Abstract
In their natural environments, moderately halophilic bacteria are confronted not only with high salinities but also with low oxygen tensions due to the high salinities. The growth of H. halophilus is strictly aerobic. To analyze the dependence of respiration on the NaCl concentration and oxygen availability of the medium, resting cell experiments were performed. The respiration rates were dependent on the NaCl concentration of the growth medium, as well as on the NaCl concentration of the assay buffer, indicating regulation on the transcriptional and the activity level. Respiration was accompanied by the generation of an electrochemical proton potential (Δμ̃H+) across the cytoplasmic membrane whose magnitude was dependent on the external pH. Genes encoding proteins involved in respiration and Δμ̃H+ generation, such as a noncoupled NADH dehydrogenase (NDH-2), complex II, and complex III, were identified in the genome. In addition, genes encoding five different terminal oxidases are present. Inhibitor profiling revealed the presence of NDH-2 and complex III, but the nature of the oxidases could not be resolved using this approach. Expression analysis demonstrated that all the different terminal oxidases were indeed expressed, but by far the most prominent was cta, encoding cytochrome caa3 oxidase. The expression of all of the different oxidase genes increased at high NaCl concentrations, and the transcript levels of cta and qox (encoding cytochrome aa3 oxidase) also increased at low oxygen concentrations. These data culminate in a model of the composition and variation of the respiratory chain of H. halophilus.
INTRODUCTION
Moderately halophilic bacteria require NaCl for growth and have the capability to grow over a wide range of NaCl concentrations, ranging from marine concentrations to near saturation, with similar growth rates. This demonstrates not only the mere NaCl requirement but an extraordinary capacity to adjust the metabolism to a wide range of different external salinities (1). In recent years, Halobacillus halophilus has become a model system to study the adaptation of Gram-positive moderately halophilic bacteria to the changing salinities in their environments. Its genome sequence has been determined, and a genetic system has been developed (2, 3). The most obvious challenge that a halophile is confronted with is to establish turgor pressure. H. halophilus accumulates molar concentrations of chloride, along with compatible solutes (4), in a highly regulated process: at intermediate NaCl concentrations, glutamine and glutamate are the major osmolytes, whereas proline becomes predominant at higher salinities (5). In addition to the salinity-dependent regulation of osmolyte synthesis, there is a second, growth phase-dependent regulation: the cellular proline content is reduced at the end of the exponential growth phase, while ectoine is synthesized instead (6). The regulatory events involved in maintaining a proper solute pool size are not well characterized, but chloride is required for glutamate and glutamine production (4).
Although turgor adjustment is an obvious challenge for moderate halophiles, the adaptation of other cellular processes is equally important. Genome-wide expression analyses in nonhalophilic bacteria, such as Escherichia coli (7) and Bacillus subtilis (8, 9), or the methanogenic archaeon Methanosarcina mazei (10), revealed upregulation of many different cellular functions, including protective pathways, such as solute transport and biosynthesis, import of phosphate, export of Na+, and upregulation of pathways for modification of DNA and cell surface architecture. In its natural environment, the salt marshes, exposure to sunlight creates an additional stress for H. halophilus that is combated by carotenoids (11). Exposure to sunlight also leads to evaporation of water, which has two important consequences: first, the salinity increases, and second, since the solubility of gases is dependent on the salinity, the consequence is a reduced availability of oxygen at high salinities. The concentration of dissolved oxygen at 3 M NaCl is reduced by 62% compared to the concentration at 0.5 M NaCl at 20°C (12, 13), and the reduction is even greater at higher temperatures. Despite the obvious correlations between salinity and oxygen availability, little is known about how these parameters affect respiration and bioenergetics in moderate halophiles. Therefore, we have studied the bioenergetics of H. halophilus, the composition of its respiratory chain, and its regulation.
MATERIALS AND METHODS
Organism and cultivation.
H. halophilus (DSMZ 2266T) was routinely grown in nutrient broth (NB-Mg2+ medium) containing 0.03 M magnesium sulfate. The final concentration of NaCl varied depending on the experiment (values are given in the text). The pH was adjusted to 7.8 with NaOH. H. halophilus was cultivated aerobically and shaken on a rotary shaker at 30°C. Growth was monitored by determining the optical density of the cultures at 578 nm (OD578). In addition to the NB-Mg2+ medium, glucose minimal medium (G10 medium) was used for anaerobic conditions. This medium contained 1 M NaCl, 50 mM glucose, 37 mM NH4Cl, 36 μM FeSO4, 100 mM Tris base, 3 mM K2HPO4, yeast extract (0.1 g · liter−1), DSM 141 vitamin solution (1 ml · liter−1), and DSM 79 artificial seawater (250 ml · liter−1). For anaerobic cultivation, both media were gassed with N2-CO2 (80:20, vol/vol). The pH of the G10 medium was adjusted to 7.8 with H2SO4. The growth conditions were strictly anaerobic under N2-CO2 (80:20, vol/vol) in 16-ml Hungate tubes (Ochs, Bovenden, Germany) containing 5 ml medium. Growth was monitored by determining the OD578 of the cultures using a Genesys 10 photometer (Spectronic Instruments, USA) designed for Hungate tubes. All data points given reflect the means of triplicate tubes of one experiment that was performed at least two times. Trimethylamine-N-oxide (TMAO), dimethyl sulfoxide (DMSO), or NO3− was used as the potential electron acceptor, with glucose as the carbon and energy source. Potassium acetate and succinate were used as electron donors, with Na2SO4 or Fe(NO3)3 as the electron acceptor. Unless otherwise noted, every electron acceptor and donor was used in a concentration of 50 mM.
Preparation of cell suspensions.
For preparation of cell suspensions, NB-Mg2+ medium with 1.0, 2.0, or 3.0 M NaCl was inoculated (5%) with exponentially growing cells that had been pregrown at the same salinity. The cells were harvested in the mid-exponential growth phase (OD578 of 0.5 to 0.7) by centrifugation (25 min at 12,800 × g in an Avanti JA-10 rotor; Beckmann Coulter, USA) and washed once with 0.05 M Tris buffer (pH 7.8) containing 0.05 M MgSO4 and NaCl as used for growth. The cell pellet was resuspended in the same buffer to an OD578 of 50 and stored on ice until use. The protein concentration of the cell suspension was determined according to the Bradford method (14), with bovine serum albumin as the standard.
For preparation of the concentrated cell suspensions used to determine the bioenergetic parameters, NB-Mg2+ was inoculated (1%) from cultures maintained in the same medium. Fresh cell suspensions were prepared for each experiment. Cells in the late logarithmic growth phase were harvested by centrifugation and washed once with 0.05 M Tris-HCl buffer, pH 7.5, containing 0.66 M Na2SO4 and 0.05 M MgSO4. The cell pellet was resuspended in the same buffer to a concentration of 12 to 18 mg protein · ml−1 and stored on ice until use. The protein concentration of the cell suspension was determined according to the Bradford method (14), with bovine serum albumin as a standard.
Determination of the ATP content.
The ATP contents of cell suspensions were determined using a luciferin-luciferase assay (15). Aliquots (0.5 ml) of the concentrated cell suspension were added to 9.5-ml volumes of 0.05 M Tris-HCl buffer, pH 7.5, containing 0.05 M MgSO4 and 1 M NaCl. The cell suspensions were shaken at room temperature. At specific time points, 0.5-ml samples were taken and directly transferred into Eppendorf tubes containing 0.2 ml of ice-cold 3 M perchloric acid. After incubation of the samples on ice for 2 h, the pH was increased to 7.4 by the addition of 30 μl saturated K2CO3 solution and 0.2 ml 0.4 M TES [N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid]-NaOH buffer, pH 7.4. The KClO4 formed was removed by centrifugation. The supernatants were kept at 4°C until ATP determination. Amounts of 10 μl of the supernatants were transferred into luma cuvettes (Celsis Lumac, Landgraaf, Netherlands) containing 250 μl of the assay buffer (20 mM glycylglycine, 5 mM sodium arsenate, and 4 mM MgSO4, pH 8.0). The reaction was started by the addition of 20 μl of a luciferin-luciferase preparation (Sigma, Deisenhofen, Germany). The cuvettes were rapidly mixed and introduced into a Biocounter (Celsis Lumac, Landgraaf, Netherlands). The amount of light emitted is proportional to the ATP content of the assay, which was determined using a calibration curve between 0 to 40 pmol ATP.
Measurements of Δψ and ΔpH.
The transmembrane electrical field (Δψ) was estimated from the distribution of radioactively labeled tetraphenylphosphonium ions ([3H]TPP+) (16, 17). Amounts of 0.5 ml of the concentrated cell suspensions were added to 9.5 ml of 0.05 M Tris-HCl buffer, pH 7.5, containing 0.05 M MgSO4, sodium salts as indicated, and 1 μCi of [3H]TPP+Br− (final concentration, 3.2 μM). At this concentration, TPP+ did not affect the respiration and viability of the cells (data not shown). At specific time points, 0.5-ml samples were withdrawn from the cell suspension and transferred to Eppendorf tubes containing 0.2 ml silicon oil (ρ = 1.065 to 1.095, depending on the salt concentration of the buffer used). The cells were separated from the buffer by centrifugation through silicon oil as described previously (17). An amount of 50 μl of the supernatant was transferred into a scintillation vial that contained 0.5 ml of 3 M NaOH. The remainder of the supernatant and the silicon oil was removed by suction. The tip of the Eppendorf tube with the pellet was cut off with a razor blade and transferred into a second scintillation vial containing 0.5 ml of 3 M NaOH. The vials were vigorously shaken, and after an incubation time of 12 h at 37°C, the cells were completely hydrolyzed. After the addition of 5 ml Rotiszint ecoplus (Roth, Karsruhe, Germany), the levels of radioactivity were determined in a liquid scintillation counter type 2100 TR (Packard, Dreieich, Germany).
Δψ was calculated from the distribution of [3H]TPP+ using the Nernst equation (17). The internal and total water spaces of H. halophilus under these conditions were determined earlier (18). The accumulation of [3H]TPP+ was corrected for unspecific binding to the cells. The radioactivity found in the pellet after treating cells with 4% butanol for 1 h at 37°C was defined as unspecific binding.
ΔpH was calculated from the distribution of the weak base methylamine, but at pH values lower than 7.0, [14C]benzoic acid was used as a marker. The treatment of the cells was exactly as described above for the measurement of Δψ except that, instead of [3H]TPP+, 10 μCi 3H2O and 1 μCi [14C]methylamine (final concentration, 1.9 μM) were added to the cell suspensions. The ΔpH was calculated as described previously (17). The transmembrane electrochemical proton gradient (Δμ̃H+) was calculated according to the equation Δμ̃H+ = Δψ − 59 × ΔpH.
Measurement of oxygen uptake.
An oxygen electrode (Rank Brothers, Bottisham, Cambridge, United Kingdom) connected to a chart recorder was used to measure oxygen uptake. Before the experiment was started, the air-saturated buffer (0.05 M Tris, pH 7.8, 0.05 M MgSO4, and NaCl as indicated below) was incubated for 5 min at 30°C. The reaction vessel contained 2.9 ml of the buffer and 100 μl of the cell suspension or the membrane suspension. The sample was stirred throughout the experiment using a small magnetic stirrer. Inhibitors like rotenone, HDQ [1-hydroxy-2-dodecyl-4(1H)quinolone], and antimycin A were dissolved in ethanol and used in different concentrations. Sodium azide, potassium cyanide, and zinc chloride were dissolved in water, also in different concentrations.
Isolation of cytoplasmic membrane.
Cell suspensions prepared as described above were supplemented with lysozyme (100 mg/ml, final concentration) for 15 min at 30°C. After the addition of 0.5 mM phenylmethylsulfonyl fluoride (PMSF), cells were disrupted in a French pressure cell. Cell debris was removed by centrifugation (30 min at 20,500 × g in an Avanti JA-25.50 rotor; Beckmann, USA), and the resulting cell extract was centrifuged at 170,000 × g and 4°C for 1.5 h (rotor 70 Ti, Optima L-100K centrifuge; Beckmann, USA). The pellet was homogenized in buffer (0.05 M Tris, pH 7.8, 0.05 M MgSO4, and NaCl as used for growth) and then centrifuged a second time at 170,000 × g and 4°C for 1.5 h. The washed membranes were resuspended in 1 ml of the same buffer to an average protein concentration of 25 to 40 mg/ml.
Real-time PCR analysis.
For real-time PCR analysis, H. halophilus cells were harvested in the early exponential growth phase (OD578 of 0.15 to 0.3). Isolation of RNA, cDNA synthesis, and qPCR were done as described before (19). Amplification of cbaB, qoxA, cydA, ctaC, and ythA fragments was achieved with primers RT_cbaB_fw, RT_cbaB_rev, RT_qoxA_fw, RT_qoxA_rev, RT_cydA_fw, RT_cydA_rev, RT_ctaC_fw, RT_ctaC_rev, RT_ythA_fw, and RT_ythA_rev. Data analysis was accomplished using the threshold cycle (2−ΔΔCT) method (20). Real-time PCR analysis was done with three independent physiological parallels to ensure statistical relevance.
Chemicals.
HDQ was kindly provided by W. Bohne (University of Göttingen), and antimycin A, rotenone, and sodium azide by B. Ludwig (University of Frankfurt). Potassium cyanide and zinc chloride were purchased from Merck (Darmstadt, Germany).
RESULTS
The metabolism of H. halophilus is strictly aerobic.
To address the question of whether H. halophilus can grow in the absence of oxygen by anaerobic respiration or fermentation, experiments were performed under strictly anaerobic conditions (21, 22). When cells were incubated anaerobically in NB-Mg2+ medium, the optical density doubled 2.5 times within the first 12 h, from 0.012 to 0.073. Although very poor, this increase in optical density was accompanied by a decrease in the pH of about 1.25 units, demonstrating a physiological activity in the absence of oxygen. The same was observed in G10 minimal medium with glucose as the carbon source, but in a time frame of 200 h (data not shown). Growth under fermentative conditions was not supported by proline, lysine, arginine, alanine, ornithine, glutamate, arabinose, lactose, glucuronic acid, pyruvate, glucose, or Casamino Acids as carbon sources or in the presence of alternative electron acceptors, such as TMAO, DMSO, nitrate, sulfate, or Fe3+, and glucose, acetate, or succinate as electron donors. In addition, no growth was observed after inoculating from a culture grown anaerobically, and thus, the low increase in optical density observed was assigned to endogenous assimilation. These data demonstrate that H. halophilus has a strictly aerobic metabolism.
H. halophilus has a respiratory chain that leads to the generation of an electrochemical proton gradient across the cytoplasmic membrane.
The growth of H. halophilus is dependent not only on Cl− but also on Na+ (18, 23). The latter may be involved in pH regulation, as observed in other halophiles (24), but could potentially also be used as a coupling ion for ATP synthesis. Respiration catalyzed by whole cells was independent of the external pH, tested from 6.0 to 9.0, although growth had a sharp pH optimum at 7.5. Respiration was not influenced by the anion used and was identical in buffer containing 1 M NaCl, NaNO3, NaBr, Na glutamate, Na gluconate, or Na2SO4, showing that the nature of the anion was not important for activity. Na+ could be replaced by K+, indicating that Na+ is not a coupling ion for a respiratory enzyme. In line with this argument is that a Na+-translocating Nqr is not encoded by the genome, the F1Fo ATP synthase subunit c does not have a signature for Na+ binding (2), and respiration is stimulated not by Na+ ionophores but by protonophores. These data are consistent with the hypothesis that respiration in H. halophilus leads to the generation of an electrochemical proton potential.
To determine the magnitude of the electrochemical proton potential, cells were grown at 1.0 M NaCl in NB-Mg2+ medium, harvested, and washed, and cell suspensions were prepared. An aliquot of the concentrated cell suspension was added to buffer with a composition as indicated in Table 1. When the external pH (pHe) was increased from 6.6 to 8.9, the internal pH (pHi) increased from 6.5 to 8.0 (Table 1). The ΔpH was inverse (acidic inside) at all external pH values and approached a value close to 1 at an external pH of 8.9. Therefore, like other alkalitolerant/alkaliphilic organisms, H. halophilus is able to maintain a relatively neutral internal pH over a wide range of external pH values. The ΔΨ was fairly constant at pH 6.6 to 8.9 and ranged between −190 and −230 mV. The intracellular ATP content was highest at pHe values optimal for growth and declined slightly at suboptimal pHe values. The bioenergetic parameters were identical in chloride- or sulfate-containing buffers.
Table 1.
ΔpH, Δψ, Δμ̃H+, and ATP content in cell suspensions of H. halophilus at different external pH valuesa
| pHe | Mean value ± SD for: |
||||
|---|---|---|---|---|---|
| pHi | ΔpH | Δψ (mV) | Δμ̃H+ (mV) | ATP (nmol/mg) | |
| 6.6 | 6.52 ± 0.07 | −0.08 ± 0.07 | −194 ± 9 | −190 ± 13 | 1.5 ± 0.1 |
| 7.4 | 7.24 ± 0.06 | −0.19 ± 0.06 | −201 ± 9 | −190 ± 13 | 3.8 ± 0.2 |
| 8.0 | 7.41 ± 0.12 | −0.59 ± 0.12 | −218 ± 4 | −183 ± 11 | 3.2 ± 0.3 |
| 8.5 | 7.86 ± 0.08 | −0.64 ± 0.08 | −220 ± 5 | −182 ± 10 | 2.8 ± 0.2 |
| 8.9 | 7.98 ± 0.02 | −0.96 ± 0.02 | −188 ± 12 | −133 ± 13 | 1.3 ± 0.3 |
Cell suspensions were incubated in 0.05 M Tris-HCl, pH 7.5, containing 0.05 M MgSO4 and 1 M NaCl. The bioenergetics parameters were determined as described in Materials and Methods. pHe, external pH; pHi, internal pH.
Respiration in H. halophilus is salinity dependent.
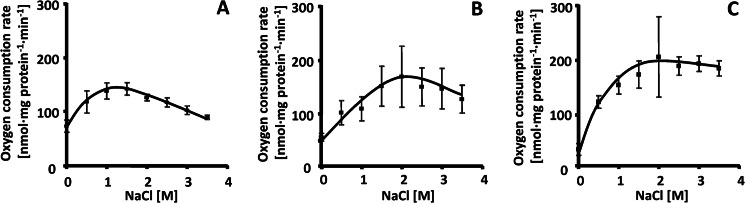
After we had determined a strictly aerobic metabolism of H. halophilus and its basic bioenergetic parameters, we analyzed the dependence of respiration on the salinity of the medium. This was the more interesting since the solubility of oxygen decreases with increasing salinity (12). To determine whether increasing salinities have an effect on the respiration of whole cells of H. halophilus, cells were grown in NB-Mg2+ medium in the presence of 1.0, 2.0, or 3.0 M NaCl to the exponential growth phase, harvested, and washed, and cell suspensions (OD578 of 50) were prepared. An aliquot of the concentrated cell suspension was added to buffer containing NaCl in concentrations ranging from 0 to 3.5 M, and the respiration rates were measured using an oxygen electrode. As can be seen from the data in in Figure 1, the respiration rates were clearly dependent on the salinity of the buffer. Different correlations were observed: first, maximal respiration rates were observed at the NaCl concentration used to grow the cells (cells grown at 3 M NaCl did not have a sharp maximum but a plateau at 2 to 3.5 M NaCl). Second, the higher the NaCl concentration of the growth medium, the lower was the activity at low salt. For example, cells grown at 1 M NaCl and incubated in the absence of NaCl still had ca. 50% of the maximal activity. This value decreased to 10% for cells grown at 3 M NaCl. Third, the higher the NaCl concentrations in the growth medium, the higher were the respiration rates. Taken together, these data demonstrate that H. halophilus adjusts its respiration depending on the salinity in its environment. These findings imply that H. halophilus may have alternative electron transport chains or alternative enzymes for certain respiratory complexes. Therefore, the genome of H. halophilus was searched for genes encoding electron transfer complexes.
Fig 1.
Effect of salinity on the respiration of H. halophilus. Cells were cultivated in NB-Mg2+ medium in the presence of 1.0 M NaCl (A), 2.0 M NaCl (B), or 3.0 M NaCl (C) to an OD578 of about 0.4. After harvesting and washing, the cells were directly resuspended in air-saturated buffer (50 mM Tris, 50 mM MgSO4, pH 7.8) containing NaCl concentrations as indicated. One hundred microliters of the cell suspension was incubated in 2.9 ml of the air-saturated buffer that also contained 50 mM glucose, and the oxygen consumption was measured with a Clark oxygen electrode. Error bars show standard deviations.
Genes encoding respiratory complexes.
Inspection of the genome sequence (2) for known respiratory enzyme complexes revealed that, apparently, a complex I is not encoded in the genome of H. halophilus. Instead, genes encoding a noncoupled NADH dehydrogenase, NDH-2 (Hbhal_3901), are present. The three subunits of complex II are apparently encoded by the genes sdhC, sdhA, and sdhB (Hbhal_3710 to 3712). From there, the electrons are probably channeled to complex III, a cytochrome bc1 complex encoded by the qcrABC (Hbhal_3267 to Hbhal_3269) cluster of H. halophilus. The deduced proteins are very similar to the corresponding subunits from B. subtilis and have the same predicted cofactors. Interestingly, the genome of H. halophilus has the inventory to potentially encode five different cytochrome oxidases. These were identified based on sequence comparisons (E values < e−30) and cofactor content. Two are cytochrome c oxidases. ctaABCDEFG (Hbhal_4053 and Hbhal_2834 to Hbhal_2839) encode a cytochrome caa3 oxidase, whereas cbaA (Hbhal_4958) and cbaB (Hbhal_4959) may encode a cytochrome c oxidase of the b(o/a3) type. Three potential quinol oxidases are encoded by qoxABCD (Hbhal_3694 to Hbhal_3697), cydAB (Hbhal_2799 to Hbhal_2800), and ythAB (Hbhal_1233 to Hbhal_1234) of H. halophilus.
Inhibitor profiling of respiratory enzyme complexes.
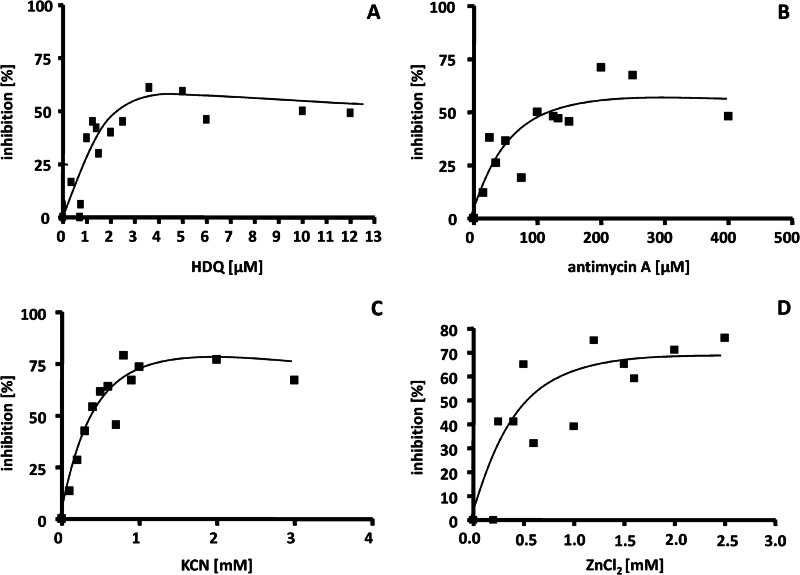
To verify the presence of the different respiratory complexes in H. halophilus as proposed by the genome sequence, membranes were prepared and electron transfer from NADH to oxygen was measured using the oxygen electrode. Different inhibitors were used to screen for the presence of the different complexes in the respiratory chain of H. halophilus. NDH-2 carries out the same redox reaction as complex I (NDH-1) but is insensitive to rotenone, an inhibitor of complex I, and sensitive to HDQ, a high-affinity inhibitor for membrane-bound NDH-2 (25). As expected, respiration as catalyzed by washed membranes of H. halophilus was insensitive to rotenone (up to 250 μM) but inhibited by HDQ with a 50% inhibitory concentration (IC50) of 1.6 μM (Fig. 2A). Antimycin, an inhibitor of complex III, inhibited respiration of H. halophilus with an IC50 of 59 μM (Fig. 2B). Specific inhibitors for the alternative oxidases are not known, but cyanide and azide are typical inhibitors for cytochrome c oxidases. KCN also inhibited the respiration of H. halophilus (Fig. 2C), with maximal inhibition reached at about 1.5 mM. Azide was also a potent inhibitor of respiration; maximal inhibition was observed at about 75 mM (data not shown). The bd oxidase of Geobacillus stearothermophilus was inhibited by ZnCl2 (26). ZnCl2 also inhibited respiration in H. halophilus membranes, with 50% inhibition at 0.55 mM (Fig. 2D). Finally, these data demonstrate the presence of NDH-2 and complex III but do not allow discrimination between the different terminal oxidases.
Fig 2.
Inhibition of NADH-dependent respiration. H. halophilus was grown in NB-Mg2+medium in the presence of 1.0 M NaCl to the exponential growth phase (OD578 of about 0.6 to 0.8). After harvesting and washing the cells, membranes were isolated. One hundred microliters of the membranes (protein concentration, 32 mg/ml) was incubated in 2.9 ml buffer (50 mM Tris, 50 mM MgSO4, pH 7.8) containing HDQ (A), antimycin A (B), KCN (C), or ZnCl2 in the concentrations indicated.
Expression analyses of alternative oxidase genes.
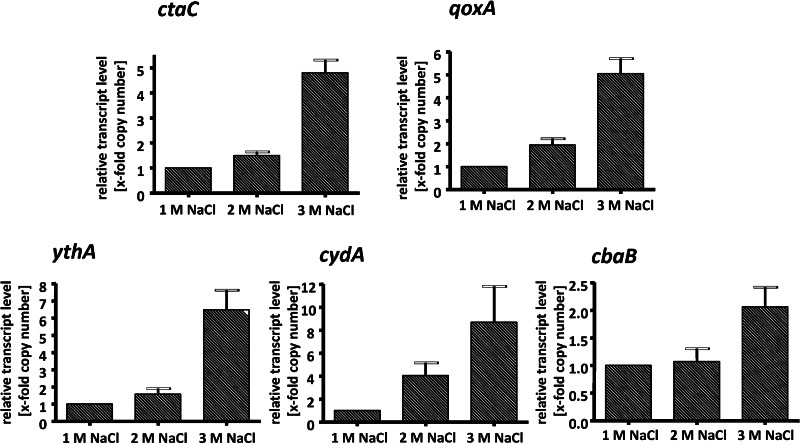
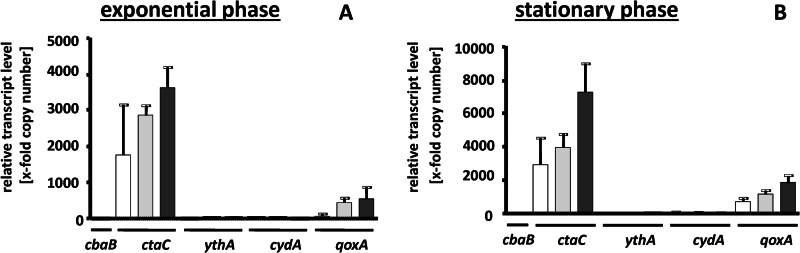
Apparently, H. halophilus has several genes encoding different oxidases that could not be differentiated by inhibitor profiling. To analyze whether the identified genes are expressed, cells were grown in NB-Mg2+ medium in the presence of 1.0, 2.0, or 3.0 M NaCl to the exponential growth phase. RNA was isolated and transcribed into cDNA as described previously (19), and the cDNA was then used for quantitative real-time PCR analyses with primers detecting one representative gene for each alternative oxidase. The results depicted in Figure 3 show that all genes are expressed, but the transcription level for every gene shows a strong dependence on the salinity of the medium. In every case, the highest transcript levels were observed at 3 M NaCl. The multiplication factor of induction was lowest for cbaB (2-fold) and highest for cydA (9-fold). Next, we analyzed the transcript levels of the different oxidases relative to each other. Interestingly, the transcript levels for ythA, cbaB, and cydA were up to 4,000-fold lower than the level of ctaC and 2,000-fold lower than the level of qoxA. Compared to ctaB as the standard, the ctaC levels were 8,000-fold higher and the qoxA levels 2,000-fold higher. This was independent of the growth phase and the salinity used (Fig. 4). Again, the expression levels were salinity dependent and highest at 3 M NaCl but not dependent on the growth phase. These data indicate that cytochrome caa3 oxidase is the predominant oxidase under the conditions tested.
Fig 3.
Effect of salinity on the expression of different terminal oxidase genes. H. halophilus was grown in NB-Mg2+medium in the presence of 1.0, 2.0, and 3.0 M NaCl to the exponential growth phase. RNA was isolated and transcribed into cDNA as described previously (19). The cDNA was then used for quantitative real-time PCR analyses. Real-time PCR analysis was done with three independent physiological parallels to ensure statistical relevance. The x-fold copy numbers were determined using the value of the cbaB sample as the reference. Error bars show standard deviations.
Fig 4.
Effect of salinity on the expression of different terminal oxidase genes. H. halophilus was grown in NB-Mg2+ medium in the presence of 1.0 (white bars), 2.0 (light gray bars), and 3.0 M NaCl (dark gray bars) to the exponential (A) or stationary (B) growth phase. RNA was isolated and transcribed into cDNA as described previously (19). The cDNA was then used for quantitative real-time PCR analyses. Real-time PCR analysis was done with three independent physiological parallels to ensure statistical relevance. The x-fold copy numbers were determined using the value of the cbaB sample as the reference. Error bars show standard deviations.
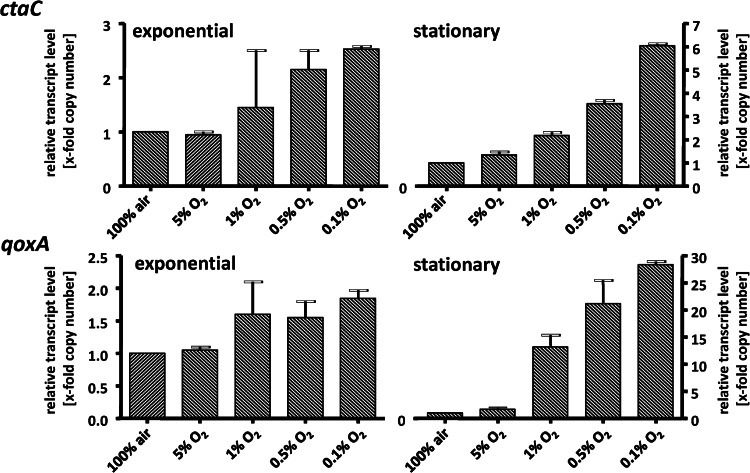
When cells were grown at different oxygen concentrations, the same overall expression pattern was observed: the transcript levels for ctaC and qoxA were highest. However, the transcript levels of both genes were oxygen regulated, with only a small effect in exponential-phase cells but a much more pronounced effect in stationary phase (Fig. 5). The ctaC and qoxA levels increased with decreasing oxygen concentration, up to 6-fold for ctaC and even 28-fold for qoxA. This pronounced effect in the stationary phase is also reflected in the relative ratio of ctaC to qoxA. Under air, ctaC dominated 550-fold over qoxA, but decreasing oxygen concentrations led to a decrease of the ctaC/qoxA ratio down to 0.3. These data demonstrate that the caa3 oxidase (cta) is the predominant oxidase under air and the aa3 oxidase (qox) dominates under oxygen limitation.
Fig 5.
Effects of growth phase and oxygen concentration on the expression of ctaC and qoxA. H. halophilus was cultivated in 1.2-liter flasks in NB-Mg2+ medium in the presence of 1.0 M NaCl and different oxygen concentrations ranging from 0.1% to 5.0%. Cells were harvested in the exponential and stationary growth phases. RNA was isolated and transcribed into cDNA as described previously (19). The cDNA was then used for quantitative real-time PCR analyses. Real-time PCR analysis was done with three independent physiological parallels to ensure statistical relevance. The x-fold copy numbers were determined using the value of the ythA sample as the reference. Error bars show standard deviations.
DISCUSSION
The moderately halophilic bacterium H. halophilus has to cope with changes of pH in its environment. The capacity to tolerate slightly alkaline pH values is reflected by its capacity to regulate its internal pH, which was kept fairly constant. Under alkaline conditions, the ΔpH was increased (by ≈1 unit), whereas the Δψ was kept constant. Thus, the electrochemical proton potential (Δμ̃H+) dropped from −190 mV to −130 mV. The rotor subunit (subunit c) of the F1Fo ATP synthase of H. halophilus does not contain a Na+ binding motif, indicating that it uses H+ as the coupling ion. Furthermore, we did not obtain any indication that respiration is coupled to Na+ transport. Therefore, the respiratory chain is presumably H+ motive and the Δμ̃H+ determined must be still high enough to drive ATP synthesis under these conditions. It is also interesting to note that Cl− has no effect on the basic bioenergetic parameters in H. halophilus, although the growth of H. halophilus is strictly Cl− dependent and a Cl− modulon has been described that regulates a number of different cellular processes (4).
At increasing salinities, the cell requires more energy to combat salt stress by synthesizing or taking up solutes and extruding Na+ from the cytoplasm. This is consistent with our finding that respiration increases with the salinity of the medium. Since increasing salinities are accompanied by decreasing oxygen concentrations, we looked for regulatory effects exerted by O2 availability. The expression of genes encoding terminal oxidases, cta and qox, was upregulated in the presence of low oxygen (and at stationary growth phase), indicating O2 stress under these conditions. However, since the metabolism of H. halophilus is strictly respiratory, the upregulation of these oxidases may just prolong survival at very low oxygen concentrations by enabling respiration at reduced oxygen tensions.
While to our knowledge, the effect of changing salinities on the expression of different oxidases has not been studied so far, several publications reported regulated oxidase gene expression in response to changing oxygen concentrations in other aerobic and facultative aerobic bacteria. The composition of the respiratory chain plays a decisive role during the adaptation of aerobic and facultative aerobic bacteria to environmental or developmental changes (27). In general, the last step of this pathway is characterized by the four-electron reduction of dioxygen to two water molecules, catalyzed by terminal oxidases (28). E. coli has two different terminal oxidases, cytochrome bo3 and cytochrome bd. The former is used under aerobic growth conditions, and the latter is induced under microaerobic conditions (29, 30). Furthermore, the obligately aerobic, nitrogen-fixing bacterium Azotobacterium vinelandii has a cytochrome bo3 oxidase and a cytochrome bd oxidase. The latter oxidase has a very high affinity to oxygen and, therefore, protects the oxygen-labile nitrogenase by keeping the oxygen level low (31). Moreover, B. subtilis synthesizes alternative electron transport chains with three or four terminal oxidases under aerobic growth conditions (32, 33). H. halophilus obviously uses a similar strategy to combat increasing salinity and the accompanying changes in O2 tension. The expression data indicate that cytochrome caa3 and aa3 oxidases are the predominant oxidases in H. halophilus, with caa3 levels being higher than aa3 levels. The same was observed for G. stearothermophilus (34), but the contrary was observed in B. subtilis (35). In Bacillus cereus, the caa3 and aa3 oxidases were differentially expressed in vegetative and sporulating cells. The caa3 oxidase was found in both types of cells but was 2-fold upregulated in sporulating cells. On the other hand, caa3 was not found in sporulating cells. This expression profile corresponds to the affinities of the oxidases to oxygen: the aa3 oxidase has a higher affinity than the caa3 for O2 (36). The same may be true for H. halophilus, since the relative ratios of ctaC/qoxA mRNA levels decreased with decreasing oxygen concentration and the qoxA mRNA became predominant.
To summarize, the regulatory chain of H. halophilus has a noncoupled NADH dehydrogenase, a complex II and a complex III, the two predominant alternative oxidases caa3 and aa3, and a proton-motive ATP synthase (Fig. 6). Both oxidases are upregulated in response to increasing salinities and decreasing oxygen concentrations, but the dominance of the low-affinity caa3 oxidase is displaced by the high-affinity aa3 oxidase under high-salt and oxygen limitation conditions.
Fig 6.
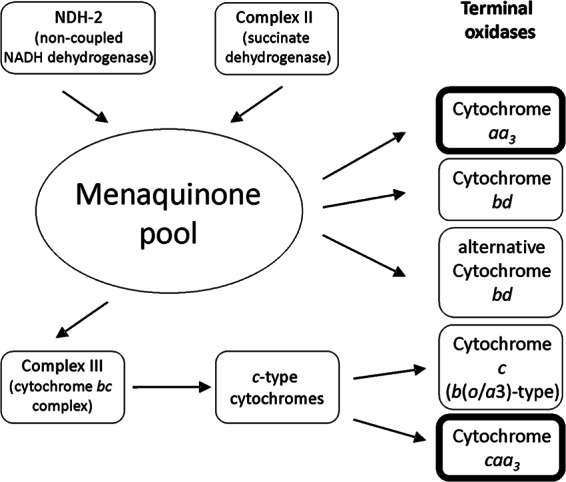
Composition of the respiratory chain of H. halophilus. The chain contains a noncoupled NDH-2 and complexes II and III converted by the menaquinone pool. Electrons are delivered to different oxidases. The two major oxidases, cytochrome aa3 (qoxABCD) and cytochrome caa3 (ctaA to -G), are highlighted. For the roles of the different oxidases, see the text.
ACKNOWLEDGMENTS
This work was supported by a grant from the Deutsche Forschungsgemeinschaft.
We are grateful to B. Ludwig, Frankfurt, Germany, for advice and stimulating discussions and to H. D. Osiewacz, Frankfurt, Germany, for the loan of the oxygen electrode.
Footnotes
Published ahead of print 12 April 2013
REFERENCES
- 1. Ventosa A, Nieto JJ, Oren A. 1998. Biology of moderately halophilic aerobic bacteria. Microbiol. Mol. Biol. Rev. 62:504–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Saum S, Pfeiffer F, Palm P, Rampp M, Schuster S, Müller V, Oesterhelt D. 14 May 2012. Chloride and organic osmolytes: a hybrid strategy to cope with elevated salinities by the moderately halophilic, chloride-dependent bacterium Halobacillus halophilus. Environ. Microbiol. [Epub ahead of print.] doi: 10.1111/j.1462-2920.2012.02770.x [DOI] [PubMed] [Google Scholar]
- 3. Köcher S, Averhoff B, Müller V. 2011. Development of a genetic system for the moderately halophilic bacterium Halobacillus halophilus: generation and characterization of mutants defect in the production of the compatible solute proline. Environ. Microbiol. 13:2122–2131 [DOI] [PubMed] [Google Scholar]
- 4. Saum SH, Müller V. 2008. Regulation of osmoadaptation in the moderate halophile Halobacillus halophilus: chloride, glutamate and switching osmolyte strategies. Saline Systems 4:4 doi: 10.1186/1746-1448-4-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Saum SH, Müller V. 2007. Salinity-dependent switching of osmolyte strategies in a moderately halophilic bacterium: glutamate induces proline biosynthesis in Halobacillus halophilus. J. Bacteriol. 189:6968–6975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Saum SH, Müller V. 2008. Growth phase-dependent switch in osmolyte strategy in a moderate halophile: ectoine is a minor osmolyte but major stationary phase solute in Halobacillus halophilus. Environ. Microbiol. 10:716–726 [DOI] [PubMed] [Google Scholar]
- 7. Weber A, Jung K. 2002. Profiling early osmostress-dependent gene expression in Escherichia coli using DNA macroarrays. J. Bacteriol. 184:5502–5507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hahne H, Mader U, Otto A, Bonn F, Steil L, Bremer E, Hecker M, Becher D. 2010. A comprehensive proteomics and transcriptomics analysis of Bacillus subtilis salt stress adaptation. J. Bacteriol. 192:870–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Steil L, Hoffmann T, Budde I, Völker U, Bremer E. 2003. Genome-wide transcriptional profiling analysis of adaptation of Bacillus subtilis to high salinity. J. Bacteriol. 185:6358–6370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pflüger K, Ehrenreich A, Salmon K, Gunsalus RP, Deppenmeier U, Gottschalk G, Müller V. 2007. Identification of genes involved in salt adaptation in the archaeon Methanosarcina mazei Gö1 using genome-wide gene expression profiling. FEMS Microbiol. Lett. 277:79–89 [DOI] [PubMed] [Google Scholar]
- 11. Köcher S, Breitenbach J, Müller V, Sandmann G. 2009. Structure, function and biosynthesis of carotenoids in the moderately halophilic bacterium Halobacillus halophilus. Arch. Microbiol. 191:95–104 [DOI] [PubMed] [Google Scholar]
- 12. Sherwood JE, Stagnitti F, Kokkinn MJ. 1991. Dissolved oxygen concentrations in hypersaline waters. Limnol. Oceanogr. 36:235–250 [Google Scholar]
- 13. Sherwood JE, Stagnitti F, Kokkinn MJ, Williams WD. 1992. A standard table for predicting equilibrium dissolved oxygen concentrations in salt lakes dominated by sodium chloride. Int. J. Salt Lake Res. 1:1–6 [Google Scholar]
- 14. Bradford MM. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 15. Kimmich GA, Randles J, Brand JS. 1975. Assay of picomole amounts of ATP, ADP and AMP using the luciferase enzyme system. Anal. Biochem. 69:187–206 [DOI] [PubMed] [Google Scholar]
- 16. Blaut M, Gottschalk G. 1984. Coupling of ATP synthesis and methane formation from methanol and molecular hydrogen in Methanosarcina barkeri. Eur. J. Biochem. 141:217–222 [DOI] [PubMed] [Google Scholar]
- 17. Rottenberg H. 1979. The measurement of membrane potential and ΔpH in cells, organelles and vesicles. Methods Enzymol. 55:547–569 [DOI] [PubMed] [Google Scholar]
- 18. Roeßler M, Müller V. 1998. Quantitative and physiological analyses of chloride dependence of growth of Halobacillus halophilus. Appl. Environ. Microbiol. 64:3813–3817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Saum SH, Sydow JF, Palm P, Pfeiffer F, Oesterhelt D, Müller V. 2006. Biochemical and molecular characterization of the biosynthesis of glutamine and glutamate, two major compatible solutes in the moderately halophilic bacterium Halobacillus halophilus. J. Bacteriol. 188:6808–6815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 21. Hungate R. 1969. A roll tube method for cultivation of strict anaerobes. Methods Microbiol. 3:117–131 [Google Scholar]
- 22. Bryant MP. 1972. Commentary on the Hungate technique for culture of anaerobic bacteria. Am. J. Clin. Nutr. 25:1324–1328 [DOI] [PubMed] [Google Scholar]
- 23. Claus D, Fahmy F, Rolf HJ, Tosunoglu N. 1983. Sporosarcina halophila sp. nov., an obligate, slightly halophilic bacterium from salt marsh soils. Syst. Appl. Microbiol. 4:496–506 [DOI] [PubMed] [Google Scholar]
- 24. Krulwich TA. 1983. Na+/H+ antiporters. Biochim. Biophys. Acta 726:245–264 [DOI] [PubMed] [Google Scholar]
- 25. Eschemann A, Galkin A, Oettmeier W, Brandt U, Kerscher S. 2005. HDQ (1-hydroxy-2-dodecyl-4(1H)quinolone), a high affinity inhibitor for mitochondrial alternative NADH dehydrogenase: evidence for a ping-pong mechanism. J. Biol. Chem. 280:3138–3142 [DOI] [PubMed] [Google Scholar]
- 26. Sakamoto J, Koga E, Mizuta T, Sato C, Noguchi S, Sone N. 1999. Gene structure and quinol oxidase activity of a cytochrome bd-type oxidase from Bacillus stearothermophilus. Biochim. Biophys. Acta 1411:147–158 [DOI] [PubMed] [Google Scholar]
- 27. Richardson DJ. 2000. Bacterial respiration: a flexible process for a changing environment. Microbiology 146:551–571 [DOI] [PubMed] [Google Scholar]
- 28. Anraku Y. 1988. Bacterial electron transport chains. Annu. Rev. Biochem. 57:101–132 [DOI] [PubMed] [Google Scholar]
- 29. Gennis RB, Stewart V. 1996. Respiration, p 217–286 In Neidhardt FC, Curtiss R, III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE. (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed, vol 1 ASM Press, Washington, DC [Google Scholar]
- 30. Unden G, Bongaerts J. 1997. Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim. Biophys. Acta 1320:217–234 [DOI] [PubMed] [Google Scholar]
- 31. Poole RK, Hill S. 1997. Respiratory protection of nitrogenase activity in Azotobacter vinelandii—roles of the terminal oxidases. Biosci. Rep. 17:303–317 [DOI] [PubMed] [Google Scholar]
- 32. Winstedt L, Yoshida K, Fujita Y, von Wachenfeldt C. 1998. Cytochrome bd biosynthesis in Bacillus subtilis: characterization of the cydABCD operon. J. Bacteriol. 180:6571–6580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. von Wachenfeldt C, Hederstedt L. 1992. Molecular biology of Bacillus subtilis cytochromes. FEMS Microbiol. Lett. 79:91–100 [DOI] [PubMed] [Google Scholar]
- 34. Sone N, Fujiwara Y. 1991. Effects of aeration during growth of Bacillus stearothermophilus on proton pumping activity and change of terminal oxidases. J. Biochem. 110:1016–1021 [DOI] [PubMed] [Google Scholar]
- 35. Winstedt L, von Wachenfeldt C. 2000. Terminal oxidases of Bacillus subtilis strain 168: one quinol oxidase, cytochrome aa3 or cytochrome bd, is required for aerobic growth. J. Bacteriol. 182:6557–6564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Garcia-Horsman JA, Barquera B, Escamilla JE. 1991. Two different aa3-type cytochromes can be purified from the bacterium Bacillus cereus. Eur. J. Biochem. 199:761–768 [DOI] [PubMed] [Google Scholar]