Abstract
The risk to human health of the annual sandhill crane (Grus canadensis) migration through Nebraska, which is thought to be a major source of fecal pollution of the central Platte River, is unknown. To better understand potential risks, the presence of Campylobacter species and three fecal indicator bacterial groups (Enterococcus spp., Escherichia coli, and Bacteroidetes) was assayed by PCR from crane excreta and water samples collected during their stopover at the Platte River, Nebraska, in 2010. Genus-specific PCR assays and sequence analyses identified Campylobacter jejuni as the predominant Campylobacter species in sandhill crane excreta. Campylobacter spp. were detected in 48% of crane excreta, 24% of water samples, and 11% of sediment samples. The estimated densities of Enterococcus spp. were highest in excreta samples (mean, 4.6 × 108 cell equivalents [CE]/g), while water samples contained higher levels of Bacteroidetes (mean, 5.1 × 105 CE/100 ml). Enterococcus spp., E. coli, and Campylobacter spp. were significantly increased in river water and sediments during the crane migration period, with Enterococcus sp. densities (∼3.3 × 105 CE/g) 2 to 4 orders of magnitude higher than those of Bacteroidetes (4.9 × 103 CE/g), E. coli (2.2 × 103 CE/g), and Campylobacter spp. (37 CE/g). Sequencing data for the 16S rRNA gene and Campylobacter species-specific PCR assays indicated that C. jejuni was the major Campylobacter species present in water, sediments, and crane excreta. Overall, migration appeared to result in a significant, but temporary, change in water quality in spring, when there may be a C. jejuni health hazard associated with water and crops visited by the migrating birds.
INTRODUCTION
Every year, millions of birds migrate across national and intercontinental borders in response to factors such as food availability, seasonal climate change, and the need for habitats in which to nest and raise offspring (1). Waterfowl migration has a positive economic impact due to recreational activities, such as hunting and bird watching. On the other hand, some waterfowl are known vectors of zoonotic pathogens and antibiotic resistance genes (2–5), which they can acquire via coprophagy or by ingestion of contaminated water (6). When roosting in high numbers, waterfowl may cause water quality deterioration, and therefore, humans and animals in contact with such water risk higher incidences of illness (7, 8). From a public health risk perspective, Salmonella, Campylobacter, Listeria, and Yersinia are among the genera of human bacterial pathogens most often detected in waterfowl (9). Of all human bacterial pathogens, Campylobacter species, such as Campylobacter jejuni, Campylobacter coli, and Campylobacter lari from gulls, Canada geese, and whistling swans, have been the most frequently documented in bird feces (10–13).
Besides their potential role in pathogen transmission, waterfowl migrations can also cause significant increases in fecal indicator bacteria (FIB) in recreational waters that can result in beach closures. For example, the number of Escherichia coli bacteria in two reservoir lakes in Scotland was related to the number of roosting gulls (14). In another study, waterfowl, such as ducks, geese, and swans, were considered one of the major sources of fecal coliforms in Buttermilk Bay, MA, although the loading of fecal coliforms varied seasonally with bird species and bird population density (15). In western Sydney, Australia, a recreational lake was closed for several years until a risk assessment demonstrated that there was a relatively low risk from the waterfowl during dry weather (16).
The North American migration of sandhill cranes (Grus canadensis) is linked to the roosting of over 500,000 birds along the central Platte River in Nebraska (17). The Platte River has numerous beneficial uses for people in central Nebraska, such as crop irrigation, fishing, and as a source of water for hydroelectric power plants and drinking. When migrating to the Platte River, sandhill cranes feed in nearby corn fields and then move to the Platte River or surrounding wetlands to roost at night. While sandhill cranes and other waterfowl are suspected to have an impact on the microbial water quality of the Platte River during their annual migration, there is little documentation on FIB and the possible public health risk. Nonetheless, studies in Canada and Alaska have shown a link between crane excretion of C. jejuni (5, 12, 18) and human disease (7). Additionally, the human health significance of Salmonella in hooded cranes (Grus monacha) and whitenaped cranes (Grus vipio) in Japan (19, 20) and Mycobacterium avium in sandhill cranes (21) is unclear.
Hundreds of thousands of sandhill cranes use the Central Flyway (Fig. 1), and a recent food-borne outbreak (7) implies that these birds may also represent a risk at other migration stopovers. However, the risks are poorly understood due to the lack of data on C. jejuni carriage during crane migration. Hence, the main objective of this study was to determine the occurrence and quantity of Campylobacter species and fecal indicators in the excreta of sandhill cranes and comigrating snow geese roosting along the Platte River and in stream water and sediments associated with the major roosting zone. This information is not only needed to inform managers about FIB impacts, but also provides data to allow the development of quantitative microbial risk assessment models (16, 46).
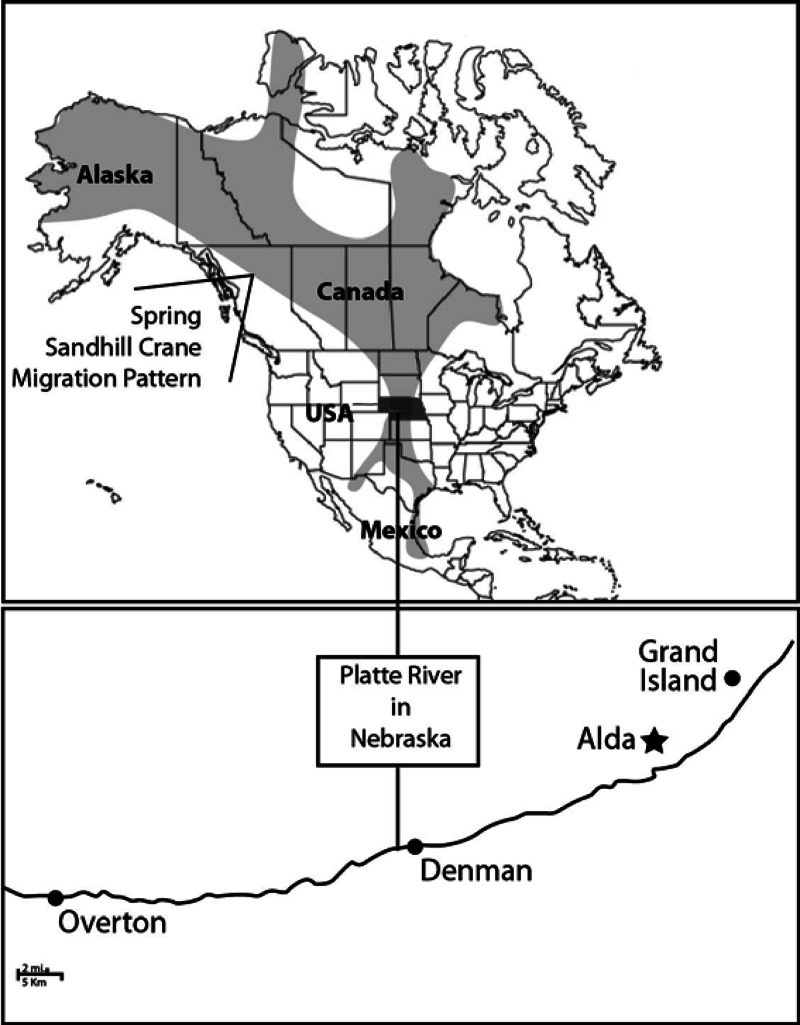
Fig 1.
Map of the migration area and flyway of sandhill cranes and sampling sites for excreta (Alda), water (Overton, Denman, and Grand Island), and sediments (Alda). The upper map was adapted, with permission, from a map created by the Crane Trust using USGS data.
MATERIALS AND METHODS
Sample collection and processing and DNA extraction.
Water samples were collected in triplicate from January to May 2010 on 14 different dates from three locations within the Platte River (Overton, Denman, and Grand Island, NE) (Fig. 1). During the period of bird migration, the field parameters—temperature (5.7 to 9.3°C), pH (8.33 to 8.63), turbidity (5.3 to 21.5 nephelometric turbidity units [NTU]), and specific conductivity (771 to 1,134 μS · cm−1)—were measured from a subset of the samples collected with an Isco refrigerated autosampler (Teledyne Isco, Lincoln, NE). Sandhill crane numbers were also estimated by watching the migrating crane population. For the purpose of quality assurance and quality control, on each water-sampling day, two field water blanks, consisting of ultrapure water from the U.S. Geological Survey (USGS) National Water Quality Laboratory, were poured from a jug into a sample bottle at the field site using the same sampling protocol that was used for environmental samples. The field blanks (n = 26, to monitor for any experiment contamination) and all other samples were processed together. Additionally, sediment samples (coarse river sands, approximately 2 mm in diameter) were collected in triplicate on the same days (n = 9) as the water samples at approximately 11:00 a.m. The location for sediment samples was from a roosting area near Alda, NE, close to the Grand Island sampling site (Fig. 1). Sandhill crane (n = 64) and associated snow goose (Chen caerulescens) (n = 22) excreta samples were collected at locations near the sediment-sampling site on the grounds of the Platte River Whooping Crane Maintenance Trust near Alda, NE, soon after defecation and immediately placed into sterile plastic bags and stored in ice coolers for overnight shipment to the U.S. Environmental Protection Agency (EPA) research facility (Cincinnati, OH).
For each sample of bird excreta, subsamples were used for culture-based studies and molecular analyses. Approximately 1 gram of excreta was transferred to sterile conical tubes containing 3 ml phosphate-buffered saline (PBS) and enriched for Campylobacter spp. Specifically, 1 ml of the fecal-PBS solution was transferred to Bolton broth (CM0983; Oxoid, Basingstoke, United Kingdom) and incubated under microaerophilic conditions (5% O2, 10% CO2, 85% N2) at 42°C for 72 h. After 72 h, an aliquot (1 ml) of the enriched cultures was harvested via centrifugation at 15,000 × g for 10 min, and DNA was extracted using the MasterPure DNA Purification Kit following the manufacturer's instructions (Epicenter Biotechnologies, Madison, WI). For excreta and water samples, DNA extracts were further purified using the Genomic DNA Clean and Concentrator kit as recommended (Zymo Research Corp., Irvine, CA). Final DNA extracts were eluted in 100 μl molecular-grade water, and DNA concentrations were estimated with a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, Inc., Wilmington, DE). We have previously described this protocol and validated it for various aquatic biofilm samples, demonstrating the removal of PCR inhibition (J. Lu, T. L. Gerke, H. Y. Buse, and N. J. Ashbolt, submitted for publication). Specifically, the procedures for various sample types prior to DNA extraction were as follows. Crane excreta samples (0.3 g) were disrupted by beating with 0.1-mm silica beads (MP Biomedicals LLC, Solon, OH) in 300 μl T&C lysis buffer (Epicenter Biotechnologies, Madison, WI) for 60 s, using a Mini Bead Beater (BioSpec Products, Inc., Bartlesville, OK), followed by DNA extraction and purification. Sediment samples (5.0 g) were transferred to conical tubes containing 50 ml of PBS buffer and dispersed three times at 70% amplitude for 30 s using a Misonix Sonicator S-4000 (Qsonica, LLC, Newtown, CT). The sediment samples were centrifuged at 1,000 × g for 5 min. A subsample of the supernatant (5 ml) was processed for Campylobacter species enrichment as described above. The rest of the sample was transferred to three 15-ml conical tubes and centrifuged at 4,500 × g for 15 min. The resulting pellets were pooled in one microtube for DNA extraction, but without the additional (Zymo purification kit) purification step. Water samples (100 ml) were filtered through polycarbonate membranes (0.40-μm pore size; 47-mm diameter; GE Water and Process Technologies, Trevose, PA). The membranes were aseptically cut into four pieces according to an optimized protocol from previous studies (22, 23), placed into tubes containing lysing solution, and processed using silica bead disruption, and DNA was extracted and further purified as described for the excreta samples. Additionally, water samples (5 ml) were enriched for Campylobacter spp. and processed as described above.
Quantitative PCR.
Quantitative PCR (qPCR) assays were used to detect Campylobacter spp. (24), Enterococcus spp. (25), Bacteroidetes (26), and E. coli (27), with the modifications described by Lu et al. (28) for the samples from crane excreta, water, and sediments. Additionally, a putative crane-specific qPCR assay (29) targeting Catellicoccus-like bacteria was used to detect sandhill crane excreta in water and sediment. The cited primer sequences are listed in Table S1 in the supplemental material. Target cells in the extracts were quantified as spiked cell equivalents (CE). The detection limits and standard curves for Campylobacter spp. were obtained using standardized procedures we developed (J. Lu, T. L. Gerke, H. Y. Buse, and N. J. Ashbolt, submitted for publication), i.e., determining standard curves by spiking a strain with targeted sequences into matrices originated from sources the same as or similar to the samples assayed. Specifically, five crane excreta and water samples and four sediments that previously tested negative for C. jejuni were pooled and used as C. jejuni-spiking matrices. Serial dilutions of C. jejuni from 108 to 101 were prepared in PBS and spiked into 9 (8 dilutions and 1 negative control) 1-ml triplicate samples of crane excreta-PBS buffer, sediment-PBS buffer, or river water, and DNA was isolated as described above. For quantifying fecal indicators, serial dilutions (108 to 101) of E. coli K-12 MG1655 and Enterococcus faecalis ATCC 29212 following overnight culture and Bacteroides thetaiotaomicron (ATCC 29741) cells within BioBalls (BioBall Custom HighDose10K; BTF, North Ryde, Australia) were prepared and spiked into nine (eight dilutions and one negative control) 1-ml filter-sterilized matrix samples, and DNA was isolated as described above. For the putative crane excreta marker, duplicate serial dilutions of crane fecal DNA (10−8 to 10−12 g/reaction) were used to generate standard curves as described previously (30). The quality assurance control for PCR was set up as a DNA standard control in the first row of the plate and a no-template control for each row of each 96-well plate assayed. To analyze qPCR fluorescent signals and obtain threshold cycle (Ct) values and cell equivalent numbers, manual setting of threshold and baseline parameters for the run was undertaken. To test qPCR inhibition, each DNA extract was assayed neat, after a 10-fold dilution in T&C lysis buffer (Epicenter Biotechnologies), and with the addition of the TaqMan Exogenous Internal Positive Control Reagents (a VIC-labeled probe) manufactured by ABI (Applied Biosystems). When any inhibition was identified, another DNA extraction with a further purification step was conducted. Furthermore, the final quantity of CE for each qPCR run was obtained by analyzing the standard control against samples. The lowest number of gene copies that was detected consistently in the standard curves was considered the qPCR limit of detection. Results below the detection limit were treated as zero. The qPCR efficiency was calculated using the following equation: efficiency percent = 100 × [10(−1/slope) − 1] (Applied Biosystems).
PCR and cloning.
A Campylobacter genus-specific PCR assay was used to determine the presence of campylobacters (30). Species-specific PCR assays targeting the mapA (31), ceuE (32), and 16S-23S rRNA gene internal transcribed spacer (ITS) (33) were used to detect C. jejuni, C. coli, and C. lari, respectively, when a sample was positive at the genus level. DNAs from the following bacterial strains were used as controls for all PCRs performed: C. jejuni LMG 8842, C. coli ATCC 33559, and C. lari ATCC 35221. For a range of excreta, water, and sediment samples positive in the Campylobacter genus-specific PCR, amplicons were cloned into pCR4.1 TOPO (Invitrogen, Carlsbad, CA) to aid in further identification. In addition, positive samples for the mapA assay were also cloned into pCR4.1 TOPO and sequenced as described by Lu et al. (28). Six Campylobacter genus-specific PCR clone libraries were generated: one snow goose fecal library (collected on 15 March 2010 [n = 2]); three sandhill crane fecal-clone libraries, by pooling PCR products from different crane excreta (collected on 15 March [n = 2], 18 March [n = 4], and 5 April [n = 6] 2010); one water clone library, by pooling a positive PCR product from each site (collected on 18 March 2010); and one sediment clone library from one sample (collected on 18 March 2010). To further confirm the presence of C. jejuni in crane excreta, a mapA PCR library was generated from pooled PCR products collected on three different sample dates (15 March [n = 2], 18 March [n = 4], and 5 April [n = 6] 2010). Raw sequences were processed with Sequencher 4.9 software (Gene Codes, Ann Arbor, MI) for editing, comparing, and alignment, using protocols, homology searches, a chimera check, and phylogenetic analysis, as previously described (28). Phylogenetic analysis based on mapA sequences was done using MEGA 4.1 (34). In order to investigate other possible sources of contamination, additional PCR assays were conducted to amplify cattle-specific (CF128) and human-specific (HF183) Bacteroidetes members in the sediment and water samples (35). A general Bacteroidales 16S rRNA gene PCR assay (Bac32f/Bac708r) was used to amplify Bacteroidales from nine water samples (35). After excluding chimeric sequences, a total of 851 Bacteroidales sequences were obtained and used for constructing a phylogenetic tree with 27 Bacteroidales sequences from the companion study (29).
Data analysis and sequence submission.
For qPCR data management and analysis, Microsoft Excel 2003 and SAS Systems version 8.2 (SAS, Cary, NC) were used. Statistical analysis included Pearson's correlation of qPCR quantity between Campylobacter spp. and fecal indicators and between sandhill crane markers and fecal indicators, and a t test with unequal sample sizes on all the qPCR signals between the pre- or postmigration and crane migration periods in water and sediment samples.
Nucleotide sequence accession numbers.
Representative Campylobacter species and Bacteroidetes sequences from cloning experiments were deposited in GenBank with accession numbers JQ693816 to JQ693898 and KC149757 to KC149853, respectively.
RESULTS
C. jejuni isolates from excreta, water, and sediment samples.
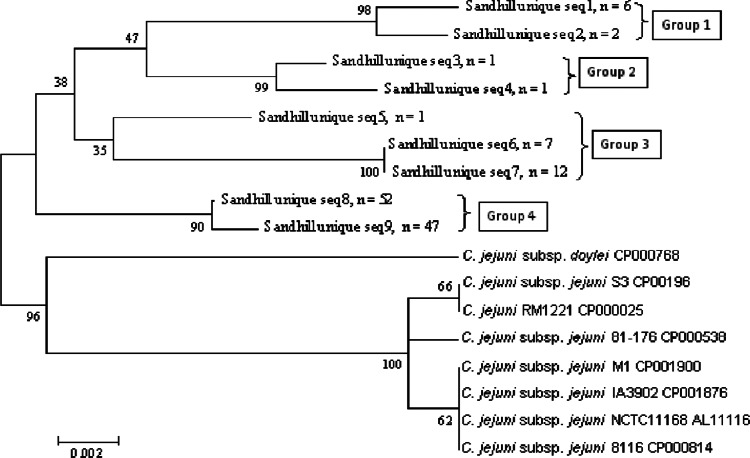
A total of 583 sequences were generated from the 12 crane excreta samples positive for the Campylobacter species 16S rRNA gene (Table 1), and 99% were homologous to C. jejuni subsp. jejuni (strain ICDCCJ07001; >99% sequence identity), with the remaining 1% identified as Campylobacter canadensis (strain EF621902; >99% sequence identity) and Campylobacter sp. strain MIT 07-5158 (taxonomy ID, 862209; <95% sequence identity) (Table 1). For the other three libraries cloned from snow goose excreta, water, and sediment, all 116 of the sequences were homologous to the same reference strain (C. jejuni subsp. jejuni strain ICDCCJ07001; >99% sequence identity), which was different from the C. jejuni strain LMG 8842 used as a positive control. The presence of C. jejuni was supported by the detection of the C. jejuni mapA gene in the samples that were positive in the genus-specific assay and by the fact that C. coli- and C. lari-specific assays were negative in all the samples tested. Moreover, mapA fecal-clone sequences were closely related to those of the C. jejuni subsp. jejuni S3 mapA gene (i.e., 95 to 97% sequence identity). Phylogenetic analysis of the mapA gene sequences indicated four sequence groups out of the nine operational taxonomic units (OTUs) at 99% identity, but all of them were significantly different from known strains isolated from different sources (Fig. 2).
Table 1.
BLAST results of the clone libraries of Campylobacter spp. for the samples of Sandhill crane excreta, snow goose excreta, water, and sediment
| Sample | No. of clones | Bacterial reference strains | Accession no. | Identity (%) |
|---|---|---|---|---|
| Crane excreta (16S rRNA) | 579 | C. jejuni subsp. jejuni ICDCCJ07001 | CP002029 | >99 |
| 2 | Campylobacter sp. MIT 07-5158 | HM211852 | <95 | |
| 2 | C. canadensis L285 | EF621902 | >99 | |
| Crane excreta (mapA) | 130 | C. jejuni subsp. jejuni S3, ICDCCJ07001, IA3902 | CP001960 | 95–97 |
| Snow goose feces (16S rRNA) | 20 | C. jejuni subsp. jejuni ICDCCJ07001 | CP002029 | >99 |
| Water (16S rRNA) | 52 | C. jejuni subsp. jejuni ICDCCJ07001 | CP002029 | >99 |
| Sediment (16S rRNA) | 44 | C. jejuni subsp. jejuni ICDCCJ07001 | CP002029 | >99 |
Fig 2.
Unrooted neighbor-joining tree of mapA gene sequences (seq) obtained from sandhill crane excreta clone libraries. Sequences (length = 644; minimum match percentage ≥ 99%) were aligned with Sequencher 4.9, and a bootstrap consensus tree was created with MEGA 4.1. Bar represents 2‰ estimated phylogenetic divergence. Bootstrap values (500 replicates) are shown at the nodes.
Campylobacter spp. were detected in 48% of crane excreta (see Table S2 in the supplemental material) (n = 64), 9% of snow goose excreta (see Table S3 in the supplemental material) (n = 22), 24% of water samples (see Table S4 in the supplemental material) (n = 45), and 11% of sediment samples (see Table S5 in the supplemental material) (n = 11). Generally, similar results were obtained using DNA extracted either directly from samples or from enrichment cultures (see Table S2 to S5 in the supplemental material), implying limited culture bias (36) associated with the presence/absence detection approach (Campylobacter genus-specific PCR assay) use in this study, possibly attributable to the presence of only one dominant species. For some samples, direct DNA extracts showed weak positive, but not in enrichments, indicating a likelihood of inadequate addition of sample material to the enrichments. Based on the 16S rRNA gene clone libraries derived from composite water (n = 3) and sediment (n = 1) samples, all sequences were nearly identical to that of C. jejuni subsp. jejuni (Table 1, strain ICDCCJ07001; >99% sequence identity), which was present in the crane excreta samples, as mentioned above. In contrast, negative results were obtained for all field blanks (n = 26).
Impact of crane excreta on Platte River water quality.
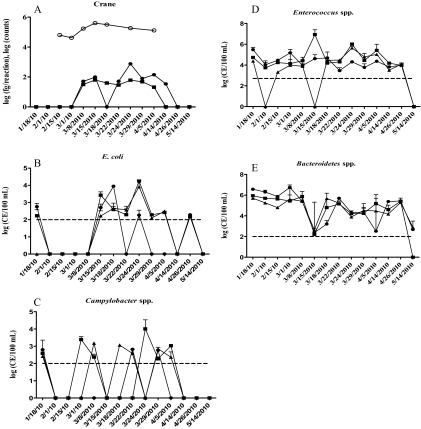
The efficiency of Campylobacter qPCR for three different samples (excreta, water, and sediment) ranged from 90% to 98% (see Table S6 in the supplemental material), which was within normal range (100% ± 10%), according to the instructions for the Applied Biosystems 7900HT Fast Real-Time PCR System. The detection limits of Campylobacter qPCR in the three sample matrices were all about 100 CE ml−1, which was the same level as in pure water (see Table S6 in the supplemental material), indicating the protocol described for DNA purification was also valid for this study. On average, excreta, water, and sediment samples contained 5.0 × 103 CE/g, 431 CE/100 ml, and 30 CE/g of Campylobacter, respectively. The mean Campylobacter estimates for the three water sites were 1.6 × 103, 1.4 × 103, and 3.0 × 102 CE/100 ml at Overton, Denman, and Grand Island, respectively. According to the observed bird presence in the river (1 March to 5 April is the main migration period) (Fig. 3B), mean densities (Table 2) for each site were significantly different during and after migration (P < 0.0001) in the three locations and during and before migration at Denman and Grand Island (P < 0.001). The occurrence of Campylobacter spp. detected during crane presence was 75%, 63%, and 25% at Denman, Grand Island, and Overton, respectively. The variation of Campylobacter density was consistent with observed bird counts and the putative crane marker (Fig. 3A and B). For example, high signals of the crane marker were measured on March 8, 15, 22, and 24 at Denman and Grand Island, coinciding with peaks in Campylobacter detection. Overall, the Campylobacter CE values were significantly higher during crane presence (1.1 × 103 CE/100 ml) than before (1.9 × 102 CE/100 ml) or after (below the detection limit) the migration period.
Fig 3.
Variations in the relative abundances of E. coli (B), Campylobacter spp. (C), Enterococcus spp. (D), and Bacteroidetes (E) corresponding to crane numbers (A) observed in the field (○) and crane marker signals detected at Overton (●), Denman (■), and Grand Island (▲) in central Platte River water samples before (January-February), during (March-5 April), and after (14 April-May) sandhill crane migration. Note that in panel A the data for crane assays performed before and after the migration dates were not detectible. The dashed lines in panels B and C indicate the detection limits of qPCR for each targeted organism group.
Table 2.
Average CE of Campylobacter spp. and fecal indicator bacteria estimated using qPCR
| Location | No. of sampling times | Sample period (2010) | Campylobacter spp. [CE/100 ml (SD)] | Bacteroidetes [103 CE/100 ml (SD)] | Enterococcus spp. [103 CE/100 ml (SD)] | E. coli [CE/100 ml (SD)] |
|---|---|---|---|---|---|---|
| Overton | 3 | Jan.-15 Feb. | 348 (602) | 2,223 (1,552) | 122 (177) | 203 (352) |
| 8 | 1 Mar.-5 Apr. | 398 (906) | 913(2090) | 56(76) | 643(1531) | |
| 4 | 13 Apr.-24 May | 0a | 181 (159) | 6 (5) | 64 (110) | |
| Denman | 3 | Jan.-15 Feb. | 130 (226) | 609 (248) | 27 (24) | 56 (98) |
| 8 | 1 Mar.-5 Apr. | 2,384 (4,829) | 293 (335) | 1,577 (3,839) | 2,698 (6,137) | |
| 4 | 13 Apr.-24 May | 0 | 102 (136) | 8 (8) | 50 (86) | |
| Grand Island | 3 | Jan.-15 Feb. | 105 (181) | 252 (229) | 10 (12) | 0 |
| 8 | 1 Mar.-5 Apr. | 551 (580) | 197 (221) | 156 (200) | 1,401 (3,359) | |
| 4 | 13 Apr.-24 May | 0 | 74 (112) | 6 (8) | 51 (88) | |
| Sediment | 6 | 15 Mar.-5 Apr. | 46 (73) | 6 (8) | 49 (48) | 3,268 (7,899) |
| 3 | 13 Apr.-24 May | 0 | 3 (4) | 2 (2) | 0 |
0, below the detection limit.
The efficiency of fecal indicator qPCR ranged from 90% to 118%, and the detection limits varied from 100 to 520 CE ml−1 for different fecal indicator qPCR assays (see Table S7 in the supplemental material), which were used to estimate the detectible quantity of samples. The mean densities of Enterococcus spp., E. coli, and Bacteroidetes were estimated at 4.6 × 108, 6.9 × 107, and 1.4 × 105 CE per gram of crane excreta, respectively. For the water samples, Bacteroidetes densities were the highest among the range of FIB tested (5.1 × 105 CE/100 ml), followed by Enterococcus spp. (3.5 × 105 CE/100 ml) and E. coli (9.3 × 102 CE/100 ml). Enterococcus spp. and E. coli reached their peaks during the presence of cranes in the surrounding area, especially from 15 to 24 March (Fig. 3C and D), while the lowest densities were observed before the birds' arrival, except Enterococcus spp. at Overton, and after the birds' departure (Fig. 3C and Table 2). Specifically, the CE values were significantly higher during crane presence than after that period for Enterococcus spp. (P < 0.0001) and E. coli (P = 0.014) and during crane presence than before that period for Enterococcus spp. (P = 0.020) and E. coli (P = 0.028). There was a significant positive relationship between the Campylobacter spp. and Enterococcus spp. (R2 = 0.312; P = 0.012) in crane excreta. The putative crane marker was detected only during the bird migration time at Denman and Grand Island. Interestingly, there were significant correlations of the crane fecal marker signals with Enterococcus spp. both in the excreta (R2 = 0.512; P = 0.022) and in water (R2 = 0.325; P = 0.036) and with E. coli in water (R2 = 0.468; P = 0.002). The agreement between the putative crane marker and Enterococcus spp. in excreta and water indicated that the increase in the river water Enterococcus spp. was most likely predominantly caused by the migrating cranes and partially by snow geese. Importantly, during the crane migration period, Enterococcus spp. appeared to be indicative of crane excreta contamination in surface waters. According to Pearson correlation analysis, there were significant correlations between Bacteroidetes and the other three bacterial groups (Campylobacter spp., R2 = 0.339, P = 0.032; E. coli, R2 = 0.383, P = 0.019; Enterococcus spp., R2 = 0.841, P < 0.0001) before the crane migration, but there was no significant correlation between Bacteroidetes and any of the other three bacterial groups during the crane migration period. To compare the change in water quantity between the presence and absence of cranes, the four bacterial group means are reported in Table 2. Generally, means were higher in water during crane migration, with Enterococcus sp. densities in sediments (∼3.3 × 105 CE/g) 2 to 4 orders of magnitude higher than densities of Bacteroidetes (4.9 × 103 CE/g), E. coli (2.2 × 103 CE/g), and Campylobacter spp. (46 CE/g). Additionally, the highest bacterial estimates in the sediments were also associated with the cranes' presence (Table 2; see Fig. S1 in the supplemental material). To further explore other potential fecal sources, Bacteroidetes-like sequences cloned from water samples (n = 851) were compared with those cloned from crane excreta (n = 27) and the sequences retrieved from GenBank. The phylogenetic analysis for the comparison (see Fig. S2 in the supplemental material) indicated that the Bacteroidetes-like sequences from water were only about 7.4%, 8.2%, and 1.9% similar to those from crane excreta, human feces, and cattle feces, respectively, whereas the majority of Bacteroidetes-like sequences (82.5%) were most likely from unidentified sources. To investigate human and cattle sources further, human and cattle markers targeting Bacteroidetes were also estimated for the water samples (see Table S4 in the supplemental material), and the PCR results indicated that the occurrence of human and cattle markers did not increase during, before, or after migration.
DISCUSSION
C. jejuni appears to be the dominant Campylobacter species in sandhill crane excreta (48% occurrence), which is consistent with previous studies reported for the migration of birds in Canada and Alaska (7, 12). Further, the results indicated C. jejuni was diverse at the strain level, but that diversity cannot be resolved using 16S rRNA gene sequences (37). Previously, Pacha et al. (12) isolated Campylobacter strains from 81% of sandhill crane samples, all of which were later identified as C. jejuni. Similarly, Gardner et al. (7) also isolated C. jejuni from all 14 sandhill crane fecal samples examined, although C. canadensis was also detected. Because the central Platte River in Nebraska is the most important stopover area for the sandhill crane (17), it is reasonable to assume that the waterfowl carried the bacteria during their migration north through Canada to Alaska, although genotyping studies are needed to confirm this assumption. Other migrating waterfowl species have also been reported to have a high prevalence of C. jejuni. For example, Pacha et al. (12) isolated Campylobacter spp. from 73% of samples from wild duck (four species) and 5% of samples from Canada geese (Branta canadensis), and all the isolates were identified as C. jejuni. In other studies, 42% of samples from Canada geese (11) and 44% of samples from ducks (38) were isolated as C. jejuni. In addition to sandhill cranes in the current study, snow geese, as they comigrate with sandhill cranes, also appeared to share C. jejuni, but at a lower rate of occurrence (5%) than in sandhill crane excreta, coincident with the presence in Canada geese reported by Pacha et al. (12). Hence, given the close proximity of the two bird species, it would be valuable to follow migrating snow geese to see if their exposure to sandhill cranes was the source of their acquired C. jejuni; as inferred by Waldenström et al. (39), when migratory birds share feeding locations, they have similar habitat preferences and migration patterns. The ecological character of sandhill cranes, a longer life span (40), and an omnivorous diet may subject them to harboring Campylobacter. In contrast, snow geese have shorter life spans, are herbivores, and feed side by side at water edges. If the sandhill cranes are the primary carriers, that could explain the lower level of Campylobacter occurrence in snow geese (5%), which are vulnerable to crane contamination in shallow waters, which were identified in the current study to harbor C. jejuni.
Previous studies have shown that C. jejuni strains are diverse and source specific. Waldenström et al. (13) detected diverse C. jejuni strains from wild birds and indicated that the different host species largely carried their own campylobacters and that cross-species transmission was rare. Gardner et al. (7) isolated C. jejuni from 14 individual crane fecal samples showing 26 unique pulsed-field gel electrophoresis (PFGE) pattern combinations (up to 7 different genotypes) in PulseNet (41). In the current study, the mapA gene assay was used to identify C. jejuni. This assay was first characterized by Stucki et al. (31), who showed that the MapA protein was present in all of the C. jejuni strains tested and absent in C. coli and related campylobacters and hence was considered C. jejuni specific, although there was high variation of mapA gene sequences among the isolates of C. jejuni. The mapA sequences in the current study were 3 to 5% different from those of known C. jejuni strains and may therefore imply that new strains were present in the crane excreta. The four different groups (9 OTUs at 99% identity) of C. jejuni sequences identified using the mapA gene assay also indicate they may be diverse and unique C. jejuni strains.
Champion et al. (42) examined 70 C. jejuni strains associated with human disease by genotyping; surprisingly, the majority (56%) were in the nonlivestock clade, and they suggested that most C. jejuni infections might be from nonlivestock (and possibly nonagricultural) sources. On a similar line, C. jejuni strains from wild European starlings (Sturnus vulgaris) (43) were shown to be a source of human and farm animal infection but distinct from poultry or other human disease isolates. A recent outbreak of campylobacteriosis was related to only a limited range of sandhill crane fecal isolates (7), which were very similar to those from patients infected by C. jejuni. Whether our mapA sequences of C. jejuni were different strains from those found in Alaska (7) or from those found in other animals and whether the C. jejuni strains in our study were infectious to humans require further investigation to determine and should form an important part of future risk assessments of crane migration.
16S rRNA gene-based Campylobacter species sequences showed consistent results among crane excreta, water, and sediments in the central Platte River, Nebraska; i.e., they were 99% identical to the same bacterial strain, C. jejuni subsp. jejuni ICDCCJ07001 (Table 1). Further, peaks of Campylobacter qPCR signals in water (and also in sediment) corresponded to the high presence of the crane population, and there were significant differences in Campylobacter qPCR signals between the premigration and migration periods in water (Fig. 3) and sediments (see Fig. S1 in the supplemental material), suggesting that Campylobacter bacteria shed by sandhill cranes had a significant impact on river water and sediment quality. In addition to this possible primary source, there also appear to be other sources, such as humans, cattle, and snow geese, of Campylobacter contamination of Platte River water. For the former sources, the evidence was as follows: (i) Bacteroidetes markers for humans and cattle were detected during the investigation, (ii) sequences of Bacteroidetes specific to humans and cattle were identified from water samples, and (iii) during the absence of cranes, the three bacterial groups were positively related to Bacteroidetes. As for the last source, in 2010, snow geese migrated to the Platte River during the same period as sandhill cranes but left before late March and were identified at all three locations. As discussed above, the snow goose excreta were shed at the riverside and in the water, and the same type of C. jejuni was detected from the excreta. Although sandhill crane marker was not detected at Overton, sandhill cranes and snow geese were observed around crop fields and the riverside; therefore, the qPCR signals of Campylobacter spp. detected at a lower level at the site could be from mixed sources from waterfowl and mammals, as human and cattle markers were detected (see Table S4 in the supplemental material).
The impact of crane excreta on Platte River water quality was also quantified by qPCR analysis of fecal indicator bacteria (i.e., Enterococcus spp., E. coli, and Bacteroidetes) and the sandhill crane fecal marker. Both E. coli and Enterococcus levels seemed significantly impacted, increasing approximately 11 to 90 times during the migration period compared to before and after migration (see Table S8 in the supplemental material). Several studies have shown that waterfowl contributed a substantial amount of fecal coliforms to surface recreational waters (44, 45). The positive correlation of sandhill crane marker signals with E. coli and Enterococcus in water and excreta strongly suggests that the quantity fluctuations of these bacterial groups over the migration period were heavily influenced by the crane fecal loadings. The other known sources, like humans and cattle, did not significantly increase in occurrence compared to the crane, according to Bacteroidetes-based human- and cattle-targeted PCR results. On the other hand, neither cranes nor, possibly, snow geese appeared to significantly impact Bacteroidetes quantity in water (based on qPCR signals between putative crane markers and Bacteroidetes spp. [R2 = 0.064]). This lack of Bacteroidetes impact was consistent with other lines of evidence. The first was the lack of detectable Bacteroidetes marker in crane excreta, and only 3.2% of the 16S rRNA gene library sequences (n = 1,151) were similar to those of Bacteroidetes members (29). Second, most Bacteroidetes-like sequences (>82.5%) might be from unidentified sources in water rather than from crane excreta (7.4%) or other known sources (human and cattle). Therefore, the large quantity of Bacteroidetes organisms in water was more likely from other, unidentified source(s) (see Fig. S2 in the supplemental material). It should be noted that the quantity of Bacteroidetes organisms was also smaller during than before migration (Fig. 3A and E). In managing pathogen risks in this section of the Platte River, it is important to mitigate both human (septage) and cattle manure runoff, as both have been determined to contribute more human-infective pathogens (when normalized for E. coli or Enterococcus species densities) than bird excreta with similar Campylobacter occurrences (as identified in sandhill cranes in the current study) (46, 47). Also, while Bacteroidetes markers have been used to identify human and cattle fecal contamination, the group may not be useful for evaluating waterfowl fecal contamination (35, 47). Finally, while both viable and dead cells are detected, the interpretion of qPCR results in risk management decision making provides a conservative estimate that is suitable for an initial-screening-level assessment (36).
Summary.
Overall, sandhill crane migration via the central Platte River in Nebraska resulted in a significant increase in fecal indicator bacteria, especially E. coli and Enterococcus spp. in surface water and sediments, along with C. jejuni. Although public health risks associated with sandhill cranes and other waterfowl surrounding those areas are unknown, the increase in environmental C. jejuni isolates suggests that risks may be higher than previously estimated for other waterfowl, such as seagulls (16, 25). Furthermore, the contamination by C. jejuni could be a risk to tourists and consumers/handlers of crops through irrigation and via local drinking water consumption.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dennis Lye, Katharine O'Connell, and Asja Korajkic (U.S. EPA, Cincinnati, OH) and Matt Moser (USGS, Lincoln, NE) for sampling and technical support.
Although this work was reviewed by the EPA and approved for publication, it does not necessarily reflect official agency policy. The mention of products or trade names does not constitute a recommendation for use.
Footnotes
Published ahead of print 12 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03990-12.
REFERENCES
- 1. Boyle WA, Conway CJ. 2007. Why migrate? A test of the evolutionary precursor hypothesis. Am. Nat. 169:344–359 [DOI] [PubMed] [Google Scholar]
- 2. Alderisio KA, DeLuca N. 1999. Seasonal enumeration of fecal coliform bacteria from the feces of ring-billed gulls (Larus delawarensis) and Canada Geese (Branta canadensis). Appl. Environ. Microbiol. 65:5628–5630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dolejská M, Bierošová B, Kohoutová L, Literák I, Čížek A. 2009. Antibiotic-resistant Salmonella and Escherichia coli isolates with integrons and extended-spectrum beta-lactamases in surface water and sympatric black-headed gulls. J. Appl. Microbiol. 106:1941–1950 [DOI] [PubMed] [Google Scholar]
- 4. Graczyk TK, Majewska AC, Schwab KJ. 2008. The role of birds in dissemination of human waterborne enteropathogens. Trends Parasitol. 24:55–59 [DOI] [PubMed] [Google Scholar]
- 5. Hoar BM, Whiteside DP, Ward L, Douglas Inglis G, Morck DW. 2007. Evaluation of the enteric microflora of captive whooping cranes (Grus americana) and Sandhill cranes (Grus canadensis). Zoo Biol. 26:141–153 [DOI] [PubMed] [Google Scholar]
- 6. Broman T, Palmgren H, Bergstrom S, Sellin M, Waldenström J, Danielsson-Tham ML, Olsen B. 2002. Campylobacter jejuni in black-headed gulls (Larus ridibundus): prevalence, genotypes, and influence on C. jejuni epidemiology. J. Clin. Microbiol. 40:4594–4602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gardner TJ, Fitzgerald C, Xavier C, Klein R, Pruckler J, Stroika S, McLaughlin JB. 2011. Outbreak of campylobacteriosis associated with consumption of raw peas. Clin. Infect. Dis. 53:26–32 [DOI] [PubMed] [Google Scholar]
- 8. Medema GJ, Baher M, Schnets FM.1997. Survival of Cryptosporidium parvum, Escherichia coli, faecal streptococci, and Clostridium perfringens in river water: influence of temperature and autochthonous microorganisms. Water Sci. Technol. 35:249–252 [Google Scholar]
- 9. Reed KD, Meece JK, Henkel JS, Shukla SK. 2003. Birds, migration and emerging zoonoses: West Nile virus, Lyme disease, influenza A and enteropathogens. Clin. Med. Res. 1:5–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ahmed W, Hodgers L, Sidhu JPS, Toze S. 2012. Fecal indicators and zoonotic pathogens in household drinking water taps fed from rainwater tanks in southeast Queensland, Australia. Appl. Environ. Microbiol. 78:219–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fallacara DM, Monahan CM, Morishita TY, Bremer CA, Wack RF. 2004. Survey of parasites and bacterial pathogens from free-living waterfowl in zoological settings. Avian Dis. 48:759–767 [DOI] [PubMed] [Google Scholar]
- 12. Pacha RE, Clark GW, Williams EA, Carter AM. 1988. Migratory birds of central Washington as reservoirs of Campylobacter jejuni. Can. J. Microbiol. 34:80–82 [DOI] [PubMed] [Google Scholar]
- 13. Waldenström J, On SL, Ottvall R, Hasselquist D, Olsen B. 2007. Species diversity of campylobacters in a wild bird community in Sweden. J. Appl. Microbiol. 102:424–432 [DOI] [PubMed] [Google Scholar]
- 14. Benton C, Khan F, Monaghan P, Richards WN, Sheddon CB. 1983. Contamination of a major water supply by gulls (Larus spp.): a study of the problem and remedial action taken. Water Res. 17:789–798 [Google Scholar]
- 15. Valiela I, Alber M, LaMontagne M. 1991. Fecal coliform loadings and stocks in Buttermilk Bay, Massachusetts and management implications. Environ. Manag. 15:659–674 [Google Scholar]
- 16. Roser DJ, Davies CM, Ashbolt NJ, Morison P. 2006. Microbial exposure assessment of an urban recreational lake: a case study of the application of new risk-based guidelines. Water Sci. Technol. 54:245–252 [DOI] [PubMed] [Google Scholar]
- 17. Krapu GL, Facey DE, Fritzell EK, Johnson DH. 1984. Habitat use by migrant sandhill cranes in Nebraska. J. Wildl. Manage. 48:407–417 [Google Scholar]
- 18. Inglis GD, Hoar BM, Whiteside DP, Morck DW. 2007. Campylobacter canadensis sp. nov., from captive whooping cranes in Canada. Int. J. Syst. Evol. Microbiol. 57:2636–2644 [DOI] [PubMed] [Google Scholar]
- 19. Homan Y, Muroga N, Taharaguchi S, Chuma T, Takase K, Shioya K, Mohri S. 2005. Salmonella Typhimurium isolated from the feces of wild cranes from the Izumi plain, Kagoshima. J. Jpn. Vet. Med. Assoc. 58:411–414 [Google Scholar]
- 20. Kitadai N, Ninomiya N, Murase T, Obi T, Takase K. 2010. Salmonella isolated from the feces of migrating cranes at the Izumi Plain (2002–2008): serotype, antibiotic sensitivity and PFGE type. J. Vet. Med. Sci. 72:939–942 [DOI] [PubMed] [Google Scholar]
- 21. Thoen CO, Himes EM, Barrett RE. 1977. Mycobacterium avium serotype 1 infection in a Sandhill crane (Grus canadensis). J. Wildl. Dis. 13:40–42 [DOI] [PubMed] [Google Scholar]
- 22. Urakawa H, Martens-Habbena W, Stahl DA. 2010. High abundance of ammonia-oxidizing Archaea in coastal waters, determined using a modified DNA extraction method. Appl. Environ. Microbiol. 76:2129–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yan PY, Hou HS, Chen T, Zhang SH, Sun WJ. 2012. Methods for extraction of microorganism DNA from glacier surface snow. Sci. Cold Arid Reg. 4:0484–0489 [Google Scholar]
- 24. Lund M, Nordentoft S, Pedersen K, Madsen M. 2004. Detection of Campylobacter spp. in chicken fecal samples by real-time PCR. J. Clin. Microbiol. 42:5125–5132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Haugland RA, Siefring SC, Wymer LJ, Brenner KP, Dufour AP. 2005. Comparison of Enterococcus measurements in freshwater at two recreational beaches by quantitative polymerase chain reaction and membrane filter culture analysis. Water Res. 39:559–568 [DOI] [PubMed] [Google Scholar]
- 26. Dick LK, Field KG. 2004. Rapid estimation of numbers of fecal Bacteroidetes by use of a quantitative PCR assay for 16S rRNA genes. Appl. Environ. Microbiol. 70:5695–5697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ludwig W, Schleifer KH. 2000. How quantitative is quantitative PCR with respect to cell counts? Syst. Appl. Microbiol. 23:556–562 [DOI] [PubMed] [Google Scholar]
- 28. Lu J, Ryu H, Santo Domingo JW, Griffith JF, Ashbolt N. 2011. Molecular detection of Campylobacter spp. in California gull (Larus californicus) excreta. Appl. Environ. Microbiol. 77:5034–5039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ryu R, Lu J, Vogel J, Elk M, Chavez-Ramirez F, Ashbolt N, Santo Domingo J. 2012. Development of a novel crane-specific qPCR assay and molecular characterization of bacterial populations in Sandhill crane (Grus canadensis) and snow goose (Chen caerulescens). Appl. Environ. Microbiol. 78:4338–4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Linton D, Owen RJ, Stanley J. 1996. Rapid identification by PCR of the genus Campylobacter and of five species enteropathogenic for man and animals. Res. Microbiol. 147:707–718 [DOI] [PubMed] [Google Scholar]
- 31. Stucki URS, Joachim F, Nicolet J, Burnens AP. 1995. Identification of Campylobacter jejuni on the basis of a species gene that encodes a membrane protein. J. Clin. Microbiol. 33:855–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gonzalez I, Grant KA, Richardson PT, Park SF, Collins MD. 1997. Specific identification of the enteropathogens Campylobacter jejuni and Campylobacter coli using a PCR test based on the ceuE gene encoding a putative virulence determinant. J. Clin. Microbiol. 35:759–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Khan IUH, Edge TA. 2007. Development of a novel triplex PCR assay for the detection and differentiation of thermophilic species of Campylobacter using 16 S–23 S rDNA internal transcribed spacer (ITS) region. J. Appl. Microbiol. 103:2561–2569 [DOI] [PubMed] [Google Scholar]
- 34. Tamura K, Dudley KJ, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 35. Bernhard AE, Field KG. 2000. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl. Environ. Microbiol. 66:4571–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Klein M, Brown L, Ashbolt NJ, Stuetz RM, Roser DJ. 2011. Inactivation of indicators and pathogens in cattle feedlot manures and compost as determined by molecular and culture assays. FEMS Microbiol. Ecol. 77:200–210 [DOI] [PubMed] [Google Scholar]
- 37. Williams LK, Sait LC, Cogan TA, Jørgensen F, Grogono-Thomas R, Humphrey TJ. 2012. Enrichment culture can bias the isolation of Campylobacter subtypes. Epidemiol. Infect. 140:1227–1235 [DOI] [PubMed] [Google Scholar]
- 38. Gorkiewicz G, Feierl G, Schober C, Dieber F, Köfer J, Zechner R, Zechner EL. 2003. Species-specific identification of campylobacters by partial 16S rRNA gene sequencing. J. Clin. Microbiol. 41:2537–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Waldenström J, Broman T, Carlsson I, Hasselquist D, Achterberg RP, Wagenaar JA, Olsen B. 2002. Prevalence of Campylobacter jejuni, Campylobacter lari, and Campylobacter coli in different ecological guilds and taxa of migrating birds. Appl. Environ. Microbiol. 68:5911–5917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cramp S, Simmons KFL. (ed). 1990. The birds of the Western Palearctic, vol III Oxford University Press, Oxford, United Kingdom [Google Scholar]
- 41. Gerner-Smidt P, Stroika SG, Fitzgerald C. 2008. National molecular subtyping network for food-borne bacterial disease surveillance in the United States, p 277–285 In Nachamkin I, Szymanski CM, Blaser MJ. (ed), Campylobacter, 3rd ed ASM Press, Washington, DC [Google Scholar]
- 42. Champion OL, Gaunt MW, Gundogdu O, Elmi A, Witney AA, Hinds J, Dorrell N, Wren BW. 2005. Comparative phylogenomics of the food-borne pathogen Campylobacter jejuni reveals genetic markers predictive of infection source. Proc. Natl. Acad. Sci. U. S. A. 102:16043–16048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Colles FM, McCarthy ND, Howe JC, Devereux CL, Gosler AG, Maiden MC. 2009. Dynamics of Campylobacter colonization of a natural host, Sturnus vulgaris (European starling). Environ. Microbiol. 11:258–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hussong D, Damaré JM, Limpert RJ, Sladen WJ, Weiner RM, Colwell RR. 1979. Microbial impact of Canada geese (Branta canadensis) and whistling swans (Cygnus columbianus columbianus) on aquatic ecosystems. Appl. Environ. Microbiol. 37:14–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Standridge JH, Delfino JJ, Kleppe LB, Butler R. 1979. Effect of waterfowl (Anas platyrhynchos) on indicator bacteria populations in a recreational lake, Madison, Wisconsin. Appl. Environ. Microbiol. 38:547–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schoen ME, Ashbolt NJ. 2010. Assessing pathogen risk to swimmers at non-sewage impacted recreational beaches. Environ. Sci. Technol. 44:2286–2291 [DOI] [PubMed] [Google Scholar]
- 47. Soller JA, Schoen ME, Bartrand T, Ravenscroft JE, Ashbolt NJ. 2010. Estimated human health risks from exposure to recreational waters impacted by human and non-human sources of faecal contamination. Water Res. 44:4674–4691 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.