Abstract
The genetic diversity of Vibrio vulnificus isolates from clinical and environmental sources originating from the Baltic Sea region was evaluated by multilocus sequence typing (MLST), and possible relationships between MLST clusters, potential genotypic and phenotypic traits associated with pathogenicity, and source of isolation were investigated. The studied traits included genotyping of polymorphic loci (16S rRNA, vcg, and pilF), presence/absence of potential virulence genes, including nanA, nab, and genes of pathogenicity regions, metabolic features, hemolytic activity, resistance to human serum, and cytotoxicity to human intestinal cells. MLST generated 35 (27 new) sequence types and divided the 53 isolates (including four reference strains) into two main clusters, with cluster I containing biotype 1 and 2 isolates of mainly environmental origin and cluster II containing biotype 1 isolates of mainly clinical origin. Cluster II isolates were further subdivided into two branches. Branch IIB included isolates from recent cases of wound infections that were acquired at the German Baltic Sea coastline between 2010 and 2011 and isolates from seawater samples of the same regions isolated between 1994 and 2010. Comparing the MLST data with the results of genotyping and phenotyping showed that strains of MLST cluster II possess a number of additional pathogenicity-associated traits compared to cluster I strains. Rapid microbiological methods such as matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry combined with typing of selected virulence-associated traits (e.g., serum resistance, mannitol fermentation, nanA, and pathogenicity region XII) could be used for risk assessment purposes regarding V. vulnificus strains isolated from the Baltic Sea region.
INTRODUCTION
Vibrio vulnificus is a potent bacterial pathogen present in coastal waters worldwide and is found preferentially in waters with moderate salinity. It can cause serious wound infections with lethal outcome and is also responsible for cases of death caused by consumption of contaminated seafood. In the United States, particularly oysters contaminated with V. vulnificus have been reported to be responsible for deadly infections (1–3). The severity of disease is strongly influenced by the health condition of exposed individuals. Immunocompromised people and persons with underlying diseases resulting in elevated serum iron levels are especially at high risk. In the case of primary septicemia, mortality rates greater than 50% have been reported, and for wound infections, approximately rates of 25% have been reported (4). Environmental factors, such as warm water and moderate salinity, are known to favor the multiplication of the pathogen. Therefore, the effect of global warming on seawater temperatures has aroused concerns that infections caused by V. vulnificus will increase in numbers (5–8). However, despite the frequent occurrence of the pathogen, the number of cases reported is relatively low, indicating that not all strains of V. vulnificus are equally virulent (9, 10).
The Baltic Sea is an intracontinental sea and a low-salinity marine ecosystem. At the same time it shows an increased rate of warming of surface water, especially in summers, which is distinctly higher than that of most other ocean seas. This led in some very hot summers (e.g., 1994, 2002, 2003, and 2006) to a significant number of reports of wound infection after contact with Baltic Sea water due to bathing or strolling along the shore line (11–16). During the summer of 2006, 67 Vibrio sp. infections were reported from the whole Baltic Sea region (8). Health authorities in the state of Mecklenburg-Vorpommern, Germany, have repeatedly detected V. vulnificus in nine out of 10 investigated German bathing locations since 2006 (15, 17). Locally acquired food-borne infections have not yet been reported in Germany, probably because oyster and mussel production does not play a role in the Baltic Sea and mandatory notification of suspected V. vulnificus infections does not exist.
The species V. vulnificus displays a high degree of intraspecies diversity and includes strains with various virulence potentials (4). Although the majority of strains are virulent in animal models (18), several investigations showed genetic divergence among strains from clinical and environmental origins. Phylogenetic and comparative genomic analyses grouped V. vulnificus strains into three major lineages: clade/cluster 1 includes biotype 1 (BT1) strains as well as biotype 2 (BT2) strains isolated mainly from the environment, while clade/cluster 2 strains include the majority of human clinical isolates of biotype 1 (19–21); a third lineage is comprised of strains that are biotype 3 human pathogens but seem to be regionally restricted to Israel (4). Recently, Broza et al. defined a fourth phylogenetic group, termed clade A, based on simple sequence repeats and multilocus sequence typing (MLST) (22).
Several studies developed rapid methods to distinguish clinical (C-type) from environmental (E-type) strains based on variations in the sequences of the 16S rRNA gene (23–25), of the virulence-correlated gene (vcg) (10, 26, 27), and of the pilF gene, encoding a protein required for pilus type IV assembly (1, 28). Other key genes that may differentiate between the genomes of clinical and environmental genotypes were recently identified using a comparative genome analysis approach (29), and Kim et al. identified further potential virulence factors by comparative transcriptomic analyses between clinical and environmental isolates using a cDNA microarray (30). Thiaville et al. analyzed the virulence properties of 33 clinical and 36 environmental isolates of V. vulnificus biotype 1 from the United States in a mouse model and found that higher virulence was correlated with, but not exclusive to, the clinical genotype (31). While several studies examined the distribution and characteristics of this species from tropical regions and the United States, studies on pathogenic and nonpathogenic V. vulnificus strains from northern temperate waters are scarce.
In the present study, we performed a detailed comparative analysis of genotypic and phenotypic traits known to be involved in or indicative for virulence of a diverse selection of V. vulnificus Baltic Sea strains. So far, V. vulnificus cases have been relatively rare in Germany, and thus a more systematic collection of strains from the Baltic Sea started only in the last few years. From the Vibrio strain collection of the Robert Koch Institute (RKI) and from samplings from the local authorities, 49 Baltic Sea isolates originating from wound infections and from environmental sources sampled between 1994 and 2011 and four reference strains were investigated.
MATERIALS AND METHODS
Bacterial strains.
A total of 49 V. vulnificus isolates from the Baltic Sea region and four reference strains (ATCC 33149, ATCC 27562, MO6-24, and CMCP6) were selected. In the study, all available Baltic Sea clinical strains (n = 19) present in our strain collection were investigated, while environmental strains (n = 30) were chosen to represent the period from 1990 to the present. A detailed list of isolates is shown in Table S1 in the supplemental material. Most strains originated from the Vibrio strain collection of the Robert Koch Institute, Berlin (RKI), which is now located at the Federal Institute for Risk Assessment (BfR), or were collected by the Landesamt für Gesundheit und Soziales, Rostock, Germany (LAGuS). All Baltic Sea clinical strains were isolated from patients with wound infections and other nonintestinal infections after contact with seawater (recreational activities or fish handling). Four reference strains from different sources were used for comparison. Species identities of all strains were confirmed by biochemical assays (32) and species PCR as described by Bauer and Roervik (33).
MLST.
Multilocus sequence typing (MLST) analyses were performed on 10 housekeeping genes from both chromosomes as described by Bisharat et al. (19, 34). PCR products were purified using the QIAquick PCR purification kit (Qiagen, Hilden, Germany) and sequenced (both strands) through a sequencing service (Qiagen, Hilden, Germany). Chromatograms were imported, assembled, and trimmed using BioNumerics (version 6.6.4; Applied Maths, Sint-Martens-Latem, Belgium). The alleles at each locus were compared to the PUBMLST database (35). Different allele numbers were given to each distinct allele sequence within a locus, and a distinct sequence type (ST) number was attributed to each distinct profile of alleles by the administrator of the PUBMLST database. Novel allele sequences and novel allelic profiles (sequence types) were submitted to the PUBMLST database. Phylogenetic and molecular evolutionary analyses of individual gene loci and concatenated gene sequences were conducted using BioNumerics and MEGA version 5.1 (36). The DnaSP software package, version 5.10 (37), was used to estimate DNA sequence variation parameters among multialigned sequences, including total number of mutations (Eta), number of segregating polymorphic sites (S), nucleotide diversity (pi), average number of nucleotide differences (k), and mutation rate (θG). Split-tree generation for individual loci, concatenated sequences, and the pairwise homoplasy test (phi) for recombination were done using the SplitsTree 4.0 software (38).
Intact-cell MALDI-TOF MS.
Sample preparation for whole-cell matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) was performed as described previously (39). In brief, colonies were directly transferred from Mueller-Hinton (MH) agar plates onto a 384-position ground steel target plate (Bruker Daltonics, Bremen, Germany) using a sterile pipette tip and immediately mixed with 1 μl of 25-mg/ml sinapinic acid (Bruker Daltonics) in 50% acetonitrile (Sigma-Aldrich, Taufkirchen, Germany), supplemented with 0.6% trifluoroacetic acid (Roth, Karlsruhe, Germany). The matrix/sample spots were crystallized by air drying. All mass spectra were acquired with an Ultraflex II MALDITOF/TOF MS (Bruker Daltonics) equipped with an all-solid-state Smartbeam laser Nd:YAG laser and operated at 100 Hz in the positive linear mode (delay, 100 ns; voltage, 25 kV; mass range, 2.2 to 20 kDa) under the control of Flexcontrol software version 3.0 (Bruker Daltonics). The spectra were obtained by averaging up to 7,000 laser shots acquired at a fixed laser power, which had been set to the minimum laser power necessary for ionization of selected samples before starting the analyses. Spectra were recorded in automatic mode. All spectra were externally calibrated by using the standard calibration mixture, protein calibration standard I (Bruker Daltonics). Sets of potentially cluster-discriminating biomarker ions were identified by visual inspection of mass spectra.
PCR genotyping.
Genomic DNA was extracted using the RTP bacterial DNA kit from Stratec Molecular, Berlin, Germany. PCRs were performed using a Mastercycler EP gradient (Eppendorf, Hamburg, Germany) in a volume of 25 μl with 1× PCR buffer (2 mM MgCl2), 0.2 mM each deoxynucleoside triphosphate (dNTP), 0,2 μM each primer, and 1.5 U DreamTaq DNA polymerase (Fermentas, St. Leon-Rot, Germany). Real-Time PCR experiments were performed using an Applied Biosystems 7500 real-time PCR System (Applied Biosystems, Foster City, CA). The PCR primers, target genes, and amplicon sizes used are shown in Table 1. Sequencing reactions (MLST genes, vcg, and pilF) were performed commercially at Eurofins MWG Operon (Ebersberg, Germany). Primers were synthesized by Metabion International AG (Martinsried, Germany).
Table 1.
Primers and probes used for PCR and sequencing
| Primer | Gene target or designation | Sequence (5′ to 3′)c | Amplicon size(s) (bp) | Reference and/or source |
|---|---|---|---|---|
| vcg-typeC-F | Virulence-correlated gene clinical allele | AGCTGCCGATAGCGATCT | 26 | |
| vcg-typeE-F | Virulence-correlated gene environmental allele | CTCAATTGACAATGATCT | 26 | |
| vcg-typeC/E-R | Virulence-correlated gene | CGCTTAGGATGATCGGTG | 277 | 26 |
| vcg-seq-Fb | vcg, partial | TGTCAAACGACATCGAAACAA | This study | |
| vcg-seq-Rb | vcg, partial | CTTAGGATGATCGGTGTAAAGG | 380 | This study |
| VVA1612F | Region XII, flanking region | ACCCTGATCGTTGGCTACTC | 21 | |
| VVA1613R | Region XII | GGAGCGGTGTGATGGTGTTG | 2,257 | 21 |
| VVA1625F | Region XII | CGGTCTGTGGTTTATCG | 21 | |
| VVA1625R | Region XII | TCGTTTCCAGTCGTCAC | 1,822 | 21 |
| VVA1634 F | Region XII | TGACACCCAACCTAGACCAC | 21 | |
| VVA1634R | Region XII | ATTGATGCCAACCTGAG | 1,364 | 21 |
| VVA1636F | Region XII | TGTCCACGACTTGAACACG | 21 | |
| VVA1637R | Region XII, flanking region | AACATCAACCAGCGAGTCGAAC | 1,547 | 21 |
| VVA1612bF | Region XII, flanking region | TGTGGAGAGCGGCAAGATCAAG | 21 | |
| VVA1637R | Region XII, flanking region | AACATCAACCAGCGAGTCGAAC | 1,200 | 21 |
| Vvu16S51-Fa | 16S rRNA gene | CAAGTCGAGCGGCAGCA | 25 | |
| Vvu16S221-Ra | 16S rRNA gene | TCCTGACGCGAGAGGCC | 171 | 25 |
| Vvu16SA-Pa (2091859) | 16S rRNA gene type A allele | 6-FAM-TGATAGCTTCGGCTCAA-MGBNFQ | Probe | 25 |
| Vvu16SB-Pa (2091860) | 16S rRNA gene type B allele | VIC-CCCGTAGGCATCATGC-MGBNFQ | Probe | 25 |
| Vv-pilF-Fb | Type IV pilus assembly protein pilF gene | CGATTGGTAGGCAATAGAC | 28 | |
| Vv-pilF-Rb | Type IV pilus assembly protein pilF gene | GCAACTCAACCTCAAGACG | 917 | 28 |
| Vv-pilF-F2b | Type IV pilus assembly protein pilF gene | AGGCCAAGCACAAAAAGATCC | This study | |
| Vv-pilF-R2b | Type IV pilus assembly protein pilF gene | TTCACCAGCGCCACATTACC | This study | |
| vllY-F | Hemolysin vllY gene | TATGTGGGATGGCATTTCGAG | 44 | |
| vllY-R | Hemolysin vllY gene | CAACATCCGACTTCGCTAGGC | 237 | 44 |
| hlyIII-F | Hemolysin III hlyIII gene | CCCAACAAGGCTATCGACCAA | 44 | |
| hlyIII-R | Hemolysin III hlyIII gene | GGTCACACACGGGTTAGGCAT | 318 | 44 |
| nanA-F | Sialic acid catabolism cluster | TKATCGCCGCTCCYCATACA | This study | |
| nanA-R | Sialic acid catabolism cluster | GCAACGCCACCGTATTCAAC | 745 | 45 |
| Mann Hemo F | Hemolysin upstream of mannitol operon | ACATTTGACGGATGAGCAG | 357; 1,821 | 49 |
| Mann Hemo R | Hemolysin upstream of mannitol operon | TCCCAGACAAACAGATGATG | 357 | 49 |
| Mann TRAP F | TRAP-type mannitol transport system | CGCTGAAGAAATGTCAAACG | 361 | 49 |
| Mann TRAP R | TRAP-type mannitol transport system | ACGCATTTTCAACCCTTT | 361; 1,821 | 49 |
| Man IIA F | Mannitol fermentation operon | GATGTTGGTGAACAACTTCTCTGC | 49 | |
| Man IIA R | Mannitol fermentation operon | TCTGAAGCCTGTTGGATGCC | 243 | 49 |
| VVA0081F | Ribosomal protein S12 methylthiotransferase | CGTTTACGCCAAACCAAACC | This study, 21 | |
| VVA0081R | Ribosomal protein S12 methylthiotransferase | ACCAATAGCTCTTTCACCCC | 637 | This study, 21 |
| VVA0303F | Phospholipase/hemolysin | CACCAACACATCACAAAACG | This study, 21 | |
| VVA0303R | Phospholipase/hemolysin | GTTGAACTCGACGATCTTCG | 661 | This study, 21 |
| VVA0320F | Zn hydrolase | GTGTAAGGCACCACTTCAAG | This study, 21 | |
| VVA0320R | Zn hydrolase | GCAAAAGCTGGGCAAAAAC | 723 | This study, 21 |
| VVA0331F | RTX toxin | GCCGTTAACATTGACGATCC | This study, 21 | |
| VVA0331R | RTX toxin | CGCAAGGCTGCTTAAATCTTC | 756 | This study, 21 |
| VV0316F | YJ-like nab1 allele | GGCCACCCCTTCAATTGAG | 47 | |
| VV0316R | YJ-like nab1 allele | GTCGCATACACAACCGTGG | 435 | 47 |
| VV0312F | YJ-like nab2 allele | CGACGAAGCACTGGCGTTTAA A | 47 | |
| VV0312R | YJ-like nab2 allele | GCTCGAGCATCTCCCAATACT | 986 | 47 |
| VV10803F | CM-like nab1 allele | TTATCGGCGACAAGGTGA | 47 | |
| VV10803R | CM-like nab1 allele | ATCCATTACATAGGCAAATATG | 346 | 47 |
| VV10808F | CM-like nab2 allele | TATTCGTTTAGCCAAACAGTTGA | 47 | |
| VV10808R | CM-like nab2 allele | CCACTTCATCCCAACGCGTT | 902 | 47 |
Used for real-time PCR.
Used for gene sequencing.
6-FAM, 6-carboxyfluorescein; MGBNFQ, minor groove-binding nonfluorescent quencher.
Mannitol fermentation.
V. vulnificus strains were streaked onto LB agar plates and incubated at 37°C for 24 h. Five milliliters of mannitol fermentation broth (1% mannitol, 0.0075% bromothymol blue, 1% peptone, 0.5% NaCl, pH 7.4) was inoculated using an inoculating needle, incubated at 37°C, and examined for mannitol fermentation after 24 h and 5 days.
Cytotoxicity assay.
Caco-2 cells were cultivated in Dulbecco's modified Eagle medium (DMEM) (with 4.5 g/liter glucose) supplemented with 10% fetal calf serum (FCS), 1 mM sodium pyruvate, 4 mM glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. Cells (2.5 × 104 cells in 100 μl) were seeded in 96-well plates (TPP, Trasadingen, Switzerland) and incubated at 37°C with 5% CO2 for 22 h to reach a confluent monolayer. Before the assay, cells were washed once with DMEM without phenol red. Bacterial cells grown in 20 ml brain heart infusion broth (BHIB) for 16 h at 37°C and 200 rpm were harvested at 5,000 × g for 10 min, and supernatants were filtered (0.2 μm). Filtered culture supernatants (200 μl) were added to Caco-2 cells and incubated at 37°C with 5% CO2 for 2 h. Plates were centrifuged at 275 × g for 4 min, and 50 μl of the supernatants was transferred into a new plate. For determining cytotoxicity, lactate dehydrogenase (LDH) activity in the supernatant was determined using the Cytotox96 kit (Promega) according to the manufacturer's protocol. Additionally, the induced morphological changes of the Caco-2 cells were examined microscopically after 4 h. Experiments were performed at least twice.
Hemolytic activity of cells and culture supernatants.
Twenty milliliters of BHIB was inoculated with the appropriate volume of V. vulnificus overnight cultures to give an optical density at 600 nm (OD600) of 0.1 and incubated at 37°C and 200 rpm for 2 to 4 h. After reaching the late logarithmic phase at an OD600 of 1.6, cells were harvested by centrifugation (5.000 × g, 10 min, 4°C), washed twice, and resuspended in the harvested volume of ice-cold phosphate-buffered saline (PBS). Culture supernatants were filtered (0.2 μm), and 500 μl of filtered culture supernatants or 1:10-diluted cells in PBS was mixed with the same volume of 4% human erythrocyte suspensions in PBS (blood donation service of the German Red Cross, Berlin-Dahlem, Germany) and incubated for 2 h at 37°C on a tilting shaker. Preparations were centrifuged at 2,500 rpm and 4°C for 10 min. Release of hemoglobin into the supernatant was determined by measuring the absorbance at 570 nm. Erythrocyte suspensions were incubated with 500 μl of 2% Triton X-100 in PBS or BHIB and with sterile PBS or BHIB to determine the maximum and spontaneous hemoglobin release, respectively. Hemolytic activity (percent hemolysis) was calculated with the following equation: hemolysis (%) = 100 × (A570 sample − A570 spontaneous)/(A570 maximum − A570 spontaneous). Selected supernatants were heat treated at 56°C for 30 min for control purposes and showed complete loss of hemolytic activity. All experiments were performed three times. Hemolytic activity was rated positive for values of >20% in Table 5. For Fig. 2, strains were judged as hemolytic when supernatants or cells (or both) were positive.
Table 5.
Genotypic and phenotypic characterization of V. vulnificus strains used in this study
| Strain | Biotype | MLST cluster | Mannitol fermentatione | Genotyping |
Sequence typing |
Human serum resistancef | Hemolytic activity in: |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 16S rRNA type | vcg type | PRXII | nanA | nab1 and nab2 | MLST type | pilF type | Supernatant | Cells | |||||
| VN-0010 | 1 | I | + | A | E | +a | − | CM-like | 43 | E | Res | + | + |
| VN-0016 | 1 | I | + | A | E | +a | − | YJ-like | 103 | C | Res | + | + |
| VN-0092 | 1 | IIB | + | B | E | + | + | YJ-like | 118 | C | Res | + | + |
| VN-0094 | 1 | IIA | + | B | C | + | + | CM-like | 110 | C | Res | + | + |
| VN-0095 | 1 | IIA | + | B | C | + | + | CM-like | 110 | C | Res | + | + |
| VN-0096 | 1 | I | + | A | E | +a | + | YJ-like | 111 | E | Int | + | − |
| VN-0097 | 2 | I | − | A | E | − | − | YJ-like | 112 | Cc | Sen | + | + |
| VN-0098 | 2 | I | − | A | E | − | − | YJ-like | 112 | Cc | Int | + | + |
| ATCC 33149 | 2 | I | − | A | E | − | − | YJ-like | 112 | Cc | Sen | + | + |
| VN-0100 | 1 | IIB | − | B | E | + | + | CM-like | 128 | C | Res | − | − |
| VN-0101 | 1 | I | − | A | E | − | − | YJ-like | 113 | E | Int | + | + |
| VN-0102 | 1 | I | − | AB | E | − | − | − | 114 | E | Int | − | + |
| VN-0103 | 1 | I | − | A | E | − | − | CM-like | 100 | E | Res | − | + |
| VN-0104 | 1 | IIB | − | B | E | + | + | CM-like | 115 | C | Res | − | − |
| VN-0105 | 1 | I | + | A | E | +a | + | YJ-like | 116 | E | Res | + | + |
| VN-0108 | 1 | I | − | A | E | − | + | YJ-like | 117 | E | Res | + | − |
| VN-0112 | 1 | IIB | + | B | E | + | + | YJ-like | 118 | C | Res | + | + |
| VN-0119 | 1 | IIB | + | B | E | + | − | YJ-like | 130 | C | Int | − | − |
| VN-0120 | 1 | IIA | + | B | C | + | + | − | 119 | C | Sen | + | + |
| ATCC 27562 | 1 | I | − | AB | E | − | − | − | 3 | E | Res | + | + |
| VN-0125 | 2 | I | − | A | E | − | − | YJ-like | 112 | Cc | Sen | + | + |
| VN-0126 | 1 | I | + | A | E | +a | − | CM-like | 120 | E | Res | + | + |
| VN-0127 | 1 | I | − | A | E | − | + | YJ-like | 121 | E | Res | + | + |
| VN-0128 | 1 | IIA | + | B | C | + | + | CM-like | 110 | C | Res | − | + |
| VN-0129 | 1 | IIA | + | B | C | + | + | CM-like | 110 | C | Res | − | + |
| VN-0130 | 1 | I | + | A | E | +a | − | CM-like | 43 | E | Res | + | + |
| VN-0131 | 1 | I | + | A | E | +a | + | CM-like | 122 | E | Res | − | + |
| VN-0132 | 1 | I | + | A | E | +a | − | CM-like | 43 | E | Res | + | + |
| VN-0133 | 1 | I | + | A | E | +a | − | CM-like | 131 | E | Res | + | + |
| VN-0143 | 2 | I | − | A | E | − | − | YJ-like | 112 | Cc | Int | + | + |
| VN-0204 | 2 | I | − | A | E | − | − | YJ-like | 112 | Cc | Int | + | + |
| VN-0205 | 1 | I | − | A | E | − | − | CM-like | 65 | E | Sen | − | + |
| VN-0206 | 1 | I | + | A | E | +a | − | YJ-like | 132 | E | Sen | + | + |
| VN-0207 | 1 | IIB | − | AB | E | + | + | YJ-like | 123 | C | Res | − | − |
| VN-0208 | 1 | I | − | A | E | − | − | YJ-like | 105 | C | Res | + | + |
| VN-0209 | 1 | I | − | A | E | − | − | − | 124 | C | Int | − | + |
| VN-0227 | 1 | IIB | − | B | E | +b | − | − | 125 | C | Int | − | − |
| VN-0235 | 1 | I | − | A | E | − | − | CM-like | 133 | E | Res | + | + |
| VN-0239 | 1 | I | − | A | E | − | − | CM-like | 133 | E | Res | + | + |
| VN-0243 | 1 | I | − | A | E | − | − | YJ-like | 107 | E | Int | + | + |
| VN-0251 | 1 | I | − | A | E | − | − | YJ-like | 44 | E | Sen | + | + |
| VN-0260 | 1 | I | − | A | E | − | − | YJ-like | 126 | E | Int | − | − |
| VN-0264 | 1 | I | − | A | E | − | − | YJ-like | 108 | E | Res | − | + |
| VN-0266 | 1 | I | − | A | E | − | − | YJ-like | 113 | E | Sen | + | + |
| VN-0270 | 1 | I | − | A | E | − | − | CM-like | 133 | E | Res | − | + |
| VN-0274 | 1 | I | − | A | E | − | − | YJ-like | 44 | E | Int | + | + |
| VN-0275 | 1 | I | − | A | E | − | − | CM-like | 133 | E | Res | + | + |
| VN-0276 | 1 | I | − | A | E | − | − | YJ-like | 134 | E | Int | + | + |
| VN-0277 | 1 | I | − | A | E | − | − | YJ-like | 127 | E | Res | − | − |
| VN-0280 | 1 | I | − | A | E | − | − | − | 109 | E | Res | + | + |
| MO6-24 | 1 | IIA | + | Bd | Cd | + | + | CM-like | NAg | Cd | Res | + | + |
| CMCP6 | 1 | IIA | + | Bd | Cd | + | + | CM-like | NA | Cd | Res | + | + |
| VN-0288 | 1 | IIB | − | B | E | + | − | CM-like | 135 | C | Res | + | − |
PCR positive for all tested genes of PRXII except VVA1634.
PCR positive for all tested genes of PRXII except VVA1625.
Slightly different pilF type C in biotype 2 strains (see the text).
Sequence deduced from published genome.
Mannitol fermentation tested biochemically and by presence of mannitol fermentation operon (PCR).
Res, resistant; Int, intermediate; Sen, sensitive.
NA, not assigned.
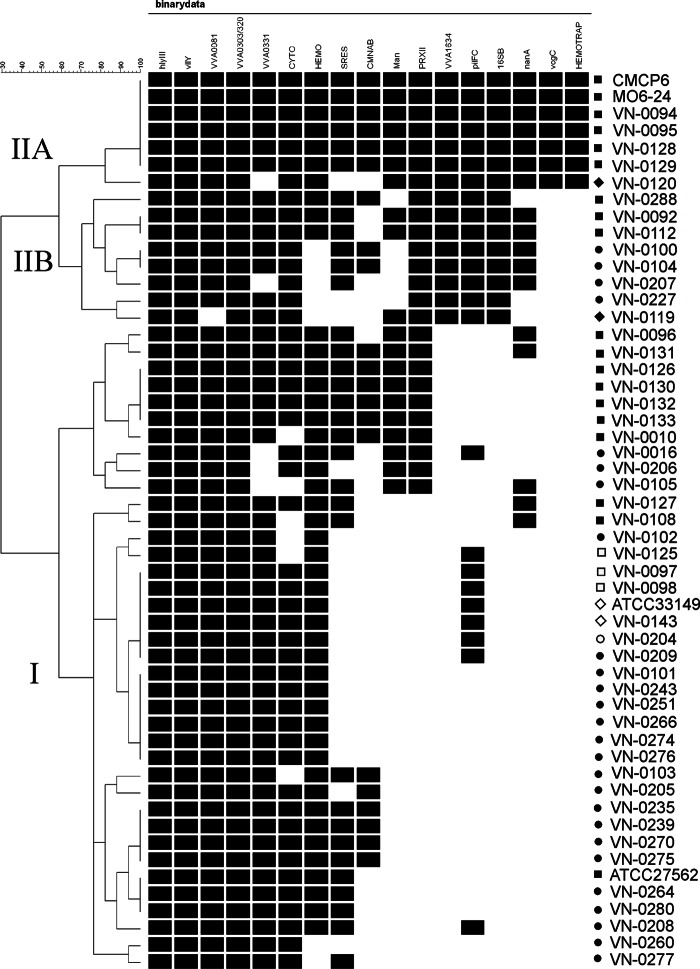
Fig 2.
Grouping of isolates based on virulence-associated traits. Similarity patterns were determined by cluster analysis by complete linkage using simple matching of binary data (BioNumerics version 6.6.4; Applied Maths, Sint-Martens-Latem, Belgium). IIA, IIB, and I, cluster designation taken from MLST analysis. ■, human; ●, environmental (seawater); ◆, seafood (mussel); □, human (BT2); ○, environmental (BT2); ♢, diseased eel (BT2).
Serum resistance.
A colorimetric serum sensitivity assay as described by Moll et al. with slight modifications was employed, which allows high-throughput screening of bacterial resistance to human serum (40, 41). Bacterial cells were transferred from sheep blood (SB) agar plates to wells of a sterile 96-well plate (Brand, Wertheim, Germany) containing 0.2 ml LB broth per well and incubated overnight at 37°C. A microplate replicator (Boekel Scientific, Feasterville, PA) was used to transfer bacteria from the overnight cultures to a new 96-well plate containing 0.1 ml LB broth. Plates were incubated for 5 h at 37°C, and bacteria were transferred to new 96-well plates containing 100 μl peptone glucose broth (1% glucose, 0.0075% bromthymol blue, 1% peptone, 0.5% NaCl, pH 7.4) with different concentrations of human serum (0, 10, 20, 40, 60, and 80% pooled serum obtained from healthy volunteers). Plates were incubated at 37°C for 24 h, and serum resistance was determined by examining the color change from blue to yellow, indicating bacterial growth. Experiments were performed at least three times. Strains of Escherichia coli K-12 and E. coli K-12(pKT107) carrying the pKT107 serum resistance plasmid (40) were used as negative and positive controls, respectively. Isolates that showed growth in the presence of 60 to 80% human serum were classified as serum resistant. Isolates that grew only in the presence of 20 to 40% and 0 to 10% human serum were classified as intermediate resistant and sensitive, respectively. For Fig. 2 (binary data) all strains that showed strong resistance (growth in 60 to 80% human serum) were rated resistant.
Grouping of isolates based on virulence-associated traits.
Similarity patterns based on virulence-associated phenotypic and genotypic traits were determined by cluster analysis by complete linkage using simple matching of binary data (BioNumerics version 6.6.4; Applied Maths, Sint-Martens-Latem, Belgium).
Nucleotide sequence accession number.
The pilF sequence of the ATCC 33149 has been deposited in GenBank (accession number HF568870).
RESULTS
Multilocus sequence typing.
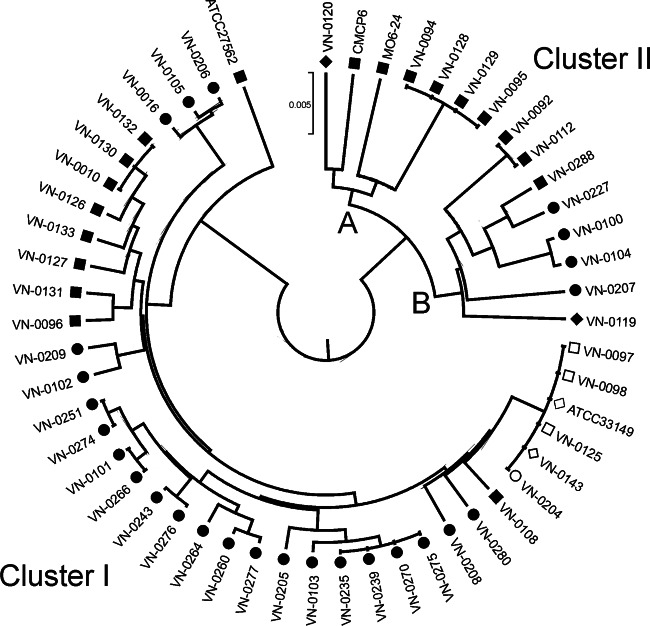
All isolates were successfully subtyped at 10 loci using multilocus sequence typing as described by Bisharat et al. (19, 34), with the exception of the two genome-sequenced reference strains, MO6-24 and CMCP6. Sequence data were concatenated in the order of loci used to define the allelic profile to produce a sequence (4,326 bp in length) for each strain. To evaluate genetic relationships among strains, phylogenetic clustering based on concatenated sequence information of all 10 gene fragments was investigated with the minimum evolution (ME) algorithm using MEGA v 5.1 (Fig. 1). The resulting dendrogram showed a clear separation into two divergent clusters, I and II, which is in accordance with other studies (19, 21, 42). Most of the environmental strains from the Baltic Sea region belonging to biotype 1 (22 of 28) fall into cluster I, which also contains some clinical strains and all biotype 2 strains. Interestingly, cluster II is further split into two branches, A and B. Branch A contains mainly older human isolates from the Baltic Sea (Denmark, 1994) and human isolates from overseas (United States and Asia), while all but one of the newer human isolates from Germany investigated in our study fall into branch IIB. Thirty-five different STs were obtained among the 49 Baltic Sea isolates, indicating a high degree of genotypic diversity. Comparison of MLST data to data for 188 sequence types in the PUBMLST database revealed that 27 new STs had been identified. The DnaSP program was used to estimate DNA sequence variation parameters in concatenated sequences (Table 2). Nucleotide diversity and mutation rates were higher (2-fold) for cluster II isolates (pi = 0.0036; θG = 14.25) than for cluster I isolates (pi = 0.0018; θG = 7.96), as already observed by Bisharat et al. (19). Generally, nucleotide diversities and mutation rates were higher for chromosome 1 alleles than for chromosome 2 alleles. The phi test for the concatenated sequences of the 49 isolates gave a P value of 0, indicating statistically significant evidence for recombination. As determined by the phi test for individual loci, three of the 10 MLST loci showed statistically significant evidence for recombination (glp, pyrC, and metG). When these three loci were analyzed separately for cluster I and cluster II isolates, significant evidence for recombination was indicated only for cluster II isolates. Lack of evidence for recombination was detected in the remaining seven loci (Table 3). Split decomposition was used to visualize the relationship of the isolates and to visualize the impact of recombination in each locus (see Fig. S1 in the supplemental material).
Fig 1.
Population structure of Vibrio vulnificus isolates from the Baltic Sea region determined using concatenated MLST sequences of 10 housekeeping genes (MEGA version 5.1). Clinical BT1 reference strains ATCC 27562, CMCP6, and MO6-24 and one biotype 2 strain from a Japanese eel (ATCC 33149) were included for comparison. The evolutionary history was inferred using the unweighted-pair group method using average linkages (UPGMA), and evolutionary distances were computed using the maximum-composite-likelihood method. ■, human; ●, environmental (seawater); ◆, seafood (mussel); □, human (BT2); ○, environmental (BT2); ♢, diseased eel (BT2).
Table 2.
Analysis of V. vulnificus DNA sequence polymorphisms using the DnaSP program
| Genes | No. of: |
Nucleotide diversity (pi)a | Avg no. of nucleotide differences (k)a | Mutation rate (θG)a | ||
|---|---|---|---|---|---|---|
| Sites | Mutations (Eta)a | Segregating polymorphic sitesa | ||||
| All | 4,326 | 71.19 (308) | 67.96 (294) | 0.0038 (0.01644) | 16.41 (71.1118) | 15.62 (67.59) |
| Chromosome 1 | 2,301 | 61.28 (141) | 59.54 (137) | 0.0061 (0.01410) | 14.10 (32.4493) | 13.45 (30.94) |
| Chromosome 2 | 2,025 | 82.47 (167) | 77.53 (157) | 0.0094 (0.01909) | 19.09 (38.6625) | 18.09 (36.65) |
| All (cluster 1 isolates) | 4,326 | 38.6 (167) | 37.22 (161) | 0.0018 (0.0079) | 9.02 (39.03) | 7.96 (34.44) |
| All (cluster 2 isolates) | 4,326 | 43.9 (190) | 43.23 (187) | 0.0036 (0.01576) | 15.76 (68.19) | 14.25 (61.23) |
Data are normalized for number of sites and expressed per 1,000 bases. Raw values are shown in parentheses.
Table 3.
Level of recombination using the pairwise homoplasy test (PHI) implemented in Splits Tree software
| Gene(s) | Statistically significant evidence for recombination | P value |
|---|---|---|
| All | Yes | 0 |
| Chromosome 1 | Yes | 0 |
| Chromosome 2 | Yes | 2.62E−12 |
| dtdS (chromosome 2) | No | 0.7863 |
| glp (chromosome 1) | Yes | 2.13E−5 |
| Cluster I | No | 0.05173 |
| Cluster II | Yes | 2.82E−7 |
| gyrB (chromosome 1) | No | 0.5056 |
| lysA (chromosome 2) | No | 1.439E−6 |
| mdh (chromosome 1) | No | 0.2934 |
| metG (chromosome 1) | Yes | 2.83E−4 |
| Cluster I | No | 0.01959 |
| Cluster II | Yes | 0.01595 |
| pntA (chromosome 2) | No | 0.9509 |
| pyrC (chromosome 2) | Yes | 0.02787 |
| Cluster I | No | 1 |
| Cluster II | Yes | 0.08041 |
| tnaA (chromosome 2) | No | 0.9051 |
| purM (chromosome 1) | No | 0.7035 |
Intact-cell MALDI-TOF MS.
We recently evaluated matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) as a rapid tool to identify Vibrio at the species level (39). Here we investigated the potential of MALDI-TOF MS for discrimination of major lineages of Vibrio vulnificus revealed by MLST and biotyping. Following routine species identification using MALDI-TOF MS, spectra were carefully inspected for sets of subcluster- or branch-identifying biomarker ions. To further analyze the respective biomarker ions, we made use of the steadily growing genome sequence databases and applied theoretical masses of biomarker proteins derived from translated gene sequences as references (Table 4). Based on our subset of isolates, biotype 2 strains could be discriminated from biotype 1 strains using a biomarker ion at m/z 6,196 that corresponds to ribosomal protein 32 (RL32). In contrast, biotype 1 strains displayed a peak for RL32 at m/z 6,168, corresponding to a K-to-R exchange in the respective protein sequence. Similarly, putative biomarker ions indicative of cluster II strains (C-type strains), e.g., ribosomal protein L25 (m/z 10,276), integration host factor subunit alpha (IHFA) (m/z 10,705), and sigma 54 modulation protein (m/z 11,052) could be identified. To further discriminate branch IIA from IIB, possible branch-identifying biomarker ions were observed at, among others, m/z 10,607 (integration host factor subunit beta [IHFB]), m/z 9,477 (RS17), m/z 11,100 (RL24), and m/z 8,070 (RL31) The discriminative value of these putative biomarkers has to be further evaluated using extended sets of isolates from different sources.
Table 4.
Selected subcluster- or branch-identifying biomarkers observed by MALDI-TOF mass spectrometric analysis of V. vulnificus isolates from different MLST lineages
| Tentative protein identity | Group-specific peaks |
Peaks present in multiple groups |
|||||
|---|---|---|---|---|---|---|---|
| Exptl mass (avg m/z) | Predicted avg mass [(M + H)+], Da | Subcluster/branch specificity | Exptl mass (avg m/z) | Predicted avg mass [(M + H)+], Da | Presence in cluster/branch | Amino acid exchange | |
| 50S ribosomal protein L32 | 6,196 | 6,195.9 (Met loss) | BT 2 isolates | 6,168 | 6,167.9 (Met loss) | Most BT 1 isolates | K→R |
| Integration host factor subunit alpha | 10,705 | 10,705.2 (Met loss) | Cluster II isolates | 10,806 | NAa | Most cluster I isolates | NA |
| Putative sigma 54 modulation protein | 11,052 | 11,052.5 | Most cluster II isolates | 11,067 | 11,066.5 | Most cluster I isolates | T→S |
| Integration host factor subunit beta | 10,607 | 10,607.1 | Cluster IIA isolates | 10,592 | 10,593.0 | Cluster IIB and cluster I isolates | V→I |
| 30S ribosomal protein S17 | 9,477 | 9,477.1 (Met loss) | Cluster IIA isolates | 9,420 | NA | Cluster IIB and many cluster I isolates | NA |
| 50S ribosomal protein L24 | 11,100 | 11,100.9 (Met loss) | Cluster IIA isolates | 11,086 | NA | Cluster IIB and most cluster I isolates | NA |
| NA | 7,393 | NA | Cluster IIA isolates | 7,365 | NA | Cluster IIB and most cluster I isolates | NA |
| 50S ribosomal protein L31 | 8,070 | 8,070.2 | Cluster IIB isolates | 8,084 | 8,084.2 | Most cluster IIA and cluster I isolates | E→D |
NA, not assigned.
Detection of virulence-associated genes by PCR.
Detection of virulence genes and typing of polymorphic DNA sequences were carried out via PCR using published primers or sequencing of PCR products (Table 1). Types were designated either clinical (C) or environmental (E) in accordance to published nomenclature based on PCR assays targeting sequence polymorphisms in the 16S rRNA (A→E; B→C) and vcg genes and by sequencing the pilF gene (25, 26, 28). In case of the 16S rRNA gene, all strains of cluster II possessed allele B (only VN-0207 is of type AB) and all cluster I strains were type A, with the exceptions of ATCC 27562 and VN-0102, which possessed both alleles (type AB). The vcg type C allele was found exclusively in cluster IIA. Analysis of the 16S rRNA and vcg gene also revealed that 10 (53%) and 13 (68%) of the 19 clinical BT1 isolates were of the respective E type. While 10 (53%) of the clinical BT1 isolates showed pilF type E, all cluster II strains and only three environmental strains of cluster I possessed pilF type C (Table 5).
Five PCR assays described by Cohen et al. were performed to check for the presence of the 33-kb pathogenicity region XII (PRXII) (21): four PCR assays amplify parts of the genes encoding two putative chondroitinase AC lyases (genes VVA1613 and VVA1636), a putative arylsulfatase A (gene VVA1634), and a putative methyl-accepting chemotaxis protein (gene VVA1625), whereas another PCR proves complete absence of the region by targeting the flanking regions of PRXII (genes VVA1612 and VVA1637). Strains were considered positive for PRXII when at least three of the four genes on PRXII could be amplified and no amplicon was detected with PCR primers VVA1612F and VVA1637R. Eighty-four percent (16 of 19) of all BT1 clinical strains were positive for PRXII, whereas only 32% (9 of 28) of the BT1 environmental strains contained this region. Interestingly, the presence of the pathogenicity region XII was proven in all cluster II strains by detecting all four genes of PRXII, with the exception of VN-0227, which showed no amplification of VVA1625. In contrast, BT1 strains of cluster I (only 10 out of 32 positive, including 7 clinical isolates) contained a different PRXII in which one gene (VVA1634) could not be detected with the primers used (VN-0010, VN-0016, VN-0096, VN-0105, VN-0126, VN-0130, VN-0131, VN-0132, VN-0133, and VN-0206).
Furthermore, oligonucleotide primers for three potential virulence genes located in a 53-kb region of chromosome II (21) were designed and applied in PCR assays: open reading frames (ORFs) VVA0303, encoding a putative thermolabile hemolysin, VVA0320, encoding a zinc-dependent hydrolase, and VVA0331, encoding a 489-kDa RTX-like protein (43). These results are not displayed in Table 5 because all strains were positive for VVA0303 and VVA0320 and only strains VN-0016, VN-0105, VN-0120, VN-0206, and VN-0207 were negative for VVA0331. Another suspected pathogenicity region (21) spanning VVA0080 to VVA0186 was investigated by targeting ORF VVA0081, encoding a ribosomal protein S12 methylthiotransferase. As all strains were positive for VVA0081 with the exception of strain VN-0119, the results are also omitted from Table 5. Similar results were obtained for the hemolysin genes hlyIII and vllY (44), as each strain possessed both genes.
The strains were further examined for the presence of the virulence-associated gene nanA of the sialic acid catabolism region (45, 46), which could be detected in 12 of 19 (63%) clinical and in only 5 of 28 (18%) environmental BT1 isolates. All cluster II strains were nanA positive except three cluster IIB strains, while most (84%) of the BT1 cluster I strains were nanA negative. Furthermore, allele types of the nab1 and nab2 genes were determined, which are responsible for biosynthesis of nonulosonic acids (NulOs) such as sialic acids (47). Sixty-eight percent (13 of 19) of the clinical BT1 isolates possessed CM-like alleles of the nab1 and nab2 genes, and one strain was not typeable. Nearly half of the environmental isolates (46%) also showed one of these genotypes. Six out of seven cluster IIA isolates possessed CM-like alleles of the nab1 and nab2 genes, whereas this was the case for only three of the cluster IIB isolates.
Mannitol fermentation.
Mannitol fermentation has recently been correlated with clinical genotypes (vcg type C and 16S rRNA type B), and mannitol transport and fermentation genes were found to be predominantly present in the C genotype (48, 49). Of the 47 biotype 1 isolates in our study, mannitol fermentation was observed in 15 of 19 (79%) isolates from human cases, but in only 5 of 28 (18%) isolates from environmental sources (Table 5). Interestingly, the five mannitol-positive environmental isolates were also positive for pathogenicity region XII. The ability to ferment mannitol was in complete accordance with the presence of the mannitol fermentation operon, which was determined by PCR analysis of the gene encoding the IIA domain of the mannitol phosphotransferase system. All isolates were also tested for the presence and orientation of two genes that are immediately upstream of the mannitol fermentation operon in the three published V. vulnificus genomes, a putative hemolysin gene (Mann Hemo) and a TRAP-type mannitol transport gene (Mann TRAP), as described by Froelich and Oliver (49). Although these two genes could be detected in each of the strains, their orientation being identical to the published genomes was a specific feature of the vcg type C strains of cluster IIA.
Cytotoxicity against Caco-2 cells.
To investigate the virulence of V. vulnificus isolates to human intestinal epithelial cells, Caco-2 cells were exposed to supernatants of V. vulnificus liquid cultures. Supernatants of all tested strains showed strong cytopathic effects on Caco-2 cells, inducing either cell lysis, cell rounding, or other morphological changes (data not shown). Most of the strains induced lysis of 80 to 100% of the cells, which was determined by LDH activity released into the medium. A few strains (VN-0010, VN-0102, VN-0103, VN-0105, VN-0108, and VN-0125) caused no lysis according to the LDH measurement but induced strong morphological changes of the cells, which were detected by microscopic examination.
Hemolysis assay.
Of the 47 biotype 1 isolates in our study, significant hemolytic activity of culture supernatants (>20%) was observed in 15 of 19 (79%) isolates from human cases and 15 out of 28 (54%) isolates from environmental sources. Three isolates from clinical cases that displayed no hemolytic activity in culture supernatant were positive when viable cells were used in the assay. Seven of the 28 (25%) BT1 environmental strains (including all five cluster IIB strains) showed no hemolytic activity when supernatants or cells were tested (Table 5; see Fig. S2 in the supplemental material). Heat inactivation of culture supernatants of 13 randomly chosen isolates (VN-0010, VN-0095, VN-0096, VN-0101, VN-0108, VN-0112, VN-0120, VN-0126, VN-0204, VN-0260, VN-0280, ATCC 33149, and MO6-24) completely abolished hemolytic activity.
Human serum resistance.
Of the 47 biotype 1 isolates in our study, strong resistance to human serum (growth in 60 to 80% human serum) was observed in 18 of 19 (95%) isolates from human cases. One clinical isolate (VN-0096) showed intermediate serum resistance (growth in 20 to 40% human serum). Fourteen of the 28 environmental isolates (50%) also displayed strong serum resistance, nine (32%) displayed intermediate serum resistance, and five (18%) isolates were classified as serum sensitive (growth in 0 to 10% human serum). All results are included in Table 5.
Biotype 2 isolates.
For comparison we included a subset of isolates of biotype 2. Strains of this biotype are regarded as eel pathogens but have also been obtained from human infections. We studied four Danish isolates from 1994 (three clinical strains from fisherman after handling eels [VN-0097, VN-0098, and VN-0125] and one environmental isolate [VN-0204]), one isolate from a diseased eel isolated in Sweden (VN-0143), and the reference strain ATCC 33149 from a diseased eel in Japan. These isolates turned out to be phylogenetically highly related (all ST 112) despite their different origins. All biotype 2 isolates were E type with respect to the vcg and 16S rRNA genes, were negative for nanA and pathogenicity region XII, were positive for hlyIII, vllY, VVA0303, VVA0320, and VVA0331, and possessed YJ-like alleles of the nab1 and nab2 genes. The isolates were not able to ferment mannitol and showed no or only intermediate resistance to human serum but displayed comparably high hemolytic activities. A pilF gene variant of the C type that was identical in all biotype 2 strains was found by sequencing the complete pilF gene.
DISCUSSION
This study was carried out as the public awareness of Vibrio infections has increased in Germany due to sporadic deaths caused by contact with contaminated Baltic Sea water. Climate warming is thought to drive bacterial waterborne infectious diseases, and the seasonal environmental temperature changes observed in the Baltic Sea region correlate with the emergence of Vibrio infections (6, 7, 15). Forecasts indicate a further increase of infections, as the effects of climate warming on surface temperatures is more pronounced at higher latitudes (6–8, 50). MLST grouping of Vibrio vulnificus isolates revealed the existence of two major phylogenetic clusters (19, 21). Though the MLST clusters I and II do not show a clear distinction between clinical and environmental strains, it was found that in cluster II, clinical strains of biotype 1 are overrepresented (19). When applying the MLST typing scheme to a diverse selection of isolates from the Baltic Sea region (Fig. 1), a comparable topology was obtained regarding two phylogenetic lineages. Cluster I contained Baltic Sea biotype 2 strains (three clinical and two environmental) which possessed the same sequence type (ST) as the reference strain from Japan (ATCC 33149). Of the biotype 1 strains, nine clinical isolates and 22 environmental isolates were grouped into cluster I, while seven clinical and six environmental strains belonged to cluster II. The phylogenetic tree analyses indicated that cluster II splits into two subclusters, branches IIA and IIB. Most of the investigated clinical Baltic Sea strains were isolated before 2000, and only four isolates (VN-0092, VN-0108, VN-0112, and VN-0288) were from recent years (2010 and 2011). It is remarkable that the latter isolates do not cluster with the older strains. Three of the recently isolated strains are grouped in branch IIB, and one strain (VN-0108) belongs to cluster I; however, it is clearly separated from all other clinical biotype 1 strains of cluster I. As VN-0108 was isolated from a patient suffering from a coinfection with a Vibrio cholerae non-O1, O139 strain, its pathogenic potential is unclear and will not be discussed in detail. The MLST analyses of the clinical strains from the early 1990s revealed that they are separated in branch IIA and in cluster I; however, all strains showed closely related sequence types within the respective group.
Due to its relatedness to the C-type strains of cluster IIA from Asian and American sources, which have been reported to possess higher virulence potential (19–21, 47) and to be more likely to cause lethal systemic infection with more severe indicators of virulence (31), branch IIB could represent a currently clinically relevant clonal lineage confined to the Baltic Sea and perhaps European coasts. Only four more STs of strains isolated in European countries that fall into this branch, which was formerly designated branch D of cluster II by Bisharat et al. (19), were present in the PUBMLST database (two clinical isolates [ST57, Spain, 2001; ST61, Sweden, 1997] and two environmental isolates [ST14, Denmark, 1996; ST45, Germany, 1995]). Therefore, we focused on the comparison of branch IIB strains to the remaining strains using a panel of virulence-related genotypic and phenotypic traits. Cluster analysis based on these data displayed in a binary table (1/0) resulted in a grouping of the Baltic Sea isolates in two clusters which largely correspond to MLST clusters I and II (Fig. 2). The two genome-sequenced C-type strains CMCP6 and MO6-24 grouped with cluster IIA Baltic Sea strains, which are clearly separated from cluster IIB strains. Most of the clinical strains of cluster I form a separate branch immediately below the IIB strains. The majority of the environmental strains together with the biotype 2 strains are displayed in the bottom part of the dendrogram.
Two genetic traits, gene VVA1634 of pathogenicity region XII (PRXII) and the 16S rRNA type B, were present exclusively in cluster II strains and could be used to discriminate the two clusters. In agreement with Cohen et al. (21), PRXII was not restricted to cluster II. Interestingly, in all 10 BT1 isolates of cluster I with PRXII (7 clinical strains), gene VVA1634, encoding a putative arylsulfatase, was either missing or altered and could not be detected in these isolates with the PCR assay. PRXII showed a good correlation with the isolation source of BT1 strains, as 84% of clinical isolates and only 32% of environmental isolates possessed this region, supporting the potential role of PRXII-encoded gene products mediating sulfate reduction, oligopeptide transport, and chondroitinase or aryl sulfatase activities for pathogenicity of V. vulnificus (20, 21, 29).
Differentiation of MLST clusters IIA and IIB is possible, as all cluster IIA strains possessed the clinical associated vcg type C, whereas the newly described Baltic Sea isolates belonging to cluster IIB possessed the environmental vcg type E, irrespective of their isolation source. Furthermore, the genetic arrangement of the putative hemolysin and TRAP transporter genes (HEMOTRAP), similar to the published genomes (49), was also present exclusively in cluster IIA. All cluster II strains possessed pilF type C, which has previously been correlated with pathogenicity (1, 28) and was only rarely found in cluster I BT1 strains. In contrast to the study of Roig et al. (28), in which nearly all clinical isolates possessed the pilF type C allele, more than half of the clinical strains in our strain collection possessed pilF type E. Altogether, our data on the distribution of type C and E alleles of the pilF, 16S rRNA, and vcg genes among environmental and clinical isolates from the Baltic Sea region support the opinion of Thiaville et al. that E-type strains can be pathogenic and should not be classified as avirulent per se (31).
The mannitol fermentation operon and the nanA gene were present in most cluster II strains but could also be detected in some cluster I strains. A correlation with MLST cluster II (designated lineage 1) was already described for nanA (45). Both markers showed a significant correlation with the isolation source and may be relevant for virulence, since they can promote a selective advantage by enabling the use of alternative carbon and nitrogen sources (48, 51). The nanA gene may be of particular importance, as it is a component of the sialic acid catabolism (SAC) cluster, whose gene products confer the ability of enteropathogens or commensals to metabolize sialic acid components of mucins in the human intestine (52), and was already found to be essential for pathogenicity of V. vulnificus (46).
Whereas in MLST clusters I and IIB almost equal percentages of CM-like and “not typeable” alleles of the nab1 and nab2 genes versus YJ-like alleles were found, all cluster IIA isolates possessed CM-like or “not typeable” alleles. According to Lewis et al. (47) the latter two alleles are associated with high or intermediate NulO expression, while YJ-like alleles are associated with lower expression. In addition, they demonstrated a higher proportion of CM-like alleles in clinical lineage I isolates (corresponding to cluster IIA) and speculated that NulO expression contributed to the virulence of these strains by participating in host-pathogen interactions (47). PCR analyses of several other suspected virulence genes (VVA0081, VVA0303, VVA0320, VVA0331, hlyIII, and vllY) revealed that they are too frequently present to be useful for a discrimination of strains, as was already shown for hlyIII and vllY by Wong et al. (44).
Cytotoxic and hemolytic properties were exhibited by most of the strains irrespective of their origin, as described by others (53, 54). However, it is also worth mentioning that all clinical isolates showed hemolytic activity in testing of supernatants or intact cells or both. Serum resistance was found in 95% of the clinical strains irrespective of their phylogenetic origin, but 50% of the environmental strains also grew in 60 to 80% serum. This is in accordance with the results reported by Roig et al., who observed serum resistance in more than 90% of clinical isolates and ca. 40% of nonclinical isolates and postulated serum resistance as a prerequisite for pathogenicity (28).
Altogether, cluster IIA constitutes a rather homogenous lineage, since all strains were able to ferment mannitol, showed hemolytic activity, possessed the pilF C, 16S rRNA B, and vcg C alleles, and were positive for PRXII and the nanA gene. Furthermore, most of the strains showed strong serum resistance and the CM-like alleles of the nab1 and nab2 genes. In contrast, cluster IIB strains showed a higher genetic diversity with a less defined pattern of virulence-associated traits. MLST lineages correlate with some virulence-associated genotypes; however, clinical strains are distributed in all lineages.
Epidemiological surveillance and risk assessment of potential virulent Vibrio vulnificus strains require rapid and effective analytical tools, as bacteria of this species are widespread along the German Baltic Sea coastline and raise to significant counts (>105 CFU · liter−1) in hot summers (55). As the MLST scheme is laborious, requiring the sequence analysis of 10 conserved genes, we evaluated intact-cell MALDI-TOF MS (ICMS) analysis, which allows rapid species identification of Vibrio spp. (39) but could also have the potential for discrimination of bacteria below the species level. This approach revealed potentially discriminating biomarker sets for biotype 2 strains as well as for the identification of the cluster II and cluster IIB isolates, which will be further evaluated using extended subsets of isolates. Likewise, the 16S rRNA B and vcg C alleles could serve as a marker for cluster II and specifically cluster IIA, respectively. Gene polymorphisms are valuable markers to recognize strains belonging to MLST clusters which include a high proportion of clinical strains. For assessment of potential pathogenicity, the presence/absence of pathogenicity region XII, the mannitol fermentation operon, or nanA can be studied quickly by simple PCR assays. As traditional biochemical typing is still performed by routine laboratories, mannitol fermentation could also be used for risk assessment of potentially pathogenic strains (48). These rapid tests may be complemented by human serum resistance assays that allow easy and high-throughput screening of large V. vulnificus strain collections.
The clustering based on the binary data suggests that the probability for strains to be highly virulent is lower for strains clustering at the bottom of the diagram (Fig. 2). Presumably, strains of cluster I which test negatively for growth in 60 to 80% human serum, pathogenicity region XII, mannitol fermentation, or nanA are likely to possess less pathogenic potential. On the other hand, environmental isolates that are phylogenetically related to clinical strains and already possess a number of virulence-associated traits might become virulent, e.g., by horizontal transfer of additional virulence genes or adaptations in response to selective forces favoring the evolution of pathogenic clones.
Further studies will focus on the investigation of differences concerning the virulence potential of environmental and clinical cluster II strains. Genome sequencing of selected cluster IIB strains is currently in process, and further investigations will include knockout strategies with selected genes, for example, components of pathogenicity region XII, to compare mutants and wild-type strains in animal models.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the Federal Ministry of Education and Research (VibrioNet, BMBF grant 01KI1015A).
Footnotes
Published ahead of print 29 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00477-13.
REFERENCES
- 1. Baker-Austin C, Lemm E, Hartnell R, Lowther J, Onley R, Amaro C, Oliver JD, Lees D. 2012. pilF polymorphism-based real-time PCR to distinguish Vibrio vulnificus strains of human health relevance. Food Microbiol. 30:17–23 [DOI] [PubMed] [Google Scholar]
- 2. Daniels NA. 2011. Vibrio vulnificus oysters: pearls and perils. Clin. Infect. Dis. 52:788–792 [DOI] [PubMed] [Google Scholar]
- 3. Haq SM, Dayal HH. 2005. Chronic liver disease and consumption of raw oysters: a potentially lethal combination—a review of Vibrio vulnificus septicemia. Am. J. Gastroenterol. 100:1195–1199 [DOI] [PubMed] [Google Scholar]
- 4. Jones MK, Oliver JD. 2009. Vibrio vulnificus: disease and pathogenesis. Infect. Immun. 77:1723–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Paz S, Bisharat N, Paz E, Kidar O, Cohen D. 2007. Climate change and the emergence of Vibrio vulnificus disease in Israel. Environ. Res. 103:390–396 [DOI] [PubMed] [Google Scholar]
- 6. Martinez-Urtaza J, Bowers JC, Trinanes J, DePaola A. 2010. Climate anomalies and the increasing risk of Vibrio parahaemolyticus and Vibrio vulnificus illnesses. Food Res. Int. 43:1780–1790 [Google Scholar]
- 7. Baker-Austin C, Stockley L, Rangdale R, Martinez-Urtaza J. 2010. Environmental occurrence and clinical impact of Vibrio vulnificus and Vibrio parahaemolyticus: a European perspective. Environ. Microbiol. Rep. 2:7–18 [DOI] [PubMed] [Google Scholar]
- 8. Baker-Austin C, Trinanes JA, Taylor NGH, Hartnell R, Siitonen A, Martinez-Urtaza J. 2012. Emerging Vibrio risk at high latitudes in response to ocean warming. Nat. Clim. Change 3:73–77 [Google Scholar]
- 9. Strom MS, Paranjpye RN. 2000. Epidemiology and pathogenesis of Vibrio vulnificus. Microb. Infect. 2:177–188 [DOI] [PubMed] [Google Scholar]
- 10. Rosche TM, Binder EA, Oliver JD. 2010. Vibrio vulnificus genome suggests two distinct ecotypes. Environ. Microbiol. Reports 2:128–132 [DOI] [PubMed] [Google Scholar]
- 11. Kuhnt-Lenz K, Krengel S, Fetscher S, Heer-Sonderhoff A, Solbach W. 2004. Sepsis with bullous necrotizing skin lesions due to Vibrio vulnificus acquired through recreational activities in the Baltic Sea. Eur. J. Clin. Microbiol. Infect. Dis. 23:49–52 [DOI] [PubMed] [Google Scholar]
- 12. Dalsgaard A, Frimodt-Moeller N, Bruun B, Hoei L, Larsen JL. 1996. Clinical manifestations and molecular epidemiology of Vibrio vulnificus infections in Denmark. Eur. J. Clin. Microbiol. Infect. Dis. 15:227–232 [DOI] [PubMed] [Google Scholar]
- 13. Sieffert M, Stolle A. 2002. Nachweis und Differenzierung von Vibrio spp. in Krusten- und Schalentieren. Bundesgesundheitsbl. 6:507–513 [Google Scholar]
- 14. Stephan R, Knabner D. 1996. Vibrio vulnificus-Erste Nachweise in Deutschland. Bundesgesundheitbl. 6:209–212 [Google Scholar]
- 15. Frank C, Littmann M, Alpers K, Hallauer J. 2006. Vibrio vulnificus wound infections after contact with Baltic Sea, Germany. Euro Suveill. 11:3024–3025 [DOI] [PubMed] [Google Scholar]
- 16. Ruppert J, Panzig B, Guertler L, Hinz P, Schwesinger G, Felix SB, Friesecke S. 2004. Two cases of severe sepsis due to Vibrio vulnificus wound infection acquired in the Baltic Sea. Eur. J. Clin. Microbiol. Infect. Dis. 23:912–915 [DOI] [PubMed] [Google Scholar]
- 17. Böer S, Hauk G, Duty O, Luden K, Heinemeyer Brennholt E-AN. 2012. Pathogenic Vibrio species in German coastal waters of the North Sea and the Baltic Sea—a comparison. In German Fed. Inst. Hydrol. Int. Symp. Veranstaltungen 4/2012:36-42 doi: 10.5675/BfG_Veranst_2012.4 [DOI] [Google Scholar]
- 18. DePaola A, Nordstrom JL, Dalsgaard A, Forslund A, Oliver JD, Bates T, Bourdage KL, Gulig PA. 2003. Analysis of Vibrio vulnificus from market oysters and septicemia cases for virulence markers. Appl. Environ. Microbiol. 69:4006–4011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bisharat N, Cohen DI, Maiden MC, Crook DW, Peto T, Harding RM. 2007. The evolution of genetic structure in the marine pathogen, Vibrio vulnificus. Infect. Genet. Evol. 7:685–693 [DOI] [PubMed] [Google Scholar]
- 20. Gulig PA, Crecy-Lagard VD, Wright C, Walts B, Telonis-Scott M, McIntyre LM. 2010. SOLiD sequencing of four Vibrio vulnificus genomes enables comparative genomic analysis and identification of candidate clade-specific virulence genes. BMC Genomics 11:512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cohen ALV, Oliver JD, DePaola A, Feil EJ, Boyd EF. 2007. Emergence of a virulent clade of Vibrio vulnificus and correlation with the presence of a 33-kilobase genomic island. Appl. Environ. Microbiol. 73:5553–5565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Broza YY, Raz N, Lerner L, Danin-Poleg Y, Kashi Y. 2012. Genetic diversity of the human pathogen Vibrio vulnificus: a new phylogroup. Int. J. Food Microbiol. 153:436–443 [DOI] [PubMed] [Google Scholar]
- 23. Aznar R, Ludwig W, Amann RI, Schleifer KH. 1994. Sequence determination of rRNA genes of pathogenic Vibrio species and whole-cell identification of Vibrio vulnificus with rRNA-targeted oligonucleotide probes. Int. J. Syst. Bacteriol. 44:330–337 [DOI] [PubMed] [Google Scholar]
- 24. Nilsson WB, Paranjype RN, DePaola A, Strom MS. 2003. Sequence polymorphism of the 16S rRNA gene of Vibrio vulnificus is a possible indicator of strain virulence. J. Clin. Microbiol. 41:442–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vickery MCL, Nilsson WB, Strom MS, Nordstrom JL, DePaola A. 2007. A real-time PCR assay for the rapid determination of 16S rRNA genotype in Vibrio vulnificus. J. Microbiol. Methods 68:376–384 [DOI] [PubMed] [Google Scholar]
- 26. Rosche TM, Yano Y, Oliver JD. 2005. A rapid and simple PCR analysis indicates there are two subgroups of Vibrio vulnificus which correlate with clinical or environmental isolation. Microbiol. Immunol. 49:381–389 [DOI] [PubMed] [Google Scholar]
- 27. Sanjuan E, Fouz B, Oliver JD, Amaro C. 2009. Evaluation of genotypic and phenotypic methods to distinguish clinical from environmental Vibrio vulnificus strains. Appl. Environ. Microbiol. 75:1604–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Roig FJ, Sanjuan E, Llorens A, Amaro C. 2010. pilF polymorphism-based PCR to distinguish Vibrio vulnificus strains potentially dangerous to public health. Appl. Environ. Microbiol. 76:1328–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Morrison SS, Williams T, Cain A, Froelich B, Taylor C, Baker-Austin C, Verner-Jeffreys D, Hartnell R, Oliver JD, Gibas CJ. 2012. Pyrosequencing-based comparative genome analysis of Vibrio vulnificus environmental isolates. PLoS One 7:e37553 doi: 10.1371/journal.pone.0037553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim IH, Kim BS, Lee KS, Kim IJ, Son JS, Kim KS. 2011. Identification of virulence factors in Vibrio vulnificus by comparative transcriptomic analyses between clinical and environmental isolates using cDNA microarray. J. Microbiol. Biotechnol. 21:1228–1235 [DOI] [PubMed] [Google Scholar]
- 31. Thiaville PC, Bourdage KL, Wright AC, Farrell-Evans M, Garvan CW, Gulig PA. 2011. Genotype is correlated with but does not predict virulence of Vibrio vulnificus biotype 1 in subcutaneously inoculated, iron dextran-treated mice. Infect. Immun. 79:1194–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Abbott SL, Janda JM, Johnson JA, Farmer JJ., III 2007. Vibrio and related organisms, p 723–733 In Murray PR, Baron EJ, Jorgensen JH, Landry ML, Pfaller MA. (ed), Manual of clinical microbiology, 9th ed ASM Press, Washington, DC [Google Scholar]
- 33. Bauer A, Roervik LM. 2007. A novel multiplex PCR for the identification of Vibrio parahaemolyticus, Vibrio cholerae and Vibrio vulnificus. Lett. Appl. Microbiol. 45:371–375 [DOI] [PubMed] [Google Scholar]
- 34. Bisharat N, Cohen DI, Harding RM, Falush D, Crook DW, Peto T, Maiden MC. 2005. Hybrid Vibrio vulnificus. Emerg. Infect. Dis. 11:30–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jolley KA, Chan MS, Maiden MCJ. 2004. mlstdbNet—distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics 5:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452 [DOI] [PubMed] [Google Scholar]
- 38. Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254–267 [DOI] [PubMed] [Google Scholar]
- 39. Dieckmann R, Strauch E, Alter T. 2010. Rapid identification and characterization of Vibrio species using whole-cell MALDI-TOF mass spectrometry. J. Appl. Microbiol. 109:199–211 [DOI] [PubMed] [Google Scholar]
- 40. Moll A, Manning PA, Timmis KN. 1980. Plasmid-determined resistance to serum bactericidal activity: a major outer membrane protein, the traT gene product, is responsible for plasmid-specified serum resistance in Escherichia coli. Infect. Immun. 28:359–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Moll A, Cabello F, Timmis KN. 1979. Rapid assay for the determination of bacterial resistance to the lethal activity of serum. FEMS Microbiol. Lett. 6:273–276 [Google Scholar]
- 42. Gonzalez-Escalona N, Whitney B, Jaykus LA, DePaola A. 2007. Comparison of direct genome restriction enzyme analysis and pulsed-field gel electrophoresis for typing of Vibrio vulnificus and their correspondence with multilocus sequence typing data. Appl. Environ. Microbiol. 73:7494–7500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chou LF, Peng HL, Yang YC, Kuo MC, Chang HY. 2009. Localization and characterization of VVA0331, a 489-kDa RTX-like protein, in Vibrio vulnificus YJ016. Arch. Microbiol. 191:441–450 [DOI] [PubMed] [Google Scholar]
- 44. Wong HC, Liu SH, Chen MY. 2005. Virulence and stress susceptibility of clinical and environmental strains of Vibrio vulnificus isolated from samples from Taiwan and the United States. J. Food Prot. 68:2533–2540 [DOI] [PubMed] [Google Scholar]
- 45. Lubin JB, Kingston JJ, Chowdhury N, Boyd EF. 2012. Sialic acid catabolism and transport gene clusters are lineage specific in Vibrio vulnificus. Appl. Environ. Microbiol. 78:3407–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jeong HG, Man HO, Byoung SK, Min YL, Ho JH, Sang HC. 2009. The capability of catabolic utilization of N-acetylneuraminic acid, a sialic acid, is essential for Vibrio vulnificus pathogenesis. Infect. Immun. 77:3209–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lewis AL, Lubin JB, Argade S, Naidu N, Choudhury B, Boyd EF. 2011. Genomic and metabolic profiling of nonulosonic acids in Vibrionaceae reveal biochemical phenotypes of allelic divergence in Vibrio vulnificus. Appl. Environ. Microbiol. 77:5782–5793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Drake SL, Whitney B, Levine JF, DePaola A, Jaykus LA. 2010. Correlation of mannitol fermentation with virulence-associated genotypic characteristics in Vibrio vulnificus isolates from oysters and water samples in the gulf of Mexico. Foodborne Pathog. Dis. 7:97–101 [DOI] [PubMed] [Google Scholar]
- 49. Froelich B, Oliver JD. 2011. Orientation of mannitol related genes can further differentiate strains of Vibrio vulnificus possessing the vcgC allele. Adv. Stud. Biol. 3:151–160 [Google Scholar]
- 50. Oberbeckmann S, Fuchs BM, Meiners M, Wichels A, Wiltshire KH, Gerdts G. 2012. Seasonal dynamics and modeling of a Vibrio community in coastal waters of the North Sea. Microb. Ecol. 63:543–551 [DOI] [PubMed] [Google Scholar]
- 51. Vimr ER, Kalivoda KA, Deszo EL, Steenbergen SM. 2004. Diversity of microbial sialic acid metabolism. Microbiol. Mol. Biol. Rev. 68:132–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Almagro-Moreno S, Boyd EF. 2009. Sialic acid catabolism confers a competitive advantage to pathogenic Vibrio cholerae in the mouse intestine. Infect. Immun. 77:3807–3816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Johnson DE, Calia FM. 1981. Hemolytic reaction of clinical and environmental strains of Vibrio vulnificus. J. Clin. Microbiol. 14:457–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Oliver JD, Wear JE, Thomas MB, Warner M, Linder K. 1986. Production of extracellular enzymes and cytotoxicity by Vibrio vulnificus. Diagn. Microbiol. Infect. Dis. 5:99–111 [DOI] [PubMed] [Google Scholar]
- 55. Hauk G, Duty O, Littmann M. 2010. Vibrio vulnificus in der Ostsee-klinische “Ausgangsfälle.” Messstellen und Messdaten. (Pathogene Vibrionen in der marinen Umwelt.) In German Fed. Inst. Hydrol. Int. Symp. Veranstaltungen 3/2010:23–30 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.