Abstract
AlfB and AlfC α-l-fucosidases from Lactobacillus casei were used in transglycosylation reactions, and they showed high efficiency in synthesizing fucosyldisaccharides. AlfB and AlfC activities exclusively produced fucosyl-α-1,3-N-acetylglucosamine and fucosyl-α-1,6-N-acetylglucosamine, respectively. The reaction kinetics showed that AlfB can convert 23% p-nitrophenyl-α-l-fucopyranoside into fucosyl-α-1,3-N-acetylglucosamine and AlfC at up to 56% into fucosyl-α-1,6-N-acetylglucosamine.
TEXT
Fucosyl-oligosaccharides (FUS) are coming to be of great interest due to their presence in human milk and their antiadhesive activity against pathogens. This last property is characterized by blocking the attachment of intestinal pathogens to host cells due to FUS structural similarity with cell-surface glycoconjugate receptors (1–4). In addition, genes encoding α-l-fucosidases (EC 3.2.1.51) have recently been found in genomes of bifidobacteria (5, 6) and lactobacilli (7), which raises issues regarding the role of these enzymes in the metabolism of fucose-containing structures and opens up the possibility of using FUS as novel prebiotics (8). To fully establish the biological function of FUS as antiadhesins and prebiotics, extensive studies are required, for which synthesis of sufficient amounts would be necessary. Chemical synthesis of FUS such as ABH and Lewis blood group antigens has long been performed, but it is a tedious and expensive process since it requires multiple protection and deprotection steps to achieve the desired selectivity (9, 10). The enzymatic synthesis of FUS is an alternative to chemical methods, and it offers the advantage of forming specific glycosidic linkages in the presence of other reactive functional groups. There are two types of enzymes that can carry out fucosylation: fucosyltransferases and fucosidases. Fucosyltransferases present high specificity toward the acceptor and do not hydrolyze the product (11, 12). However, those enzymes are generally difficult to express and they require an expensive sugar nucleotide or a complex multienzymatic system for the regeneration (11, 13). In contrast, oligosaccharides have been largely produced using the transglycosylation activity of glycosidases and engineered glycosynthases (14, 15). Only a few works describe the synthesis of FUS by transglycosylation using α-l-fucosidases; however, they cover a wide range of sources such as mammalian tissues (16), fungi (17), and bacteria (18, 19). The transglycosylation activity of α-l-fucosidases is generally moderate compared with the hydrolysis activity, although it is variable and depends on the origin of the enzymes (20).
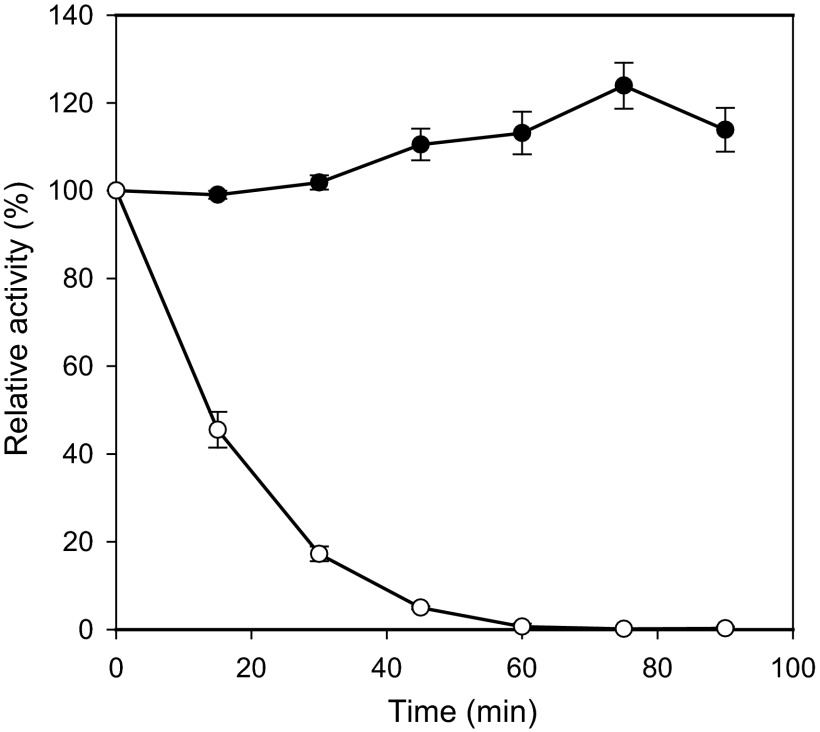
We have previously isolated three α-l-fucosidases (AlfA, AlfB, and AlfC) from Lactobacillus casei that cleaved α-linked l-fucose from natural FUS (7). AlfB and AlfC showed high specific activity on fucosyl-α-1,3-N-acetylglucosamine (Fuc-α-1,3-GlcNAc) and fucosyl-α-1,6-N-acetylglucosamine (Fuc-α-1,6-GlcNAc), respectively. These substrate specificities led us in the present work to explore the likely use of L. casei α-l-fucosidases to synthesize those FUS. AlfB and AlfC were expressed in Escherichia coli as 6×(His)-tagged proteins and purified to near homogeneity as previously described (7). Transfucosylation activity of 6×(His)AlfB and 6×(His)AlfC was analyzed in 100 mM sodium phosphate buffer at pH 7.0. The reactions mixtures contained 50 mM p-nitrophenyl-α-l-fucopyranoside (pNP-fuc) as the donor and 200 mM N-acetylglucosamine (GlcNAc) as the acceptor. The mixtures were heated at 100°C to overcome the poor solubility of pNP-fuc and then cooled to the reaction temperature of 42°C. The reactions were started by the addition of 50 U/ml AlfB or 100 U/ml AlfC (1 U was defined as the amount of enzyme able to release 1 μmol of pNP in 1 h). Double the amount of AlfC compared to AlfB was needed to achieve the maximum disaccharide synthesis, due to the lower stability for AlfC, which is completely inactivated after 60 min at 42°C (Fig. 1). Samples (10 μl) were heated at 100°C for 3 min to stop the reaction, dried, and dissolved in 500 μl of D2O. The reaction products were analyzed by nuclear magnetic resonance (NMR) spectroscopy. NMR spectra were recorded at 313 K using a Bruker Avance Ultrashield Plus 600 spectrometer equipped with a 5-mm TCI cryoprobe. Chemical shifts were referenced using trimethylsilyl propionate (TSP) as an internal reference. One-dimensional (1D) 1H and 1H, 13C heteronuclear single quantum coherence–heteronuclear multiple-bond correlation (HSQC/HMBC) experiments were acquired for the reaction mixtures and for each of the substrates and products. Typical parameters for the 2D experiments were as follows: for HSQC, 256 and 2,048 points in F1 and F2, respectively (48 transients each); and for HMBC, 512 and 2,048 points in F1 and F2, respectively (64 transients each). NMR spectra were processed using the program Topspin 1.3 (Bruker GmbH, Karlsruhe, Germany). The structural analysis of the reaction products showed that the enzymes AlfB (Fig. 2) and AlfC (Fig. 3) generate by their transglycosylation activity the disaccharides Fuc-α-1,3-GlcNAc and Fuc-α-1,6-GlcNAc, respectively. No byproducts other than the hydrolyzed fucose and pNP were detected in the reactions. This is the first reported example of synthesis of Fuc-α-1,6-GlcNAc by transglycosylation. The synthesis of Fuc-α-1,3-GlcNAc has been previously described by using an α-l-fucosidase partially purified from a culture broth of Penicillium multicolor (17). However, the substrate specificity demonstrated for this enzyme suggests that a mixing of transient disaccharides occurred in the transfucosylation reaction. Both disaccharides synthesized here are important molecules that form part of glycoconjugates present at mucosal surfaces. Fuc-α-1,3-GlcNAc forms part of the Lewis X antigen core, which is found in many human glycoproteins at mucosal surfaces (21) and also forms part of human milk oligosaccharides (22). Fuc-α-1,6-GlcNAc is part of the core sugar in protein N-glycosylation also at mucosal surfaces (23).
Fig 1.
Stability of the α-l-fucosidases AlfB (●) and AlfC (○). The enzymes (10 μg/ml) were incubated in 100 mM sodium phosphate buffer at pH 7.0 and 42°C, and the residual activities were determined at different time intervals with 2 mM p-nitrophenyl α-l-fucopyranoside as the substrate. The initial activity was defined as 100%. The data presented are mean values from three replicate experiments. The errors bars represent the standard deviations.
Fig 2.
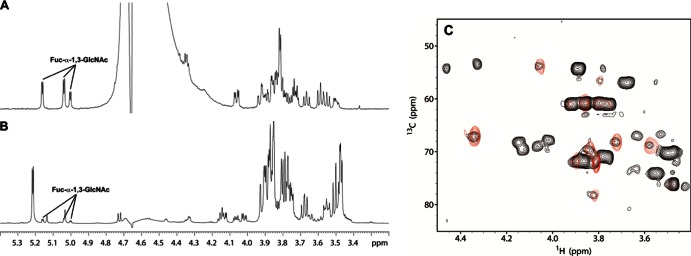
(A and B) Partial 1D 1H NMR spectra (600 MHz, 313 K) covering the anomeric region of commercial fucosyl-α-1,3-N-acetylglucosamine (Fuc-α-1,3-GlcNAc) (A) and the transfucosylation reaction with AlfB (B). (C) 1H, 13C-HSQC of Fuc-α-1,3-GlcNAc (red) and the reaction mixture (black).
Fig 3.
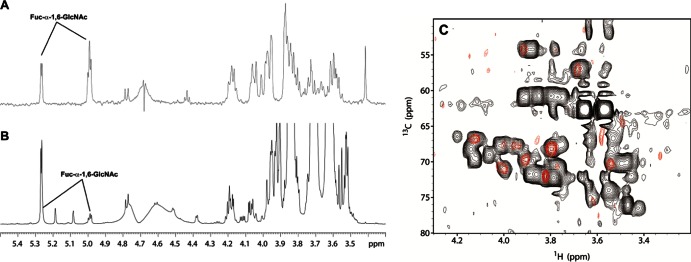
(A and B) Partial 1D 1H NMR spectra (600 MHz, 313 K) covering the anomeric region of commercial fucosyl-α-1,6-N-acetylglucosamine (Fuc-α-1,6-GlcNAc) (A) and the transfucosylation reaction with AlfC (B). (C) 1H, 13C-HSQC of Fuc-α-1,6-GlcNAc (red) and the reaction mixture (black).
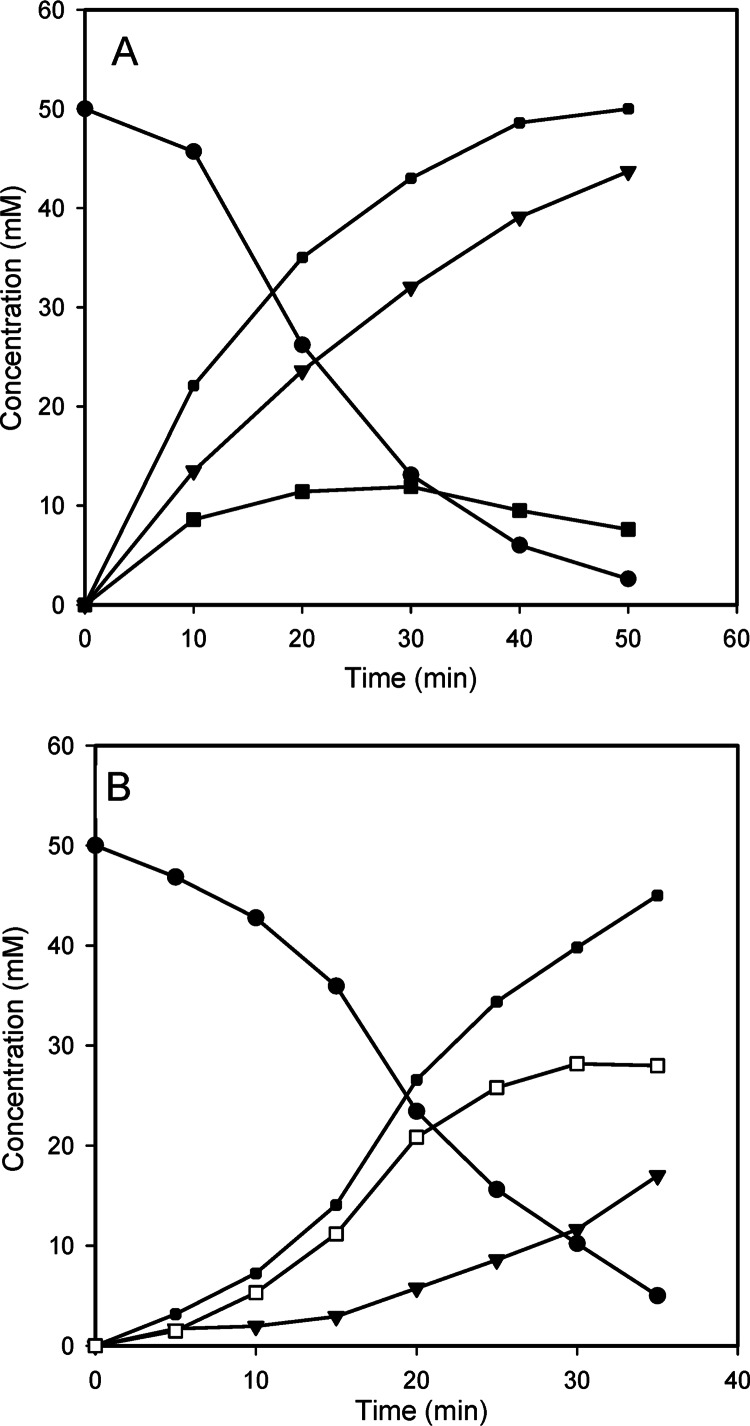
In order to analyze the transfucosylation kinetics of the AlfB and AlfC enzymes, a time course analysis was performed. 1D 1H spectra were collected for several reaction time points, with 64 transients, 32 K points, and a recovery time of 5 s, using water presaturation to suppress the residual HOD signal. Compound yields at each time point were determined from the integration of characteristic nonoverlapped proton signals for each component present in the reaction mixture. The total integral was normalized and the concentration of each molecule calculated using as a reference the added TSP (100 μM). The transglycosylation/hydrolysis ratio was 7.7-fold higher for AlfC than for AlfB (Table 1). This resulted in maximum transient yields of approximately 23% for AlfB and 56% for AlfC with respect to the added pNP-fuc (Fig. 4). The efficiency of transfucosylation is largely dependent on the type of enzyme. Thus, α-l-fucosidases from some bifidobacteria are completely devoid of transfucosylation activity unless they are mutated at specific amino acid sites (19). The reaction efficiency of AlfB was similar to the efficiencies of other α-l-fucosidases found in the literature, which ranged from 3% to 30% (18, 20, 24). Interestingly, the 56% AlfC yield is the maximum percentage described for a wild-type α-l-fucosidase, showing that this enzyme has very high transfucosylation activity which resembles that of engineered α-l-fucosidases; i.e., yields of pNP-fucosyl-α-1,2-galactose that ranged from 28% to 65% compared to 7% with wild-type enzyme have been obtained with mutants of an α-l-fucosidase from Thermotoga maritima (18). We have recently reported that probiotic bacterium L. casei strain BL23 can use Fuc-α-1,3-GlcNAc as a carbon source and that AlfB is necessary for this. However, Fuc-α-1,6-GlcNAc cannot be metabolized by L. casei (25). This suggests that, in spite of the high specific activity of AlfC on Fuc-α-1,6-GlcNAc (13.6 μmol fucose liberated/min · mg protein, compared to 27.5 μmol fucose liberated from Fuc-α-1,3-GlcNAc/min · mg protein for AlfB [7]), this disaccharide is not the natural substrate of AlfC. This could explain the elevated capacity of AlfC to synthesize Fuc-α-1,6-GlcNAc.
Table 1.
Transglycosylation and hydrolysis activities of AlfB and AlfC
| Enzyme | Transglycosylation (μmol/min/mg protein) | Hydrolysis (μmol/min/mg protein) | Vtrans/Vhyda |
|---|---|---|---|
| AlfB | 0.95 | 1.50 | 0.63 |
| AlfC | 12.79 | 2.63 | 4.86 |
Tranglycolylation/hydrolysis (Vtrans/Vhyd) ratios were calculated from the initials rates of formation of transglycosylation products over the initial rates of hydrolysis.
Fig 4.
Time course of the substrate consumption and the product formation by transfucosylation catalyzed by α-l-fucosidases AlfB (A) and AlfC (B). ●, p-nitrophenyl-α-l-fucopyranoside; ◆, p-nitrophenol; ▼, l-fucose; ■, fucosyl-α-1,3-N-acetylglucosamine; □, fucosyl-α-1,6-N-acetylglucosamine. Data from a representative experiment are shown.
In this report, we have shown that the high transglycosylation activity of AlfB and AlfC enables the enzymatic synthesis of two important fucosyldisaccharides. The use of glycosidases from probiotic species is within the rational strategies for efficient prebiotic oligosaccharide production, because all the machinery necessary for uptake and degradation is already present in these bacteria and also constitutes a tool for the synthesis of FUS to be used as antiadhesin ingredients.
ACKNOWLEDGMENTS
This work was financed by funds of the Spanish Ministry for Science and Innovation (MICINN)/FEDER through projects AGL2010-18696 and Consolider Fun-c-Food CSD2007-00063 and of the Valencian government through projects GV/2011/079 and ACOMP/2012/030. J.R.-D. was supported by a JAE-doc contract from CSIC/FSE.
Footnotes
Published ahead of print 29 March 2013
REFERENCES
- 1. Chessa D, Winter MG, Jakomin M, Baumler AJ. 2009. Salmonella enterica serotype Typhimurium Std fimbriae bind terminal α(1,2) fucose residues in the cecal mucosa. Mol. Microbiol. 71:864–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Morrow AL, Ruiz-Palacios GM, Jiang X, Newburg DS. 2005. Human-milk glycans that inhibit pathogen binding protect breast-feeding infants against infectious diarrhea. J. Nutr. 135:1304–1307 [DOI] [PubMed] [Google Scholar]
- 3. Newburg DS. 2009. Neonatal protection by an innate immune system of human milk consisting of oligosaccharides and glycans. J. Anim. Sci. 87:26–34 [DOI] [PubMed] [Google Scholar]
- 4. Ruiz-Palacios GM, Cervantes LE, Ramos P, Chavez-Munguia B, Newburg DS. 2003. Campylobacter jejuni binds intestinal H(O) antigen (Fucα1, 2Galβ1, 4GlcNAc), and fucosyloligosaccharides of human milk inhibit its binding and infection. J. Biol. Chem. 278:14112–14120 [DOI] [PubMed] [Google Scholar]
- 5. Ashida H, Miyake A, Kiyohara M, Wada J, Yoshida E, Kumagai H, Katayama T, Yamamoto K. 2009. Two distinct alpha-l-fucosidases from Bifidobacterium bifidum are essential for the utilization of fucosylated milk oligosaccharides and glycoconjugates. Glycobiology 19:1010–1017 [DOI] [PubMed] [Google Scholar]
- 6. Sela DA, Garrido D, Lerno L, Wu S, Tan K, Eom HJ, Joachimiak A, Lebrilla CB, Mills DA. 2012. Bifidobacterium longum subsp. infantis ATCC 15697 alpha-fucosidases are active on fucosylated human milk oligosaccharides. Appl. Environ. Microbiol. 78:795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rodríguez-Díaz J, Monedero V, Yebra MJ. 2011. Utilization of natural fucosylated oligosaccharides by three novel alpha-l-fucosidases from a probiotic Lactobacillus casei strain. Appl. Environ. Microbiol. 77:703–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yu ZT, Chen C, Kling DE, Liu B, McCoy JM, Merighi M, Heidtman M, Newburg DS. 2013. The principal fucosylated oligosaccharides of human milk exhibit prebiotic properties on cultured infant microbiota. Glycobiology 23:169–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kameyama A, Ishida H, Kiso M, Hasegawa A. 1991. Total synthesis of sialyl Lewis X. Carbohydr. Res. 209:c1–c4 [DOI] [PubMed] [Google Scholar]
- 10. Kretzschmar G, Stahl W. 1998. Large scale synthesis of linker-modified sialyl LewisX, LewisX and N-acetyllactosamine. Tetrahedron 54:6341–6358 [Google Scholar]
- 11. Albermann C, Piepersberg W, Wehmeier UF. 2001. Synthesis of the milk oligosaccharide 2′-fucosyllactose using recombinant bacterial enzymes. Carbohydr. Res. 334:97–103 [DOI] [PubMed] [Google Scholar]
- 12. Murray BW, Takayama S, Schultz J, Wong CH. 1996. Mechanism and specificity of human alpha-1,3-fucosyltransferase V. Biochemistry 35:11183–11195 [DOI] [PubMed] [Google Scholar]
- 13. Lee WH, Han NS, Park YC, Seo JH. 2009. Modulation of guanosine 5′-diphosphate-D-mannose metabolism in recombinant Escherichia coli for production of guanosine 5′-diphosphate-l-fucose. Bioresour. Technol. 100:6143–6148 [DOI] [PubMed] [Google Scholar]
- 14. Bojarová P, Kren V. 2009. Glycosidases: a key to tailored carbohydrates. Trends Biotechnol. 27:199–209 [DOI] [PubMed] [Google Scholar]
- 15. Shaikh FA, Withers SG. 2008. Teaching old enzymes new tricks: engineering and evolution of glycosidases and glycosyl transferases for improved glycoside synthesis. Biochem. Cell Biol. 86:169–177 [DOI] [PubMed] [Google Scholar]
- 16. Murata T, Morimoto S, Zeng X, Watanabe S, Usui T. 1999. Enzymatic synthesis of alpha-l-fucosyl-N-acetyllactosamines and 3′-O-alpha-l-fucosyllactose utilizing alpha-l-fucosidases. Carbohydr. Res. 320:192–199 [DOI] [PubMed] [Google Scholar]
- 17. Ajisaka K, Fujimoto H, Miyasato M. 1998. An alpha-l-fucosidase from Penicillium multicolor as a candidate enzyme for the synthesis of α(1→3)-linked fucosyl oligosaccharides by transglycosylation. Carbohydr. Res. 309:125–129 [DOI] [PubMed] [Google Scholar]
- 18. Osanjo G, Dion M, Drone J, Solleux C, Tran V, Rabiller C, Tellier C. 2007. Directed evolution of the alpha-l-fucosidase from Thermotoga maritima into an alpha-l-transfucosidase. Biochemistry 46:1022–1033 [DOI] [PubMed] [Google Scholar]
- 19. Sakurama H, Fushinobu S, Hidaka M, Yoshida E, Honda Y, Ashida H, Kitaoka M, Kumagai H, Yamamoto K, Katayama T. 2012. 1,3-1,4-α-l-Fucosynthase that specifically introduces Lewis a/x antigens into type-1/2 chains. J. Biol. Chem. 287:16709–16719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Berteau O, Bielicki J, Kilonda A, Machy D, Anson DS, Kenne L. 2004. α-l-Fucosidases: exoglycosidases with unusual transglycosylation properties. Biochemistry 43:7881–7891 [DOI] [PubMed] [Google Scholar]
- 21. King MJ. 1994. Blood group antigens on human erythrocytes—distribution, structure and possible functions. Biochim. Biophys. Acta 1197:15–44 [DOI] [PubMed] [Google Scholar]
- 22. Kobata A. 2010. Structures and application of oligosaccharides in human milk. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 86:731–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moran AP, Gupta A, Joshi L. 2011. Sweet-talk: role of host glycosylation in bacterial pathogenesis of the gastrointestinal tract. Gut 60:1412–1425 [DOI] [PubMed] [Google Scholar]
- 24. Eneyskaya EV, Kulminskaya AA, Kalkkinen N, Nifantiev NE, Arbatskii NP, Saenko AI, Chepurnaya OV, Arutyunyan AV, Shabalin KA, Neustroev KN. 2001. An alpha-l-fucosidase from Thermus sp. with unusually broad specificity. Glycoconj. J. 18:827–834 [DOI] [PubMed] [Google Scholar]
- 25. Rodríguez-Díaz J, Rubio-del-Campo A, Yebra MJ. 2012. Lactobacillus casei ferments the N-acetylglucosamine moiety of fucosyl-α-1,3-N-acetylglucosamine and excretes l-fucose. Appl. Environ. Microbiol. 78:4613–4619 [DOI] [PMC free article] [PubMed] [Google Scholar]