Abstract
Microbial cells often serve as an evolutionary battlefield for different types of mobile genetic elements, such as viruses and plasmids. Here, we describe the isolation and characterization of two new archaeal plasmids which share the host with the spindle-shaped Thermococcus prieurii virus 1 (TPV1). The two plasmids, pTP1 and pTP2, were isolated from the hyperthermophilic archaeon Thermococcus prieurii (phylum Euryarchaeota), a resident of a deep-sea hydrothermal vent located at the East Pacific Rise at 2,700-m depth (7°25′24 S, 107°47′66 W). pTP1 (3.1 kb) and pTP2 (2.0 kb) are among the smallest known plasmids of hyperthermophilic archaea, and both are predicted to replicate via the rolling-circle mechanism. The two plasmids and the virus TPV1 do not have a single gene in common and stably propagate in infected cells without any apparent antagonistic effect on each other. The compatibility of the three genetic elements and the high copy number of pTP1 and pTP2 plasmids (50 copies/cell) might be useful for developing new genetic tools for studying hyperthermophilic euryarchaea and their viruses.
INTRODUCTION
Mobile genetic elements (MGE), such as viruses, plasmids, and transposons, which collectively represent the mobilome, are omnipresent companions of cellular organisms from all three domains of life. It is becoming increasingly clear that universal suffering of cellular organisms is not the only outcome of the MGE-host interactions; instead, MGE embody a powerful engine driving the evolution, adaptation, and pathogenicity of their cellular hosts (1–3). Such patterns are perhaps most apparent in prokaryotes, bacteria and archaea, where MGE are very abundant and have been studied extensively. One of the least understood facets of the prokaryotic mobilome is the interaction between different types of MGE (e.g., viruses and plasmids) replicating within the same host (4). Comparative genomic analyses have revealed that there is a substantial overlap in the genomic content between different types of MGE (4–7), and even the transition from one type of element to another (e.g., from a plasmid to a virus and vice versa) appears to be permitted and likely has occurred on multiple occasions during the course of evolution (8–10). Furthermore, due to their abundance, various MGE are often forced to compete for the host cell (11), and this competition leads to coevolution of MGE not only with the host but also with the rival MGE as well.
Exploration of the archaeal mobilome has provided valuable insights into the nature and diversity of interactions between archaeal viruses and plasmids (4, 12–14). Sequences of more than 100 archaeal plasmids and 50 archaeal viruses have been obtained so far. Notably, distinction between these two types of MGE based on the genome sequence alone is not always straightforward (15–17); in some cases, entities originally considered to represent plasmids eventually have been demonstrated to actually represent temperate viruses (18). Studies on the mobilome of hyperthermophilic archaea belonging to the phylum Crenarchaeota revealed that interaction between different viruses and plasmids often extends beyond gene exchange; both neutral and antagonistic relationships have been observed. For example, different members of the pRN-like plasmid family (12) interact with the spindle-shaped Sulfolobus shibatae virus 1 (SSV1)-like fuselloviruses in radically different ways. Plasmid pXZ1 and virus SSV4 can be stably propagated together without imposing any prominent negative effect on each other (19). In contrast, upon superinfection of plasmid-containing Sulfolobus cells with fusellovirus SSV2, plasmids pSSVi and pSSVx, but not pXZ1, hijack the virion morphogenesis machinery of the virus and are encapsidated into virus-like particles (20, 21). This parasitic strategy ensures that the plasmids are disseminated throughout the cellular population in a virus-like fashion. A more aggressive interaction with a resident plasmid is displayed by the filamentous lipothrixvirus Acidianus filamentous virus 1 (AFV1) (22). The infection of the Acidianus host with AFV1 leads to the exclusion of the episomal form of the conjugative plasmid pAH1, which is otherwise stably maintained in the host cell population. More generally, it has been reported that screening of over 300 Sulfolobus isolates for the presence of extrachromosomal elements did not reveal a single isolate that contained both a virus and a conjugative plasmid (22), suggesting that such virus-plasmid incompatibility is a widespread phenomenon in at least some hyperthermophilic archaea.
Much less is known about virus-plasmid interactions in hyperthermophiles from the other major phylum of Archaea, the Euryarchaeota. In this phylum, plasmids have been isolated from hyperthermophilic organisms belonging to three different orders, the Methanococcales (genus Methanocaldococcus) (23), Archaeoglobales (24), and Thermococcales (25). However, viruses have been isolated only from members of the latter order (26, 27). We have previously observed a close genetic relationship between a group of Thermococcales plasmids (7, 28) and the spindle-shaped Pyrococcus abysii virus 1 (PAV1) (26, 29), with more than half of the viral genome being composed of genes that have homologues in plasmids (7). Consequently, it was proposed that PAV1-like viruses emerged as a result of recombination between a plasmid and a virus, which donated genetic determinants for genome propagation and virion formation. Indeed, the second recently isolated virus infecting Thermococcales, Thermococcus prieurii virus 1 (TPV1) (27), shares with PAV1 only two genes encoding putative structural proteins, but it does not carry the ones common to PAV1 and the plasmids.
TPV1 was originally isolated from T. prieurii (27), a euryarchaeon residing in deep-sea hydrothermal vents of the East Pacific Rise (30), but was subsequently shown to infect a number of other Thermococcus species (31). Like PAV1, TPV1 possesses a spindle-shaped virion and contains a circular double-stranded DNA genome (27). The virus is not lytic, and its production is stimulated by UV irradiation. Interestingly, besides the TPV1 genome, two additional extrachromosomal elements were observed in infected T. prieurii cells (27). However, the nature of these elements remained unclear. To better understand the interplay between viruses and plasmids of hyperthermophilic archaea, we set out to characterize the two MGE sharing the host with the virus TPV1. Here, we report on their isolation and sequence analysis.
MATERIALS AND METHODS
Strains and growth conditions.
Thermococcus prieurii strain Bio-pl-0405IT2 (JCM16307T; Japan Collection of Microorganisms designation) was isolated from a hydrothermal chimney sample collected from the East Pacific Rise at 2,700-m depth at the Sarah Spring area (7°25′24 S, 107°47′66 W). Cells were cultured in Ravot medium as previously described (26, 27), with minor modifications as detailed below.
The medium contained, per liter of distilled water, the following: 1 g NH4Cl, 0.2 g MgCl2×6H2O, 0.1 g CaCl2×2H2O, 0.1 g KCl, 0.83 g CH3COONa×2H2O, 20 g NaCl, 5 g yeast extract, 5 g tryptone, 3.45 g piperazine-N,N′-bis (2-ethanesulfonic acid) (PIPES), and 0.001 g resazurin. The pH was adjusted to 7 before autoclaving. After autoclaving, 5 ml of 6% (wt/vol) K2HPO4 solution and 5 ml of 6% (wt/vol) KH2PO4 solution, separately sterilized, were added. The medium was dispensed (50 ml) into 100-ml sterile vials and completed by adding 1% (wt/vol) previously sterilized elemental sulfur. Anaerobiosis was obtained by applying vacuum to the medium and saturating it with N2. Finally, a sterile solution of Na2S×9H2O (final concentration, 0.05% [wt/vol]) was added to reduce the medium. The medium, inoculated to a final concentration of 1%, was incubated at 85°C.
DNA extraction and sequencing.
Total DNA from T. prieurii was prepared as previously described (26, 32). Covalently closed circular DNA was extracted from cells in exponential growth phase by the alkaline lysis method (26, 33). Plasmids designated here pTP1 and pTP2 were completely digested with restriction endonucleases HindIII and SmaI, respectively, and all of the fragments obtained were cloned in the corresponding sites of pUC18 to obtain an overlapping clone library of pTP1 and pTP2 genomes. Sequencing reactions were carried out with the BigDye Terminator kit (Applied Biosystems) and analyzed at the Platforme Biogenouest (Roscoff, France; http://www.sb-roscoff.fr/plateformes-techniques/genomique-sbr.html) on an ABI Prism 3100 genetic analyzer. Each insert was sequenced from both ends using the M13 forward and M13 reverse primers. The sequences were trimmed and assembled using the SeqMan Lasergene 8.0 program (DNASTAR, Inc., Madison, WI) with both strands completely sequenced and with a 3-fold coverage.
Sequence analysis and annotation.
Glimmer (34), GenMark (35), and RBS finder (36) were used to identify open reading frames (ORFs). The amino acid sequence of each ORF was searched against the NCBI nonredundant protein database (February 2013) using PSI-BLAST (37) and compared to the database of known protein structures using HHPred (38). Membrane-spanning regions were predicted using the TMHMM program (39). The Ka/Ks ratio, namely, the ratio of the number of nonsynonymous substitutions per nonsynonymous site (Ka) to the number of synonymous substitutions per synonymous site (Ks) (40), was estimated using a web-based Ka/Ks calculation tool at http://services.cbu.uib.no/tools/kaks.
Plasmid copy number determination.
Real-time PCR was used to determine the copy number of plasmid genomes in the host cell using an ABI Prism 7500 sequence detection system (Applied Biosystems). The method is based on the difference in threshold cycle values (ΔΔCT) of amplicons targeting the plasmids and an amplicon targeting the single-copy gene of 16S rRNA carried by the host chromosome and used as an internal standard. Total and plasmid DNAs were obtained as described previously (27). Primers were designed with Primer Express software, version 3.0. The following primer pairs were used for gene amplification: for the partial 16S rRNA gene, forward primer 5′-CGTGCGGTTTAATTGGATTCA-3′ and reverse primer 5′-ACCTTCAGGCTGGCCTTCA-3′; for pTP1 DNA, forward primer 5′-CCCGTAGCGAGTCGTCTGAT-3′ and reverse primer 5′-GCGCGCAGACAACACTATGA-3′; and for pTP2 DNA, forward primer 5′-CGAAAGATGGCAAGAACAAGGT-3′ and reverse primer 5′-TCACGGCGAACAAGAACGA-3′. In all three cases, the fragments were identically sized.
Total DNA was mixed with 12.5 μl SYBR green mix for quantitative PCR (qPCR), 100 nM forward primer, and 100 nM reverse primer. Samples were incubated at 95°C for 10 min, followed by 60 cycles of amplification (at 95°C for 15′ and at 60°C for 1 min), and then cooled to 4°C. A standard curve was obtained using 10-fold serial dilutions of DNA, ranging from 1 to 10−6. Samples were tested in triplicate. The PCR products amplified had identical melting points (about 86°C) and were analyzed by agarose gel electrophoresis.
ΔΔCT, used to calculate the relative plasmid copy number, is the difference in the mean CT (threshold cycle) value of the amplicon targeting single-copy 16S rRNA genes and the mean CT value of the amplicon(s) targeting the plasmid DNAs.
Nucleotide sequence accession numbers.
The complete sequences of pTP1 and pTP2 were deposited in GenBank under accession numbers KC617920 and KC617921, respectively.
RESULTS
Isolation and characterization of plasmids pTP1 and pTP2.
Agarose gel electrophoresis of the extrachromosomal DNA extracted from TPV1-infected T. prieurii cells revealed that in addition to the viral genome, two smaller elements were present (27). The latter were isolated and digested with several type II restriction endonucleases (REases), including HindIII, EcoRI, EcoRV, and SmaI. The analysis showed that the elements represent covalently closed circular DNA molecules of approximately 2 and 3 kb. The DNA was susceptible to digestion with both methylation-insensitive and methylation-sensitive (e.g., SmaI) REases.
The complete nucleotide sequences of both DNA molecules were subsequently determined. The sizes were found to be consistent with those defined using restriction analysis: the larger element, designated pTP1 (for T. prieurii plasmid 1), consists of 3,126 bp, while the smaller one, pTP2, is 2,038 bp long. Six-frame in silico translation of pTP1 and pTP2 sequences suggested that each plasmid contains 5 open reading frames (ORFs) (Fig. 1). The G+C content of pTP1 and pTP2 was found to be 42.5 and 41.7%, respectively. These value are considerably lower than those of TPV1 (49.9%) and the T. prieurii chromosome (53.6%) (27) but are similar to the G+C content of other small plasmids of Thermococcales, namely, pTN1 (3.6 kb; 45.5% G+C) from T. nautilus (41), pGT5 (3.4 kb; 43.5% G+C) from Pyrococcus abysii (42), and pRT1 (3.4 kb; 43% G+C) from Pyrococcus sp. strain JT1 (43). pTN1 and pGT5 encode homologous replication proteins (Reps) and replicate via a rolling-circle (RC) mechanism (41, 42). The predicted Rep protein of pRT1, on the other hand, does not display the canonical order of RC-Rep motifs (44) and the protein is not related to the RC-Reps of pTN1 and pGT5 (43, 45). The mechanism of pRT1 replication thus remains unresolved.
Fig 1.

Circular genome maps of the two plasmids isolated from Thermococcus prieurii. Predicted genes are depicted with arrows indicating the direction of transcription. The putative genes for the rolling-circle replication initiation proteins are shown in black.
pTP1 is a unique recombinant variant related to the pTN1-like rolling-circle plasmids.
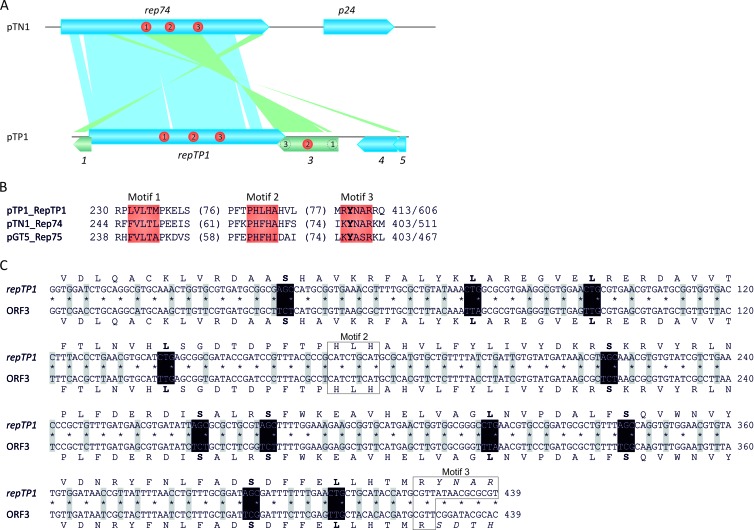
Three of the five predicted pTP1 ORFs (Table 1) display sequence similarity to the RC-Rep, Rep74, from plasmid pTN1 (Fig. 2A), but only one of them is predicted to encode a full-length Rep74-like protein, which we designate RepTP1. All three conserved motifs typical of RC-Rep proteins of the superfamily II (SFII; with one catalytic Tyr residue in motif 3) (44) are readily identifiable in RepTP1 (Fig. 2B). The two other ORFs, ORF1 and ORF3, match the central and 3′-distal regions, respectively, of the pTN1 rep74 (Fig. 2A and Table 1). The similarity between ORF3 and rep74 extends throughout the region encompassing the three signature motifs. However, there is a deletion of motif 1 in ORF3, while the catalytic Tyr in motif 3 is changed to a Ser residue. Interestingly, ORF1 and ORF3 are separated by a repTP1 gene and are in the opposite orientation with respect to repTP1. These observations suggest that ORF1 and ORF3 once constituted a single ORF encoding an RC-Rep of the ancestral pTP1. The gene was subsequently split in the course of recombination between two ancestral pTP1 plasmids, leading to integration of a new RC-Rep gene copy into the preexisting one (Fig. 2A). Notably, the inverse and nested orientation of the repTP1 gene suggests that the incoming rep was provided in trans rather than emerging in the course of duplication of the original gene copy.
Table 1.
Annotation of plasmid pTP1
| ORF | Position (strand) | Start/stop codon | Length (aa) | Function/feature | Homologue (GenBank accession no.) | Identity (%); E value |
|---|---|---|---|---|---|---|
| 1 | 5…178 (−) | CTG/TGA | 58 | RC-Rep fragment | Thermococcus nautilus plasmid pTN1, Rep74 (YP_001351689) | 15/37 (41%); 3e−03 |
| 2 (repTP1) | 158…1975 (+) | ATG/TAA | 606 | RC-Rep | Thermococcus nautilus plasmid pTN1, Rep74 (YP_001351689) | 172/488 (35%); 4e−81 |
| 3 | 1896…2468 (−) | ATG/TAA | 191 | RC-Rep fragment | Thermococcus nautilus plasmid pTN1, Rep74 (YP_001351689) | 64/179 (36%); 5e−22 |
| 4 | 2646…3008 (−) | GTG/TAG | 121 | TMDa (2×) | ||
| 5 | 2977…3090 (−) | GTG/TAA | 38 | TMD (1×) |
TMD, transmembrane domain. The numbers in parentheses denote the number of predicted TMDs.
Fig 2.
pTP1 and its relationship to the T. nautilus plasmid pTN1. (A) Alignment of the linearized genome maps of pTN1 (top) and pTP1 (bottom). Homologous regions of pTP1 and pTN1, which are transcribed in the same direction, are connected by blue shading. pTP1 ORFs (1 and 3) derived from the ancestral disrupted rolling-circle replication initiation (RC-Rep) gene are colored green and are connected to the homologous regions in pTN1 using green shading. Positions of the three conserved motifs (1 to 3) characteristic of RC-Rep proteins are indicated with red circles. Key residues of motifs 1 and 3 are mutated/absent in the gene product of ORF3, and the corresponding regions are indicated by empty circles. (B) Alignment of the three conserved motifs of superfamily II (SFII) RC-Reps encoded by plasmids of Thermococcales. The limits of the depicted motifs are indicated by the residue positions on each side of the alignment; the total lengths of the proteins are also provided. The numbers in parentheses indicate the distance between the motifs. The single catalytic Tyr residue, a distinguishing feature of SFII RC-Reps, is highlighted in boldface. (C) Detailed comparison of the homologous regions of repTP1 and ORF3 from plasmid pTP1. Nucleotides and corresponding protein sequences are shown for both genes. The third position of each codon is indicated by an asterisk. Gray shading highlights all of the changes in the third position of the corresponding codon in the two sequences. Codons (for Leu and Ser) that have been changed in the two other positions, while preserving the same amino acid sequence, are indicated with black shading. Two of the three conserved motifs of RC-Rep proteins that are present in the depicted region are boxed. Note that identity between RepTP1 and ORF abruptly stops at the second position of the codon for the catalytic Tyr residue in motif 3.
The scenario described above predicts that the incoming and the resident RC-Rep gene copies were identical. To verify this hypothesis, we compared the sequences of RepTP1 and ORF3. The two turned out to be identical at the protein level over the 142-amino-acid (aa) region (note that identity abruptly stops at the catalytic Tyr residue in motif 3; Fig. 2C). This observation argues against the possibility that the two Rep genes (fragmented and full length) are derived from heterologous plasmids. Strikingly, however, when the corresponding nucleotide sequences were compared, we found that there is only 75% identity (322/428) between them (Fig. 2C). Closer examination of the pairwise nucleotide sequence alignment revealed that all mutations are silent, with the vast majority of them occurring at the third position of the codons; 81 out of 142 codons had mutations in the third position. In addition, 11 codons (6 for Ser and 5 for Leu) also displayed changes in other positions (Fig. 2C). This observation is consistent with the fact that Ser and Leu represent two of the three amino acids (the third one is Arg) for which the genetic code is most degenerated: each can be encoded by six different codons. Since ORF3 (and ORF1) apparently represents a fragment of a disrupted Rep gene, we expected that it will be evolving under neutral (in the case of the inactivated gene) or positive (in the case of an interfering gene product) selection. Instead, the result presented above clearly indicates that ORF3 evolves under strong purifying selection. Indeed, the Ka/Ks ratio for the ORF3-RepTP1 gene pair was estimated to be 0.036 (Ka/Ks ratios below 1 indicate purifying selection [40]). Thus, despite the lack of motifs 1 and 3, the product of ORF3 is likely to be a functional player in pTP1 propagation, and biochemical studies are needed to reveal its exact role(s). As mentioned above, ORF1 shares sequence similarity with the 3′-distal region of rep74 from pTN1 but not with the equivalent region of repTP1. Consistent with this, the C-terminal regions of RepTP1 and Rep74 are not homologous. Thus, it appears that the original Rep gene of pTP1, which was subsequently disrupted by introduction of repTP1, was more similar to rep74 of pTN1 than the current repTP1 gene is.
pTP2 is unrelated to other plasmids of Thermococcales.
With 2,038 bp, pTP2 is the smallest known plasmid of hyperthermophilic archaea. Sequence analysis revealed that it is not related to pTP1 or, in fact, to any other plasmid of Thermococcales; not a single gene is shared between pTP2 and the other plasmids. Nevertheless, similar to pTP1, pTP2 is likely to replicate using the RC mechanism. ORF3 of pTP2 encodes a putative protein (RepTP2) of 252 aa which shares significant sequence similarity with RC-Reps encoded by diverse archaeal MGE (Table 2), including Archaeoglobus profundus plasmid pGS5 (24), putative Methanococcus voltae A3 provirus (MVV) (46), and a group of pleomorphic haloarchaeal viruses (47). Sequence analysis of RepTP2 showed that, unlike RepTP1, it possesses two conserved Tyr residues in motif 3 (Fig. 3) and should be considered a member of SFI of RC-Reps. RepTP2 is considerably shorter than its homologues in other archaeal MGE (252 aa versus 430 to 700 aa; Fig. 3). The size difference is mainly due to the lack of an N-terminal domain (∼200 aa) in RepTP2. Notably, the N-terminal extensions in other RepTP2-like proteins are very divergent, to the point that N-terminal regions of Reps from haloarchaeal MGE do not share identifiable similarity with the corresponding sequences from methanococcal or methanosarcinal RC-Reps. We hypothesize that an ORF preceding and overlapping the repTP2 gene encodes a functional counterpart of the N-terminal domains observed in other RepTP2-like proteins. However, the validity of this prediction remains to be verified by biochemical studies of the pTP2 proteins.
Table 2.
Annotation of plasmid pTP2
| ORF | Position (strand) | Start/stop codon | Length (aa) | Function/featurea | Homologue(s) (GenBank accession no.) | Identity (%); E value |
|---|---|---|---|---|---|---|
| 1 | 38…418 (+) | ATG/TGA | 127 | Putative transcriptional regulator, wHTH fold | Distant similarity to wHTH proteins of the Rf2 family; HHpred hit to 1stz, probability of 89.4 | |
| 2 | 418…861 (+) | GTG/TGA | 148 | |||
| 3 (repTP2) | 791…1546 (+) | ATG/TGA | 252 | RC-Rep | Methanococcus voltae A3 (YP_003706702); Methanococcus voltae A3 provirus MVV (YP_003707078) | 97/232 (42); 4e−52; 79/264 (30); 8e−21 |
| Halorubrum pleomorphic virus 1 (YP_002791886) | 63/219 (29); 5e−11 | |||||
| 4 | 1580…1711 (−) | ATG/TGA | 44 | TMD (1×) | ||
| 5 | 1814…1960 (+) | ATG/TAG | 49 | SpoVT/AbrB-like transcriptional regulator; swapped-hairpin fold | Methylocystis sp. strain ATCC 49242 (ZP_08073517); HHpred hits to 1mvf (MazE), probability of 97.4, and1yfb (AbrB), probability of 93 | 13/41 (32); 0.32 |
TMD, transmembrane domain (the number in parentheses denotes the number of predicted TMDs); wHTH, winged helix turn helix.
Fig 3.
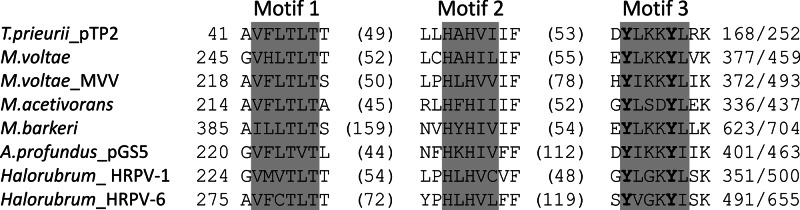
Alignment of the three conserved motifs of SFI rolling-circle replication proteins (RC-Reps) encoded by various archaeal MGE with those of RepTP2 from plasmid pTP2. The limits of the depicted motifs are indicated by the residue positions on each side of the alignment; the total lengths of the proteins are also provided. The numbers in parentheses indicate the distance between the motifs. The two catalytic Tyr residues, a distinguishing feature of SFI RC-Reps, are highlighted in boldface. GenBank accession numbers for the depicted proteins are the following: Methanococcus voltae A3, YP_003706642; M. voltae A3 provirus MVV, YP_003707078; Methanosarcina acetivorans C2A, NP_617505; Methanosarcina barkeri, YP_305744; Archaeoglobus profundus plasmid pGS5, YP_002995766; Halorubrum pleomorphic virus 1 (HRPV-1), YP_002791886; and HRPV-6, YP_005454285.
Besides repTP2, putative functions could be inferred for two other pTP2 ORFs (Table 2). HHpred analysis suggests that ORF1 and ORF5 encode DNA-binding proteins that possibly are involved in transcription regulation. The product of ORF1 is predicted to possess a winged helix-turn-helix motif, while the one of ORF5 belongs to a family of SpoVT/AbrB-like transcriptional regulators displaying the swapped-hairpin fold (Table 2). Notably, an SpoVT/AbrB-like protein is also encoded by TPV1 (27); however, it does not share significant sequence similarity with the pTP2 protein.
pTP1 and pTP2 stably propagate in TPV1-infected cells.
We have previously shown that in infected T. prieurii cells, the TPV1 genome is present at 20 copies per chromosome (27). Interestingly, qPCR analysis showed that pTP1 and pTP2 are considerably more abundant than TPV1 genome in infected cells; both plasmids were found to be present in 50 copies per chromosome. This number is similar to those reported for numerous bacterial RC plasmids (48) but is much higher than the copy number estimated for the pTN1-based Escherichia coli-T. kodakaraensis shuttle vector pLC70, which was found to be present in just 3 copies per chromosome (49).
To gain insights into the possible interaction between the two plasmids and TPV1, T. prieurii, with a doubling time of 23 min, was subjected to serial passages in liquid cultures (8 times, corresponding to approximately 187 generations), and the resultant culture was streaked on a plate and examined for the presence of the three extrachromosomal elements. Both plasmids and the virus TPV1 were stably maintained in T. prieurii cells during the course of the experiment, with all colony clones analyzed testing positive for the presence of all three MGE. However, unlike the case of crenarchaeal fuselloviruses, which can package and transfer resident plasmids in a virus-like fashion (20, 21), TPV1 apparently does not offer such service to pTP1 and pTP2. TPV1 virions produced by T. prieurii could infect T. kodakaraensis, T. barophilus, T. celer, and T. gorgonarius (27), but the interspecies plasmid transfer was not observed. Consistent with this, neither pTP1 nor pTP2 was observed in DNA preparations extracted directly from TPV1 viral particles (27).
DISCUSSION
In this study, we have characterized two cryptic plasmids of T. prieurii. Our results indicate that the two plasmids and the spindle-shaped virus TPV1 stably propagate within T. prieurii cells. Such stable coexistence is reminiscent of that described for certain pRN-like plasmids and fuselloviruses of Sulfolobus (19). Moreover, comparative genome analysis did not reveal a single gene that would be shared between either of the two plasmids and the virus. This is in contrast to the extensive gene content overlap between the medium-sized pTN2-like plasmids of Thermococcales and PAV1 (7), the only other virus known to infect Thermococcales (29, 31); however, none of these PAV1 plasmid genes was found in TPV1. Nevertheless, MGE other than pTN2-like plasmids appear to have contributed to shaping of the TPV1 genome (27).
Similar to many other medium-sized euryarchaeal viruses and plasmids (50), TPV1 encodes a replicative minichromosome maintenance (MCM) helicase and is likely to replicate via a theta mechanism (27). pTP1 and pTP2, on the other hand, apparently utilize the rolling-circle mode of replication. The two plasmids encode proteins belonging to two distinct superfamilies (SF) of RC-Reps. Whereas SFII Reps were previously found in plasmids of Thermococcales (41, 42), MGE encoding SFI RC proteins have not been observed in this euryarchaeal order. Our analysis has uncovered a unique recombination event which occurred in the evolution of pTP1. As a result of this recombination, an ancestral copy of the RC-Rep gene has been disrupted by the incoming copy of a homologous RC-Rep gene. What the mechanism of such integration could have been is currently unclear. Remarkably, a relationship between RepTP1-like RC-Reps and bacterial transposases has been noticed previously (41). Thus, it is possible that, akin to the transposition reaction of rolling-circle transposons, the integration was catalyzed by RepTP1 itself. Another unexpected finding was that ORF3, the larger fragment of the disrupted RC-Rep of pTP1, is evolving under strong purifying selection (Fig. 2A and C), indicating that its product retained some function in plasmid reproduction. Biochemical studies will be crucial for rationalizing these perplexing observations.
Although constantly growing, the genetic toolkit for Thermococcales is still limited. Especially modest is the choice of replicative expression vectors (51, 52). Indeed, all of them are currently based on the small RC plasmid pTN1 of T. nautilus (41); thus, they share the same origin of replication (49). The two plasmids described in this study encode unrelated Rep proteins and are naturally compatible. Similarly, the genome replication cassette of TPV1 may be also used to construct an even more versatile array of compatible vectors for genetic manipulations. Furthermore, the high copy number of the two plasmids (50 copies/cell) may prove to be a useful property in developing new protein expression systems in Thermococcales. The availability of such genetic tools would undoubtedly propel the studies on hyperthermophilic euryarchaeal viruses and their interaction with the Thermococcus hosts.
ACKNOWLEDGMENTS
This work was supported by the ANR project Thermovésicules (ANR-12-BSV3-OO23-01) (to P.F. and C.G.), a MENRT grant (to A.G.), and the Foundation for Research on Biodiversity (DIVVIR 2009–2012).
Footnotes
Published ahead of print 12 April 2013
REFERENCES
- 1. Feschotte C, Pritham EJ. 2007. DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 41:331–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Frost LS, Leplae R, Summers AO, Toussaint A. 2005. Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3:722–732 [DOI] [PubMed] [Google Scholar]
- 3. Sobecky PA, Hazen TH. 2009. Horizontal gene transfer and mobile genetic elements in marine systems. Methods Mol. Biol. 532:435–453 [DOI] [PubMed] [Google Scholar]
- 4. Krupovic M, Prangishvili D, Hendrix RW, Bamford DH. 2011. Genomics of bacterial and archaeal viruses: dynamics within the prokaryotic virosphere. Microbiol. Mol. Biol. Rev. 75:610–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Koonin EV, Senkevich TG, Dolja VV. 2006. The ancient virus world and evolution of cells. Biol. Direct 1:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Krupovic M, Bamford DH. 2007. Putative prophages related to lytic tailless marine dsDNA phage PM2 are widespread in the genomes of aquatic bacteria. BMC Genomics 8:236 doi: 10.1186/1471-2164-8-236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Krupovic M, Gonnet M, Hania WB, Forterre P, Erauso G. 2013. Insights into dynamics of mobile genetic elements in hyperthermophilic environments from five new Thermococcus plasmids. PLoS One 8:e49044 doi: 10.1371/journal.pone.0049044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Krupovic M. 2012. Recombination between RNA viruses and plasmids might have played a central role in the origin and evolution of small DNA viruses. Bioessays 34:867–870 [DOI] [PubMed] [Google Scholar]
- 9. Krupovic M, Ravantti JJ, Bamford DH. 2009. Geminiviruses: a tale of a plasmid becoming a virus. BMC Evol. Biol. 9:112 doi: 10.1186/1471-2148-9-112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ravin NV. 2011. N15: the linear phage-plasmid. Plasmid 65:102–109 [DOI] [PubMed] [Google Scholar]
- 11. Jalasvuori M. 2012. Vehicles, replicators, and intercellular movement of genetic information: evolutionary dissection of a bacterial cell. Int. J. Evol. Biol. 2012:874153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lipps G. 2006. Plasmids and viruses of the thermoacidophilic crenarchaeote Sulfolobus. Extremophiles 10:17–28 [DOI] [PubMed] [Google Scholar]
- 13. Peng X, Garrett RA, She Q. 2012. Archaeal viruses—novel, diverse and enigmatic. Sci. China Life Sci. 55:422–433 [DOI] [PubMed] [Google Scholar]
- 14. Pina M, Bize A, Forterre P, Prangishvili D. 2011. The archeoviruses. FEMS Microbiol. Rev. 35:1035–1054 [DOI] [PubMed] [Google Scholar]
- 15. Dyall-Smith ML, Pfeiffer F, Klee K, Palm P, Gross K, Schuster SC, Rampp M, Oesterhelt D. 2011. Haloquadratum walsbyi: limited diversity in a global pond. PLoS One 6:e20968 doi: 10.1371/journal.pone.0020968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Holmes ML, Pfeifer F, Dyall-Smith ML. 1995. Analysis of the halobacterial plasmid pHK2 minimal replicon. Gene 153:117–121 [DOI] [PubMed] [Google Scholar]
- 17. Roine E, Kukkaro P, Paulin L, Laurinavičius S, Domanska A, Somerharju P, Bamford DH. 2010. New, closely related haloarchaeal viral elements with different nucleic acid types. J. Virol. 84:3682–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang Z, Liu Y, Wang S, Yang D, Cheng Y, Hu J, Chen J, Mei Y, Shen P, Bamford DH, Chen X. 2012. Temperate membrane-containing halophilic archaeal virus SNJ1 has a circular dsDNA genome identical to that of plasmid pHH205. Virology 434:233–241 [DOI] [PubMed] [Google Scholar]
- 19. Peng X. 2008. Evidence for the horizontal transfer of an integrase gene from a fusellovirus to a pRN-like plasmid within a single strain of Sulfolobus and the implications for plasmid survival. Microbiology 154:383–391 [DOI] [PubMed] [Google Scholar]
- 20. Arnold HP, She Q, Phan H, Stedman K, Prangishvili D, Holz I, Kristjansson JK, Garrett R, Zillig W. 1999. The genetic element pSSVx of the extremely thermophilic crenarchaeon Sulfolobus is a hybrid between a plasmid and a virus. Mol. Microbiol. 34:217–226 [DOI] [PubMed] [Google Scholar]
- 21. Wang Y, Duan Z, Zhu H, Guo X, Wang Z, Zhou J, She Q, Huang L. 2007. A novel Sulfolobus non-conjugative extrachromosomal genetic element capable of integration into the host genome and spreading in the presence of a fusellovirus. Virology 363:124–133 [DOI] [PubMed] [Google Scholar]
- 22. Basta T, Smyth J, Forterre P, Prangishvili D, Peng X. 2009. Novel archaeal plasmid pAH1 and its interactions with the lipothrixvirus AFV1. Mol. Microbiol. 71:23–34 [DOI] [PubMed] [Google Scholar]
- 23. Bult CJ, White O, Olsen GJ, Zhou L, Fleischmann RD, Sutton GG, Blake JA, FitzGerald LM, Clayton RA, Gocayne JD, Kerlavage AR, Dougherty BA, Tomb JF, Adams MD, Reich CI, Overbeek R, Kirkness EF, Weinstock KG, Merrick JM, Glodek A, Scott JL, Geoghagen NS, Venter JC. 1996. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science 273:1058–1073 [DOI] [PubMed] [Google Scholar]
- 24. López-García P, Forterre P, van der Oost J, Erauso G. 2000. Plasmid pGS5 from the hyperthermophilic archaeon Archaeoglobus profundus is negatively supercoiled. J. Bacteriol. 182:4998–5000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Soler N, Gaudin M, Marguet E, Forterre P. 2011. Plasmids, viruses and virus-like membrane vesicles from Thermococcales. Biochem. Soc. Trans. 39:36–44 [DOI] [PubMed] [Google Scholar]
- 26. Geslin C, Le Romancer M, Erauso G, Gaillard M, Perrot G, Prieur D. 2003. PAV1, the first virus-like particle isolated from a hyperthermophilic euryarchaeote, “Pyrococcus abyssi.” J. Bacteriol. 185:3888–3894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gorlas A, Koonin EV, Bienvenu N, Prieur D, Geslin C. 2012. TPV1, the first virus isolated from the hyperthermophilic genus Thermococcus. Environ. Microbiol. 14:503–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Soler N, Marguet E, Cortez D, Desnoues N, Keller J, van Tilbeurgh H, Sezonov G, Forterre P. 2010. Two novel families of plasmids from hyperthermophilic archaea encoding new families of replication proteins. Nucleic Acids Res. 38:5088–5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Geslin C, Gaillard M, Flament D, Rouault K, Le Romancer M, Prieur D, Erauso G. 2007. Analysis of the first genome of a hyperthermophilic marine virus-like particle, PAV1, isolated from Pyrococcus abyssi. J. Bacteriol. 189:4510–4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gorlas A, Alain K, Bienvenu N, Isaac S, Geslin C. 25 January 2013. Thermococcus prieurii sp. nov., a novel hyperthermophilic archaeon isolated from a deep-sea hydrothermal vent at the East Pacific Rise. Int. J. Syst. Evol. Microbiol. [Epub ahead of print.] doi: 10.1099/ijs.0.026419-0 [DOI] [PubMed] [Google Scholar]
- 31. Gorlas A, Geslin C. 2013. A simple procedure to determine the infectivity and host range of viruses infecting anaerobic and hyperthermophilic microorganisms. Extremophiles 17:349–355 [DOI] [PubMed] [Google Scholar]
- 32. Charbonnier F, Erauso G, Barbeyron T, Prieur D, Forterre P. 1992. Evidence that a plasmid from a hyperthermophilic archaebacterium is relaxed at physiological temperatures. J. Bacteriol. 174:6103–6108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Birnboim HC, Doly J. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Besemer J, Borodovsky M. 2005. GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 33:W451–W454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Suzek BE, Ermolaeva MD, Schreiber M, Salzberg SL. 2001. A probabilistic method for identifying start codons in bacterial genomes. Bioinformatics 17:1123–1130 [DOI] [PubMed] [Google Scholar]
- 37. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Söding J. 2005. Protein homology detection by HMM-HMM comparison. Bioinformatics 21:951–960 [DOI] [PubMed] [Google Scholar]
- 39. Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580 [DOI] [PubMed] [Google Scholar]
- 40. Hurst LD. 2002. The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet. 18:486. [DOI] [PubMed] [Google Scholar]
- 41. Soler N, Justome A, Quevillon-Cheruel S, Lorieux F, Le Cam E, Marguet E, Forterre P. 2007. The rolling-circle plasmid pTN1 from the hyperthermophilic archaeon Thermococcus nautilus. Mol. Microbiol. 66:357–370 [DOI] [PubMed] [Google Scholar]
- 42. Erauso G, Marsin S, Benbouzid-Rollet N, Baucher MF, Barbeyron T, Zivanovic Y, Prieur D, Forterre P. 1996. Sequence of plasmid pGT5 from the archaeon Pyrococcus abyssi: evidence for rolling-circle replication in a hyperthermophile. J. Bacteriol. 178:3232–3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ward DE, Revet IM, Nandakumar R, Tuttle JH, de Vos WM, van der Oost J, DiRuggiero J. 2002. Characterization of plasmid pRT1 from Pyrococcus sp. strain JT1. J. Bacteriol. 184:2561–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ilyina TV, Koonin EV. 1992. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 20:3279–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gonnet M, Erauso G, Prieur D, Le Romancer M. 2011. pAMT11, a novel plasmid isolated from a Thermococcus sp. strain closely related to the virus-like integrated element TKV1 of the Thermococcus kodakaraensis genome. Res. Microbiol. 162:132–143 [DOI] [PubMed] [Google Scholar]
- 46. Krupovic M, Bamford DH. 2008. Archaeal proviruses TKV4 and MVV extend the PRD1-adenovirus lineage to the phylum Euryarchaeota. Virology 375:292–300 [DOI] [PubMed] [Google Scholar]
- 47. Senčilo A, Paulin L, Kellner S, Helm M, Roine E. 2012. Related haloarchaeal pleomorphic viruses contain different genome types. Nucleic Acids Res. 40:5523–5534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rasooly A, Rasooly RS. 1997. How rolling circle plasmids control their copy number. Trends Microbiol. 5:440–446 [DOI] [PubMed] [Google Scholar]
- 49. Santangelo TJ, Cubonova L, Reeve JN. 2008. Shuttle vector expression in Thermococcus kodakaraensis: contributions of cis elements to protein synthesis in a hyperthermophilic archaeon. Appl. Environ. Microbiol. 74:3099–3104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Krupovic M, Gribaldo S, Bamford DH, Forterre P. 2010. The evolutionary history of archaeal MCM helicases: a case study of vertical evolution combined with hitchhiking of mobile genetic elements. Mol. Biol. Evol. 27:2716–2732 [DOI] [PubMed] [Google Scholar]
- 51. Atomi H, Imanaka T, Fukui T. 2012. Overview of the genetic tools in the Archaea. Front. Microbiol. 3:337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hileman TH, Santangelo TJ. 2012. Genetics techniques for Thermococcus kodakarensis. Front. Microbiol. 3:195. [DOI] [PMC free article] [PubMed] [Google Scholar]