Abstract
We investigated the relationship between the genetic diversity of indigenous soybean-nodulating bradyrhizobia and their geographical distribution in the United States using nine soil isolates from eight states. The bradyrhizobia were inoculated on three soybean Rj genotypes (non-Rj, Rj2Rj3, and Rj4). We analyzed their genetic diversity and community structure by means of restriction fragment length polymorphisms of PCR amplicons to target the 16S-23S rRNA gene internal transcribed spacer region, using 11 USDA Bradyrhizobium strains as reference strains. We also performed diversity analysis, multidimensional scaling analysis based on the Bray-Curtis index, and polar ordination analysis to describe the structure and geographical distribution of the soybean-nodulating bradyrhizobial community. The major clusters were Bradyrhizobium japonicum Bj123, in the northern United States, and Bradyrhizobium elkanii, in the middle to southern regions. Dominance of bradyrhizobia in a community was generally larger for the cluster belonging to B. elkanii than for the cluster belonging to B. japonicum. The indigenous American soybean-nodulating bradyrhizobial community structure was strongly correlated with latitude. Our results suggest that this community varies geographically.
INTRODUCTION
The United States is the world's largest producer and consumer of soybeans (Glycine max [L.] Merr.), which are utilized for food products, feedstuff, and biofuel feedstock. Soybean is a legume that forms root nodules after infection with soybean-nodulating rhizobia, which perform symbiotic nitrogen fixation by taking up atmospheric nitrogen (as ammonia) through the root nodules. The major soybean-nodulating rhizobia are Bradyrhizobium japonicum, Bradyrhizobium elkanii, and Sinorhizobium (Ensifer) fredii (1–5). In addition, Bradyrhizobium yuanmingense, Bradyrhizobium liaoningense, Sinorhizobium xinjiangense, and Mesorhizobium tianshanense have been classified as soybean-nodulating rhizobia (6–11).
Inoculation of soybean with bradyrhizobia can improve nitrogen fixation, resulting in increased soybean yield. However, the efficiency of the inoculum may be poor if the inoculum cannot compete with indigenous soybean-nodulating rhizobia in the soil or cannot establish an efficient symbiosis with the host plants. To solve this problem, it will be necessary to understand the ecology of indigenous soybean-nodulating rhizobia in terms of their genetic diversity, geographical distribution, compatibility with the host soybean, and the environmental factors associated with the localization and dominance of the rhizobial strains in the soil.
Saeki et al. (12) investigated the genetic diversity and geographical distribution of indigenous soybean-nodulating rhizobia isolates from five sites in Japan (Hokkaido, Fukushima, Kyoto, Miyazaki, and Okinawa) by analyzing PCR restriction fragment length polymorphisms (PCR-RFLP) of the 16S-23S rRNA gene internal transcribed spacer (ITS) region. They reported that a geographical distribution of indigenous bradyrhizobia varied from northern to southern Japan. Furthermore, Saeki et al. (13) reported that the distribution of soybean-nodulating rhizobia in Japan was strongly correlated with latitude (r2 = 0.924). The representative clusters of the isolated bradyrhizobia changed from those of B. japonicum strains USDA 123, 110, and 6T to B. elkanii strain USDA 76T, moving from northern to southern Japan (14). Adhikari et al. (15) revealed the genetic diversity of soybean-nodulating bradyrhizobia in relation to climate depending on altitude and soil properties, such as soil pH, in Nepal. Other researchers found that S. fredii strains were dominant in the alkaline soils of Vietnam and Okinawa, Japan (16, 17). These results suggest that a relationship exists between the geographic distribution of indigenous soybean-nodulating rhizobia, soil temperature (and its variations due to latitude and altitude), and soil pH.
The ability of a soybean plant to host bradyrhizobia depends on the characteristics of nodulation regulatory genes (Rj genes), and the Rj genotypes rj1, Rj2, Rj3, Rj4, and non-Rj, which lacks these genes, have been confirmed to exist in nature (18–21). Previous experimental results have also demonstrated that the community structure of soybean-nodulating bradyrhizobia depends on the host soybean Rj genotype and on the soybean cultivar, and it varies with cultivation temperature even in an identical soil sample (22, 23).
The United States is the world's biggest soybean producer, and soybean cultivars are grown at latitudes similar to those of the soybean production areas in Japan. Understanding the geographical distribution of soybean-nodulating rhizobia in the United States therefore would provide important knowledge about bradyrhizobial ecology and insights into appropriate inoculation techniques for soybean-nodulating rhizobia with high nitrogen fixation ability. Thus, in this study, we examined the relationship between the genetic diversity and geographical distribution of indigenous soybean-nodulating bradyrhizobia by isolating these organisms using three Rj genotype soybean cultivars from nine soil samples of American soybean fields. Our objectives were to investigate the genetic diversity, community structure, and geographic distribution of the bradyrhizobia by means of PCR-RFLP analysis of the 16S-23S rRNA gene ITS region. In addition, we described the community structure and geographical distribution of the bradyrhizobia using several tools from mathematical ecology.
MATERIALS AND METHODS
Soil samples.
We obtained soil samples for isolation of soybean-nodulating bradyrhizobia from nine experimental fields and farm fields in eight American states (Michigan, Ohio, Kentucky, North Carolina, Alabama, Georgia, Florida, and Louisiana) in August 2010 (Table 1). Three samples (each at least 200 cm3) were obtained from each field, to a depth of 10 cm, after removal of the surface litter. The samples were homogenized to produce a single composite sample. The Alabama soil sample was collected from two separate soybean fields, which we designated Alabama1 and Alabama2. Table 1 summarizes the location, soil pH, electrical conductivity (EC), total carbon (C) and total nitrogen (N) contents, and C/N ratio at these sites.
Table 1.
Soil sample sites for analysisa
| Sampling site | Soil group | Latitude and longitude | Change in latitude | pH (H2O) | EC (dS m−1) | C (%) | N (%) | C/N |
|---|---|---|---|---|---|---|---|---|
| Michigan (MI) | Loam | 43.05°N, −82.53°W | 12.84 | 7.68 | 0.15 | 2.03 | 0.18 | 11.3 |
| Ohio (OH) | Silt loam | 40.78°N, −81.93°W | 10.56 | 6.34 | 0.10 | 0.97 | 0.09 | 10.9 |
| Kentucky (KY) | Silt loam | 38.93°N, −86.47°W | 6.71 | 6.14 | 0.10 | 2.05 | 0.19 | 11.1 |
| North Carolina (NC) | Sandy loam | 35.79°N, −78.69°W | 5.57 | 5.23 | 0.06 | 1.37 | 0.12 | 11.9 |
| Alabama1 (AL1) | Loamy sand | 32.59°N, −85.49°W | 2.37 | 5.77 | 0.07 | 1.16 | 0.09 | 13.6 |
| Alabama2 (AL2) | Loamy sand | 32.59°N, −85.48°W | 2.37 | 5.18 | 0.04 | 0.91 | 0.06 | 15.2 |
| Georgia (GA) | Loamy sand | 31.48°N, −83.52°W | 1.26 | 5.70 | 0.03 | 0.45 | 0.03 | 17.3 |
| Florida (FL) | Coarse sand | 30.68°N, −85.31°W | 0.46 | 5.58 | 0.02 | 0.40 | 0.01 | 35.9 |
| Louisiana (LA) | Clay | 30.22°N, −91.10°W | 0 | 5.52 | 0.05 | 0.94 | 0.09 | 10.6 |
Identification of soil group refers to the following website: http://websoilsurvey.nrcs.usda.gov/app/WebSoilSurvey.aspx.
Isolation of the indigenous bradyrhizobia.
To isolate indigenous soybean-nodulating bradyrhizobia, we used three soybean cultivars of three Rj genotypes, Bragg as non-Rj, CNS as Rj2Rj3, and Hill as Rj4, and planted each soybean cultivar in 1-liter culture pots (n = 3 plants per cultivar). The culture pots were filled with vermiculite containing N-free nutrient solution (24) at 40% (vol/vol) water content and then were autoclaved at 121°C for 20 min. Soybean seeds were sterilized by being soaked in 70% ethanol for 30 s and in a dilute sodium hypochlorite solution (0.25% available chlorine) for 3 min. They were then washed with sterile distilled water. A soil sample (2 to 3 g) was placed in the vermiculite at a depth of 2 to 3 cm, the soybean seeds were sown on the soil, and then the pot weight was measured. The plants were grown for 4 weeks in a growth chamber (day, 28°C for 16 h; night, 23°C for 8 h), and sterile distilled water was supplied weekly until it reached the initial pot weight.
After 4 weeks, 23 to 24 nodules were randomly collected from the soybean roots and sterilized by soaking them in 70% ethanol for 3 min and in a diluted sodium hypochlorite solution (0.25% available chlorine) for 30 min; they were then washed with sterile distilled water. Each nodule was homogenized in sterile distilled water, streaked onto a yeast extract-mannitol agar (YMA) (25) plate medium, and incubated for 5 to 7 days in the dark at 28°C. To determine the genus of the isolates, a single colony was streaked onto YMA plate medium containing 0.002% (wt/wt) bromothymol blue (26) and incubated as described above. After incubation, each isolate was maintained on YMA slant medium at 4°C for further analysis. Sixy-nine to 72 isolates per soil sample were used to represent the soybean-nodulating bradyrhizobial community; we obtained a total of 645 isolates from the nine soybean fields and used them in the diversity analysis and multidimensional scaling (MDS) analysis described below.
Representative isolates in each operational taxonomic unit (OTU) of the dendrogram were confirmed for their nodulation capability on host soybean by inoculation test. Each isolate was cultured in yeast extract-mannitol broth culture (25) for 6 days at 28°C, and the cultures were then diluted with sterile distilled water to approximately 106 cells ml−1. The soybean seeds were sown into 500-ml prepared culture pots without soil, as described above, and inoculated with a 1-ml aliquot of each isolate per seed, with two or three replicates. We assessed nodule formation after 3 weeks in a growth chamber under the conditions described above.
PCR-RFLP analysis of the 16S-23S rRNA gene ITS region.
For DNA extraction, we cultured each isolate in 1.5 ml of HEPES-morpholineethanesulfonic acid (MES) medium (27) supplemented with 0.1% l-arabinose (28) for 5 days at 28°C. Total DNA was extracted from the isolates using BL extraction buffer, as described previously (22), based on the method reported by Hiraishi et al. (29). As reference strains, we used B. japonicum USDA strains 4, 6T, 38, 110, 115, 123, 124, and 135 and B. elkanii USDA strains 46, 76T, and 94 (30). Total DNAs of the reference strains were extracted by means of the same procedure as that used for the isolates.
The PCR amplification for the 16S-23S rRNA gene ITS region was carried out using Ex Taq DNA polymerase (TaKaRa Bio, Otsu, Shiga, Japan) and a previously developed ITS primer set (BraITS-F, 5′-GACTGGGGTGAAGTCGTAAC-3′; BraITS-R, 5′-ACGTCCTTCATCGCCTC-3′) (12). The PCR cycle consisted of a prerun at 94°C for 5 min, 30 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 1 min, with a final postrun extension at 72°C for 10 min.
The RFLP analysis of the ITS region was investigated using the restriction enzymes HaeIII, HhaI, MspI, and XspI (TaKaRa Bio). A 5-μl aliquot of the PCR product was digested with the restriction enzymes at 37°C for 16 h in a 20-μl reaction mixture. The restriction fragments were separated on 3 or 4% agarose gels by means of electrophoresis and visualized with ethidium bromide.
Cluster analysis of the indigenous soybean-nodulating rhizobia.
The fragment sizes on the electrophoresis gels were measured using a 50-bp reference ladder marker (TaKaRa Bio) and the fragment sizes from the sequences of the reference strains. All reproducible fragments longer than 50 bp were used for the cluster analysis (described below), and some irreproducible fragments were excluded. The genetic distance between pairs of isolates (D) was determined using the equation DAB = 1 − [2NAB/(NA + NB)], where NAB represents the number of RFLP bands shared by strains A and B and NA and NB represent the numbers of RFLP bands found only in strains A and B, respectively (31, 32). The cluster analysis was carried out using the unweighted pair-group method using average linkages (UPGMA). The dendrograms were constructed using version 3.69 of the PHYLIP software (J. Felsenstein, University of Washington, Seattle, WA; http://evolution.genetics.washington.edu/phylip.html).
Diversity analysis for the bradyrhizobial communities.
To estimate the diversity of the bradyrhizobial communities in the United States that we isolated from the host soybeans, we used the Shannon-Wiener diversity index (13, 33, 34), H′ = −∑ Pi ln Pi, where Pi is the dominance of dendrogram cluster i, defined as ni/N, where N is the total number of isolates (n = 69 or 72) and ni is the total number of tested isolates belonging to dendrogram cluster i. We also calculated the alpha diversity (H′α), beta diversity (H′β), and gamma diversity (H′γ) to estimate the differences in the bradyrhizobial communities between pairs of soil samples (35, 36). The H′α index represents a weighted average of the diversity indices of two bradyrhizobial communities, the H′β index represents the differences between the bradyrhizobial communities from two soil samples (i.e., differences between sites), and the H′γ index represents the diversity of the total isolate communities from the two soil samples (n = 141 or 144). The relationship among these indices is H′β = H′γ − H′α.
We also estimated the differences among the compositions of the bradyrhizobial communities by comparing the ratio of beta to gamma diversity (H′β/H′γ), taking into consideration the difference in gamma diversity in each pairwise comparison of bradyrhizobial communities.
Multidimensional scaling analysis, cluster analysis, and polar ordination analysis. To describe the characteristics of the bradyrhizobial communities and the differences among field sample sites, we performed an MDS analysis based on the Bray-Curtis similarity measure. The Bray-Curtis similarity measure has a robust monotonic relationship with ecological distance and a robust linear relationship with ecological distance until large values of the distance. Thus, the Bray-Curtis similarity measure (BC) is one of the indices that best reflect the properties between communities (37). The Bray-Curtis similarity measure was calculated using the equation BCAB = ∑ ΣnA − nBΣ/∑ (nA + nB), where BCAB is the dissimilarity between communities A and B and nA and nB represent the total number of strains in a particular cluster for communities A and B, respectively (38, 39). The three-dimensional MDS analysis and the UPGMA analysis based on the Bray-Curtis similarity measure were conducted using version 2.15.1 of the R software (http://www.r-project.org/). Furthermore, UPGMA analysis was conducted as described above for all communities as OTUs with Bray-Curtis similarity to obtain the objective index for the clustering of MDS plots.
To determine the relative distances among the diversities of the bradyrhizobial communities based on the three-dimensional MDS plots as a function of latitude (°N), we calculated the Euclidean distances between the bradyrhizobial communities. The distances between the MDS plot were calculated using the coordinates on the x, y, and z axes as the Euclidean distance (Ed) using the equation EdAB = (ΣXA − XBΣ2 + ΣYA − YBΣ2 + ΣZA − ZBΣ2)1/2, where EdAB is the linear distance between communities A and B in the MDS plot and XA and XB, YA and YB, and ZA and ZB represent the x, y, and z coordinates of communities A and B, respectively. The distances from each pole were converted into percent differences, D1 and D2, from the two polar communities (i.e., the Michigan and Louisiana sites, which were considered to have a 100% difference).
Polar ordination was conducted to determine the coordinates of communities on the axis between communities of the Michigan and Louisiana sites using the Pythagorean theorem (13, 35, 40). We constructed simultaneous equations from the trigonometric figure using the Pythagorean theorem as described previously (13). Parameter Pd represents the polar difference (%) from the 0% pole (the bradyrhizobial community of Louisiana) and is calculated as Pd = (L2 + D12 − D22)/2L, where D1 and D2 are the percent differences between a particular bradyrhizobial community and the communities at Louisiana and Michigan, respectively (40). The parameter L represents the total length of the polar axis (i.e., 100%).
RESULTS
Isolation of indigenous soybean-nodulating bradyrhizobia.
We obtained 23 or 24 isolates from each Rj soybean genotype and a total of 645 indigenous soybean-nodulating bradyrhizobia that could be used for further analysis. The indigenous bradyrhizobia isolated from each soil sample plus host soybean cultivar combination were labeled using a combination of the site abbreviation, an abbreviation for the three cultivars (B, Bragg; C, CNS; and H, Hill), and the number of the isolate (1-23 or 1-24) (e.g., for Michigan, MIB1-23, MIC1-23, and MIH1-23). The YMA cultures of all isolates turned blue as a result of the presence of bromothymol blue, indicating that all isolates belonged to the genus Bradyrhizobium (2). Representative isolates indicated their nodulation capability on the host soybean by the inoculation test.
PCR-RFLP analysis of the 16S-23S rRNA gene ITS region.
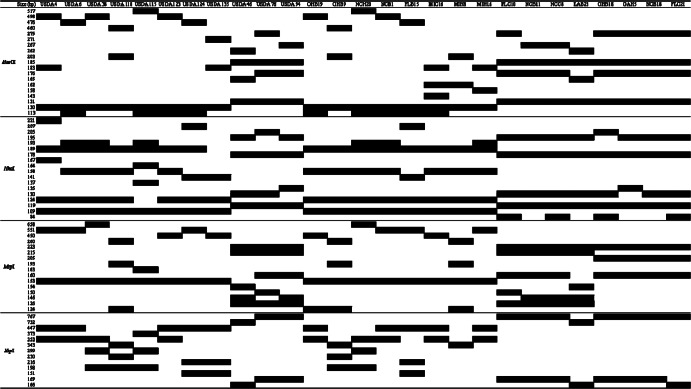
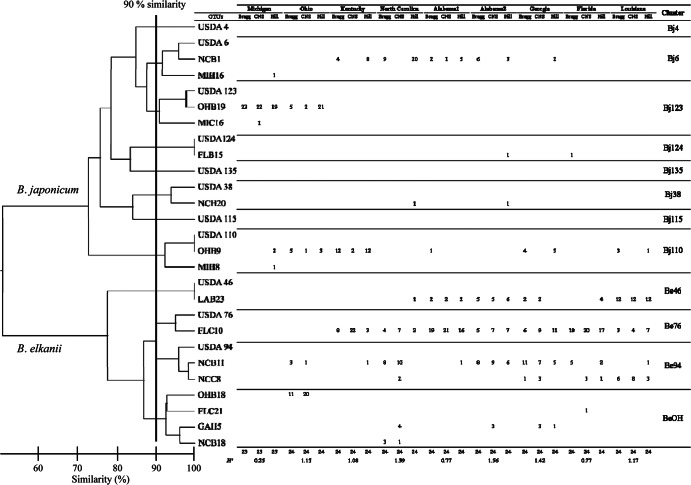
PCR products of the amplified 16S-23S rRNA gene ITS region of isolates were digested using four restriction enzymes, and the restriction fragments were separated by gel electrophoresis. Figure 1 provides a schematic representation of the restriction fragment patterns for each enzyme. The dendrogram was constructed based on the differences in fragment size and pattern shown in Fig. 1. Figure 2 presents the results of the cluster analysis based on PCR-RFLP data. We detected a total of 24 OTUs containing 11 reference strains (Fig. 2). The maximum similarity between the OTUs and the reference strains was 90%, and it occurred between OTUs USDA 76 and USDA 94. These results were then applied as the criterion for distinguishing clusters in the dendrogram, which produced 12 clusters, 11 of which included 1 of the reference strains. All indigenous bradyrhizobia were classified into 9 clusters: Bj6, Bj38, Bj110, Bj123, Bj124, Be46, Be76, Be94, and BeOH (Fig. 2). Three of the clusters included only a single reference strain and no indigenous bradyrhizobia. Bj6, Bj38, Bj110, Bj123, and Bj124 showed RFLP patterns identical or similar to those of B. japonicum strains USDA 6T, 38, 110, 115, 123, and 124, respectively. Be46, Be76, and Be94 showed RFLP patterns identical or similar to those of B. elkanii strains USDA 46, 76T, and 94, respectively. Cluster BeOH consisted of isolates from five sites and was independent from those of the reference strains.
Fig 1.
Schematic representation of RFLP patterns of the 16S-23S rRNA gene ITS region. Reference strains and representative isolates are indicated: USDA 4, 6, 38, 110, 115, 123, 124, and 135 represent Bradyrhizobium japonicum USDA 4, 6T, 38, 110, 115, 123, 124, and 135, respectively, and USDA 46, 76, and 94 represent Bradyrhizobium elkanii USDA 46, 76T, and 94, respectively. MIC, MIH, OHB, NCB, NCC, NCH, GAH, FLB, FLC, and LAB represent Michigan-CNS, Michigan-Hill, Ohio-Bragg, North Carolina-Bragg, North Carolina-CNS, North Carolina-Hill, Georgia-Hill, Florida-Bragg, Florida-CNS, and Louisiana-Bragg, respectively.
Fig 2.
Dendrogram of the 16S-23S rRNA gene ITS region of indigenous soybean-nodulating bradyrhizobia and Bradyrhizobium USDA reference strains. The similarity between Bradyrhizobium elkanii USDA 76T and 94 (which was 90%) was applied as the criterion to differentiate the clusters. Clusters are indicated on the right. The diversity index (H′) was calculated using the following equation: H′ = −Σ Pi ln Pi.
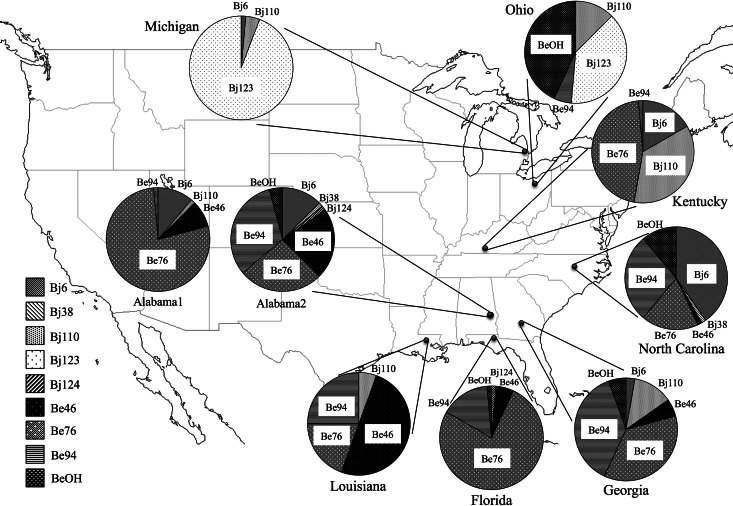
Figure 3 shows the geographical distribution of the soybean-nodulating bradyrhizobial isolates that belonged to each cluster. The isolates belonging to Bj123 were isolated from northern regions (Michigan and Ohio), whereas those belonging to Bj6 were found in the middle regions (Kentucky, North Carolina, and Alabama), and those belonging to Be46 were found in southern regions (Georgia, Florida, and Louisiana). Furthermore, the southern sites were dominated by B. elkanii isolates belonging to clusters Be46, Be76, and Be94. The dominance of B. elkanii exceeded the dominance of B. japonicum in the middle to southern regions (Fig. 2 and 3).
Fig 3.
Distribution of clusters and the population ratio of indigenous soybean-nodulating bradyrhizobia in the United States (map from CraftMAP [http://www.craftmap.box-i.net]).
Diversity analysis for the indigenous bradyrhizobial communities.
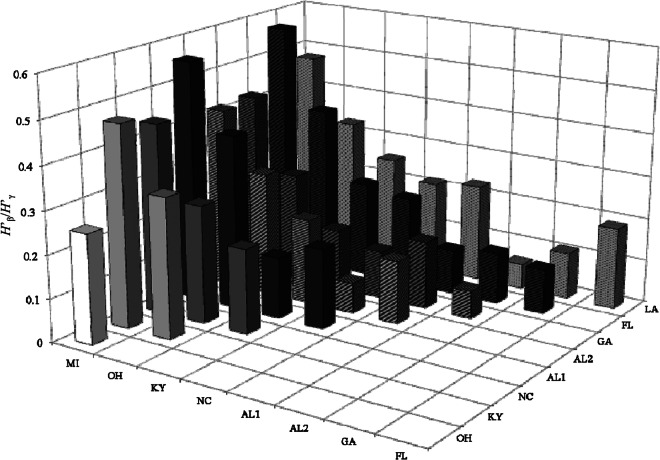
The differences in the indigenous bradyrhizobial communities among the nine sample sites were estimated based on the H′β/H′γ ratio. The diversity index (H′) values are indicated at the bottom of Fig. 2, and the beta diversity ratio (H′β/H′γ) at each field site is shown in Fig. 4. As shown in Fig. 2, in the indigenous American bradyrhizobial communities, Alabama2 had the highest H′ value (1.56), whereas Michigan had the lowest diversity index (0.25). The values of H′β/H′γ were largest for comparisons of the northern regions (Michigan and Ohio) to the other field sites (Fig. 4).
Fig 4.
Ratio of beta diversity to gamma diversity (H′β/H′γ) among pairs of soil sampling sites.
Multidimensional scaling analysis, UPGMA analysis, and polar ordination analysis.
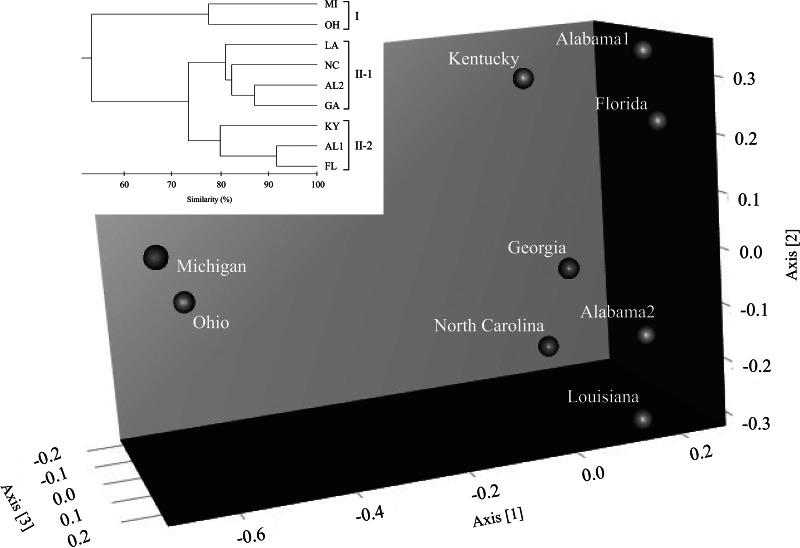
Figure 5 shows the results of the MDS analysis. The MDS plots showed three groups of bradyrhizobial communities. Based on UPGMA analysis, group I comprised Michigan and Ohio, group II-1 comprised North Carolina, Alabama2, Georgia, and Louisiana, and group II-2 comprised Kentucky, Alabama1, and Florida. Based on results shown in Fig. 3, the dominant region of the B. japonicum cluster belonged to group I, the region with high diversity in the B. elkanii cluster belonged to group II-1, and the dominant region for the Be76 cluster belonged to group II-2.
Fig 5.
Plots of indigenous soybean-nodulating bradyrhizobial communities of the soil sampling sites by 3-dimensional scaling analysis based on the Bray-Curtis index and dendrogram from UPGMA analysis. The dendrogram was applied as the objective index for grouping of MDS plots.
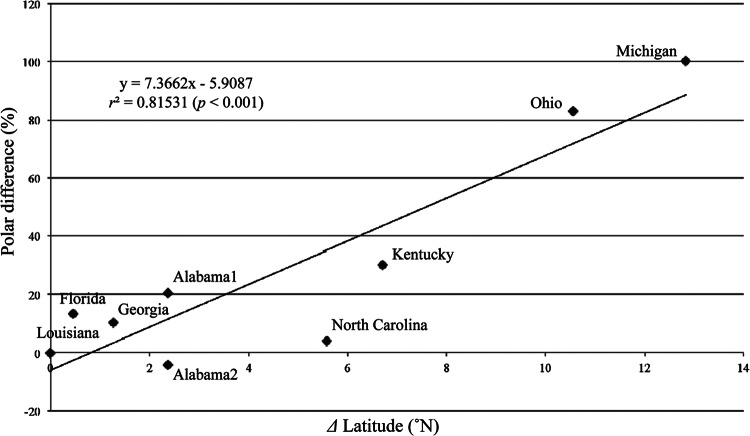
Figure 6 shows the results of the polar ordination analysis based on the percent differences of the bradyrhizobial communities from each pole and the differences in the latitudes of the sample sites. The results of the polar ordination indicate that the transition in the composition of the indigenous soybean-nodulating bradyrhizobial communities was strongly and significantly related to latitude (r2 = 0.815, F = 30.9, P < 0.001).
Fig 6.
Relationship between indigenous soybean-nodulating bradyrhizobial community and latitude of the soil-sampling site.
DISCUSSION
The American soils from which we isolated soybean-nodulating bradyrhizobia in the present study were acidic to slightly alkaline (pH 5.18 to 7.68; Table 1), and the cluster analysis based on PCR-RFLP data shows that strains of both B. japonicum and B. elkanii were isolated from nine field soils in the United States (Fig. 2). The major clusters from the sample soils were Bj123 in the northern region and Be46, Be76, and Be94 in the middle to southern regions (Fig. 3). Bj6 and Bj110 were moderately dominant in the middle regions. On the whole, the number of clusters of B. elkanii strains was larger than the number of clusters of B. japonicum strains (Fig. 2 and 3). Cluster Be76 was especially dominant in most sample soils in the middle to southern regions. Keyser et al. (41) investigated the distribution of Bradyrhizobium serogroups, which were determined using rabbit antisera prepared against 15 serotype strains in 12 states (Arkansas, Delaware, Florida, Kansas, Louisiana, Minnesota, Mississippi, New Jersey, North Carolina, Pennsylvania, South Carolina, and South Dakota) and found that the dominant serogroup was serogroup 31, which belongs to B. elkanii USDA 31 (21.5%), followed by serogroup 123, which belongs to B. japonicum USDA 123 (13.6%), and serogroup 76, which belongs to B. elkanii USDA 76T (10.2%). Interestingly, this report also indicated that B. elkanii was more dominant than B. japonicum, which confirms the results of the PCR-RFLP analysis of the 16S-23S rRNA gene ITS region in the present study; in addition, the distribution of the major serogroups generally agreed with the distribution of the major clusters in the present study. Additionally, Fuhrmann (42) investigated the diversity and symbiotic effectiveness of indigenous soybean-nodulating bradyrhizobia isolated from 18 locations in Delaware using serological, morphological, rhizobitoxine, and hydrogenase phenotypes, and they revealed that the dominant serogroup of indigenous bradyrhizobia was serogroup 94, which belongs to B. elkanii USDA 94 (17.5%), followed by serogroup 6, which belongs to B. japonicum USDA 6T (10.3%), serogroup 122, which belongs to B. japonicum USDA 122 (8.6%), and serogroup 76, which belongs to B. elkanii USDA 76T (5.8%). On the other hand, the grouping of the Bradyrhizobium USDA strains by sequence analysis and PCR-RFLP targeted to the 16S-23S rRNA gene ITS region used in this study revealed that B. japonicum USDA 110 and USDA 122 and B. elkanii USDA 31 and USDA 76T belong to the same clusters (30, 43). Therefore, to distinguish between B. japonicum USDA 110 and USDA 122 and between B. elkanii USDA 31 and USDA 76T, a characterization method based on gene markers such as those for housekeeping genes and/or single-nucleotide polymorphism of the ITS region will be necessary, because some of the isolates belonging to the Bj110 and Be76 cluster in the present study may be included in the serogroup of B. japonicum USDA 122 and B. elkanii USDA 31.
Our diversity analysis (Fig. 2) revealed that the sampling site with the highest diversity index (H′) was Alabama2 (1.56), and that Michigan had the lowest diversity index (0.25). The value of H′β/H′γ was largest for the comparison of the two northern regions (Michigan and Ohio) to the other regions (Fig. 4). This indicates that the bradyrhizobial communities of soybean-nodulating bradyrhizobia differed greatly between the northern region and the other regions (Table 2 and Fig. 4). The Bj123 cluster was only dominant in the northern region (Michigan and Ohio), and the BeOH cluster that was dominant in Ohio may have affected the difference among the communities.
Table 2.
Alpha, beta, and gamma diversity indices for each soil sample site pair
| Index | MI |
OH |
KY |
KY |
NC |
AL1 |
AL2 |
GA |
FL |
|||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MI-OH | MI-KY | MI-NC | MI-AL1 | MI-AL2 | MI-GA | MI-FL | MI-LA | OH-KY | OH-NC | OH-AL1 | OH-AL2 | OH-GA | OH-FL | OH-LA | KY-NC | KY-AL1 | KY-AL2 | KY-GA | KY-FL | KY-LA | NC-AL1 | NC-AL2 | NC-GA | NC-FL | NC-LA | AL1-AL2 | AL1-GA | AL1-FL | AL1-LA | AL2-GA | AL2-FL | AL2-LA | GA-FL | GA-LA | FL-LA | |
| H′α | 0.71 | 0.68 | 0.84 | 0.52 | 0.92 | 0.85 | 0.52 | 0.72 | 1.12 | 1.27 | 0.96 | 1.36 | 1.28 | 0.96 | 1.16 | 1.24 | 0.92 | 1.32 | 1.25 | 0.93 | 1.13 | 1.08 | 1.48 | 1.41 | 1.08 | 1.28 | 1.16 | 1.09 | 0.77 | 0.97 | 1.49 | 1.17 | 1.37 | 1.09 | 1.29 | 0.97 |
| H′β | 0.24 | 0.60 | 0.66 | 0.65 | 0.67 | 0.63 | 0.69 | 0.66 | 0.54 | 0.48 | 0.65 | 0.54 | 0.44 | 0.60 | 0.56 | 0.30 | 0.15 | 0.33 | 0.20 | 0.26 | 0.39 | 0.24 | 0.11 | 0.18 | 0.28 | 0.35 | 0.20 | 0.20 | 0.09 | 0.29 | 0.10 | 0.16 | 0.08 | 0.13 | 0.16 | 0.23 |
| H′γ | 0.95 | 1.28 | 1.50 | 1.17 | 1.59 | 1.48 | 1.21 | 1.38 | 1.65 | 1.75 | 1.61 | 1.90 | 1.72 | 1.56 | 1.73 | 1.54 | 1.08 | 1.65 | 1.45 | 1.19 | 1.51 | 1.32 | 1.59 | 1.58 | 1.37 | 1.63 | 1.36 | 1.29 | 0.86 | 1.25 | 1.59 | 1.32 | 1.45 | 1.22 | 1.45 | 1.20 |
The results of the geographical distribution analysis using polar ordination (Fig. 6) show a strong correlation between the latitude of the samples and the community structure (r2 = 0.815). However, the correlation between the community structure in Japan and latitude was higher (r2 = 0.924) (13). The major clusters of indigenous soybean-nodulating bradyrhizobia in Japan were regularly distributed, with clusters in the order of B. japonicum strains USDA 123, USDA 110, and USDA 6T and B. elkanii strain USDA 76T moving from northern to southern regions, and was generally dominated by B. japonicum strains (12, 14). On the other hand, in the geographical distribution of the American soybean-nodulating bradyrhizobia, the B. elkanii clusters were more dominant than the B. japonicum clusters (Fig. 3), and the dominance of the Bj110 cluster in the middle regions was lower than that in Japan. Furthermore, the American and Japanese agricultural systems differ greatly, including different climates and soil types; most American agriculture is irrigated aerobic cultivation, whereas a major portion of Japanese agriculture is flooded rice cultivation on alluvial soils, although the aerobic cultivation is also major in soils such as Andosol. The difference in dominance of the Bj110 cluster between the United States and Japan might be affected by these factors. These results are one reason why the coefficient of determination for the relationship between community structure and latitude was lower in the United States than in Japan. However, since the transitions of Bj123, Bj110, Bj6, and Be clusters are in common with those of Japan, the present results clearly indicate that the indigenous soybean-nodulating bradyrhizobial community in the United States varies with latitude.
In the United States, the dominance of localized B. elkanii strains in the soil was high. Minamisawa et al. (44) investigated the preferential nodulation of soybean cultivars, a wild soybean progenitor (Glycine soja), and siratro (Macroptilium atropurpureum) by B. japonicum and B. elkanii strains and found that B. japonicum and B. elkanii preferentially nodulated G. max and M. atropurpureum, respectively, whereas both bradyrhizobial species formed nodules on G. soja with similar efficiency. Marr et al. (45) reported that Amphicarpaea bracteata performed nodule formation with B. japonicum and B. elkanii and performed nitrogen fixation with B. elkanii, though A. bracteata will not be the original host for B. elkanii. Furthermore, Tang et al. (46) investigated the microevolution and origins of Bradyrhizobium populations in eastern North America associated with soybean and native legumes (A. bracteata and Desmodium canadense) using genetic characterization by multilocus sequence typing of six core (housekeeping) gene sequences and two symbiotic gene sequences, and the results suggested that soybean-nodulating bacteria associated with native legumes represent a novel source of ecologically adapted bacteria for soybean inoculation. In addition, the Rj4 soybean genotype is known to produce weak nodulation with certain B. elkanii strains (e.g., USDA 61 and USDA 94) (47, 48). In fact, infection by bradyrhizobia belonging to the Be94 cluster isolated from Bragg (non-Rj) and CNS (Rj2Rj3) cultivars in Ohio and North Carolina, which produced high dominance of B. japonicum, was inhibited by the soybean cultivar with the Rj4 genotype (Fig. 2). These results suggest that the interaction between host plants and rhizobia involved both host specificity and host preference. Further analysis of phenotypic traits of isolates, such as compatibility and preference with Rj genotypes, and genetic diversity of other genes of isolates, such as nodulation genes and housekeeping genes, must be conducted for elucidation of ecological, evolutionary, and phylogenetic relationships.
Siratro is a major pasture legume that is cultivated in the tropics and subtropics, including parts of Australia, South and Central America, and some Pacific islands (49). Additionally, A. bracteata is an annual legume that distributes widely in eastern North America (50, 51). The presence of these legumes that have compatibility for nodulation with B. elkanii might contribute strongly to the high dominance of B. elkanii in North America. We expect that further research on the competition between inoculated and indigenous bradyrhizobia and the environmental adaptability of bradyrhizobia will lead to the establishment of improved inoculation techniques based on the use of the most effective bradyrhizobium for a given host and region. The knowledge of the geographical distribution of indigenous soybean-nodulating bradyrhizobia provided by the present study will help to guide that future research.
ACKNOWLEDGMENTS
This study was supported mainly by a Japan Society for the Promotion of Science KAKENHI grant (Grants-in-Aid for Scientific Research [C], no. 22580068) and partially by a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan (Genomics for Agricultural Innovation SOY-2001).
Footnotes
Published ahead of print 5 April 2013
REFERENCES
- 1. Chen WX, Yan GH, Li LJ. 1988. Numerical taxonomic study of fast-growing soybean rhizobia and a proposal that Rhizobium fredii be assigned to Sinorhizobium gen. nov. Int. J. Syst. Bacteriol. 38:392–397 [Google Scholar]
- 2. Jordan DC. 1982. Transfer of Rhizobium japonicum Buchanan 1980 to Bradyrhizobium gen. nov., a genus of slow-growing, root nodule bacteria from leguminous plants. Int. J. Syst. Bacteriol. 32:136–139 [Google Scholar]
- 3. Kuykendall LD, Saxena B, Cevine TE, Udell SE. 1992. Genetic diversity in Bradyrhizobium Jordan 1982 and a proposal for Bradyrhizobium elkanii sp. nov. Can. J. Microbiol. 38:501–505 [Google Scholar]
- 4. Scholla MH, Elkan HG. 1984. Rhizobium fredii sp.nov. a fast-growing species that effectively nodulates soybeans. Int. J. Syst. Bacteriol. 34:484–486 [Google Scholar]
- 5. Young JM. 2003. The genus name Ensifer Casida 1982 takes priority over Sinorhizobium Chen et al. 1988, and Sinorhizobium morelense Wang et al. 2002 is a later synonym of Ensifer adhaerens Casida 1982. Is the combination ‘Sinorhizobium adhaerens ’ (Casida 1982) Willems et al. 2003 legitimate? Request for an opinion. Int. J. Syst. Evol. Microbiol. 53:2107–2110 [DOI] [PubMed] [Google Scholar]
- 6. Chen W, Wang E, Wang S, Li Y, Chen X, Li Y. 1995. Characteristics of Rhizobium tianshanense sp. nov., a moderately and slowly growing root nodule bacterium isolated from an arid saline environment in Xinjiang, People's Republic of China. Int. J. Syst. Bacteriol. 45:153–159 [DOI] [PubMed] [Google Scholar]
- 7. Peng GX, Tan ZY, Wang ET, Reinhold-Hurek B, Chen WF, Chen WX. 2002. Identification of isolates from soybean nodules in Xinjiang region as Sinorhizobium xinjiangense and genetic differentiation of S. xinjiangense from Sinorhizobium fredii. Int. J. Syst. Evol. Microbiol. 52:457–462 [DOI] [PubMed] [Google Scholar]
- 8. Tan ZY, Xu XD, Wang ET, Gao JL, Martinez-Romero E, Chen WX. 1997. Phylogenetic and genetic relationships of Mesorhizobium tianshanense and related rhizobia. Int. J. Syst. Bacteriol. 47:874–879 [DOI] [PubMed] [Google Scholar]
- 9. Vinuesa P, Rojas-Jiménez K, Contreras-Moreira B, Mahna SK, Prasad BN, Moe H, Selvaraju SB, Thierfelder H, Werner D. 2008. Multilocus sequence analysis for assessment of the biogeography and evolutionary genetics of four Bradyrhizobium species that nodulate soybeans on the Asiatic continent. Appl. Environ. Microbiol. 74:6987–6996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xu ML, Ge C, Cui Z, Li J, Fan H. 1995. Bradyhizobium liaoningense sp. nov., isolated from the root nodules of soybeans. Int. J. Syst. Bacteriol. 45:706–711 [DOI] [PubMed] [Google Scholar]
- 11. Yao YZ, Kan FL, Wang ET, Wei GH, Chen X. 2002. Characterization of rhizobia that nodulate legume species of the genus Lespedeza and description of Bradyrhizobium yuanmingense sp. nov. Int. J. Syst. Evol. Microbiol. 52:2219–2230 [DOI] [PubMed] [Google Scholar]
- 12. Saeki Y, Aimi N, Tsukamoto S, Yamakawa T, Nagatomo Y, Akao S. 2006. Diversity and geographical distribution of indigenous soybean-nodulating bradyrhizobia in Japan. Soil Sci. Plant Nutr. 52:418–426 [Google Scholar]
- 13. Saeki Y, Minami M, Yamamoto A, Akao S. 2008. Estimation of the bacterial community diversity of soybean-nodulating rhizobia isolated from Rj-genotype soybean. Soil Sci. Plant Nutr. 54:718–724 [Google Scholar]
- 14. Saeki Y. 2011. Characterization of soybean-nodulating rhizobial communities and diversity, p 163–184 In Aleksandra S. (ed), Soybean—molecular aspects of breeding. Intech, Rijeka, Croatia [Google Scholar]
- 15. Adhikari D, Kaneto M, Itoh K, Suyama K, Pokharel BB, Gaihre YK. 2012. Genetic diversity of soybean-nodulating rhizobia in Nepal in relation to climate and soil properties. Plant Soil 357:131–145 [Google Scholar]
- 16. Saeki Y, Kaneko A, Hara T, Suzuki K, Yamakawa T, Nguyen MT, Nagatomo Y, Akao S. 2005. Phylogenetic analysis of soybean-nodulating rhizobia isolated from alkaline soils in Vietnam. Soil Sci. Plant Nutr. 51:1043–1052 [Google Scholar]
- 17. Suzuki K, Oguro H, Yamakawa T, Yamamoto A, Akao S, Saeki Y. 2008. Diversity and distribution of indigenous soybean-nodulating rhizobia in the Okinawa Islands, Japan. Soil Sci. Plant Nutr. 54:237–246 [Google Scholar]
- 18. Caldwell BE. 1966. Inheritance of a strain-specific ineffective nodulation in soybean. Crop Sci. 6:427–428 [Google Scholar]
- 19. Vest G. 1970. Rj3—a gene conditioning ineffective nodulation in soybean. Crop Sci. 10:34–35 [Google Scholar]
- 20. Vest G, Caldwell EB. 1972. Rj4—a gene conditioning ineffective nodulation in soybean. Crop Sci. 12:692–693 [Google Scholar]
- 21. Williams LF, Lynch LD. 1954. Inheritance of a non-nodulating character in the soybean. Agron. J. 46:28–29 [Google Scholar]
- 22. Minami M, Yamakawa T, Yamamoto A, Akao S, Saeki Y. 2009. Estimation of nodulation tendency among Rj-genotype soybeans using the bradyrhizobial community isolated from an Andosol. Soil Sci. Plant Nutr. 55:65–72 [Google Scholar]
- 23. Shiro S, Yamamoto A, Umehara Y, Hayashi M, Yoshida N, Nishiwaki A, Yamakawa T, Saeki Y. 2012. Effect of Rj genotype and cultivation temperature on the community structure of soybean-nodulating bradyrhizobia. Appl. Environ. Microbiol. 78:1243–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Saeki Y, Akagi I, Takaki H, Nagatomo Y. 2000. Diversity of indigenous Bradyrhizobium strains isolated from three different Rj-soybean cultivars in terms of randomly amplified polymorphic DNA and intrinsic antibiotic resistance. Soil Sci. Plant Nutr. 46:917–926 [Google Scholar]
- 25. Vincent JM. 1970. A manual for the practical study of the root-nodule bacteria. International Biological Program. Blackwell Scientific, Oxford, United Kingdom [Google Scholar]
- 26. Keyser HH, Bohlool BB, Hu TS, Weber DF. 1982. Fast-growing rhizobia isolated from root nodules of soybean. Science 215:1631–1632 [DOI] [PubMed] [Google Scholar]
- 27. Cole MA, Elkan GH. 1973. Transmissible resistance to penicillin G, neomycin & chloramphenicol in Rhizobium japonicum. Antimicrob. Agents Chemother. 4:248–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sameshima R, Iwasa T, Sadowsky MJ, Hamada T, Kasai H, Shutsrirung A, Mitsui H, Minamisawa K. 2003. Phylogeny and distribution of extra-slow-growing Bradyrhizobium japonicum harboring high copy numbers of RSα, RSβ and IS1631. FEMS Microbiol. Ecol. 44:191–202 [DOI] [PubMed] [Google Scholar]
- 29. Hiraishi A, Kamagata Y, Nakamura K. 1995. Polymerase chain reaction amplification and restriction fragment length polymorphism analysis of 16S rRNA genes from methanogens. J. Ferment. Bioeng. 79:523–529 [Google Scholar]
- 30. Saeki Y, Aimi N, Hashimoto M, Tsukamoto S, Kaneko A, Yoshida N, Nagatomo Y, Akao S. 2004. Grouping of Bradyrhizobium USDA strains by sequence analysis of 16S rDNA and 16S–23S rDNA internal transcribed spacer region. Soil Sci. Plant Nutr. 50:517–525 [Google Scholar]
- 31. Nei M, Li HW. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. U. S. A. 76:5269–5273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sakai M, Futamata H, Kim SJ, Matuguchi T. 1998. Effect of soil salinity on population structure of fluorescent pseudomonads in spinach rhizosphere. Soil Sci. Plant Nutr. 44:701–705 [Google Scholar]
- 33. MacArthur RH. 1965. Patterns of species diversity. Biol. Rev. 40:510–533 [Google Scholar]
- 34. Pielou EC. 1969. Ecological diversity and its measurement. An introduction to mathematical ecology, p 221–235 Wiley Interscience, New York, NY [Google Scholar]
- 35. Kobayashi S. 1995. Multivariate analysis of biological communities. Soju Shobo, Tokyo, Japan [Google Scholar]
- 36. Whittaker RH. 1972. Evolution and measurement of species diversity. Taxon 21:213–251 [Google Scholar]
- 37. Faith PD, Minchin PR, Belbin L. 1987. Compositional dissimilarity as a robust measure of ecological distance. Vegetatio 69:57–68 [Google Scholar]
- 38. Bray JR, Curtis TJ. 1957. An ordination of the upland forest communities of southern Wisconsin. Ecol. Monogr. 27:325–349 [Google Scholar]
- 39. Michie GM. 1982. Use of the Bray-Curtis similarity measure in cluster analysis of foraminiferal date. Math. Geol. 14:661–667 [Google Scholar]
- 40. Whittaker RH. 1967. Gradient analysis of vegetation. Biol. Rev. 49:207–264 [DOI] [PubMed] [Google Scholar]
- 41. Keyser HH, Weber DF, Uratsu SL. 1984. Rhizobium japonicum serogroup and hydrogenase phenotype distribution in 12 states. Appl. Environ. Microbiol. 47:613–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fuhrmann J. 1990. Symbiotic effectiveness of indigenous soybean bradyrhizobia as related to serological, morphological, rhizobitoxine, and hydrogenase phenotypes. Appl. Environ. Microbiol. 56:224–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. van Berkum P, Fuhrmann JJ. 2000. Evolutionary relationships among the soybean bradyrhizobia reconstructed from 16S rRNA gene and internally transcribed spacer region sequence divergence. Int. J. Syst. Evol. Microbiol. 50:2165–2172 [DOI] [PubMed] [Google Scholar]
- 44. Minamisawa K, Onodera S, Tanimura Y, Kobayashi N, Yuhashi K, Kubota M. 1997. Preferential nodulation of Glycine max, Glycine soja and Macroptilium atropurpureum by two Bradyrhizobium species japonicum and elkanii. FEMS Microbiol. Ecol. 24:49–56 [Google Scholar]
- 45. Marr DL, Devine TE, Parker MA. 1997. Nodulation restrictive genotypes of Glycine and Amphicarpaea: a comparative analysis. Plant Soil 189:181–188 [Google Scholar]
- 46. Tang J, Bromfield ESP, Rodrigue N, Cloutier S, Tambong JT. 2012. Microevolution of symbiotic Bradyrhizobium populations associated with soybeans in east North America. Ecol. Evol. 2:2943–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Devine ET, Kuykendall LD. 1996. Host genetic control of symbiosis in soybean (Glycine max L.). Plant Soil. 186:173–187 [Google Scholar]
- 48. Devine ET, Kuykendall LD, O'Neill JJ. 1990. The Rj4 allele in soybean represses nodulation by chlorosis-inducing bradyrhizobia classified as DNA homology group II by antibiotic resistance profiles. Theor. Appl. Genet. 80:33–37 [DOI] [PubMed] [Google Scholar]
- 49. Shaw HN, Whiteman PC. 1977. Siratro—a success story in breeding a tropical pasture legume. Trop. Grassl. 11:7–14 [Google Scholar]
- 50. Parker MA. 1991. Local genetic differentiation for disease resistance in a selfing annual. Biol. J. Linn. Soc. 42:337–349 [Google Scholar]
- 51. Turner BL, Fearing OS. 1964. A taxonomic study of the genus Amphicarpaea (Leguminosae). Southwest. Nat. 9:207–218 [Google Scholar]