispensable during specific stages of transcriptional elongation in Saccharomyces cerevisiae.
The eukaryotic RNA polymerases are multisubunit enzymes that transcribe DNA to produce a wide variety of RNAs. RNA polymerase II (RNAPII)1 is responsible for the production of messenger RNA, which occurs through an elaborate and highly regulated transcription cycle (1). During each phase of the RNAPII transcription cycle, accessory proteins are recruited to carry out key co-transcriptional processes. These co-transcriptional processes include chromatin modification, chromatin remodeling, and mRNA processing. It has been well established that the individual subunits of RNAPII can alter the recruitment of accessory factors during transcription. This is especially true for the largest subunit of RNAPII referred to as Rpb1. Rpb1 contains a specialized C-terminal domain (CTD) that is the target of multiple post-translational modifications such as phosphorylation and proline isomerization. Specifically, the CTD is phosphorylated at serine 2, serine 5, and serine 7 in the hepta-peptide repeat 1YSPTSPS7, which is present as tandem repeats (2–5). It has been postulated that dynamic changes in the modification of the CTD dictate recruitment of accessory factors during all phases of transcription (6). However, the other subunits of RNAPII also play a role in the recruitment of accessory factors during transcription, increasing the potential diversity of protein-protein interactions that occur during each round of transcription.
Many of the other 11 subunits of RNAPII, termed Rpb2–12, have been shown to mediate RNAPII interactions with regulatory proteins. Rpb9, for instance, has been shown to mediate interactions between RNAPII and the largest subunit of initiation factor TFIIE (Tfa1) (7). Rpb9 is also required for the interaction between RNAPII and the initiation/elongation factor TFIIF (7, 8). Rpb5 interacts with the Rsc4 subunit of the chromatin remodeling complex RSC (9). Disruption of the Rpb5-Rsc4 interaction has been shown to cause decreases in RNAPII transcription, illustrating the importance of this interaction in the regulation of RNAPII function perhaps through the regulation of chromatin structure (9, 10). Rpb2 interacts with Ssu72, a CTD phosphatase that dephosphorylates a population of serine 5-phosphorylated RNAPII (11, 12). The well described heterodimer of Rpb4 and Rpb7 (Rpb4/7) has also been shown to interact with a number of accessory proteins. This includes interactions between Rpb4/7 and the head of the Mediator complex suggesting a potential role for the heterodimer during transcriptional initiation (13). The C terminus of Rpb4 has been shown to be required for the interaction between the serine 2-specific CTD phosphatase Fcp1 and RNAPII in Saccharomyces cerevisiae, Schizosaccharomyces pombe, and Drosophila melanogaster (14–16). Both S. pombe and S. cerevisiae Rpb7 have been shown to interact with the termination factor Nrd1, which also binds to the serine 2-phosphorylated RNAPII CTD (17).
Previous studies have shown that Rpb4/7 dissociates from S. cerevisiae RNAPII when RPB4 is deleted or when isolated RNAPII is subjected to mild denaturing conditions or nondenaturing gel electrophoresis (18–21). However, a biological role for the dissociation of Rpb4/7 has not been discovered. To address the role of Rpb4/7 during the transcription cycle, multiple groups have used chromatin immunoprecipitation (ChIP) experiments to determine whether Rpb4/7 dissociates from RNAPII during transcription in both yeast and human cells (22–25). Each study concluded that Rpb4/7 associates with promoters and throughout coding regions during RNAPII transcription. However, because ChIP experiments measure the average occupancy of DNA interacting proteins from a large population of cells, these data do not conclusively show that a small percentage of RNAPII complexes lacking Rpb4/7 do not exist in S. cerevisiae.
Our current work has identified a group of known RNAPII-interacting proteins that associate with a 10-subunit form of RNAPII lacking the Rpb4/7 heterodimer. In agreement with previous studies, analysis of isolated RNAPII using quantitative proteomics revealed consistent association of Rpb4/7 with the majority of cellular RNAPII. However, we have found that the levels of Rpb4/7 are decreased in RNAPII complexes isolated using a subset of tagged elongation factors for purification. Upon further characterization of these isolated complexes, we have found that the loss of Rpb4/7 correlates with increased phosphorylation of the Rpb1 CTD at both serine 2 and serine 5. These data suggest that Rpb4/7 can dissociate from RNAPII as a consequence of specific elongation factor interactions during transcription.
EXPERIMENTAL PROCEDURES
Yeast Strains
The genotype and source of all strains used in this study are provided in the supplemental material. Note that all epitope tags (TAP and FLAG) were integrated at the C terminus of the endogenous chromosomal locus for each tagged protein as described previously (26, 27). Insertion of the epitope tag at the chromosomal copy of each gene results in endogenous level expression of each tagged protein from its own promoter.
TAP Purification
Tandem affinity purifications were performed as described previously (26, 27). In brief, TAP strains were grown to A600 = 3–4 and then pelleted by centrifugation. Yeast cell pellets were resuspended in TAP lysis buffer and frozen in liquid nitrogen followed by grinding in a Waring blender. The resulting lysate was thawed and then subjected to DNase I and heparin sulfate treatment to solubilize chromatin-associated complexes. As a result, our TAP purifications contain both soluble and chromatin-associated RNAPII. After solubilization, the lysate was cleared by centrifugation and then incubated overnight with IgG-Sepharose. After extensive washing, IgG-Sepharose resin was resuspended in TEV cleavage buffer, and AcTEV protease was added, and the slurry was allowed to incubate at 4°C overnight to facilitate site-specific cleavage of the protein A portion of the TAP tag resulting in elution of the protein complexes from IgG-Sepharose. The slurry was then pipetted into an empty 30-ml Econoprep column, and the proteins were separated from the IgG-Sepharose by gravity flow. The elutions were then diluted in calmodulin binding buffer, and additional CaCl2 was added to a final concentration of 2 mm CaCl2. The samples were subsequently incubated with calmodulin-Sepharose resin to allow for binding of the protein complexes followed by elution with calmodulin elution buffer containing 2 mm EGTA.
Mass Spectrometry
Mass spectrometry analysis of isolated protein complexes was performed as described previously (26, 27). For this study, TAP elutions were subjected to TCA precipitation followed by digestion with endoproteinase Lys-C and trypsin. The digested peptides were pressure-loaded onto a three-phase MudPIT column containing Aqua C18 and Luna SCX resins (Phenomenex) as described previously. Twelve-step MudPIT was performed with increasing concentrations of ammonium acetate pulsed at the beginning of the step followed by a 100-min acetonitrile gradient. All data were acquired on an LTQ with the exception of the Cdc73-TAP data, which was acquired on an LTQ-Velos.
Quantitative Proteomics Analyses
Peptide sequences were matched to the obtained spectra using Proteome Discoverer 1.3 (Thermo) using SEQUEST (proteomicsresource.washington.edu/sequest.php) version 1.3.0.339 as the matching algorithm. The following parameters were used: (i) enzyme/trypsin; (ii) two missed cleavages allowed; (iii) peptide tolerance, 2.0 daltons; (iv) MS/MS tolerance, 0.8 daltons; (v) fixed modification, +57 daltons on cysteine; (vi) variable modifications, +16 daltons on methionine. Spectra were searched against a FASTA database containing 5819 S. cerevisiae protein sequences (downloaded from NCBI build from April 26, 2011) and 281 common contaminant proteins, including human keratins, immunoglobins, and proteolytic enzymes. The resulting peptide spectral matches (PSMs) were then subjected to false discovery rate calculation using Scaffold 3 (www.proteomesoftware.com/products/scaffold/) with the retained matches having a false discovery rate of less than or equal to 1%. Individual peptides were also required to have a peptide identification probability of 95% and a protein identification probability of 95% such that peptides/proteins with lower probabilities were excluded from the final dataset. Proteins were also required to be detected by at least two peptides so proteins that were identified by only one peptide were not included in the final dataset. All peptide matching information including XCorr values and peptide sequence information are available upon request as a Scaffold viewer file and are provided in text format as supplemental Tables 1–3. Peptides that could be assigned to multiple proteins were attributed to the protein identified with the most evidence (such as unique matching peptides). In addition, all peptides that could be assigned to multiple proteins contain the accession numbers for the other potential matching proteins in supplemental Tables 1–3, in the column “Other Proteins.” Using the PSMs that passed all filtering criteria, the NSAF value of proteins in each individual TAP were calculated as described previously (28, 29). NSAF values from each purification were then compared with a mock TAP, which was performed in the parental untagged strain BY4741. Proteins that had NSAF values at least 2-fold higher than the mock purification were considered as enriched in the TAP purification and were used for further analyses (supplemental Table 5). Hierarchical clustering was performed as described previously using either the raw number of PSMs or the NSAF value for each identified protein of interest (27, 30, 31).
Comparison of the Relative Abundance of Each RNA Polymerase II Subunit
To allow for the direct comparison of the subunit composition of RNA polymerase II complexes isolated through different tagged subunits, we performed complex normalized spectral abundance calculations by only considering the PSM values for the 12 subunits of RNA polymerase II (cNSAFRPB). Equation 1 is given for this calculation,
 |
Western Blot Analyses
Three to six liters of TAP-tagged and Rpb7–3×FLAG-containing strains were grown to A600 ≈4.0 and then pelleted by centrifugation. Cell pellets were lysed as described for the TAP purifications, and protein complexes were isolated by incubation with IgG-Sepharose followed by TEV cleavage. At this point, SDS-PAGE loading buffer was added to the TEV cleavage products, and the protein samples were boiled for 5 min. The loading volume of each TAP-isolated complex was adjusted to allow for equivalent levels of Rpb3 to be loaded in all samples for comparison purposes. Three 5-fold varying concentrations of Rpb3-TAP Rpb7–3×FLAG and Rpb7–3×FLAG samples were loaded for reference. Samples were separated on 10–20% gradient Tris-glycine precast CriterionTM gels (Bio-Rad) and transferred onto nitrocellulose for blotting. The antibodies used for Western blotting have been described previously (26). Protein quantity from replicate Western blots was determined by densitometry using ImageQuant software.
RESULTS
Analysis of RNAPII Dynamics Using Quantitative Proteomics
To determine whether Rpb4/7 interact with RNAPII during all phases of transcription, we purified RNAPII and a number of known RNAPII-associated proteins that function during specific steps in the transcription cycle. The tagged proteins (baits) used for purification are indicated on the top of the heat maps shown in Fig. 1. To purify both soluble and DNA-associated RNA polymerase complexes, yeast extracts were treated with heparin and DNase I to solubilize the chromatin-associated complexes prior to purification. Following purification, the protein mixtures were analyzed using MudPIT and quantitative proteomics where the NSAF of the proteins within each sample was calculated allowing for the comparison of the relative enrichment of proteins across various preparations (27, 28, 32). The data from each of these analysis steps are provided as supplemental Tables 1–5. Hierarchical clustering was performed using a Ward algorithm and Pearson correlation as described previously, using the raw number of PSMs or the NSAF values of the 25 most enriched proteins (Fig. 1, A and B, respectively) (27, 33). The cluster analysis revealed separation in the dendrogram between the specific subunits of RNAPII and known RNAPII-associated proteins using either the number of PSMs or the NSAF values (Fig. 1).
Fig. 1.

Multiple RNAPII-associated proteins interact with an RNAPII that lacks Rpb4 and Rpb7. The protein that was TAP-tagged and used as the bait for protein purification is shown at the top of A and B. Proteins identified by mass spectrometry are shown down the right side of A and B. A, hierarchical cluster performed using the total number of PSMs identified. PSM values for each protein in each purification are shown in a yellow to black color scale according to the legend shown to the right of the figure. B, hierarchical cluster performed using the NSAF value for each protein of interest. NSAF values for each protein in each purification are shown in a yellow to black color scale according to the NSAF legend shown to the right of the figure. The order of the proteins shown was determined by hierarchical clustering using the Ward algorithm as described previously (27, 31). The relationship between each protein to the right is shown in a dendrogram shown to the left of the figure. The identified proteins separate into two major branches within the dendrogram that represent RNAPII subunits (top branch) and RNAPII-associated proteins (bottom branch).
All 12 subunits of RNAPII were identified when using any of the RNAPII-specific subunits as bait (Rpb3, Rpb11, and Rpb7; see Fig. 1). As reported previously, the most enriched RNAPII-interacting proteins were Spt4/Spt5 (also known as DSIF) and Tfg1/Tfg2/Taf14 (the known subunits of TFIIF) (27). All 12 subunits of RNAPII were also observed when Spt4-, Tfg1-, or Cdc73-TAP were used for purification. These proteins are part of the DSIF, TFIIF, and PAF complexes, respectively, and have previously been shown to interact with RNAPII at multiple phases of the transcription cycle (34–39). The dendrogram to the left of Fig. 1, A and B, clearly delineate the known subunits of DSIF, TFIIF, and PAF. DSIF and TFIIF, which were reproducibly identified using RNAPII-specific baits for purification, have been shown to interact with the globular domain of RNAPII and interact with RNAPII regardless of the phosphorylation state of the CTD (7, 8, 40, 41). DSIF was also identified in TFIIF purifications (Tfg1-TAP), implying that Spt4/5 and TFIIF can interact with RNAPII simultaneously.
Surprisingly, purifications of RNAPII using Rtr1- or Set2-TAP as the bait protein resulted in the isolation of a 10-subunit RNAPII lacking the Rpb4/7 heterodimer (Fig. 1). This observation is intriguing because Rtr1 and Set2 are both known to be involved in the regulation of transcriptional elongation and both interact with hyperphosphorylated forms of RNAPII (3, 26, 42–45). The GTPase Npa3 was also found by MudPIT using Rtr1-TAP as the bait. It has previously been shown that the human homolog of Npa3 (known as GPN1) also co-purifies with the human homolog of Rtr1 (RPAP2) (46). Yeast Npa3 has been implicated in the regulation of RNAPII transcription and nuclear import of RNAPII (47). However, in previously published Npa3 purifications only the RNAPII subunits Rpb1, Rpb2, Rpb3, and Rpb5 were detected by mass spectrometry (47, 48). To determine whether Npa3 associates with a 10-subunit or 12-subunit RNAPII, we performed MudPIT analysis of Npa3-TAP purifications. As shown in Fig. 1, Npa3-TAP purification resulted in the identification of a 10-subunit RNAPII lacking Rpb4/7. Intriguingly, although Npa3 has been implicated in the nuclear import of RNAPII from the cytoplasm, Npa3-TAP purifications also contained Spt5, the two largest subunits of TFIIF, and the CTD phosphatase Rtr1 (Fig. 1) (46, 49–51). Co-purification of Npa3 and proteins that play a role in RNAPII transcription indicates that Npa3 may also play a role in the regulation of RNAPII function in the nucleus.
Multiple purifications of Set2-TAP resulted in the identification of the recently described ubiquitin E3 ligase Asr1 (Fig. 1 and data not shown). Asr1 has been proposed to regulate the subunit composition of RNAPII through ubiquitination of Rpb1 or Rpb2 that causes dissociation of the Rpb4/7 heterodimer (52). To confirm the finding that Asr1 interacts with RNAPII lacking Rpb4/7 and to further investigate the interaction between Asr1 and Set2, we performed Asr1-TAP purifications and analyzed them by MudPIT. As shown in Fig. 1, Asr1 purifications contain Set2, and the two proteins cluster together on the dendrogram indicating that the E3 ligase may interact with Set2 in vivo (Fig. 1). In addition, we confirmed that Asr1-TAP purifications do not contain Rpb4/7 as shown previously (52). Surprisingly, analysis of Asr1-TAP also identified Rtr1 and Npa3 indicating that Asr1, Rtr1, Npa3, and Set2 could be recruited during a specific stage of RNAPII elongation.
Understanding the biological role of Rpb4/7 dissociation from RNAPII is of great interest. Although multiple studies concluded that the majority of Rpb4/7 remains associated with RNAPII during all phases of the transcription cycle using ChIP-based experiments, recent studies have concluded that a free Rpb4/7 heterodimer may play an important role in coupling RNAPII transcription to translation of the RNA in the cytoplasm (53–55). To quantitate the relative abundance of the free Rpb4/7 heterodimer and to further characterize Rtr1-associated RNAPII, we performed additional quantitative proteomic analysis of Rpb3-, Rpb7-, Rpb11-, and Rtr1-TAP samples. As shown in Fig. 2 and supplemental Table 6, Rpb7-TAP purifications do not show a significant increase in the amount of Rpb4 or Rpb7 when compared with Rpb3- and Rpb11-TAP samples. These data indicate there may not be a significant amount of free Rpb4/7 heterodimer present in log-phase yeast. The only reproducibly significant difference in spectral abundance identified between these TAP purifications is the decrease in the association of Rpb4 and Rpb7 in Rtr1-TAP purifications, which showed a significant decrease when compared with the amount of Rpb4/7 detected in the Rpb3-, Rpb11-, and Rpb7-TAP purifications (Fig. 2B).
Fig. 2.
Analysis of biological replicate Rpb3-, Rpb7-, Rpb11-, and Rtr1-TAP samples indicates that there is not a significant level of non-RNAPII associated Rpb4/7 in total yeast lysate. A, average plus or minus one standard deviation of cNSAF values obtained from MudPIT analyses of three biological replicates of each purification as indicated. B, summary of p values obtained from Student's t test analysis of the cNSAF values are shown from comparing the levels of the RNAPII subunits Rpb3, Rpb4, and Rpb7 across each purification as indicated.
RNAPII Lacking Rpb4/7 Is Enriched in Hyperphosphorylated CTD
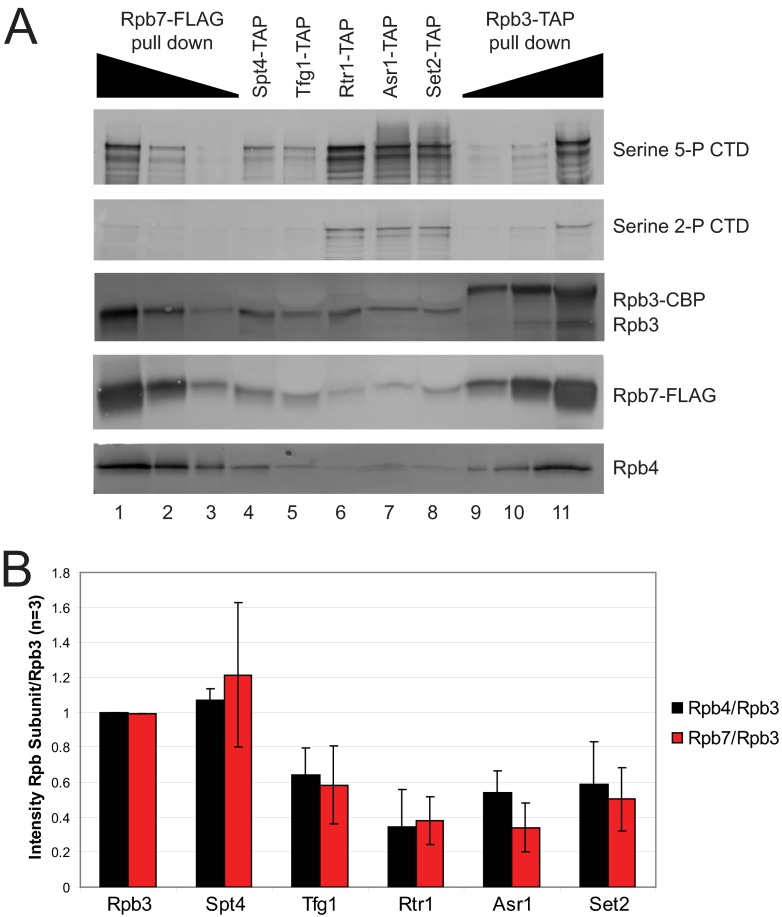
To confirm that the Rpb4/7 heterodimer is depleted in specific purified complexes, we performed Western blot analysis of the indicated TAP-purified samples (Fig. 3). Because we were unable to find a suitable Rpb7 antibody, endogenous RPB7 was FLAG-tagged via homologous recombination in all the strains for comparison. As shown in Fig. 3A, Rtr1-, Asr1-, and Set2-TAP samples were depleted in Rpb4 and Rpb7 in agreement with the MudPIT results (Fig. 3, lanes 6–8). Quantitation of replicate Western blots revealed that Rpb4/7 were depleted by ∼50% (Fig. 3B). To monitor the modification state of the CTD, we also performed Western blots using monoclonal antibodies against serine 5 and serine 2 CTD phosphorylation. Using this approach, we found that Rtr1-, Asr1-, and Set2-TAP samples contained increased levels of serine 5- and serine 2-phosphorylated CTD when compared with all other samples tested (Fig. 3A, compare lanes 6–8 to lanes 1–5 and 9–11). These data support previous work that has established that Rtr1, Asr1, and Set2 are recruited at a specific stage of RNAPII elongation in a manner dependent on the phosphorylation state of the CTD (26, 42, 44, 52, 56, 57). Interestingly, we also observed a lack of serine 2-phosphorylated CTD in the Rpb7-FLAG-isolated RNAPII as compared with Rpb3-TAP-isolated samples (Fig. 3A, compare lanes 9–11 to lanes 1–3).
Fig. 3.
Purifications that lack Rpb4 and Rpb7 are enriched in RNAPII with a hyperphosphorylated CTD. A, Western blot analysis of specific TAP-purified samples as indicated at the top of the figure. Increasing and decreasing concentrations of the Rpb7-FLAG (lanes 1–3) and Rpb3-TAP (lanes 9–11) immunoprecipitants varied by 5-fold across wells. The antibody used for visualization of each row of Western blots is indicated to the right of the figure. Lane numbers are given below the figure for comparison purposes. B, quantitation by densitometry of the levels of Rpb4 or Rpb7 when normalized to the levels of Rpb3 (n = 3). The purification to which each set of bars refers is listed at the bottom of the graph.
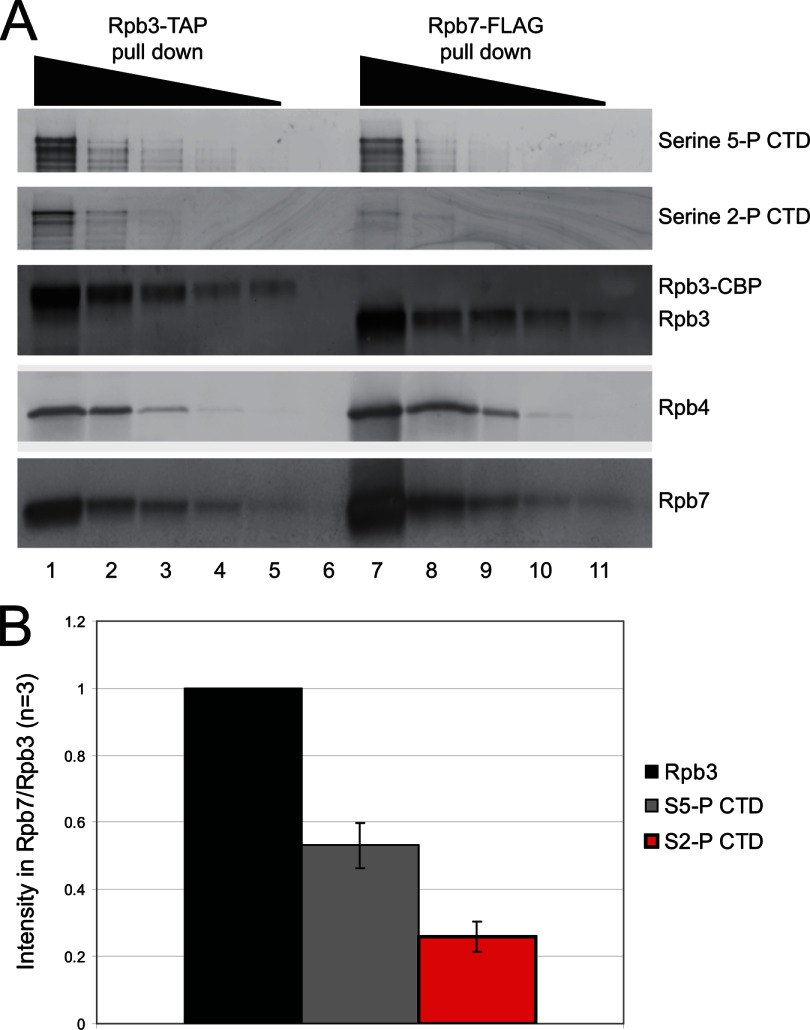
To determine whether there is a mutually exclusive relationship between serine 2 phosphorylation and the presence of Rpb4/7, we performed Western blot analysis on RNAPII isolated through either Rpb7 or Rpb3. To improve quantitation by Western blotting, we loaded 5-fold decreasing concentrations of each immunoprecipitant as shown in Fig. 4A. We observed a decreased amount of both serine 5- and serine 2-phosphorylated CTD in Rpb7-FLAG immunoprecipitants as compared with the levels of Rpb3 (Fig. 4A, compare lanes 1–5 against lanes 7–11). Replicate Western blots were quantitated by densitometry and revealed a 50% decrease in serine 5-phosphorylated CTD and a 70% decrease in serine 2-phosphorylated CTD in Rpb7-FLAG immunoprecipitants. These data clearly show that Rpb4/7 dissociate from the other 10 subunits of RNAPII in a manner that is dependent on the phosphorylation state of the CTD.
Fig. 4.
Rpb7-FLAG immunoprecipitants are depleted in serine 2- and serine 5-phosphorylated RNAPII. A, Western blot analysis of decreasing concentrations of the Rpb7-FLAG (lanes 1–6) and Rpb3-TAP (lanes 7–11) immunoprecipitants varied by 5-fold steps between wells. The antibody used for visualization of each row of Western blots is indicated to the right of the figure. Lane numbers are given below the figure for comparison purposes. B, quantitation by densitometry of the levels of serine 5- (S5-P CTD) or serine 2 (S2-P CTD)-phosphorylated Rpb1 in each purification normalized to the levels of Rpb3 (n = 3).
DISCUSSION
In addition to modification of the CTD, alterations in the subunit composition or modification state of the 12 RNAPII subunits have the potential to act as a specific regulatory signal during the regulation of transcription. The known heterodimer of Rpb4 and Rpb7 has been suggested to dissociate from the core of RNAPII; however, multiple studies using ChIP-based analyses have not been able to discover an example of Rpb4/7 dissociation during the RNAPII transcription cycle (22–25). In an effort to study the dynamics of RNAPII during transcriptional elongation, we have characterized multiple RNAPII-containing purifications using quantitative proteomics and found that Rtr1, Npa3, Set2, and Asr1 interact with hyperphosphorylated RNAPII that lacks the Rpb4/7 heterodimer. These studies clearly show that changes in the subunit composition of RNAPII may serve as an additional regulatory step during transcriptional elongation.
Although we have identified multiple RNAPII-containing purifications that are depleted specifically in Rpb4/7, we did not observe a significant amount of free Rpb4/7 using quantitative proteomics analyses. If a population of free Rpb4/7 did exist in the cell in significant amounts, we would expect to observe a statistically significant increase in the levels of Rpb4/7 in Rpb7-TAP purifications (Fig. 2). We did, however, observe a slight increase in the standard deviation of Rpb4 and Rpb7 levels in Rpb7-TAP as compared with Rpb3- and Rpb11-TAP purifications. It is possible that this increase in variability is observed specifically in Rpb7-TAP samples as a result of Rpb4/7 dissociation from RNAPII. Even after considering this possibility, our data clearly show that there is not a large population of free Rpb4/7 heterodimers suggesting that the dissociation of Rpb4/7 from RNAPII is short lived. Another possibility is that a ubiquitin ligase like Asr1 (52) may mark Rp4 and Rpb7 for degradation by the proteasome.
Previous studies have clearly shown that Set2, Rtr1, and Asr1 are enriched within the gene body using ChIP in yeast (26, 43, 52). The human homolog of Rtr1, RPAP2, has also been shown to localize to actively transcribed genes and to bind to hyperphosphorylated RNAPII (45). Human Set2 has also been shown to bind to hyperphosphorylated RNAPII specifically indicating that these interactions are conserved from yeast to humans (57). Our findings suggest that as serine 2 CTD phosphorylation increases, the association of Rpb4/7 decreases, and it is well established that serine 2 CTD phosphorylation increases within the coding region of RNAPII target genes in S. cerevisiae (1, 2, 58). However, it is not currently known if the dissociation of Rpb4/7 or serine 2 CTD phosphorylation is a prerequisite to the recruitment of Rtr1, Npa3, and/or Set2. Previous work by Daulny et al. (52) has shown that the E3 ligase Asr1 is capable of binding to RNAPII that either contains or lacks Rpb4/7. In our studies, Western blot analysis of Rtr1-, Asr1-, and Set2-isolated RNAPII complexes revealed that each complex was capable of interacting with Rpb4/7 to some extent, although at reduced levels relative to those seen in Spt4- or Tfg1-TAP (Fig. 3). These data suggest that even in the context of these elongation-specific protein interactions, Rpb4/7 dissociation may occur rapidly or in a gene-specific manner.
The observation that Rpb7-FLAG-isolated complexes are depleted in serine 2-phosphorylated RNAPII suggests that this CTD modification may play a role in the dissociation of Rpb4/7. Previously characterized functions of Rpb4/7 have implicated the heterodimer in additional regulation of transcriptional initiation and termination (14–16) (17). An additional explanation for the lack of serine 2 CTD phosphorylation in Rpb7-isolated RNAPII may be explained by the observation that the C terminus of Rpb4 is required for the interaction between the serine 2-specific CTD phosphatase Fcp1 and RNAPII (14–16). Under this circumstance, it is possible that the dissociation of Rpb4/7 is necessary to maintain the level of serine 2 phosphorylation through the 3′ end of specific genes. This could occur through prevention or reduction of Fcp1 recruitment until a specific stage in transcriptional elongation or termination during which Rpb4/7 is able to reassociate with RNAPII.
Supplementary Material
Acknowledgments
We thank the members of the Mosley, Washburn, and Workman laboratories for critical feedback during this project and during the preparation of this manuscript.
Footnotes
* This work was supported, in whole or in part, by National Institutes of Health Grant R01GM099714 (to A.L.M.). This work was also supported by the Stowers Institute for Medical Research (to M.P.W. and J.L.W.).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- RNAPII
- RNA polymerase II
- CTD
- C-terminal domain
- TEV
- tobacco etch virus
- PSM
- peptide spectral match
- MudPIT
- multidimensional protein identification technology
- NSAF
- normalized spectral abundance factor.
REFERENCES
- 1. Buratowski S. (2009) Progression through the RNA polymerase II CTD cycle. Mol. Cell 36, 541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Komarnitsky P., Cho E. J., Buratowski S. (2000) Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14, 2452–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kim M., Suh H., Cho E. J., Buratowski S. (2009) Phosphorylation of the yeast Rpb1 C-terminal domain at serines 2, 5, and 7. J. Biol. Chem. 284, 26421–26426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chapman R. D., Heidemann M., Albert T. K., Mailhammer R., Flatley A., Meisterernst M., Kremmer E., Eick D. (2007) Transcribing RNA polymerase II is phosphorylated at CTD residue serine 7. Science 318, 1780–1782 [DOI] [PubMed] [Google Scholar]
- 5. Egloff S., O'Reilly D., Chapman R. D., Taylor A., Tanzhaus K., Pitts L., Eick D., Murphy S. (2007) Serine 7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science 318, 1777–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Buratowski S. (2003) The CTD code. Nat. Struct. Biol. 10, 679–680 [DOI] [PubMed] [Google Scholar]
- 7. Van Mullem V., Wery M., Werner M., Vandenhaute J., Thuriaux P. (2002) The Rpb9 subunit of RNA polymerase II binds transcription factor TFIIE and interferes with the SAGA and elongator histone acetyltransferases. J. Biol. Chem. 277, 10220–10225 [DOI] [PubMed] [Google Scholar]
- 8. Ziegler L. M., Khaperskyy D. A., Ammerman M. L., Ponticelli A. S. (2003) Yeast RNA polymerase II lacking the Rpb9 subunit is impaired for interaction with transcription factor IIF. J. Biol. Chem. 278, 48950–48956 [DOI] [PubMed] [Google Scholar]
- 9. Soutourina J., Bordas-Le Floch V., Gendrel G., Flores A., Ducrot C., Dumay-Odelot H., Soularue P., Navarro F., Cairns B. R., Lefebvre O., Werner M. (2006) Rsc4 connects the chromatin remodeler RSC to RNA polymerases. Mol. Cell. Biol. 26, 4920–4933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Parnell T. J., Huff J. T., Cairns B. R. (2008) RSC regulates nucleosome positioning at pol II genes and density at pol III genes. EMBO J. 27, 100–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dichtl B., Blank D., Ohnacker M., Friedlein A., Roeder D., Langen H., Keller W. (2002) A role for SSU72 in balancing RNA polymerase II transcription elongation and termination. Mol. Cell 10, 1139–1150 [DOI] [PubMed] [Google Scholar]
- 12. Krishnamurthy S., He X., Reyes-Reyes M., Moore C., Hampsey M. (2004) Ssu72 Is an RNA polymerase II CTD phosphatase. Mol. Cell 14, 387–394 [DOI] [PubMed] [Google Scholar]
- 13. Cai G., Imasaki T., Yamada K., Cardelli F., Takagi Y., Asturias F. J. (2010) Mediator head module structure and functional interactions. Nat. Struct. Mol. Biol. 17, 273–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kimura M., Suzuki H., Ishihama A. (2002) Formation of a carboxyl-terminal domain phosphatase (Fcp1)/TFIIF/RNA polymerase II (pol II) complex in Schizosaccharomyces pombe involves direct interaction between Fcp1 and the Rpb4 subunit of pol II. Mol. Cell. Biol. 22, 1577–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tombácz I., Schauer T., Juhász I., Komonyi O., Boros I. (2009) The RNA pol II CTD phosphatase Fcp1 is essential for normal development in Drosophila melanogaster. Gene 446, 58–67 [DOI] [PubMed] [Google Scholar]
- 16. Kamenski T., Heilmeier S., Meinhart A., Cramer P. (2004) Structure and mechanism of RNA polymerase II CTD phosphatases. Mol. Cell 15, 399–407 [DOI] [PubMed] [Google Scholar]
- 17. Mitsuzawa H., Kanda E., Ishihama A. (2003) Rpb7 subunit of RNA polymerase II interacts with an RNA-binding protein involved in processing of transcripts. Nucleic Acids Res. 31, 4696–4701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dezélée S., Wyers F., Sentenac A., Fromageot P. (1976) Two forms of RNA polymerase B in yeast. Proteolytic conversion in vitro of enzyme BI into BII. Eur. J. Biochem. 65, 543–552 [DOI] [PubMed] [Google Scholar]
- 19. Edwards A. M., Kane C. M., Young R. A., Kornberg R. D. (1991) Two dissociable subunits of yeast RNA polymerase II stimulate the initiation of transcription at a promoter in vitro. J. Biol. Chem. 266, 71–75 [PubMed] [Google Scholar]
- 20. Choder M., Young R. A. (1993) A portion of RNA polymerase II molecules has a component essential for stress responses and stress survival. Mol. Cell. Biol. 13, 6984–6991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McKune K., Richards K. L., Edwards A. M., Young R. A., Woychik N. A. (1993) RPB7, one of two dissociable subunits of yeast RNA polymerase II, is essential for cell viability. Yeast 9, 295–299 [DOI] [PubMed] [Google Scholar]
- 22. Cojocaru M., Jeronimo C., Forget D., Bouchard A., Bergeron D., Côte P., Poirier G. G., Greenblatt J., Coulombe B. (2008) Genomic location of the human RNA polymerase II general machinery: evidence for a role of TFIIF and Rpb7 at both early and late stages of transcription. Biochem. J. 409, 139–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jasiak A. J., Hartmann H., Karakasili E., Kalocsay M., Flatley A., Kremmer E., Strässer K., Martin D. E., Söding J., Cramer P. (2008) Genome-associated RNA polymerase II includes the dissociable Rpb4/7 subcomplex. J. Biol. Chem. 283, 26423–26427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Verma-Gaur J., Rao S. N., Taya T., Sadhale P. (2008) Genomewide recruitment analysis of Rpb4, a subunit of polymerase II in Saccharomyces cerevisiae, reveals its involvement in transcription elongation. Eukaryot. Cell 7, 1009–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Runner V. M., Podolny V., Buratowski S. (2008) The Rpb4 subunit of RNA polymerase II contributes to cotranscriptional recruitment of 3′ processing factors. Mol. Cell. Biol. 28, 1883–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mosley A. L., Pattenden S. G., Carey M., Venkatesh S., Gilmore J. M., Florens L., Workman J. L., Washburn M. P. (2009) Rtr1 is a CTD phosphatase that regulates RNA polymerase II during the transition from serine 5 to serine 2 phosphorylation. Mol. Cell 34, 168–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mosley A. L., Sardiu M. E., Pattenden S. G., Workman J. L., Florens L., Washburn M. P. (2011) Highly reproducible label free quantitative proteomic analysis of RNA polymerase complexes. Mol. Cell. Proteomics 10, M110.000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zybailov B., Mosley A. L., Sardiu M. E., Coleman M. K., Florens L., Washburn M. P. (2006) Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J. Proteome Res. 5, 2339–2347 [DOI] [PubMed] [Google Scholar]
- 29. Paoletti A. C., Parmely T. J., Tomomori-Sato C., Sato S., Zhu D., Conaway R. C., Conaway J. W., Florens L., Washburn M. P. (2006) Quantitative proteomic analysis of distinct mammalian Mediator complexes using normalized spectral abundance factors. Proc. Natl. Acad. Sci. U.S.A. 103, 18928–18933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sardiu M. E., Florens L., Washburn M. P. (2009) Evaluation of clustering algorithms for protein complex and protein interaction network assembly. J. Proteome Res. 8, 2944–2952 [DOI] [PubMed] [Google Scholar]
- 31. Sardiu M. E., Gilmore J. M., Carrozza M. J., Li B., Workman J. L., Florens L., Washburn M. P. (2009) Determining protein complex connectivity using a probabilistic deletion network derived from quantitative proteomics. PLoS One 4, e7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang Y., Wen Z., Washburn M. P., Florens L. (2009) Effect of dynamic exclusion duration on spectral count based quantitative proteomics. Anal. Chem. 81, 6317–6326 [DOI] [PubMed] [Google Scholar]
- 33. Sardiu M. E., Cai Y., Jin J., Swanson S. K., Conaway R. C., Conaway J. W., Florens L., Washburn M. P. (2008) Probabilistic assembly of human protein interaction networks from label-free quantitative proteomics. Proc. Natl. Acad. Sci. U.S.A., 105, 1454–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pokholok D. K., Hannett N. M., Young R. A. (2002) Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell 9, 799–809 [DOI] [PubMed] [Google Scholar]
- 35. Chambers R. S., Wang B. Q., Burton Z. F., Dahmus M. E. (1995) The activity of COOH-terminal domain phosphatase is regulated by a docking site on RNA polymerase II and by the general transcription factors IIF and IIB. J. Biol. Chem. 270, 14962–14969 [DOI] [PubMed] [Google Scholar]
- 36. Tan S., Aso T., Conaway R. C., Conaway J. W. (1994) Roles for both the RAP30 and RAP74 subunits of transcription factor IIF in transcription initiation and elongation by RNA polymerase II. J. Biol. Chem. 269, 25684–25691 [PubMed] [Google Scholar]
- 37. Squazzo S. L., Costa P. J., Lindstrom D. L., Kumer K. E., Simic R., Jennings J. L., Link A. J., Arndt K. M., Hartzog G. A. (2002) The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 21, 1764–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wade P. A., Werel W., Fentzke R. C., Thompson N. E., Leykam J. F., Burgess R. R., Jaehning J. A., Burton Z. F. (1996) A novel collection of accessory factors associated with yeast RNA polymerase II. Protein Expr. Purif. 8, 85–90 [DOI] [PubMed] [Google Scholar]
- 39. Shi X., Finkelstein A., Wolf A. J., Wade P. A., Burton Z. F., Jaehning J. A. (1996) Paf1p, an RNA polymerase II-associated factor in Saccharomyces cerevisiae, may have both positive and negative roles in transcription. Mol. Cell. Biol. 16, 669–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Klein B. J., Bose D., Baker K. J., Yusoff Z. M., Zhang X., Murakami K. S. (2011) RNA polymerase and transcription elongation factor Spt4/5 complex structure. Proc. Natl. Acad. Sci. U.S.A. 108, 546–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lindstrom D. L., Hartzog G. A. (2001) Genetic interactions of Spt4-Spt5 and TFIIS with the RNA polymerase II CTD and CTD modifying enzymes in Saccharomyces cerevisiae. Genetics 159, 487–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li B., Howe L., Anderson S., Yates J. R., 3rd, Workman J. L. (2003) The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 278, 8897–8903 [DOI] [PubMed] [Google Scholar]
- 43. Krogan N. J., Kim M., Tong A., Golshani A., Cagney G., Canadien V., Richards D. P., Beattie B. K., Emili A., Boone C., Shilatifard A., Buratowski S., Greenblatt J. (2003) Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 23, 4207–4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gibney P. A., Fries T., Bailer S. M., Morano K. A. (2008) Rtr1 is the Saccharomyces cerevisiae homolog of a novel family of RNA polymerase II-binding proteins. Eukaryot. Cell 7, 938–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Egloff S., Zaborowska J., Laitem C., Kiss T., Murphy S. (2012) Ser7 phosphorylation of the CTD recruits the RPAP2 Ser5 phosphatase to snRNA genes. Mol. Cell 45, 111–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jeronimo C., Forget D., Bouchard A., Li Q., Chua G., Poitras C., Thérien C., Bergeron D., Bourassa S., Greenblatt J., Chabot B., Poirier G. G., Hughes T. R., Blanchette M., Price D. H., Coulombe B. (2007) Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol. Cell 27, 262–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Staresincic L., Walker J., Dirac-Svejstrup A. B., Mitter R., Svejstrup J. Q. (2011) GTP-dependent binding and nuclear transport of RNA polymerase II by Npa3 protein. J. Biol. Chem. 286, 35553–35561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gavin A. C., Bösche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J. M., Michon A. M., Cruciat C. M., Remor M., Höfert C., Schelder M., Brajenovic M., Ruffner H., Merino A., Klein K., Hudak M., Dickson D., Rudi T., Gnau V., Bauch A., Bastuck S., Huhse B., Leutwein C., Heurtier M. A., Copley R. R., Edelmann A., Querfurth E., Rybin V., Drewes G., Raida M., Bouwmeester T., Bork P., Seraphin B., Kuster B., Neubauer G., Superti-Furga G. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141–147 [DOI] [PubMed] [Google Scholar]
- 49. Uetz P., Giot L., Cagney G., Mansfield T. A., Judson R. S., Knight J. R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Qureshi-Emili A., Li Y., Godwin B., Conover D., Kalbfleisch T., Vijayadamodar G., Yang M., Johnston M., Fields S., Rothberg J. M. (2000) A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403, 623–627 [DOI] [PubMed] [Google Scholar]
- 50. Carre C., Shiekhattar R. (2011) Human GTPases associate with RNA polymerase II to mediate its nuclear import. Mol. Cell. Biol., 31, 3953–3962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Calera M. R., Zamora-Ramos C., Araiza-Villanueva M. G., Moreno-Aguilar C. A., Peña-Gómez S. G., Castellanos-Terán F., Robledo-Rivera A. Y., Sánchez-Olea R. (2011) Parcs/Gpn3 is required for the nuclear accumulation of RNA polymerase II. Biochim. Biophys. Acta 1813, 1708–1716 [DOI] [PubMed] [Google Scholar]
- 52. Daulny A., Geng F., Muratani M., Geisinger J. M., Salghetti S. E., Tansey W. P. (2008) Modulation of RNA polymerase II subunit composition by ubiquitylation. Proc. Natl. Acad. Sci. U.S.A. 105, 19649–19654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Harel-Sharvit L., Eldad N., Haimovich G., Barkai O., Duek L., Choder M. (2010) RNA polymerase II subunits link transcription and mRNA decay to translation. Cell 143, 552–563 [DOI] [PubMed] [Google Scholar]
- 54. Goler-Baron V., Selitrennik M., Barkai O., Haimovich G., Lotan R., Choder M. (2008) Transcription in the nucleus and mRNA decay in the cytoplasm are coupled processes. Genes Dev. 22, 2022–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lotan R., Goler-Baron V., Duek L., Haimovich G., Choder M. (2007) The Rpb7p subunit of yeast RNA polymerase II plays roles in the two major cytoplasmic mRNA decay mechanisms. J. Cell Biol. 178, 1133–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Selitrennik M., Duek L., Lotan R., Choder M. (2006) Nucleocytoplasmic shuttling of the Rpb4p and Rpb7p subunits of Saccharomyces cerevisiae RNA polymerase II by two pathways. Eukaryot. Cell 5, 2092–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Vojnic E., Simon B., Strahl B. D., Sattler M., Cramer P. (2006) Structure and carboxyl-terminal domain (CTD) binding of the Set2 SRI domain that couples histone H3 Lys36 methylation to transcription. J. Biol. Chem. 281, 13–15 [DOI] [PubMed] [Google Scholar]
- 58. Li M., Phatnani H. P., Guan Z., Sage H., Greenleaf A. L., Zhou P. (2005) Solution structure of the Set2-Rpb1 interacting domain of human Set2 and its interaction with the hyperphosphorylated C-terminal domain of Rpb1. Proc. Natl. Acad. Sci. U.S.A. 102, 17636–17641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ahn S. H., Kim M., Buratowski S. (2004) Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell 13, 67–76 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.