Abstract
The esterases and lipases from the α/β hydrolase superfamily exhibit an enormous sequence diversity, fold plasticity, and activities. Here, we present the comprehensive sequence and biochemical analyses of seven distinct esterases and lipases from the metagenome of Lake Arreo, an evaporite karstic lake in Spain (42°46′N, 2°59′W; altitude, 655 m). Together with oligonucleotide usage patterns and BLASTP analysis, our study of esterases/lipases mined from Lake Arreo suggests that its sediment contains moderately halophilic and cold-adapted proteobacteria containing DNA fragments of distantly related plasmids or chromosomal genomic islands of plasmid and phage origins. This metagenome encodes esterases/lipases with broad substrate profiles (tested over a set of 101 structurally diverse esters) and habitat-specific characteristics, as they exhibit maximal activity at alkaline pH (8.0 to 8.5) and temperature of 16 to 40°C, and they are stimulated (1.5 to 2.2 times) by chloride ions (0.1 to 1.2 M), reflecting an adaptation to environmental conditions. Our work provides further insights into the potential significance of the Lake Arreo esterases/lipases for biotechnology processes (i.e., production of enantiomers and sugar esters), because these enzymes are salt tolerant and are active at low temperatures and against a broad range of substrates. As an example, the ability of a single protein to hydrolyze triacylglycerols, (non)halogenated alkyl and aryl esters, cinnamoyl and carbohydrate esters, lactones, and chiral epoxides to a similar extent was demonstrated.
INTRODUCTION
Esterases and lipases from the α/β hydrolase family have received considerable attention, because they are widely distributed within the microbial communities operating in most of environments where they have important physiological functions (1) and because they are one of the most important groups of biocatalysts for biotechnological applications (2–4). Upon searching the list of genes using Pfam (the protein family database [5]) from the approximately 140 metagenomic projects in various stages of sequencing on the GOLD website (Genomes OnLine Database; http://www.genomesonline.org/) and the available sequences of esterases and lipases, more than 72,000 predicted esterases/lipases of the α/β hydrolase superfamily were retrieved, which revealed the richness of uncultured biodiversity (6), to provide wide collections of such biocatalysts. This is one of the largest protein families with available sequences. In relation to the cultivation-independent methods used to identify them, it should be highlighted that sequence-based metagenomics only provide the presumptive compositional and functional blueprint represented in the community genome (7, 8), but at the same time, this method causes serious problems regarding both sequencing errors (9) and the erroneous assignment of substrate specificity (10). In contrast, the activity-directed techniques have been shown to provide a direct view of known or new protein families and functionalities (for examples, see references 11–22, and 23). Whatever the platform used (i.e., either gene cloning from sequence resources or naïve activity screens), in many cases, enzyme properties have been shown to be modulated by ecosystem characteristics (24, 25).
From the whole set of sequences available, only a limited number of metagenomically derived esterases and lipases have been experimentally characterized (see Table S1 in the supplemental material), and only 27% of them (or approximately 60) have been shown to possess characteristics likely needed for industrial operations. In this respect, useful features, such as broad substrate specificity complemented with high activity levels under a wide range of thermal and pH conditions and salt concentrations, stability in organic solvents, and enantio- and stereoselectivity, are widely used for defining the potential application of esterases and lipases in biotechnology settings (4); however, not all characterized enzymes match these criteria (for example, see reference 26). Table S1 in the supplemental material provides an exhaustive list (with biochemical characteristics) of more than 200 different esterases/lipases from the α/β hydrolase superfamily which have been identified by metagenomic methods from various environments, including soils, compost piles, landfill leachate, bioreactors, activated sludges, marine water, and sediment samples (including tidal flat sediments, deep sea samples, and water columns) and freshwater samples (including drinking water, pond water, rivers, and hot springs).
In the present work, we identified and successfully cloned, expressed, purified, and characterized seven esterases/lipases from the α/β hydrolase superfamily via an activity-centered metagenome analysis from a microbial community of an evaporite karstic lake (Lake Arreo) in northwest Spain (42°46′N, 2°59′W; altitude, 655 m). Our findings point to the importance of evaporite karstic lakes as rich resources for novel low-temperature- and salt-adapted α/β hydrolases that may be useful for biotechnological applications because of their remarkable and broad substrate profiles. The substrate spectrum of newly isolated enzymes was compared to that of commercially available preparations, and structural models were also used to suggest molecular features that allow for the acceptance of different sets of substrates. Additionally, our work provided further insights into the placing of esterases/lipases at Lake Arreo by using genome linguistics, sequence similarity, and activity phenotyping.
MATERIALS AND METHODS
Materials, strains, and esterase/lipase preparations.
Chemicals, biochemicals, and solvents were purchased from Sigma Chemical Co. (St. Louis, MO), Aldrich (Oakville, Canada), or Fluka (Oakville, Canada) and were of pro-analysis quality. The oligonucleotides used for DNA amplification and sequencing were synthesized by Sigma Genosys Ltd. (Pampisford, Cambs, United Kingdom) and are provided in Table S2 in the supplemental material. The nickel-nitrilotriacetic acid (Ni-NTA) His · Bind chromatographic media were from Sigma Chemical Co. (St. Louis, MO). The Escherichia coli EPI300-T1R strain (Epicentre Biotechnologies; Madison, WI), used for the fosmid library construction and screening, and E. coli GigaSingles for the gene cloning and BL21(DE3) for the expression using the pET-46 Ek/LIC vector (Novagen, Darmstadt, Germany), were cultured and maintained according to the recommendations of the suppliers and standard protocols (27). Commercial esterase/lipase preparations Novozym 735 (lipase CalA from Candida antarctica), Novozym CALB L (lipase CalB from C. antarctica), Lipolase 100L (lipase from Thermomyces lanuginosa), and Lipozyme RM–Novozym 388L (lipase from Rhizomocur miehei) were provided by Novozymes A/S (Bagsvaerd, Denmark). Lipase from Alcaligenes sp. was kindly donated by Meito Sangyo Co. (Japan). Lipase from Rhizopus oryzae was obtained and prepared as described elsewhere (28).
Selection of hydrolases derived from the evaporite karstic lake (Lake Arreo) fosmid library.
A large-insert pCCFOS1 fosmid library was created from the DNA of a microbial community inhabiting sediment samples from an evaporite karstic lake (Lake Arreo). Sediment sampling (10 cm deep) was carried out in February 2007 with a sterilized 50-ml polypropilene Falcon tube in the eastern shallow part of the lake (see Fig. S1 in the supplemental material). The sample was maintained cold and dark during transportation to the laboratory, where it was frozen at −20°C until processing. Total DNA (5.2 μg DNA/g sediment) was extracted using the G'NOME DNA isolation kit (Qbiogene, Heidelberg, Germany). Purified and size-fractioned DNA was ligated into the pCCFOS fosmid vector and further cloned in E. coli EPI300-T1R according to the instructions of Epicentre Biotechnologies (WI) and a procedure described earlier (11). Fosmid clones (40,000; average insert size, 29.7 kbp) harboring approximately 1 Gbp of community genomes were arrayed and grown in 384-well microtiter plates containing Luria-Bertani (LB) medium with chloramphenicol (12.5 μg/ml) and 15% (vol/vol) glycerol and stored at −80°C.
Approximately 11,520 clones, plated onto small (12.5- by 12.5-cm) petri plates (each containing 96 clones) with LB agar containing chloramphenicol (12.5 μg/ml) and the induction solution (Epicentre Biotechnologies, WI), as recommended by the supplier to induce a high fosmid copy number, were screened with α-naphthyl acetate under previously described conditions (29). The positive clones were selected and their DNA inserts sequenced with a Roche 454 GS FLX Ti sequencer (454 Life Sciences, Branford, CT) at Life Sequencing SL (Valencia, Spain). Upon completion of sequencing, the reads were assembled to generate nonredundant metasequences using Newbler GS De Novo Assembler v.2.3 (Roche). GeneMark software (30) was employed to predict potential protein-coding regions (open reading frames [ORFs] with ≥20 amino acids) from the sequences of each assembled contig, and deduced proteins were screened via BLASTP and PSI-BLAST (31). Sequences are available at NCBI under accession number SRA059294.
Cloning, expression, and purification of selected proteins.
The cloning, expression, and purification of selected His6-tagged proteins in the Ek/LIC 46 vector and E. coli BL21 were performed as described elsewhere (32), except that protein expression was performed using 1.0 mM isopropyl-β-d-galactopyranoside for 16 h at 16°C. After purification using a Ni-NTA His · Bind resin (Sigma Chemical Co., St. Louis, MO), protein solutions were extensively dialyzed with 20 mM HEPES buffer (pH 7.0) by ultrafiltration through low-adsorption, hydrophilic, 10,000-nominal-molecular-weight-limit-cutoff membranes (regenerated cellulose; Amicon, Madrid, Spain) and stored at −86°C until use. Purity was assessed as >95% using SDS-PAGE, performed on 12% (vol/vol) acrylamide gels as described by Laemmli (33), in a Bio-Rad Mini protein system. Protein concentrations were determined according to Bradford (34).
Biochemical assays.
If not otherwise stated, hydrolase reactions using p-nitrophenyl (pNP) esters were performed in a microplate reader (Synergy HT multimode microplate reader; BioTek) as described previously (32), with minor modifications. Briefly, reaction mixtures contained 2 μg pure enzyme and 0.8 mM pNP ester (from a 20 mM stock solution in acetone) in 20 mM HEPES buffer, pH 7.0, at 30°C, in a total volume of 190 μl. Hydrolase-specific activity using other structurally diverse esters was also determined using p-nitrophenol as a pH indicator as described elsewhere (35), with small modifications. Briefly, specific activity was calculated by adding 2 μg pure enzyme (a smaller amount of enzyme [0.4 μg] was used for LAE6) and 2 mM substrate from a 100 mM ester stock solution (in acetonitrile) in 2 mM N,N-bis(2-hydroxyethyl)-2-aminoethanesulfonic acid (BES) buffer (pH 7.2) containing 0.45 mM p-nitrophenol and 2.5% acetonitrile (vol/vol), at 30°C, in a total volume of 150 μl. In all assays, the reactions were monitored every 2 min by spectrophotometrically measuring the absorbance of p-nitrophenol at 410 nm (for pNP esters) or 404 nm (for substrates other than pNP esters) during 15 min (except for LAE6, for which a 10-min incubation time was used). Under our experimental conditions, the absorption coefficient for p-nitrophenol was measured as 15,200 M−1 · cm−1. One unit of enzyme activity was defined as the amount of enzyme hydrolyzing 1 μmol of substrate in 1 min under the assay conditions. In all cases, three independent experiments were performed, and graphs were plotted using mean values; the standard deviations were less than 5%. Kinetic parameters were calculated by using a conventional Lineweaver and Burk model, at 30°C, as outlined above, in 96-well microtiter plates where each well contained 0.388 to 0.882 μM enzyme solution and 0 to 100 mM substrate.
The optimal pH, temperature, and salt concentration were determined using pNP butyrate (0.8 mM final concentration) as the substrate; pH values between 4.0 and 9.0, temperatures between 4 and 70°C, and sodium chloride concentrations ranging from 0.1 to 1.2 M were tested. All of the following buffers were tested at 20 mM: sodium citrate (pH 4.0 to 4.5), sodium acetate (pH 5.0 to 6.0), 2-(N-morpholino)ethanesulfonic acid (pH 5.5 to 6.0), HEPES (pH 7.0 to 8.0), piperazine-N,N′-bis(ethanesulfonic acid) (pH 6.0 to 7.0), K/Na phosphate (pH 7.5), Tris-HCl (pH 8.5), and glycine (pH 9.0 to 9.5). The pH was always adjusted at 25°C. The pH and temperature profiles were obtained at 30°C and pH 7.0, respectively. The optimal anion concentration was obtained at 30°C and pH 7.0. Under our experimental conditions, the absorption coefficients for p-nitrophenol were measured for each indicated temperature and pH and ranged from 132 (for pH 4.0) to 28,381 (for pH 9.5) M−1 · cm−1. These values were considered for calculating activity for each pH and temperature, as described above.
Oligonucleotide usage pattern analysis.
DNA sequences of contigs were searched for oligonucleotide compositional similarity against all sequenced bacterial chromosomes, plasmids, and phages by using the GOHTAM web tool (36). Frequencies of tetranucleotides for compositional genome comparison were calculated as described earlier (37).
Three-dimensional (3D) model analysis.
Suitable protein structures to be used as templates for modeling were searched using the PSIPRED server (http://bioinf.cs.ucl.ac.uk/psipred/). The Swiss Model server (http://swissmodel.expasy.org/) was used to generate the enzyme models. DeepView-Swiss-PdbViewer (http://spdbv.vital-it.ch/) was used to analyze the structures and generate the figures.
Sequence read accession number.
The sequence reads determined in the course of this work are available at NCBI under accession number SRA059294.
RESULTS AND DISCUSSION
Study site description.
Lake Arreo (42°46′N, 2°59′W; altitude, 655 m) is one of the deepest karstic lakes on the Iberian Peninsula (maximum depth, 24.8 m) (38, 39) (see Fig. S1 in the supplemental material). Chemically, the lake is subsaline, with an electrical conductivity range of 703 to 1,727 μS/cm during the last 20 years. Most of the sediment showed anoxic conditions, except for a thin surface layer, which has a Ca-(Mg)-(Na)-SO4-HCO3-(Cl) ionic composition (40, 41). The main water physicochemical data from the sampling point were the following: conductivity, 1.079 μS/cm; temperature, 6.9°C; dissolved oxygen content, 11.5 mg/liter (101%); pH 8.0. The mean values of major ionic concentrations (mg/liter) are the following: sulfates (354.9), bicarbonates (269.1), chloride (99.6), calcium (188.4), magnesium (37.5), sodium (51.52), and potassium (14.08).
Anoxic sediment samples (10 cm deep) were taken, and total DNA was purified. Size-fractionated DNA was ligated into the pCCFOS fosmid vector and further cloned in E. coli EPI300-T1R to create a fosmid clone library harboring approximately 1 Gbp of community genomes.
Metagenome library screening and sequence analysis.
Approximately 11,520 fosmid clones from the metagenomic library of Lake Arreo sediment (nearly 342 Mbp of metagenome DNA) were screened using plate-based screens for hydrolytic activity against α-naphthyl acetate, a model esterase substrate (29). Ten positive clones (incidence of positive clones, 1/1,152) were identified. Fosmid DNA isolated from each clone was mixed in equal amounts and sequenced as a pool using a Roche GS FLX DNA sequencer (1/16 plate), which produced 77,841 reads with an average length per read of 454.69 bp. Accordingly, a total of 38.5 Mbp of raw DNA sequences were obtained, which were assembled into 735,686 bp (19 contigs with lengths ranging from 1,854 to 43,416 bp), with an average GC content of 58.75%. A total of 378 open reading frames were identified, which were analyzed and compared to the sequences available in the databases. Seven out of 10 identified genes encoding the predicted esterases from the α/β hydrolase superfamily (named LAE1 to LAE7) were expressed and produced in soluble form in E. coli BL21(DE3). The corresponding gene products were purified (see Fig. S2 in the supplemental material), and their activities were characterized using 15 model substrates and an array of 86 additional structurally diverse esters (see Fig. S3 in the supplemental material).
Sequence analysis of α/β hydrolases from Lake Arreo.
Analysis of sequences of the Lake Arreo proteins showed that they all belong to the α/β hydrolase superfamily, with amino acid sequence identity ranging from 31 to 57%. As determined by Matcher (EMBOSS package), among Lake Arreo α/β hydrolases, LAE4 and LAE7 were the most similar enzymes (71.4% sequence identity), whereas LAE5 and LAE6 were the most different at the sequence level (18.2% identity) (see Table S3 in the supplemental material). The LAE1 protein was structurally most similar to thermophilic esterase from Thermotoga maritima (31%; Protein Data Bank [PDB] code 3DOH_A [42]) and α/β hydrolase from Agrobacterium tumefaciens (24%; PDB code 2r8b); LAE2 was most similar to GDSL-like brain platelet-activating factor acetylhydrolases (SGNH_hydrolase subfamily) (31%, PDB code 1FXW [43], and 28%, PDB code 1ES9 [44]); LAE3 was most similar to thermostable esterase/lipase from uncultured bacterium (36%; PDB code 3V9A_A) and hormone-sensitive esterase (35%; PDB code 3fak [45]); LAE4 was most similar to esterase from Pyrobaculum calidifontis (25%; PDB code 2YH2_A [46]) and bacterial α/β hydrolase (19%; PDB code 3k2i); LAE5 was most similar to the GDSL family lipolytic protein (SGNH_hydrolase subfamily) from Alicyclobacillus acidocaldarius subsp. acidocaldarius Dsm 446 (54%; PDB code 3RJT_B); LAE6 was most similar to thermophilic and thermostable carboxylesterase from the metagenomic library (41%; PDB code 2c7b [47]); and LAE7 was most similar to GDSL-like brain platelet-activating factor acetylhydrolase (SGNH_hydrolase subfamily) (32%; PDB code 1ES9 [44]).
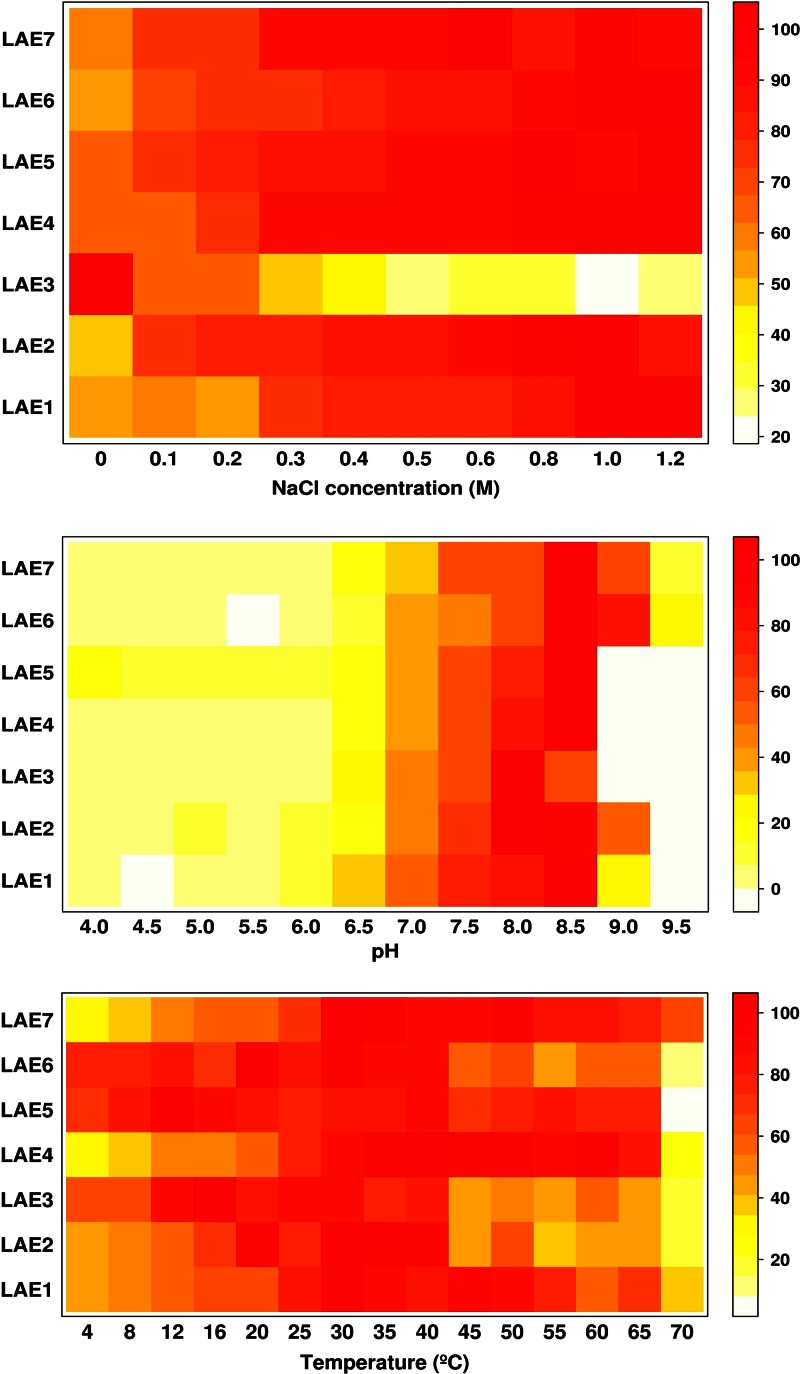
The deduced molecular masses and estimated pI values of Lake Arreo α/β hydrolases range from 24,328.83 to 34,002.96 Da and from 4.78 to 10.04, respectively. Suitable templates have been found for each enzyme, and 3D models of the corresponding protein structures are represented in Fig. S4 in the supplemental material. The analysis of the seven enzymes, which can be classified in the microbial lipase/esterase families described by Arpigny and Jaeger (48), allows the distinction of three structural groups. The first group (LAE2, LAE5, and LAE7) are GDSL esterases/lipases (49). A characteristic feature of GDSL enzymes is that the serine-containing motif is close to the protein N terminus. However, whereas the presumptive nucleophile of LAE5 corresponds to Ser15, that of LAE2 and LAE7 correspond to Ser143 and Ser130, respectively. The structural analysis of the LAE2 and LAE7 enzymes shows that they are bimodular enzymes in which the α/β hydrolase fold of the esterase catalytic unit is preceded at an N-terminal position by a 100- to 120-amino-acid module whose structure could not be modeled due to the absence of suitable templates. A remarkable feature of this module is the presence of a collagen-like sequence with a glycine/proline-rich stretch (GPGGPGGPRGGGFGAPPTPPGP in LAE2). These sequences, which are found in diverse bacterial proteins, are considered to be signatures of phage origin (50); this agrees with the analysis of DNA fragments using a genome linguistics approach (see below). Whatever the case, the fact that the catalytic Ser located closer to the N terminus in LAE5 than in LAE2 and LAE7 could make the catalytic site more accessible to substrates; this was further confirmed (see below) by showing that LAE5 accepted a higher number of substrates and by showing that although all three enzymes hydrolyzed pNP acetate and pNP propionate, LAE5 did prefer pNP propionate, while LAE2 and LAE7 preferred shorter pNP acetate (Table 1). Additionally, the fact that LAE5 retained circa 70% of the activity at 4°C, while LAE2 and LAE7 retained only 42 and 28%, respectively (Fig. 1 and see below), suggests a link between the differences in active center location and optimization of activity at low temperatures. Further structural analysis is in progress to confirm this hypothesis. The second group corresponds to common esterases/lipases with typical α/β hydrolase folds and GxSxG motif (LAE1, LAE4, and LAE6). LAE1 belongs to family I and LAE4 and LAE6 to family VI; with a molecular mass in the range 23 to 26 kDa, the enzymes in family VI are among the smallest esterases known, in agreement with the theoretical molecular mass of LAE4, which is the smallest (24 kDa) among the Lake Arreo esterases. The third group, represented by LAE3, belongs to the hormone-sensitive lipase/esterase (HSL) type (51) and also has a GxSxG motif. It is thought that HSL hydrolases retained high activity at low temperature, although sequence analysis indicates that temperature adaptation is not responsible for such sequence conservation (48); the fact that LAE3 did show maximal activity at 16°C (Fig. 1) agrees with this hypothesis.
Table 1.
Specific activities of the wild-type α/β hydrolases from Lake Arreo against a set of model α-naphthyl, pNP, and triacylglycerol ester substrates
| Substrate | Activitya (U/mg) |
||||||
|---|---|---|---|---|---|---|---|
| LAE1 | LAE2 | LAE3 | LAE4 | LAE5 | LAE6 | LAE7 | |
| α-Naphthyl acetate | ND | 111.04 | ND | 1.63 | 83.36 | 124.8 | 99.22 |
| α-Naphthyl propionate | 3.58 | 16.01 | ND | 8.34 | 24.35 | 55.1 | 11.73 |
| α-Naphthyl butyrate | 3.36 | 0.92 | 0.74 | 3.93 | 1.23 | 28.3 | 1.21 |
| pNP acetate | 0.133 | 5.57 | 23.80 | 0.05 | 1.23 | 34.3 | 0.64 |
| pNP propionate | 0.365 | 0.29 | 0.84 | 0.15 | 2.05 | 135.5 | 0.13 |
| pNP butyrate | 1.072 | 0.27 | 0.29 | 0.11 | 0.01 | 105.2 | 0.02 |
| pNP octanoate | 0.129 | ND | 0.04 | ND | ND | 2.6 | ND |
| pNP decanoate | 0.028 | ND | 0.02 | ND | ND | 1.0 | ND |
| pNP laurate | ND | ND | ND | ND | ND | 0.2 | ND |
| Triacetin | 0.015 | 1.07 | 0.01 | 1.01 | 1.14 | 18.7 | 1.19 |
| Tripropionin | 0.080 | ND | 0.01 | 2.33 | 0.02 | 14.9 | 0.39 |
| Tributyrin | 0.005 | ND | 0.09 | 0.83 | 0.00 | 11.3 | 0.14 |
| Tricaprin | 0.001 | ND | ND | 0.06 | 0.00 | 5.5 | ND |
| Tricaprylin | ND | ND | ND | ND | ND | 3.4 | ND |
| Trilaurin | ND | ND | ND | ND | ND | ND | ND |
For α-naphthyl and triacylglycerol esters, reaction mixtures contained 2 μg pure enzyme (a lower amount of enzyme [0.4 μg] was used for LAE6), 2 mM substrate, 0.45 mM p-nitrophenol, and 2.5% (vol/vol) acetonitrile in 2 mM BES buffer (pH 7.2), at 30°C, in a total volume of 150 μl. For pNP esters, reaction mixtures contained the same amount of enzymes and 0.8 mM pNP ester in 20 mM HEPES buffer (pH 7.0), at 30°C, in a total volume of 190 μl. In all cases, the reactions were monitored every 2 min during 15 min, and the absorbance of p-nitrophenol at 410 nm (for pNP esters) or 404 nm (for substrates other than pNP esters) was measured. In all cases, 1 U of enzyme activity was defined as the amount of enzyme hydrolyzing 1 μmol of substrate in 1 min under the assay conditions. Three independent experiments were performed for each parameter, and mean values are given; the standard deviations were less than 5%. ND, no activity detected under the experimental conditions assayed.
Fig 1.
Effect of NaCl concentration (upper), pH (middle), and temperature (lower) on the hydrolytic activity of Lake Arreo enzymes. The heat map colors represent the relative percentages of specific activity (U/mg) compared to the maximum (100%) within each enzyme, using pNP butyrate as the substrate. The effect of NaCl concentration was measured in 20 mM HEPES buffer (pH 7.0) at 30°C and varied from 0 to 1.2 M; the pH dependence was tested in the range of pH 4.0 to 9.5 at 30°C in 20 mM buffers; the temperature dependence in the range of 4 to 70°C at pH 7.0 was also determined. Reaction conditions were as described in Table 1 and Materials and Methods.
According to sequence and 3D model analyses (see Fig. S4 in the supplemental material), the catalytic triads were tentatively identified: Ser166, Glu217, and His249 (in LAE1), Ser140, Glu292, and His295 (in LAE2), Ser143, Glu237, and His267 (in LAE3), Ser144, Asp248, and His281 (in LAE4), Ser15, Asp192, and His195 (in LAE5), Ser161, Asp256, and His286 (in LAE6), and Ser130, Glu282, and His285 (in LAE7).
Optimization of activity and anion stimulation of esterases/lipases from Lake Arreo.
As shown in Fig. 1, purified proteins showed high hydrolytic activity against the model esterase substrate pNP butyrate at 16 to 40°C and pH 8.0 to 8.5, with the hydrolases LAE3, LAE5, and LAE6 being the most active at temperatures as low as 4°C, as they retained from 64 to 78% of the activity compared to the activity level at the optimal temperatures. Hydrolytic activities were stimulated (1.5 to 2.2 times) by the addition of NaCl to the reaction mixture (0.1 to 1.2 M NaCl) for all enzymes but one (LAE3), which was inhibited from 1.6 (at 0.1 M) to 3.8 (at 1.2 M) times. This is consistent with the fact that the Lake Arreo basin was formed by dissolved evaporates; thus, it represents a subsaline environment (52). The fact that all but one of the esterases was activated by NaCl indicates that enzyme properties resembled habitat-specific characteristics, and that activation by chloride is common for enzymes from Lake Arreo. Similar observations have been reported for the cold-active and anion-activated carboxyl esterase OLEI01171 from the oil-degrading marine bacterium Oleispira antarctica (53).
Using model substrates that included 3 α-naphthyl, 6 pNP, and 6 triacylglycerol esters and according to specific activity (U/mg) determinations (Table 1), we observed that LAE6 did show the highest capacity to accept longer esters (up to tricaprin, pNP laurate, and α-naphthyl butyrate), which is in line with the highest lipase character of this enzyme compared to other Lake Arreo α/β hydrolases. pNP esters were preferred substrates for LAE3, whereas LAE1, LAE2, LAE4, LAE5, and LAE7 preferentially hydrolyzed α-naphthyl esters. LAE6 showed similar activity for both substrates. In all cases, the activity toward triacylglycerols was significantly lower (from 3.5-fold for LAE4 to 100-fold for LAE2 compared to α-naphthyl esters). Overall, LAE6 (approximately 125 U/mg for α-naphthyl acetate) was the most active enzyme, whereas LAE3 showed the lowest activity (0.74 U/mg with α-naphthyl butyrate) (Table 1).
Substrate fingerprinting of esterases/lipases from Lake Arreo.
The substrate range of the purified esterases was characterized using a battery of 86 different esters (see Fig. S3 in the supplemental material) that included 25 halogenated alkyl and aryl esters, 34 alkyl esters, 12 aryl esters, 10 hydroxycinammic esters, 2 epoxides, 1 lactone, and 2 carbohydrate esters, using a colorimetric method in which p-nitrophenol was used as a pH indicator (35). The fingerprints of LAE1 to LAE7 are shown in Fig. 2 and 3, which showed that 52 out of the 86 (or 61%) esters used in the present study were accepted as substrates (for substrates not hydrolyzed by any of the enzymes, see Fig. S3 in the supplemental material).
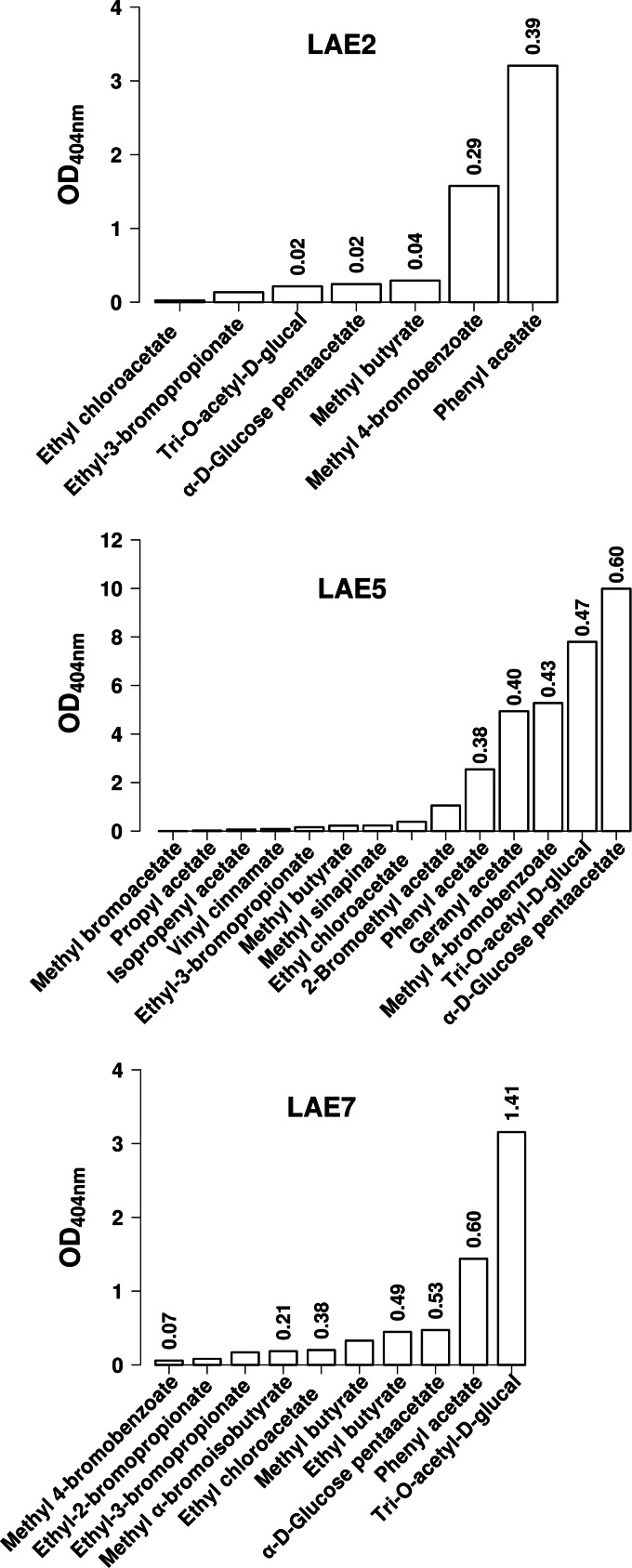
Fig 2.
Substrate profiles of the wild-type LAE2, LAE5, and LAE7 α/β hydrolases from Lake Arreo against a set of structurally diverse ester substrates. LAE2, LAE5, and LAE7 were the α/β hydrolases characterized by a restricted substrate spectrum. Reactions (30°C and pH 7.2) were performed as described for the α-naphthyl and triacylglycerol esters (Table 1) and were monitored during 15 min, and the absorbance of p-nitrophenol at 404 nm was recorded and plotted. For best and/or representative substrates, specific activities (U/mg) were calculated as described in Table 1 and Materials and Methods and are shown on the top of bars. In all cases, three independent experiments were performed for each parameter, and graphs were plotted using mean values; the standard deviations were less than 5%.
Using the ester library substrates (Fig. 2 and 3), we observed that LAE6 exhibits the broader substrate spectrum, being able to hydrolyze 43 substrates, followed by LAE3 (28 substrates), LAE4 (27 substrates), LAE1 (26 substrates), and, to lesser extents, LAE5 (14 esters), LAE7 (10 esters), and LAE2 (7 esters). The majority of compounds were accepted by at least two or more enzymes; however, 14 substrates were LAE6 specific (Fig. 3), including ethyl (S)-(−)-4-chloro-3-hydroxybutyrate, (+)-methyl-(R)-2-chloropropionate, methyl α-bromophenylacetate, methyl α-bromoisobutyrate, ethyl octanoate, vinyl decanoate, ethyl benzoate, vinyl benzoate, methyl cinnamate, methyl trans-cinnamate, isobutyl cinnamate, phenethyl cinnamate, (R)-(−)-glycidyl butyrate, (S)-(+)-glycidyl butyrate, and γ-butyrolactone, and the flavor terpene compound geranyl acetate was accepted as the substrate only by LAE5 (Fig. 2). Specific activity (U/mg) determinations were performed for the best representative substrates (Fig. 2 and 3). At 30°C and pH 7.2, methyl-2-chloro-3-hydroxypropionate (for LAE1, 0.28 U/mg), phenyl acetate (for LAE2, 0.39 U/mg), tri-O-acetyl-d-glucal (for LAE3, 0.33 U/mg; for LAE7, 1.41 U/mg), methyl 4-bromobenzoate (for LAE4, 1.01 U/mg), α-d-glucose penta-acetate (for LAE5, 0.60 U/mg), and (S)-(+)-glycidyl butyrate (for LAE6, 17.45 U/mg) were the preferred substrates.
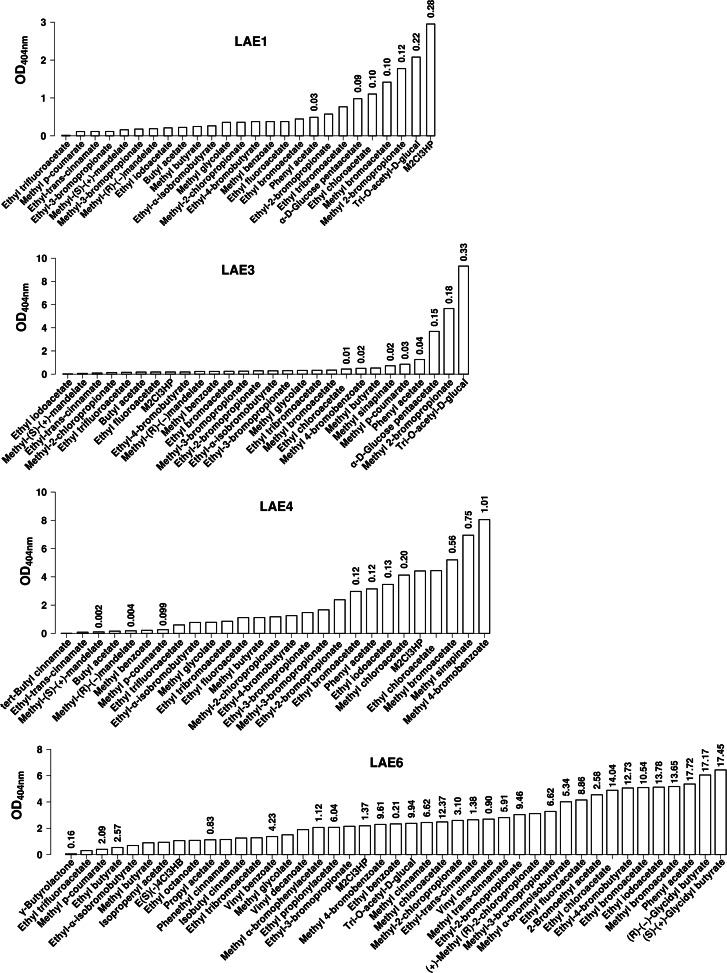
Fig 3.
Substrate specificity of the wide-substrate-spectrum LAE1, LAE3, LAE4, and LAE6 α/β hydrolases from Lake Arreo against a set of structurally diverse ester substrates. LAE1, LAE3, LAE4, and LAE6 were the hydrolases characterized by the widest substrate spectrum. Reaction conditions (30°C and pH 7.2) and activity parameter determinations were as described in Table 1 and the legend to Fig. 2. M2Cl3HP, methyl-2-chloro-3-hydroxypropionate.
The ability to hydrolyze nonhalogenated and halogenated (including those containing bromo-, chloro-, fluor-, and iodo-) alkyl and aryl esters was demonstrated for all enzymes (Fig. 2 and 3). Of special significance is that long alkyl and aryl esters with vinyl and isoprenyl substituents, such as vinyl decanoate, vinyl benzoate, and isoprenyl acetate, were mainly hydrolyzed by LAE6 (up to 4.23 U/mg) (Fig. 3); thus, this enzyme may be applied in transesterification reactions using vinyl esters, unlike the other enzymes from Lake Arreo. In addition, LAE1 was the only enzyme that did not accept aromatic halogenated esters, such as methyl 4-bromobenzoate (Fig. 3), although it was able to hydrolyze the nonhalogenated substrate (methyl benzoate). As shown in Fig. 2 and 3, four enzymes exhibited the capacity to accept the carbohydrate ester α-d-glucose penta-acetate (LAE2, LAE3, LAE5, and LAE7), and all but one (LAE4) showed activity for tri-O-acetyl-glucal. Finally, four enzymes were able to hydrolyze one or several cinnamate esters (LAE1, LAE3, LAE4, and LAE5), four accept p-coumarate esters (LAE1, LAE3, LAE4, and LAE6), and LAE3 and LAE5 were the only ones accepting methyl sinnapinate esters; no activity toward methyl ferulate was observed for any of the enzymes. Of special significance is that LAE4 (Fig. 3) was the only enzyme that showed activity on cinnamate esters of tertiary alcohols, such as tert-butyl cinnamate, although it was unable to accept primary and secondary alcohol substituents, such as methyl or isobutyl substituents. Further, LAE6 exhibited activity toward cinnamate esters with methyl, ethyl, isobutyl, vinyl, and the large aromatic phenethyl substituents, for which low or now activity was detected for the majority of other esterases/lipases (Fig. 3).
In relation to the enantioselective character of Lake Arreo hydrolases, we further observed that LAE1, LAE3, and LAE4 did show the capacity to hydrolyze both enantiomers of methyl mandelate, whereas LAE6 was the only one accepting both enantiomers of glycidyl butyrate (Fig. 2 and 3); other chiral esters, such as menthyl, neomenthyl, or lactate esters, were not substrates for any of the enzymes. According to the Quick E assay (34) and kcat/Km determinations for separate enantiomers (see Table S4 in the supplemental material), apparent enantiomeric ratios (Eapp values) were calculated. At 30°C and pH 7.2, Eapp varied from 816 (for LAE3) to 15 (for LAE6), 8.3 (for LAE1), and 2.0 (for LAE4). LAE1 and LAE3 did show enantiopreference for methyl-(R)-(−)-mandelate, whereas LAE4 preferred methyl-(S)-(+)-mandelate and LAE6 preferred (R)-(−)-glycidyl butyrate. In addition, LAE6 further exhibited activity toward γ-butyrolactone, although the activity for this substrate was 108-fold lower than that of (S)-(+)-glycidyl butyrate, which was the best substrate (Fig. 3).
Our results indicate that esterases/lipases from Lake Arreo are characterized by high activities and different substrate profiles and enantioselectivities; therefore, they are potentially useful for various biotechnological applications. The differences in hydrolytic capacities (using the half-saturation [Michaelis] coefficient [Km], the catalytic rate constant [kcat], the catalytic efficiency [kcat/Km] values, and/or specific activities; see Table S4 in the supplemental material) with distinct acid and alcohol substituents of different lengths and nature further indicate differences in active sites. Having said that, it should be highlighted that, compared to previously reported esterases/lipases (see Table S1 in the supplemental material), some of the substrate profiles described here are biologically relevant, such as the ability of a single esterase/lipase such as LAE6 to hydrolyze triacylglycerols, halogenated and nonhalogenated alkyl and aryl esters, cinnamoyl and carbohydrate esters, lactones, and chiral epoxides to a similar extent, which has not been previously described. Enzymes LAE1 and LAE2, which belong to the same contig (i.e., are produced by the same bacterium; see below), have similar optimal parameters (pH 8.0 to 8.5, 30°C, and up to 1.2 M NaCl; Fig. 1) but have quite distinct substrate profiles (Fig. 3 and 4). Thus, only 6 out of 27 substrates were hydrolyzed by both enzymes, with LAE1 possessing the broader substrate spectrum; these distinct functionalities suggest complementary metabolic and ecological capacities in vivo, together with different biotechnological capacities.
Fig 4.
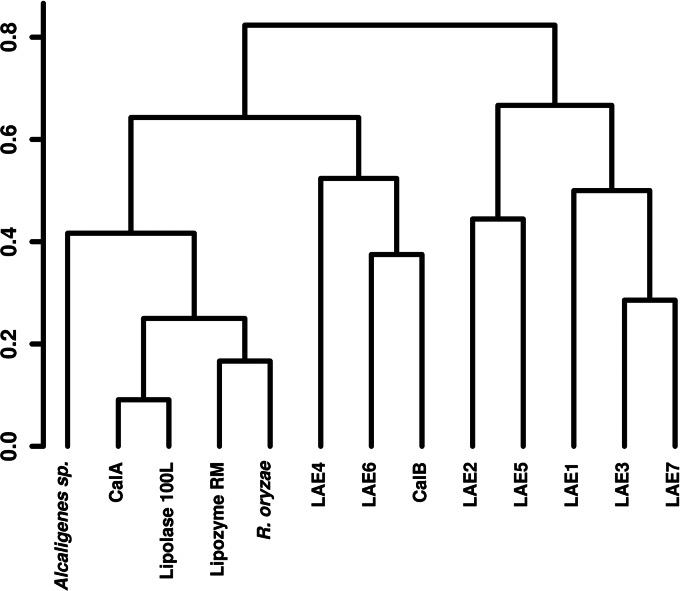
Clustering of substrate spectrum profile of Lake Arreo enzymes and commercial preparations, applying the Pearson's correlation to calculate the distances. Hierarchical clustering was derived from binomial distribution based on the presence or absence of activity against particular substrates (activity data not shown), measured as described in Table 1, using 2-μg protein extracts.
Comparison to commercial esterases and lipases.
Lipases and esterases are one of the most important classes of hydrolytic enzymes in industrial settings (2, 54). There is a wide range of available preparations that can be tested for particular applications and for comparative studies (3, 4, 55); in this context, it is interesting to evaluate and compare the substrate spectrum of newly isolated enzymes to that of commercially available and most common esterases and lipases. Here, we used five esterase/lipase commercial preparations, namely, Novozym 735 (lipase CalA from Candida antarctica), Novozym CALB L (lipase CalB from C. antarctica), Lipolase 100L (lipase from Thermomyces lanuginosa), Lipozyme RM-Novozym 388L (lipase from Rhizomocur miehei), and lipase from Alcaligenes sp. Lipase from Rhizopus oryzae, obtained and prepared as described elsewhere (28), was also added to the study.
The clustering analysis, generated from a binomial distribution based on the presence or absence of activity (at 30°C and pH 7.2) against the set of 101 different esters used here (Fig. 4), suggested that six of the Arreo Lake α/β hydrolases were functionally closer to each other than to the other preparations which clustered together. Lipases from Rhizopus oryzae and Rhizomocur miehei also showed similar profiles and clustered together with lipases from T. lanuginosa and CalA from C. antarctica. Interestingly, the lipase LAE6 clusters more closely with the lipase CALB L from C. antarctica, with both forming a separate group, which further indicates that both enzymes possess similar substrate spectra; this is of special significance, as Novozym CALB L is one of the most common commercially available lipases (Novozymes A/S, Bagsvaerd, Denmark). However, some minor differences were observed: (i) LAE6 hydrolyzed tri-O-acetyl-d-glucal (9.9 U/mg) and methyl 4-bromobenzoate (9.6 U/mg), for which no activity with CalB was detected, and (ii) CalB hydrolyzed methyl 2-bromopropionate (0.74 U/mg), for which negligible activity was found for LAE6. This suggests structural factors as determinants for substrate specificity when comparing both lipases.
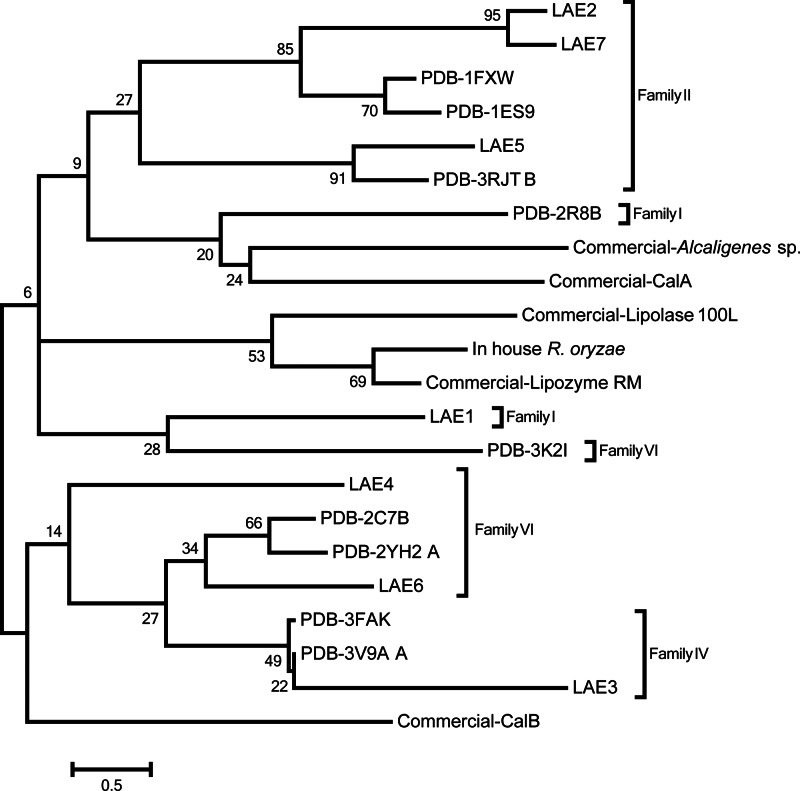
To analyze the sequence similarity of known commercial and newly discovered esterases/lipases belonging to different families, a dendrogram was created in MEGA5 (56) by the maximum-likelihood algorithms (Fig. 5). The dendrogram was further used to evaluate whether the placing of the different enzymes in the phylogenetic tree was related to their placing in the activity profile-based tree (Fig. 4). The sequences of reference and commercial esterases were obtained from the NCBI and PDB databases. The protein sequences were aligned by MUSCLE (57) and edited manually in JalView 2.5.1 (58), correlating with the alignment quality histogram. Due to an extreme diversity of selected sequences of enzymes, some of which were bacterial (PDB and Lake Arreo sequences) and others fungal (5 out of 6 sequences of commercial enzymes), the final alignment was reduced to 161 amino acid residues by removal of ambiguously aligned regions constituting 70% of the initial alignment. Analysis of the alignment by MEGA5 suggested WAG+G as the best evolutionary model. Robustness of the dendrogram was analyzed by the Bootstrap analysis based on 100 replicates of the initial data set. As shown in Fig. 4 and 5, we made the following observations: (i) commercial lipases CalA, Lipolase 100L, Lipozyme RM, Alcaligenes sp., and R. oryzae formed a separate cluster at the sequence level, as was also found for the activity clustering; (ii) similarly, lipases/esterases CalB, LAE4, and LAE6 clustered together at both the sequence and activity levels, which is of special significance considering the extreme diversity of selected sequences; (iii) LAE1, LAE2, LAE5, and LAE7 formed a separate group from LAE4 and LAE6 at both levels; and (iv) no clear correlation between sequence and activity was observed for LAE3. Accordingly, under the experimental setting applied here, the results suggest that, to some extent, an association between the phylogenetic/sequence positioning and the activity relationships exists.
Fig 5.
Dendrogram of protein sequence similarity relationships between newly identified and reference esterases/lipases. Enzyme families are depicted according to the Arpigny and Jaeger classification (48).
Analysis of metagenomic DNA fragments using genome linguistics approach.
Compositional similarity between the metagenomic fragments and the sequences of sequenced bacterial genomes and plasmids was analyzed by the comparison of frequencies of tetranucleotides in DNA sequences. A comprehensive analysis of the GOHTAM and BLAST analysis results is shown in the supplemental material. Five studied metagenomic DNA fragments containing genes encoding α/β hydrolases (excluding the LAE6-containing contig 19, which was too short for the compositional analysis) shared a significant level of tetranucleotide usage pattern similarity that indicated their origination from related organisms, which may be of Burkholderia/Ralstonia (Betaproteobacteria) and/or Rhizobium/Methylobacterium (Alphaproteobacteria) lineages. A search against the GOHTAM database showed compositional similarities to multiple bacterial genomes, plasmids, and phages. The protein blast of contig-encoded proteins against plasmid proteins also showed many hits but no gene syntenies. The DNA fragment (contig 2) containing LAE1 and LAE2 enzymes is most similar to several plasmids from Cupriavidus and Ralstonia, and it shares one major facilitator superfamily (MFS_1) gene and an integrase fragment with a genomic island from Ralstonia pickettii. Accordingly, we can conclude that this fragment most likely is a mobilome associated with either a plasmid or an integrated genomic island of a betaproteobacterium related to Cupriavidus and Ralstonia. Several genes of this fragment share similarity with the corresponding genes of Arthrobacter aurescens TC1 plasmid TC2 (NC_008713) and Shewanella baltica OS155 plasmid pSbal01 (NC_009035). The DNA fragment (contig 3) containing LAE3 enzyme shows some compositional similarity to alphaproteobacterial Methylobacterium plasmids, and it shares several genes with Nitrobacter hamburgensis X14 plasmid 1 (NC_007959). It also contains a gene for transposase IS4. The DNA fragment (contig 4) containing LAE7 enzyme resembles betaproteobacterial Burkholderia phages and contains phage major capsid proteins. The DNA fragment (contig 6) containing LAE4 enzyme has weak but consistent compositional similarity to alphaproteobacterial Rhizobium plasmids and multiple blast hits against Rhizobium leguminosarum bv. trifolii WSM1325 plasmid pR132504 (NC_012852), Rhizobium leguminosarum bv. trifolii WSM1325 plasmid pR132505 (NC_012854), Rhizobium etli CFN 42 plasmid p42c (NC_007764), and Azospirillum sp. strain B510 plasmid pAB510f (NC_013860). The DNA fragment (contig 7) containing LAE5 enzyme shows weak compositional similarity to the gammaproteobacterial Azotobacter genome and alphaproteobacterial Azospirillum plasmids and weak but persistent blast hits against several bacterial plasmids. The DNA fragment (contig 19) containing LAE6 enzyme is too short for the compositional analysis, but its genes are similar to those in the alphaproteobacterial Sphingomonas sp. strain KA1 plasmid pCAR3 (NC_008308). According to this analysis, DNA fragments containing Lake Arreo esterases may be plasmids or plasmid-borne genomic islands from proteobacteria, although contig 4, containing LAE7, most likely is a prophage.
Taken together, in the present study an esterase-driven assay, based on a well-established screen with α-naphthyl acetate, was used to identify hydrolytic activity in 10 E. coli clones. These clones harbor DNA plasmid fragments from a microbial community from an evaporite karstic lake (Lake Arreo). Lake Arreo is one of the few relatively deep (maximum depth [zmax] = 24 m) karstic lakes in Spain, and it developed in gypsum formations. Our study of α/β hydrolases mined from Lake Arreo suggests that they display habitat-specific characteristics, and that cold-adapted or psychrophilic proteobacteria occupy this ecological niche, with average temperatures ranging from 4.7 to 19.8°C, as they produced low-temperature active and anion-activated enzymes with unusual substrate specificities. The production of highly promiscuous hydrolases with broad substrate profiles may have important metabolic and ecological implications, such as that the function of these enzymes lies in substrate scavenging in low-temperature and substrate-poor karstic environments. A more comprehensive structural survey of enzymes from Lake Arreo, currently in progress, will determine whether or not this hypothesis is correct. Having said that, recent developments for the production of esterase and lipase improved commercial preparations have been achieved, and many factors account for the cost-effective utilization of esterases and lipases (2). This study provides experimental evidence that enzymes from evaporite bacterial metagenomes are of great interest for biotechnological processes, because they are salt-tolerant and are active at low temperatures and against a broad constellation of structurally diverse esters. This should be further evaluated and analyzed with commercial preparations under application conditions in a plethora of reactions according to the substrate specificity reported here. For example, preliminary tests confirmed that LAE6 preparations had performed well for esterification and transesterification reactions of sucrose with methyl and vinyl esters in several organic solvents; also, LAE3 preparations have been found preliminarily to be suitable for the production of enantiomers, such as (R)-mandelic acid, for synthetic purposes. Finally, this study may open research avenues into comparative catalysis models and structural-functional studies and confirms the necessity of isolating and characterizing new enzymes from environmental metagenomes.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge the financial support provided by the Spanish Ministry of Economy and Competitiveness (project CSD2007-00005), the European Community project MAGICPAH (FP7-KBBE-2009-245226), the European Regional Development Fund (ERDF), and the Government of Canada through Genome Canada, Ontario Genomics Institute, and Ontario Research Fund (2009-OGI-ABC-1405 and ORF-GL2-01-004). M.-E.G. thanks the CSIC for a JAE fellowship.
We thank Javier Tamames for his excellent support for taxonomic analysis. We thank Francisco J. Plou for his excellent support in the use of commercial lipase preparations used in the present study.
Footnotes
Published ahead of print 29 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00240-13.
REFERENCES
- 1. McQueen DA, Schottel JL. 1987. Purification and characterization of a novel extracellular esterase from pathogenic Streptomyces scabies that is inducible by zinc. J. Bacteriol. 169:1967–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Casas-Godoy L, Duquesne S, Bordes F, Sandoval G, Marty A. 2012. Lipases: an overview. Methods Mol. Biol. 861:3–30 [DOI] [PubMed] [Google Scholar]
- 3. Fan X, Niehus X, Sandoval G. 2012. Lipases as biocatalyst for biodiesel production. Methods Mol. Biol. 861:471–483 [DOI] [PubMed] [Google Scholar]
- 4. Hudlicky T, Reed JW. 2009. Applications of biotransformations and biocatalysis to complexity generation in organic synthesis. Chem. Soc. Rev. 38:3117–3132 [DOI] [PubMed] [Google Scholar]
- 5. Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, Studholme DJ, Yeats C, Eddy SR. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138–D141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Desai N, Antonopoulos D, Gilbert JA, Glass EM, Meyer F. 2012. From genomics to metagenomics. Curr. Opin. Biotechnol. 23:72–76 [DOI] [PubMed] [Google Scholar]
- 7. Galperin MY. 2008. Sorting out the mix in microbial genomics. Environ. Microbiol. 10:3187–3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sharma VK, Kumar N, Prakash T, Taylor TD. 2010. MetaBioME: a database to explore commercially useful enzymes in metagenomic datasets. Nucleic Acids Res. 38:D468–D472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gomez-Alvarez V, Teal TK, Schmidt TM. 2009. Systematic artifacts in metagenomes from complex microbial communities. ISME J. 3:1314–1317 [DOI] [PubMed] [Google Scholar]
- 10. Fernández-Arrojo L, Guazzaroni ME, López-Cortés N, Beloqui A, Ferrer M. 2010. Metagenomic era for biocatalyst identification. Curr. Opin. Biotechnol. 21:725–733 [DOI] [PubMed] [Google Scholar]
- 11. Beloqui A, Nechitaylo TY, López-Cortés N, Ghazi A, Guazzaroni ME, Polaina J, Strittmatter AW, Reva O, Waliczek A, Yakimov MM, Golyshina OV, Ferrer M, Golyshin PN. 2010. Diversity of glycosyl hydrolases from cellulose-depleting communities enriched from casts of two earthworm species. Appl. Environ. Microbiol. 76:5934–5946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Beloqui A, Pita M, Polaina J, Martínez-Arias A, Golyshina OV, Zumárraga M, Yakimov MM, García-Arellano H, Alcalde M, Fernández VM, Elborough K, Andreu JM, Ballesteros A, Plou FJ, Timmis KN, Ferrer M, Golyshin P-N. 2006. Novel polyphenol oxidase mined from a metagenome expression library of bovine rumen: biochemical properties structural analysis and phylogenetic relationships. J. Biol. Chem. 281:22933–22942 [DOI] [PubMed] [Google Scholar]
- 13. Beloqui A, Polaina J, Vieites JM, Reyes-Duarte D, Torres R, Golyshina OV, Chernikova TN, Waliczek A, Aharoni A, Yakimov MM, Timmis KN, Golyshin PN, Ferrer M. 2010. Novel hybrid esterase-haloacid dehalogenase enzyme. Chembiochem 11:1975–1978 [DOI] [PubMed] [Google Scholar]
- 14. Ekkers DM, Cretoiu MS, Kielak AM, Elsas JD. 2012. The great screen anomaly—a new frontier in product discovery through functional metagenomics. Appl. Microbiol. Biotechnol. 93:1005–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ellrott K, Jaroszewski L, Li W, Wooley JC, Godzik A. 2010. Expansion of the protein repertoire in newly explored environments: human gut microbiome specific protein families. PLoS Comput. Biol. 6:e1000798 doi: 10.1371/journal.pcbi.1000798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ferrer M, Golyshina OV, Chernikova TN, Khachane AN, Martins Dos Santos VA, Yakimov MM, Timmis KN, Golyshin PN. 2005. Microbial enzymes mined from the Urania deep-sea hypersaline anoxic basin. Chem. Biol. 12:895–904 [DOI] [PubMed] [Google Scholar]
- 17. Gabor E, Niehaus F, Aehle W, Eck J. 2012. Zooming in on metagenomics: molecular microdiversity of subtilisin Carlsberg in soil. J. Mol. Biol. 418:16–20 [DOI] [PubMed] [Google Scholar]
- 18. Lei Y, Peng H, Wang Y, Liu Y, Han F, Xiao Y, Gao Y. 2012. Preferential and rapid degradation of raw rice starch by an α-amylase of glycoside hydrolase subfamily GH13_37. Appl. Microbiol. Biotechnol. 94:1577–1584 [DOI] [PubMed] [Google Scholar]
- 19. Nagarajan S. 2012. New tools for exploring “old friends-microbial lipases.” Appl. Biochem. Biotechnol. 168(5):1163–1196 [DOI] [PubMed] [Google Scholar]
- 20. Park SY, Shin HJ, Kim GJ. 2011. Screening and identification of a novel esterase EstPE from a metagenomic DNA library. J. Microbiol. 49:7–14 [DOI] [PubMed] [Google Scholar]
- 21. Pham VD, Palden T, DeLong EF. 2007. Large-scale screens of metagenomic libraries. J. Vis. Exp. 4:201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vieites JM, Ghazi A, Beloqui A, Polaina Jreu Golyshina JMOV, Nechitaylo TY, Waliczek A, Yakimov MM, Golyshin PN, Ferrer M. 2010. Inter-conversion of catalytic abilities in a bifunctional carboxyl/feruloyl-esterase from earthworm gut metagenome. Microb. Biotechnol. 3:48–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yooseph S, Sutton G, Rusch DB, Halpern AL, Williamson SJ, Remington K, Eisen JA, Heidelberg KB, Manning G, Li W, Jaroszewski L, Cieplak P, Miller CS, Li H, Mashiyama ST, Joachimiak MP, van Belle C, Chandonia JM, Soergel DA, Zhai Y, Natarajan K, Lee S, Raphael BJ, Bafna V, Friedman R, Brenner SE, Godzik A, Eisenberg D, Dixon JE, Taylor SS, Strausberg RL, Frazier M, Venter JC. 2007. The Sorcerer II Global Ocean Sampling expedition: expanding the universe of protein families. PLoS Biol. 5:e16 doi: 10.1371/journal.pbio.0050016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Beier K, Jones CM, Mohit V, Hallin S, Betrtilsson S. 2011. Global phylogenography of chitinase genes in aquatic metagenomes. Appl. Environ. Microbiol. 77:1101–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chu X, He H, Guo C, Sun B. 2008. Identification of two novel esterases from a marine metagenomic library derived from South China Sea. Appl. Microbiol. Biotechnol. 80:615–625 [DOI] [PubMed] [Google Scholar]
- 26. Elend C, Schmeisser C, Leggewie C, Babiak P, Carballeira JD, Steele HL, Reymond JL, Jaeger KE, Streit WR. 2006. Isolation and biochemical characterization of two novel metagenome-derived esterases. Appl. Environ. Microbiol. 72:3637–3645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed, p 6.22 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 28. Guillén M, Benaiges MD, Valero F. 2011. Comparison of the biochemical properties of a recombinant lipase extract from Rhizopus oryzae expressed in Pichia pastoris with a native extract. Biochem. Eng. J. 54:117–123 [Google Scholar]
- 29. Reyes-Duarte D, Ferrer M, García-Arellano H. 2012. Functional-based screening methods for lipases, esterases, and phospholipases in metagenomic libraries. Methods Mol. Biol. 861:101–113 [DOI] [PubMed] [Google Scholar]
- 30. Lukashin AV, Borodovsky M. 1998. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 26:1107–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ferrer M, Ghazi A, Beloqui A, Vieites JM, López-Cortés N, Marín-Navarro J, Nechitaylo TY, Guazzaroni ME, Polaina J, Waliczek A, Chernikova TN, Reva ON, Golyshina OV, Golyshin PN. 2012. Functional metagenomics unveils a multifunctional glycosyl hydrolase from the family 43 catalysing the breakdown of plant polymers in the calf rumen. PLoS One 7:e38134 doi: 10.1371/journal.pone.0038134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 34. Bradford MM. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 35. Janes LE, Löwendahl C, Kazlauskas RJ. 1998. Rapid quantitative screening of hydrolases using pH indicators. Finding enantioselective hydrolases. Chem. Eur. J. 4:2317–2324 [Google Scholar]
- 36. Ménigaud S, Mallet L, Picord G, Churlaud C, Borrel A, Deschavanne P. 2012. GOHTAM: a website for genomic origin of horizontal transfers, alignment and metagenomics. Bioinformatics 28:1270–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ganesan H, Rakitianskaia AS, Davenport CF, Tümmler B, Reva ON. 2008. The SeqWord Genome Browser: an online tool for the identification and visualization of atypical regions of bacterial genomes through oligonucleotide usage. BMC Bioinformatics 9:333 doi: 10.1186/1471-2105-9-333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Corella JP, El Amrani A, Sigró J, Morellón M, Rico E, Valero-Garcés BL. 2011. Recent evolution of Lake Arreo, northern Spain: influences of land use change and climate. J. Paleolimnol. 46:469–485 [Google Scholar]
- 39. Rico E, Chicote A, González ME, Montes M. 1995. Batimetría y análisis morfométrico del Lago Arreo (N. España). Limnética 11:55–58 [Google Scholar]
- 40. Chicote A. 2004. Limnología y ecología microbiana de un lago kárstico evaporítico: el lago de Arreo (Norte de España). Ph.D. thesis Universidad Autónoma de Madrid, Madrid, Spain [Google Scholar]
- 41. González-Mozo ME, Chicote A, Rico Montes EC. 2000. Limnological characterizetion of an evaporite karstic lake in Spain (Lake Arreo). Limnética 18:91–98 [Google Scholar]
- 42. Levisson M, Sun L, Hendriks S, Swinkels P, Akveld T, Bultema JB, Barendregt A, van den Heuvel RH, Dijkstra BW, van der Oost J, Kengen SW. 2009. Crystal structure and biochemical properties of a novel thermostable esterase containing an immunoglobulin-like domain. J. Mol. Biol. 385:949–962 [DOI] [PubMed] [Google Scholar]
- 43. Sheffield PJ, McMullen TW, Li J, Ho YS, Garrard SM, Derewenda U, Derewenda ZS. 2001. Preparation and crystal structure of the recombinant α1/α2 catalytic heterodimer of bovine brain platelet-activating factor acetylhydrolase Ib. Protein Eng. 14:513–519 [DOI] [PubMed] [Google Scholar]
- 44. McMullen TW, Li J, Sheffield PJ, Aoki J, Martin TW, Arai H, Inoue K, Derewenda ZS. 2000. The functional implications of the dimerization of the catalytic subunits of the mammalian brain platelet-activating factor acetylhydrolase (Ib). Protein Eng. 13:865–871 [DOI] [PubMed] [Google Scholar]
- 45. Nam KH, Kim M, Kim S, Priyadarshi A, Lee WH, Hwang KY. 2009. Structural and functional analysis of a novel EstE5 belonging to the subfamily of hormone-sensitive lipase. Biochem. Biophys. Res. Commun. 379:553–556 [DOI] [PubMed] [Google Scholar]
- 46. Hotta Y, Ezaki S, Atomi H, Imanaka T. 2002. Extremely stable and versatile carboxylesterase from a hyperthermophilic archaeon. Appl. Environ. Microbiol. 68:3925–3931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Byun J, Rhee J, Kim ND, Yoon J, Kim D, Koh E, Oh J, Cho H. 2007. Crystal structure of hyperthermophilic esterase EstE1 and the relationship between its dimerization and thermostability properties. BMC Struct. Biol. 7:47 doi: 10.1186/1472-6807-7-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Arpigny JL, Jaeger KE. 1999. Bacterial lipolytic enzymes: classification and properties. Biochem. J. 343:177–183 [PMC free article] [PubMed] [Google Scholar]
- 49. Akoh CC, Lee GC, Liaw YC, Huang TH, Shaw JF. 2004. GDSL family of serine esterases/lipases. Prog. Lipid Res. 43:534–552 [DOI] [PubMed] [Google Scholar]
- 50. Ghosh N, McKillop TJ, Jowitt TA, Howard M, Davies H, Holmes DF, Roberts IS, Bella J. 2012. Collagen-like proteins in pathogenic E. coli strains. PLoS One 7:e37872 doi: 10.1371/journal.pone.0037872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mandrich L, Merone L, Manco G. 2009. Structural and kinetic overview of the carboxylesterase EST2 from Alicyclobacillus acidocaldarius: a comparison with the other members of the HSL family. Protein Pept. Lett. 16:1189–1200 [DOI] [PubMed] [Google Scholar]
- 52. Martín-Rubio M, Rodriguez-Lazaro J, Anadón P, Robles F, Utrilla R, Vázquez A. 2005. Factors affecting the distribution of recent lacustrine ostracoda from the Caicedo de Yuso-Arreo Lake (Western Ebro Basin, Spain). Palaeogeogr. Palaeoclimatol. Palaeoecol. 225:118–133 [Google Scholar]
- 53. Lemak S, Tchigvintsev A, Petit P, Flick R, Singer AU, Brown G, Evdokimova E, Egorova O, Gonzalez CF, Chernikova TN, Yakimov MM, Kube M, Reinhardt R, Golyshin PN, Savchenko A, Yakunin AF. 2012. Structure and activity of the cold-active and anion-activated carboxyl esterase OLEI01171 from the oil-degrading marine bacterium Oleispira antarctica. Biochem. J. 4452:193–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Biely P. 2012. Microbial carbohydrate esterases deacetylating plant polysaccharides. Biotechnol. Adv. 30:1575–1588 [DOI] [PubMed] [Google Scholar]
- 55. Goswami D, Basu JK, De S. 2013. Lipase applications in oil hydrolysis with a case study on castor oil: a review. Crit. Rev. Biotechnol. 33:81–96 [DOI] [PubMed] [Google Scholar]
- 56. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113 doi: 10.1186/1471-2105-5-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. 2009. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.