Abstract
Plant-derived aromatic compounds are important components of the dissolved organic carbon pool in coastal salt marshes, and their mineralization by resident bacteria contributes to carbon cycling in these systems. Members of the roseobacter lineage of marine bacteria are abundant in coastal salt marshes, and several characterized strains, including Sagittula stellata E-37, utilize aromatic compounds as primary growth substrates. The genome sequence of S. stellata contains multiple, potentially competing, aerobic ring-cleaving pathways. Preferential hierarchies in substrate utilization and complex transcriptional regulation have been demonstrated to be the norm in many soil bacteria that also contain multiple ring-cleaving pathways. The purpose of this study was to ascertain whether substrate preference exists in S. stellata when the organism is provided a mixture of aromatic compounds that proceed through different ring-cleaving pathways. We focused on the protocatechuate (pca) and the aerobic benzoyl coenzyme A (box) pathways and the substrates known to proceed through them, p-hydroxybenzoate (POB) and benzoate, respectively. When these two substrates were provided at nonlimiting carbon concentrations, temporal patterns of cell density, gene transcript abundance, enzyme activity, and substrate concentrations indicated that S. stellata simultaneously catabolized both substrates. Furthermore, enhanced growth rates were observed when S. stellata was provided both compounds simultaneously compared to the rates of cells grown singly with an equimolar concentration of either substrate alone. This simultaneous-catabolism phenotype was also demonstrated in another lineage member, Ruegeria pomeroyi DSS-3. These findings challenge the paradigm of sequential aromatic catabolism reported for soil bacteria and contribute to the growing body of physiological evidence demonstrating the metabolic versatility of roseobacters.
INTRODUCTION
The structural diversity of aromatic compounds in the environment is influenced by the various mechanisms that produce them, including naturally occurring abiotic (1) and biotic (2, 3) processes as well as those of anthropogenic origin (4). Regardless of the source, the dissolved organic carbon pool containing aromatic compounds in nature is typically structurally heterogeneous (5, 6). This is an important consideration for microbial degradation and has received significant attention in studies examining the catabolism of aromatic compound mixtures classified as environmental pollutants (7). Considerably less attention has been focused on microbial physiology of mixtures of naturally occurring aromatic compounds, such as those derived from lignin, the structural component of vascular plants (8).
Microbial mineralization of aromatic compounds plays an important role in global carbon cycling and bioremediation. Bacterial aromatic catabolism is described as “catabolic funneling” where upper (also called peripheral) pathways transform a diverse suite of aromatic compounds into one of a limited number of intermediates that are then subject to ring cleavage (9). The β-ketoadipate pathway is one such pathway and is a paradigm for aerobic catabolism of plant-derived aromatics (10, 11). In organisms possessing one or both its branches, peripheral pathways generate dihydroxylated intermediates, either catechol or protocatechuate. Alternative aerobic ring-cleaving mechanisms are also present in bacteria, including epoxidation of coenzyme A (CoA)-thioesterified aromatics, as occurs in the benzoyl-CoA (box) pathway for benzoate degradation (12). These catabolic pathways are typically under tight transcriptional regulation (13, 14) and subject to catabolite repression (15–17).
Previous studies reveal that substrate preferences are the norm when bacterial strains are presented with a mixture of aromatic compounds (9). This phenomenon has been best characterized for selected soil-derived bacteria provided mixtures of benzoate and p-hydroxybenzoate (POB) (18–20). Each of these compounds is ultimately processed through parallel ring cleavage branches of the β-ketoadipate pathway, referred to as the catechol (cat) and protocatechuate (pca) branches (21), respectively. The hierarchical nature of substrate utilization profiles has been mechanistically explained as cross-regulation and typically involves transcriptional regulation by pathway metabolites or regulatory proteins (for examples, see references 22 and 23). However, the extent to which similar hierarchies exist in environmentally relevant microbes is not yet clear. Furthermore, as environmental bacteria are dependent upon growth substrate pools that are highly heterogeneous in composition (24, 25), mixed-substrate studies may provide the foundation for a better understanding of bacterial catabolism in nature.
The Roseobacter lineage of marine bacteria is numerically abundant and active in the world's oceans (26, 27). Group members are most dominant in coastal environments, including salt marshes heavily influenced by lignocellulosic vascular plant material (28). Roseobacter ubiquity and success in the oceans have been attributed, in part, to their ability to use a large repertoire of growth substrates, including aromatic compounds (29–31). Genome analyses have identified several ring-cleaving pathways in roseobacters, including the box and pca pathways (30). Yet it is unknown whether roseobacters show evidence of substrate preference when presented with mixtures of aromatic compounds representative of compounds derived from vascular plants abundant in coastal marine habitats.
MATERIALS AND METHODS
Media and growth conditions.
A marine basal medium (MBM) containing 1.5% (wt/vol) sea salts (Sigma-Aldrich, St. Louis, MO) with 225 nM K2HPO4, 13.35 μM NH4Cl, 71 mM Tris-Cl (pH 7.5), 68 μM Fe-EDTA, trace metals, and vitamins was used to culture Ruegeria pomeroyi DSS-3 and Sagittula stellata E-37 at 30°C (32). Variovorax paradoxus EPS was also cultured at 30°C in M9 basal medium (33). All growth experiments used cells preconditioned on 3.5 or 7 mM acetate at early stationary phase to match the carbon concentration of the new medium. Initial inocula were ≤1% transfer volume (∼105 cells ml−1). Benzoic acid and p-hydroxybenzoic acid were obtained from Sigma-Aldrich, and sodium acetate was obtained from Fisher Scientific (Waltham, MA). All glassware was combusted minimally for 6 h at 450°C to remove trace carbon, and negative (non-carbon-amended) controls of cells in basal medium were also performed.
Nucleic acid isolation.
For RNA preservation and isolation, approximately 108 cells were pelleted at 5,000 × g for 5 min, resuspended in 1 ml of RNAlater (Ambion, Austin, TX), and preserved for 1 h at room temperature. RNAlater was removed by aspiration following centrifugation at 6,000 × g for 5 min, and the cells were flash frozen in liquid N2 and stored at −70°C until processed. Cells were lysed by agitation in the presence of low-binding 200μm zirconium beads (OPS Diagnostics, L.L.C., Lebanon, NJ), and RNA was extracted using the RNeasy minikit (Qiagen, Valencia, CA). Genomic DNA was removed using the vigorous Turbo DNase (4 U) (Ambion) method as described in the product manual. Nucleic acids were quantified and purity was assessed with an ND-1000 spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE). Reverse transcription was carried out in 60-μl volumes containing 180 ng RNA, 600 U Moloney murine leukemia virus (M-MLV) reverse transcriptase (Invitrogen, Carlsbad, CA), 500 μg ml−1 random hexamers (Promega, Madison, WI), 120 U RNaseOUT (Invitrogen), 500 μM deoxynucleoside triphosphates (dNTPs) (Promega), and 10 mM dithiothreitol (DTT; Invitrogen). Initially, the random hexamers in the presence of dNTPs were annealed to the RNA for 5 min at 65°C, followed by an immediate transfer to ice to maintain the binding interaction. Next, to protect the mRNA and increase full-length cDNA yields, DTT and RNaseOUT were added and incubated for 2 min at 37°C. Finally, cDNAs were generated from the RNA by M-MLV reverse transcriptase by activation for 10 min at 25°C followed by 50 min of synthesis at 37°C. The enzymes were denatured and inactivated for 15 min with 70°C, and the remaining cDNA was stored at −20°C.
Gene transcription assays.
Reverse transcription-quantitative PCR (RT-qPCR) was used to assess relative gene expression. Transcripts diagnostic of the benzoyl-CoA pathway (boxA) and the protocatechuate branch of the β-ketoadipate pathway (pcaH) were measured and normalized to the expression of three reference genes (alaS, map, and rpoC). Primers were designed for each of these 5 genes and are shown in Table S1 in the supplemental material. All primer sets were optimized for qPCR using the following method. In 25-μl qPCR volumes, a matrix of forward and reverse primer concentrations (final concentrations ranging from 100 to 1,500 nM) was used along with a fixed concentration of E-37 genomic DNA (2.5 × 105 genomes reaction−1) and 1× SYBR Premix Ex Taq (Perfect Real Time) (TaKaRa Bio Inc., Otsu, Japan). The qPCR amplification included an initial 95°C denaturation for 15 min, followed by 40 cycles of amplification (95°C denaturation for 45 s, 58°C annealing for 45 s, and 72°C elongation and fluorescence detection for 15 s) and a final 72°C extension for 5 min. A melting curve from 50°C to 100°C read every 1°C was performed after each reaction to ensure that only a single product melted around 90°C. Within each primer set matrix, the combination of forward and reverse primer concentrations that yielded the lowest quantification cycle (Cq) was used in the subsequent RT-qPCR.
The 25-μl qPCR volumes consisted of 15% cDNA template, 577 μM oligonucleotide primers, and SYBR Premix Ex Taq (Perfect Real Time) (TaKaRa). Each cDNA template sample was amplified with primer sets to quantitate alaS, boxA, map, pcaH, and rpoC. Technical qPCR triplicates were performed for each of the five primer sets used to amplify the three biological triplicates at five time points. The cycling conditions were also the same as described for the qPCR optimization. Non-reverse-transcribed aliquots were also performed as negative controls to ensure that the RT-qPCR measurements represented cDNA concentration and that other nucleic acids' Cq values were negligible (>5 difference). Reference genes for normalization remained unchanged during the sampling time points and have been successfully applied as reference genes to another roseobacter in our laboratory (34). All normalized boxA and pcaH RT-qPCR data were relativized to their basal expression of E-37 cells grown on 7 mM acetate according to calculations described by Hellemans et al. (35). Technical replicate errors were propagated with a truncated first-order Taylor series expansion.
PcaHG enzyme assays.
Approximately 1010 cells were washed in a 4°C solution containing 1.5% sea salts (Sigma-Aldrich) and 50 mM Tris-acetate (pH 7.5). Rinsed and pelleted cells were suspended in 396 μl Bugbuster protein extraction reagent (Novagen, Inc., Madison, WI). Lysozyme (4 μl) was added (to a final concentration of 0.1 ng ml−1), and the cells were incubated at 30°C for 15 min. After the cell debris was removed via centrifugation (21,000 × g for 30 min at 4°C), crude cell lysates were assayed for protocatechuate 3,4-dioxygenase (PcaHG, EC 1.13.11.3) activity by measuring the kinetic loss of protocatechuate at 290 nm with a DU 800 UV-visible (UV-Vis) spectrophotometer (Beckman Coulter, Inc., Brea, CA) (36). Briefly, the 1-ml assays included 400 μM protocatechuate and 50 mM Tris-acetate (pH 7.5) along with ≥3 different volumes of lysate tested below the saturation point. Protein concentrations were determined with the Coomassie Plus protein assay reagent kit (Thermo Scientific Pierce, Rockford, IL) (37). The specific activity for each sample was calculated using the change in molar extinction coefficients (Δε) of εprotocatechuate and εβ-carboxymuconolactone (2,280 cm−1 M−1). E-37 cells grown solely on 7 mM acetate and on 2 mM protocatechuate served as negative and positive controls.
HPLC-PDA analysis of substrate concentrations.
Aromatic substrates in the spent media were separated with a Waters 2695 high-performance liquid chromatography (HPLC) instrument containing a reverse-phase 3.9- by 150-mm Novak-Pak C18 column (Waters Corp., Milford, MA) coupled to a Waters 2996 photodiode array (PDA) detector. For the spent MBM, an isocratic elution of 0.8 ml min−1 at 25°C with the mobile phase containing 30% MeCN and 0.07% phosphoric acid produced distinct peaks for benzoate (3.62 min) and POB (2.12 min). The same separation conditions were used for M9 spent medium, with the exception of increased (2.5%) phosphoric acid. A 10-point serial dilution curve of authentic standards was used to determine the concentration of each compound at their λmax values in the MBM solution, which were 230.3 and 256.2 nm for benzoate and POB, respectively. The peak area of each eluate was calculated with ApexTrack's integration tool using the Empower 2 Pro software package (Waters) for each of the technical (HPLC-PDA machine) triplicates performed on each sample. Linear regression of the temporally paired substrate concentrations was used to assess the statistical correlation between the catabolic/disappearance rate of each substrate using SigmaPlot 11.0 (Systat Software, Inc., Chicago, IL).
Ash-free dry mass (AFDM) measurements.
E-37 cells grown at 30°C with agitation at 200 rpm in 250-ml baffled flasks containing 100 ml 14 mM carbon in MBM were used to estimate the total carbon biomass yields on different growth substrates. Cells grown to early stationary phase on 2 mM benzoate, on 2 mM POB, and on 1 mM benzoate plus 1 mM POB were harvested by centrifugation at 6,000 × g for 10 min. To capture any cells that did not pellet, supernatants were passed through prewashed and preweighed fiberglass (GF/F) filters with a nominal (0.7-μm) pore size (Whatman Ltd., Maidstone, ME). Cell pellets were resuspended in 1 ml spent medium and filtered onto their respective GF/F filters. All biomass was dried on the filters for 24 h at 60°C, after which their masses were measured. The dried organic material was then combusted for 4 h at 450°C. The total carbon biomass for each population was determined, taking into account the combusted filter masses. Three biological replicates were performed for each growth substrate, and a one-way analysis of variance (ANOVA) was performed with Tukey's post hoc tests in SigmaPlot 11.0. One-tailed Student's t tests were performed with MS Excel 2010 (Microsoft Corp., Redmond, WA).
Genome analyses.
Using the NCBI genome database (http://blast.ncbi.nlm.nih.gov/), co-occurrences of box and pca pathways were identified in cultured taxa. The following protein sequences from organisms with experimental data were used as tBLASTn queries: BenA (benzoate 1,2-dioxygenase alpha subunit; YP_046122.1) and BenB (benzoate 1,2-dioxygenase beta subunit; YP_046123.1), CatA (catechol 1,2-dioxygenase; YP_046127.1) and CatB (muconate cycloisomerase I; YP_046131.1), BoxB (benzoyl-CoA 2,3-epoxidase; Q9AIX7.1) and BoxC (2,3-epoxybenzoyl-CoA dihydrolase; Q84HH6.1), PcaG (protocatechuate 3,4-dioxygenase alpha subunit; YP_046376.1) and PcaH (protocatechuate 3,4-dioxygenase beta subunit; YP_046375.1), and AraC-type PobR (transcriptional regulator; YP_299213.1) and IclR-type PobR (transcriptional regulator; YP_046382.1).
RESULTS
Sagittula stellata E-37, the representative roseobacter selected for these studies, is a coastal seawater strain that can catabolize a variety of plant-derived sugars as well as aromatic compounds representative of lignin breakdown products (32, 38, 39). Furthermore, it has been shown to selectively attach to lignocellulose particles as well as transform and partially mineralize synthetic lignin (32). Its genome contains six ring-cleaving pathways (aerobic benzoyl-CoA [box], gentisate [gtd], homogentisate [hmg], homoprotocatechuate [hpa], phenylacetate [paa], and protocatechuate [pca]) (30), making it an especially attractive model system to examine mixed-substrate growth during aromatic-compound catabolism. In order to facilitate comparisons with previous studies conducted with soil bacteria, we focused our efforts here on the structurally similar growth substrates benzoate and p-hydroxybenzoate (POB).
Catabolic funneling in Sagitulla stellata E-37.
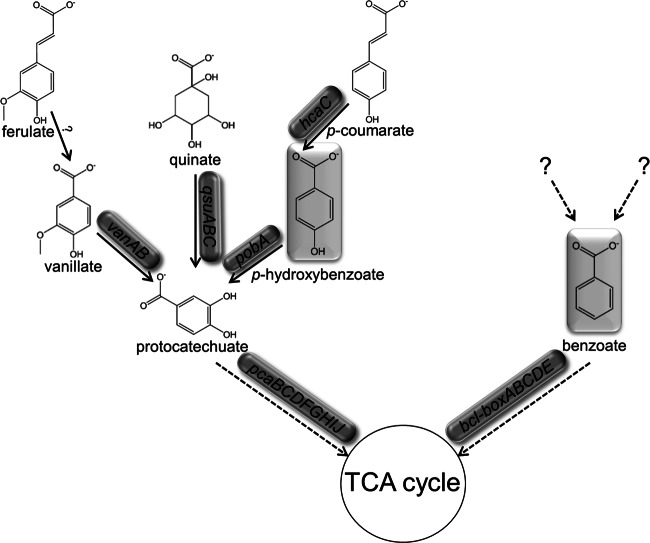
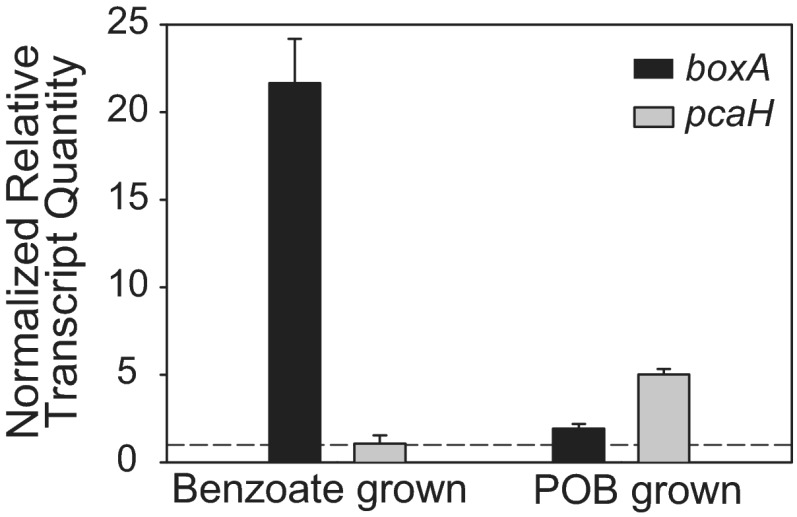
Before addressing the primary issue of mixed-substrate growth, it was first useful to have a broader understanding of aromatic-compound catabolism in E-37. Experimental studies with roseobacters are largely restricted to the protocatechuate (pca) branch of the β-ketoadipate pathway. Activity of the ring-cleaving enzyme protocatechuate 3,4-dioxygenase (PcaHG) has been previously shown to be inducible by growth on POB in E-37 and other roseobacters (39). Furthermore, pobA, the gene encoding a hydroxylase that mediates the conversion of POB to protocatechuate (pca), appears to be coordinately expressed with the pca genes in E-37 and other roseobacters (38). To address which additional aromatic substrates capable of supporting growth are processed via the pca pathway, PcaHG enzyme assays were performed on extracts of E-37 grown on different substrates, only one of which (p-coumarate) is predicted to generate POB as a catabolic intermediate. Basal-level activity (<0.04 μmol pca min−1 μg protein−1) was observed for cells grown on acetate, benzoate, and phenylacetate, suggesting that the pca pathway is not induced when these growth substrates are provided. Conversely, specific activity of PcaHG was detected in cells grown on ferulate, p-coumarate, p-hydroxybenzoate, protocatechuate, quinate, and vanillate (>0.50 μmol pca min−1 μg protein−1), suggesting that all of these compounds are catabolized via the pca pathway (Fig. 1). In some cases, these data are supported by the presence of gene homologs to known catabolic enzymes from soil bacteria (Table 1 and Fig. 1). It is not yet clear what, if any, compounds other than benzoate are degraded through the box pathway. Transcriptional assays of pcaH were also performed on E-37 grown solely on acetate, benzoate, or POB. pcaH transcripts were significantly higher (P ≤ 0.01) in POB-grown cells than in those grown on either acetate or benzoate. These findings corroborate the PcaHG enzyme assay data (Fig. 2 and Table 1).
Fig 1.
Substrates proceeding through the protocatechuate branch of the β-ketoadipate and the aerobic benzoyl-CoA pathway in Sagittula stellata E-37. Gene designations are based on sequence homology to characterized proteins. Question marks indicate uncharacterized genes or substrates.
Table 1.
Phenotypic and genetic evidence for aromatic-compound catabolism in S. stellata E-37
| Carbon substrate | Concn(s) (mM) | Growthb | PcaHG sp actc | Catabolic gene(s) in E-37 | Locus tag(s)f |
|---|---|---|---|---|---|
| Acetate | 3.5, 7, and 25 | + | 0.017 ± 0.008 | TCA cycle (gltA, acnA, icd, sucABCD, shdABCD, fumC, and mdh) | SSE37_06879, SSE37_25138, SSE37_06274, SSE37_11324, SSE37_11329, SSE37_11294, SSE37_11309, SSE37_11244, SSE37_11229, SSE37_11254, SSE37_11249, SSE37_05510, SSE37_11289 |
| Benzoate | 0.5, 1, and 2 | + | 0.030 ± 0.016 | bcl-boxDCBAE | SSE37_24404, SSE37_24409, SSE37_24419, SSE37_24424, SSE37_24439, SSE37_24444 |
| Caffeate | 1, 2, and 3 | − | ND | hcaABC | SSE37_01000, SSE37_01050, SSE37_12324 |
| Catechol | 1, 2, and 3 | − | ND | xylE | SSE37_12149 |
| Chlorogenate | 1, 2, and 3 | − | ND | hcaG, hcaABC, and pobA | SSE37_18822, SSE37_01000, SSE37_01050, SSE37_12324, SSE37_18837 |
| Ferulate | 2 | + | (+)d | Unke and vanAB | SSE37_02815, SSE37_00265 |
| Gentisate | 1, 2, and 3 | − | ND | nagILK | SSE37_02745g, SSE37_02750, SSE37_02755 |
| Homogentisate | NDa | ND | ND | hmgAB and hmgC | SSE37_24544, SSE37_24549, SSE37_14734 |
| Homoprotocatechuate | ND | ND | ND | hpaBC and hpaDEF | SSE37_00435, SSE37_00425, SSE37_23379, SSE37_23374, SSE37_23369 |
| p-Coumarate | 2 | + | 1.205 ± 0.702 | hcaC and pobA | SSE37_12324, SSE37_18837 |
| p-Hydroxybenzoate | 0.5, 1, and 2 | + | 0.477 ± 0.051 | pobA | SSE37_18837 |
| Phenylacetate | 2 | + | 0.005 ± 0.017 | paaABCDEZ and paaIJK | SSE37_00390, SSE37_00385, SSE37_00380, SSE37_00375, SSE37_00370, SSE37_00360, SSE37_15758, SSE37_15753, SSE37_15748 |
| Protocatechuate | 2 | + | 0.632 ± 0.141 | pcaCHG, pcaDB, pcaIJ, and pcaF | SSE37_18842, SSE37_18847, SSE37_18852, SSE37_21470, SSE37_21460, SSE37_23014, SSE37_23019, SSE37_16533 |
| Quinate | 2 | + | 0.694 ± 0.140 | qsuBCD | SSE37_03985, SSE37_06077, SSE37_03980 |
| Salicylate | 1, 2, and 3 | − | ND | salA | SSE37_01015 |
| Vanillate | 2 | + | (+) | vanAB | SSE37_02815, SSE37_00265 |
ND, not determined.
Growth was assessed by monitoring the optical density at 540 nm of cultures in MBM.
PcaHG activity was scored as “−” or “+” if the specific activity was <0.04 or >0.50 μmol pca min−1 μg protein−1, respectively.
“(+)” indicates that specific activity was not quantified but levels were >0.50 μmol pca min−1 μg protein−1.
Unk, gene(s) involved in the pathway is unknown.
Locus tags unique to the E-37 genome are listed in respective order of the catabolic genes.
A disparately located nagI duplicate exists (SSE37_02425).
Fig 2.
boxA and pcaH transcript abundance for E-37 grown solely on POB (2 mM) or benzoate (2 mM). cDNA copy numbers were relativized to those from cells grown on an equimolar C concentration of acetate (7 mM). The horizontal dashed line represents unchanged expression. Error bars represent the standard error of the mean for each biological triplicate.
Growth on benzoate induces boxA expression in Sagitulla stellata E-37.
Like many alphaproteobacteria, roseobacters lack the catechol (cat) branch of the β-ketoadipate pathway that is a well-described route for benzoate degradation in other taxa. Instead, E-37 contains the complete complement of box genes encoding enzymes that convert benzoate to the tricarboxylic acid (TCA) cycle intermediates acetyl-CoA and succinyl-CoA (12). Growth on benzoate as a sole carbon source has been demonstrated previously in E-37 (32), but experimental observations linking the box pathway to this physiology have not yet been performed. In this study, we observed increased abundance of boxA transcripts when the strain was grown on benzoate relative to cells grown solely on acetate or POB, indicating that E-37 uses the benzoyl-CoA pathway for benzoate catabolism (Fig. 2). These results also suggest that the box pathway is strongly inducible and therefore likely subject to transcriptional regulation. The approximately 5-fold difference in transcript abundance between boxA and pcaH when the organism is grown on benzoate and POB, respectively, may be the result of a combination of factors, including differences in promoter regulation and strength, as well as transcript stability. Given these differences, transcript abundance for the mixed-substrate experiments described below is expressed relative to those obtained for cells grown on each substrate alone.
Simultaneous catabolism of benzoate and p-hydroxybenzoate.
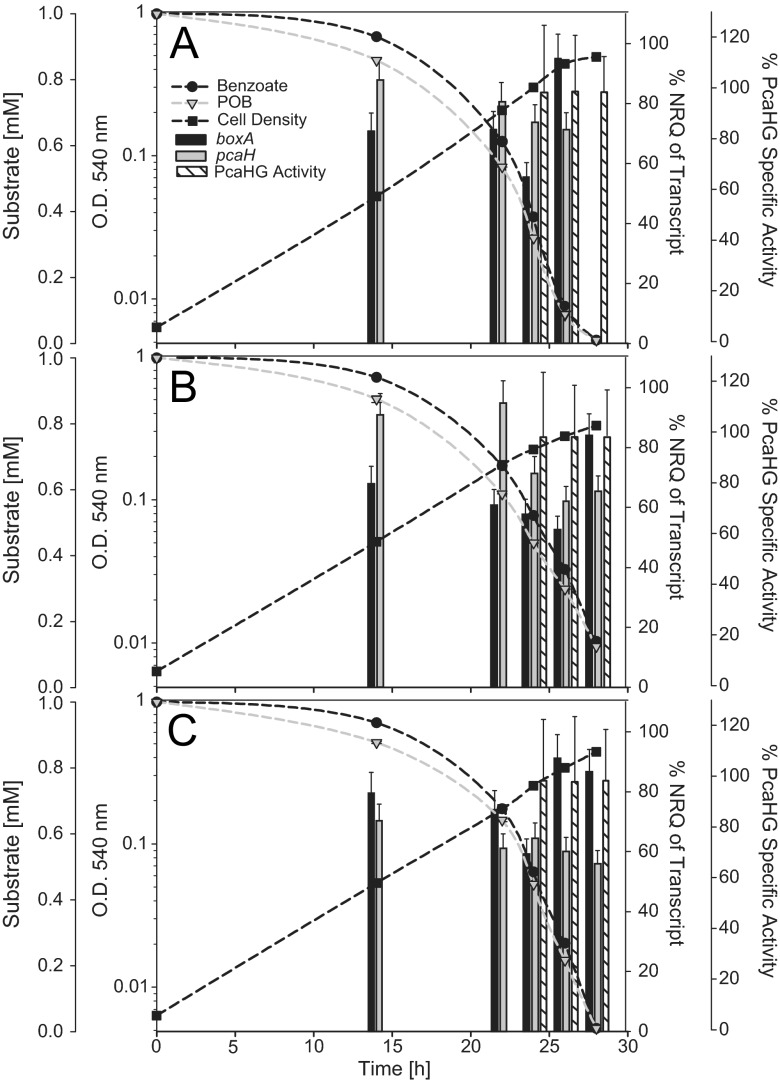
In order to assess whether there was a preferential use of either benzoate or POB under nonlimiting carbon conditions, E-37 was provided a mixture of the two compounds at equal concentrations (1 mM each). Cell density, transcript abundance, PcaHG enzyme activity, and substrate concentration were monitored throughout the growth cycle for each of three biological replicates (Fig. 3). Collectively, these data provided no evidence for substrate preference. Replicate growth curves were monophasic, and while both boxA and pcaH transcript abundances were, on average, 58 to 95% of that found in cultures grown on either substrate alone, the normalized relative quantities of both transcript abundances were significantly higher than basal levels and remained high even as substrate concentrations fell. Furthermore, PcaHG specific activities remained unchanged over time and were equivalent to that found in POB-grown cells. Over the course of the experiment, benzoate and POB were removed from the growth medium at approximately the same rate (see Fig. S1 in the supplemental material). However, substrate concentrations in early-logarithmic-phase cultures indicate that POB is removed from the medium at a slightly higher rate than benzoate during the initial growth phase. The rates of disappearance for benzoate and for POB during simultaneous catabolism were not significantly different from disappearance rates when E-37 was grown on equimolar carbon concentrations of each substrate individually (data not shown).
Fig 3.
Simultaneous catabolism of benzoate (1 mM) and POB (1 mM) by E-37. Panels A to C represent three biological replicates. Data for all three biological replicates are shown in separate panels due to slight variation in growth phase and sample time points among parallel cultures. The boxA and pcaH transcript abundances are expressed as percentages of those when E-37 was grown solely on benzoate and POB, respectively. Error bars represent the standard errors of the means (n = 3 per gene). PcaHG specific activity is also expressed as the percentage of activity obtained from POB-grown cells; error bars represent standard deviations of at least 3 replicates. Standard deviations of the technical variation (n = 3) for benzoate and POB concentrations are smaller than the symbol. NRQ, normalized relative quantity; O.D., optical density.
Simultaneous aromatic catabolism confers enhanced growth rates.
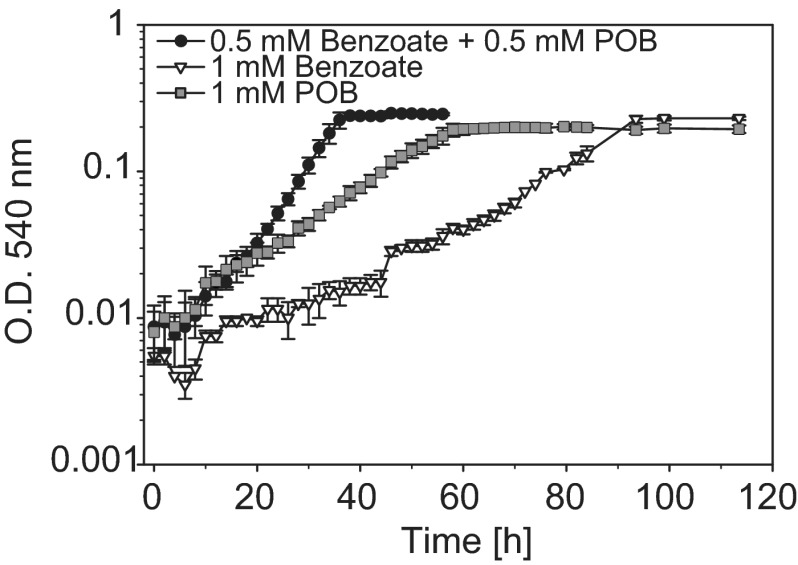
Growth kinetics were monitored to compare the physiology of cells simultaneously catabolizing benzoate and POB to that of cells grown on each substrate individually at the same carbon concentrations. The growth rate for E-37 grown on both compounds simultaneously (μ = 0.129) was significantly higher (P < 0.001) than for cells grown on benzoate (μ = 0.048) and POB (μ = 0.048) alone (Fig. 4). Interestingly, there was a difference in cell yield when this strain was grown on equimolar concentrations of substrates that differ by a single hydroxyl group. Early-stationary-phase biomass yields of 100-ml cultures of E-37 grown on POB (69.13 ± 2.28 mg C) were significantly lower (P < 0.015) than those of benzoate-grown cells (79.57 ± 4.90 mg C), while the mixed-substrate-grown cells yielded an intermediate value (74.27 ± 4.89 mg C). One explanation for the observed differences in biomasses of cultures grown on equimolar carbon concentrations is that there is a difference in the ATP yield for the substrates. In fact, theoretical energy calculations suggest that more ATP is made per molecule of benzoate proceeding through box than ATP per molecule of POB proceeding through pob-pca (see Table S2 in the supplemental material).
Fig 4.
Growth responses to benzoate (1 mM), POB (1 mM), and a mixture of benzoate (0.5 mM) and POB (0.5 mM) by S. stellata E-37. Error bars for O.D. represent the standard deviations of biological replicates (n = 3) for each.
Genome analysis and growth assays with other strains.
To ascertain whether a diagnostic genetic signature might be evident in bacteria demonstrating the simultaneous-catabolism phenotype, a bioinformatics analysis of bacterial genomes containing the box and pca pathways was undertaken. Searches indicate that all putative box operons contain a gene, typically designated boxR, with high homology to an XRE-type transcriptional regulator, suggesting functional similarity among the bacteria harboring this pathway (data not shown). Conversely, gene synteny and genetic regulators for POB catabolism are much less conserved. A focused analysis of bacterial genomes that contain both the box and pca pathways (as of January 2013) revealed significant variation in the organization of genes for POB catabolism (pobA and the pca genes) and their transcriptional regulators (see Fig. S2 in the supplemental material). To date, colocalization of pobA with the pca genes is unique to roseobacters, and pobA transcription appears to be coordinated with that of the pca genes and under the control of a LysR-type regulator, called PcaQ (38). Outside of the Roseobacter clade, gene organization indicates that pobA transcription is mediated by the activity of an adjacently located gene, pobR, encoding a regulatory protein belonging to either the AraC or IclR family (40–42). Of the strains analyzed, simultaneous-catabolism studies have been performed only for Cupriavidus necator JMP134, which possesses an AraC-type PobR protein and exhibits a substrate preference for benzoate over POB (19).
Given the unique pobA gene organization identified in roseobacters, we hypothesized that the absence of a PobR homolog facilitates the growth phenotypes described here. As a first step in exploring this hypothesis, we performed additional mixed-substrate growth experiments with the bacterium Variovorax paradoxus EPS, which contains both the box and pca pathways and has an IclR-type pobR, as well as another roseobacter, Ruegeria pomeroyi DSS-3, that lacks a pobR homolog (see Fig. S2 in the supplemental material). It was first confirmed that both organisms could grow on the two substrates individually (data not shown). In mixed-substrate experiments, simultaneous catabolism of benzoate and POB was observed in DSS-3; however, preferential consumption of POB was observed in EPS (see Fig. S3 in the supplemental material).
DISCUSSION
When a bacterium is provided a mixture of growth-supporting compounds, the utilization profile is generally substrate concentration dependent. Under nonlimiting (replete) carbon concentrations, sequential utilization of substrates by bacteria is typically observed, with the substrate supporting the highest growth rate receiving preference (43). This response has been well studied for bacteria provided a mixture of two sugars (43) but has also been demonstrated with substrate mixtures from other compound classes, including organic acids and aromatic compounds (for examples, see references 44 and 45). Control mechanisms that are responsible for substrate utilization hierarchies include substrate transport into the cell (46, 47), transcriptional regulation (48, 49), or posttranslational modification of an enzyme(s) (50). However, these regulatory controls are often relieved in substrate-limiting concentrations, when mixed-substrate use is essential for growth (for examples, see references 51 and 52). In fact, as oligotrophic conditions dominate the microbial landscape (53), it is possible that substrate preferences are the exception rather than the rule in most natural environments (54). However, we demonstrate here that simultaneous catabolism of two aromatic compounds processed through separate ring-cleaving pathways in roseobacter representatives occurs under carbon-replete conditions (C/N/P ratio of >10,000:59:1). Furthermore, we show that this metabolic versatility leads to an increased growth rate, which may contribute to the competitiveness of these organisms in natural systems, particularly in coastal salt marshes, in which aromatic compounds are a significant component of the dissolved organic carbon pool (55). Consistent with this hypothesis is the observation that roseobacters are one of the most abundant groups of bacteria associated with decaying Spartina alterniflora (56), a primary source of lignin-derived aromatic compounds to these coastal systems (57).
The cross-regulatory mechanisms resulting in substrate preference when strains are provided mixtures of POB and benzoate are complex and difficult to predict from genome sequences alone. Variations in the regulatory proteins and gene organization of each pathway can result in differences in substrate preferences. For example, PobR proteins belong to one of two families (AraC and IclR) and activate pobA transcription in response to the inducer POB (19, 40). Yet the regulatory mechanisms that dictate repression of pobA transcription in the presence of benzoate vary among the organisms possessing different PobR representatives. For example, in the soil bacterium Cupriavidus necator JMP134, benzoate serves as a structurally similar anti-inducer to prevent AraC-type PobR-mediated expression of pobA (19). Conversely, in the soil bacterium Acinetobacter baylyi ADP1, benzoate does not directly modulate the activity of the IclR-type PobR protein found in this strain (40). Instead, repression of the pca catabolic genes, whose products are necessary for complete degradation of POB, appears to be primarily mediated by the activities of regulatory proteins of benzoate and catechol catabolic gene loci (BenM and CatM) that upon binding an intermediate of catechol catabolism (cis, cis muconate) repress transcription of the pca operon, which includes a gene encoding a POB permease (18, 58). Our observation that V. paradoxus EPS preferentially catabolizes POB over benzoate in mixed-substrate experiments is consistent with the notion that the activities of IclR-type PobR proteins are not directly and negatively influenced by benzoate. However, additional studies are needed to confirm this model and to better understand the underlying regulatory mechanisms that lead to the novel phenotype of POB preference over benzoate evidenced in this strain. The unique pobA-pca gene organization found in roseobacters is suggestive of a more simplified regulatory scheme for pobA, which may be manifested in the simultaneous-catabolism phenotype described here and may indicate that POB serves as an important substrate in the environmental niches that roseobacters occupy. Furthermore, most roseobacter genomes contain multiple, potentially competing ring-cleaving pathways for the degradation of aromatic compounds (59), raising the possibility that the simultaneous-catabolism phenotype demonstrated here is representative of group members' utilization of a broader class of aromatic compounds.
While the specific mechanism(s) that facilitates enhanced growth on mixed substrates is not clear, the answer may lie at the cell membrane. Prior studies have suggested that distinct and dedicated transport mechanisms contribute to enhanced growth during mixed-substrate use of glucose and sucrose (60–62). In these previous studies, catabolic enzyme activities were unchanged in mixed-substrate relative to single-substrate experiments. The PcaHG activities reported for cells in different phases of the growth curve are in accordance with this conclusion (Fig. 3), although the specific transporters of POB and benzoate in E-37 are not yet known. In other taxa, PcaK has been shown to transport POB (63), and BenK and BenP transport benzoate (64, 65). However, homologs to the genes for these proteins are absent in the E-37 genome (GenBank accession number AAYA00000000), as well as those of other roseobacters containing the box and pca pathways. This suggests that an alternative transport system for these substrates exists in lineage members. No putative transport genes are found in the local vicinity (10 kb) of the pobA-pca operon in E-37. However, a candidate system for benzoate transport is found directly adjacent to the box operon (SSE37_24379 and SSE37_24389), which putatively encodes a dicarboxylate TRAP transporter. A thorough investigation of the POB and benzoate transport systems is needed to confirm that enhanced growth by strains simultaneously utilizing these compounds is due to the presence and activities of separate, noncompeting transport systems for these substrates. An alternative explanation for the observed physiology is downhill diffusion as a result of metabolism.
In the coastal salt marshes in which roseobacters are abundant, the dissolved organic carbon pool is highly aromatic and heterogeneous in structure and in distribution (55). A successful ecological strategy for bacteria in such environments is metabolic versatility, a strategy exemplified by cultivated roseobacter representatives (for examples, see reference 28). The simultaneous catabolism of growth substrates demonstrated here further illustrates the flexible metabolic characteristic of this abundant group of marine bacteria.
Supplementary Material
ACKNOWLEDGMENTS
We thank Paul M. Orwin of California State University, San Bernardino, for providing us with the Variovorax paradoxus EPS strain. We are also grateful to James J. Daleiden and Joseph J. Bozell for technical assistance and use of their HPLC-PDA.
C.A.G. and A.B. were supported as part of the Center for Direct Catalytic Conversion of Biomass to Biofuels (C3Bio), an Energy Frontier Research Center funded by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, award number DE-SC0000997.
Footnotes
Published ahead of print 5 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00405-13.
REFERENCES
- 1. Wang Y, Yang J, Lee OO, Dash S, Lau SCK, Al-Suwailem A, Wong TYH, Danchin A, Qian P-Y. 2011. Hydrothermally generated aromatic compounds are consumed by bacteria colonizing in Atlantis II Deep of the Red Sea. ISME J. 5:1652–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boerjan W, Ralph J, Baucher M. 2003. Lignin biosynthesis. Annu. Rev. Plant Biol. 54:519–546 [DOI] [PubMed] [Google Scholar]
- 3. Higuchi T. 1990. Lignin biochemistry: biosynthesis and biodegradation. Wood Sci. Technol. 24:23–63 [Google Scholar]
- 4. Wakeham SG, Schaffner C, Giger W. 1980. Polycyclic aromatic hydrocarbons in Recent lake sediments—I. Compounds having anthropogenic origins. Geochim. Cosmochim. Acta 44:403–413 [Google Scholar]
- 5. Christian JR, Anderson TR. 2002. Modeling DOM biogeochemistry, p 717–755 In Hansell DA, Carlson CA. (ed), Biogeochemistry of marine dissolved organic matter. Academic Press, San Diego, CA [Google Scholar]
- 6. Crawford DL, Pometto AL, Crawford RL. 1983. Streptomyces viridosporus: isolation and characterization of a new polymeric lignin degradation intermediate. Appl. Environ. Microbiol. 45:898–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alvarez PJJ, Vogel TM. 1991. Substrate interactions of benzene, toluene, and para-xylene during microbial degradation by pure cultures and mixed culture aquifer slurries. Appl. Environ. Microbiol. 57:2981–2985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kirk TK, Farrell RL. 1987. Enzymatic “combustion”: the microbial degradation of lignin. Annu. Rev. Microbiol. 41:465–505 [DOI] [PubMed] [Google Scholar]
- 9. Fuchs G, Boll M, Heider J. 2011. Microbial degradation of aromatic compounds—from one strategy to four. Nat. Rev. Microbiol. 9:803–816 [DOI] [PubMed] [Google Scholar]
- 10. Harwood CS, Parales RE. 1996. The beta-ketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol. 50:553–590 [DOI] [PubMed] [Google Scholar]
- 11. Stanier RY, Ornston LN. 1973. The beta-ketoadipate pathway. Adv. Microb. Physiol. 9:89–151 [PubMed] [Google Scholar]
- 12. Rather LJ, Knapp B, Haehnel W, Fuchs G. 2010. Coenzyme A-dependent aerobic metabolism of benzoate via epoxide formation. J. Biol. Chem. 285:20615–20624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Romero-Steiner S, Parales RE, Harwood CS, Houghton JE. 1994. Characterization of the pcaR regulatory gene from Pseudomonas putida, which is required for the complete degradation of p-hydroxybenzoate. J. Bacteriol. 176:5771–5779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wheelis ML, Ornston LN. 1972. Genetic control of enzyme induction in the β-ketoadipate pathway of Pseudomonas putida: deletion mapping of cat mutations. J. Bacteriol. 109:790–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mazzoli R, Pessione E, Giuffrida MG, Fattori P, Barello C, Giunta C, Lindley ND. 2007. Degradation of aromatic compounds by Acinetobacter radioresistens S13: growth characteristics on single substrates and mixtures. Arch. Microbiol. 188:55–68 [DOI] [PubMed] [Google Scholar]
- 16. Dal S, Steiner I, Gerischer U. 2002. Multiple operons connected with catabolism of aromatic compounds in Acinetobacter sp. strain ADP1 are under carbon catabolite repression. J. Mol. Microbiol. Biotechnol. 4:389–404 [PubMed] [Google Scholar]
- 17. Cánovas JL, Stanier RY. 1967. Regulation of the enzymes of the beta-ketoadipate pathway in Moraxella calcoacetica. I. General aspects. Eur. J. Biochem. 1:289–300 [DOI] [PubMed] [Google Scholar]
- 18. Brzostowicz PC, Reams AB, Clark TJ, Neidle EL. 2003. Transcriptional cross-regulation of the catechol and protocatechuate branches of the beta-ketoadipate pathway contributes to carbon source-dependent expression of the Acinetobacter sp. strain ADP1 pobA gene. Appl. Environ. Microbiol. 69:1598–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Donoso RA, Pérez-Pantoja D, González B. 2011. Strict and direct transcriptional repression of the pobA gene by benzoate avoids 4-hydroxybenzoate degradation in the pollutant degrader bacterium Cupriavidus necator JMP134. Environ. Microbiol. 13:1590–1600 [DOI] [PubMed] [Google Scholar]
- 20. Gaines GL, Smith L, Neidle EL. 1996. Novel nuclear magnetic resonance spectroscopy methods demonstrate preferential carbon source utilization by Acinetobacter calcoaceticus. J. Bacteriol. 178:6833–6841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ornston LN, Stanier RY. 1966. Conversion of catechol and protocatechuate to beta-ketoadipate by Pseudomonas putida. I. Biochemistry. J. Biol. Chem. 241:3776–3786 [PubMed] [Google Scholar]
- 22. Siehler SY, Dal S, Fischer R, Patz P, Gerischer U. 2007. Multiple-level regulation of genes for protocatechuate degradation in Acinetobacter baylyi includes cross-regulation. Appl. Environ. Microbiol. 73:232–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bleichrodt FS, Fischer R, Gerischer UC. 2010. The beta-ketoadipate pathway of Acinetobacter baylyi undergoes carbon catabolite repression, cross-regulation and vertical regulation, and is affected by Crc. Microbiology 156:1313–1322 [DOI] [PubMed] [Google Scholar]
- 24. del Giorgio PA, Davis J. 2003. Patterns in dissolved organic matter lability and consumption across aquatic ecosystems, p 399–424 In Findlay SEG, Sinsabaugh RL. (ed), Aquatic ecosystems: interactivity of dissolved organic matter. Academic Press, San Diego, CA [Google Scholar]
- 25. McCarthy M, Hedges J, Benner R. 1996. Major biochemical composition of dissolved high molecular weight organic matter in seawater. Mar. Chem. 55:281–297 [Google Scholar]
- 26. Suzuki MT, Preston CM, Chavez FP, DeLong EF. 2001. Quantitative mapping of bacterioplankton populations in seawater: field tests across an upwelling plume in Monterey Bay. Aquat. Microb. Ecol. 24:117–127 [Google Scholar]
- 27. Selje N, Simon M, Brinkhoff T. 2004. A newly discovered Roseobacter cluster in temperate and polar oceans. Nature 427:445–448 [DOI] [PubMed] [Google Scholar]
- 28. Moran MA, Belas R, Schell MA, González JM, Sun F, Sun S, Binder BJ, Edmonds J, Ye W, Orcutt B, Howard EC, Meile C, Palefsky W, Goesmann A, Ren Q, Paulsen I, Ulrich LE, Thompson LS, Saunders E, Buchan A. 2007. Ecological genomics of marine roseobacters. Appl. Environ. Microbiol. 73:4559–4569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mou X, Sun S, Edwards RA, Hodson RE, Moran MA. 2008. Bacterial carbon processing by generalist species in the coastal ocean. Nature 451:708–711 [DOI] [PubMed] [Google Scholar]
- 30. Newton RJ, Griffin LE, Bowles KM, Meile C, Gifford S, Givens CE, Howard EC, King E, Oakley CA, Reisch CR, Rinta-Kanto JM, Sharma S, Sun SL, Varaljay V, Vila-Costa M, Westrich JR, Moran MA. 2010. Genome characteristics of a generalist marine bacterial lineage. ISME J. 4:784–798 [DOI] [PubMed] [Google Scholar]
- 31. Rocker D, Brinkhoff T, Grüner N, Dogs M, Simon M. 2012. Composition of humic acid-degrading estuarine and marine bacterial communities. FEMS Microbiol. Ecol. 80:45–63 [DOI] [PubMed] [Google Scholar]
- 32. González JM, Mayer F, Moran MA, Hodson RE, Whitman WB. 1997. Sagittula stellata gen. nov., sp. nov., a lignin-transforming bacterium from a coastal environment. Int. J. Syst. Bacteriol. 47:773–780 [DOI] [PubMed] [Google Scholar]
- 33. Jamieson WD, Pehl MJ, Gregory GA, Orwin PM. 2009. Coordinated surface activities in Variovorax paradoxus EPS. BMC Microbiol. 9:124 doi: 10.1186/1471-2180-9-124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cude WN, Mooney J, Tavanaei AA, Hadden MK, Frank AM, Gulvik CA, May AL, Buchan A. 2012. Production of the antimicrobial secondary metabolite indigoidine contributes to competitive surface colonization in the marine roseobacter Phaeobacter sp. strain Y4I. Appl. Environ. Microbiol. 78:4771–4780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. 2007. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8:R19 doi: 10.1186/gb-2007-8-2-r19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stanier RY, Ingraham JL. 1954. Protocatechuic acid oxidase. J. Biol. Chem. 210:799–808 [PubMed] [Google Scholar]
- 37. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 38. Buchan A, Neidle EL, Moran MA. 2004. Diverse organization of genes of the beta-ketoadipate pathway in members of the marine Roseobacter lineage. Appl. Environ. Microbiol. 70:1658–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Buchan A, Collier LS, Neidle EL, Moran MA. 2000. Key aromatic-ring-cleaving enzyme, protocatechuate 3,4-dioxygenase, in the ecologically important marine Roseobacter lineage. Appl. Environ. Microbiol. 66:4662–4672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. DiMarco AA, Averhoff B, Ornston LN. 1993. Identification of the transcriptional activator pobR and characterization of its role in the expression of pobA, the structural gene for p-hydroxybenzoate hydroxylase in Acinetobacter calcoaceticus. J. Bacteriol. 175:4499–4506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. DiMarco AA, Ornston LN. 1994. Regulation of p-hydroxybenzoate hydroxylase synthesis by PobR bound to an operator in Acinetobacter calcoaceticus. J. Bacteriol. 176:4277–4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Quinn JA, McKay DB, Entsch B. 2001. Analysis of the pobA and pobR genes controlling expression of p-hydroxybenzoate hydroxylase in Azotobacter chroococcum. Gene 264:77–85 [DOI] [PubMed] [Google Scholar]
- 43. Harder W, Dijkhuizen L. 1982. Strategies of mixed substrate utilization in microorganisms. Philos. Trans. R. Soc. Lond. B Biol. Sci. 297:459–480 [DOI] [PubMed] [Google Scholar]
- 44. Zylstra GJ, Olsen RH, Ballou DP. 1989. Cloning, expression, and regulation of the Pseudomonas cepacia protocatechuate 3,4-dioxygenase genes. J. Bacteriol. 171:5907–5914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hugouvieux-Cotte-Pattat N, Köhler T, Rekik M, Harayama S. 1990. Growth-phase-dependent expression of the Pseudomonas putida TOL plasmid pWW0 catabolic genes. J. Bacteriol. 172:6651–6660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kamogawa A, Kurahash K. 1967. Inhibitory effect of glucose on the growth of a mutant strain of Escherichia coli defective in glucose transport system. J. Biochem. 61:220–230 [DOI] [PubMed] [Google Scholar]
- 47. Nichols NN, Harwood CS. 1995. Repression of 4-hydroxybenzoate transport and degradation by benzoate: a new layer of regulatory control in the Pseudomonas putida beta-ketoadipate pathway. J. Bacteriol. 177:7033–7040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ampé F, Lindley ND. 1995. Acetate utilization is inhibited by benzoate in Alcaligenes eutrophus: evidence for transcriptional control of the expression of acoE coding for acetyl coenzyme A synthetase. J. Bacteriol. 177:5826–5833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fujihara H, Yoshida H, Matsunaga T, Goto M, Furukawa K. 2006. Cross-regulation of biphenyl- and salicylate-catabolic genes by two regulatory systems in Pseudomonas pseudoalcaligenes KF707. J. Bacteriol. 188:4690–4697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zwaig N, Lin ECC. 1966. Feedback inhibition of glycerol kinase, a catabolic enzyme in Escherichia coli. Science 153:755–757 [DOI] [PubMed] [Google Scholar]
- 51. Heinaru E, Viggor S, Vedler E, Truu J, Merimaa M, Heinaru A. 2001. Reversible accumulation of p-hydroxybenzoate and catechol determines the sequential decomposition of phenolic compounds in mixed substrate cultivations in pseudomonads. FEMS Microbiol. Ecol. 37:79–89 [Google Scholar]
- 52. van der Kooij D, Oranje JP, Hijnen WAM. 1982. Growth of Pseudomonas aeruginosa in tap water in relation to utilization of substrates at concentrations of a few micrograms per liter. Appl. Environ. Microbiol. 44:1086–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Poindexter JS. 1981. Oligotrophy. Fast and famine existence. Adv. Microb. Ecol. 5:63–89 [Google Scholar]
- 54. Egli T. 2010. How to live at very low substrate concentration. Water Res. 44:4826–4837 [DOI] [PubMed] [Google Scholar]
- 55. Moran MA, Hodson RE. 1990. Contributions of degrading Spartina alterniflora lignocellulose to the dissolved organic carbon pool of a salt marsh. Mar. Ecol. Prog. Ser. 62:161–168 [Google Scholar]
- 56. Buchan A, Newell SY, Butler M, Biers EJ, Hollibaugh JT, Moran MA. 2003. Dynamics of bacterial and fungal communities on decaying salt marsh grass. Appl. Environ. Microbiol. 69:6676–6687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Moran MA, Hodson RE. 1994. Dissolved humic substance of vascular plant origin in a coastal marine environment. Limnol. Oceanogr. 39:762–771 [Google Scholar]
- 58. Gerischer U, Segura A, Ornston LN. 1998. PcaU, a transcriptional activator of genes for protocatechuate utilization in Acinetobacter. J. Bacteriol. 180:1512–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Buchan A, González JM. 2010. Roseobacter, p 1336–1343 In Timmis KN. (ed), Handbook of hydrocarbon and lipid microbiology. Springer-Verlag, Berlin, Germany [Google Scholar]
- 60. Wood AP, Kelly DP. 1977. Heterotrophic growth of Thiobacillus A2 on sugars and organic acids. Arch. Microbiol. 113:257–264 [DOI] [PubMed] [Google Scholar]
- 61. Wood AP, Kelly DP. 1982. Kinetics of sugar transport by Thiobacillus A2. Arch. Microbiol. 131:156–159 [Google Scholar]
- 62. Wood AP, Kelly DP. 1982. Mechanisms of sugar transport by Thiobacillus A2. Arch. Microbiol. 131:160–164 [Google Scholar]
- 63. Harwood CS, Nichols NN, Kim MK, Ditty JL, Parales RE. 1994. Identification of the pcaRKF gene cluster from Pseudomonas putida—involvement in chemotaxis, biodegradation, and transport of 4-hydroxybenzoate. J. Bacteriol. 176:6479–6488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Thayer JR, Wheelis ML. 1976. Characterization of a benzoate permease mutant of Pseudomonas putida. Arch. Microbiol. 110:37–42 [DOI] [PubMed] [Google Scholar]
- 65. Collier LS, Nichols NN, Neidle EL. 1997. benK encodes a hydrophobic permease-like protein involved in benzoate degradation by Acinetobacter sp. strain ADP1. J. Bacteriol. 179:5943–5946 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.