Abstract
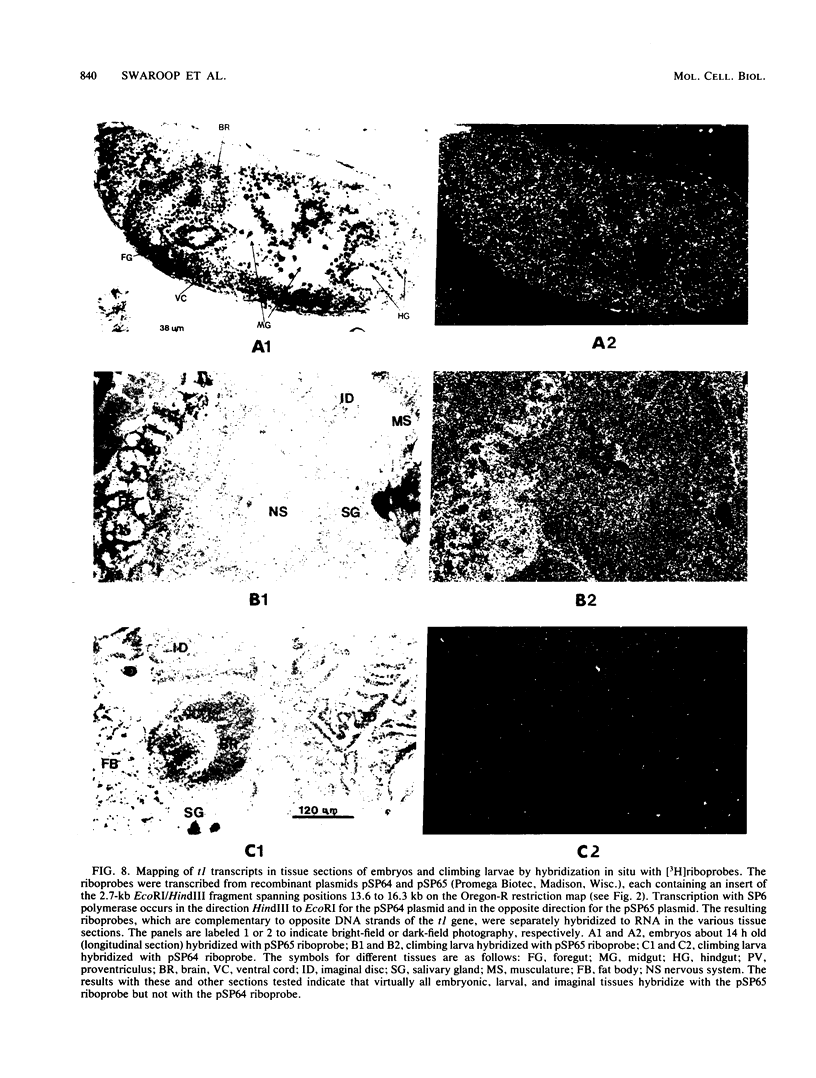
The Glued locus of Drosophila melanogaster is genetically defined as the functional unit which is affected by the dominant Glued mutation Gl. Genomic DNA was cloned from the region of the Glued locus, at 70C2 on chromosome 3, by using a P element insertion in the region as a molecular marker. Three genes encoding polyadenylated transcripts were detected within a 30-kilobase span of the cloned DNA. The gene nearest the P element insertion site was identified as a Glued gene on the basis of alterations in its DNA and encoded transcript associated with the Gl mutation and with reversions of Gl which eliminate the dominant effect by inactivation of the mutant allele. Expression of the wild-type Gl+ gene is temporally regulated during development; the amount of the encoded transcript is highest in the embryonic stage, decreasing in the first and second larval instars, and then increasing in the third instar and pupal stages. There is a maternal contribution of the Gl+ transcript to the embryo, which probably accounts for the maternal lethal effect of Glued mutations on early development. In situ hybridizations of Gl+ DNA to RNA in tissue sections showed that the Gl+ transcript is present in virtually all tissues of the embryo, late larva, and pupa. The general distribution of this transcript is consistent with genetic evidence indicating that the Glued locus controls a generally essential cell function (P. J. Harte and D. R. Kankel, Genetics 101:477-501, 1982). Different Glued mutations produce distinct phenotypic effects, including adults with severe visual defects, larvae lacking imaginal discs, and early lethality. These diverse mutant phenotypes are discussed in terms of quantitative changes in the Glued function. Closely adjacent to Gl+ is another gene which is transcribed in a divergent direction and expressed coordinately with Gl+ throughout Drosophila development. It remains to be determined whether this gene is also involved with the Glued function.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akam M. E. The location of Ultrabithorax transcripts in Drosophila tissue sections. EMBO J. 1983;2(11):2075–2084. doi: 10.1002/j.1460-2075.1983.tb01703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S., Muskavitch M. A., Yedvobnick B. Molecular cloning of Notch, a locus affecting neurogenesis in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1977–1981. doi: 10.1073/pnas.80.7.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garabedian M. J., Hung M. C., Wensink P. C. Independent control elements that determine yolk protein gene expression in alternative Drosophila tissues. Proc Natl Acad Sci U S A. 1985 Mar;82(5):1396–1400. doi: 10.1073/pnas.82.5.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garen A., Miller B. R., Paco-Larson M. L. Mutations affecting functions of the Drosophila gene glued. Genetics. 1984 Aug;107(4):645–655. doi: 10.1093/genetics/107.4.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garen S. H., Kankel D. R. Golgi and genetic mosaic analyses of visual system mutants in Drosophila melanogaster. Dev Biol. 1983 Apr;96(2):445–466. doi: 10.1016/0012-1606(83)90182-3. [DOI] [PubMed] [Google Scholar]
- Garfinkel M. D., Pruitt R. E., Meyerowitz E. M. DNA sequences, gene regulation and modular protein evolution in the Drosophila 68C glue gene cluster. J Mol Biol. 1983 Aug 25;168(4):765–789. doi: 10.1016/s0022-2836(83)80074-6. [DOI] [PubMed] [Google Scholar]
- Hafen E., Levine M., Garber R. L., Gehring W. J. An improved in situ hybridization method for the detection of cellular RNAs in Drosophila tissue sections and its application for localizing transcripts of the homeotic Antennapedia gene complex. EMBO J. 1983;2(4):617–623. doi: 10.1002/j.1460-2075.1983.tb01472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harte P. J., Kankel D. R. Genetic analysis of mutations at the Glued locus and interacting loci in Drosophila melanogaster. Genetics. 1982 Jul-Aug;101(3-4):477–501. doi: 10.1093/genetics/101.3-4.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell M. G., Kidwell J. F., Sved J. A. Hybrid Dysgenesis in DROSOPHILA MELANOGASTER: A Syndrome of Aberrant Traits Including Mutation, Sterility and Male Recombination. Genetics. 1977 Aug;86(4):813–833. doi: 10.1093/genetics/86.4.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer-Safer P. R., Levine M., Ward D. C. Immunological method for mapping genes on Drosophila polytene chromosomes. Proc Natl Acad Sci U S A. 1982 Jul;79(14):4381–4385. doi: 10.1073/pnas.79.14.4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M., Garen A., Lepesant J. A., Lepesant-Kejzlarova J. Constancy of somatic DNA organization in developmentally regulated regions of the Drosophila genome. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2417–2421. doi: 10.1073/pnas.78.4.2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Hardison R. C., Lacy E., Lauer J., O'Connell C., Quon D., Sim G. K., Efstratiadis A. The isolation of structural genes from libraries of eucaryotic DNA. Cell. 1978 Oct;15(2):687–701. doi: 10.1016/0092-8674(78)90036-3. [DOI] [PubMed] [Google Scholar]
- Meyerowitz E. M., Kankel D. R. A genetic analysis of visual system development in Drosophilia melanogaster. Dev Biol. 1978 Jan;62(1):112–142. doi: 10.1016/0012-1606(78)90096-9. [DOI] [PubMed] [Google Scholar]
- Plough H H, Ives P T. Induction of Mutations by High Temperature in Drosophila. Genetics. 1935 Jan;20(1):42–69. doi: 10.1093/genetics/20.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ready D. F., Hanson T. E., Benzer S. Development of the Drosophila retina, a neurocrystalline lattice. Dev Biol. 1976 Oct 15;53(2):217–240. doi: 10.1016/0012-1606(76)90225-6. [DOI] [PubMed] [Google Scholar]
- Searles L. L., Jokerst R. S., Bingham P. M., Voelker R. A., Greenleaf A. L. Molecular cloning of sequences from a Drosophila RNA polymerase II locus by P element transposon tagging. Cell. 1982 Dec;31(3 Pt 2):585–592. doi: 10.1016/0092-8674(82)90314-2. [DOI] [PubMed] [Google Scholar]
- Shearn A., Garen A. Genetic control of imaginal disc development in Drosophila. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1393–1397. doi: 10.1073/pnas.71.4.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearn A., Rice T., Garen A., Gehring W. Imaginal disc abnormalities in lethal mutants of Drosophila. Proc Natl Acad Sci U S A. 1971 Oct;68(10):2594–2598. doi: 10.1073/pnas.68.10.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A. C., Rubin G. M. Transposition of cloned P elements into Drosophila germ line chromosomes. Science. 1982 Oct 22;218(4570):341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- Stewart M., Murphy C., Fristrom J. W. The recovery and preliminary characterization of X chromosome mutants affecting imaginal discs of Drosophila melanogaster. Dev Biol. 1972 Jan;27(1):71–83. doi: 10.1016/0012-1606(72)90113-3. [DOI] [PubMed] [Google Scholar]
- Swaroop A., Paco-Larson M. L., Garen A. Molecular genetics of a transposon-induced dominant mutation in the Drosophila locus Glued. Proc Natl Acad Sci U S A. 1985 Mar;82(6):1751–1755. doi: 10.1073/pnas.82.6.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West R. W., Jr, Yocum R. R., Ptashne M. Saccharomyces cerevisiae GAL1-GAL10 divergent promoter region: location and function of the upstream activating sequence UASG. Mol Cell Biol. 1984 Nov;4(11):2467–2478. doi: 10.1128/mcb.4.11.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]







