Abstract
The proteome of the ropy strain Bifidobacterium animalis subsp. lactis A1dOxR, compared to that of its nonropy isogenic strain, showed an overproduction of a protein involved in rhamnose biosynthesis. Results were confirmed by gene expression analysis, and this fact agreed with the high rhamnose content of the ropy exopolysaccharide.
TEXT
The ability to synthesize an extracellular layer of carbohydrates, or exopolysaccharides (EPS), is a common trait in bacteria, such as those members of the microbiota inhabiting our gut (1, 2), including Bifidobacterium and Lactobacillus (3–5). For instance, some strains of these human commensal genera are able to produce complex EPS under laboratory conditions. These polymers, built on repeating units of different monosaccharides, are also known as heteropolysaccharides (6, 7). Recently, EPS-producing bacteria have received particular attention due to the presumptive implication of the polymers in the cross talk between bacteria and host (8–11). However, there is limited information as to why only some specific EPS are able to interact with human cells (12–14). Previous results from our group, also supported by other literature data, have shown that some physicochemical characteristics of EPS could be correlated with their immune-modulating capability. Specifically, it seems that polymers with high molecular weights (HMWs) are able to act as suppressors of the immune response (15). Part of the results supporting this hypothesis were obtained with a model of three isogenic EPS-producing Bifidobacterium animalis subsp. lactis strains, A1, A1dOx, and A1dOxR, the last being able to synthesize an HMW EPS fraction with an immunosuppressive profile (13).
This strain, A1dOxR, also named IPLA-R1, displays a characteristic “ropy” phenotype, denoted by the formation of a long filament when a loop is introduced into the colony. Interestingly, this ropy character was not observed in two related (isogenic) EPS-producing strains, A1 and A1dOx, which also lack the production of the HMW EPS fraction; in fact, it seems that all three strains synthesize a low-molecular-weight (LMW) EPS fraction (Table 1). Additionally, the monosaccharides building the EPS produced by the three strains are the same (glucose, galactose, and rhamnose), but they are present at different ratios (16). Furthermore, the LMW EPS from the two isogenic strains did not elicit a suppression of the immune response (13). Therefore, it seems that both the ability to suppress the immune response and the ropy phenotype in strain A1dOxR may be related to the synthesis of the HMW polymer. In this regard, it has been indicated that the ability of certain EPS-producing bacteria to confer ropiness to a fermented product is directly related to the molecular weight of its polymer. That is, ropy strains have EPS of high molecular weights, whereas nonropy strains produce polymers of smaller molecular weights (17, 18). In the current work, we have tried to gain insight into some molecular and physiological aspects of the occurrence of the ropy phenotype in the B. animalis subsp. lactis strain A1dOxR.
Table 1.
Growth parameters of strainsa
| Strain | Strain phenotype | EPS MW distribution | Mean ± SD |
||||
|---|---|---|---|---|---|---|---|
| OD600 | No. of log CFU ml−1 | pH | Total organic acids (mM) | Ratio of acetic acid to lactic acid | |||
| A1 | Nonropy | LMW | 7.97 ± 0.11 | 9.42 ± 0.05 | 4.36 ± 0.01 | 116.8 ± 15.8 | 1.32 ± 0.04 |
| A1dOx | Nonropy | LMW | 8.60 ± 0.11 | 9.39 ± 0.05 | 4.39 ± 0.01 | 110.93 ± 14.5 | 1.32 ± 0.07 |
| A1dOxR | Ropy | LWM + HMW | 8.27 ± 0.13 | 9.58 ± 0.01 | 4.41 ± 0.01 | 106.65 ± 16.96 | 1.29 ± 0.08 |
Measured after 24 h of incubation in MRS broth plus 0.25% l-cysteine (MRSC) cultured with the exopolysaccharide (EPS)-producing strains Bifidobacterium animalis subsp. lactis A1, A1dOx, and A1dOxR. Abbreviations: MW, molecular weight; LMW, low molecular weight; HMW, high molecular weight.
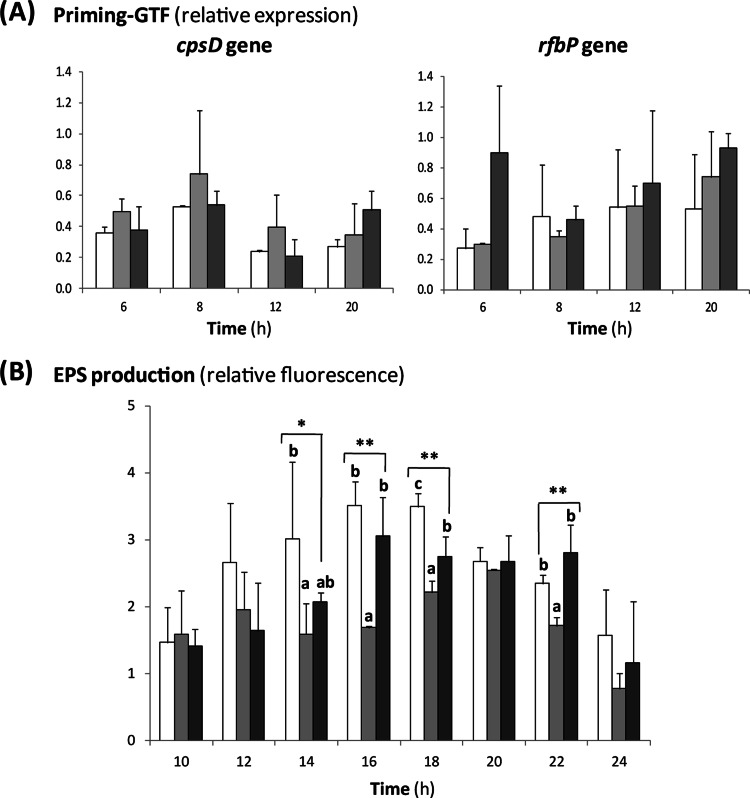
The strains A1 (parental, isolated from a fermented dairy product), A1dOx (adapted to OxGall by exposure of strain A1 to increasing concentrations of these bile salts), and A1dOxR (a derivative of A1dOx, which, after consecutive generations in the absence of bile, spontaneously acquired a ropy phenotype) were grown in MRSC (MRS [Difco, BD Biosciences, San Diego, CA] containing 0.25% l-cysteine [Sigma Chemical Co., St. Louis, MO]) as previously described (13). As an initial approach, several physiological parameters were determined during the growth of the strains in order to determine potential differences among them. Growth curves were performed for 24 h at 37°C under anaerobic conditions in at least two biological replicates, and measurements in each replicate were done in duplicate (see the supplemental material). The evolution of the optical density at 600 nm (OD600) and counts were parallel in the three bifidobacteria, and statistical differences were not detected in the specific growth rates (μmax) among the strains (μmax = 0.58 ± 0.01, 0.57 ± 0.02, and 0.56 ± 0.01 for A1, A1dOx, and A1dOxR, respectively). In addition, the acidification rates, determined by monitoring the pH decrease and by analyzing the production of total (acetic plus lactic plus formic) organic acids, were similar in the three strains. The final values for these parameters reached after 24 h of incubation are collected in Table 1. Regarding EPS synthesis, the expression levels of the two priming glycosyltransferase genes (cpsD and rfbP) located within the B. animalis subsp. lactis eps cluster did not show any significant variation among the three strains (Fig. 1A). This suggests that the amounts of EPS synthesized by the three strains were rather similar. Indeed, the production of total EPS, quantified by means of fluorescent-dye-conjugated lectins (19), showed slight differences among the three strains (Fig. 1B). This variability may be related to the different sensitivities of this lectin, which is specific for the detection of α-mannopyranosyl and α-glucopyranosyl residues and can detect the monosaccharides present at different ratios in the three polymers (16). Nevertheless, all previous facts suggest that the levels of glucose turnover in metabolic pathways, such as glycolysis and EPS biosynthesis, were not significantly different among the three strains and, therefore, could not explain the occurrence of the ropy phenotype of strain A1doxR.
Fig 1.
(A) Relative expression of the cpsD and rfbP genes, encoding the priming glycosyltransferase (GTF) CpsD and RfbP in the EPS-producing strains B. animalis subsp. lactis A1 (white bars), A1dOx (light-gray bars), and A1dOxR (dark-gray bars) during growth in MRSC medium. Within each strain, the quantitative-PCR result for each gene was compared with that for the reporter gene recA, encoding recombinase A, and finally to the basal (time zero [hours]) expression level. No statistical differences, analyzed by means of analysis of variance (ANOVA) tests, were detected among strains at each incubation point. (B) EPS production, expressed as the relative fluorescence emitted after EPS labeling with concanavalin A conjugated with Alexa Fluor 488, by the strains B. animalis subsp. lactis A1 (white bars), A1dOx (light-gray bars), and A1dOxR (dark-gray bars) during growth in MRSC medium. Values of fluorescence emitted were corrected for the number of bifidobacteria obtained at each sampling point, and finally, fluorescence was calculated relative to that at the initial point (0 h). Results represent average values from three biological replicates, which were analyzed by means of one-way ANOVA tests (*, P < 0.05; **, P < 0.01). Bars that are not labeled with the same letter are significantly different (P < 0.05) according to the mean-comparison least significant difference test.
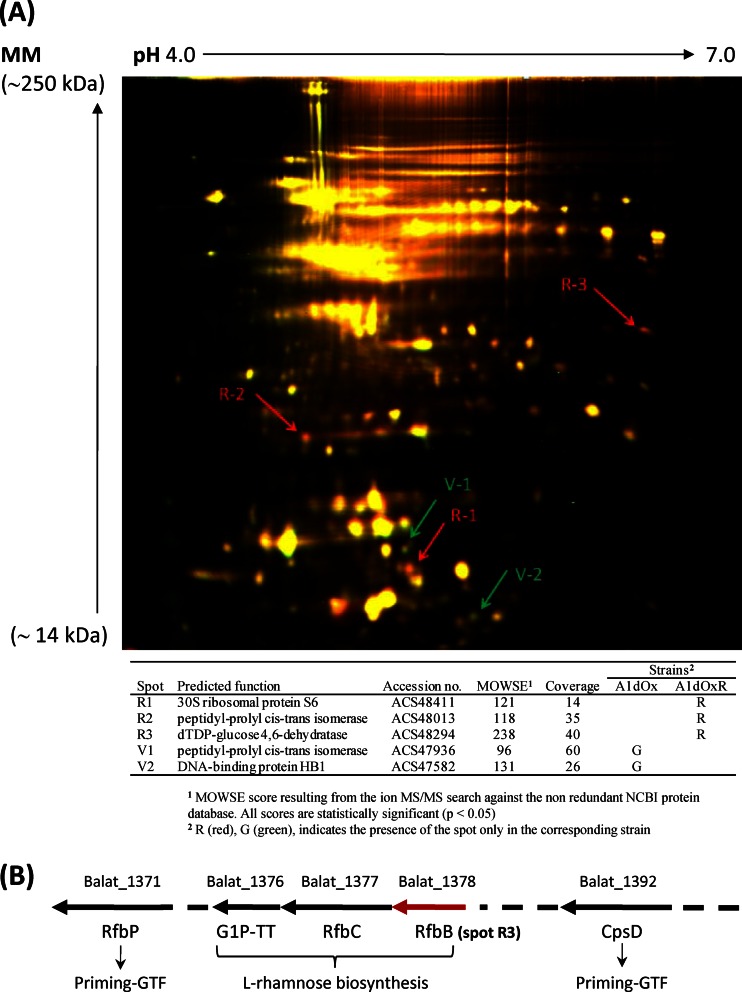
In a step forward, we also wanted to check whether differences at the molecular level could be detected among the three isogenic strains. For this purpose, a proteomic analysis was carried out with samples collected in the same growth phase (∼6 h; OD600, ∼1). Cultures were broken by sonication, and proteins from the cell debris-free supernatants were precipitated by methanol-chloroform (3:1, vol/vol) and finally frozen until use (see the supplemental material). The protein extracts were analyzed by two-dimensional difference in gel electrophoresis (2D-DIGE) by comparing strain A1 with strain A1dOx and strain A1dOx with strain A1dOxR. The proteomes of the nonropy strains (A1 and A1dOx) did not show apparent differences (data not shown); however, the comparison of A1dOx with its ropy derivative A1dOxR evidenced a few proteins that differed between them (Fig. 2A). One of these proteins (spot R3 in Fig. 2A) was present in the extracts of strain A1dOxR but not in those of A1dOx; it was annotated in the NCBI protein database as dTDP-d-glucose 4,6-dehydratase (RfbB; accession no. COG1088), and it is involved in the biosynthesis of l-rhamnose in Gram-negative bacteria (20). Remarkably, this enzyme is encoded by the first gene (Balat_1378) out of the three detected for this biosynthetic pathway inside the eps cluster in B. animalis subsp. lactis (Fig. 2B). Then, specific primers (see Table S1 in the supplemental material) were designed to check the expression of these three genes by quantitative reverse transcriptase PCR in our strains. From this analysis, we determined that the ropy A1dOxR strain overexpressed the Balat_1376, Balat_1377, and Balat-1378 genes by 5.01 (±0.62)-, 4.57 (±1.2)-, and 7.16 (±1.17)-fold, respectively, relative to their expression by AldOxR's parental strain, A1dOx. Similar levels of overexpression were detected in the two intergenic regions of the cluster (5.60 ± 0.25 and 8.61 ± 0.41 for intergenic regions Balat_1376 to _1377 and Balat_1377 to _1378, respectively), suggesting an operon structure. Thus, it seems that the rhamnose biosynthetic pathway was activated in strain A1dOxR. Indeed, the monosaccharide composition, determined by gas chromatography-mass spectrometry (GC-MS) technology as previously described (7), of the EPS purified from both strains showed a higher percentage of rhamnose in the polymer A1dOxR (34.6% ± 2.6%) than that detected in the polymer A1dOx (24.3% ± 0.8%). This fact may also explain the high (50%) rhamnose content found in the repeating-unit structure of the HMW EPS fraction synthesized exclusively by the ropy A1dOxR strain, which was determined in a previous work by nuclear magnetic resonance (NMR) (21). The three genes of the rhamnose biosynthesis pathway present in the eps cluster and the flanking regions of this cluster were sequenced in the three strains, but no single nucleotide polymorphisms (SNPs) were detected among them. This suggests that a pleiotropic effect of a mutation(s) located somewhere else in the chromosome, or a loss in the transcriptional control of the operon, may be the basis of the changes in expression of these genes in the ropy strain. On the other hand, a second protein (spot V2 in Fig. 2A) showing homology to the DNA-binding protein HB1 was detected only in the nonropy strain A1dOx. This protein belongs to the DNA BII family and is able to bind and bend DNA, thus acting as an architectural factor that may mediate many cellular processes (22). No variations in the sequences of the corresponding genes, or in their expression, were detected between the two strains under study. Additionally, as far as we know, there is currently no evidence of genetic EPS regulation by this type of protein in Gram-positive bacteria. Finally, the eps cluster from strain A1dOxR, which was sequenced in a previous work (21), showed a structural organization that was identical, with a few nucleotide changes but no deletions or insertions, to that found in other available B. animalis subsp. lactis genomes (23). This fact is not surprising due to the scarce genetic variability detected in the whole genomes of strains of this subspecies (24).
Fig 2.
(A) Representative overlay image of a 2D-DIGE gel containing proteins extracted at the mid-exponential growth phase (∼6 h) from the B. animalis subsp. lactis strains A1dOx (dyed green) and A1dOxR (dyed red). The yellow protein spots showed no differences in abundance between the two strains, whereas those in red were more abundant in strain A1dOxR and those in green were more abundant in strain A1dOx. The numbered spots were excised from a duplicated 2D-PAGE gel stained with Gelcode Blue Safe, and the proteins were identified by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) MS, as indicated in the table. MM, molecular mass; MOWSE, molecular weight search. (B) Partial physical map of the eps cluster in B. animalis subsp. lactis DSM10140 (type strain), showing some genes of interest in this work, namely, Balat_1392, galactosyltransferase (CpsD); Balat_1978, dTDP-d-glucose 4,6-dehydratase (RfbB); Balat_1977, dTDP-4-dehydro-rhamnose 3,5-epimerase (RfbC); Balat_1976, dTDP-glucose pyrophosphorylase or glucose-1-phosphate thymidylyltransferase (G1P_TT_ short); and Balat_1971, UDP-phosphate sugar phosphotransferase (RfbP).
In short, in this study, we have found an association between the high rhamnose content of the HMW EPS synthesized by strain A1dOxR and the overexpression of the rhamnose biosynthesis genes located within its eps cluster. Further work will focus on the analysis of the genomes of these three isogenic strains and the SNP variations among them in order to identify the molecular bases of the ropy phenotype.
Supplementary Material
ACKNOWLEDGMENTS
This work was financed by the Spanish Ministry of Science and Innovation (MICINN) and FEDER European Union funds through project AGL2009-09445. C. Hidalgo-Cantabrana acknowledges his FPI fellowship and B. Sánchez his Juan de la Cierva postdoctoral contract with MICINN (currently the Ministry of Economy and Competitiveness).
We are grateful to Isabel Cuesta (IPLA-CSIC) for her excellent technical assistance in the high-performance liquid chromatography analysis.
Footnotes
Published ahead of print 12 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00633-13.
REFERENCES
- 1. Coyne MJ, Chatzidaki-Livanis M, Paoletti LC, Comstock LE. 2008. Role of glycan synthesis in colonization of the mammalian gut by the bacterial symbiont Bacteroides fragilis. Proc. Natl. Acad. Sci. U. S. A. 105:13099–13104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Patrick S, Blakely GW, Houston S, Moore J, Abratt VR, Bertalan M, Cerdeño-Tárraga M, Quail MA, Corton N, Corton C, Bignell A, Barron A, Clark L, Bentley SD, Parkhill J. 2010. Twenty-eight divergent polysaccharide loci specifying within- and amongst-strain capsule diversity in three strains of Bacteroides fragilis. Microbiology 156:3255–3269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lebeer S, Vanderleyden J, De Keersmaecker SCJ. 2010. Host interactions of probiotic bacteria surface molecules: comparison with commensal and pathogens. Nat. Rev. Microbiol. 8:171–184 [DOI] [PubMed] [Google Scholar]
- 4. Turroni F, Ventura M, Buttó LF, Duranti S, O'Toole PW, O'Connell-Motherway M, van Sinderen D. 2013. Molecular dialogue between the human gut microbiota and the host: a Lactobacillus and Bifidobacterium perspective. Cell. Mol. Life Sci. doi: 10.1007/s0018-013-1318-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ventura M, Turroni F, O'Connell-Motherway M, MacSharry J, van Sinderen D. 2012. Host-microbe interactions that facilitate gut colonization by commensal bifidobacteria. Trends Microbiol. 20:467–476 [DOI] [PubMed] [Google Scholar]
- 6. Ruas-Madiedo P, Moreno JA, Salazar N, Delgado S, Mayo B, Margolles A, de los Reyes-Gavilán CG. 2007. Screening of exopolysaccharide-producing Lactobacillus and Bifidobacterium strains isolated from the human intestinal microbiota. Appl. Environ. Microbiol. 73:4385–4388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Salazar N, Prieto A, Leal JA, Mayo B, Bada-Gancedo JC, de los Reyes-Gavilán CG, Ruas-Madiedo P. 2009. Production of exopolysaccharides by Lactobacillus and Bifidobacterium strains of human origin, and metabolic activity of the producing bacteria in milk. J. Dairy Sci. 92:4158–4168 [DOI] [PubMed] [Google Scholar]
- 8. Denou E, Pridmore RD, Berger B, Panoff JM, Arigoni F, Brüssow H. 2008. Identification of genes associated with the long-gut-persistence phenotype of the probiotic Lactobacillus johnsonii strain NCC522 using a combination of genomic and transcriptomic analysis. J. Bacteriol. 190:3161–3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fanning S, Hall LJ, Cronin M, Zomer A, MacSharry J, Goulding D, O'Connell-Motherway M, Shanahan F, Nally K, Dougan G, van Sinderen D. 2012. Bifidobacterial surface-exopolysaccharide facilitates commensal-host interaction through immune modulation and pathogen protection. Proc. Natl. Acad. Sci. U. S. A. 109:2108–2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lebeer S, Claes IJJ, Verhoeven TLA, Vanderleyden J, De Keersmaecker SCJ. 2011. Exopolysaccharides of Lactobacillus rhamnosus GG form a protective shield against innate immune factors in the intestine. Microb. Biotechnol. 4:368–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, Mazmanian SK. 2011. The Toll-like receptor 2 pathway established colonization by a commensal of the human microbiota. Science 322:974–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bleau C, Monges A, Rashidan K, Laverdure J-P, Lacroix M, Van Calsteren M-R, Millette M, Savard R, Lamontagne L. 2010. Intermediate chains of exopolysaccharides from Lactobacillus rhamnosus RW-9595M increase IL-10 production by macrophages. J. Appl. Microbiol. 108:666–675 [DOI] [PubMed] [Google Scholar]
- 13. López P, Monteserín DC, Gueimonde M, de los Reyes-Gavilán CG, Margolles A, Suárez A, Ruas-Madiedo P. 2012. Exopolysaccharide-producing Bifidobacterium strains elicit different in vitro responses upon interaction with human cells. Food Res. Int. 46:99–107 [Google Scholar]
- 14. Nikolic M, López P, Strahinic I, Suárez A, Kojic M, Fernández-García M, Topisirovic L, Golic N, Ruas-Madiedo P. 2012. Characterization of the exopolysaccharide (EPS)-producing Lactobacillus paraplantarum BGCG11 and its non-EPS producing derivative strains as potential probiotics. Int. J. Food Microbiol. 158:155–162 [DOI] [PubMed] [Google Scholar]
- 15. Hidalgo-Cantabrana C, López P, Gueimonde M, de los Reyes-Gavilán CG, Suñarez A, Margolles A, Ruas-Madiedo P. 2012. Immune modulation capability of exopolysaccharides synthesised by lactic acid bacteria and bifidobacteria. Probiotics Antimicrob. Proteins 4:227–237 [DOI] [PubMed] [Google Scholar]
- 16. Ruas-Madiedo P, Medrano M, Salazar N, de los Reyes-Gavilán CG, Pérez PF, Abraham AG. 2010. Exopolysaccharides produced by Lactobacillus and Bifidobacterium strains abrogate in vitro the cytotoxic effect of bacterial toxins on eukaryotic cells. J. Appl. Microbiol. 109:2079–2086 [DOI] [PubMed] [Google Scholar]
- 17. Faber EJ, Zoon P, Kemerling JP, Vliegenthart JF. 1998. The exopolysaccharides produced by Streptococcus thermophilus Rs and Sts have the same repeating unit but differ in viscosity of their milk cultures. Carbohydr. Res. 310:269–276 [DOI] [PubMed] [Google Scholar]
- 18. Petry S, Furlan S, Waghorne E, Saulnier L, Cerning J, Maguin E. 2003. Comparison of the thickening properties of four Lactobacillus delbrueckii subsp. bulgaricus strains and physicochemical characterization of their exopolysaccharides. FEMS Microbiol. Lett. 221:285–291 [DOI] [PubMed] [Google Scholar]
- 19. Ruas-Madiedo P, Gueimonde M, Arigoni F, de los Reyes-Gavilán CG, Margolles A. 2009. Bile affects the synthesis of exopolysaccharides by Bifidobacterium animalis. Appl. Environ. Microbiol. 75:1204–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Allard ST, Giraud MF, Whitfiled C, Graninger M, Messner P, Naismith JH. 2001. The crystal structure of dTDP-d-glucose 4,6-dehydratase (RmlB) from Salmonella enterica serovar Typhimurium, the second enzyme in the dTDP-1-rhamnose pathway. J. Mol. Biol. 307:283–295 [DOI] [PubMed] [Google Scholar]
- 21. Leivers S, Hidalgo-Cantabrana C, Robinson G, Margolles A, Ruas-Madiedo P, Laws AP. 2011. Structure of the high molecular weight exopolysaccharide produced by Bifidobacterium animalis subsp. lactis IPLA-R1 and sequence analysis of its putative eps cluster. Carbohydr. Res. 346:2710–2717 [DOI] [PubMed] [Google Scholar]
- 22. Swinger KK, Rice PA. 2004. IHF and HU: flexible architects of bent DNA. Curr. Opin. Struct. Biol. 14:28–35 [DOI] [PubMed] [Google Scholar]
- 23. Ruas-Madiedo P, Sánchez B, Hidalgo-Cantabrana C, Margolles A, Laws AP. 2012. Exopolysaccharides from lactic acid bacteria and bifidobacteria, p 125–152 In Hui YH, Evranuz EO. (ed), Handbook of animal-based fermented food and beverage technology, 2nd ed CRC Press, Boca Raton, FL [Google Scholar]
- 24. Briczinski EP, Loquasto JR, Barragou R, Dudley EG, Roberts AM, Roberts RF. 2009. Strain-specific genotyping of Bifidobacterium animalis subsp. lactis by using single-nucleotide polymorphisms, insertions, and deletions. Appl. Environ. Microbiol. 75:7501–7508 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.