Abstract
CSF-1 mRNA 3′UTR contains multiple unique motifs, including a common microRNA (miRNA) target in close proximity to a noncanonical G-quadruplex and AU-rich elements (AREs). Using a luciferase reporter system fused to CSF-1 mRNA 3′UTR, disruption of the miRNA target region, G-quadruplex, and AREs together dramatically increased reporter RNA levels, suggesting important roles for these cis-acting regulatory elements in the down-regulation of CSF-1 mRNA. We find that nucleolin, which binds both G-quadruplex and AREs, enhances deadenylation of CSF-1 mRNA, promoting CSF-1 mRNA decay, while having the capacity to increase translation of CSF-1 mRNA. Through interaction with the CSF-1 3′UTR miRNA common target, we find that miR-130a and miR-301a inhibit CSF-1 expression by enhancing mRNA decay. Silencing of nucleolin prevents the miRNA-directed mRNA decay, indicating a requirement for nucleolin in miRNA activity on CSF-1 mRNA. Downstream effects followed by miR-130a and miR-301a inhibition of directed cellular motility of ovarian cancer cells were found to be dependent on nucleolin. The paradoxical effects of nucleolin on miRNA-directed CSF-1 mRNA deadenylation and on translational activation were explored further. The nucleolin protein contains four acidic stretches, four RNA recognition motifs (RRMs), and nine RGG repeats. All three domains in nucleolin regulate CSF-1 mRNA and protein levels. RRMs increase CSF-1 mRNA, whereas the acidic and RGG domains decrease CSF-1 protein levels. This suggests that nucleolin has the capacity to differentially regulate both CSF-1 RNA and protein levels. Our finding that nucleolin interacts with Ago2 indirectly via RNA and with poly(A)-binding protein C (PABPC) directly suggests a nucleolin-Ago2-PABPC complex formation on mRNA. This complex is in keeping with our suggestion that nucleolin may work with PABPC as a double-edged sword on both mRNA deadenylation and translational activation. Our findings underscore the complexity of nucleolin's actions on CSF-1 mRNA and describe the dependence of miR-130a- and miR-301a-directed CSF-1 mRNA decay and inhibition of ovarian cancer cell motility on nucleolin.
mRNA degradation and translation are intimately coupled processes in which RNA-binding proteins (RBPs)1 and miRNAs interact with cis-acting regulatory elements in mRNA-untranslated regions (UTRs). miRNAs usually elicit translation repression and mRNA decay (1–4), a complex process that involves multiple components, including argonaute (Ago) proteins. Ago2, as part of the miRNA-induced silencing complex (miRISC), triggers rapid mRNA degradation by accelerating a two-phase deadenylation process (5). In general, longer 3′UTRs contain a higher density of miRNA-binding sites increasing the chances of being regulated by miRNA (6).
In addition to miRNA target sequences (6), the mRNA 3′UTR confers multiple layers of post-transcriptional regulation as it can contain other unique motifs such as a G-quadruplex (7), and AU-rich elements (AREs), for protein binding. Several reports indicate that RBPs, which bind to mRNA 3′UTR, can modulate miRNA-directed repression of target mRNA. For instance, miRNA-directed translational repression and mRNA decay can be relieved by RBPs (8–10). RBPs can also enhance miRNA-directed mRNA decay and translational suppression (11–13). In this regard, importin 8 binds Ago2 and can enhance the association efficiency between miRNPs and target mRNAs (11). The RNA-binding protein TRIM32 binds Ago1, thereby increasing the activity of Let-7 miRNA (12).
The cytokine CSF-1 activates the CSF-1 tyrosine kinase receptor, encoded by the proto-oncogene c-fms. Overexpression of CSF-1/c-fms is associated with poor prognosis of epithelial ovarian cancer patients and enhances invasive and metastatic behavior (14, 15). The longest CSF-1 transcript (3,939 nt) gives rise to the major form of secreted CSF-1 (16, 17) in ovarian cancer. The 2,172-nt 3′UTR of the major 3,939-nt CSF-1 mRNA contains several unique motifs, including a common miRNA binding target we described (18), AREs (19, 20), and a noncanonical G-quadruplex (this report), which are involved in post-transcriptional regulation.
Nucleolin is a multifunctional protein that we describe in this report to bind the G-quadruplex and AREs in CSF-1 3′UTR mRNA. It is involved in ribosome biogenesis for rRNA transcription and processing as well as pre-ribosome assembly in the nucleolus (21). Even though nucleolin is highly localized in the nucleolus (22, 23), its presence is also found in the cytoplasm and on the cell surface (24). In the cytoplasm, nucleolin binds both strategic mRNAs and proteins. Nucleolin, which contains three major domains for RNA or protein binding, is known to post-transcriptionally regulate mRNA stability (25, 26) and also to interact with mRNA 3′UTRs and proteins for translational regulation (27, 28).
The G-quadruplex is a noncanonical tetrahelix formed from guanine-rich sequences, which is found both in the promoter region of the gene (29) and in mRNA 5′- and 3′UTRs (7). G-quadruplex DNA structures have a role in regulating telomere maintenance. Although the roles for G-quadruplex RNA structures are not fully understood, those found in the 5′UTR are involved in internal ribosome entry site-dependent translation initiation (30) as well as translational repression (31). Nucleolin has been described to bind G-rich elements in coding and noncoding regions (32). However, little is known about the effects of interactions of nucleolin, and RBPS in general, with the mRNA 3′UTR G-quadruplex.
AREs are well studied, typically found in mRNA 3′UTR, and are known to dictate mRNA decay (33). The seed for miR-16 is complementary to ARE, and miR-16 is involved in ARE-mediated mRNA degradation (34), requiring an ARE-binding protein tristetraproline, which interacts with Ago/eIF2C proteins. Nucleolin has previously shown an affinity for AREs on bcl-2 and Bcl-XL mRNAs (26, 35).
In this report, we show that disruption of all three cis-acting regulatory elements, including G-quadruplex, AREs, and the common miRNA target region in the CSF-1 mRNA 3′UTR, dramatically increases luciferase reporter RNA levels. This suggests important roles for these cis-acting regulatory elements in the down-regulation of CSF-1 mRNA. Our studies show that nucleolin's actions on CSF-1 mRNA are complex, with nucleolin enhancing CSF-1 mRNA deadenylation, thereby promoting miRNA-directed CSF-1 mRNA decay, while activating translation of CSF-1 mRNA. We offer a model of how nucleolin may work with PABPC as a double-edged sword on both mRNA deadenylation and translational activation.
EXPERIMENTAL PROCEDURES
Cell Culture and Protein Fractionation
Human epithelial ovarian cancer cell lines (Hey and SKOV3) that constitutively express high levels of CSF-1 were maintained in DMEM/F-12 (Mediatech) supplemented with 10% fetal bovine serum. Bix3 human epithelial ovarian cancer cells that express low levels of CSF-1 (36) were also maintained in DMEM/F-12 supplemented with 10% fetal bovine serum. Nuclear and cytoplasmic protein extract was prepared from cells using NE-PER nuclear and cytoplasmic extraction reagents (Pierce). Total cellular protein extract was prepared from cells using RIPA buffer (25 mm Tris-HCl, pH 7.6, 150 mm NaCl, 1% Nonidet P-40, 1% sodium deoxycholate, 0.1% SDS) and protease inhibitor mixture set 1 at 1:100 dilution (Calbiochem). Protein concentrations were determined by BCA assay (Pierce).
Identification of Nucleolin as a G-quadruplex-binding Protein
CSF-1 mRNA 3′UTR G-quadruplex tetrahelix-binding proteins were purified from Hey cell lysates. The lysates were applied to a Sephacryl S-400 gel and eluted with 250 mm NaCl. Active fractions were identified by UV cross-linking, pooled together, and applied to a heparin-agarose resin to enrich for RBPs. After the heparin column, active fractions were identified by UV cross-linking and pooled together. Active fractions were then applied to the biotin-labeled G-quadruplex RNA-bound streptavidin resin. Bound proteins were eluted by 8 m guanidine-HCl and separated by SDS-PAGE. The ∼105-kDa band was excised and sequenced by LC-MS/MS analysis (University of Arizona Cancer Center, Proteomics Core).
Identification of Nucleolin as an ARE-binding Protein
32P-labeled CSF-1 3′UTR 144-nt RNA, which contains three AREs, was UV cross-linked with Hey cell lysates. RNase A was added to the reaction and incubated at 37°C for 1 h. The reaction was resolved on an IEF gel for the first dimension and a 12% SDS-polyacrylamide gel for the second dimension and autoradiographed. The protein spots in the 2-D gel were excised and sequenced by LC-MS/MS analysis (University of Arizona Cancer Center, Proteomics Core).
LC-MS/MS for Nucleolin Identification
Protein bands excised from SDS-PAGE were in-gel-digested with trypsin (10 μg/ml) at 37°C overnight (56). Using a linear quadrupole ion trap ThermoFinnigan LTQ mass spectrometer (San Jose, CA) equipped with a Michrom Paradigm MS4 HPLC, a SpectraSystems AS3000 autosampler, and a nanoelectrospray source, LC-MS/MS analyses were done. Peptides were eluted from a 15-cm pulled tip capillary column (100 μm inner diameter × 360 μm outer diameter; 3–5-μm tip opening) packed with a 7-cm Vydac C18 (5 μm, 300 Å pore size, Hesperia, CA), using a 0–70% gradient of solvent B (98% methanol, 2% water, 0.5% formic acid, 0.01% trifluoroacetic acid) over a 60-min period at a flow rate of 350 nl/min. The LTQ electrospray positive mode spray voltage was 1.6 kV, and the capillary temperature was 180°C. Dependent data scanning was performed by the Xcalibur version 1.4 software (57) with a default charge of 2, an isolation width of 1.5 atomic mass units, an activation amplitude of 35%, activation time of 30 ms, and a minimal signal of 100 ion counts. Globally dependent data were set as follows: reject mass width of 1.5 atomic mass units; dynamic exclusion enabled; exclusion mass width of 1.5 atomic mass units; repeat count of 1; repeat duration of 1 min; and exclusion duration of 5 min. Scan event series included one full scan with a mass range of 350–2000 Da, followed by three dependent MS/MS scans of the most intense ion. Tandem MS spectra of peptides were analyzed with SEQUESTTM (version 27, revision 12, ThermoFinnigan, San Jose, CA), a program that allows the correlation of experimental tandem MS data with theoretical spectra generated from known protein sequences (58). The peak list for the search was generated by Bioworks 3.1. Parent peptide mass error tolerance was set at 2.0 atomic mass units, and fragment ion mass tolerance was set at 1.0 atomic mass unit during the search. The criteria used for a preliminary positive peptide identification are the same as described previously, namely a dCn score >0.08 and Xcorr >1.8 for +1, Xcorr >2.5 for +2, and Xcorr >3.5 for +3 charged peptide precursor ions (59). Peptides matching proteins of interest were confirmed by visual examination of the spectra. All spectra were searched against a human protein database downloaded from IPI October 30, 2009 (ipihuman version 3.65), based on semitryptic peptides between 700 and 4,500 atomic mass units with up to two missed cleavages. At the time of the search, the ipihuman version 3.65 protein database contained 86,379 entries. The results were also validated using X!Tandem, another search engine (60), and visualized with Scaffold (version Scaffold_3_00_02, Proteome Software Inc., Portland, OR), a program that relies on various search engine results (i.e. SEQUESTTM, X!Tandem, MASCOT) and uses Bayesian statistics to reliably identify more spectra (61, 62). X!Tandem identifications required at least −log(expect scores) of greater than 3.0. Protein identifications were accepted if they contained at least two identified peptides. Oxidation of methionine and iodoacetamide derivative of cysteine was specified in SEQUESTTM and X!Tandem as variable modifications. The protein false discovery rate was calculated at 0.1% by the Scaffold software using the ProteinProfit-based probabilities ((sum of probabilities of proteins/sum of maximum probabilities for each protein) = A; 1.00 − + = B·100 = false discovery rate).
UV-Cross-linking and Label Transfer of Nucleolin with 3′UTR CSF-1 RNA
UV cross-linking of nucleolin was performed as described previously (37). Synthesis of the 32P-labeled riboprobe was performed according to the manufacturer's instructions (Ambion). RNAs of different regions of CSF-1 3′UTR labeled with [32P]ATP to the same specific activity were incubated with 0.5 μg of nucleolin or 0.5 μg of maltose-binding protein (MBP). To analyze protein-RNA cross-links, reaction mixtures were electrophoresed on an SDS-polyacrylamide gel, and radioactive bands were detected by phosphorimager after 24 h.
RNA Electrophoretic Mobility Shift Assay (EMSA)
RNA EMSA was performed as described previously (18).
Immunoblot Analysis
Proteins were extracted from Bix3, Hey, or SKOV3 cells and loaded (50–60 μg of protein) per lane on a 4–15% SDS-polyacrylamide gel, electrophoresed, and transferred to an Immune-Blot PVDF Membrane (Bio-Rad). Membranes were probed with CSF-1 mAb (ab66236, Abcam), nucleolin mAb (sc-17826, Santa Cruz Biotechnology), or GFP mAb (sc-9996, Santa Cruz Biotechnology). Immunoblot processing and ECL protein detection were performed according to the manufacturer's instructions using SuperSignal West Pico (Pierce) and HRP-conjugated 2° Abs (Santa Cruz Biotechnology).
Production and Purification of Recombinant Nucleolin
Human nucleolin cDNA cloned in the pMAL-c2X vector was expressed in Escherichia coli for recombinant nucleolin protein production according to the manufacturer's protocol (New England Biolabs).
DNA Constructs
cDNA fragments corresponding to the nucleolin acidic stretches, RRM domain, and/or RGG repeats were cloned into the XhoI and BamHI sites of pEGFP-C1 (Clontech). After transfection into human ovarian cancer cells, immunoblots of cell lysates were performed with anti-nucleolin and anti-EGFP antibodies to confirm that the respective proteins were overexpressed.
Reporter Constructs for Luciferase Analysis
Both wild type and mutant 3′UTR CSF-1 2,172 nt (1,768–3,939) were cloned into the 3′UTR of pMirTarget vector (Origene).
Dual-Luciferase Activity Analysis
SKOV3 and Bix3 cells were cultured in DMEM/F-12 as described above. For luciferase analysis, SKOV3 cells were transiently transfected using FuGENE HD (Roche Applied Science) with Luc-CSF-1–3′UTR-WT or Luc-CSF-1–3′UTR-Mut and Renilla control plasmid. Dual-Luciferase activity assays were performed 48 h after transfection according to the manufacturer's directions (Promega). Luciferase and GAPDH mRNA levels were measured by qRT-PCR.
mRNP Immunoprecipitation
IP of endogenous RNA-protein complexes was done by the protocol described previously (37). Briefly, cells were fixed in 1% formaldehyde. Cellular proteins were extracted using RIPA buffer. For nucleolin IP, 5 μg of mouse monoclonal anti-human nucleolin antibody (sc-17826, Santa Cruz Biotechnology) was used. A reaction containing normal mouse IgG (Sigma-Aldrich) served as a negative control. One-quarter of the beads was washed and boiled for Western blot analyses. The rest were used for mRNA extraction of the immunoprecipitates, followed by RNase-free DNase I (Fermentas) and proteinase K (Ambion) digestions, phenol/chloroform extraction, and ethanol precipitation. qRT-PCR was used to assess the abundance of nucleolin-associated CSF-1 or Bcl-xL mRNA, as described above. Actin mRNA was served as a loading control. The oligomer pairs used for the amplification of PCR products are listed in supplemental Table S1.
Size-exclusion Chromatography
Formaldehyde cross-linked SKOV3 cell lysates were size-fractionated in a Sephacryl 400-HR column. Five hundred-μl fractions were collected for immunoblot analysis and qRT-PCR.
Gain-of-Function and Loss-of-Function Assay
Plasmids encoding a control shRNA or shRNAs directed against nucleolin were purchased from Origene. The shRNAs correspond to coding region nucleotides 553–581 (5′-AAGAATGCCAAGAAGGAAGACAGTGATGA-3′), 769–796 (5′-GCTATGGAGACTACACCAGCCAAAGGAA-3′), 1141–1168 (5′-AATGATCTTGCTGTTGTGGATGTCAGAA-3′), and 2242–2270 (5′-AAGCCACAAGGAAAGAAGACGAAGTTTGA-3′), respectively, of human nucleolin (GenBankTM NM_005381). An shRNA containing a noneffective 29-nt GFP sequence (TR30003, Origene) was used as a negative control (Empty). For RNAi, 5 × 106 cells were transfected with 10 μg of shRNA plasmid using FuGENE HD (Roche Applied Science) according to the manufacturer's instructions. Transfected cells were maintained in culture medium for 2 days to permit knockdown before assays.
For nucleolin overexpression, nucleolin-pEGFP-C1 was used. SKOV3 and Bix3 cells at 70% confluence in 6-well plates were transfected with 4 μg of plasmids. The overexpression effects were monitored 48 h after transfection by qRT-PCR and Western blot analyses.
For miRNA overexpression, either pEP-has-miR-130a or pEP-has-miR-301a (Cell Biolabs) was used. pEP-miR-Null was used as an empty vector control.
For miRNA inhibition, 40 nm hsa-miR-130a inhibitor or hsa-miR-301a inhibitor (Exiqon) was used. miRCURY LNATM microRNA inhibitor negative control (Exiqon) was used as a negative control.
Analysis of CSF-1 mRNA Half-life
To determine CSF-1 or reporter mRNA half-life in human ovarian cancer cells, actinomycin-D (Act-D) chase experiments were performed with 3 μg/ml Act-D (Sigma-Aldrich) added to inhibit new transcription. For cells undergoing transfection of various constructs, the experiments were performed 2 days after transfection. Cells were harvested at 2, 2.5, 3, 3.5, 4, 4.5, 5, and 6 h after Act-D treatment; total cellular RNA was extracted using TRIzol (Invitrogen), and CSF-1 mRNA or luciferase RNA and GAPDH or Renilla mRNA levels were analyzed by qRT-PCR. CSF-1 or luciferase mRNA half-lives were calculated after qRT-PCR and normalized to GAPDH mRNA; values were plotted, and the time period required for a given transcript to decrease to one-half of the initial abundance was calculated. GAPDH mRNA was not affected by either nucleolin or miR-130a or -301a, and it has a long half-life (>18 h). Three independent experiments were performed.
Polyribosome Profile Assay
Polysome profiling was described previously (37). A sucrose gradient was constituted by adding 2 ml of 47, 37, 27, 17, and 7% sucrose solution (75 mm KCl, 20 mm Tris-HCl, pH 7.4, 1.5 mm MgCl2, 50 μg/ml cycloheximide) from bottom to top in a polyallomer centrifuge tube (Beckman). Transfected SKOV3 cells were cultured in two 10-cm Petri dishes to 75–80% confluence. Before harvest, cells were treated with 100 μg/ml cycloheximide for 15 min and then washed two times with cold PBS containing 100 μg/ml cycloheximide. Three mg of protein samples were loaded onto the top of the sucrose gradient and centrifuged at 39,000 rpm at 4°C for 2 h using a Beckman L7-55 ultracentrifuge. After centrifugation, gradient samples were fractionated 600 μl per fraction, and absorbance was monitored at λ = 254 nm. The RNAs of each fraction were extracted using TRIzol (Invitrogen). For analysis of the relative distributions of CSF-1 and GAPDH mRNA in polyribosome gradients, Ct values from individual fractions 1–10 were each subtracted from the Ct value from either fraction 1 for CSF-1 and GAPDH mRNAs, as fraction 1 had the largest Ct values (that is the lowest CSF-1 and GAPDH mRNA abundances). The resulting ΔCt numbers were converted into fold differences. The abundance of each mRNA as a percentage of the total from all 10 fractions was then calculated. Because GAPDH mRNA is not a binding target of nucleolin (35), its abundance was used as a control for estimation of relative distribution of CSF-1 mRNA in polyribosome gradients after nucleolin knockdown or overexpression. For IB, equal volumes (40 μl) of each fraction were electrophoresed through 4–15% SDS-polyacrylamide gels.
Ligation-mediated poly(A) test was done according to Murray and Schoenberg (38) with modifications. Total RNA was isolated by TRIzol. For cDNA synthesis, 5′-phosphorylated oligo(dT)16 and oligo(dT) primer/adapter (CGG GGG ATC CGC GGT TTT TTT TTT) were annealed and synthesized by Superscript II reverse transcriptase (Invitrogen). cDNA was diluted 10-fold by H2O and used in PCRs with target-specific primers. PCR products were separated in 2% agarose gel, blotted to nylon membrane, and hybridized by either digoxigenin-labeled luciferase, CSF-1, or GAPDH DNAs. Signal was detected by anti-digoxigenin antibody (Abcam), and x-ray film was exposed.
Transwell Motility Assay
After 2-days of transfection, 5 × 104 cells were seeded in 1% NuSerumTM in the top chamber of 24-well inserts with uncoated 8-μm pore membranes (BD Biosciences). The bottom chamber contained 20% FBS and 12.5 μg/ml fibronectin as chemoattractants. Six hours after seeding, the top chamber cells were wiped off completely with Q-tips, and both top and bottom chambers were washed with cold PBS, and then the membrane was dried and frozen at −80°C for 30 min. Cells that have migrated through the membrane into the bottom chamber were quantified by the lysis method using CyQuant cell proliferation assay kit (Invitrogen) as per the manufacturer's protocol. Percent motility toward a chemoattractant in the bottom chamber under various conditions was calculated by dividing the number of cells in the bottom chamber by the number of cells loaded in the upper chamber.
Statistical Analyses
Data are depicted as means ± S.D. or means ± S.E. from at least three independent experiments. Exact n values are provided in the figure legends. The unpaired two-way t test and one-way analysis of variance test were performed using SigmaStat (Jandel Scientific Corp.). p < 0.05 was considered statistically significant. The Pearson product moment correlation test was performed using SigmaStat (Jandel Scientific Corp.) for correlation analysis between miRNA and CSF-1 expression levels.
RESULTS
CSF-1 mRNA Expression Is Down-regulated by Three Elements within Its 3′UTR
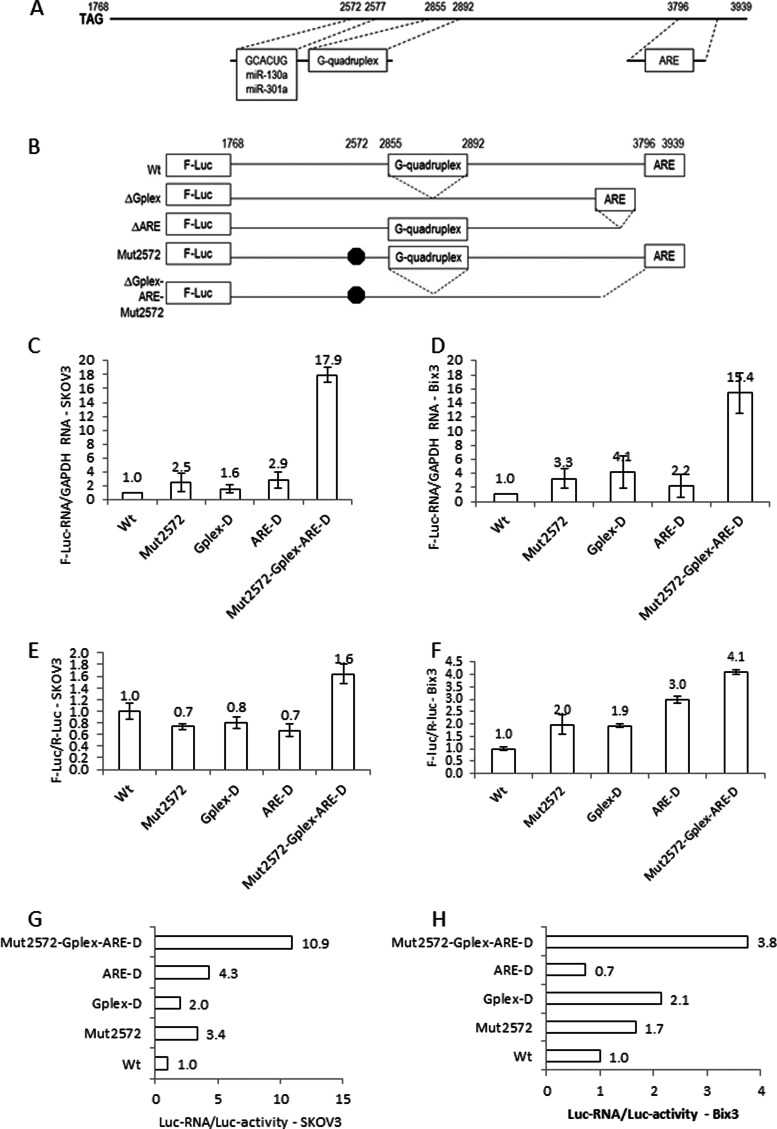
Exon 10 in the CSF-1 gene encodes the mRNA 3′UTR, which is 2,172 nt-long excluding the poly(A+) tail (Fig. 1A). In CSF-1 mRNA 3′UTR, both AU-rich elements (AREs) (3796–3939) and a common miRNA target sequence (2572–2577) exist, which we have previously described (18, 19, 20). In this report, we will describe the presence of a putative noncanonical G-quadruplex (2855G3N2G3N6G3N9G3N2G3N1G32892) sequence in the CSF-1 mRNA 3′UTR. To obtain insight into the mechanism by which these cis-acting regulatory elements regulate the expression of CSF-1 in ovarian cancer cells, we generated constructs of luciferase RNA fused to CSF-1 mRNA 3′UTR with point mutations in the common miRNA target and/or deletion of G-quadruplex and/or AREs to disrupt the function of these elements (Fig. 1B). We found that either single disruption or combination disruption of all three cis-acting regulatory sequences together showed more pronounced effects on enhancing luciferase RNA levels than luciferase activities in SKOV3 cells (Fig. 1, C and E). Disruption of all three cis-acting regulatory sequences resulted in a 17.9-fold increase (p = <0.001) in luciferase RNA levels contrasted to a 1.6-fold increase (p = <0.001) in luciferase activity compared with the wild type construct (Fig. 1, C and E). A similar effect was observed in Bix3 cells, in which disruption of all three cis-acting regulatory sequences resulted in a 15.4-fold increase (p = <0.001) in luciferase RNA level contrasted to a 4.1-fold increase (p = <0.001) in luciferase activity (Fig. 1, D and F). The ratio of normalized luciferase RNA to the corresponding normalized luciferase activity is 10.9, by mutation of all three cis-acting regulatory sequences in SKOV3 cells (Fig. 1G). A similar effect was also observed in Bix3 cells (Fig. 1H), with a 3.76-fold difference observed, favoring a primary effect at the RNA level. Mutations in the pairwise combinations of two cis-acting elements did not show as great of an effect as mutations in all three elements (supplemental Fig. S1), indicating that the three elements act together to repress CSF-1 mRNA expression.
Fig. 1.
miRNA target, G-quadruplex, and ARE work together to down-regulate CSF-1 mRNA. A, CSF-1 mRNA 3′UTR showing the miR-130a and miR-301a target, G-quadruplex, and ARE. B, schematic diagram of G-quadruplex (2855–2892) and ARE (3796–3939) deletion mutants and miRNA target mutant (2572) ligated with firefly luciferase (F-Luc) RNA. C, SKOV3. D, Bix3 cells, firefly luciferase RNA levels were normalized to those of the GAPDH mRNA and set to 1 in cells expressing the wild type sequence. E, SKOV3. F, Bix3 ovarian cancer cells, firefly luciferase activities were normalized to those of the Renilla luciferase (R-Luc) activities. n = 3. Mean ± S.D. is presented. In each experiment (C–F), there was a significant difference between the effects of the mutant targeting all three motifs (G-quadruplex, ARE, and common miRNA target) and wild type (p < 0.001). G, SKOV3. H, Bix 3 cells, the normalized firefly luciferase RNA values were divided by the corresponding normalized firefly luciferase activities for each condition. These ratios were set to 1 in cells expressing wild type sequence.
Identification of Nucleolin as a CSF-1 mRNA 3′UTR Interacting Protein
Because 3′UTRs can regulate mRNA stability and translation by interacting with different RBPs and miRNAs, we hypothesized that several RBPs and miRNAs may be involved in CSF-1 mRNA stabilization and/or translation. We have previously identified GAPDH as one protein that stabilizes CSF-1 mRNA through its 3′UTR AREs, in ovarian cancer cells (19, 20).
In the CSF-1 mRNA 3′UTR, the putative G-quadruplex sequence with six repeats of “GGG ” is located 279 nt downstream from the common miRNA target sequence (Fig. 1A). Both CD and NMR analyses indicate that this region (2855–2892) in CSF-1 mRNA 3′UTR can form a noncanonical G-quadruplex for protein binding (supplemental Fig. S2, A and B). Based on this observation, we decided to identify the RBPs that bind this G-quadruplex. Through serial biochemical purifications, we enriched for CSF-1 mRNA G-quadruplex-binding proteins from extracts of Hey epithelial ovarian cancer cells (supplemental Fig. S2, C and D). We used a synthetic 127-nt RNA sequence, including a 38-nt G-quadruplex sequence (2855–2892) of CSF-1 mRNA 3′UTR to capture G-quadruplex RBPs. After the bound proteins were resolved by SDS-PAGE and silver-stained, there were at least three protein bands that were potential CSF-1 mRNA G-quadruplex RBPs (supplemental Fig. S3A). The most abundant protein (∼105 kDa) was excised, and five peptides were identified as nucleolin by LC-MS/MS sequencing analysis (accession number gi|55956788, 11.4% sequence coverage, supplemental Fig. S4).
Because the majority of AREs are clustered in a 144-nt sequence (3796–3939) in CSF-1 mRNA 3′UTR (Fig. 1A), and GAPDH protein was found to interact with this ARE-rich 144 nt in ovarian cancer cells (19, 20), a synthetic 144-nt RNA sequence of CSF-1 mRNA 3′UTR was used to search for RBPs from the Hey cell lysates. After UV cross-linking RBPs with 32P-labeled 144-nt RNA, the proteins were resolved by two-dimensional PAGE (supplemental Fig. S3B). We found a single spot (∼105 kDa, pI = ∼4.8) that was a potential ARE-binding protein. Interestingly, nucleolin was the protein identified as the CSF-1 mRNA 3′UTR ARE-binding protein (accession number gi|55956788, 3.6% sequence coverage, supplemental Fig. S4). Nucleolin has been reported in the post-transcriptional regulation of several mRNAs, including Bcl-XL mRNA (39) by binding AREs in mRNA 3′UTRs.
Because nucleolin is simultaneously identified as a CSF-1 mRNA 3′UTR G-quadruplex- and ARE-binding protein in vitro, and CSF-1 can be post-transcriptionally regulated, we hypothesized that nucleolin may regulate CSF-1 expression by binding to the 3′UTR of CSF-1 mRNA. To test whether nucleolin directly associates with CSF-1 mRNA in ovarian cancer cells, IP assay using anti-human nucleolin monoclonal antibody was performed. The association of CSF-1 mRNA with nucleolin was determined by isolating RNA from the IP material and subjecting it to qRT-PCR analysis. As shown in Fig. 2, A and B, the CSF-1 mRNA was dramatically enriched in nucleolin IP samples compared with control IgG IP samples in Hey cell lysates. The association of CSF-1 mRNA with nucleolin was 9.9-fold higher than that seen in control IgG IP reaction (Fig. 2B). Actin mRNA does not interact with nucleolin, and amplification of actin mRNA served to monitor the evenness of sample input. Association of CSF-1 mRNA with nucleolin was also observed in SKOV3 epithelial ovarian cancer cells (SKOV3 cells, supplemental Fig. S5). Nucleolin binding to Bcl-XL mRNA (34) served as a positive binding control, with the association of Bcl-XL mRNA with nucleolin being 3.4-fold higher than that seen in the control IgG IP reaction (Fig. 2A).
Fig. 2.
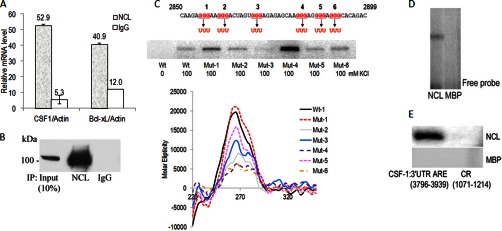
Nucleolin binds to the G-quadruplex and ARE in CSF-1 mRNA 3′UTR in vitro. A and B, IP of nucleolin-RNA complexes from Hey cell lysates. IP assays were carried out using anti-human nucleolin mAb or IgG. The presence of nucleolin in the IP materials was monitored by IB. qRT-PCR measurements of CSF-1 mRNA in nucleolin IP show direct interaction between nucleolin and CSF-1 mRNA. Bcl-xL mRNA in nucleolin IP serves as a positive control. The mean ± S.D. of CSF-1 mRNA normalized for actin mRNA is depicted (n = 3). Actin mRNA was set to equal 1. C, in vitro binding assay by UV cross-linking shows recombinant nucleolin binds to the G-quadruplex (WT) and mutant sequences (Mut) in the CSF-1 RNA 3′UTR 50-nt sequence in the presence of 100 mm KCl. In the absence of KCl, no binding of wild type (Wt) sequence to recombinant nucleolin was detected. Mutation in region 3 GGG to UUU (Mut-3) abrogated nucleolin binding. CD spectra showing the G-quadruplex formation in CSF-1 RNA 3′UTR (WT, Mut-1, and Mut-5) in the presence of 100 mm KCl. CD spectra of Mut-2, -3, -4, and -6 show formation of unknown structures. D, EMSA shows recombinant nucleolin associates with the CSF-1 3′UTR G-quadruplex RNA. The 32P-labeled CSF-1 3′UTR G-quadruplex riboprobe (48 nt, 2,850–2,897 nt) from 3′UTR associated with nucleolin and shifted in native PAGE. MBP, used as a negative control, was not associated with CSF-1 3′UTR G-quadruplex RNA. E, in vitro binding assay by UV cross-linking shows nucleolin binds to the ARE in the CSF-1 RNA 3′UTR 144-nt sequence. No binding of CSF-1 mRNA coding region (CR) to recombinant nucleolin was detected. MBP, used as a negative control, was not bound to CSF-1 RNA.
Nucleolin Binds the Putative Noncanonical G-Quadruplex in the CSF-1 mRNA 3′UTR with High Specificity in Vitro
To determine whether the binding of nucleolin to the G-quadruplex in CSF-1 mRNA 3′UTR was specific, we performed a UV cross-linking and label transfer analyses by incubating recombinant nucleolin with radiolabeled 48-mer G-quadruplex RNA from CSF-1 mRNA 3′UTR (Fig. 2C). Our results show that nucleolin can bind to the G-quadruplex RNA from CSF-1 mRNA 3′UTR with high specificity in the presence of 100 mm KCl. In the absence of KCl, nucleolin could not bind to the G-quadruplex sequence, suggesting dependence of nucleolin binding of G-quadruplex on the presence of the K+ ion (Fig. 2C, 1st lane). Electrophoretic mobility shift assay confirmed the binding activity of nucleolin to G-quadruplex RNA (Fig. 2D). These results support that the G-quadruplex in CSF-1 mRNA 3′UTR could serve as a binding target of nucleolin.
To confirm that the G-quadruplex contains part of the binding site for nucleolin, mutations in the G-quadruplex sequence were generated, and a nucleolin binding assay was performed by UV cross-linking. In the G-quadruplex sequence, six regions that collectively encompass “GGG ” (regions 1–6) were chosen for mutation analysis. Mutations of GGG to “UUU ” in region 3 (Mut-3) abrogated nucleolin binding, suggesting importance of GGG in region 3 for nucleolin binding (Fig. 2C). Interestingly, mutations of GGG to UUU in region 4 (Mut-4) showed better binding activity by nucleolin than wild type G-quadruplex sequence (Fig. 2C). All other mutations except Mut-3 have binding activities by nucleolin. However, CD analysis revealed all mutations except Mut-1 disrupted the G-quadruplex structure and formed unknown secondary structures (Fig. 2C). RNA folding analysis predicts that both wild type and all mutants except Mut-3 can form a large hairpin loop structure (WT and Mut-1, -2, and -6) or two hairpin loops (Mut-4 and -5) (supplemental Fig. S6). The small hairpin loop structure predicted in Mut-3 may not be recognized by nucleolin. The data suggest that binding of nucleolin may favor large hairpin loops as well as the G-quadruplex. This observation is supported by the finding that nucleolin recognizes and binds a pre-formed RNA stem-loop (39).
Nucleolin Binds to the AREs in CSF-1 mRNA 3′UTR in Vitro
To confirm that nucleolin can bind to the ARE-containing 144 nt in CSF-1 mRNA 3′UTR, we used UV cross-linking and label transfer assays, with maltose-binding protein (MBP) serving as a negative control (Fig. 2E). A riboprobe made from the 144-nt sequence bound recombinant nucleolin (Fig. 2E). In contrast, nucleolin did not bind sequences from the coding region of CSF-1 mRNA. The data suggest that nucleolin-binding sites are contained in part within the 144 nt of AU-rich CSF-1 mRNA 3′UTR.
Effects of Nucleolin on CSF-1 mRNA Stability and Translation
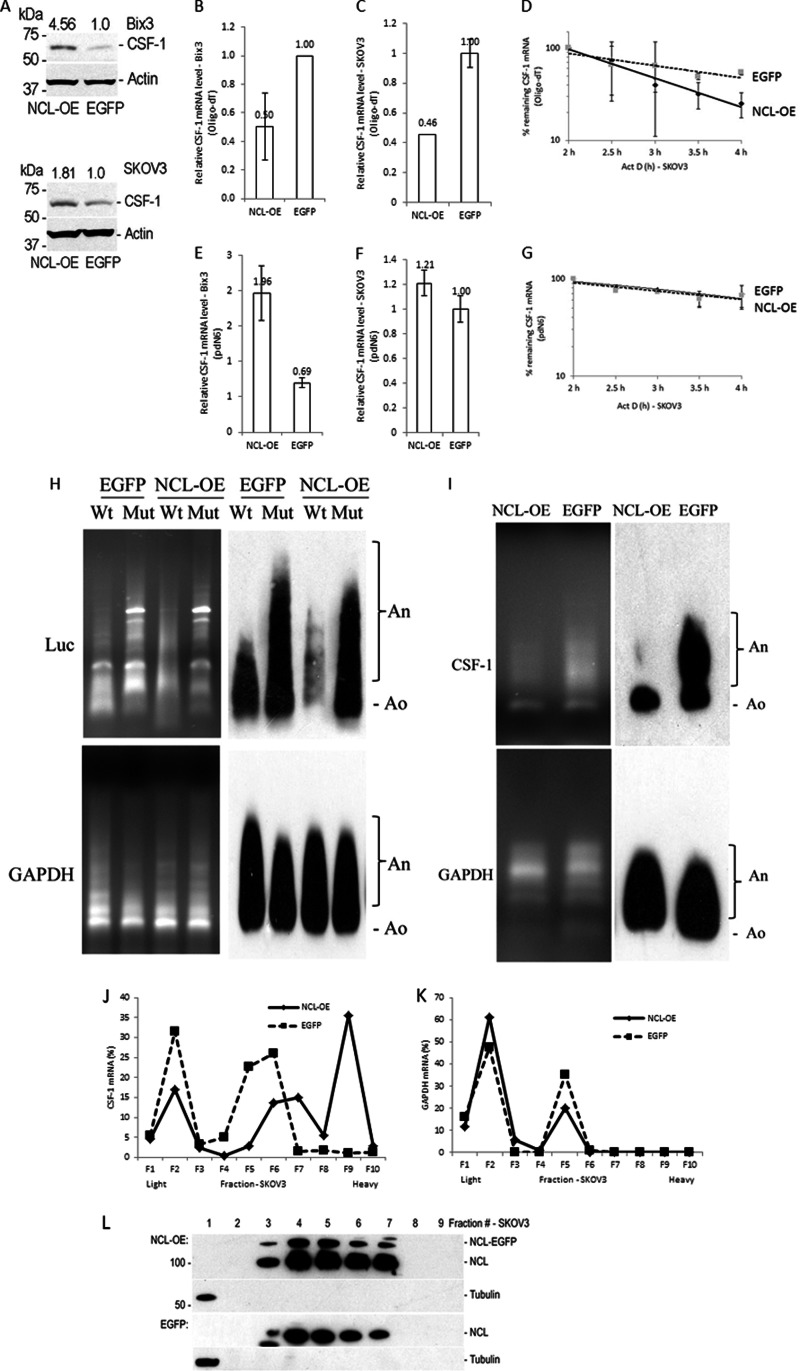
Because our data indicate that nucleolin interacts with both G-quadruplex (2855–2892) and AREs (3769–3939) in CSF-1 mRNA 3′UTR (Fig. 2), we studied the role of nucleolin on CSF-1 mRNA and protein expression. Initially we observed that overexpression of nucleolin increased the CSF-1 protein level by 4.6-fold in Bix3 cells and 1.8-fold in SKOV3 cells (Fig. 3A). However, surprisingly, overexpression of nucleolin decreased steady-state levels of CSF-1 mRNA by 34–50% in Bix3 and SKOV3 cells (Fig. 3, B and C). To further confirm the nucleolin effects, CSF-1 mRNA stability was determined in nucleolin overexpressed (NCL-OE) SKOV3 cells. Four hours after Act-D treatment, ∼25% CSF-1 mRNA remained in the nucleolin overexpressed group compared with ∼54.1% in the control group (EGFP, Fig. 3D). There was a trend in the decrease of the CSF-1 mRNA half-life from ∼4.3 to ∼2.8 h, by overexpression of nucleolin.
Fig. 3.
Nucleolin enhances deadenylation of CSF-1 mRNA but increases CSF-1 mRNA translation. A, nucleolin overexpression increases CSF-1 protein levels in Bix3 and SKOV3 ovarian cancer cells. B and C, when the cDNA was synthesized by oligo(dT)18, nucleolin overexpression decreases CSF-1 mRNA levels in Bix3 and SKOV3 ovarian cancer cells. D, CSF-1 mRNA stability is decreased by nucleolin overexpression when cDNA was synthesized by oligo(dT)18 in SKOV3 cells. E, when the cDNA was synthesized by random primer (pdN6), nucleolin overexpression increases CSF-1 mRNA levels in Bix3 cells. F and G, in SKOV3 cells, when the cDNA was synthesized by random primer (pdN6), there was no significant effect of nucleolin overexpression on CSF-1 mRNA or mRNA stability. This suggests an effect of nucleolin on CSF-1 translation in these cells. B–G, n = 3. Mean ± S.D. is presented. Deadenylation assay of luciferase RNA fused with CSF-1 mRNA 3′UTR sequences (H) or endogenous CSF-1 mRNA, by ligation-mediated poly(A) test (I) is shown. J, nucleolin abundance affects the distribution of CSF-1 mRNA on polysomes. SKOV3 cells were transfected with either nucleolin-EGFP or EGFP. Polysome profiles were prepared with the sucrose density gradient ultracentrifugation. Relative distributions of CSF-1 mRNA. K, GAPDH mRNA in polysome gradients after nucleolin overexpression. L, relative distributions of nucleolin and β-tubulin were obtained by IB analyses of the polysome gradient. Fractions 1 and 2, unbound RNPs (Unb.); fractions 3 and 4, monosomes (Mono.); and fractions 5–10, low and high molecular weight polysomes (LMW and HMW).
Because oligo(dT)18 was used in cDNA synthesis (Fig. 3, B--D), reverse transcription was repeated using random primers (pdN6) instead, which does not require the poly(A+) tail sequence for priming (Fig. 3, E–G). With this approach, overexpression of nucleolin increased steady-state levels of CSF-1 mRNA by 1.96-fold in Bix3 cells (Fig. 3E), more in keeping with the enhancement of CSF-1 protein levels by nucleolin. However, there were no significant differences in either steady-state CSF-1 mRNA levels or mRNA stability between the two groups in SKOV3 cells (Fig. 3, F and G). These data suggest that nucleolin may have a role in CSF-1 mRNA deadenylation, while also having the ability to enhance CSF-1 mRNA and protein levels.
We therefore studied whether nucleolin enhances deadenylation of CSF-1 mRNA, using the ligation-mediated poly(A) test assay (63). Luciferase RNA fused to CSF-1 mRNA 3′UTR with or without point mutations in the common miRNA target, and deletion of G-quadruplex and AREs (Fig. 1B) was cotransfected with the nucleolin overexpression plasmid in SKOV3 cells (Fig. 3H). Overexpression of nucleolin enhanced deadenylation of the wild type RNA sequence, but it did not enhance deadenylation of mutant RNA (Fig. 3H). As a control, nucleolin did not influence the deadenylation of GAPDH mRNA. Overexpression of nucleolin also enhanced deadenylation of endogenous CSF-1 mRNA without effects on GAPDH mRNA (Fig. 3I). This suggests that nucleolin enhances deadenylation of CSF-1 mRNA, via interaction with one or more of the three regulatory motifs in CSF-1 mRNA 3′UTR.
We also studied whether nucleolin can affect translation of CSF-1 mRNA in SKOV3 cells. After cycloheximide treatment to arrest polyribosome migration, polysome profiles of cytoplasmic lysates from SKOV3 cells were generated (Fig. 3, J–L). Overexpression of nucleolin did not affect the distribution of nucleolin in polysome profiles (Fig. 3L). Most of the nucleolin cosedimented with polysomes, where cytosolic β-tubulin was detected with free RNP. In SKOV3 cells transfected with the control empty vector (pEGFP), 94.3% of CSF-1 mRNA cosedimented with light polysomes and free RNPs (Fig. 3J, fractions 1–6). Upon overexpression of nucleolin, 58.9% of CSF-1 mRNA cosedimented with heavy polysomes instead (Fig. 3J, fractions 7–10). There were no changes in GAPDH mRNA distribution by nucleolin overexpression (Fig. 3K). We also silenced nucleolin by shRNA in SKOV3 and Bix3 cells; however, there were no significant changes in CSF-1 mRNA distribution in nucleolin-silenced cells (data not shown).
Taken together, these results indicate that nucleolin regulation of CSF-1 mRNA is complex. On the one hand, nucleolin enhances deadenylation of CSF-1 mRNA. On the other hand, nucleolin can enhance translation of CSF-1 mRNA, in keeping with its paradoxical effect on enhancement of CSF-1 protein levels.
miR-130a and miR-301a Down-regulate Reporter Expression by Binding to the Common miRNA Target
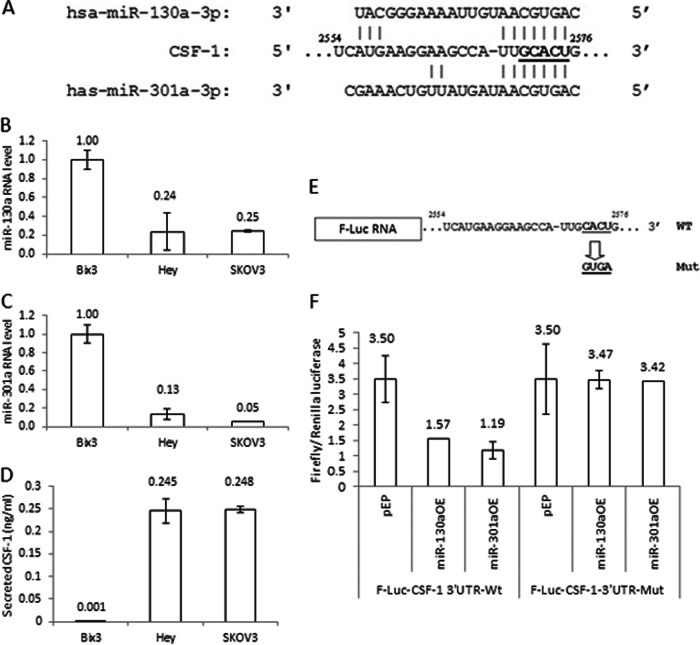
Bioinformatics analysis revealed that a putative miRNA target sequence (2572–2577) for miR-130a and miR-301a is located in CSF-1 mRNA 3′UTR (Fig. 4A). This is the only target sequence for these miRNAs predicted in CSF-1 mRNA 3′UTR. We have described this target sequence as a common target sequence for several miRNAs in ovarian cancer cells (44). The miRNA target sequence 2572GCACUG2577 in CSF-1 mRNA 3′UTR was previously identified as matching the seeds of 68% of the miRNA population in mouse embryonic stem cells (50). In addition, miR-130a was previously reported to down-regulate CSF-1 mRNA expression in A2780 ovarian cancer cells (40). In Hey and SKOV3 cells, we find that the levels of miR-130a and miR-301a RNAs are lower than in Bix3 cells. (Fig. 4, B and C). In contrast, the secreted CSF-1 protein level is significantly higher in Hey and SKOV3 cells compared with Bix3 ovarian cancer cells (Fig. 4D) Expression of miR-130a and miR-301a are both inversely correlated with CSF-1 protein expression (correlation coefficients = −1.000, p = 0.014 for miR-130a; and −0.998, p = 0.042 for miR-301a) (Fig. 4, B–D). These associations suggest that miR-130a and miR-301a may down-regulate CSF-1 expression.
Fig. 4.
miR-130a and miR-301a repress CSF-1 expression by binding a common target in CSF-1 mRNA 3[prime]UTR. A, miR-130a and miR-301a target the same region (2554–2576) in CSF-1 mRNA 3′UTR. B and C, miR-130a and miR-301a RNA levels are high in Bix3 cells compared with Hey and SKOV3 cells. D, secreted CSF-1 protein levels detected by ELISA. There is little secreted CSF-1 protein in Bix3 cells compared with Hey and SKOV3 cells. E, firefly luciferase (F-Luc) RNA ligated with either wild type full-length CSF-1 mRNA 3′UTR or one containing target point mutations of the common target were generated. F, SKOV3 ovarian cancer cells were transfected with the indicated F-Luc-CSF-1 3′UTR-WT or point mutant (F-Luc-CSF-1 3′UTR-Mut) constructs. Relative luciferase activity is the ratio of the activities of Firefly and the Renilla luciferases. n = 3. Mean ± S.D. is presented.
To study the activity of miRNAs on CSF-1 mRNA 3′UTR, the miRNA target sequence (2573CACU2576) in CSF-1 mRNA 3′UTR was mutated to “GUGA ” and cloned into the 3′end of the luciferase reporter vector (Fig. 4E). These constructs were transfected into SKOV3 cells that express basal levels of miR-130a and miR-301a (Fig. 4, B and C). Overexpression of either miR-130a or miR-301a in SKOV3 cells transfected with CSF-1 mRNA 3′UTR wild type construct decreased luciferase activity by 50% (p = 0.016) and 65% (p = 0.004), respectively (Fig. 4F). In contrast, when transfected with the CSF-1 mRNA 3′UTR mutant construct instead, no significant difference was observed by overexpression of either miR-130a or miR-301a in SKOV3 cells, compared with the vector control. This suggests that both miRNAs down-regulate reporter expression by interacting with this target (2573CACU2576) in CSF-1 mRNA 3′UTR.
miR-130a and miR-301a Down-regulate Endogenous CSF-1 Expression
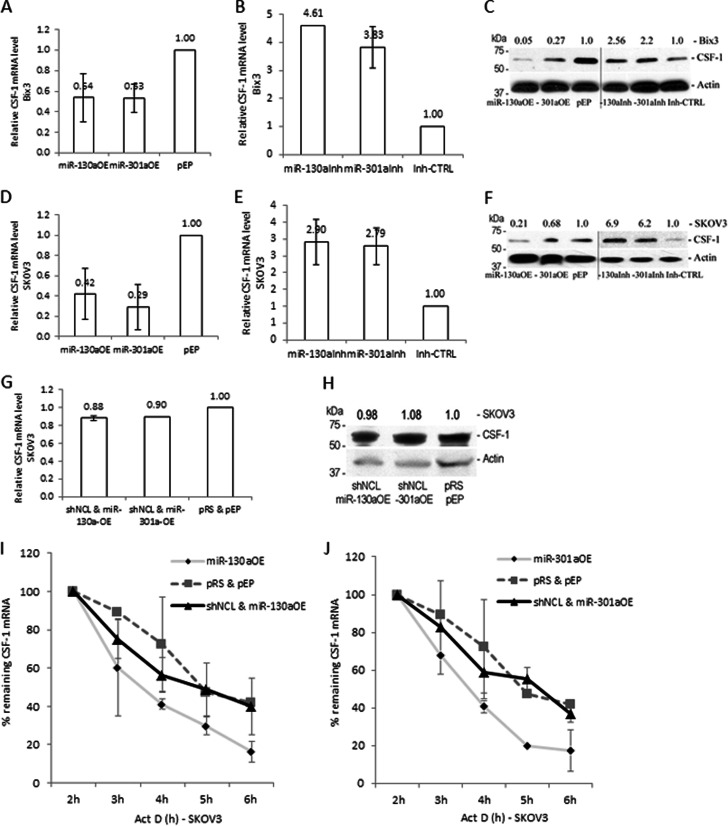
We have shown that overexpression of miR-130a or miR-301a down-regulates reporter activity by binding the common miRNA target sequence (Fig. 4). To determine the influence of these miRNAs on the expression of endogenous CSF-1 mRNA and protein, each miRNA was overexpressed in Bix3 and SKOV3 cells (Fig. 5, A and D). Overexpression of either miR-130a or miR-301a decreased the level of CSF-1 mRNA by 46–47% compared with pEP-miR-Null negative control vector-transfected Bix3 cells (pEP) (Fig. 5A) and 58–71% compared with the negative control vector transfected SKOV3 cells (Fig. 5D). At the same time, the CSF-1 protein levels decreased by 73–95% and by 32–79%, compared with the negative control vector transfected Bix3 or SKOV3 cells, respectively (Fig. 5, C and F). Thus, both miRNAs have negative effects on CSF-1 mRNA and protein levels.
Fig. 5.
miRNA-directed CSF-1 mRNA decay is dependent on nucleolin. A and D, overexpression of miR-130a and miR-301a reduced CSF-1 mRNA levels in Bix3 (p = 0.026 and 0.024, respectively) and in SKOV3 cells (p = 0.021 and 0.01, respectively). B and E, inhibition of miR-130a and miR-301a increased CSF-1 mRNA levels in Bix3 and SKOV3 cells. C and F, immunoblot analysis of CSF-1 protein in either miR-130a or miR-301a overexpressed or inhibited Bix3 and SKOV3 cells. G and H, silencing of nucleolin in miR-130a or miR-301a overexpressed cells did not decrease CSF-1 mRNA and protein levels in SKOV3 cells. I and J, at 2 days after transfection, Bix3 and SKOV3 cells were treated with Act-D, and the half-life of CSF-1 mRNA was measured by qRT-PCR. I, half-life of CSF-1 mRNA was ∼4.3 h. Overexpression of miR-130a reduced the half-life to ∼2.4 h. Silencing of nucleolin in miR-130a-overexpressed cells recovered the half-life to ∼3.4 h. J, overexpression of miR-301a reduced the half-life of CSF-1 mRNA to ∼2.6 h. Silencing of nucleolin in miR-301a overexpressed cells recovered the half-life to ∼3.4 h. n = 3. Mean ± S.D. is presented.
In contrast, silencing of miR-130a and miR-301a by inhibitors (miR-130aInh and miR-301aInh) increased the levels of CSF-1 mRNA by 4.6- and 3.8-fold (both p < 0.001) compared with the miRNA inhibitor negative control-transfected Bix3 cells (Inh-CTRL) (Fig. 5B), and by 2.9- and 2.8-fold (p = 0.009, 0.012, respectively) compared with the Inh-CTRL-transfected SKOV3 cells (Fig. 5E). At the same time, the CSF-1 protein levels increased by 2.6- and 2.2-fold compared with the Inh-CTRL-transfected Bix3 cells (Fig. 5C), and 6.9- and 6.2-fold compared with the Inh-CTRL-transfected SKOV3 cells (Fig. 5F). These data confirm that miR-130a and miR-301a suppress CSF-1 expression both at the mRNA and protein levels.
Down-regulation of CSF-1 Expression by miR-130a and miR-301a Requires Nucleolin
Because the target sequence (2572–2577) for miR-130a and miR-301a binding and the G-quadruplex sequence (2855–2892) for nucleolin binding is located in close proximity (279 nt apart), we determined the influence of nucleolin on miR-130a- and miR-301a-directed activities on regulation of CSF-1 expression by cotransfecting nucleolin shRNA with either the miR-130a or miR-301a overexpression plasmid. Silencing of nucleolin abolished the inhibitory effect of each miRNA on CSF-1 expression, such that CSF-1 mRNA and protein levels were comparable with vector control-transfected SKOV3 cells (Fig. 5, G and H). This result suggests that activities of miR-130a and miR-301a require nucleolin that binds to both G-quadruplex and AREs in CSF-1 mRNA 3′UTR.
Regulation of CSF-1 mRNA Stability by miR-130a and miR-301a in Ovarian Cancer Cells, miR-130a and miR-301a Decrease the Half-Life of CSF-1 mRNA
Because SKOV3 cells overexpressing miR-130a and miR-301a showed a decrease in steady-state level of CSF-1 mRNA compared with empty vector-transfected cells (Fig. 5, A and D), the CSF-1 mRNA half-life in miR-130a- or miR-301a-overexpressed SKOV3 cells was determined. The stability of the CSF-1 transcript was influenced by miR-130a or miR-301a abundance; CSF-1 mRNA half-life was decreased from ∼4.3 to ∼2.4 h, by overexpression of miR-130a (Fig. 5I), and from ∼4.3 to ∼2.6 h by overexpression of miR-301a (Fig. 5J). Taken together, these results (Figs. 4 and 5) suggest the destabilizing influence of miR-130a and miR-301a on target mRNAs, including those that encode CSF-1 mRNA.
Silencing of Nucleolin Blocks the CSF-1 mRNA Destabilizing Effects of miR-130a and miR-301a
Because the miRNA effect on down-regulation of steady-state CSF-1 mRNA appears to depend on nucleolin (Fig. 5, G and H), CSF-1 mRNA half-life was measured in nucleolin silenced by shRNA in either miR-130a or miR-301a overexpressed SKOV3 cells. We found that down-regulation of nucleolin blocks the miR-130a- or miR-301a-destabilizing effect on CSF-1 mRNA; CSF-1 mRNA half-life approached that of control conditions, in the absence of nucleolin in miRNA overexpressed SKOV3 cells (Fig. 5, I and J). Taken together, our data support that miR-130a and miR-301a require nucleolin to enhance CSF-1 mRNA decay.
Inhibition of Ovarian Cancer Cell Motility in Vitro by miR-130a and miR-301a Depends on Nucleolin
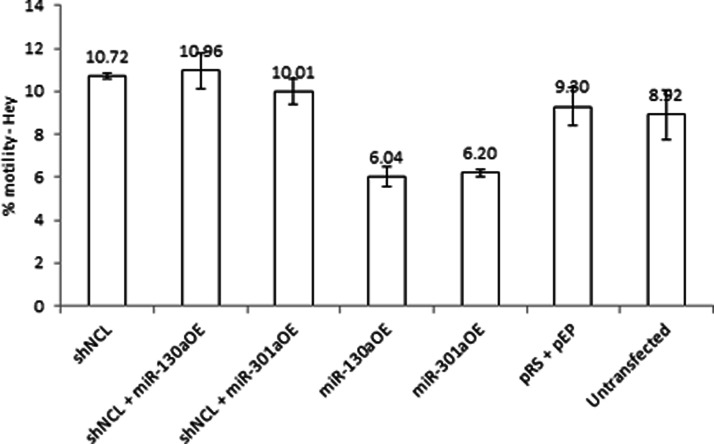
CSF-1 promotes ovarian cancer cell motility, invasiveness, and metastasis (15, 36). To assess whether there may be a downstream change in cellular behavior as a consequence of nucleolin's mediation of miRNA-directed CSF-1 down-regulation, the chemo-attractant-directed motility of Hey cells was studied in vitro. These ovarian cancer cells produce high level of secreted CSF-1 protein and express low levels of miR-130a and miR-301a (Fig. 4). The motility of the Hey cells overexpressing miR-130a or miR-301a was studied in comparison with the pEP-miR-Null negative control vector-transfected Hey cells. CSF-1 protein levels were confirmed under these conditions to be as described previously (Fig. 5). Overexpression of either miR-130a or miR-301a significantly inhibited Hey cell motility by 31–35% compared with the control vector (pRS + pEP)-transfected Hey cells (p = 0.016 and 0.014, respectively; Fig. 6). This finding of inhibition of motility by overexpressing miRNA is in line with their effect on inhibiting CSF-1 expression.
Fig. 6.
miR-130a and miR-301a inhibit in vitro motility of ovarian cancer cells, an effect that is dependent on nucleolin. After transfection with the indicated construct(s), Hey cells were plated on an 8-μm pore membrane for a 6-h directed motility assay. Mean ± S.E. is depicted. n = 4.
We performed in vitro loss-of-function analyses by silencing nucleolin in Hey cells and then studying their effects on miRNA-directed inhibition of motility. Alteration of nucleolin alone was first studied on changes in motility in Hey cells with low levels of miR-130a and -301a (Fig. 6), and no significant effect was found compared with the control vector-transfected Hey cells (p = 0.163, Fig. 6). However, when these conditions were studied in combination with overexpressing miR-130a or miR-301a in Hey cells, we found that silencing of nucleolin in miR-130a- or miR-301a-overexpressed Hey cells blocked the miRNA inhibitory effects on cell motility, leading to a level of motility similar to that of the control vector-transfected cells (p = 0.221, 0.53, respectively, Fig. 6; shNCL+miR-130a-OE or shNCL+miR-301a-OE versus control vector alone). These results confirm a nucleolin requirement for miRNA-130a or miR-301a directed inhibition of ovarian cancer cell motility.
Nucleolin as a Double-edged Sword for CSF-1 Expression
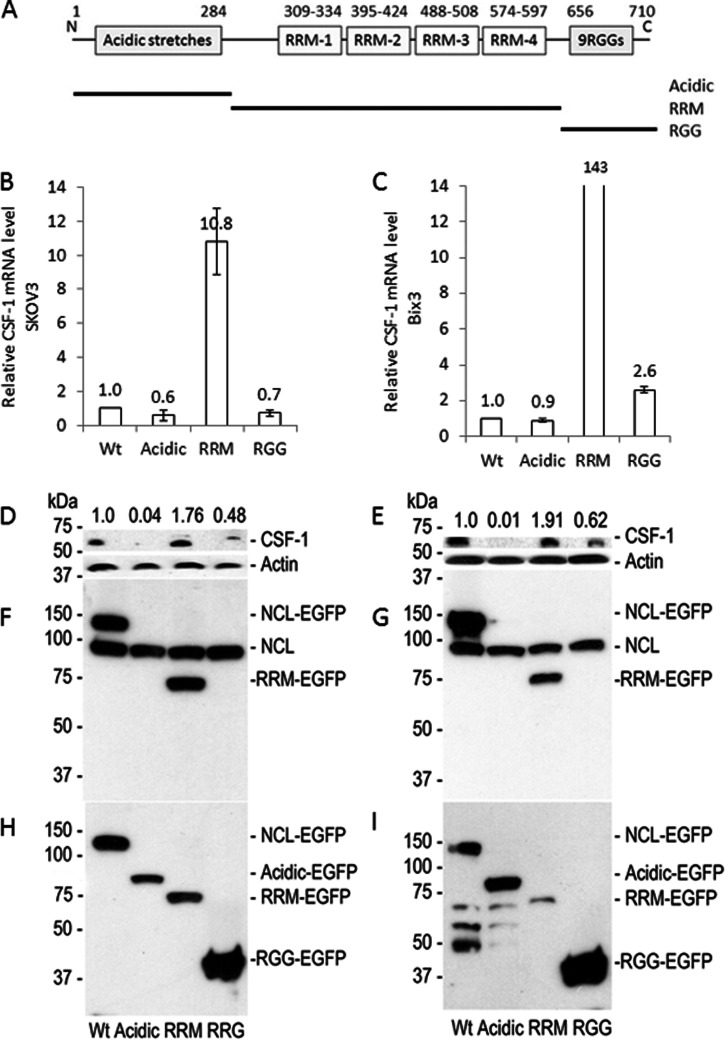
Because nucleolin enhances deadenylation of CSF-1 mRNA, and thereby miRNA-directed CSF-1 mRNA decay, but increases translation in ovarian cancer cells (Fig. 3), we hypothesized that nucleolin has two opposite functions, one for mRNA deadenylation and the other for translational activation. Nucleolin contains four acidic stretches (rich in glutamic acid and aspartic acid) in the N-terminal region, which is known to interact with other proteins (28), four RRMs in the middle domain for RNA-binding (41), and nine RGG repeats in C-terminal region, which is known to interact with other proteins as well as the RNA G-quadruplex (21, 42–44).
To investigate how these individual domains in nucleolin may work on the regulation of CSF-1 expression, each of these individual domains was overexpressed in SKOV3 and Bix3 cells (Fig. 7). Overexpression of the RRM domain alone increased CSF-1 mRNA levels by 10.8-fold in SKOV3 and 143-fold in Bix3 cells compared with wild type nucleolin (Fig. 7, B and C). CSF-1 protein levels were also increased by 1.8-fold in SKOV3 and 1.9-fold in Bix3 cells (Fig. 7, D and E). In contrast, overexpression of either the acidic domain or RGG repeats decreased CSF-1 protein levels (Fig. 7, D and E). The effect is most dramatic with overexpression of the acidic domain, which decreased CSF-1 protein levels by 96–99% in SKOV3 and Bix3 cells, respectively, with little effects on CSF-1 mRNA levels (Fig. 7, B and C).
Fig. 7.
Nucleolin domains differentially regulate CSF-1 mRNA and protein expression. A, schematic diagram of nucleolin functional domains. Expression constructs containing wild type nucleolin or each domain-tagged by EGFP were generated. Wild type nucleolin or the individual functional domain was transiently transfected in SKOV3 (B, D, F, and H) and Bix3 (C, E, G, and I) ovarian cancer cells. In both cells (B and C), introduction of the RRM domain increased CSF-1 mRNA levels dramatically. In contrast, the presence of acidic or RGG domains did not influence CSF-1 mRNA levels significantly. n = 3. Mean ± S.D. is presented. D and E, introduction of acidic and RGG domains decreased, whereas the RRM domain increased CSF-1 protein levels. F and G, immunoblot of transiently transfected SKOV3 and Bix3 ovarian cancer cells by nucleolin Ab. Endogenous nucleolin is shown at ∼110 kDa. Only endogenous nucleolin and RRM domain can be detected by the nucleolin Ab, which is raised by the RRM domain epitope. H and I, immunoblot of transiently transfected SKOV3 and Bix3 ovarian cancer cells by EGFP Ab.
We show that the RRM domain in the middle region enhances CSF-1 mRNA significantly and protein levels moderately. In contrast, both the N-terminal acidic stretches and the C-terminal RGG repeats have the capacity to down-regulate CSF-1 protein levels, suggesting that these two domains are negative regulators of CSF-1. These intriguing observations add to the growing evidence that nucleolin has the capacity to differentially regulate CSF-1 mRNA and protein levels, likely depending on the context. The exact function of the different domains of nucleolin on regulation of CSF-1 mRNA has yet to be fully understood.
Nucleolin, Ago2, and PABPC May Exist Together as a Complex Containing CSF-1 mRNA in Ovarian Cancer Cells, and Nucleolin Interacts with Ago2 Indirectly via RNA
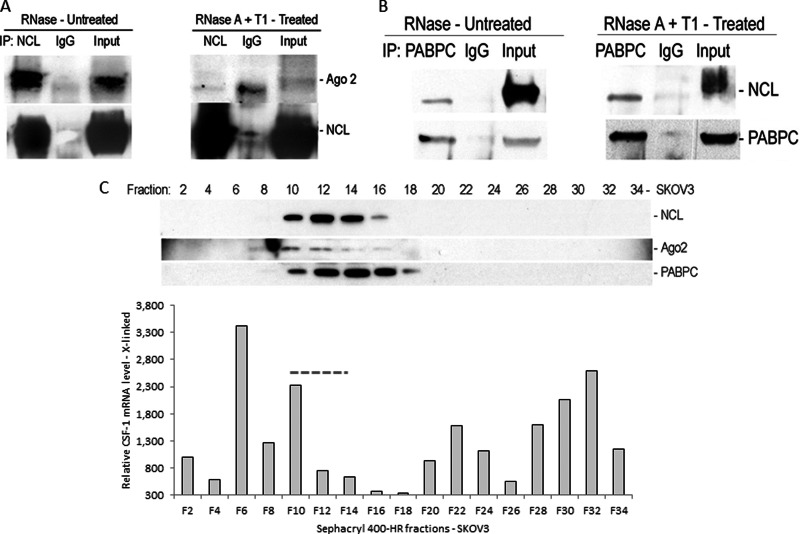
Because both the G-quadruplex and common miRNA target sequence are in close proximity (Fig. 1A) and nucleolin is required for miR-130a and miR-301a activities on CSF-1 mRNA (Fig. 5), the association of nucleolin with Ago2 as part of the miRISC complex was determined by a co-IP assay. Association of nucleolin with Ago2 via RNA was verified in the cytoplasmic fractions of Hey cells (Fig. 8A). In the presence of RNase inhibitor in the IP reaction, nucleolin and Ago2 coimmunoprecipitated. Including RNase A and T1 in the IP reaction led to the disappearance of Ago2 from the nucleolin-containing complex, indicating that the interaction between nucleolin and Ago2 is mediated by RNA.
Fig. 8.
Interaction of nucleolin with Ago2 and PABPC. A, co-IP assay from Hey cell lysates. Co-IP assay was carried out using anti-human nucleolin mAb or IgG. The presence of nucleolin and Ago2 in the IP materials was monitored by IB. Nucleolin and Ago2 are coimmunoprecipitated in the presence of RNase inhibitor. In contrast, in the presence of RNase A and T1, nucleolin and Ago2 are not coimmunoprecipitated, indicating an indirect interaction between nucleolin and Ago2 via mRNA. B, co-IP assay from Hey cell lysates. Co-IP assay was carried out using anti-human PABPC mAb or IgG. Nucleolin and PABPC are coimmunoprecipitated in either presence of RNase inhibitor or RNase A and T1, indicating a direct interaction between PABPC and nucleolin. C, size-exclusion chromatography of SKOV3 cell lysates. Nucleolin, Ago2, and PABPC are eluted together in fractions 10–16, in which CSF-1 mRNA is present.
Nucleolin Interacts Directly with PABPC
The N-terminal region of PABPC interacts with the translation initiation factor eIF4G to form an “mRNP closed loop ” for translational activation (45–48). In contrast, the C-terminal domain of PABPC interacts with TNRC6 (human counterpart of Drosophila GW182), a core component of miRISC (49), and attracts the Pan2-Pan3 and CCR4-NOT-CAF1 deadenylase complex to the 3′end of the mRNA (50). PABPC, which binds the poly(A+) tail of mRNA, enhances miRISC association with mRNAs resulting in translational repression followed by mRNA deadenylation (51). Because we have shown that nucleolin enhances mRNA deadenylation, but can also increase translation (Fig. 3), we tested whether nucleolin and PABPC interact in the cytoplasmic fractions of Hey cell lysates by a co-IP assay. Nucleolin and PABPC coimmunoprecipitated either in the presence of RNase inhibitor or RNase A and T1 in the IP reaction (Fig. 8B). This suggests that there is a direct interaction between nucleolin and PABPC.
Associations of nucleolin with Ago2 and PABPC were also observed by gel filtration chromatography (Fig. 8C). Total cellular extracts of SKOV3 cells were size-fractionated by Sephacryl 400-HR. Ago2, PABPC, and nucleolin are present in the same fractions with CSF-1 mRNA (Fig. 8C, fractions 10–14). Together, these results suggest an indirect interaction between nucleolin and Ago2, a member of the miRISC complex, via mRNA, a direct interaction between nucleolin and PABPC, and formation of a nucleolin-Ago2-PABPC complex on CSF-1 mRNA 3′UTR.
DISCUSSION
Post-transcriptional Regulation of CSF-1 by Its cis-Acting Regulatory Elements in the 3′UTR
In this study, we found an unexpected role for the RNA-binding protein nucleolin in the control of miRNA activity for both CSF-1 mRNA deadenylation and decay, and inhibition of ovarian cancer cell motility. From an evolutionary point of view, a long 3′UTR could contain more cis-acting regulatory elements than a short 3′UTR. Exon 10 of the CSF-1 gene encodes 2,172-nt 3′UTR, which is more than half of the 3,939-nt CSF-1 major transcript. Our results indicate CSF-1 mRNA 3′UTR contains several functional cis-acting regulatory elements (Fig. 1), including a common miRNA target region conserved between human, mouse, and rat. Previously, we predicted using bioinformatics analysis that at least 14 miRNAs likely target this region “2572–2577 ” in CSF-1 mRNA 3′UTR, and we showed that by interacting with this common target, miR-128 and miR-152 down-regulate CSF-1 mRNA and protein expression and ovarian cancer cell behavior (18). Here, we report that nucleolin is required for the activities of two different miRNAs (miR-130a and miR-301a), which also interact with the common miRNA target region (Fig. 5). Our data therefore suggest that nucleolin requirement may be generalized for all miRNAs targeting this common miRNA target region, situated close to the noncanonical G-quadruplex we describe here as binding to nucleolin. Moreover, to our knowledge, this may be the first report to date of nucleolin mediating miRNA-directed mRNA deadenylation, and cancer cell behavior.
Supporting this concept, we found by both co-IP and size-fractionation chromatography that Ago2, a component of miRISC, coimmunoprecipitates with nucleolin along with RNA (Fig. 8A). Ago2 is also coeluted with nucleolin and CSF-1 mRNA (Fig. 8C). These results strongly support that formation of the nucleolin-Ago2 complex with CSF-1 mRNA 3′UTR could induce mRNA decay via the activities of miRNA.
In this report, we describe a noncanonical G-quadruplex sequence in CSF-1 mRNA 3′UTR (Fig. 1), and we identified nucleolin to favor binding this G-quadruplex as well as large hairpin loop structures (Fig. 2). Fragile X mental retardation protein is an RBP, whose RGG peptide domain binds G-quadruplex RNA by recognizing the RNA duplex-quadruplex junction (44, 52). Nucleolin, which also has nine repeats of RGG domain in its C terminus, could bind the G-quadruplex RNA on this basis. Our finding is in line with a recent report of nucleolin binding to mRNA targets with G-rich sequences (32).
We have previously studied the interaction of GAPDH with AREs of terminal CSF-1 mRNA 3′UTR, in the regulation of CSF-1 expression (19, 20). Our results indicate nucleolin also binds AREs in CSF-1 mRNA 3′UTR (Fig. 2D). Nucleolin's affinity for AREs has been previously described (26, 35). The central globular domain of nucleolin contains four RRMs that bind to AREs (26) and RNA stem-loop structures (53). Binding of nucleolin to both the G-quadruplex and AREs within CSF-1 mRNA 3′UTR support a key role for nucleolin in the regulation of CSF-1 mRNA.
Finally, we show that the three elements within CSF-1 mRNA 3′UTR (the miRNA common target, G-quadruplex, and AREs) work together to promote mRNA decay (Fig. 1). Disruption of these three cis-acting regulatory elements in a CSF-1 mRNA 3′UTR reporter construct results in a more pronounced increase in the level of reporter RNA than on the level of reporter activity. We interpret this result to mean that disruption of these three cis-acting regulatory sequences disrupts both nucleolin and miRNA binding, which is required for miRISC complex formation on the common miRNA target sequence. Stable binding of nucleolin to CSF-1 mRNA 3′UTR may require both the G-quadruplex and AREs, because deletion of either G-quadruplex or AREs, along with mutation of the miRNA common target, did not show the same degree of reporter up-regulation as did disruption of all three elements together (supplemental Fig. S1).
Nucleolin Enhances mRNA Deadenylation but Activates Translation
In vivo gain-of-function assay of nucleolin supports the influence of nucleolin on CSF-1 mRNA (Fig. 3). Overexpression of nucleolin decreased the CSF-1 mRNA level when the reverse transcription was primed by oligo(dT)18, i.e. when detecting poly(A+) mRNA. In contrast, when the mRNA was reverse-transcribed by random primer (pdN6), which does not require the poly(A+) sequence for priming, nucleolin overexpression increased CSF-1 mRNA levels in Bix3 ovarian cancer cells, correlating with an increase in CSF-1 protein levels. We confirm that nucleolin enhances deadenylation of both reporter RNA and endogenous CSF-1 mRNA (Fig. 3, H and I). Our studies of miRNA-directed destabilization of CSF-1 mRNA (Fig. 5) also were performed by studying poly(A+) mRNA, suggesting miRNA-directed deadenylation, in keeping with other reports (1, 54). This suggests that nucleolin mediation of miRNA activities on CSF-1 mRNA may be via its ability to deadenylate CSF-1 mRNA.
Because we found no significant effect of nucleolin overexpression on CSF-1 mRNA when random priming was used in another ovarian cancer cell line, SKOV3, containing low levels of miR-130a and miR-301a (Fig. 3F), we used these cells to show that nucleolin overexpression has the capacity to enhance translational activity of CSF-1 mRNA instead, correlating with an increase in CSF-1 protein levels (Fig. 3, J–L). Thus, we envision this double-edged sword effect as at times formation of nucleolin-miRISC initiating mRNA deadenylation (Fig. 9A), and at the other times, nucleolin with its association with PABPC (Fig. 9B) helping the formation of an mRNP closed loop for translational activation. The initiating factors that enable nucleolin to function more in mRNA deadenylation versus translational activation are unknown at this time.
Fig. 9.
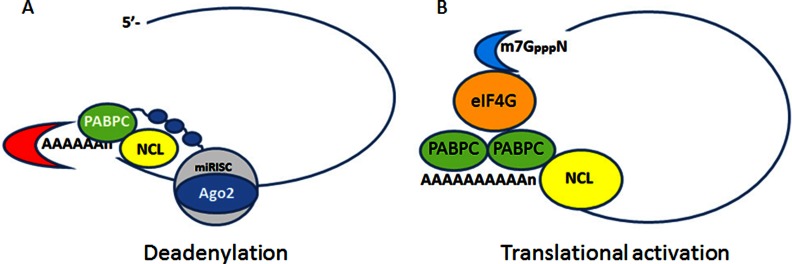
Model for post-transcriptional regulation of CSF-1 mRNA and protein expression. A, formation of nucleolin-Ago2-PABPC complex induces deadenylation of CSF-1 mRNA. B, nucleolin in association with PABPC activates translation by formation of the cap-dependent mRNP closed loop.
Nucleolin as a Double-edged Sword in the Post-transcriptional Regulation of CSF-1
We found that the RRM domain substantially increases the level of CSF-1 mRNA (Fig. 7). It is possible that the RRM domain alone may be sufficient to bind and stabilize the CSF-1 mRNA, without engaging the miRISC apparatus for mRNA decay. Binding of CSF-1 mRNA by the RRM domain could protect the mRNA from degradation by blocking nuclease activity and helping to form an mRNP closed loop for translational activation (45). This could explain the substantial increase in the steady-state CSF-1 mRNA level; however, we observed only a small effect on increasing protein levels (Fig. 7). Thus, although the RRM domain may help to form an mRNP closed loop by binding to CSF-1 mRNA, there are likely other rate-limiting factor(s) for translation activation.
Our work suggests that both N-terminal acidic stretches and C-terminal RGG repeats work as negative regulators for CSF-1 protein expression. It is possible that the N-terminal acidic stretches and the C-terminal RGG repeats may compete with endogenous nucleolin depleting the available proteins for translation machinery formation, because both domains are involved in protein-protein interactions (21, 42). In addition, both domains may deplete Ago2, which is a component of the miRISC complex for mRNA decay. Depletion of Ago2 in the miRISC complex may prevent further CSF-1 mRNA decay. However, the exact function of these domains in this context is unknown and remains a conjecture at this time.
Interestingly, we observed that PABPC coimmunoprecipitates with nucleolin (Fig. 8B). PABPC has two opposite functions in mRNA decay and translation (45), much as we are finding for nucleolin. PABPC binds to eIF4G to form a “cap-dependent mRNP closed loop ” for translational activation (45). In contrast, PABPC also interacts with GW182, a component of miRISC, and attracts deadenylase complex for 3′end deadenylation (49, 51, 55). We propose that nucleolin may work together with PABPC in this context (Fig. 9).
In summary, our work proposes a model where, by binding the G-quadruplex and AREs in CSF-1 mRNA 3′UTR, nucleolin mediates the actions of miR-130a and miR-301a on repressing CSF-1 levels by enhancing CSF-1 mRNA deadenylation and decay. Downstream consequences of these interactions are observed in our studies of human ovarian cancer cell motility, where the miRNA inhibition of cellular motility is only observed in the presence of nucleolin. In other contexts, however, nucleolin may increase CSF-1 translation by activating the formation of an mRNP closed loop (45), enhancing CSF-1 protein levels.
Our findings suggest that nucleolin's role as a regulator of CSF-1 mRNA is complex, and our observations have opened up many channels of investigations. Nucleolin may serve as a model for how some regulators may function to regulate gene expression.
Supplementary Material
Footnotes
* This work was supported, in whole or in part, by National Institutes of Health Grant CA60665. This work was also supported by the Rodel Foundation, the Arizona Cancer Center (to S.K.C.), and the Proteomics and Biometry Shared Services (supported by National Institutes of Health Grant CA023074 to the University of Arizona Cancer Center).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- NCL
- nucleolin
- ARE
- AU-rich element
- RRM
- RNA recognition motif
- PABPC
- poly(A)-binding protein 1
- RBP
- RNA-binding protein
- RGG
- arginine-glycine-glycine motif
- qRT-PCR
- quantitative real time-PCR
- Ab
- antibody
- EGFP
- enhanced GFP
- nt
- nucleotide
- miRNA
- microRNA
- IP
- immunoprecipitation
- MBP
- maltose binding protein
- miRISC
- miRNA-induced silencing complex
- RNP
- ribonucleoprotein
- Act-D
- actinomycin-D
- IB
- immunoblot
- mRNP
- Messenger Ribonucleoprotein.
REFERENCES
- 1. Djuranovic S., Nahvi A., Green R. (2012) miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 336, 237–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Filipowicz W., Bhattacharyya S. N., Sonenberg N. (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 9, 102–114 [DOI] [PubMed] [Google Scholar]
- 3. Filipowicz W., Jaskiewicz L., Kolb F. A., Pillai R. S. (2005) Post-transcriptional gene silencing by siRNAs and miRNAs. Curr. Opin. Struct. Biol. 15, 331–341 [DOI] [PubMed] [Google Scholar]
- 4. Eulalio A., Huntzinger E., Izaurralde E. (2008) Getting to the root of miRNA-mediated gene silencing. Cell 132, 9–14 [DOI] [PubMed] [Google Scholar]
- 5. Chen C. Y., Zheng D., Xia Z., Shyu A.-B. (2009) Ago-TNRC6 triggers microRNA-mediated decay by promoting two deadenylation steps. Nat. Struct. Mol. Biol. 16, 1160–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stark A., Brennecke J., Bushati N., Russell R. B., Cohen S. M. (2005) Animal microRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell 123, 1133–1146 [DOI] [PubMed] [Google Scholar]
- 7. Huppert J.-L., Bugaut A., Kumari S., Balasubramanian S. (2008) G-quadruplexes: the beginning and end of UTRs. Nucleic Acids Res. 36, 6260–6268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bhattacharyya S. N., Habermacher R., Martine U., Closs E. I., Filipowicz W. (2006) Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125, 1111–1124 [DOI] [PubMed] [Google Scholar]
- 9. Kundu P., Fabian M. R., Sonenberg N., Bhattacharyya S. N., Filipowicz W. (2012) HuR protein attenuates miRNA-mediated repression by promoting miRISC dissociation from the target RNA. Nucleic Acids Res. 40, 5088–5100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kedde M., Strasser M. J., Boldajipour B., Oude Vrielink J. A., Slanchev K., le Sage C., Nagel R., Voorhoeve P. M., van Duijse J., Ørom U. A., Lund A. H., Perrakis A., Raz E., Agami R. (2007) RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell 131, 1273–1286 [DOI] [PubMed] [Google Scholar]
- 11. Weinmann L., Höck J., Ivacevic T., Ohrt T., Mütze J., Schwille P., Kremmer E., Benes V., Urlaub H., Meister G. (2009) Importin 8 is a gene silencing factor that targets argonaute proteins to distinct mRNAs. Cell 136, 496–507 [DOI] [PubMed] [Google Scholar]
- 12. Schwamborn J. C., Berezikov E., Knoblich J. A. (2009) The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell 136, 913–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Galgano A., Forrer M., Jaskiewicz L., Kanitz A., Zavolan M., Gerber A. P. (2008) Comparative analysis of mRNA targets for human PUF-family proteins suggests extensive interaction with the miRNA regulatory system. PLoS One 3, e3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chambers S. K. (2009) Role of CSF-1 in progression of epithelial ovarian cancer. Future Oncol. 5, 1429–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Toy E. P., Azodi M., Folk N. L., Zito C. M., Zeiss C. J., Chambers S. K. (2009) Enhanced ovarian cancer tumorigenesis and metastasis by the macrophage colony-stimulating factor. Neoplasia 11, 136–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Price L. K., Choi H. U., Rosenberg L., Stanley E. R. (1992) The predominant form of secreted colony stimulating factor-1 is a proteoglycan. J. Biol. Chem. 267, 2190–2199 [PubMed] [Google Scholar]
- 17. Suzu S., Ohtsuki T., Makishima M., Yanai N., Kawashima T., Nagata N., Motoyoshi K. (1992) Biological activity of a proteoglycan form of macrophage colony-stimulating factor and its binding to type V collagen. J. Biol. Chem. 267, 16812–16815 [PubMed] [Google Scholar]
- 18. Woo H. H., Laszlo C., Greco S., Chambers S. K. (2012) Regulation of colony stimulating factor-1 expression and ovarian cancer cell behavior in vitro by miR-128 and miR-152. Mol. Cancer 11(58), doi:10.1186/1476-4598-11-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhou Y., Yi X., Stoffer J. B., Bonafe N., Gilmore-Hebert M., McAlpine J., Chambers S. K. (2008) The multifunctional protein glyceraldehyde-3-phosphate dehydrogenase is both regulated and controls colony-stimulating factor-1 messenger RNA stability in ovarian cancer. Mol. Cancer Res. 6, 1375–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bonafé N., Gilmore-Hebert M., Folk N. L., Azodi M., Zhou Y., Chambers S. K. (2005) Glyceraldehyde-3-phosphate dehydrogenase binds to the AU-rich 3′-untranslated region of colony-stimulating factor-1 (CSF-1) messenger RNA in human ovarian cancer cells: possible role in CSF-1 posttranscriptional regulation and tumor phenotype. Cancer Res. 65, 3762–3771 [DOI] [PubMed] [Google Scholar]
- 21. Ginisty H., Sicard H., Roger B., Bouvet P. (1999) Structure and functions of nucleolin. J. Cell Sci. 112, 761–772 [DOI] [PubMed] [Google Scholar]
- 22. Orrick L. R., Olson M. O., Busch H. (1973) Comparison of nucleolar proteins of normal rat liver and Novikoff hepatoma ascites cells by two-dimensional polyacrylamide gel electrophoresis. Proc. Natl. Acad. Sci. U.S.A. 70, 1316–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sirri V., Urcuqui-Inchima S., Roussel P., Hernandez-Verdun D. (2008) Nucleolus: the fascinating nuclear body. Histochem. Cell Biol. 129, 13–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Destouches D., El Khoury D., Hamma-Kourbali Y., Krust B., Albanese P., Katsoris P., Guichard G., Briand J. P., Courty J., Hovanessian A. G. (2008) Suppression of tumor growth and angiogenesis by a specific antagonist of the cell-surface expressed nucleolin. PLoS One 3, e2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ishimaru D., Zuraw L., Ramalingam S., Sengupta T. K., Bandyopadhyay S., Reuben A., Fernandes D. J., Spicer E. K. (2010) Mechanism of regulation of bcl-2 mRNA by nucleolin and A+U-rich element-binding factor 1 (AUF1). J. Biol. Chem. 285, 27182–27191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Miniard A. C., Middleton L. M., Budiman M. E., Gerber C. A., Driscoll D. M. (2010) Nucleolin binds to a subset of selenoprotein mRNAs and regulates their expression. Nucleic Acids Res. 38, 4807–4820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Takagi M., Absalon M. J., McLure K. G., Kastan M. B. (2005) Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell 123, 49–63 [DOI] [PubMed] [Google Scholar]
- 28. Chen J., Guo K., Kastan M. B. (2012) Interactions of nucleolin and ribosomal protein L26 (RPL26) in translational control of human p53 mRNA. J. Biol. Chem. 287, 16467–16476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Qin Y., Hurley L. H. (2008) Structures, folding patterns, and functions of intramolecular DNA G-quadruplexes found in eukaryotic promoter regions. Biochimie 90, 1149–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bonnal S., Schaeffer C., Créancier L., Clamens S., Moine H., Prats A.-C., Vagner S. (2003) A single internal ribosome entry site containing a G quartet RNA structure drives fibroblast growth factor 2 gene expression at four alternative translation initiation codons. J. Biol. Chem. 278, 39330–39336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Beaudoin J.-D., Perreault J. P. (2010) 5′-UTR G-quadruplex structures acting as translational repressors. Nucleic Acids Res. 38, 7022–7036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Abdelmohsen K., Tominaga K., Lee E. K., Srikantan S., Kang M.-J., Kim M. M., Selimyan R., Martindale J. L., Yang X., Carrier F., Zhan M., Becker K. G., Gorospe M. (2011) Enhanced translation by nucleolin via G-rich elements in coding and non-coding regions of target mRNAs. Nucleic Acids Res. 39, 8513–8530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bakheet T., Frevel M., Williams B. R., Greer W., Khabar K. S. (2001) ARED: human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional repertoire of encoded proteins. Nucleic Acids Res. 29, 246–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jing Q., Huang S., Guth S., Zarubin T., Motoyama A., Chen J., Di Padova F., Lin S.-C., Gram H., Han J. (2005) Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell 120, 623–634 [DOI] [PubMed] [Google Scholar]
- 35. Zhang J., Tsaprailis G., Bowden G. T. (2008) Nucleolin stabilizes Bcl-XL messenger RNA in response to UVA irradiation. Cancer Res. 68, 1046–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chambers S. K., Wang Y., Gertz R. E., Kacinski B. M. (1995) Macrophage colony-stimulating factor mediates invasion of ovarian cancer cells through urokinase. Cancer Res. 55, 1578–1585 [PubMed] [Google Scholar]
- 37. Woo H.-H., Yi X., Lamb T., Menzl I., Baker T., Shapiro D. J., Chambers S. K. (2011) Posttranscriptional suppression of proto-oncogene c-fms expression by vigilin in breast cancer. Mol. Cell. Biol. 31, 215–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Murray E. L., Schoenberg D. R. (2008) Assays for determining poly(A) tail length and the polarity of mRNA decay in mammalian cells. Methods Enzymol. 448, 483–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bouvet P., Allain F. H., Finger L. D., Dieckmann T., Feigon J. (2001) Recognition of pre-formed and flexible elements of an RNA stem-loop by nucleolin. J. Mol. Biol. 309, 763–775 [DOI] [PubMed] [Google Scholar]
- 40. Sorrentino A., Liu C.-G., Addario A., Peschle C., Scambia G., Ferlini C. (2008) Role of microRNAs in drug-resistant ovarian cancer cells. Gynecol. Oncol. 111, 478–486 [DOI] [PubMed] [Google Scholar]
- 41. Johansson C., Finger L. D., Trantirek L., Mueller T. D., Kim S., Laird-Offringa I. A., Feigon J. (2004) Solution structure of the complex formed by the two N-terminal RNA-binding domains of nucleolin and a pre-rRNA target. J. Mol. Biol. 337, 799–816 [DOI] [PubMed] [Google Scholar]
- 42. Bouvet P., Diaz J.-J., Kindbeiter K., Madjar J.-J., Amalric F. (1998) Nucleolin interacts with several ribosomal proteins through its RGG domain. J. Biol. Chem. 273, 19025–19029 [DOI] [PubMed] [Google Scholar]
- 43. Hanakahi L. A., Bu Z., Maizels N. (2000) The C-terminal domain of nucleolin accelerates nucleic acid annealing. Biochemistry 39, 15493–15499 [DOI] [PubMed] [Google Scholar]
- 44. Phan A. T., Kuryavyi V., Darnell J. C., Serganov A., Majumdar A., Ilin S., Raslin T., Polonskaia A., Chen C., Clain D., Darnell R. B., Patel D. J. (2011) Structure-function studies of FMRP RGG peptide recognition of an RNA duplex-quadruplex junction. Nat. Struct. Mol. Biol. 18, 796–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Huntzinger E., Izaurralde E. (2011) Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 12, 99–110 [DOI] [PubMed] [Google Scholar]
- 46. Imataka H., Gradi A., Sonenberg N. (1998) A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 17, 7480–7489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wakiyama M., Imataka H., Sonenberg N. (2000) Interaction of eIF4G with poly(A)-binding protein stimulates translation and is critical for Xenopus oocyte maturation. Curr. Biol. 10, 1147–1150 [DOI] [PubMed] [Google Scholar]
- 48. Burgess H. M., Gray N. K. (2010) mRNA-specific regulation of translation by poly(A)-binding proteins. Biochem. Soc Trans. 38, 1517–1522 [DOI] [PubMed] [Google Scholar]
- 49. Fabian M. R., Mathonnet G., Sundermeier T., Mathys H., Zipprich J. T., Svitkin Y. V., Rivas F., Jinek M., Wohlschlegel J., Doudna J. A., Chen C. Y., Shyu A. B., Yates J. R., 3rd, Hannon G. J., Filipowicz W., Duchaine T. F., Sonenberg N. (2009) Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol. Cell 35, 868–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zekri L., Huntzinger E., Heimstädt S., Izaurralde E. (2009) The silencing domain of GW182 interacts with PABPC1 to promote translational repression and degradation of microRNA targets and is required for target release. Mol. Cell. Biol. 29, 6220–6231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Moretti F., Kaiser C., Zdanowicz-Specht A., Hentze M. W. (2012) PABP and the poly(A) tail augment microRNA repression by facilitated miRISC binding. Nat. Struct. Mol. Biol. 19, 603–608 [DOI] [PubMed] [Google Scholar]
- 52. Bensaid M., Melko M., Bechara E. G., Davidovic L., Berretta A., Catania M. V., Gecz J., Lalli E., Bardoni B. (2009) FRXAE-associated mental retardation protein (FMR2) is an RNA-binding protein with high affinity for G-quartet RNA forming structure. Nucleic Acids Res. 37, 1269–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Finger L. D., Trantirek L., Johansson C., Feigon J. (2003) Solution structures of stem-loop RNAs that bind to the two N-terminal RNA-binding domains of nucleolin. Nucleic Acids Res. 31, 6461–6472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bazzini A. A., Lee M. T., Giraldez A. J. (2012) Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 336, 233–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Huntzinger E., Braun J. E., Heimstädt S., Zekri L., Izaurralde E. (2010) Two PABPC1-binding sites in GW182 proteins promote miRNA-mediated gene silencing. EMBO J. 29, 4146–4160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 57. Andon N. L., Hollingworth S., Koller A., Greenland A. J., Yates J. R., 3rd, Haynes P. A. (2002) Proteomic characterization of wheat amyloplasts using identification of proteins by tandem mass spectrometry. Proteomics 2, 1156–1168 [DOI] [PubMed] [Google Scholar]
- 58. Eng J. K., Mccormack A. L., Yates J. R. (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 [DOI] [PubMed] [Google Scholar]
- 59. Cooper B., Eckert D., Andon N. L., Yates J. R., Haynes P. A. (2003) Investigative proteomics: identification of an unknown plant virus from infected plants using mass spectrometry. J. Am. Soc. Mass Spectrom. 14, 736–741 [DOI] [PubMed] [Google Scholar]
- 60. Craig R., Beavis R. C. (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 61. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 62. Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.