Abstract
blaSHV genes from Escherichia coli and Salmonella enterica isolates from chicken (n = 19) and pork (n = 1) were identified as blaSHV-2 (n = 5) or blaSHV-2a (n = 15). Eighteen were on plasmids of the incI1 (n = 15), incP (n = 2), and incFIB (n = 1) incompatibility groups. These plasmids were all transferable by conjugation between E. coli and S. enterica.
TEXT
Although SHV enzymes remain among the major extended-spectrum β-lactamases (ESBLs) in bacteria from humans (1), data on their prevalence in bacteria from food animals are sparse (2), particularly in North America. Broad-spectrum cephalosporin resistance seen in bacteria from farm animals in North America, and Canada in particular, has been dominated by cephamycinase resistance determinants (3, 4). However, in addition to numerous blaCMY-2-positive isolates, we recently detected Escherichia coli and Salmonella enterica isolates carrying blaSHV genes in Canadian farm animals. Therefore, the objective of this study was to investigate blaSHV and associated plasmids from these E. coli and S. enterica isolates, obtained by the Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS) in 2006 to 2007 (5, 6), to provide a baseline for comparison with isolates from later years and of human origin.
PCR screening for blaSHV (7) was performed on 675 E. coli isolates (448, 187, and 40 isolates of chicken, porcine, and bovine origins, respectively) and 205 Salmonella enterica isolates (142, 63 isolates of chicken and porcine origins, respectively) resistant to ampicillin recovered by CIPARS in 2006 and 2007 from retail meat and abattoir cecal samples across Canada. Eighteen E. coli isolates (2.7%) of chicken (n = 17) and porcine (n = 1) origins and two S. enterica isolates (0.9%) of chicken origin were positive. These 20 isolates were screened for the presence of blaTEM and blaCMY-2 (7). One Salmonella isolate (SA18) was positive for blaTEM. The isolates were serotyped at the Laboratory for Food-Borne Zoonoses (LFZ; Guelph, Ontario, Canada) and tested in duplicate on different days by broth microdilution for susceptibility to a panel of 27 antimicrobial agents, including 16 β-lactams (8). Each isolate belonged to a different serotype. The two S. enterica isolates had the highest MICs for most β-lactams (Table 1).
Table 1.
β-Lactam MICs in the original E. coli and Salmonella isolates and in E. coli DH10B transformants and Salmonella Typhimurium LB500 transconjugants
| β-Lactama | MIC (μg/ml) forb: |
|||||||
|---|---|---|---|---|---|---|---|---|
| Original isolates |
Transformants |
Original isolates |
E. coli transformants | Salmonella Typhimurium transconjugants | ||||
| blaSHV-2 | blaSHV-2a | blaSHV-2 | blaSHV-2a | E. coli | Salmonella | |||
| CZO | 20 (≤8–32) | 16 (≤8–32) | 16 (≤8–32) | 32 (16–32) | 16 (≤8–32) | 32 (32–32) | 32 (≤8–32) | 32 (≤8–32) |
| FEP | ≤1 (≤1–≤1) | ≤1 (≤1–2) | ≤1 (≤1–2) | ≤1 (≤1–≤1) | ≤1 (≤1–≤1) | 2 (2–8) | ≤1 (≤1–2) | ≤1 (≤1–2) |
| CTX | 0.75 (0.25–4) | 1 (0.25–8) | 0.75 (0.25–1) | 2 (1–2) | 1 (0.25–4) | 8 (4–16) | 1 (0.25–2) | 4 (0.5–4) |
| FOX | ≤4 (≤4–16) | ≤4 (≤4–8) | 8 (≤4–8) | 8 (≤4–16) | ≤4 (≤4–16) | ≤4 (≤4–≤4) | 8 (≤4–16) | ≤4 (≤4–≤4) |
| CPD | 4 (1–16) | 4 (1–>32) | 6 (4–8) | 12 (4–32) | 4 (1–16) | 32 (32–64) | 8 (4–32) | 16 (4–32) |
| CAZ | 0.5 (0.25–1) | 0.5 (0.25–2) | 1 (0.25–2) | 2 (1–2) | 0.5 (0.25–2) | 4 (2–8) | 2 (0.5–2) | 2 (0.5–4) |
| CCV | ≤0.12 (≤0.12–≤0.12) | ≤0.12 (≤0.12–0.25) | 0.25 (0.25–0.25) | 0.25 (0.25–0. 5) | ≤0.12 (≤0.12–0.25) | 0.25 (≤0.12–0.5) | 0.25 (≤0.12–0.5) | 0.25 (≤0.12–0.25) |
| CRO | ≤1 (≤1–4) | ≤1 (≤1–8) | ≤1 (≤1–2) | 2 (≤1–4) | ≤1 (≤1–4) | 8 (4–16) | 1.5 (≤1–4) | 4 (≤1–8) |
CZO, cefazolin; FEP, cefepime; CTX, cefotaxime; FOX, cefoxitin; CPD, cefpodoxime; CAZ, ceftazidime; CCV, ceftazidime-clavulanic acid; CRO, ceftriaxone.
Numbers in parentheses represent the MIC ranges (μg/ml) observed in each respective group. Data for ampicillin, amoxicillin-clavulanic acid, piperacillin-tazobactam, cephalothin, cefotaxime-clavulanic acid, and carbapenems (imipenem and meropenem) are not shown because the MICs were monomorphic and outside the testing range.
Plasmid preparations from each original isolate (Qiagen plasmid midikit; Qiagen, Hilden, Germany) were electroporated into ElectroMAX E. coli DH10B cells (Invitrogen, Carlsbad, CA). Transformants were recovered with the plasmid preparations from 18 isolates on Mueller-Hinton agar containing 50 μg/ml ampicillin or 25 μg/ml tetracycline. However, despite repeated attempts, transformants were not recovered with plasmid preparations of two E. coli isolates. Southern blot hybridizations (9) of total DNA (DNeasy blood and tissue kit; Qiagen) and of plasmid preparations with a digoxigenin-labeled blaSHV probe (PCR DIG probe synthesis kit; Roche, Mannheim, Baden-Württemberg, Germany) confirmed the blaSHV from these isolates was chromosomal. DNA sequencing (described below) showed that these two isolates carried blaSHV-2a, and they were not investigated further. The blaSHV genes from the remaining 18 isolates were carried on plasmids of sizes ranging from approximately 95 kbp to 200 kbp.
DNA sequencing (Laboratory Services, University of Guelph) with a combination of PCR and primer walking and comparison with DNA sequences in NCBI's GenBank (10) and with SHV amino acid sequences (http://www.lahey.org/studies/webt.asp#SHV) was used to identify blaSHV variants. Five and 13 plasmids carried blaSHV-2 and blaSHV-2a, respectively. All blaSHV-2 and blaSHV-2a genes were identical to GenBank accession no. AF148851 and X53817, respectively (11), and matched the SHV-2 and SHV-2a amino acid sequences in the Lahey Clinic database. The median MICs of the original isolates for β-lactams were generally identical for SHV-2a and SHV-2. However, they were 1 dilution higher for SHV-2a than for SHV-2 with most β-lactams when transformants were compared (Table 1). Thus, the slight differences in β-lactam resistance phenotypes provided by the substitution between SHV-2 and SHV-2a and in their promoter regions may be detectable when expressed under a standardized genetic background, but not readily in natural bacterial populations.
As the two S. enterica isolates had median MICs at least 2 dilution steps higher than the E. coli isolates for most β-lactams (Table 1), blaSHV promoter regions of the two S. enterica isolates (SA17 and SA18) and of two E. coli isolates (EC7 and EC15) were characterized by PCR (12). The promoter regions for blaSHV-2 for E. coli EC7 and Salmonella SA18 were identical in sequence, and so were blaSHV-2a promoter sequences for E. coli EC15 and Salmonella SA17, thus suggesting the observed difference in MICs between bacterial host species was not due to promoter variants affecting SHV expression levels.
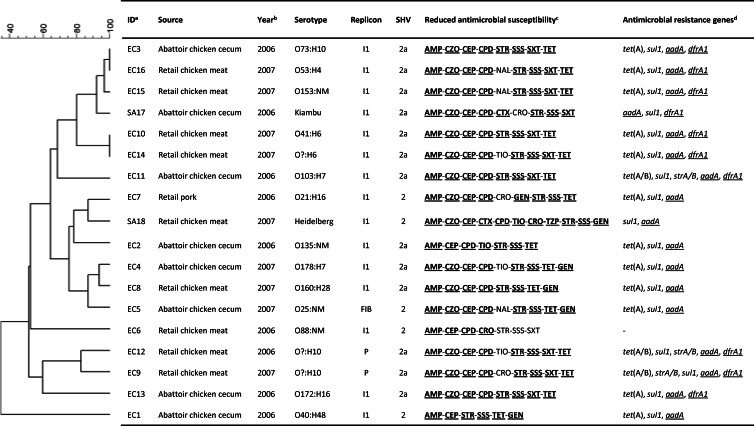
Although some minor variation resulting in seven antimicrobial susceptibility profiles was observed, transformants were similar overall, with most blaSHV plasmids associated with resistance to sulfonamides, tetracycline, streptomycin, and occasionally trimethoprim or gentamicin (Fig. 1). PCR screening (7) of transformants confirmed the presence of associated resistance genes (Fig. 1) for sulfonamides (n = 17), tetracycline (n = 15), and streptomycin (n = 17). PCR screening for intI1 and intI2 genes (13) indicated that 17 of the 18 blaSHV plasmids carried class 1 integrons. Sequencing of PCR products (14) confirmed integron variable regions of 1,200 bp (n = 10) and 900 bp (n = 7), consisting of dfrA1 for trimethoprim resistance plus aadA1 for streptomycin resistance or aadA1 alone (Fig. 1). The presence of these additional resistance determinants suggests a potential for coselection and persistence of blaSHV plasmids in the absence of cephalosporin use. The plasmids belonged to three different replicon types (2), but the vast majority were of the incI1 type (Fig. 1). EcoRI restriction analysis of transformant plasmids was conducted following standard protocols (3) and analyzed using BioNumerics v5.1 software (Applied Maths, Austin, TX). Only partial correlations between plasmid clusters, replicon types, antimicrobial susceptibility, integron size, antimicrobial resistance genes, and blaSHV variants were visible (Fig. 1), suggesting numerous recombinations in the history of these plasmids. However, the S. enterica isolates and some E. coli isolates shared highly related replicon type I1 plasmids, and two pairs of E. coli isolates (EC3 and EC16 and EC10 and EC14) from spatially and temporally distinct sources carried indistinguishable I1 plasmids (Fig. 1). This suggests the ability of blaSHV plasmids to move across bacterial populations and from commensal E. coli to Salmonella. To confirm this, the 2 S. enterica and 13 E. coli isolates were conjugated with a nalidixic acid-resistant derivative of S. enterica serovar Typhimurium strain LB5000 (15) by overnight mating in Luria-Bertani (LB) broth using a 1:1 ratio of donor and recipient cells. Transconjugants were selected using LB agar containing 50 μg/ml ampicillin and 50 μg/ml nalidixic acid. Three E. coli isolates (EC5, EC15, and EC16) were resistant to nalidixic acid and could not be tested. The blaSHV plasmids of all 15 isolates tested were transferable by conjugation. Efficiencies of 7.36 × 10−2 and 1.41 × 10−6 per donor, respectively, were recorded for the transfer of the blaSHV plasmid between two randomly selected E. coli isolates (EC2, replicon type I1, and EC9, replicon type P) and S. enterica serovar Typhimurium LB5000. Overall, these results demonstrate widespread horizontal transfer of blaSHV plasmids, in particular of replicon type incI1, rather than clonal spread of a few SHV-2- and SHV-2a-producing strains. Furthermore, these plasmids appear to move easily between E. coli and S. enterica.
Fig 1.
EcoRI restriction analysis of blaSHV plasmids and characteristics of blaSHV isolates. The scale on the top left of the figure indicates the percentage of similarity for the EcoRI restriction profiles of the blaSHV plasmids. Footnotes: a, isolate identification number (all isolates are E. coli, except SA17 and SA18, which are Salmonella enterica); b, year isolate was collected; c, antimicrobial for which the original isolates' MICs were in the intermediate or resistant range; d, antimicrobial resistance genes detected in transformants. Genes that were shown to be part of a class 1 integron variable region are underlined. Antimicrobial abbreviations: AMP, ampicillin; CEP, cephalothin; CPD, cefpodoxime; CRO, ceftriaxone; CTX, cefotaxime; CZO, cefazolin; GEN, gentamicin; NAL, nalidixic acid; SSS, sulfonamides; STR, streptomycin; SXT, sulfonamide-trimethoprim; TET, tetracycline; TIO, ceftiofur; TZP, piperacillin-tazobactam. Reduced susceptibilities that were transferred to the respective transformants are in boldface and underlined.
Previous reports have suggested that lower cephalosporin MIC values should be used for E. coli than for S. enterica to trigger investigations of the presence of ESBLs (16). Although not extensive enough for statistical evaluation, the results from the present study strongly support these findings in the case of blaSHV. The two S. enterica isolates had median MICs at least 2 dilution steps higher than those of the E. coli isolates for cefotaxime, ceftriaxone, ceftazidime, and cefpodoxime and 1 dilution step higher for most other β-lactams (Table 1). Similar trends were visible when comparing the blaSHV plasmids in the S. Typhimurium LB5000 host with the E. coli DH10B host or with the original E. coli isolates, whereas transfer from the original S. enterica isolate into S. Typhimurium LB5000 resulted in minor changes only (Table 1).
In conclusion, although their prevalence was low (3.2% and 0.4% of ampicillin-resistant isolates from chicken and porcine origins, respectively), the detection of plasmid-borne blaSHV-2 and blaSHV-2a in bacteria from Canadian food animals and retail meat with a high potential for horizontal transfer between E. coli and S. enterica is a public health concern. It is certainly not a coincidence that the first blaSHV characterized in a Canadian non-Typhi Salmonella isolate of human origin was also a blaSHV-2a (17). Despite their high mobility, the vast majority of blaSHV plasmids were found in bacteria originating from chicken. Further testing of 12 E. coli isolates from 2010 (9 chicken and 2 bovine isolates and 1 porcine isolate) and of two Salmonella isolates from 2008 (S. enterica serovar Kiambu from chicken) and 2010 (S. enterica serovar Derby from swine) from the CIPARS collection confirmed that blaSHV-2a (n = 11) and blaSHV-2 (n = 2) are still persisting in bacteria from food animals in Canada (P. Boerlin, G. Chalmers, and L. Martin, unpublished data), in particular from chickens. Concerns have been expressed previously regarding use of broad-spectrum cephalosporins and the presence of cephamycinases in E. coli and Salmonella isolates from chickens (4). The data presented here suggest that these concerns may also extend to the selection of ESBLs, such as blaSHV.
ACKNOWLEDGMENTS
This project was funded by the Public Health Agency of Canada.
We thank the abattoir and retail components of CIPARS, the LFZ OIE Reference Laboratory for Salmonellosis, and the LFZ CIPARS laboratories, Guelph, Ontario, Canada, and St. Hyacinthe, Quebec, Canada. Special thanks goes to Emily Weir, Laura Martin, Kim Ziebell, Gabhan Chalmers, and Charles Shearer for technical assistance with this study.
Footnotes
Published ahead of print 5 April 2013
REFERENCES
- 1. Bush K. 2008. Extended-spectrum beta-lactamases in North America, 1987–2006. Clin. Microbiol. Infect. 14(Suppl 1):134–143 [DOI] [PubMed] [Google Scholar]
- 2. Carattoli A. 2008. Animal reservoirs for extended spectrum beta-lactamase producers. Clin. Microbiol. Infect. 14(Suppl 1):117–123 [DOI] [PubMed] [Google Scholar]
- 3. Martin LC, Weir EK, Poppe C, Reid-Smith RJ, Boerlin P. 2012. Characterization of blaCMY-2 plasmids in Salmonella and Escherichia coli isolates from food animals in Canada. Appl. Environ. Microbiol. 78:1285–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dutil L, Irwin R, Finley R, Ng LK, Avery B, Boerlin P, Bourgault AM, Cole L, Daignault D, Desruisseau A, Demczuk W, Hoang L, Horsman GB, Ismail J, Jamieson F, Maki A, Pacagnella A, Pillai DR. 2010. Ceftiofur resistance in Salmonella enterica serovar Heidelberg from chicken meat and humans, Canada. Emerg. Infect. Dis. 16:48–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Government of Canada 2009. Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS) 2006. Public Health Agency of Canada, Guelph, Ontario, Canada [Google Scholar]
- 6. Government of Canada 2010. Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS) 2007. Public Health Agency of Canada, Guelph, Ontario, Canada [Google Scholar]
- 7. Kozak GK, Boerlin P, Janecko N, Reid-Smith RJ, Jardine C. 2009. Antimicrobial resistance in Escherichia coli isolates from swine and wild small mammals in the proximity of swine farms and in natural environments in Ontario, Canada. Appl. Environ. Microbiol. 75:559–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clinical and Laboratory Standards Institute 2007. Performance standards for antimicrobial susceptibility testing; seventeenth informational supplement. M100-S17 CLSI, Wayne, PA [Google Scholar]
- 9. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 10. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 11. Bradford PA. 1999. Automated thermal cycling is superior to traditional methods for nucleotide sequencing of blaSHV genes. Antimicrob. Agents Chemother. 43:2960–2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Podbielski A, Schonling J, Melzer B, Haase G. 1991. Different promoters of SHV-2 and SHV-2a beta-lactamase lead to diverse levels of cefotaxime resistance in their bacterial producers. J. Gen. Microbiol. 137:1667–1675 [DOI] [PubMed] [Google Scholar]
- 13. Sandvang D, Diggle M, Platt DJ. 2002. Translocation of integron-associated resistance in a natural system: acquisition of resistance determinants by Inc P and Inc W plasmids from Salmonella enterica Typhimurium DT104. Microb. Drug Resist. 8:151–160 [DOI] [PubMed] [Google Scholar]
- 14. Levesque C, Piche L, Larose C, Roy PH. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bullas LR, Ryu JI. 1983. Salmonella typhimurium LT2 strains which are r− m+ for all three chromosomally located systems of DNA restriction and modification. J. Bacteriol. 156:471–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Aarestrup FM, Hasman H, Veldman K, Mevius D. 2010. Evaluation of eight different cephalosporins for detection of cephalosporin resistance in Salmonella enterica and Escherichia coli. Microb. Drug Resist. 16:253–261 [DOI] [PubMed] [Google Scholar]
- 17. Mulvey MR, Soule G, Boyd D, Demczuk W, Ahmed R. 2003. Characterization of the first extended-spectrum beta-lactamase-producing Salmonella isolate identified in Canada. J. Clin. Microbiol. 41:460–462 [DOI] [PMC free article] [PubMed] [Google Scholar]