Summary
Uropathogenic Escherichia coli (UPEC) contain multiple horizontally acquired pathogenicity-associated islands (PAI) implicated in the pathogenesis of urinary tract infection. In a murine model of cystitis, type 1 pili-mediated bladder epithelial invasion and intracellular proliferation are key events associated with UPEC virulence. In this study, we examined the mechanisms by which a conserved PAI contributes to UPEC pathogenesis in acute cystitis. In the human UPEC strain UTI89, spontaneous excision of PAI IIUTI89 disrupts the adjacent leuX tRNA locus. Loss of wild-type leuX-encoded tRNA5Leu significantly delayed, but did not eliminate, FimB recombinase-mediated phase variation of type 1 pili. FimX, an additional FimB-like, leuX-independent recombinase, was also found to mediate type 1 pili phase variation. However, whereas FimX activity is relatively slow in vitro, it is rapid in vivo as a non-piliated strain lacking the other fim recombinases rapidly expressed type 1 pili upon experimental infection. Finally, we found that disruption of leuX, but not loss of PAI IIUTI89 genes, reduced bladder epithelial invasion and intracellular proliferation, independent of type 1 piliation. These findings indicate that the predominant mechanism for preservation of PAI IIUTI89 during the establishment of acute cystitis is maintenance of wild-type leuX, and not PAI IIUTI89 gene content.
Introduction
Escherichia coli primarily inhabit vertebrate gastrointestinal tracts as commensal organisms. Yet certain strains of E. coli are distinguished as pathogens, in part, by the presence of unique virulence factor genes not found in environmental or commensal strains (Groisman and Ochman, 1994). These pathogen-associated genes are often found clustered together on horizontally acquired genomic islands termed pathogenicity-associated islands (PAI) (Hacker et al., 1990). This type of horizontal gene transfer enables recipient strains to colonize unique host niches through a number of mechanisms, including host tissue invasiveness (Groisman and Ochman, 1996). Features typical of PAI include: high frequency of insertion at tRNA loci, GC content distinct from the rest of the chromosome, flanking direct repeats and presence of mobility genes (Dobrindt et al., 2004; Gal-Mor and Finlay, 2006). Hacker and colleagues have proposed that PAI become fixed into the genome by mutation over time as part of the normal adaptation process of PAI acquisition, where continued mutation and rearrangement eventually lead to genome reduction (Dobrindt et al., 2004).
Pathogenicity-associated islands are common among uropathogenic E. coli (UPEC) (Blum et al., 1995; Brzuszkiewicz et al., 2006; Chen et al., 2006; Lloyd et al., 2007), a genetically heterogeneous pathotype of E. coli isolates that are the most common cause of urinary tract infections (Foxman, 2002). However, despite their prevalence, the importance of these PAI in the urinary tract niche remains unclear. This is because many of these islands have become fixed within the genome, either losing the elements necessary for excision or becoming linked to critical genes such that excision would reduce the fitness of the strain (Hacker and Kaper, 1999). An example of the latter is found in the human pyelonephritis UPEC isolate 536, where spontaneous excision of a genomic island, PAI II536, results in deletion of two nucleotides within the adjacent leuX tRNA gene, leuXΔGC80-81. This occurs because PAI insertion into the ancestral strain duplicated the 3′ end of the leuX tRNA, leaving an intact leuX gene at one end of the PAI and a 3′ fragment of leuX at the other end. This non-coding 3′ fragment of leuX can then undergo mutation without disrupting tRNA5Leu expression and function. However, upon PAI excision via the direct repeats any mutated sequence within the 3′ end of this leuX fragment becomes incorporated into the coding leuX gene. In the case of strain 536, a two-nucleotide deletion has occurred near the 3′ end of the non-coding leuX fragment (Blum et al., 1994). These PAI excision dynamics have important clinical relevance as it has been demonstrated in vitro that fluoroquinolone antibiotic exposure can induce PAI excision and fluoroquinolone resistant UPEC isolates commonly lack PAI-encoded virulence factors (Soto et al., 2006).
Although leuX-associated PAI with synteny to PAI II536 are found among many UPEC (Bidet et al., 2005), the importance of these genomic islands, independent of leuX, in urinary tract disease pathogenesis is largely unproven. The UPEC strain UTI89, a human cystitis isolate, contains a previously uncharacterized 118.9 kb PAI, designated PAI IIUTI89, that is closely related to PAI II536 (99.1% identical over 97.3 kb) and is also found adjacent to the leuX locus. Almost all of the 124 open reading frames found on PAI IIUTI89 are unique within the UTI89 genome, and many encode putative virulence factor genes that could hypothetically potentiate the invasiveness of UPEC in the urinary tract (Chen et al., 2006). These include the genes for P pili (Roberts et al., 1994; Winberg et al., 1995), the toxins α-haemolysin (Nagy et al., 2006) and cytotoxic necrotizing factor 1 (CNF1) (Doye et al., 2002; Davis et al., 2005), the putative invasin, Hek (Srinivasan et al., 2003), and the recently identified contact-dependent inhibition of growth exoprotein, CdiA (Aoki et al., 2005). The presence of PAI IIUTI89 also directly affects the expression of virulence factors elsewhere on the chromosome by encoding transcriptional regulators such as PapB, which negatively regulates type 1 pili expression (Xia et al., 2000; Holden et al., 2006).
Type 1 pili are the major virulence factors in UPEC pathogenesis in a murine model of cystitis (Connell et al., 1996; Wright et al., 2007). The type 1 pilus-associated adhesin, FimH, mediates UPEC adherence to and invasion into the urinary bladder epithelium (urothelium) (Mulvey et al., 1998). These early events of cystitis are critical for the establishment of both acute infection and chronic persistence within quiescent intracellular reservoirs (Mulvey et al., 1998; 2001; Mysorekar and Hultgren, 2006). Type 1 pili bind with high affinity to mannosylated glycoprotein receptors present on urothelial cells, such as uroplakin Ia and α3β1 integrins (Zhou et al., 2001; Eto et al., 2007). After invasion of the urothelium, some of these bacteria proceed to replicate rapidly within the cytoplasm to form intracellular bacterial communities (IBC) (Anderson et al., 2003; Justice et al., 2004). The biofilm-like organization of the IBC requires the expression of type 1 pili (Wright et al., 2007) and appears to provide a protected niche where UPEC can more effectively multiply and survive in the face of vigorous innate immune responses (Mulvey et al., 2000; Anderson et al., 2003), which are mounted in response to UPEC infection (Mysorekar et al., 2002).
Spontaneous excision of PAI II536 and the resultant leuXΔGC80–81 mutation completely abolishes type 1 pili expression in the UPEC strain 536, and this defect is likely the primary mechanism for loss of urinary tract virulence (Blum et al., 1994). Indeed, type 1 pili expression, and therefore virulence in cystitis, is highly dependent on the leuX-encoded tRNA5Leu, which translates the rare leucine codon UUG (Ritter et al., 1995). Type 1 pili are encoded by the fim operon and their expression is phase variable, originating from the fim operon promoter, which is contained within the fimS-invertible element (Abraham et al., 1985). Two tyrosine recombinases, FimB and FimE, are known to control the orientation of the fimS-invertible region. FimB has bidirectional activity, but predominantly switches fim operon transcription from OFF to ON, while FimE mediates solely ON to OFF phase switching (Klemm, 1986). Additional FimB homologues also mediate type 1 pili phase variation in vitro or in vivo (Bryan et al., 2006; Xie et al., 2006). The dependence of type 1 pili expression on tRNA5Leu was demonstrated to be a direct result of impaired UUG codon translation within the fimB recombinase transcript in leuX mutants (Ritter et al., 1997). Therefore, the regulation of rare codon tRNA expression may provide a mechanism for the fine tuning of virulence gene expression in UPEC as they adapt to diverse niches (Saier, 1995; Piechaczek et al., 2000; Dobrindt and Hacker, 2001).
Our goal in this study was to dissect the specific mechanisms by which PAI IIUTI89 contributes to virulence in a murine model of cystitis. In particular, we hypothesized that the presence of PAI IIUTI89 genes as a whole, independent of leuX, would be important or even necessary for the early events of cystitis, namely urothelial invasion and IBC formation. We discovered that the spontaneous loss of PAI IIUTI89 in UTI89 disrupts leuX, thereby delaying but not eliminating type 1 pili phase switching, in vitro. This defect, which was restored by complementation with wild-type (wt) leuX, was a result of an apparent loss of tRNA5Leu function, but not expression. Overexpression of either FimB or FimX, a FimB homologue in UTI89, also restored normal phase switching, in vitro. However, we found that FimX is less dependent on tRNA5Leu-mediated translation than FimB and was able to mediate phase switching in UTI89ΔfimBE slowly in vitro, but rapidly in vivo in a murine model of cystitis. Finally, we showed that the absence of PAI IIUTI89 gene content did not significantly affect the early establishment of cystitis, whereas mutation of leuX led to a significant attenuation of UTI89, independent of type 1 piliation, during the early acute stages of UPEC pathogenesis.
Results
PAI IIUTI89 genes are conserved among lower urinary tract isolates
The prevalence of PAI IIUTI89-like islands among UPEC isolates from women with lower urinary tract infection (LUTI) was unknown. Therefore, we surveyed a collection of 163 UPEC isolates from adult women with LUTI by multiplex PCR for the presence of the three PAI IIUTI89 genes cnf1, hek and hlyA, which, when found concurrently, are a specific indicator of a PAI IIUTI89-like genomic island (Bidet et al., 2005). We found that the individual prevalences of these genes in our collection of clinical isolates ranged from 44% to 48% (Table S1), and that all three genes were found concurrently in 43% of isolates. These prevalences were significantly higher (P < 0.0001, Fisher's exact test) than those found in the E. coli reference collection (ECOR) of mostly commensal isolates (Ochman and Selander, 1984) in which the individual gene prevalences ranged from 8% to 17%. Only six strains (8%) in the ECOR set were positive for all three genes, and one of these was a human cystitis isolate. These findings indicate that at least 43% of these clinical isolates contain a PAI IIUTI89-like island, and suggest that PAI similar to PAI IIUTI89 are evolutionarily conserved among lower urinary tract isolates.
Wild-type leuX , a PAI IIUTI89-associated tRNA gene, is important but not essential for fimS phase switching in vitro
In contrast to UPEC strain 536ΔPAI II536, which is unable to express type 1 pili, spontaneous excision of PAI IIUTI89 does not abolish expression of type 1 pili in vitro (G. Anderson and S. Hultgren, unpubl. data). Additional spontaneous PAI IIUTI89 deletion mutants (PAI IIUTI89-) were isolated by selection on blood agar plates as described in Experimental procedures. Chromosomal sequencing of the leuX locus in six non-haemolytic mutants lacking PAI IIUTI89 revealed that each had an identical single-base-pair deletion at nucleotide 80 in the 3′ end of the leuX gene, leuXΔG80 (leuX*), as would be predicted by the genome sequence (Fig. 1). All six clones were found to express type 1 pili as determined by mannose-sensitive haemagglutination (MSHA) titres, but at a markedly delayed rate compared with wt UTI89 (data not shown). The kinetics of type 1 pili expression in one of these UTI89ΔPAI IIUTI89 clones, designated TJH1 (leuX*, PAI IIUTI89-), is shown in Fig. 2A. Reversion of the 3′ end of the leuX locus in TJH1 to the wt sequence, TJH1 COMP (leuXwt, PAI IIUTI89-), restored wt piliation (Fig. 2A). Phase assays, which amplify the fimS-invertible region and utilize a restriction endonuclease digest of the PCR product to determine the orientation of the fim operon promoter, demonstrated that the delay in type 1 pili expression in TJH1 was a result of delayed OFF to ON phase switching (Fig. 2A). A directed mutation of leuX in UTI89 mimicking that found in TJH1, UTI89 leuXΔG80 (leuX*, PAI IIUTI89+), also resulted in delayed but not absent expression of type 1 pili, and was complemented by leuX expressed in trans (Fig. 2B). These data demonstrate that the leuXΔG80 mutation causes delayed phase variation in TJH1, while loss of PAI IIUTI89-associated genes has no effect.
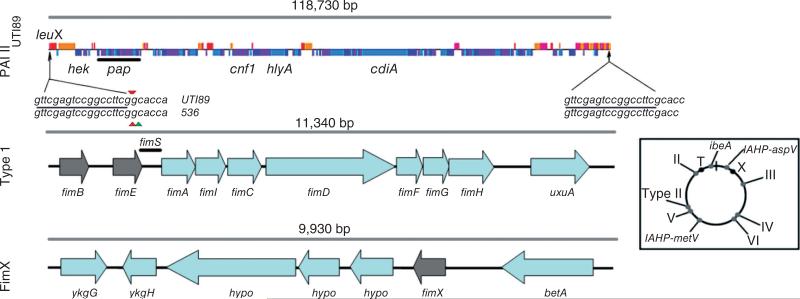
Fig. 1.
Overview of the genomic organization of PAI IIUTI89, the fim operon (‘Type 1’) and the fimX locus (‘FimX’). Open reading frames shown in the PAI IIUTI89 diagram are depicted either above or below the line, representing forward or reverse strand orientation of the genes, respectively. The direct repeat locations (arrows) are shown flanking the PAI IIUTI89 region, and the repeat sequences are shown below (underlined text). The small triangles flanking the direct repeat sequence text indicate the nucleotides deleted in the leuX gene of either UTI89 or 536 after spontaneous excision of each respective PAI II by recombination of the direct repeats. Recombinase genes in Type 1 and FimX loci are indicated in grey and ‘hypo’ indicates that the gene was annotated as a hypothetical protein. Inset: The relative chromosomal positions of these loci (‘II’, PAI IIUTI89; ‘T’, fim operon; ‘X’, fimX locus) and other UTI89 PAI are shown on the circular map at the lower right. ‘ibeA’ indicates PAI containing the invasion of brain endothelium virulence gene (Johnson et al., 2007). ‘Type II’ indicates type II secretion system island (Chen et al., 2006). PAI containing IcmF-associated homologous proteins are denoted IAHP (Das and Chaudhuri, 2003) followed by the tRNA insertion site. Roman numerals indicate similarity to PAI numbered by convention in pyelonephritis strain 536 (Dobrindt et al., 2002; Brzuszkiewicz et al., 2006).
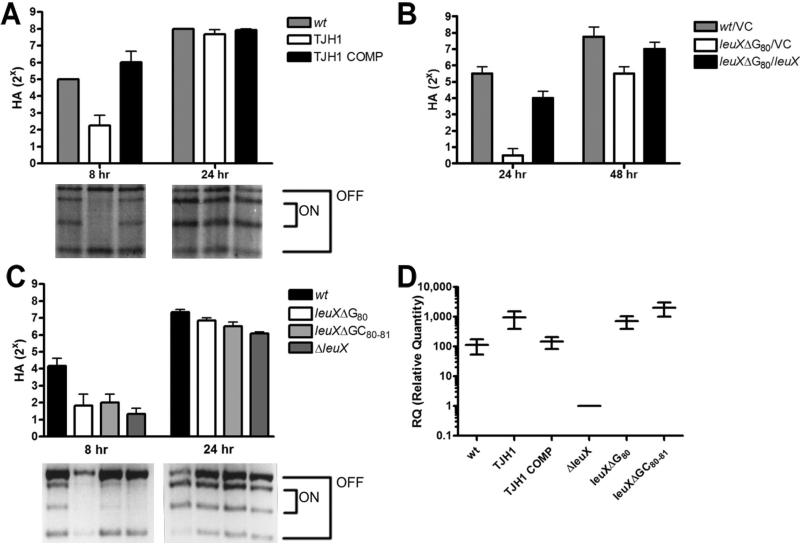
Fig. 2.
Mutation of leuX due to spontaneous excision of PAI IIUTI89 delays type 1 pili expression due to a complete loss of tRNA5Leu function.
A–C. Type 1 pili expression and fimS phase switch orientation in static broth cultures of wt UTI89 (wt) and derivatives grown at 37°C.
A. Mannose-sensitive haemagglutination (MSHA) titres (8 and 24 h) and representative phase assays of wt, TJH1 (leuX-, PAI II-) and TJH1 COMP (leuX+, PAI II-).
B. MSHA titres (24 and 48 h) of wt and UTI89 leuXΔG80, carrying either pBAD33 (VC) or pBAD-leuX (leuX) in trans.
C. MSHA titres (8 and 24 h) and representative phase assays of wt or isogenic leuX mutants. All trials in (A), (B) and (C) were performed in duplicate in three independent experiments. Error bars represent standard deviations.
D. Relative quantity (RQ) of leuX tRNA in wt or isogenic leuX mutants compared with UTI89 ΔleuX after 24 h of static growth in broth culture at 37°C. Horizontal bars represent mean values and vertical bars show RQ minimum and maximum range. qRT-PCR was performed on cDNA derived from each strain grown in three independent cultures in two independent experiments. qRT-PCR reactions were performed in duplicate.
To explain the differences in type 1 piliation phenotypes between spontaneous PAI II deletion mutants in UPEC strains 536 and UTI89, we hypothesized that a UTI89 strain with a two-nucleotide deletion in leuX mimicking that found in 536 ΔPAI II536, UTI89 leuXΔGC80-81, would be completely unable to express type 1 pili. Surprisingly, however, we found that such a mutant, as well as one with a complete leuX deletion, expressed type 1 pili with kinetics similar to UTI89 leuXΔG80 (Fig. 2C). Furthermore, quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis of these mutant strains indicated that the various single- or double-nucleotide mutants expressed higher levels of tRNA5Leu, relative to the leuX null strain, UTI89 ΔleuX (Fig. 2D). These data demonstrate that both leuXΔG80 and leuXΔGC80-81 strains continue to produce tRNA5Leu, but these mutant tRNA are apparently non-functional, resulting in delayed phase variation and type 1 piliation similar to that seen in a leuX null strain. Therefore, leuXΔG80 strains are functionally leuX null (leuX-). Furthermore, the UTI89ΔleuX, phenotype indicates that tRNA5Leu are not absolutely required for production of type 1 pili in UTI89.
FimX, a fimB homologue in UTI89, has lower UUG leucine codon usage than fimB
An alternative explanation for the ability of TJH1 (leuX-, PAI IIUTI89-) to express type 1 pili, despite leuX mutation, was the presence of additional, leuX-independent, fim recombinases. Therefore, we analysed the UTI89 genome and identified a FimB homologue, FimX (Fig. 1). FimX shares 49.1% amino acid identity to FimB and orthologues were recently described in two other extraintestinal E. coli strains (Bryan et al., 2006; Xie et al., 2006). We performed a PCR survey of a collection of 61 UPEC isolates, and found that the fimX gene was present in 83.9% of these strains including strain 536, but was not detected in the K12 strain MG1655. Interestingly, the usage of the UUG codon, which is translated by the leuX-encoded tRNA5Leu, differs significantly among the three fim recombinases in UTI89. UUG is used in 23%, 9% and 6% of the leucine codons in fimB, fimX and fimE respectively (Table S2). This codon usage in fimB is significantly higher than that found in either fimX or fimE (P < 0.05 and P < 0.01 respectively; binomial test). The leucine codon bias between fimX and fimB suggested that fimX expression may maintain the ability to phase switch in some leuX mutant strains despite poor to absent fimB translation.
Both FimB and FimX contribute to fimS phase switching in the absence of wt leuX
To test whether FimX was capable of mediating fimS phase switching, we constructed a double mutant in UTI89, lacking both fimB and fimE, that was isolated and maintained in vitro in the phase OFF position. This mutant, UTI89OFFΔfimBE, had delayed fimS phase switching from OFF to ON and type 1 piliation as measured by MSHA in vitro that resembled the kinetics of switching in TJH1 (data not shown). This similarity suggested that TJH1 (leuX-, PAI IIUTI89-) with a leuX mutation may functionally be fimB null due to the relatively high UUG codon usage of fimB. Therefore, we tested the relative contributions of FimB and FimX to fimS phase switching in the TJH1 background by constructing fimB and fimX single-and paired-deletion mutants. By 48 h of static in vitro growth, TJH1ΔfimX had a modest but statistically significant (P = 0.03, Student's t-test) decrease in MSHA compared with the parental strain, which was reflected in the amount of FimA (Fig. 3A). In contrast, TJH1ΔfimB had a marked loss of MSHA (P < 0.0001) and FimA. Interestingly, TJH1ΔfimBΔfimX continued to produce a small phase ON population and minimally detectable amount of FimA (Fig. 3A). The origin of the residual phase variation remains unknown, as FimE is thought to have only unidirectional, ON to OFF, phase activity (Klemm, 1986). Surprisingly, these data suggest that although FimX makes a significant contribution to in vitro phase switching in TJH1, FimB continues to have recombinase activity.
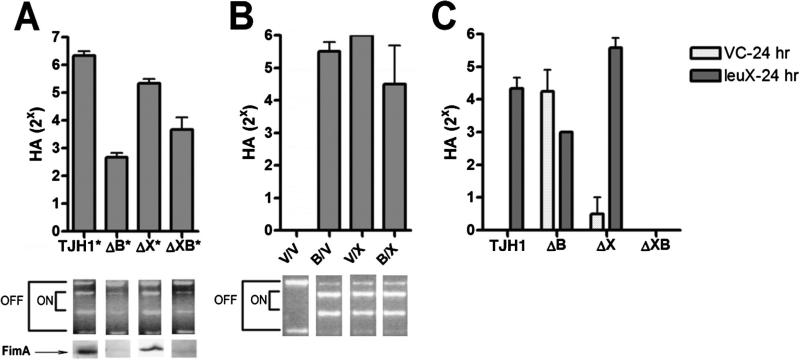
Fig. 3.
Roles of FimB and FimX recombinases in type 1 pili phase switching in vitro in TJH1.
A–C. Type 1 pili expression and fimS phase switch orientation in static broth cultures of TJH1 (leuX-, PAI II-) and derivatives grown at 37°C.
A. MSHA titres (48 h) of TJH1 and derivatives ΔB (fimB-), ΔX (fimX-) or ΔXB (fimX-fimB-). The asterisk (*) indicates each strain carried pTrc99a as a vector control. Representative phase assays and FimA immunoblot results are shown for each sample.
B. MSHA titres (24 h) of TJH1ΔXB (leuX-, PAI IIUT189-, fimX-, fimB-) carrying two of the following plasmids in trans: (i) control vectors (V), (ii) inducible fimB (B) and/or (iii) inducible fimX vector (X). Representative phase assays are shown for each sample.
C. MSHA titres (24 h) of TJH1 and derivatives with either pBAD33 (VC) or leuX in trans.
All trials in (A), (B) and (C) were performed in duplicate in three independent experiments. Error bars represent standard deviations.
To test the abilities of FimB and FimX to independently mediate phase switching activity regardless of recombinase expression levels in a leuXΔG80 background in vitro, we overexpressed either fimX or fimB in trans in TJH1ΔfimBΔfimX. Each recombinase was equally capable of restoring fimS phase switching and high MSHA in this strain after 24 h growth (Fig. 3B). This finding demonstrates that both FimB and FimX have recombinase activity when overexpressed in a leuXΔG80 mutant strain of UTI89, suggesting that increased levels of recombinase transcription are capable of overcoming the functional tRNA5Leu deficiency in this genetic background.
LeuX-encoded tRNA5Leu-dependent and -independent recombinases
To test whether FimB and FimX recombinase activities are adversely affected by leuX mutation when expressed at native levels, we complemented TJH1 fimX and fimB mutants with a plasmid expressing leuX from its native promoter. At 24 h of growth, FimB-mediated type 1 expression in TJH1ΔfimX was enhanced by leuX, provided in trans (Fig. 3C), whereas there was no positive effect on FimX-mediated phase variation in TJH1ΔfimB. These data are consistent with our hypothesis that FimB expression is more dependent on the leuX-encoded tRNA5Leu than is FimX, likely due to the differences in the UUG codon usage. However, because fimX expession in vitro appears to be inherently delayed, its contribution to overall in vitro phase variation in TJH1 still does not outweigh the role of FimB.
FimX mediates rapid phase switching of type 1 pili in vivo
FimX appears to play a lesser role than FimB in type 1 pili phase switching in vitro. We investigated the in vivo function of FimX by testing the ability of a phase OFF nonpiliated UTI89 ΔfimBΔfimE mutant (UTI89OFFΔfimBE) to infect the mouse bladder (Fig. 4A), anticipating that the mutant would have a dramatic loss of fitness. However, the phase OFF non-piliated culture was able to colonize and invade the bladder urothelium to the same level as the wt vector control, UTI89/VC (P = 0.4, Mann–Whitney test). Individual colonies (n = 10) and bacteria from an aggregate sweep of gentamicin-protected UTI89OFFΔfimBE were found to be exclusively in the phase ON orientation (data not shown). These phase ON colonies of UTI89OFFΔfimBE had high MSHA titres also, ≥ 1:28 (data not shown). These data indicate that a subpopulation of UTI89OFFΔfimBE underwent rapid fimS phase switching in vivo, produced type 1 pili and consequently invaded the urothelium. Consistent with these findings, UTI89OFFΔfimBE was able to form IBC morphologically similar to those formed by the wt parental strain at 6 h post infection (hpi). The development of high-density IBC by 6 hpi implies that the phase switch undergone by UTI89OFFΔfimBE occurred shortly following inoculation. However, UTI89OFFΔfimBE recovered from the extracellular fraction remained in the OFF orientation (n = 10 colonies). UTI89OFFΔfimBEX was found to be dramatically attenuated in its ability to infect the mouse bladder, confirming that FimX was mediating this in vivo phase switching (Fig. 4B). Accordingly, the expression of fimX in trans restored the virulence of UTI89OFFΔfimBEX.
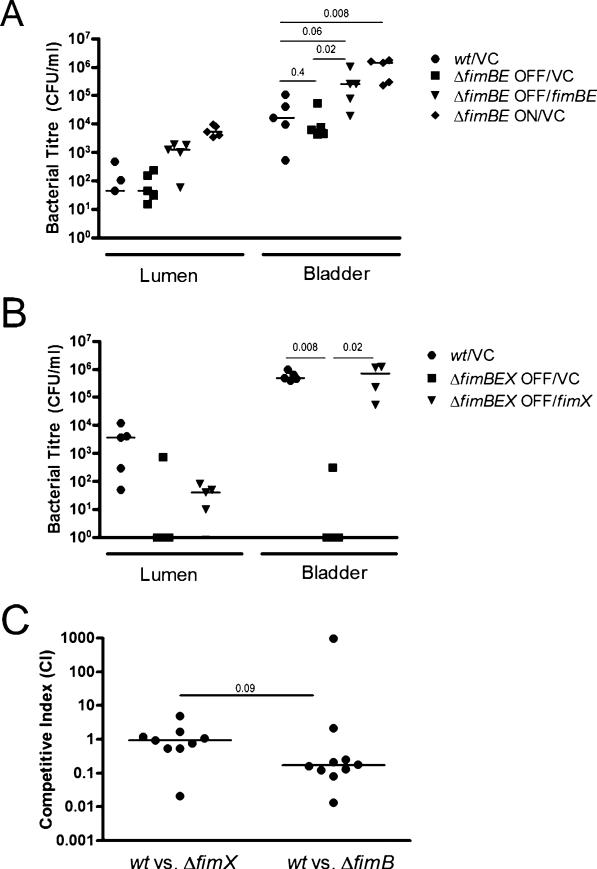
Fig. 4.
FimX-dependent phase variation of type 1 pili in vivo.
A and B. Ex vivo gentamicin protection assays were performed to distinguish extracellular and intracellular bacteria in infected C3H/HeN female mice at 6 hpi (as per Experimental procedures). Luminal contents (Lumen) are obtained from PBS washes of the removed bladder and represent relative luminal colonization. Bladder shows the cfu per ml of intracellular bacteria in the gentamicin-treated bladder homogenate.
A. Mice were transurethrally infected with 107 cfu of UTI89 with control plasmid (wt/VC), a UTI89fimB-fimE– strain phase OFF in vitro with either the control plasmid (ΔfimBE OFF/VC) or a plasmid carrying the fimB and fimE genes (ΔfimBE OFF/fimBE), or a ΔfimBE strain phase ON in vitro carrying the control plasmid (ΔfimBE ON/VC).
B. Mice were transurethrally infected with 107 cfu of wt/VC or UTI89fimB-fimE-fimX– strains phase OFF in vitro carrying either control (ΔfimBEX OFF/VC) or fimX (δfimBEX OFF/fimX) vectors in trans.
C. C3H/HeN female mice were transurethrally infected with an equal ratio of UTI89 attλ::PSSH10-1 (wt, SpecR) and either fimB or fimX null bacteria (KanR) for competitive infections. In the wt versus fimB null competitive experiment, each strain carried a plasmid with fimX under the ara promoter and grown in the presence of l-arabinose to induce phase OFF to ON changes, normally requiring native fimB in vitro. Colony-forming units (cfu) and competitive indices were scored for each strain as indicated in Experimental procedures.
A–C. Bars represent the median value for each group and P-values were calculated using the Mann–Whitney U-test.
FimX and FimB both contribute to persistence during cystitis
The ability of FimX in UTI89OFFΔfimBE to rapidly induce fimS phase switching from OFF to ON and carry out the UPEC pathogenic cycle in the mouse bladder led us to hypothesize that FimX was the dominant recombinase in vivo. This hypothesis was tested by assaying the ability of either FimX or FimB null mutants to compete with wt UTI89, in vivo. Approximately equal colony-forming units (cfu) of wt and UTI89ΔfimX or ΔfimB were co-inoculated into the bladders of C3H/HeN mice, and cfu were determined at 30 hpi. The medians of the competitive indices for each infection were 0.9 and 0.2 for ΔfimX and ΔfimB, respectively, suggesting only modest attenuation of these mutants (Fig. 4C). Furthermore, the competitive indices were not statistically different (P = 0.09, Mann–Whitney). Therefore, these findings suggest that FimB and FimX have overlapping and compensatory activities during acute cystitis.
Wild-type leuX, but not PAI IIUTI89 gene content, is necessary for full virulence of UTI89 in urothelial invasion and IBC formation
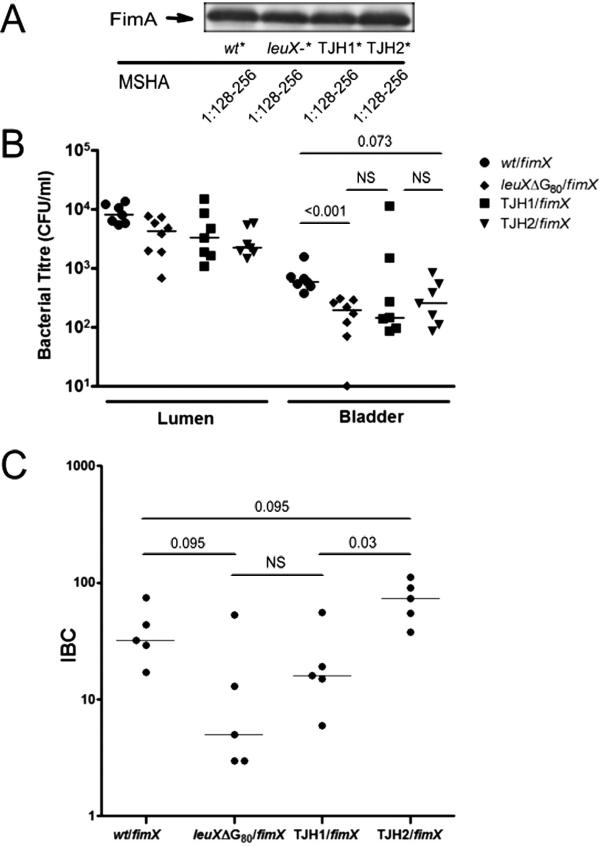
We then sought to test the individual roles of wt leuX and PAI IIUTI89 in urothelial invasion and IBC formation, key events in early acute infection. We employed the isogenic UTI89 mutants utilized in the in vitro studies, TJH1 (leuX-, PAI IIUTI89-) and UTI89 leuXΔG80 (leuX-, PAI IIUTI89+), as well as a directed PAI IIUTI89 deletion mutant that retained a wt leuX locus, called TJH2 (leuX+, PAI IIUTI89-). The growth rates of the mutants were comparable to wt in rich and minimal media (data not shown). To compensate for the differences in type 1 pili phase variation among these strains, overexpression of fimX in trans was used to direct fimS phase switching from OFF to ON, resulting in equivalent MSHA titres and FimA expression of the inoculating strains (Fig. 5A).
Fig. 5.
Wild-type leuX, not PAI IIUTI89 gene content, is necessary for full UPEC virulence during early acute events in experimental cystitis.
A–C. The individual roles of the leuX-encoded tRNA5Leu and PAI II content in early events in acute cystitis were tested in an experimental cystitis model comparing the isogenic mutants UTI89 leuXΔG80 (‘leuX–’, leuX-, PAI IIUTI89+), TJH1 (leuX-, PAI IIUTI89-) and TJH2 (leuX+, PAI IIUTI89-) with wt UTI89.
A. Immunoblot detecting FimA and MSHA in the same inocula of UTI89 (wt) and derivatives used in the experiments displayed in (B) and (C), which were performed on the same day. The asterisks (*) indicate strains carried pBAD-fimX, in trans, and were grown statically in 0.1% arabinose in LB/Cm20 for 18 h, thereby overexpressing fimX and driving type 1 piliation prior to inoculation into mice.
B. In vivo invasion of UTI89 (wt) and derivatives overexpressing fimX, measured by ex vivo gentamicin protection at 1 hpi. Luminal contents (Lumen) demonstrate the relative presence of each strain in the bladder lumen. Bladder shows the cfu present after gentamicin treatment of each bladder. These data are from a single experiment representative of three independent experiments, except for UTI89 leuXΔG80 (n = 2).
C. IBC formation by strains UTI89 and derivatives, all overexpressing fimX, at 6 hpi measured by β-galactosidase staining. These data are from a single experiment representative of two independent experiments, except for UTI89 leuXΔG80 (n = 1).
B and C: Bars represent the median value for each group and P-values were calculated using the Mann–Whitney U-test.
Urothelial invasion was determined by ex vivo gentamicin protection assays at 1 hpi (Fig. 5B). We found that UTI89 leuXΔG80 /pBAD-fimX (leuX-, PAI IIUTI89+) was consistently attenuated (P < 0.01, Mann–Whitney) in its ability to invade bladder urothelium and/or survive intracellularly. However, the gentamicin-protected bladder titres from mice infected with TJH1/pBAD-fimX (leuX-, PAI IIUTI89-) and TJH2/pBAD-fimX (leuX+, PAI IIUTI89-) were not significantly different from those infected with wt UTI89 in any experiment. In an independent control experiment, UTI89ONΔfimBE carrying either a control or fimX expression vector were shown to have equivalent invasion, suggesting that fimX had no detectable type 1 pili-independent effects in this assay (data not shown). IBC formation was assayed qualitatively by confocal microscopy and quantitatively by lacZ staining of whole mounted, fixed bladders at 6 hpi (Fig. 5C). All three mutant strains overexpressing FimX produced IBC morphologically similar to UTI89/pBAD-fimX (data not shown). However, UTI89 leuXΔG80 /pBAD-fimX (leuX-, PAI IIUTI89+) had decreased numbers of IBC compared with UTI89/pBAD-fimX (leuX+, PAI IIUTI89+; P < 0.10), and TJH1/pBAD-fimX (leuX-, PAI IIUTI89-) developed fewer IBC than TJH2/pBAD-fimX (leuX+, PAI IIUTI89-; P < 0.05). Taken together, these findings strongly indicate that wt leuX is necessary for efficient urothelial invasion and/or intracellular survival, as well as IBC formation. Surprisingly, the presence of the pathogenicity island genes, independent of leuX, did not enhance IBC formation as TJH2/pBAD-fimX (leuX+, PAI IIUTI89-) formed more IBC than UTI89/pBAD-fimX (leuX+, PAI IIUTI89+; P < 0.10), and TJH1/pBAD-fimX (leuX-, PAI IIUTI89-) formed more IBC than UTI89 leuXΔG80 /pBAD-fimX (leuX-, PAI IIUTI89+; P = 0.15).
Discussion
The acquisition of unique genes by horizontal transfer, often from other prokaryotic species, has promoted tremendous diversity within the E. coli species (Feil and Spratt, 2001), enabling many strains to evolve as successful pathotypes (Welch et al., 2002). Traditionally, if these genetic elements play a role in pathogenesis, they have been termed pathogenicity-associated islands (PAI) (Hacker et al., 1990). These islands, which are often inserted at tRNA loci, appear to become fixed within the genome over time by mutation or further recombination (Hacker and Kaper, 1999). In some cases, this fixation is mediated by disruption of the associated tRNA locus as a consequence of spontaneous excision of the island (Blum et al., 1994). Therefore, PAI gene content is not the only mechanism for island conservation in these strains. The higher prevalence of PAI IIUTI89 genes in lower urinary tract isolates when compared with commensal strains suggests that PAI IIUTI89-containing strains may be selected for in colonizing the urinary tract. However, as this correlation does not demonstrate that PAI gene content is operative in this selection, we investigated the consequences of losing this PAI in a murine model of acute cystitis. Surprisingly, we found that leuX mutation, and not loss of PAI IIUTI89 gene content, was the predominant mechanism for loss of virulence during the establishment of acute cystitis by UTI89 after spontaneous excision of this island, independent of any effect on type 1 piliation. These studies also led to the discovery that the leuX-independent fim recombinase, FimX, rapidly mediates fimS phase switching in vivo.
In this report, we show for the first time that leuX mutation, independent of its effect on type 1 pili-mediated invasion, reduces urothelial invasion and IBC formation in murine cystitis. Examination of the proteome of leuX mutants of 536 grown in minimal media demonstrated that leuX was necessary for normal expression of a small number of abundant proteins, including iron scavenging effectors (Piechaczek et al., 2000). Yet, many of the proteins that were adversely affected were not enriched in UUG codons, suggesting that the effect was at least in part mediated by undetected low abundance transcriptional regulators. LeuX is unusual among tRNA genes in that it is regulated by the stress factor RpoH (Dobrindt and Hacker, 2001). Furthermore, leuX expression was previously shown to be important for stationary-phase survival in bladder mucus (Dobrindt et al., 1998), an important extracellular phenotype of UPEC. In contrast, our studies suggest roles for tRNA5Leu during invasion, intracellular survival, and rapid intracellular proliferation. As a consequence of leuX mutation, the expression of invasins or adhesins that could potentiate the invasion process or effectors necessary for intracellular survival and IBC formation may be adversely affected. Therefore, leuX could be important for necessary bacterial stress responses initiated in both the extracellular and intracellular environments of the urinary tract.
Wild-type leuX may also be necessary for continued fimS phase switching within the developing IBC, as IBC formation is highly dependent on type 1 pili expression post invasion (Wright et al., 2007). FimS-gfp transcriptional reporter fusions responsive to Fim recombinase-mediated phase variation are predominantly inactive in early invasive bacteria (P. Seed and S. Hultgren, unpubl. data), suggesting that bacteria may initiate IBC formation starting in the phase OFF orientation. Therefore, the IBC defect due to leuXΔG80 may be at least partially due to impaired phase switching of these invasive bacteria from OFF to ON. However, leuX mutation could also be affecting the assembly of type 1 pili during rapid replication, as two proteins critical for pili formation, FimG and FimC, have unusually high UUG codon usage: 36% and 31%, respectively (Table S2) (Saulino et al., 2000).
We demonstrate that as a consequence of PAI IIUTI89 excision the recombined leuX gene acquires a single-nucleotide deletion, leuXΔG80. This mutation, independent of the presence of PAI IIUTI89, delayed but did not eliminate FimB-mediated type 1 pili phase switching from OFF to ON. Moreover, fimB overexpression restored OFF to ON phase switching in the UTI89ΔPAI IIUTI89 (leuXΔG80) mutant strain, whereas fimB overexpression was unable to restore type 1 piliation in 536ΔPAI II536 (Ritter et al., 1997). Interestingly, directed mutation of the leuX gene in UTI89 mimicking that observed with PAI II excision in 536, leuXΔGC80-81, resulted in a type 1 piliation defect similar to that seen in both leuXΔG80 and leuX null strains, despite the fact that the various leuXΔG80 and leuXΔGC80-81 mutant strains all had higher than wt levels of tRNA5Leu expression. This suggests that tRNA5Leu function is completely abrogated in the leuXΔG80 and leuXΔGC80-81 mutant strains. Thus, in contrast to strain 536, UTI89 is capable of expressing type 1 pili despite the complete absence of functional tRNA5Leu. While we have observed that wt 536 produces significantly less type 1 pili than UTI89 under optimal conditions (T. Hannan and S. Hultgren, unpubl. data), the specific differences in the requirements of each strain for tRNA5Leu to express type 1 pili may be accounted for by suppressor mutation(s) found in UTI89 but not 536. Alternatively, additional strain background factors, such as differences in the expression of the leuZ-encoded tRNA6Leu that is capable of ‘wobbling’ onto and translating the UUG codon, may also explain these differences, which merit further study.
Our studies of leuX also revealed the role of an additional Fim recombinase in uropathogenesis. Mounting evidence suggests that UPEC have acquired a battery of recombinases that have type 1 pili phase switching activity (Bryan et al., 2006; Xie et al., 2006). We found that, in contrast to its orthologue in the UPEC strain CFT073 (Bryan et al., 2006), UTI89 FimX can be the sole mediator of in vivo fimS phase switching. Furthermore, FimB and FimX are able to significantly compensate for each other during acute cystitis. However, both FimB- and FimX-mediated type 1 pili expression are sensitive to different environmental cues as was evident by comparison of in vitro and in vivo phase variation. In vitro, FimB and FimX produced rapid and slow switching, respectively. In contrast, an environmental signal in the lower urinary tract markedly affects FimX-mediated type 1 pili expression through rapid phase switching in the absence of FimB and FimE. Thus, FimX and FimB may provide important environmentally sensitive regulatory controls for type 1 pili regulation and thereby fine tune the control of type 1 pili expression in non-commensal niches.
Independent of its relationship to leuX, PAI IIUTI89 genes likely serve some function among uropathogenic E. coli strains because they are more frequently present in lower urinary tract isolates than in commensal isolates. Surprisingly, we were unable to demonstrate that PAI IIUTI89 positively affects the virulence of UPEC in urothelial invasion and IBC formation. In fact, we have shown that deletion of the entire island while leaving leuX intact may actually be beneficial to UTI89 in early intracellular stages of cystitis. This could be due to the loss of the PAI IIUTI89-encoded transcriptional regulator PapB, which negatively regulates type 1 pili expression (Xia et al., 2000). We cannot exclude that this genomic island enhances UPEC virulence in later stages of uropathogenesis such as during extracellular colonization in the face of a host immune response. For example, the ρ-GTPase-activating toxin, CNF1, is found on PAI IIUTI89 and has been demonstrated to enhance the virulence of UPEC in the urinary tract, possibly by preventing the phagocytosis of UPEC by neutrophils encountered in the extracellular environment (Rippere-Lampe et al., 2001; Davis et al., 2005). PAI IIUTI89 gene content may also be selected for in the ascension from the gut to the urinary bladder, as UPEC commonly colonize the periurethral and vaginal epithelium prior to introduction into and ascension of the urethra.
In summary, we have shown a specific role for leuX tRNA5Leu-dependent factors in the earliest stages of cystitis, namely adherence, invasion and IBC formation. The effects of leuXΔG80 mutation on invasion and IBC formation further support the hypothesis that minor codon usage could serve as a mode of regulation of virulence factor expression in vivo (Saier, 1995; Piechaczek et al., 2000). Through the elucidation of in vivo expressed tRNA5Leu-dependent factors, we anticipate the discovery of essential UPEC virulence determinants for cystitis that would be ideal targets for therapeutic inhibition.
Experimental procedures
Bacterial strains and cultivation
Strains of Escherichia coli used in this study are listed in Table S3. Bacteria were routinely cultured in Luria–Bertani (LB) broth containing, where appropriate, kanamycin 50 μg ml-1, ampicillin 100 μg ml-1 and chloramphenicol 20 μg ml-1.
Deletion strain construction
Deletion mutations were made using the red recombinase method, as previously described, using pKD4 or pKD13 as a template and the primers as listed in Table S4 (Datsenko and Wanner, 2000; Murphy and Campellone, 2003). PCR was performed with flanking primers to confirm the appropriate deletions. Antibiotic insertions were removed by transforming the mutant strains with pCP20 (Cherepanov and Wackernagel, 1995) expressing the FLP recombinase. The resultant strains subsequently had no additional antibiotic resistance compared with the parental wt strain.
Spontaneous PAI IIUTI89 deletion mutant derivation
Spontaneous PAI IIUTI89 deletion mutants were created by aerobically growing a liquid culture of UTI89 in LB broth to stationary phase at room temperature (22°C), growth conditions under which the rate of spontaneous deletion has been shown to be maximal (Middendorf et al., 2004). The bacteria were then diluted and plated onto trypticase soy agar plates containing 5% sheep blood for screening. Non-haemolytic clones were isolated and confirmed by PCR and sequencing of the scar region, using a three-primer set: PAI IIUTI89 5′ FLANK F, 5′ INT R and 3′ FLANK R.
Phase and haemagglutination assays
Phase assays were performed by a modified method of Smith et al. (Horcajada et al., 2005) using the primers PHASE 1, PHASE 2 and fim#14. The inclusion of primer fim#14 allowed PCR amplification of the fim promoter-invertible region in both wt and mutants with fimB–fimE deletions. Amplicons were digested with BstUI which singly and asymmetrically cuts within the amplicon, and the products run on a 2% TBE agarose gel. Haemagglutination assays for mannose-sensitive agglutination of guinea pig red blood cells were performed as previously described using 3% red blood cells (OD640 = ~1.9–2; Hultgren et al., 1986).
qRT-PCR
RNA was collected from 5 ml of 24 h static growth bacterial cultures in LB using disruptor beads and Trizol (Sigma-Aldrich) followed by DNase I treatment (Fisher RQ1) and RNA cleanup (Zymo Research). One microgram of RNA was reverse transcribed using random hexamers and iScript (Bio-Rad). cDNA was diluted fourfold in DEPC-treated water, and 1 μl of each dilution was used per 25 μl of qPCR reaction. Amplicons were detected using the inclusion of 1 μl per reaction (rxn-1) of a 1:3000 dilution of SYBR Green (Invitrogen) in a mixture of 1× APEX Taq polymerase buffer (Genesse Scientific), 1 U of APEX Taq polymerase, 2.5 mM MgCl2 (final) and 0.2 μM primers (IDT). Primers 16s RT L/R and leuX RT L/R were included in the 16s and leuX reaction mixtures respectively (Table S4). cDNA for 16s RNA analysis (calibrator) was performed by 1 cycle of 95°C (5 min) then 40 cycles of 95°C (10 s), 55°C (30 s), 72°C (15 s), 80°C (5 s) and plate reading. leuX cDNA analysis was performed by 1 cycle of 95°C (5 min) then 40 cycles of 95°C (10 s), 57°C (30 s), 72°C (15 s), 80°C (5 s) and plate reading. Melting curve analysis and gel electrophoresis were used to ensure amplicon homogeneity. Relative quantity (RQ) was derived by the ΔΔCt method in comparison with UTI89 ΔleuX (ABI User Bulletin #2). Reactions lacking reverse transcriptase (–RT) were performed to ensure the adequacy of DNase treatment.
PCR analysis
Bacterial chromosomal DNA from a library of clinical isolates (Garofalo et al., 2007) was isolated using the Wizard genomic purification kit (Promega). Multiplex PCR reactions to determine the presence of cnf1, hek and hlyA (primers shown in Table S4) in this library were performed as previously described (Johnson and Stell, 2000). The presence of fimX was assessed by PCR with prepared genomic DNA from clinical UPEC isolates using the primers fimX PCR PROBE #1 and #2 (Table S4).
Mouse infections
C3H/HeN mice were obtained from Harlan (Indianapolis, IN). Bacterial strains were inoculated into LB broth from freezer stock, grown aerobically to mid- to late-log phase and then subcultured 1:100 into fresh media and grown statically at 37°C for 18 h. These cultures were spun at room temperature for 7 min at 5500 r.p.m., re-suspended in PBS and then diluted to approximately 2–4 × 108 cfu ml-1. Fifty microlitres of this suspension (~1–2 × 107 cfu) was inoculated into the bladders of 6- to 8-week-old female mice by transurethral catheterization. All animal studies were approved and performed in accordance with Committee for Animal Studies at Washington University School of Medicine.
Ex vivo gentamicin protection assay to assess for invasion
The presence of intracellular bacteria in vivo in the murine bladder was assayed by a modified gentamicin-protection assay as previously described (Justice et al., 2006).
Competitive infections
The reference strain used was UTI89 attλ::PSSH10-1 (Wright et al., 2007), which has a spectinomycin resistance cassette. In the experiments involving the UTI89 ΔfimB, the reference and test strains each carried pBAD-fimX and were grown in the presence of both chloramphenicol 20 μg ml-1 and 0.1% l-arabinose to promote the in vitro expression of type 1 pili. This modification was performed to compensate for the poor in vitro phase variation and type 1 pili expression of fimB null strains. Prior studies have shown that the arabinose promoter has no appreciable activity in the lower urinary tract in the absence of induction (Justice et al., 2006). Thus, after inoculation into the murine bladder, FimX expressed from the ara promoter was expected to be diluted out due to the absence of arabinose in the bladder thus restoring the wt and ΔfimB phenotypes. Mice were inoculated transurethrally with 50 μl of a reference/test strain mixture, amounting to ~1–2 × 107 cfu per strain. Bladders were mechanically homogenized in 0.01% Triton X-100 in PBS. Dilutions of homogenates were plated on LB agar, LB kanamycin 50 μg ml-1 or LB spectinomycin 50 μg ml-1. Competitive indices were calculated as (cfu_OUTtest/cfu_OUTref)/(cfu_INtest/cfu_INref).
Visualization of whole mount bladders for IBC formation
Two approaches were used to quantify and/or examine IBC formation. In the first method, infected bladders were stretched and fixed with 3% paraformaldehyde for 45 min at room temperature in the dark. The bladders were permeabilized with PBS containing Triton X-100 0.01% for 15 min. The bladders were rinsed with sterile PBS five times. Bladders were incubated for 5 min at room temperature in a 1:1000 dilution of Topro-3 (Molecular Probes) followed by five PBS washes. The bladders were mounted on glass slides under Prolong antifade/mount (Molecular Probes) and imaged immediately on a Zeiss LSM510 laser scanning confocal microcope. Alternatively, IBC numbers were enumerated by fixation and β-galactosidase staining of whole mount bladders as previously described (Justice et al., 2006).
Statistical analysis
Statistical analyses using the non-parametric, Mann–Whitney U-test, Fisher's exact test or Student's t-test were performed using GraphPad Prism (GraphPad Software) where indicated in the main text. The binomial test was used to compare codon usage between genes using ad hoc perl scripts. Statistical significance was defined by attaining P-values ≤ 0.05.
Supplementary Material
Acknowledgements
We thank Drs Soman Abraham, Michael Caparon, Karen Dodson, Kimberly Kline, Jeffrey McKinney, Joseph St. Geme and Joseph Vogel for their insightful editorial comments and suggestions, and Gregory Anderson for providing the initial PAI IIUTI89 deletion mutant strain. This work was supported by NIH Grants K12HD00850 and K08DK07444301 (P.C.S.), F32CA108328 (I.U.M.), DK51406, AI49950, AI48689, and Office of Research on Women's Health Specialized Center of Research Grant DK64540 (S.J.H.).
Footnotes
Supplementary material
This material is available as part of the online article from: http://www.blackwell-synergy.com/doi/abs/10.1111/j.1365-2958.2007.06025.x (This link will take you to the article abstract).
Please note: Blackwell Publishing is not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Abraham JM, Freitag CS, Clements JR, Eisenstein BI. An invertible element of DNA controls phase variation of type 1 fimbriae of Escherichia coli. Proc Natl Acad Sci USA. 1985;82:5724–5727. doi: 10.1073/pnas.82.17.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson GG, Palermo JJ, Schilling JD, Roth R, Heuser J, Hultgren SJ. Intracellular bacterial biofilm-like pods in urinary tract infections. Science. 2003;301:105–107. doi: 10.1126/science.1084550. [DOI] [PubMed] [Google Scholar]
- Aoki SK, Pamma R, Hernday AD, Bickham JE, Braaten BA, Low DA. Contact-dependent inhibition of growth in Escherichia coli. Science. 2005;309:1245–1248. doi: 10.1126/science.1115109. [DOI] [PubMed] [Google Scholar]
- Bidet P, Bonacorsi S, Clermont O, De Montille C, Brahimi N, Bingen E. Multiple insertional events, restricted by the genetic background, have led to acquisition of pathogenicity island IIJ96-like domains among Escherichia coli strains of different clinical origins. Infect Immun. 2005;73:4081–4087. doi: 10.1128/IAI.73.7.4081-4087.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum G, Ott M, Lischewski A, Ritter A, Imrich H, Tschape H, Hacker J. Excision of large DNA regions termed pathogenicity islands from tRNA-specific loci in the chromosome of an Escherichia coli wild-type pathogen. Infect Immun. 1994;62:606–614. doi: 10.1128/iai.62.2.606-614.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum G, Falbo V, Caprioli A, Hacker J. Gene clusters encoding the cytotoxic necrotizing factor type 1, Prs-fimbriae and alpha-hemolysin form the pathogenicity island II of the uropathogenic Escherichia coli strain J96. FEMS Microbiol Lett. 1995;126:189–195. doi: 10.1111/j.1574-6968.1995.tb07415.x. [DOI] [PubMed] [Google Scholar]
- Bryan A, Roesch P, Davis L, Moritz R, Pellett S, Welch RA. Regulation of type 1 fimbriae by unlinked FimB- and FimE-like recombinases in uropathogenic Escherichia coli strain CFT073. Infect Immun. 2006;74:1072–1083. doi: 10.1128/IAI.74.2.1072-1083.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzuszkiewicz E, Bruggemann H, Liesegang H, Emmerth M, Olschlager T, Nagy G, et al. How to become a uropathogen: comparative genomic analysis of extraintestinal pathogenic Escherichia coli strains. Proc Natl Acad Sci USA. 2006;103:12879–12884. doi: 10.1073/pnas.0603038103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SL, Hung CS, Xu J, Reigstad CS, Magrini V, Sabo A, et al. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: a comparative genomics approach. Proc Natl Acad Sci USA. 2006;103:5977–5982. doi: 10.1073/pnas.0600938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- Connell I, Agace W, Klemm P, Schembri M, Marild S, Svanborg C. Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc Natl Acad Sci USA. 1996;93:9827–9832. doi: 10.1073/pnas.93.18.9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Chaudhuri K. Identification of a unique IAHP (IcmF associated homologous proteins) cluster in Vibrio cholerae and other proteobacteria through in silico analysis. In Silico Biol. 2003;3:287–300. [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JM, Rasmussen SB, O'Brien AD. Cytotoxic necrotizing factor type 1 production by uropathogenic Escherichia coli modulates polymorphonuclear leukocyte function. Infect Immun. 2005;73:5301–5310. doi: 10.1128/IAI.73.9.5301-5310.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrindt U, Hacker J. Regulation of tRNA Leu5-encoding gene leuX that is associated with a pathogenicity island in the uropathogenic Escherichia coli strain 536. Mol Genet Genomics. 2001;265:895–904. doi: 10.1007/s004380100486. [DOI] [PubMed] [Google Scholar]
- Dobrindt U, Cohen PS, Utley M, Muhldorfer I, Hacker J. The leuX-encoded tRNA Leu5 but not the pathogenicity islands I and II influence the survival of the uropathogenic Escherichia coli strain 536 in CD-1 mouse bladder mucus in the stationary phase. FEMS Microbiol Lett. 1998;162:135–141. doi: 10.1111/j.1574-6968.1998.tb12990.x. [DOI] [PubMed] [Google Scholar]
- Dobrindt U, Blum-Oehler G, Nagy G, Schneider G, Johann A, Gottschalk G, Hacker J. Genetic structure and distribution of four pathogenicity islands (PAI I536 to PAI IV536) of uropathogenic Escherichia coli strain 536. Infect Immun. 2002;70:6365–6372. doi: 10.1128/IAI.70.11.6365-6372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrindt U, Hochhut B, Hentschel U, Hacker J. Genomic islands in pathogenic and environmental microorganisms. Nat Rev Microbiol. 2004;2:414–424. doi: 10.1038/nrmicro884. [DOI] [PubMed] [Google Scholar]
- Doye A, Mettouchi A, Bossis G, Clement R, Buisson-Touati C, Flatau G, et al. CNF1 exploits the ubiquitin-proteasome machinery to restrict Rho GTPase activation for bacterial host cell invasion. Cell. 2002;111:553–564. doi: 10.1016/s0092-8674(02)01132-7. [DOI] [PubMed] [Google Scholar]
- Eto DS, Jones TA, Sundsbak JL, Mulvey MA. Integrin-mediated host cell invasion by type 1-piliated uropathogenic Escherichia coli. PloS Pathog. 2007;3:949–961. doi: 10.1371/journal.ppat.0030100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil EJ, Spratt BG. Recombination and the population structures of bacterial pathogens. Annu Rev Microbiol. 2001;55:561–590. doi: 10.1146/annurev.micro.55.1.561. [DOI] [PubMed] [Google Scholar]
- Foxman B. Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Am J Med. 2002;113(Suppl. 1A):5S–13S. doi: 10.1016/s0002-9343(02)01054-9. [DOI] [PubMed] [Google Scholar]
- Gal-Mor O, Finlay BB. Pathogenicity islands: a molecular toolbox for bacterial virulence. Cell Microbiol. 2006;8:1707–1719. doi: 10.1111/j.1462-5822.2006.00794.x. [DOI] [PubMed] [Google Scholar]
- Garofalo CK, Hooton TM, Martin SM, Stamm WE, Palermo JJ, Gordon JI, Hultgren SJ. Escherichia coli from urine of female patients with urinary tract infections is competent for intracellular bacterial community formation. Infect Immun. 2007;75:52–60. doi: 10.1128/IAI.01123-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman EA, Ochman H. How to become a pathogen. Trends Microbiol. 1994;2:289–294. doi: 10.1016/0966-842x(94)90006-x. [DOI] [PubMed] [Google Scholar]
- Groisman EA, Ochman H. Pathogenicity islands: bacterial evolution in quantum leaps. Cell. 1996;87:791–794. doi: 10.1016/s0092-8674(00)81985-6. [DOI] [PubMed] [Google Scholar]
- Hacker J, Kaper JB. The concept of pathogenicity islands. In: Kaper JB, Hacker J, editors. Pathogenicity Islands and Other Mobile Virulence Elements. American Society for Microbiology Press; Washington, DC: 1999. pp. 1–12. [Google Scholar]
- Hacker J, Bender L, Ott M, Wingender J, Lund B, Marre R, Goebel W. Deletions of chromosomal regions coding for fimbriae and hemolysins occur in vitro and in vivo in various extraintestinal Escherichia coli isolates. Microb Pathog. 1990;8:213–225. doi: 10.1016/0882-4010(90)90048-u. [DOI] [PubMed] [Google Scholar]
- Holden NJ, Totsika M, Mahler E, Roe AJ, Catherwood K, Lindner K, et al. Demonstration of regulatory cross-talk between P fimbriae and type 1 fimbriae in uropathogenic Escherichia coli. Microbiology. 2006;152:1143–1153. doi: 10.1099/mic.0.28677-0. [DOI] [PubMed] [Google Scholar]
- Horcajada JP, Soto S, Gajewski A, Smithson A, Jimenez de Anta MT, Mensa J, et al. Quinolone-resistant uropathogenic Escherichia coli strains from phylogenetic group B2 have fewer virulence factors than their susceptible counterparts. J Clin Micro-biol. 2005;43:2962–2964. doi: 10.1128/JCM.43.6.2962-2964.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultgren SJ, Schwan WR, Schaeffer AJ, Duncan JL. Regulation of production of type 1 pili among urinary tract isolates of Escherichia coli. Infect Immun. 1986;54:613–620. doi: 10.1128/iai.54.3.613-620.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JR, Stell AL. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis. 2000;181:261–272. doi: 10.1086/315217. [DOI] [PubMed] [Google Scholar]
- Johnson TJ, Kariyawasam S, Wannemuehler Y, Mangiamele P, Johnson SJ, Doetkott C, et al. The genome sequence of avian pathogenic Escherichia coli strain O1:K1:H7 shares strong similarities with human extraintestinal pathogenic E. coli genomes. J Bacteriol. 2007;189:3228–3236. doi: 10.1128/JB.01726-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice SS, Hung C, Theriot JA, Fletcher DA, Anderson GG, Footer MJ, Hultgren SJ. Differentiation and developmental pathways of uropathogenic Escherichia coli in urinary tract pathogenesis. Proc Natl Acad Sci USA. 2004;101:1333–1338. doi: 10.1073/pnas.0308125100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice SS, Lauer SR, Hultgren SJ, Hunstad DA. Maturation of intracellular Escherichia coli communities requires SurA. Infect Immun. 2006;74:4793–4800. doi: 10.1128/IAI.00355-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm P. Two regulatory fim genes, fimB and fimE, control the phase variation of type 1 fimbriae in Escherichia coli. EMBO J. 1986;5:1389–1393. doi: 10.1002/j.1460-2075.1986.tb04372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd AL, Rasko DA, Mobley HLT. Defining genomic islands and uropathogen-specific genes in uropathogenic Escherichia coli. J Bacteriol. 2007;189:3532–3546. doi: 10.1128/JB.01744-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middendorf B, Hochhut B, Leipold K, Dobrindt U, Blum-Oehler G, Hacker J. Instability of pathogenicity islands in uropathogenic Escherichia coli 536. J Bacteriol. 2004;186:3086–3096. doi: 10.1128/JB.186.10.3086-3096.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvey MA, Lopez-Boado YS, Wilson CL, Roth R, Parks WC, Heuser J, Hultgren SJ. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science. 1998;282:1494–1497. doi: 10.1126/science.282.5393.1494. [DOI] [PubMed] [Google Scholar]
- Mulvey MA, Schilling JD, Martinez JJ, Hultgren SJ. Bad bugs and beleaguered bladders: interplay between uropathogenic Escherichia coli and innate host defenses. Proc Natl Acad Sci USA. 2000;97:8829–8835. doi: 10.1073/pnas.97.16.8829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvey MA, Schilling JD, Hultgren SJ. Establishment of a persistent Escherichia coli reservoir during the acute phase of a bladder infection. Infect Immun. 2001;69:4572–4579. doi: 10.1128/IAI.69.7.4572-4579.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KC, Campellone KG. Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC Mol Biol. 2003;4:11. doi: 10.1186/1471-2199-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mysorekar IU, Hultgren SJ. Mechanisms of uropathogenic Escherichia coli persistence and eradication from the urinary tract. Proc Natl Acad Sci USA. 2006;103:14170–14175. doi: 10.1073/pnas.0602136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mysorekar IU, Mulvey MA, Hultgren SJ, Gordon JI. Molecular regulation of urothelial renewal and host defenses during infection with uropathogenic Escherichia coli. J Biol Chem. 2002;277:7412–7419. doi: 10.1074/jbc.M110560200. [DOI] [PubMed] [Google Scholar]
- Nagy G, Altenhoefer A, Knapp O, Maier E, Dobrindt U, Blum-Oehler G, et al. Both alpha-haemolysin determinants contribute to full virulence of uropathogenic Escherichia coli strain 536. Microbes Infect. 2006;8:2006–2012. doi: 10.1016/j.micinf.2006.02.029. [DOI] [PubMed] [Google Scholar]
- Ochman H, Selander RK. Standard reference strains of Escherichia coli from natural populations. J Bacteriol. 1984;157:690–693. doi: 10.1128/jb.157.2.690-693.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piechaczek K, Dobrindt U, Schierhorn A, Fischer GS, Hecker M, Hacker J. Influence of pathogenicity islands and the minor leuX-encoded tRNA Leu5 on the proteome pattern of the uropathogenic Escherichia coli strain 536. Int J Med Microbiol. 2000;290:75–84. doi: 10.1016/S1438-4221(00)80110-1. [DOI] [PubMed] [Google Scholar]
- Rippere-Lampe KE, O'Brien AD, Conran R, Lockman HA. Mutation of the gene encoding cytotoxic necrotizing factor type 1 (cnf1) attenuates the virulence of uropathogenic Escherichia coli. Infect Immun. 2001;69:3954–3964. doi: 10.1128/IAI.69.6.3954-3964.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter A, Blum G, Emody L, Kerenyi M, Bock A, Neuhierl B, et al. tRNA genes and pathogenicity islands: influence on virulence and metabolic properties of uropathogenic Escherichia coli. Mol Microbiol. 1995;17:109–121. doi: 10.1111/j.1365-2958.1995.mmi_17010109.x. [DOI] [PubMed] [Google Scholar]
- Ritter A, Gally DL, Olsen PB, Dobrindt U, Friedrich A, Klemm P, Hacker J. The Pai-associated leuX specific tRNA Leu5 affects type 1 fimbriation in pathogenic Escherichia coli by control of FimB recombinase expression. Mol Microbiol. 1997;25:871–882. doi: 10.1111/j.1365-2958.1997.mmi517.x. [DOI] [PubMed] [Google Scholar]
- Roberts JA, Marklund BI, Ilver D, Haslam D, Kaack MB, Baskin G, et al. The Gal(alpha 1-4)Galspecific tip adhesin of Escherichia coli P-fimbriae is needed for pyelonephritis to occur in the normal urinary tract. Proc Natl Acad Sci USA. 1994;91:11889–11893. doi: 10.1073/pnas.91.25.11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH. Differential codon usage: a safeguard against inappropriate expression of specialized genes? FEBS Lett. 1995;362:1–4. doi: 10.1016/0014-5793(95)00185-c. [DOI] [PubMed] [Google Scholar]
- Saulino ET, Bullitt E, Hultgren SJ. Snapshots of usher-mediated protein secretion and ordered pilus assembly. Proc Natl Acad Sci USA. 2000;97:9240–9245. doi: 10.1073/pnas.160070497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto SM, Jimenez de Anta MT, Vila J. Quinolones induce partial or total loss of pathogenicity islands in uropathogenic Escherichia coli by SOS-dependent or -independent pathways, respectively. Antimicrob Agents Chemother. 2006;50:649–653. doi: 10.1128/AAC.50.2.649-653.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan U, Foxman B, Marrs CF. Identification of a gene encoding heat-resistant agglutinin in Escherichia coli as a putative virulence factor in urinary tract infection. J Clin Microbiol. 2003;41:285–289. doi: 10.1128/JCM.41.1.285-289.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch RA, Burland V, Plunkett G, 3rd, Redford P, Roesch P, Rasko D, et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci USA. 2002;99:17020–17024. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winberg J, Mollby R, Bergstrom J, Karlsson KA, Leonardsson I, Milh MA, et al. The PapG-adhesin at the tip of P-fimbriae provides Escherichia coli with a competitive edge in experimental bladder infections of cynomolgus monkeys. J Exp Med. 1995;182:1695–1702. doi: 10.1084/jem.182.6.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright KJ, Seed PC, Hultgren SJ. Development of intracellular bacterial communities of uropathogenic Escherichia coli depends on type 1 pili. Cell Microbiol. 2007;9:2230–2241. doi: 10.1111/j.1462-5822.2007.00952.x. [DOI] [PubMed] [Google Scholar]
- Xia Y, Gally D, Forsman-Semb K, Uhlin BE. Regulatory cross-talk between adhesin operons in Escherichia coli: inhibition of type 1 fimbriae expression by the PapB protein. EMBO J. 2000;19:1450–1457. doi: 10.1093/emboj/19.7.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Yao Y, Kolisnychenko V, Teng CH, Kim KS. HbiF regulates type 1 Fimbriation independently of FimB and FimE. Infect Immun. 2006;74:4039–4047. doi: 10.1128/IAI.02058-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Mo W-J, Sebbel P, Min G, Neubert TA, Glockshuber R, et al. Uroplakin Ia is the urothelial receptor for uropathogenic Escherichia coli: evidence from in vitro FimH binding. J Cell Sci. 2001;114:4095–4103. doi: 10.1242/jcs.114.22.4095. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.