Abstract
Plasmid-based Escherichia coli BL21(DE3) expression systems are extensively used for the production of recombinant proteins. However, the combination of a high gene dosage with strong promoters exerts extremely stressful conditions on producing cells, resulting in a multitude of protective reactions and malfunctions in the host cell with a strong impact on yield and quality of the product. Here, we provide in-depth characterization of plasmid-based perturbations in recombinant protein production. A plasmid-free T7 system with a single copy of the gene of interest (GOI) integrated into the genome was used as a reference. Transcriptomics in combination with a variety of process analytics were used to characterize and compare a plasmid-free T7-based expression system to a conventional pET-plasmid-based expression system, with both expressing human superoxide dismutase in fed-batch cultivations. The plasmid-free system showed a moderate stress response on the transcriptional level, with only minor effects on cell growth. In contrast to this finding, comprehensive changes on the transcriptome level were observed in the plasmid-based expression system and cell growth was heavily impaired by recombinant gene expression. Additionally, we found that the T7 terminator is not a sufficient termination signal. Overall, this work reveals that the major metabolic burden in plasmid-based systems is caused at the level of transcription as a result of overtranscription of the multicopy product gene and transcriptional read-through of T7 RNA polymerase. We therefore conclude that the presence of high levels of extrinsic mRNAs, competing for the limited number of ribosomes, leads to the significantly reduced translation of intrinsic mRNAs.
INTRODUCTION
Plasmid-based expression systems have been used for the production of recombinant proteins for more than 4 decades (1, 2). They can be manipulated quickly and easily, and a variety of replicons for use in Escherichia coli have become available (3), allowing, e.g., different expression levels by using plasmids with different copy numbers (4, 5). Plasmids equipped with additional functions can be used to facilitate, for instance, coexpression of proteins assisting correct folding (6) or of tRNAs supporting transcription of rare codons (7). The dissemination of E. coli systems was further strongly supported by the availability of well-established, easy-to-use protocols from molecular manipulation to cell cultivation up to a large scale (8) and by the FDA-proven status of E. coli as a host for production of proteins for clinical use (9).
However, E. coli-based production processes are still far from optimal for exploitation of the cellular system, as the expression of heterologous proteins is performed with excessive strength, leading to a rapid exhaustion of the host cell (10) and, hence, loss of yield. One prominent example is the T7 system, combining high-copy-number plasmids with an orthogonal transcription system in the form of the T7 phage RNA polymerase, an enzyme showing an average elongation rate of 200 to 400 nucleotides (nt) s−1 (11), which is approximately 5 times the activity of the E. coli host RNA polymerase (12). The ultimate strength of the T7 system exerts extremely stressful conditions on producing cells, finally resulting in metabolic overload of the cells and reduced production periods. Even though theories on possible causes of this cellular stress response do exist (13–16), it is not fully understood to what extent these phenomena are linked to a plasmid-associated metabolic burden. Plasmid replication and expression of plasmid-encoded proteins, such as the product of the constitutively expressed antibiotic resistance gene, are responsible for an additional metabolic load which is manageable for relaxed nonproducing cells but becomes a serious problem for cells producing at high levels (17, 18). Another common problem in using plasmid-based vectors for bioprocessing is caused by the variability of the plasmid copy numbers (PCN) throughout the bioprocess. Fluctuations in the gene dosage lead to variations in expression rates of the gene of interest (GOI) and of plasmid backbone elements (e.g., the antibiotic resistance gene) and, consequently, to process instabilities. A central issue during plasmid-based expression of recombinant proteins is plasmid loss, a phenomenon tightly related to the genetic background of the host and the type of plasmid and finally resulting in a nonproducing cell population that grows at the expense of producer cells (19–22). Another challenging problem is the increase of the PCN in response to induction of recombinant gene expression. High-level protein production leads to malfunctions in the plasmid replication control mechanism. The resultant increase in the PCN induces a self-amplifying cascade of metabolic load which finally produces a fatal metabolic overburden (23). Both phenomena significantly contribute to instabilities and represent distinct limitations of plasmid-based systems in production processes. Elucidation of the reasons for adverse effects observed in plasmid-based expression systems during recombinant protein expression remains elusive due to the complexity of the interactions between plasmids and host. Although knowledge on these interactions would be essential, detailed investigations are as yet limited (24, 25). Recent years have revealed several approaches to improve T7-based expression systems, including the use of a lysozyme-expressing plasmid (pLysS or pLysE) to decrease leaky expression of genes and of a more restrictive promoter(s) for the integrated T7 RNA polymerase (e.g., rhamnose inducible) and the generation of a recA mutant derivative of BL21(DE3) called BLR(DE3). Reviews of these approaches can be found elsewhere (26, 27).
Plasmid-free T7-based expression systems in which the GOI is site-specifically integrated into a defined locus of the E. coli genome were described previously (28, 29) but have not yet been used as a reference system to reveal plasmid-related disturbances during bioprocessing. Here, the gene copy number was fixed to a single copy and the stability of these systems was significantly increased, as the GOI cannot be lost during cultivation. It has been shown that product formation rates generated in these systems are comparable to plasmid-based ones (29). Consequently, they can be used in a comparative study to assign cellular responses of plasmid-based systems after induction either to recombinant protein production or to phenomena exclusively related to plasmid systems.
In this publication, we focus on the in-depth characterization of differences in the cellular responses to recombinant gene expression of a plasmid-free and a plasmid-based expression system. Comparative transcriptomics (spotted-oligonucleotide DNA microarrays [DNA μ-arrays] and the digital nCounter technology [30]) in combination with a comprehensive number of process analytics were used to compare the plasmid-based system E. coli BL21(DE3)(pET30aSOD) and the plasmid-free system E. coli BL21(DE3)::TN7<SOD> in standard recombinant protein production processes.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Experiments were performed using E. coli BL21(DE3) (Merck KGaA, Darmstadt, Germany). BL21(DE3) was transformed with pET30aSOD, yielding BL21(DE3)(pET30aSOD). pET30aSOD was derived by subcloning of the Homo sapiens superoxide dismutase 1 gene (GenBank accession no. BT006676.1) from pET11aSOD, described elsewhere (SOD, superoxide dismutase) (31). Linear double-stranded DNA (dsDNA) cartridges, containing the expression unit from pET30a<SOD> fused to a chloramphenicol acetyltransferase resistance gene (cat), were integrated into the bacterial chromosome at the attTn7 site of E. coli BL21(DE3), which carries the pSIM6 helper plasmid, as described by Sharan et al. (32). The resulting strain was designated BL21(DE3)::TN7<T7-SOD>.
Medium, cultivation conditions, and sampling.
For cultivation of cells in a minimal medium, a 20-liter (14-liter working volume, 4-liter batch volume) computer-controlled bioreactor (MBR, Wetzikon, Switzerland), equipped with standard control units (Siemens PS7, Intellution iFIX), described elsewhere (29), was used. A fed-batch regimen with an exponential substrate feed rate was applied to provide a constant growth rate of 0.1 h−1 over 4 doubling times. The pH was maintained at a set point of 7.0 ± 0.05 by addition of 25% ammonia solution (Merck), the temperature was set to 37°C ± 0.5°C, and the dissolved oxygen level was stabilized above 30%. For inoculation, a deep-frozen (−80°C) working-cell-bank vial was thawed and 1 ml (optical density at 600 nm [OD600] = 1) was transferred aseptically with 30 ml 0.9% NaCl solution to the bioreactor. Induction of the expression system was performed by adding isopropyl-β-d-thiogalactoside (IPTG) (GERBU Biotechnik, Germany) to the reactor in a relation of 20 μmol IPTG per g calculated cell dry mass (CDM) (in total, 370 g CDM).
Sampling for the standard off-line process parameter started after one generation (7 h) in fed-batch mode. This first sample was withdrawn from the bioreactor prior to induction. After sampling, recombinant protein expression was induced and samples were withdrawn every hour. Analytical methods used for determination of optical density, CDM, plasmid copy number (PCN), product titer, viable cell number (VCN), and levels of guanosine-3′,5′-bispyrophosphate (ppGpp) are described in detail elsewhere (29, 33, 34). Samples for subsequent microarray analysis were taken between feed h 7 and 16 for the plasmid-based system and between feed h 7 and 28 for the plasmid-free system. For RNA isolation, samples were drawn directly into a 5% phenol-ethanol stabilizing solution and split into aliquots corresponding to about 3 mg CDM. The cell suspension was centrifuged for 2 min at ∼11,000 × g and 4°C. The supernatant was discarded, and the pellet was immediately frozen at −80°C.
Microarray analysis.
Microarrays comprised selective probes (approximately 70-mer oligonucleotides) for all open reading frames of the E. coli BL21 genome (custom array; Operon, Germany). The oligonucleotides were spotted onto an epoxy surface, and the mRNA expression levels of 4,537 unique genes could be measured. The experimental data (including all processing protocols) were loaded into ArrayExpress (http://www.ebi.ac.uk/microarray-as/ae/). A dye-swap design was used, and the cells in the noninduced state in each experiment were compared to cells of samples past the induction state.
Isolation of RNA.
RNA was isolated by TRIzol reagent (Invitrogen) pulping and chloroform (Sigma) extraction according to the modified protocol described by Hegde and coworkers (35). The quality of RNA was checked on a RNA1000 LabChip using an Agilent Bioanalyzer according to the protocol of the supplier (Agilent Bioanalyzer Application Guide), and the RNA concentration was determined with a NanoDrop 1000 spectrophotometer.
Reverse transcription and indirect labeling.
Ten micrograms of total RNA was labeled with either of two fluorescent dyes (Cy3 or Cy5; GE Healthcare) and then subjected to paired competitive hybridizations. Indirect labeling involves two steps: incorporation of amino-allyl dUTP by reverse transcription and then attachment of the fluorescent dyes.
Briefly, total RNA was mixed with 3 μg of random primer (hexamer) in a total volume of 15.5 μl, denatured at 65°C for 10 min, and primed during cooling prior to reverse transcription (RT). To this mixture, 0.6 μl of a 50× deoxynucleoside triphosphate (dNTP) mix (10 mM [each] dATP, dGTP, and dCTP; 4 mM dTTP; and 6 mM amino-allyl dUTP [Sigma]), 6 μl of 5× first-strand buffer, 3 μl of 0.1 M dithiothreitol (DTT) (both provided with Superscript III reverse transcriptase), 0.25 μl of RNAsin (Promega), and 2 μl of Superscript III reverse transcriptase (Invitrogen) were added. After 2 h of incubation at 42°C, 10 μl of 0.5 M EDTA and 10 μl of 1 M sodium hydroxide were added and the reaction mixture was placed at 65°C for 15 min. After cooling to room temperature, 25 μl of 1 M Tris-HCl (pH 7.5) was added, and cDNA was washed and concentrated using a Microcon YM30 centrifugal filter unit (Millipore). Finally, cDNA was dried in a Speedvac. The dried cDNA was stored at −20°C until use. The cDNA was resuspended in 4.5 μl of 0.2 M sodium carbonate buffer (pH 8.5 to 9.0) and mixed with 4.5 μl of monoreactive Cy3 or Cy5 dye that had previously been resuspended in 37 μl of dimethyl sulfoxide (DMSO). The cDNA-dye mixture was incubated for 1 h at 23°C in the dark. For dye quenching, 4.5 μl of 4 M hydroxylamine was added and incubated again for 15 min at 23°C in the dark. Labeled cDNA was mixed with 35 μl of 3 M sodium acetate (pH 5.2) and purified using a Qiagen MinElute PCR purification kit according to the manufacturer's protocol. The incorporation of Cy dye into the cDNA was measured with the NanoDrop 1000 spectrophotometer, and 200 pg of a Cy3-labeled sample was combined with 200 pg of a Cy5-labeled sample, mixed with 2 μl of salmon sperm (Invitrogen) (10 mg · ml−1 stock), and hybridized on a microarray.
Array processing.
Blocking and hybridization of the microarrays were carried out using a Tecan HS400 hybridization station. The slides were blocked with blocking buffer (4× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.5% [vol/vol] SDS, 1% bovine serum albumin [BSA]) for 1 h and then washed with water and 2× SSC containing 0.1% SDS. Combined labeled samples were dried to a volume of ∼7 μl in a Speedvac, mixed with OpArray HybSolution (Operon) to achieve a concentration of 90% and a final sample volume of 70 μl, and heated to 95°C for 3 min. Hybridization was carried out for 16 h at 50°C. After hybridization, slides were washed with 1× SSC and finally with 0.5× SSC for 1 min. Slides were dried and scanned using the Agilent microarray scanner at 10-μm resolution.
Statistical analysis of microarray data.
Resulting images were analyzed with Dapple (36), where the data were extracted. Data analysis was performed using the statistical computing environment R (37). The data were preprocessed using variance stabilization normalization (38). Differential expression estimates were calculated using limma in the Bioconductor package (39). The filtered data sets (absolute log fold change > 2) were clustered using flexclust in the R package (40), and gcExplorer (41, 42) was used for further analysis and visualization.
Verification of microarray data—NanoString nCounter gene expression system.
The Nanostring nCounter technology uses molecular barcodes and microscopic imaging to detect and count up to several hundred unique transcripts in one hybridization reaction mixture. Each color-coded barcode is attached to a single target-specific probe corresponding to a gene of interest. Barcodes hybridize directly to their corresponding target molecules and can be individually counted without the need for amplification—providing very sensitive digital data.
RNA was isolated as described above and analyzed using the NanoString nCounter gene expression system, which captures and counts individual mRNA transcripts. Advantages over existing platforms include direct measurement of mRNA expression levels without enzymatic reactions or bias, sensitivity coupled with high multiplex capability, and digital readout. The sensitivity of the NanoString nCounter gene expression system is similar to that of real-time PCR. The transcript levels for selected genes across all samples showed similar patterns of gene expression for microarray data and NanoString data (30). Due to the high transcription levels of the superoxide dismutase (SOD) gene and consequent saturation of the flow cell imaging surface, an attenuation strategy, based on competitive inhibition, was followed (see Data Set S1 in the supplemental material).
Microarray data accession numbers.
The ArrayExpress accession number for the BL21 array design is A-MARS-11. The ArrayExpress accession numbers for data from the plasmid-based and plasmid-free experiment are E-MARS-19 and E-MARS-24.
RESULTS
To characterize the plasmid-related effects on host cell metabolism during recombinant protein production, carbon-limited fed-batch cultivations in minimal medium with exponentially increasing feed amounts for each system were carried out in triplicate experiments. For the plasmid-based system, detailed analysis was limited to 9 h past induction due to the fact that this time point also represented the process optimum for the plasmid-based system in terms of product yield under the given conditions.
Fermentation characteristics, recombinant protein yield, and stress response.
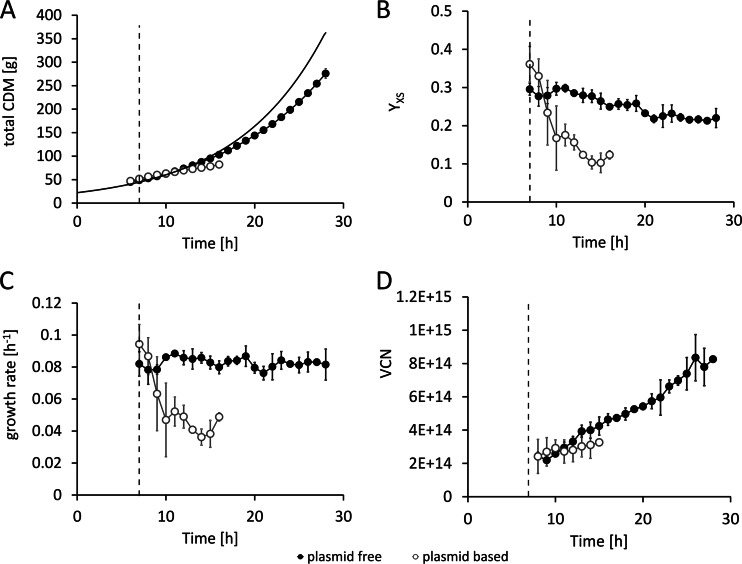
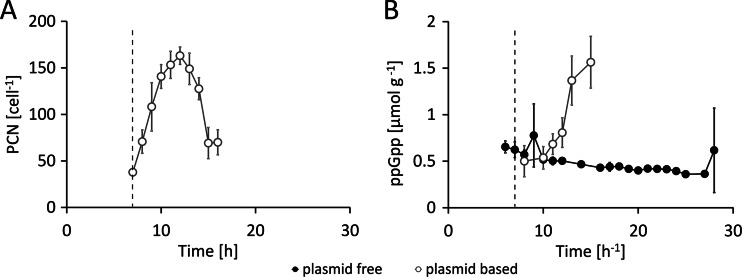
In the noninduced state, the plasmid-based system and the plasmid-free system showed similar growth behaviors; thus, replication of the pET30a<SOD> plasmid did not have a significant effect. The obtained levels of total CDM at the time point of induction were comparable in all experiments and were independent of the system used (Fig. 1A). Remarkably, the induction of recombinant gene expression with IPTG triggered totally different responses in the two systems. In the plasmid-based system, a rapid drop of the substrate yield coefficient (YX/S; g CDM/g glucose) (Fig. 1B) and, consequently, a significant (4.5-fold) decrease of the growth rate below the given rate of 0.1 h−1 were observed (Fig. 1C). In parallel, the plasmid copy number of 40 ± 4 in the noninduced state increased to 160 ± 9 within 6 h past induction followed by a steep decrease (Fig. 2A). In the plasmid-free expression system using BL21(DE3)::TN7<SOD>, the average value of YX/S after induction was decreased by a maximum of only 25%. The system was able to maintain a growth rate of approximately 0.08 h−1 throughout the whole cultivation period (Fig. 1B and C). Finally, the experiments performed with the plasmid-free system yielded a total CDM of 276 ± 10 g, whereas the plasmid-based system was able to achieve only 82 ± 3 g, due to growth cessation (9 h past induction; see Fig. 1A) caused by the metabolic overburden. This difference was also reflected in the viable cell numbers (Fig. 1D). The high metabolic load in the plasmid-based expression system in response to recombinant gene expression was demonstrated by the increased concentrations of the stress marker molecule ppGpp detected. In the plasmid-free expression system, the ppGpp concentration remained rather unaffected (Fig. 2B).
Fig 1.
Comparison of process data from the feed phase of cultivations performed with the plasmid-based [BL21(DE3)(pET30aSOD), open circles] expression system and the plasmid-free [BL21(DE3)::TN7<T7-SOD>, filled circles] expression system. Data represent determinations of the induction of recombinant gene expression conducted at feed hour 7 using 20 μmol IPTG g−1 CDM. (A) Total cell dry mass (CDM). The black line represents the calculated theoretical CDM based on a constant glucose yield coefficient of 0.3 g · g−1 · h−1. (B) Glucose yield coefficient (YX/S). (C) Growth rate. (D) Viable cell number (VCN). The dashed line indicates the time point of induction. Data shown are derived from three independent replicate experiments (n = 3).
Fig 2.
Comparison of process data from the feed phase of cultivations performed with the plasmid-based [BL21(DE3)(pET30aSOD), open circles] system and the plasmid-free [BL21(DE3)::TN7<T7-SOD>, filled circles] system. Determination of the induction of recombinant gene expression was conducted at feed hour 7. (A) Plasmid copy number. (B) Levels of the stress marker guanosin-3′,5′-tetraphosphat (ppGpp). The dashed line indicates the time point of induction. Data shown are derived from three independent replicate experiments (n = 3).
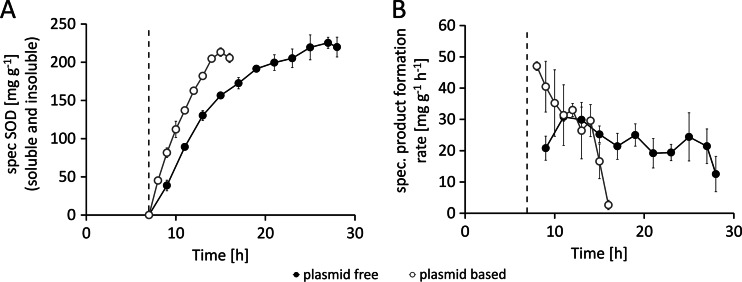
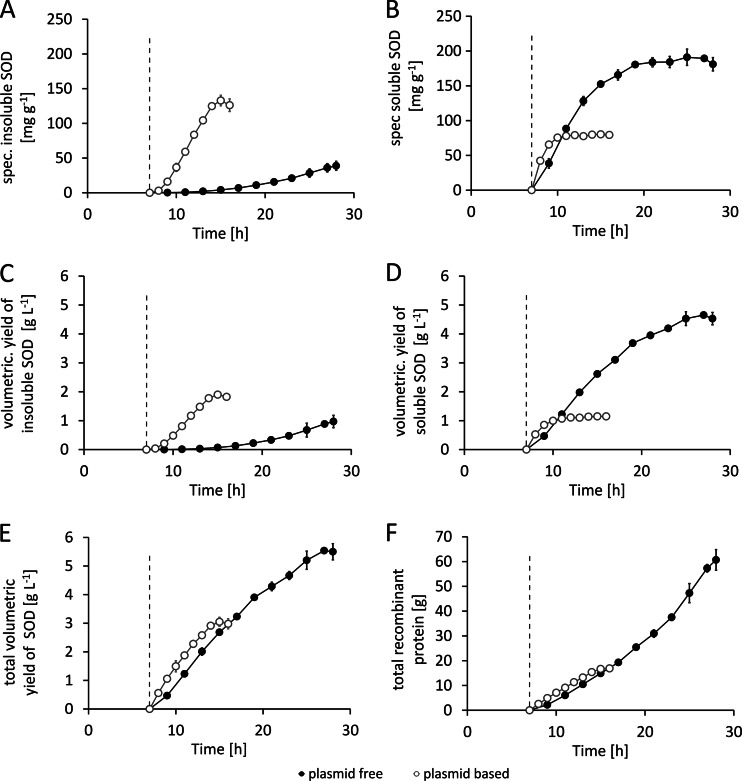
The plasmid-based system accumulated a maximum of 213 ± 6 mg · g−1 and the genome-encoded system 225 ± 7 mg · g−1 SOD per gram CDM, and yet variations in the product formation kinetics of the two systems were observed (Fig. 3A). The product formation rate (qP) in the plasmid-based system peaked at a maximum of 47 ± 1 mg · g−1 · h−1 shortly after induction and dropped to zero within 8 h. In contrast, the plasmid-free system with only a single copy of the target gene was able to achieve an average qP of 22.7 ± 5 mg · g−1 · h−1 for the whole production period of 21 h (Fig. 3B). Another important product-related issue was the influence of the individual system on the distribution of soluble versus insoluble recombinant SOD (Fig. 4A to D). During the first 2 h past induction, the plasmid-based system was able to produce the recombinant protein mainly in its soluble form; in the later stage of the process, the protein mainly accumulated as inclusion bodies. Finally, the plasmid system yielded less than 40% of the recombinant protein in a correctly folded and soluble form, whereas the genome-encoded system yielded more than 80% as soluble protein (Fig. 4B and D). The volumetric yield of 4.5 ± 0.21 g · liter−1 soluble SOD in the genome-encoded production system represents a 3.8-fold increase in the product titer under the given process conditions compared to the level seen with the plasmid-based system (Fig. 4D). With respect to the total product yields obtained, the plasmid-free system showed a 1.9-fold increase in the total volumetric yield of SOD (soluble plus insoluble protein) (Fig. 4E) and a 3.6-fold increase in the total recombinant protein yield (Fig. 4F).
Fig 3.
Comparison of product data from the feed phase of cultivations performed with the plasmid-based system [BL21(DE3)(pET30aSOD), open circles] and the plasmid-free system [BL21(DE3)::TN7<T7-SOD>, filled circles]. Determination of the induction of recombinant gene expression was conducted at feed hour 7. (A) Specific (spec) concentrations of SOD (soluble and insoluble) in mg per gram CDM. (B) Specific (spec.) product formation rates in mg per gram CDM and hour. The dashed line indicates the time point of induction. Data shown are derived from three independent replicate experiments (n = 3).
Fig 4.
Comparison of product data from the feed phase of cultivations performed with the plasmid-based system [BL21(DE3)(pET30aSOD), open circles] and the plasmid-free system [BL21(DE3)::TN7<T7-SOD>, filled circles]. Determination of the induction of recombinant gene expression was conducted at feed hour 7. (A) Specific (spec) concentrations of insoluble SOD aggregated in inclusion bodies in mg per gram CDM. (B) Specific (spec.) concentrations of soluble SOD in mg per gram CDM. (C) Volumetric yields of insoluble SOD in grams per liter. (D) Volumetric yields of soluble SOD in grams per liter. (E) Total volumetric yield (soluble plus insoluble) of SOD in grams per liter. (F) Total recombinant protein expressed in grams. The dashed line indicates the time point of induction. Data shown are derived from three independent replicate experiments (n = 3).
DNA μ-array transcription profiling experiments.
In order to investigate the impact of plasmid- and genome-encoded recombinant gene expression on host cell metabolism in more detail, genome-wide transcription profiling experiments were conducted. We used spotted-oligonucleotide DNA microarrays (DNA μ-arrays) for transcriptome analysis and the digital nCounter system (30) for the validation of DNA μ-array data and the quantification of mRNA counts for a selected subset of genes (see Data Set S1 in the supplemental material).
Analysis of genome-wide response to induction of recombinant protein expression.
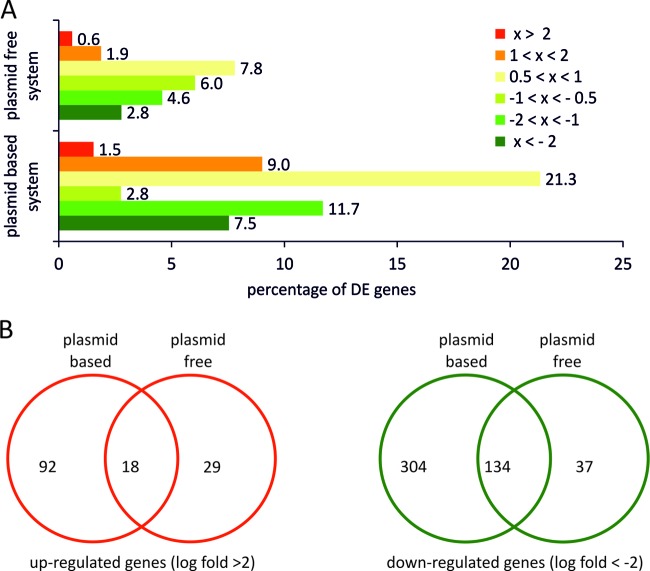
To analyze the genome-wide transcriptional response, the differentially expressed (DE) genes were categorized according to the log fold change (an absolute log fold change > 0.5 was taken as the minimum for DE). Within the first 4 h past induction, transcriptional responses were detected for both systems. The percentage of differentially expressed genes with an absolute log fold change > 0.5 from samples of both systems is shown in Fig. 5A. For BL21(DE3)(pET30aSOD), 9% of all genes showed an absolute log fold change > 2, 30% of all genes showed DE with an absolute log fold change > 1, and only 46% of genes remained unaffected in this cellular state. Further, we observed a significant reduction in the level of DE genes starting 7 h past induction. This decline of the initial transcriptional response arose in connection with the occurrence of nonproducing, plasmid-free cells that showed a different transcriptional profile, with fewer genes up- and downregulated. For BL21(DE3)::TN7<SOD>, changes in the transcriptional profile were less pronounced. Again, at 4 h past induction a steady state was reached, with 3% of the genes showing an absolute log fold change > 2, 10% of the genes with an absolute log fold change > 1, and 76% of the genes unaffected. This cellular state was maintained for the rest of the production phase without significant global changes.
Fig 5.
(A) Genome-wide overview of differentially expressed (DE) genes of the plasmid-based expression system [BL21(DE3)(pET30aSOD)] and the plasmid-free expression system [BL21(DE3)::TN7<T7-SOD>] at 4 h past induction. “x” indicates the log fold change in mRNA levels relative to the reference sample at the time point of induction. (B) Venn diagrams for up- and downregulated genes with log fold changes greater than 2 and less than −2, respectively.
For further analysis, all genes that showed an absolute log fold change > 2 were selected from both data sets and visualized using Venn diagrams (Fig. 5B). Among all 4,537 analyzed genes, 304 were significantly downregulated in the plasmid-based expression system whereas only 37 showed downregulation in the plasmid-free expression system compared to the noninduced reference system. Of the 4,537 genes, a total of 134 were downregulated in both systems. Similar behavior was observed for the upregulated genes, where 92 genes were found to be significantly upregulated in the plasmid-based system and only 29 in the plasmid-free system. A total of 18 genes were shared by the two expression systems.
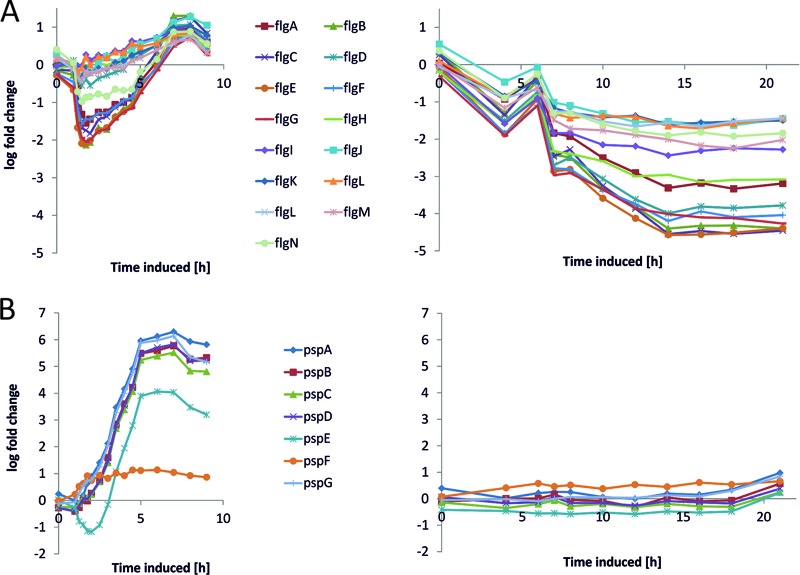
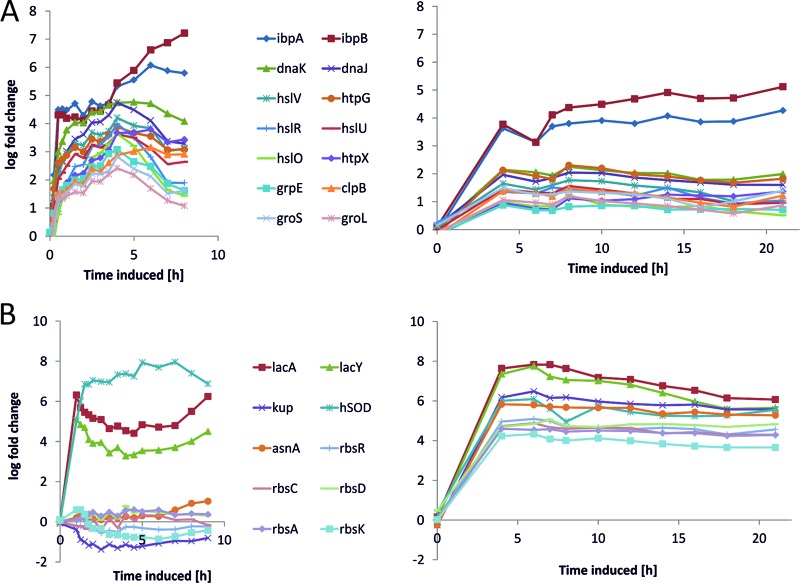
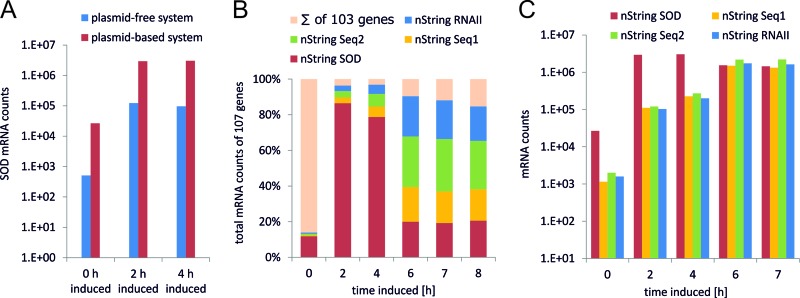
Using the set of genes with an absolute log fold change > 2, we performed a GO (Gene Ontology) term enrichment analysis using AmiGO (43). This analysis revealed that a significant proportion of genes that were downregulated only in the genome-based expression system are associated with GO term GO:0019861, the flagellum (Fig. 6A). The group of genes that were downregulated only in the plasmid-based system is associated with GO term GO:0030964, comprising respiration-related NADH dehydrogenase I-encoding genes. Top tables of DE genes revealed the genes of the phage shock protein (Psp) operon belonging to the group of genes being exclusively upregulated in BL21(DE3)(pET30aSOD) (Fig. 6B). The group of genes that were upregulated in both systems includes a significant number of chaperones and proteases involved in correct protein processing (Fig. 7A). To gain further insights, we analyzed our samples using the digital nCounter technology. We observed that the counts for the SOD probe (see Fig. 9B, nString SOD) were significantly higher (>1.3 log fold) in the case of the plasmid-based expression system (Fig. 8A). Remarkably, 97% of the total counts determined for the 108 nString probes after induction originated from plasmid-associated oligonucleotides (Fig. 8B; see also Data Set S1 in the supplemental material).
Fig 6.
(A) Transcription profiles of genes of the flg operon. (B) Genes belonging to the phage shock protein (psp gene) system (operon pspABCDE, pspF, and pspG) shown for the plasmid-based expression system (left charts) and the genome-based expression system (right charts).
Fig 7.
(A) Transcription profiles of selected heat shock proteins and chaperones shown for the plasmid-based expression system (left chart) and the genome-based expression system (right chart). (B) Selected cluster of transcription profiles from the genome-based system (right chart) and transcription profiles of these genes in the plasmid-based system (left chart).
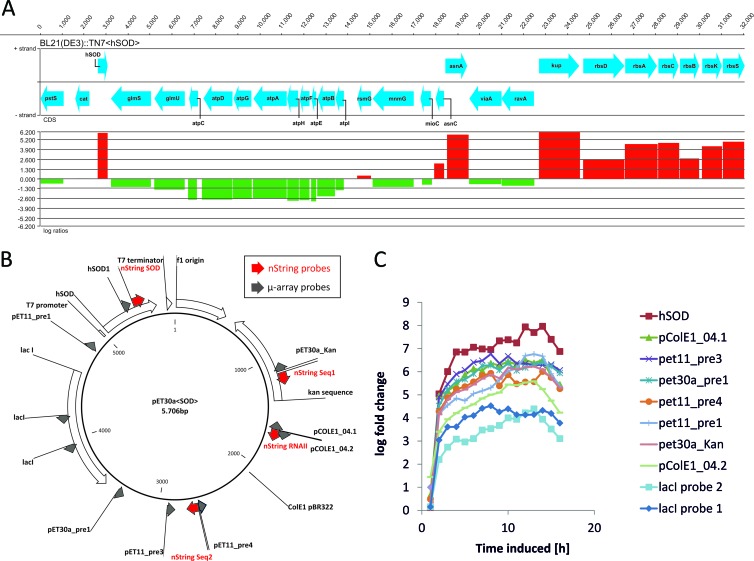
Fig 9.
(A) Log fold changes of transcript levels 4 h past induction drawn for the genomic region downstream of the attTn7 site (pstS to glmS). The genes located on the positive strand (in the same direction as the SOD gene under the control of the T7 promoter) are significantly upregulated upon induction with IPTG. (B) pET30a<SOD> plasmid map, with all plasmid-related probes. NanoString nCounter probes (nString) are shown in red; μ-array probes are shown in gray. (C) Transcription profiles of respective mRNAs in the experiments performed with the plasmid-based system.
Fig 8.
NanoString nCounter analysis results in the time course of the bioprocess. (A) Numbers of SOD mRNA molecules in samples of the plasmid-free and the plasmid-based systems. (B) Percentages of nString counts for SOD, Seq1, Seq2, and RNAII in relation to total nString counts of 107 genes. (C) nStrings counts for SOD, Seq1, and Seq2 (sequences in the plasmid backbone) and RNAII (preprimer in the plasmid ori).
Read-through transcription by the T7 RNA polymerase.
To identify groups of coregulated genes, we performed cluster analysis of the DE genes in both data sets, using the gcExplorer R package (41). We found that several genes in the plasmid-free system showed a transcription profile similar to that of the GOI transcribed via the use of T7-RNA polymerase on the plasmid system (Fig. 9A). Those genes were located downstream of the chromosomal integration site of the sod gene. An overview of the genomic region adjacent to the attTn7 integration site together with the log ratios of the corresponding genes can be found in Fig. 9A. Eight genes on the sense strand, downstream of the T7 integration site of the SOD gene, were significantly upregulated, corresponding to a region as large as 32 kbp. Our μ-array platform includes DNA probes designed specifically for the pET plasmid, and, as shown in Fig. 9B and C, all of the corresponding mRNAs were strongly upregulated (log fold change > 4). The read-through phenomenon was also confirmed with the NanoString nCounter technology for both the plasmid-based system (Fig. 8C) and the plasmid-free system (see Data Set S1 in the supplemental material).
DISCUSSION
In this work, we compared a plasmid-free expression system [BL21(DE3)::TN7<SOD>] and a plasmid-based expression system [BL21(DE3)(pET30aSOD] under production conditions in a fed-batch mode to identify plasmid-mediated effects on host metabolism during production of the recombinant protein SOD. Therefore, comprehensive data on growth characteristics, product formation, PCN, stress response, and transcription profiles were analyzed in detail.
The results clearly showed that plasmid replication itself does not trigger significant changes in cell metabolism in nonproducing cells at a growth rate of 0.1 h−1, as the two systems behaved similarly before induction. This confirms the findings of Diaz Ricci and Hernandez showing that at low growth rates the influence on cell growth is almost negligible (25). However, after induction of recombinant gene expression we observed many differences in host cell responses to recombinant gene expression. In contrast to an only moderate decrease in the growth rate of the plasmid-free system, a fundamental reduction of the growth rate in the plasmid-based system was observed (Fig. 1). Based on these significant differences, substantial variance in product formation was also expected, but we observed similar amounts of specific total SOD per gram CDM in the two systems (Fig. 3A). Interestingly, the average product formation rate in the single-copy, plasmid-free system was only 20% lower than that of the plasmid-based, multicopy system and did not cause significant adverse reactions with respect to host metabolism. Irrespective of product formation rates, the protein folding machinery of the plasmid-based system was also negatively affected, as the main fraction of SOD was accumulated in inclusion bodies. Hence, we conclude that the massive translation of the GOI observed for both systems is not the principal reason for metabolic overload in the plasmid-based system but rather that a series of superposed effects caused by the plasmid overstrain the cell metabolism shortly after induction. Among these effects are the 40-times-higher copy number of the GOI in the plasmid-based system and the thus much higher transcription rates and mRNA levels of the GOI (Fig. 8A). After induction, the PCN did increase more than 4-fold (Fig. 2A), potentiating the adverse metabolic side effects. This phenomenon can possibly be attributed to the occurrence of the uncharged tRNAs (23, 44) that do interact with the plasmid replication control mechanism and additionally trigger the stringent response (45, 46). Consequently, the high levels of ppGpp detected for the plasmid-based system may also contribute to the observed growth cessation (47). This assumption is also supported by the significant drop in PCN that we observed (Fig. 2A). This rapid decrease of PCN, within only 3 h, could have been due to (i) inhibition of DNA replication, (ii) plasmid loss due to recombination and consequent multimerization (48), or (iii) targeted degradation of the recombinant plasmid by, e.g., clustered regularly interspaced short palindromic repeat (CRISPR) endonuclease (49). The causality seems to be as follows: (i) massive translation of the GOI leads to the occurrence of uncharged tRNAs; (ii) these uncharged tRNAs do interact with the plasmid replication control mechanism, further increasing the PCN; and (iii) these elevated PCN consequently cause further increases in the mRNA levels. Finally, the cell finds itself trapped in a vicious circle in which it is forced to operate far beyond its usual cellular capacities.
Concerning the performed comparative μArray analysis, we revealed that downregulation was predominant in both systems and that the number of genes with an absolute log fold change > 2 was 2.8 times higher for the plasmid-based expression system than for the plasmid-free expression system (Fig. 5). An example of responses in the two systems that were nearly uniform but included a more pronounced response in BL21(DE3)(pET30aSOD) is the upregulation of genes encoding chaperones and proteases (Fig. 7). With regard to protein quality control, this response should support correct folding of the recombinant protein (50). The fact that SOD was mainly produced as inclusion bodies in the plasmid-based system (Fig. 4A and C) indicates that protein aggregates sequestered chaperones and proteases otherwise necessary to maintain proteostasis (51) or that translation of these mRNAs into functional proteins seems to have been disabled. In addition, protein aggregation could also be the reason for growth cessation (52–54). We also observed that the Psp operon is exclusively upregulated in BL21(DE3)(pET30aSOD). The Psp system responds to extra cytoplasmatic stress that may reduce the energy status of the cell. Expression of the psp operon is normally triggered by the occurrence of mislocalized secretins caused by the absence of specific chaperone-like pilot proteins or by defects in maintenance of the proton motive force (55). These results are in accordance with our hypothesis that the folding apparatus is severely overstrained in the plasmid-based expression system. The observation that transcription of the nuo operon is downregulated indicates an energy shortage and a reduced proton motive force, since NADH dehydrogenase transfers electrons from NADH to the respiratory chain (56). According to our data, the response of the plasmid-free system is not limited to a reduced response following the same pattern as that seen with the plasmid-based system but instead shows totally different characteristics for subsets of genes (Fig. 6). The plasmid-free expression system shuts down expression of genes associated with the assembly of the flagellum (Fig. 6A). Since synthesis of the flagella is a complex and energy-consuming process (57) and yet is not essential, it becomes obvious that the cellular response to an additional metabolic burden includes the cessation of such a dissipative process.
Additionally, we observed read-through transcription of the T7 RNA polymerase into adjacent genes located downstream of the GOI. According to our data, substantial termination was not provided upstream of the strong terminator of the rrsC-gltU-rrlC-rrfC ribosomal operon. It was reported before that the T7 terminator provides a limited termination efficiency of only 80% (58–61) due to the fact that it evolved to allow read-through in order to enable expression of T7 genes located further downstream (62). Improving the efficiency of the T7 terminator for the expression of multiple genes has been reported recently (63, 64) and might be a useful approach to avoid this effect. However, two points have to be considered. (i) The T7 terminator is not an adequate transcription termination signal for synthetic biology or metabolic engineering applications, since cross talk of the artificially introduced transcription unit, causing possible instabilities and disruption of regulons when integrated in the genome, is likely. (ii) Plasmid-based expression systems seem to be affected by the same read-through, leading to long transcripts containing, e.g., noncoding RNA structures (e.g., RNAII in the case of ColE1-like plasmids). Such sequence elements may interact with the plasmid replication mechanism, possibly explaining the increase in the PCN (Fig. 2). We therefore assume that this read-through transcription does constitute a major problem in the plasmid-based expression system where (i) rapid plasmid loss was observed upon induction and (ii) the regulatory status of the replicon is influenced, resulting in either lower or higher replication rates of the plasmid. In the case of the genome-integrated, plasmid-free expression system, this read-through transcription had no significant negative effect on the performance of the expression system.
Finally, we identified the high gene dosage and consequently massive overtranscription of the GOI in the plasmid-based expression system (Fig. 8) as one of the major factors responsible for the metabolic overburden. Thus, we assume that high concentrations of a recombinant protein within the cells are not detrimental to the host cell factory per se. Instead, the major source of the problem is the overabundance of the mRNA of the GOI (Fig. 8B). This large amount of mRNA occupies ribosomes and diverts amino acids and nucleotide precursors that would otherwise be required to maintain cellular integrity and functionality. Here, we have demonstrated that plasmid-free expression systems are applicable reference systems for investigation of host/plasmid interactions, and we conclude that they represent an attractive alternative in terms of both up- and downstream processing.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Federal Ministry of Traffic, Innovation and Technology (bmvit), the Federal Ministry of Economy, Family and Youth (BMWFJ), the Styrian Business Promotion Agency (SFG), and the Standortagentur Tirol and ZIT-Technology Agency of the City of Vienna through the COMET-Funding Program managed by the Austrian Research Promotion Agency (FFG).
The technical assistance of Florian Strobl and Norbert Auer is highly appreciated. Thanks also to Peter M. Krempl and Oana A. Tomescu from the Institute for Genomics and Bioinformatics, Graz University of Technology, for management and submission of microarray data.
We declare that we have no conflicts of interest.
J.M. participated in the design of the study, was involved in analysis and interpretation, and drafted the manuscript. T.S. performed the statistical analysis of the microarray data sets. K.M. and G.S. carried out the cultivation and μ-array experiments. M.C.-P. was involved in the off-line process analytics (ELISA, SDS-PAGE, and HPLC). All of us were involved in interpretation of data and assembly of the manuscript. G.S. conceived and directed the study and drafted the manuscript. All of us read and approved the final manuscript.
Footnotes
Published ahead of print 12 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00365-13.
REFERENCES
- 1. Cohen SN, Chang AC, Hsu L. 1972. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc. Natl. Acad. Sci. U. S. A. 69:2110–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cohen S, Chang A, Boyer H, Helling R. 1973. Construction of biologically functional bacterial plasmids in vitro. Proc. Natl. Acad. Sci. U. S. A. 70:3240–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. del Solar G, Giraldo R, Ruiz-Echevarría MJ, Espinosa M, Díaz-Orejas R. 1998. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 62:434–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Camps M. 2010. Modulation of ColE1-like plasmid replication for recombinant gene expression. Recent Pat. DNA Gene Seq. 4:58–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kittleson JT, Cheung S, Anderson JC. 2011. Rapid optimization of gene dosage in E. coli using DIAL strains. J. Biol. Eng. 5:10 doi: 10.1186/1754-1611-5-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baneyx F. 1999. Recombinant protein expression in Escherichia coli. Curr. Opin. Biotechnol. 10:411–421 [DOI] [PubMed] [Google Scholar]
- 7. Del Tito BJJ, Ward JM, Hodgson J, Gershater CJ, Edwards H, Wysocki LA, Watson FA, Sathe G, Kane JF. 1995. Effects of a minor isoleucyl tRNA on heterologous protein translation in Escherichia coli. J. Bacteriol. 177:7086–7091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Studier FW. 2005. Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 41:207–234 [DOI] [PubMed] [Google Scholar]
- 9. FDA 1982. Human insulin receives FDA approval. FDA Drug Bull. 12:18–19 [PubMed] [Google Scholar]
- 10. Hoffmann F, Rinas U. 2004. Stress induced by recombinant protein production in Escherichia coli. Adv. Biochem. Eng. Biotechnol. 89:73–92 [DOI] [PubMed] [Google Scholar]
- 11. Golomb M, Chamberlin M. 1974. Characterization of T7-specific ribonucleic acid polymerase. IV. Resolution of the major in vitro transcripts by gel electrophoresis. J. Biol. Chem. 249:2858–2863 [PubMed] [Google Scholar]
- 12. Vogel U, Jensen KF. 1994. The RNA chain elongation rate in Escherichia coli depends on the growth rate. J. Bacteriol. 176:2807–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dong H, Nilsson L, Kurland CG. 1995. Gratuitous overexpression of genes in Escherichia coli leads to growth inhibition and ribosome destruction. J. Bacteriol. 177:1497–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bentley WE, Mirjalili N, Andersen DC, Davis RH, Kompala DS. 1990. Plasmid-encoded protein: the principal factor in the “metabolic burden” associated with recombinant bacteria. Biotechnol. Bioeng. 35:668–681 [DOI] [PubMed] [Google Scholar]
- 15. Zahn K. 1996. Overexpression of an mRNA dependent on rare codons inhibits protein synthesis and cell growth. J. Bacteriol. 178:2926–2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Glick BR. 1995. Metabolic load and heterologous gene expression. Biotechnol. Adv. 13:247–261 [DOI] [PubMed] [Google Scholar]
- 17. Birnbaum SBJ. 1991. Plasmid presence changes the relative levels of many host cell proteins and ribosome components in recombinant Escherichia coli. Biotechnol. Bioeng. 37:736–745 [DOI] [PubMed] [Google Scholar]
- 18. Neidhardt FC, Ingraham JL, Schaechter M. 1990. Physiology of the bacterial cell: a molecular approach. Sinauer Associates, Sunderland, MA [Google Scholar]
- 19. Bentley WE, Kompala DS. 1990. Plasmid instability in batch cultures of recombinant bacteria. a laboratory experiment. Chem. Eng. Educ. 24:168–172 [Google Scholar]
- 20. Summers D, Sherratt D. 1984. Multimerization of high copy number plasmids causes instability: CoIE1 encodes a determinant essential for plasmid monomerization and stability. Cell 36:1097–1103 [DOI] [PubMed] [Google Scholar]
- 21. Summers DK, Beton CW, Withers HL. 1993. Multicopy plasmid instability: the dimer catastrophe hypothesis. Mol. Microbiol. 8:1031–1038 [DOI] [PubMed] [Google Scholar]
- 22. Popov M, Petrov S, Nacheva G, Ivanov I, Reichl U. 2011. Effects of a recombinant gene expression on ColE1-like plasmid segregation in Escherichia coli. BMC Biotechnol. 11:18 doi: 10.1186/1472-6750-11-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grabherr R, Nilsson E, Striedner G, Bayer K. 2002. Stabilizing plasmid copy number to improve recombinant protein production. Biotechnol. Bioeng. 77:142–147 [DOI] [PubMed] [Google Scholar]
- 24. Silva F, Queiroz JA, Domingues FC. 2012. Evaluating metabolic stress and plasmid stability in plasmid DNA production by Escherichia coli. Biotechnol. Adv. 30:691–708 [DOI] [PubMed] [Google Scholar]
- 25. Diaz Ricci JC, Hernandez ME. 2000. Plasmid effects on Escherichia coli metabolism. Crit. Rev. Biotechnol. 20:79–108 [DOI] [PubMed] [Google Scholar]
- 26. Terpe K. 2006. Overview of bacterial expression systems for heterologous protein production: from molecular and biochemical fundamentals to commercial systems. Appl. Microbiol. Biotechnol. 72:211–222 [DOI] [PubMed] [Google Scholar]
- 27. Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60–89 [DOI] [PubMed] [Google Scholar]
- 28. Marchand I, Nicholson AW, Dreyfus M. 2001. High-level autoenhanced expression of a single-copy gene in Escherichia coli: overproduction of bacteriophage T7 protein kinase directed by T7 late genetic elements. Gene 262:231–238 [DOI] [PubMed] [Google Scholar]
- 29. Striedner G, Pfaffenzeller I, Markus L, Nemecek S, Grabherr R, Bayer K. 2010. Plasmid-free T7-based Escherichia coli expression systems. Biotechnol. Bioeng. 105:786–794 [DOI] [PubMed] [Google Scholar]
- 30. Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, James JJ, Maysuria M, Mitton JD, Oliveri P, Osborn JL, Peng T, Ratcliffe AL, Webster PJ, Davidson EH, Hood L. 2008. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 26:317–325 [DOI] [PubMed] [Google Scholar]
- 31. Kramer W, Elmecker G, Weik R, Mattanovich D, Bayer K. 1996. Kinetics studies for the optimization of recombinant protein formation. Ann. N. Y. Acad. Sci. 782:323–333 [DOI] [PubMed] [Google Scholar]
- 32. Sharan SK, Thomason LC, Kuznetsov SG, Court DL. 2009. Recombineering: a homologous recombination-based method of genetic engineering. Nat. Protoc. 4:206–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cserjan-Puschmann M, Kramer W, Duerrschmid E, Striedner G, Bayer K. 1999. Metabolic approaches for the optimisation of recombinant fermentation processes. Appl. Microbiol. Biotechnol. 53:43–50 [DOI] [PubMed] [Google Scholar]
- 34. Clementschitsch F, Jurgen K, Florentina P, Karl B. 2005. Sensor combination and chemometric modelling for improved process monitoring in recombinant E. coli fed-batch cultivations. J. Biotechnol. 120:183–196 [DOI] [PubMed] [Google Scholar]
- 35. Hegde P, Qi R, Abernathy K, Gay C, Dharap S, Gaspard R, Hughes JE, Snesrud E, Lee N, Quackenbush J. 2000. A concise guide to cDNA microarray analysis. Biotechniques 29:548–550, 552,–554, 556 [DOI] [PubMed] [Google Scholar]
- 36. Buhler J, Ideker T, Haynor D. 2000. Dapple: improved techniques for finding spots on DNA microarrays. CSE Technical Report UWTR 2000-08-05. Washington University in St. Louis, St. Louis, MO: http://www.cse.wustl.edu/∼jbuhler/dapple/dapple-tr.pdf [Google Scholar]
- 37. Team RDC 2012. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 38. Huber W, von Heydebreck A, Sültmann H, Poustka A, Vingron M. 2002. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18(Suppl 1):S96–S104 [DOI] [PubMed] [Google Scholar]
- 39. Smyth GK. 2005. Limma: linear models for microarray data, p 397–420 In Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S. (ed), Bioinformatics and computational biology solutions using R and Bioconductor (statistics for biology and health). Springer-Verlag, New York, NY [Google Scholar]
- 40. Leisch F. 2006. A toolbox for K-centroids cluster analysis. Comput. Stat. Data Anal. 51:526–544 [Google Scholar]
- 41. Scharl T, Leisch F. 2009. gcExplorer: interactive exploration of gene clusters. Bioinformatics 25:1089–1090 [DOI] [PubMed] [Google Scholar]
- 42. Scharl T, Striedner G, Poetschacher F, Leisch F, Bayer K. 2009. Interactive visualization of clusters in microarray data: an efficient tool for improved metabolic analysis of E. coli. Microb. Cell Fact. 8:37 doi: 10.1186/1475-2859-8-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S, AmiGO Hub, Web Presence Working Group 2009. AmiGO: online access to ontology and annotation data. Bioinformatics 25:288–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Grabherr R, Bayer K. 2002. Impact of targeted vector design on Co/E1 plasmid replication. Trends Biotechnol. 20:257–260 [DOI] [PubMed] [Google Scholar]
- 45. Goldman E, Jakubowski H. 1990. Uncharged tRNA, protein synthesis, and the bacterial stringent response. Mol. Microbiol. 4:2035–2040 [DOI] [PubMed] [Google Scholar]
- 46. Dalebroux ZD, Swanson MS. 2012. ppGpp: magic beyond RNA polymerase. Nat. Rev. Microbiol. 10:203–212 [DOI] [PubMed] [Google Scholar]
- 47. Herman A, Wegrzyn G. 1995. Effect of increased ppGpp concentration on DNA replication of different replicons in Escherichia coli. J. Basic Microbiol. 35:33–39 [DOI] [PubMed] [Google Scholar]
- 48. Summers DK. 1991. The kinetics of plasmid loss. Trends Biotechnol. 9:273–278 [DOI] [PubMed] [Google Scholar]
- 49. Karginov FV, Hannon GJ. 2010. The CRISPR system: small RNA-guided defense in bacteria and archaea. Mol. Cell 37:7–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Baneyx F, Mujacic M. 2004. Recombinant protein folding and misfolding in Escherichia coli. Nat. Biotechnol. 22:1399–1408 [DOI] [PubMed] [Google Scholar]
- 51. Mogk A, Huber D, Bukau B. 2011. Integrating protein homeostasis strategies in prokaryotes. Cold Spring Harb. Perspect. Biol. 3:a004366 doi: 10.1101/cshperspect.a004366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Winkler J, Seybert A, Konig L, Pruggnaller S, Haselmann U, Sourjik V, Weiss M, Frangakis AS, Mogk A, Bukau B. 2010. Quantitative and spatio-temporal features of protein aggregation in Escherichia coli and consequences on protein quality control and cellular ageing. EMBO J. 29:910–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Maisonneuve E, Ezraty B, Dukan S. 2008. Protein aggregates: an aging factor involved in cell death. J. Bacteriol. 190:6070–6075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lindner AB, Madden R, Demarez A, Stewart EJ, Taddei F. 2008. Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. Proc. Natl. Acad. Sci. U. S. A. 105:3076–3081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Darwin AJ. 2005. The phage-shock-protein response. Mol. Microbiol. 57:621–628 [DOI] [PubMed] [Google Scholar]
- 56. Friedrich T. 1998. The NADH:ubiquinone oxidoreductase (complex I) from Escherichia coli. Biochim. Biophys. Acta 1364:134–146 [DOI] [PubMed] [Google Scholar]
- 57. Zhao K, Liu M, Burgess RR. 2007. Adaptation in bacterial flagellar and motility systems: from regulon members to ‘foraging’-like behavior in E. coli. Nucleic Acids Res. 35:4441–4452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Macdonald LE, Durbin RK, Dunn JJ, McAllister WT. 1994. Characterization of two types of termination signal for bacteriophage T7 RNA polymerase. J. Mol. Biol. 238:145–158 [DOI] [PubMed] [Google Scholar]
- 59. Sousa R, Patra D, Lafer EM. 1992. Model for the mechanism of bacteriophage T7 RNAP transcription initiation and termination. J. Mol. Biol. 224:319–334 [DOI] [PubMed] [Google Scholar]
- 60. Telesnitsky AP, Chamberlin MJ. 1989. Sequences linked to prokaryotic promoters can affect the efficiency of downstream termination sites. J. Mol. Biol. 205:315–330 [DOI] [PubMed] [Google Scholar]
- 61. Telesnitsky A, Chamberlin MJ. 1989. Terminator-distal sequences determine the in vitro efficiency of the early terminators of bacteriophages T3 and T7. Biochemistry 28:5210–5218 [DOI] [PubMed] [Google Scholar]
- 62. Dunn JJ, Studier FW. 1983. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J. Mol. Biol. 166:477–535 [DOI] [PubMed] [Google Scholar]
- 63. Du L, Gao R, Forster AC. 2009. Engineering multigene expression in vitro and in vivo with small terminators for T7 RNA polymerase. Biotechnol. Bioeng. 104:1189–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Du L, Villarreal S, Forster AC. 2012. Multigene expression in vivo: supremacy of large versus small terminators for T7 RNA polymerase. Biotechnol. Bioeng. 109:1043–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.