Abstract
We report the development and characterization of a novel, vendor-neutral ultra-high pressure-compatible (∼10,000 p.s.i.) LC-MS source. This device is the first to make automated connections with user-packed capillary traps, columns, and capillary emitters. The source uses plastic rectangular inserts (referred to here as cartridges) where individual components (i.e. trap, column, or emitter) can be exchanged independent of one another in a plug and play manner. Automated robotic connections are made between the three cartridges using linear translation powered by stepper motors to axially compress each cartridge by applying a well controlled constant compression force to each commercial LC fitting. The user has the versatility to tailor the separation (e.g. the length of the column, type of stationary phase, and mode of separation) to the experimental design of interest in a cost-effective manner. The source is described in detail, and several experiments are performed to evaluate the robustness of both the system and the exchange of the individual trap and emitter cartridges. The standard deviation in the retention time of four targeted peptides from a standard digest interlaced with a soluble Caenorhabditis elegans lysate ranged between 3.1 and 5.3 s over 3 days of analyses. Exchange of the emitter cartridge was found to have an insignificant effect on the abundance of various peptides. In addition, the trap cartridge can be replaced with minimal effects on retention time (<20 s).
The tremendous progress in the field of proteomics over the last decade can largely be attributed to significant advancements in mass spectrometry instrumentation (1–5). Mass spectrometers have improved in every imaginable analytical figure of merit, including the following: duty cycle, resolving power, mass measurement accuracy, sensitivity, and compatibility to various dissociation techniques (6–8). An area of advancement that is not always acknowledged is the usability of current commercial instruments. No longer is one required to be an expert in instrumentation, an electrical engineer, or an analytical chemist to operate, maintain, and acquire scientifically sound data from a mass spectrometer. This fact combined with the power of mass spectrometry to achieve absolute molecular specificity has expanded the breadth of users to experts outside the field of traditional chemistry (e.g. biology). However, in a typical proteomics experiment mass spectrometric analysis is preceded by liquid chromatography (i.e. LC-MS), which is significantly less robust.
Chromatography is a major limitation in proteomics (e.g. throughput and robustness) but an absolute necessity. Separation of analytes as a function of time accomplishes two main feats: 1) it reduces charge competition inside an ESI droplet that mitigates ion suppression; 2) it allows the mass spectrometer time to interrogate more ions. Both of these are critical to maximizing proteome coverage and identifying potential physiologically relevant species that are present at low abundance in biological samples. However, with these advantages, the robustness of chromatography suffers due to its extreme sensitivity to sample preparation, detergents, particulate (i.e. clogging), fitting fatigue, and dead volume. Several of these problems are compounded by the necessity in proteomics to operate in the nanoliter flow-rate regime. The sensitivity of electrospray ionization is inversely proportional to flow rate (9–11), and low flow rates concentrate the analyte inside the ESI droplet as species elute in a smaller peak volume. Combined, these effects of low flow rates significantly increase the sensitivity of proteomic experiments via LC-MS. Nano-flow rates require the miniaturization of all components of an LC instrument. Although this is advantageous in that it requires lower amounts of material to achieve the desired signal, it increases the susceptibility to various problems such as clogging. Other factors become increasingly important when operating at nano-flow rates, including the ability to make low dead volume connections.
As the field of proteomics continues to attract new researchers and as the seasoned proteomic laboratories move into clinical validation of putative biomarkers where thousands of analyses will be necessary, the robustness of chromatography needs improvement. Some of these factors are experimental/user-specific (e.g. good sample preparation) and are unlikely to be obviated by improved hardware design. However, there is a strong need for systems to be developed that automate connections between components, removing the variation inherently associated with manual connections. In addition, there is a need for systems where individual components can be rapidly and reproducibly exchanged independently of one another. Agilent Technologies has introduced a partial solution to this problem, referred to as HPLC-Chip Cube, in which the trapping, separation, and ESI is performed on a microfluidic chip (12, 13). The chip contains all of the components and thus has eliminated the need to make conventional finger tight fittings. In addition, because these chips can be fabricated to extremely low tolerances, the inter-chip reproducibility is exceptional. However, the device is limited in that if a single component fails (e.g. ESI tip), it often requires replacement of the whole chip, which is expensive. Eksigent Technologies developed the modular NanoFlex where trapping and separation of mixtures are performed on separate microfluidic chips, both which are separated from the ESI emitter. However, it still suffers from the high cost in purchasing replacement or additional chips. Possibly, the most noteworthy drawback of both devices is the limited versatility. Consumers are limited to a finite selection of vendor stationary phases and short set column lengths (<15 cm). These factors significantly limit both the type and performance (i.e. peak capacity) of any individual proteomic experiment. Furthermore, neither device is currently ultra-high pressure compatible (>4000 p.s.i.). Recently, Thermo Fisher Scientific introduced an integrated column and emitter device that requires the user to make a single manual connection (14). Although this device is ultra-HPLC-compatible and undoubtedly simplifies the setup, it neither allows for the exchange of individual components nor does it accept user packed capillary columns.
Herein, we describe the development and characterization of a vendor-neutral device that fills an empty “niche” in LC-MS instrumentation. The hybrid LC-MS device incorporates the advantages of the traditional setup (i.e. versatility and low cost) combined with the ability to automate connections between the individual components. The device accepts user or commercially packed capillary columns and is completely plug and play. It allows the user the ability to exchange individual components independently of one another. The device is described in detail, and several experiments are described to systematically evaluate the reproducibility of the system and the exchange of the individual components.
EXPERIMENTAL PROCEDURES
Materials
Formic acid, ammonium bicarbonate, DTT, and iodoacetamide, were obtained from Sigma-Aldrich. Proteomics grade trypsin was purchased from Promega (Madison, WI). HPLC grade acetonitrile was from Burdick & Jackson (Muskegon, MI). A Caenorhabditis elegans lysate (soluble fraction) was prepared as reported previously (15). The standard six-protein digest was obtained from Bruker-Michrom (Auburn, CA). Integra-Frit columns (IF360-75-50-N-5) and emitters (FS360-25-10-N-5) were from New Objective (Woburn, MA). Commercial LC fittings (F-152 and PK-152) were from IDEX (Lake Forest, IL).
Methods (Nano-flow LC-MS)
Splitless nano-flow chromatography was performed in the vented column configuration using a Waters NanoAcquity LC system (Waters Corp., Milford, MA). Solvents A and B were 99.9:0.1 water/formic acid and 99.9:0.1 acetonitrile/formic acid, respectively. A flow rate of 2 μl/min (98% A and 2% B) flushed sample out of a 5-μl loop and onto a self-packed capillary trap column (100 μm inner diameter × 4–5 cm). After 12 μl of wash, the six-port valve switched and closed the vent that initiated the gradient flow (250 nl/min) and data acquisition. For the lysate analyses, a 90-min analysis was used in which solvent B ramped from 2 to 34% over 60 min and from 34 to 80% over 1 min; it was held constant for 5 min, and then initial conditions were restored for the final 24 min. For the standard digest mix analyses, a 65-min run time was used in which solvent B was ramped from 2 to 34% over 40 min and from 34 to 80% over 1 min, it was held constant for 5 min and ramped down to initial conditions for 1 min; and the column was re-equilibrated at initial conditions for the remaining 18 min. Integra-Frit columns (New Objective, Woburn, MA) were packed in-house to stated lengths with either 4 μm of C12 (Phenomenex Torrance, CA), 3 μm C18 (Dr. Maisch, Ammerbuch-Entringen, Germany), 1.9 μm C18 (Dr. Maisch, Ammerbuch-Entringen, Germany) reversed-phase particles, or 5 μm Amide-80 HILIC particles (Tosoh Biosciences, King of Prussia, PA). All separations were performed with 4 μm C12 stationary phase except where noted.
Mass spectrometric analysis was performed with a Velos Pro LTQ (Thermo Fisher Scientific, San Jose, CA) or LTQ-FT-ICR ultra mass spectrometer equipped with a 7-tesla superconducting magnet (Thermo Fisher Scientific, Bremen, Germany). For the analysis with the standard protein digest, both instruments performed a full scan (m/z 400–1400) followed by four pseudo SRM scans that targeted four peptides in the mixture. For the complex lysate, the Velos Pro acquired 10 data-dependent MS2 scans per full MS1 scan for which a targeted AGC value of 30,000 charges and maximum injection time of 25 ms for MS1 was used. For MS2, a target value of 8000 charges with a maximum injection time of 80 ms was utilized. Dynamic exclusion was employed with a list size of 50 and duration of 30 s. Both instruments were calibrated following the manufacturer's protocol.
Experiment 1, Retention Time Reproducibility of System
A standard commercial six bovine protein digest was analyzed 20 times using a full scan followed by four pseudo SRM scans that targeted four known peptides in the mixture. A complex C. elegans lysate was interlaced between each standard to add complexity to the experiment. The base peak chromatograms of the standard protein digest along with the targeted analyses were used to gauge the retention time reproducibility of the system across ∼3 days of analyses. Raw files were directly imported into Skyline (16), and data were exported into R version 2.11.1 for analysis.
Experiment 2, Reproducibility of ESI-Tip Placement/Ion Abundance as a Function of Emitter Cartridge Replacement
25 injections of the standard digest were made on the same trap and column with the emitter replaced on the 6th, 11th, 16th, and 21st injection. A total of five emitters were used. These experiments allowed the assessment of peptide abundance reproducibility as a function of exchanging the emitter cartridge. Raw files were viewed in Skyline and MS1 peak areas (sum of M+0, M+1, and M+2), and RTs1 were exported into Microsoft Excel and then imported into R version 2.11.1. Peptides were identified based on accurate mass, retention time reproducibility, and isotope distributions. In addition, magnification images (×50, Dino-Lite, New Taipei City 241, Taiwan) were taken to evaluate the reproducibility of emitter placement (tip to MS capillary distance) among the different emitters.
Experiment 3, Reproducibility of Retention Time as a Function of Trap Cartridge Replacement
15 injections of the standard digest were made on the same column and emitter. The trap cartridge was exchanged on the 6th and 11th injection. A total of three different trap columns were used with measured bed lengths of 4.0, 3.9, and 4.1 cm. Each trap cartridge was first conditioned at high organic (80% B), then 50% B, and finally initial (2%B) conditions for 20 min. Prior to running the five standards, each new trap was also conditioned with a C. elegans lysate. These studies allowed the assessment of the retention time reproducibility as a function of trap cartridge replacement. Raw files were viewed in Skyline, and retention time data were exported into R version 2.11.1 for analysis.
Experiment 4, Comparison of Peak Widths between Prototype and Conventional LC-MS Setup
Nine replicate injections of a C. elegans lysate were analyzed using the prototype and a conventional LC setup. All effort was made to keep the trap (5.0 ± 0.1 cm) and column length (25.0 ± 0.1 cm) exactly the same between the two setups. Peak widths of identified peptides were extracted using an in-house program that fits a Gaussian distribution to each elution profile. The full widths at half-maximum were then calculated from these distributions and used to compare the performance between the prototype and the conventional method using finger tight fittings.
RESULTS AND DISCUSSION
Source
The device consists of a mounting bracket that interfaces the robotic source with a mass spectrometer and consumable inserts. The device accepts various kinds of “pop-in” consumable inserts, including fitting inserts, analytical column inserts, trapping column inserts, and spray emitter inserts. Leak-tight seals are then created between these inserts by a force-drive system that compresses the inserts together with a well controlled, continuous force. The force-drive system consists of electric motors, translational slides, lead screws, and an onboard microprocessor. The drive system provides a 100% linear compression of the sealing surfaces, which is in contrast to traditional fittings where components are driven together by the turning of threaded parts. This linear compression mechanism ensures no twisting of the fragile glass capillaries, and all connections are made in a fully automated manner, without the use of any tools. The sealing components are held and positioned by the robotic system ensuring the alignment of the capillary with the fittings. To avoid over or under tightening, a novel dynamic compression compensation mechanism was designed where force sensors provide constant read-back to the electronics, ensuring that the user-defined sealing force is always maintained. Upon sealing, a low dead volume connection is ensured by the spring-like mechanism that is created inside the cartridge (supplemental Fig. 1). When the actual compression force deviates from the user-defined force by ±1.0 pound (e.g. when fittings fatigue), the motors will automatically engage to re-adjust the compression force applied. This dynamic compensation mechanism eliminates the possibility of leaks/dead volume developing due to fitting fatigue. It also allows for various compression set points to be chosen via the software for situations of significantly different compression requirements. As an alternative, a second system design implements a force generation mechanism where the motors move the linear drive system a set distance and an in-line compression spring creates a constant force at a set target value ±1.0 pound. A microprocessor controls the motor direction and drive distance traveled. Again, fitting fatigue or changes in temperature are compensated by the constant applied force of the in-line spring. Both of these compression approaches remove the potential for over and under tightening that commonly occurs in practice with traditional thread-based fittings. The fine control of sealing force offered by these approaches becomes especially important for small bore capillary columns operated at ultra-HPLC (UHPLC) pressures. The compression connections formed using this device remain leak-tight to greater than 10,000 p.s.i. allowing for UPLC capabilities.
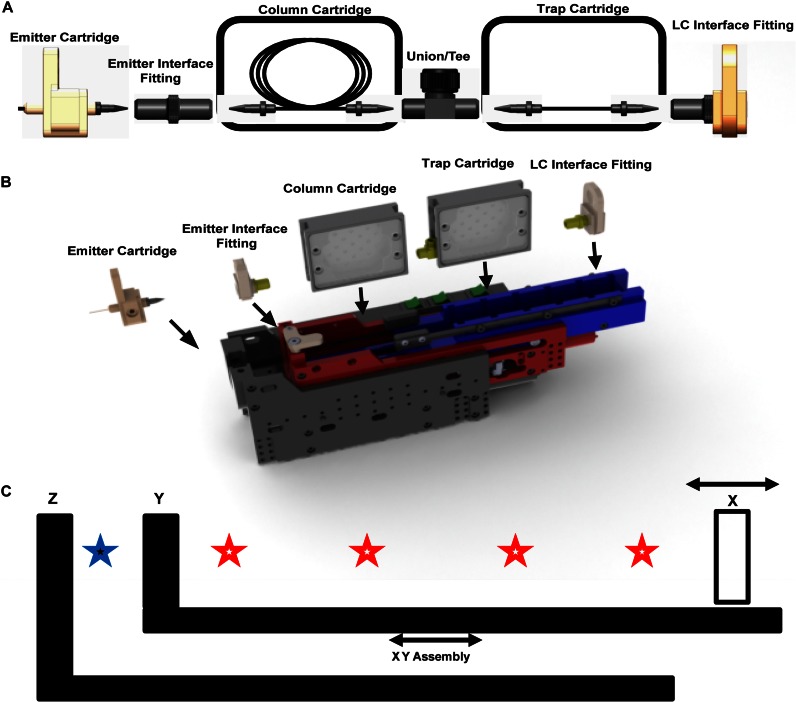
Fig. 1A displays a cross-sectional schematic of the device showing the individual components in relationship to one another. A model of the device is shown in Fig. 1B with the components floating above the figure. Here, the user would first place the fitting inserts into their respective locations in the robotic system. Then the emitter, analytical column, and trap column cartridge inserts are also placed into the device. Although Fig. 1B specifically shows a vented trap configuration (Trap-TEE-Analytical Column), the source is versatile and can accommodate various combinations of components and cartridges (e.g. single column, infusion insert, dual mixed phase analytical columns, etc.). A schematic of the two independent linear compression zones contained within the device is shown in Fig. 1C. These independent compression zones allow for the simultaneous sealing of multiple components with the first translation mechanism (XY assembly) and also an independent emitter changing in the second compression zone (Z assembly). Here, the components and cartridges placed between the movable force drive (X) and a hard stop (Y) are compressed, sealing all components simultaneously. In Fig. 1C, the locations of seals in the XY assembly are indicated by open stars. The emitter cartridge resides in location Z, acting as a hard stop for the emitter. For the emitter sealing process, the XY assembly in its entirety is driven by a second force mechanism and moves along the device's frame so that the emitter interface fitting residing in Y compresses against the emitter cartridge ferrule, held in place in Z. The seal between the emitter cartridge and the emitter interface fitting is indicated by a solid star in Fig. 1C. This allows for the column components (or any combination of components between X and Y) to remain sealed and compressed, while the emitter is exchanged or vice versa. The column cartridges ride in a track, and the fitting holder inserts and emitter cartridge are held stationary. Although all components, including the emitter, could have been designed to be compressed with a single mechanism, this dual mechanism approach allows for a rapid emitter exchange without the need for stopping flow to the column components during the change, thus saving time and disruption of the separation components. A picture of the device along with its inserts relative to a 12-inch ruler is displayed in supplemental Fig. 2. In addition, a movie of the device as it seals the individual components is shown in supplemental Fig. 3.
Fig. 1.
A, cross-sectional schematic of a vented column setup is shown. The emitter interface fitting, LC interface fitting, and emitter cartridge are stationary, although the column cartridges float in a track. B, CAD model is shown with inserts floating above. C, two independent compression zones of the device. The emitter interface fitting would be inserted in position Y, and the LC interface fitting would be inserted in position X. To form compression connections, X is driven toward Y until the pre-determined sealing force has been reached. At this point, leak-tight connections, represented by open stars, have been established between all components in the XY assembly. The emitter cartridge is stationary at location Z. To form the emitter seal, the entire XY assembly moves to compress against Z. The solid blue star represents the leak-tight compression connection made between the emitter cartridge and the emitter interface fitting. This nesting assembly design allows for emitters or columns to be exchanged independently, without affecting each other.
Experiment 1, Retention Time Reproducibility of System
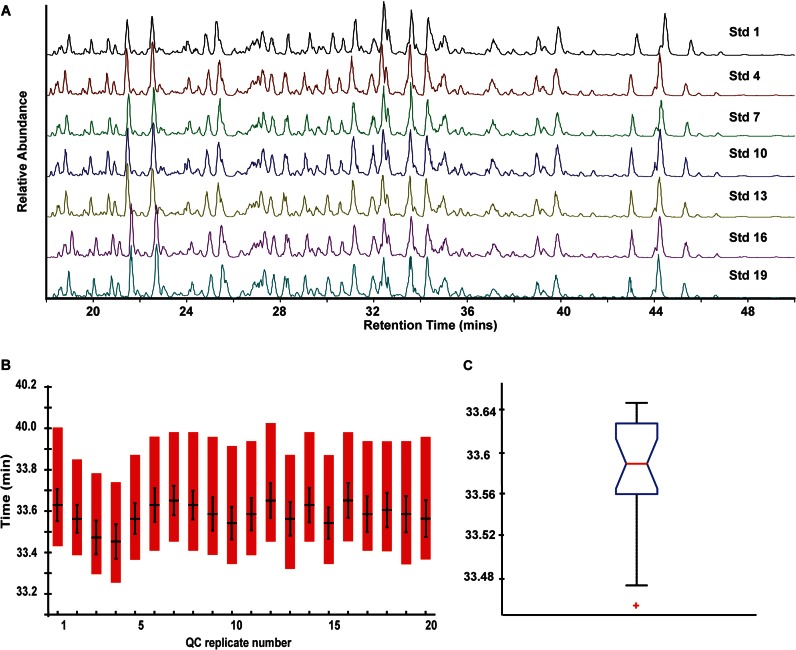
Fig. 2A displays seven representative base peak ion chromatograms of the standard protein digest stacked upon one another. Twenty digests were analyzed in total with each one interlaced with a complex lysate run. The reproducibility of the source is shown in these chromatograms as qualitatively there is little shift in the retention time between the seven analyses. The shift in the first digest (Fig. 2A, Std 1) is the most noticeable; however, we attribute this to factors associated with column conditioning. Fig. 2B displays the retention time of a single peptide (e.g. K.VLDALDSIK.T) throughout the 20 runs. The length of the red bars in Fig. 2B describes the width of the peak at base line, and the length of the black vertical bars indicates the full width at half-maximum. The horizontal black bar in Fig. 2B is the apex of the elution profile. Using a Mann-Kendall test for randomness, there is no significant trend in the retention time (p = 0.18) indicating that the distribution is free from systematic error. Fig. 2C displays a boxplot summarizing the distribution of retention times (i.e. peak's apex). The horizontal line in Fig. 2C across the box and notch represents the median of the data and the 95% confidence interval in the median, respectively. The box in Fig. 2C describes 50% of the data (1st to 3rd quartiles), and the whiskers connect the most extreme values within 1.5* interquartile ranges (IQRs) of the lower and upper quartiles. The 95% confidence interval in the median was ± 2.4 s with an interquartile range of 4.2 s over 3 days of analysis. Furthermore, the standard deviation of the four targeted peptides from these runs ranged from 3.1 to 5.2 s. These experiments illustrate the high retention time reproducibility across several days afforded by this device.
Fig. 2.
A, representative base peak chromatograms of a standard commercial digest across the 3 days of experiments illustrating the retention time reproducibility of the source. For this experiment, a standard digest was run between complex C. elegans lysates (n = 20). B, retention time reproducibility of a targeted peptide identified in the standard digest across the 20 runs. The length of the bars represents the integration widths of the peak. The vertical lines within the bars correspond to the full width at half-maximum height of the peak. The horizontal line represents the apex of the peak. C, box plot describing the distribution of the retention times (apex) of the 20 runs. The horizontal line across the box and notch represent the median of the data and the 95% confidence interval in the median, respectively. The box describes 50% of the data (1st to 3rd quartiles), and the whiskers connect the most extreme values within 1.5 IQRs of the lower and upper quartiles. The plus symbols represent outliers (>1.5 IQRs from upper and lower quartiles). For this peptide (K.VLDALDSIK.T) the IQR was 5.4 s.
Experiment 2, Reproducibility of ESI-Tip Placement/Ion Abundance as a Function of Emitter Cartridge Replacement
Next, we wanted to determine the reproducibility of exchanging the emitter in terms of tip placement, peptide retention time, and ion abundance. Because the distance of an electrospray tip to the MS inlet can affect numerous factors related to the electrospray process, including spray mode, desolvation, voltage onset, and ion loss, which in turn can have dramatic effects on ion abundance (17), it was necessary to evaluate the reproducibility of the system in its placement of the emitter. To accomplish this goal, the emitter cartridge was swapped five times, and each time a high magnification image was taken with respect to the MS inlet. This experiment was performed using the same emitter replaced five times and with using five different commercial emitters (supplemental Fig. 4). The differences in the images were indistinguishable when using the same emitter. However, small differences (∼200–300 μm) were observed in the experiment where five different emitters were used. This difference is attributed to the small tolerances in the stated length of each commercial emitter. In other words, tip placement is dependent on the length of the emitter.
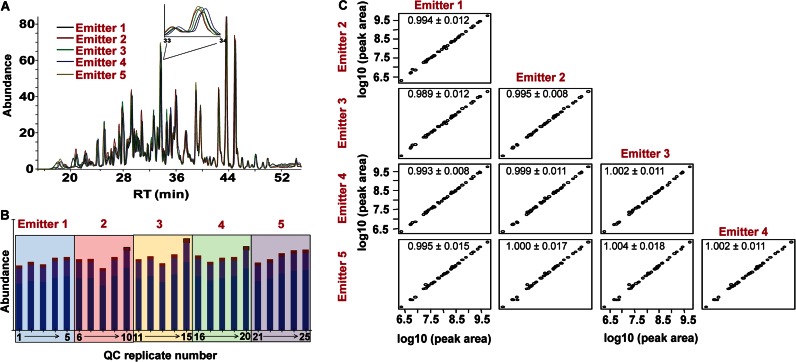
To investigate the reproducibility of emitter exchange and if the small differences in tip placement were significant in terms of ion abundance, a series of experiments were performed in which a standard digest was injected 25 times on the same trap and column cartridge with the emitter cartridge exchanged every five runs. Fig. 3A displays a qualitative view of the reproducibility in retention times and abundances of a single replicate randomly selected from each of the five emitters. The inset in Fig. 3A shows an expanded view (33–34 min) indicating very similar retention times and abundances. Fig. 3B displays the abundance of a single peptide across the five replicates for each emitter. As expected, there are slight deviations in abundance within the same emitter, possibly due to any single factor or a combination of factors, including the following: 1) variation in the amount of peptide loaded on column; 2) ESI fluctuations, and 3) differences in integration of the elution profile. However, using a one-way analysis of variance, the difference in the mean values among the emitters was not significant (p = 0.48). This indicates that one can change out the emitters reproducibly without significant effects on the ESI process and peptide abundance. To provide further evidence supporting this claim, Fig. 3C displays a draftsman plot in which the integrated MS1 intensities of various peptides (n = 43) were plotted between the different emitters. Each point represents the average of the intra-replicate (n = 5) abundances for each peptide. There is a high linearity associated with all five emitters when plotted against each other over 3 orders of dynamic range. The 95% confidence interval of the slope from the linear regression analysis for all pairwise comparisons contains the theoretical value of 1, indicating there is no statistically significant difference between the experimental and the theoretical slopes (α = 0.05). These data emphasize that the emitter cartridge can be exchanged in a highly reproducible manner with limited effects on ion abundance.
Fig. 3.
A, comparison of representative base peak chromatograms from the standard digest as a function of different emitters. The inset shows a magnified region (34–35 min) illustrating the high degree of similarity in both retention times and overall ion abundance between the various emitters. B, bar plot showing the variation in the integrated areas of the MS1 signal (sum M+0, M+1, and M+2) within and between the emitter for peptide R.GASIVEDK.L. The mean intensities were not statistically different between the emitters (p = 0.48). C, draftsman plot comparing the integrated MS1 signal intensities of various peptides (n = 43) in the standard digest between the different emitters. Each point represents the average of intra-emitter replicates. The numbers correspond to the slope and 95% confidence interval from the regression analysis.
Experiment 3, Reproducibility of Retention Time as a Function of Trap Cartridge Replacement
Because the ESI tip and trap are the components of an LC system most prone to deteriorate from extended LC-MS runs of complex samples, we wanted to investigate the reproducibility of retention time as a function of trap cartridge exchange. The final experiment investigated this metric using three different self-packed traps. The three similar length traps (4 ± 0.1 cm) were conditioned with solvent and a single complex run as described under “Methods.” Then a standard digest was injected 15 times with the same emitter and column, but the trap was exchanged after every five runs.
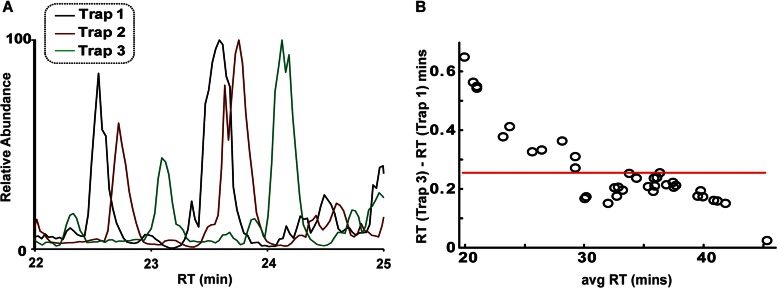
Fig. 4A shows an elution window (22–25 min) of three base peak chromatograms from representative runs among the three different trap columns. The difference in RT is most evident with the third trap as peptides eluted significantly later. The difference in peptide retention time between the two extremes (trap 3 versus trap 1) decreased as a function of retention time as shown in Fig. 4B. The largest differences were observed with hydrophilic peptides that eluted early in the gradient where much smaller differences were observed as a function of RT. The average difference in mean RT between trap 3 and trap 1 was 0.26 min and is represented as a red line in Fig. 4B. For scheduled SRM experiments where windows are typically at least 2 min wide (18, 19), the precision in the RT among the different trap cartridges is more than adequate.
Fig. 4.
A, comparison of the RT in the base peak chromatograms among the various traps from 22 to 25 min. B, plot of the differences between the RT (trap 3 to trap 1) as a function of average RT of the individual peptides (n = 37) between the two traps. Trap exchange had the most profound effect on hydrophilic peptides and much less of an effect on later eluting peptides. The average difference (red line) was ∼15.6 s.
Experiment 4, Comparison of Peak Widths between Prototype and Conventional LC-MS Setup
Finally, it was necessary to compare performance of this prototype system to using a traditional packed-tip nano-flow LC setup with finger tight fittings. For complex mixtures, the two systems perform similarly in terms of peak widths. Slightly longer peak widths at full width at half-maximum (∼1 s) were observed in the prototype system (supplemental Fig. 5), which we attribute to the inherent dead volume associated with an unpacked (i.e. open) ESI tip. Regardless, these data indicate that there is little loss of performance in terms of chromatography. The ability to make automated connections and exchange the various components in a reproducible plug and play manner offers a significant advancement in LC instrumentation for high throughput proteomics. The versatility of this device in which different column lengths, any stationary phase, and different modes of separation can be performed with ease (supplemental Fig. 6) and at relatively low cost offers significant advantages over current approaches.
CONCLUSIONS
Motivated by the difficulties in nano-flow chromatography (e.g. making reproducible low dead volume connections) and the limitations in current commercial technologies aimed at circumventing these problems (e.g. expense and versatility), we developed a vendor-neutral device that automates connections among various components and accepts user packed capillary traps, columns, and emitters in a plug and play manner. These studies have been directed at investigating the reproducibility in retention time and ion abundance of exchanging the various components within the device. The ESI emitter, a component prone to failure throughout a series of experiments, can be exchanged reproducibly in a high throughput manner without disturbing the trap and column cartridges and without significant effects on the ESI process. The trap cartridge, while showing statistically significant variation, could still be exchanged with limited effect on RT (0.26 ± 0.13 min). Because of its versatility and plug and play attributes, we feel this device will find significant use across clinical laboratories, core laboratory facilities, and even seasoned proteomic laboratories interested in high throughput. Although some practice is needed to become familiar with the device, the ease at which low dead volume connections are made is significant, and we are certain that the device decreases the technical expertise needed to perform robust nano-LC-MS measurements. Future work is ongoing to develop hardware and software to completely automate the emitter exchange system centered on real time feedback from the instrument based on heuristics calculated from peptide abundance and spray stability. The source described in this study is now marketed as the CorConneX source and is commercially available from CorSolutions (Ithaca, NY).
Supplementary Material
Footnotes
* This work was supported, in whole or in part, by National Institutes of Health Grant 5R44RR024628-04 from NCRR, Grant 8R44GM103386-04 from NIGMS, and Grant P41 GM103533. Conflict of Interest: The device described in this work is currently being marketed by CorSolutions under the product name “CorConnex LC/ESI Source.” Colleen K. Van Pelt is founder and CEO of CorSolutions and Thomas N. Corso is the Chief Technical Officer. The MacCoss laboratory has a sponsored research agreement with Thermo Fisher.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- RT
- retention time
- IQR
- interquartile range
- SRM
- Selected Reaction Monitoring.
REFERENCES
- 1. Syka J. E., Marto J. A., Bai D. L., Horning S., Senko M. W., Schwartz J. C., Ueberheide B., Garcia B., Busby S., Muratore T., Shabanowitz J., Hunt D. F. (2004) Novel linear quadrupole ion trap/FT mass spectrometer: Performance characterization and use in the comparative analysis of histone H3 post-translational modifications. J. Proteome Res. 3, 621–626 [DOI] [PubMed] [Google Scholar]
- 2. Andrews G. L., Simons B. L., Young J. B., Hawkridge A. M., Muddiman D. C. (2011) Performance characteristics of a new hybrid quadrupole time-of-flight tandem mass spectrometer (TripleTOF 5600). Anal. Chem. 83, 5442–5446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hu Q., Noll R. J., Li H., Makarov A., Hardman M., Graham Cooks R. (2005) The Orbitrap: a new mass spectrometer. J. Mass Spectrom. 40, 430–443 [DOI] [PubMed] [Google Scholar]
- 4. Olsen J. V., Schwartz J. C., Griep-Raming J., Nielsen M. L., Damoc E., Denisov E., Lange O., Remes P., Taylor D., Splendore M., Wouters E. R., Senko M., Makarov A., Mann M., Horning S. (2009) A dual pressure linear ion trap orbitrap instrument with very high sequencing speed. Mol. Cell. Proteomics 8, 2759–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Second T. P., Blethrow J. D., Schwartz J. C., Merrihew G. E., MacCoss M. J., Swaney D. L., Russell J. D., Coon J. J., Zabrouskov V. (2009) Dual-pressure linear ion trap mass spectrometer improving the analysis of complex protein mixtures. Anal. Chem. 81, 7757–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bereman M. S., Canterbury J. D., Egertson J. D., Horner J., Remes P. M., Schwartz J., Zabrouskov V., MacCoss M. J. (2012) Evaluation of front-end higher energy collision-induced dissociation on a benchtop dual-pressure linear ion trap mass spectrometer for shotgun proteomics. Anal. Chem. 84, 1533–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Williams D. K., Jr., McAlister G. C., Good D. M., Coon J. J., Muddiman D. C. (2007) Dual electrospray ion source for electron-transfer dissociation on a hybrid linear ion trap-orbitrap mass spectrometer. Anal. Chem. 79, 7916–7919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Baba T., Hashimoto Y., Hasegawa H., Hirabayashi A., Waki I. (2004) Electron capture dissociation in a radio frequency ion trap. Anal. Chem. 76, 4263–4266 [DOI] [PubMed] [Google Scholar]
- 9. Wilm M., Mann M. (1996) Analytical properties of the nanoelectrospray ion source. Anal. Chem. 68, 1–8 [DOI] [PubMed] [Google Scholar]
- 10. Luo Q., Tang K., Yang F., Elias A., Shen Y., Moore R. J., Zhao R., Hixson K. K., Rossie S. S., Smith R. D. (2006) More sensitive and quantitative proteomic measurements using very low flow rate porous silica monolithic LC columns with electrospray ionization-mass spectrometry. J. Proteome Res. 5, 1091–1097 [DOI] [PubMed] [Google Scholar]
- 11. Schmidt A., Karas M., Dülcks T. (2003) Effect of different solution flow rates on analyte ion signals in nano-ESI MS, or: when does ESI turn into nano-ESI? J. Am. Soc. Mass Spectrom. 14, 492–500 [DOI] [PubMed] [Google Scholar]
- 12. Yin H., Killeen K., Brennen R., Sobek D., Werlich M., van de Goor T. (2005) Microfluidic chip for peptide analysis with an integrated HPLC column, sample enrichment column, and nanoelectrospray tip. Anal. Chem. 77, 527–533 [DOI] [PubMed] [Google Scholar]
- 13. Fortier M.-H., Bonneil E., Goodley P., Thibault P. (2005) Integrated microfluidic device for mass spectrometry-based proteomics and its application to biomarker discovery programs. Anal. Chem. 77, 1631–1640 [DOI] [PubMed] [Google Scholar]
- 14. Kiyonami R., Ravnsborg C., Madsen O., Zabrouskov V. (2012) Easy-to-Use, Plug-and-Spray Ion Source for Robust and Reproducible Ultra High Pressure Nanoflow LC/MS. Technical Note 63546, Thermo Fisher Scientific, San Jose, CA [Google Scholar]
- 15. Bereman M. S., Egertson J. D., MacCoss M. J. (2011) Comparison between procedures using SDS for shotgun proteomic analyses of complex samples. Proteomics 11, 2931–2935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. MacLean B., Tomazela D. M., Shulman N., Chambers M., Finney G. L., Frewen B., Kern R., Tabb D. L., Liebler D. C., MacCoss M. J. (2010) Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cech N. B., Enke C. G. (2001) Practical implications of some recent studies in electrospray ionization fundamentals. Mass Spectrom. Rev. 20, 362–387 [DOI] [PubMed] [Google Scholar]
- 18. Shuford C. M., Li Q. Z., Sun Y. H., Chen H. C., Wang J., Shi R., Sederoff R. R., Chiang V. L., Muddiman D. C. (2012) Comprehensive quantification of monolignol-pathway enzymes in Populus trichocarpa by protein cleavage isotope dilution mass spectrometry. J. Proteome Res. 11, 3390–3404 [DOI] [PubMed] [Google Scholar]
- 19. Stahl-Zeng J., Lange V., Ossola R., Eckhardt K., Krek W., Aebersold R., Domon B. (2007) High sensitivity detection of plasma proteins by multiple reaction monitoring of N-glycosites. Mol. Cell. Proteomics 6, 1809–1817 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.