Abstract
Coniothyrium minitans is a sclerotial parasite of the plant-pathogenic fungus Sclerotinia sclerotiorum, and conidial production and parasitism are two important aspects for commercialization of this biological control agent. To understand the mechanism of conidiation and parasitism at the molecular level, we constructed a transfer DNA (tDNA) insertional library with the wild-type strain ZS-1. A conidiation-deficient mutant, ZS-1TN22803, was uncovered through screening of this library. This mutant could produce pycnidia on potato dextrose agar (PDA), but most were immature and did not bear conidia. Moreover, this mutant lost the ability to parasitize or rot the sclerotia of S. sclerotiorum. Analysis of the tDNA flanking sequences revealed that a peroxisome biogenesis factor 6 (PEX6) homolog of Saccharomyces cerevisiae, named CmPEX6, was disrupted by the tDNA insertion in this mutant. Targeted gene replacement and gene complementation tests confirmed that a null mutation of CmPEX6 was responsible for the phenotype of ZS-1TN22803. Further analysis showed that both ZS-1TN22803 and the targeted replacement mutants could not grow on PDA medium containing oleic acid, and they produced much less nitric oxide (NO) and hydrogen peroxide (H2O2) than wild-type strain ZS-1. The conidiation of ZS-1TN22803 was partially restored by adding acetyl-CoA or glyoxylic acid to the growth media. Our results suggest that fatty acid β-oxidation, reactive oxygen and nitrogen species, and possibly other unknown pathways in peroxisomes are involved in conidiation and parasitism by C. minitans.
INTRODUCTION
Coniothyrium minitans, a mycoparasite of the plant fungal pathogen Sclerotinia sclerotiorum, can parasitize and decay both hyphae and sclerotia of Sclerotinia spp. It also reduces the germination of sclerotia and inhibits further infection by S. sclerotiorum hyphae; therefore, its existence in crop fields may play a very important role in suppressing Sclerotinia diseases (1–5). Its antagonistic properties have made C. minitans a well-known biological control microorganism, and several formulations based on C. minitans have been developed and registered commercially (4, 6).
Coniothyrium minitans is a coelomycete fungus producing conidia in pycnidia. Understanding the conidiation of C. minitans at the molecular level may help to improve the efficiency of conidial production, which is important for the commercial use of biological control agents. Asexual reproduction of fungi which do not have a sexual stage in their life cycle is essential for survival and spread in nature; however, many previous studies on conidiation have focused on hyphomycete fungi (unenclosed conidia), such as Aspergillus nidulans, Neurospora crassa, and Magnaporthe oryzae, and little is known about conidiation in coelomycetes, except for the chestnut blight fungus Cryphonectria parasitica (7; for a review, see reference 8). Studies on the conidiation of C. minitans may improve our understanding of the general nature of asexual reproduction by coelomycetes.
Conidiation by C. minitans can be divided into five stages: hyphal growth (48 hpi), primordial formation (60 hpi), pycnidial initiation (72 hpi), pycnidial formation (84 hpi), and pycnidial maturation (96 hpi) (9, 10). Several genes and pathways regulating C. minitans conidiation have recently been elucidated, including signaling mediated by nitric oxide, cGMP, cyclic AMP (cAMP), the mitogen-activated protein (MAP) kinase cascade, and the PIF1 DNA helicase gene, as well as by many cell wall-degrading enzymes (fCWDE), such as beta-1,3-glucanase, chitinase, and so on (9–14).
The interaction between C. minitans and its host, S. sclerotiorum, also attracts extensive research interest due to the potential use of hyperparasitism-associated genes in the resistance improvement of crops against fungal diseases, especially Sclerotinia diseases. Oxalic acid, a pathogenicity factor of S. sclerotiorum, is likely to be a signal for this interaction. C. minitans produces antifungal substances with increasing activity at low pH caused by oxalic acid and produces beta-1,3-glucanase, an fCWDE, at higher pH after oxalic acid is degraded (15–17). Theoretically, the interactions between C. minitans and S. sclerotiorum are similar to that between the plant pathogen and host plants, and many similar pathways are likely to be involved in this hyperparasitism.
Peroxisomes are single-membrane-bound organelles widely existing in eukaryotic cells, and they play a pivotal role in various metabolic pathways and developmental processes of plants, fungi, and mammals (18–21). These pathways are associated with a suite of cellular functions, including detoxification of H2O2, β-oxidation of fatty acids, and utilization of amino acids (22, 23). Peroxisomes are the site for the generation of superoxide (O2 · −) and nitric oxide ( · NO) radicals in plants (24, 25), while in filamentous fungi, peroxisomes also have other specific functions. In Podospora anserina, mutations in pex2 (a peroxisome-associated gene) affected karyogamy and meiocyte formation, causing a specific block in sexual development (26). In Penicillium chrysogenum, peroxisomes are known to participate in the last step of penicillin biosynthesis (20). In Colletotrichum lagenarium and M. oryzae, peroxisomal metabolic pathways contribute to appressorial melanization, generation of appressorial turgor, and subsequent host invasion (27–29). In Alternaria alternata, peroxisomes are involved in conidiation, plant invasion, and tissue colonization (30). Peroxisomes may also play a role in the development of trap cells in the nematophagous fungus Arthrobotrys oligospora (31). Approximately 32 peroxisomal proteins for biogenesis and maintenance have been identified in yeast (32), and disrupting some of these genes in plant-pathogenic fungi, such as A. alternata, C. lagenarium, and M. oryzae, leads to loss of fungal pathogenicity (27, 28, 30).
Previously, we constructed a transfer DNA (tDNA) insertional library of C. minitans using the wild-type strain ZS-1 (33). Through a growth phenotype screening of this mutant library, we uncovered a conidiation-deficient mutant, ZS-1TN22803. The mutant formed normal hyphae but produced immature pycnidia without conidia, and it showed a phenotype similar to that of previously characterized conidiation-deficient mutants ZS-1T2029 and ZS-1T21882 (10, 11); however, unlike those two mutants, the ZS-1TN22803 mutant was unable to parasitize S. sclerotiorum. Furthermore, we identified a peroxisome biogenesis factor 6 (PEX6) homolog of Saccharomyces cerevisiae responsible for the phenotype of ZS-1TN22803. Although it is well known that the PEX proteins are involved in fungal conidiation and pathogenicity on plant or animal hosts, this is the first report on peroxisomal involvement in mycoparasitism. Here, we demonstrated the roles of PEX6 in conidiation by C. minitans and its parasitism of S. sclerotiorum, and we speculated on the possible underlying mechanisms.
MATERIALS AND METHODS
C. minitans strains and culture conditions.
The C. minitans wild-type strain ZS-1 (CCAM 041057) was isolated from garden soil at Zhushan County, Hubei Province, People's Republic of China, and the conidiation-deficient mutant ZS-1TN22803 was obtained from screening a tDNA insertional library (33, 34). Strain Ep-1PNA367 was a virulent and virus-free strain of S. sclerotiorum, derived from hypovirulent strain Ep-1PN by a single-ascospore isolation technique (35). The strains were cultured on potato dextrose agar (PDA) and broth (PDB) at 20 to 22°C and stored in PDA slants at 4°C for further use (11, 34).
Assay of growth rate, colony morphology, conidiation, and parasitic ability.
To characterize biological properties of ZS-1TN22803, growth rate, conidial production, colony morphology, and parasitic ability were examined. Hyphal extension and conidial production were examined as described by Cheng et al. (34) and Zeng et al. (14). Colony morphology was observed on PDA after incubation at 20 to 22°C for 15 days.
To assay parasitic ability, surface-sterilized sclerotia of strain Ep-1PNA367 were incubated in a conidial suspension of all C. minitans strains (106 conidia/ml) for 30 min and then transferred to sterilized moist sand in petri dishes with half of the sclerotia exposed on the surface of the sand for 30 days. The plates were sealed to retain moisture. Rot index was used to assess the parasitic activity by C. minitans according to Cheng et al. (34). To further check if sclerotia were parasitized by mutants, all conidium-treated sclerotia were surface sterilized with sodium hypochlorite solution to kill superficially growing C. minitans and washed three times with sterilized water, and then the sclerotia were placed onto PDA amended with 50 μg/ml hygromycin and incubated at 20°C for 7 days. If the sclerotia were successfully parasitized by mutants, the mutants would grow out of the sclerotia and develop colonies on hygromycin-amended PDA, since the mutants were transformed with the hygromycin resistance gene (hph).
To examine if a mutant could parasitize the hyphae of S. sclerotiorum, a method of dual culture on PDA, following Qin et al. (10), was used, with minor modifications. An agar plug of mutant ZS-1TN22803 was placed at the center of a PDA plate and incubated for 5 days, and then four mycelial discs of S. sclerotiorum were inoculated on the same plate symmetrically (discs were about 3.0 cm from the agar plug of ZS-1TN22803), and then the plates were incubated for a further 20 days. Mycelial discs were taken from the interaction zones between two colonies and transferred onto fresh PDA for further incubation to check if S. sclerotiorum emerged. Experiments were repeated three times with three replicates and with the wild-type strain ZS-1 as a control.
Observation of the pycnidial formation.
To examine the pycnidial development and fine surface structure of pycnidia of the mutants, scanning electron microscopy (SEM; Model JSM-6390/LV; NTC, Japan) was used. The mutants were cultured on cellophane membranes placed on PDA and incubated for 7 days at 20 to 22°C. The mycelia with the cellophane base then were cut into 5- by 10-mm pieces with a scalpel. Samples were fixed with 1% OsO4 for 1 h, placed on the sample holder directly, and sputter coated with platinum (model JFC-1600; NTC, Japan). The samples then were observed by SEM at an acceleration voltage of 10 kV and photographed with a digital camera (Canon Power Shot SX30 IS). The wild-type strain ZS-1 was used as a control.
DNA extraction and Southern blot analysis.
The genomic DNA of the wild-type strain ZS-1, ZS-1TN22803, and other derivative mutants was extracted according to a standard procedure (36). To estimate the tDNA insertion copy number in ZS-1TN22803, Southern blot analysis was performed by following the method described by Gong et al. (11), with some modifications. Genomic DNA of ZS-1TN22803 or ZS-1 was completely digested with SacI, which has only one recognition site on the binary vector, pTFCM, used to construct the tDNA insertional library. The nylon membrane was probed with the hygromycin resistance gene (hph) that was amplified with primer pair hph-L and hph-R (Table 1) and labeled with [α-32P]dCTP. To estimate the copy number of CmPEX6 in C. minitans, genomic DNA of ZS-1 was digested completely with ApaI or BamHI separately, neither of which has recognition sites in the DNA sequence of CmPEX6. The nylon membrane was hybridized with a 536-bp DNA fragment of CmPEX6 amplified with primer pair 22803-L and 22803-R (Table 1).
Table 1.
Primers used for vector construction and PCR
| Primer name and use | Sequence |
|---|---|
| RT-PCR | |
| 22803-L | 5′CAAAGCAGTCGCCTGGTTC3′ |
| 22803-R | 5′ATTTGGCACCGCAGGTAAGC3′ |
| I-PCR | |
| Pttrp | 5′ATGTCCTCGTTCCTGTCTGCTAATA3′ |
| LB-1 | 5′AGGGTTCCTATAGGGTTTCGCTCATG3′ |
| LB-3 | 5′ GAATTAATTCGGCGTTAATTCAGT3′ |
| Replacement vector | |
| CmPEX6 HindIII | 5′CGAAGCTTAGAACACCCCGTGGACCATCCTG 3′ |
| CmPEX6 SalI | 5′CGGTCGACAGATGGAGGCGAGGAGAAGC3′ |
| CmPEX6 XbaI | 5′GCTCTAGATATCCGCGGTTTGTTCACACACG3′ |
| CmPEX6 KpnI | 5′GGGGTACCCGAACGAGTATGTGAAATAGAGG3′ |
| Complementation vector | |
| com-HindIII | 5′GCAAGCTTATGGAAGTGCAGAACGGCACT3′ |
| com-PstI | 5′CTGCAGTCAAGAGTACAGCTCCTCGTCCC3′ |
For the analysis of CmPEX6 replacement and complemented mutants, genomic DNA of ZS-1 or the mutants were digested with HindIII. The nylon membranes were hybridized with probe P1 for CmPEX6 and P2 for hph.
Cloning and analysis of the gene disrupted by tDNA insertion.
Amplification of the flanking DNA fragments of the tDNA insertion site in ZS-1TN22803 was performed using the inverse PCR (I-PCR) technique. The genomic DNA of ZS-1TN22803 (500 ng to 1 μg) was digested with either ScaI or KpnI, precipitated with ethanol, and then resuspended in 20 μl double-distilled water (ddH2O) and mixed with 20 U of T4 DNA ligase. The reaction was allowed to proceed at 16°C for 10 to 16 h. After ethanol precipitation, the circularized DNA was resuspended in 20 μl ddH2O and used as the template for the nested PCR amplification with primer pair Pttrp and LB-1 for the primary amplification. This PCR product was then used with primer pair Pttrp and LB-3 for the secondary amplification. The primers were designed based on the sequence of vector pTFCM (Table 1). The PCR conditions used were 32 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 3 min, with a final extension at 72°C for 5 min. The PCR product of primary amplification was diluted 20-fold with ddH2O, and 1 μl of the diluted PCR product was used as the template for the secondary amplification. PCRs were carried out on a PTC-200 DNA Engine Peltier thermal cycler (Bio-Rad, USA).
RNA extraction and RT-PCR amplification.
Total RNA of fungal strains was isolated with TRIzol reagent (Invitrogen) according to the manufacturer's protocols, and potential DNA contamination was removed by DNase I treatment (RNase Free) (TaKaRa, Dalian, China). First-strand cDNA was synthesized using the RevertAid first-strand cDNA synthesis kit (MBI, Fermentas) by following the manufacturer's instructions. The total RNA of mycelia at 48, 72, 84, and 96 h postincubation on PDA was used to assess the expression pattern of CmPEX6 by reverse transcription-PCR (RT-PCR), and samples at 96 h were detected to determine the gene expression in all mutants. A 536-bp fragment of CmPEX6 was amplified by RT-PCR with gene-specific primer pair 22803-L and 22803-R (Table 1). PCR conditions used were 28 cycles of 94°C for 30 s, 57°C for 30 s, and 72°C for 3 min, with a final extension at 72°C for 5 min.
Vector construction and Agrobacterium-mediated transformation.
To assess the function of CmPEX6, a CmPEX6 replacement vector, pCMPEX-3300, and a complementary vector, pCPPE, were constructed. The replacement vector was constructed using the homologous recombination strategy. A 5′ fragment of 1.4 kb was amplified with primer pair CmPEX6 HindIII and CmPEX6 SalI (Table 1) and cloned into the same sites on pMD18-hph, resulting in the construct pMD2-hph. Subsequently, a 0.95-kb 3′ fragment was amplified with primer pair CmPEX6 XbaI and CmPEX6 KpnI (Table 1) and cloned between the same sites on pMD2-hph, resulting in the construct pNeoP3300III. After that, the neomycin resistance gene cassette (neo) was ligated into the XbaI recognition site of pNeoP330III as the second negative selectable marker, resulting in the gene replacement vector pCMPEX-3300, which ensures that the replacement transformants would be hygromycin resistant and neomycin sensitive. To construct the complementary vector, CmPEX6 cDNA was amplified by RT-PCR with primer pair com-HindIII and com-PstI (Table 1) and cloned into the same sites of pCIT vector, which contained the constitutive PtrpC promoter and terminator. Finally, the cDNA of CmPEX6 was digested with XhoI and cloned into pNeoP3300, resulting in the CmPEX6 complementary vector pCPPE.
The transformation of C. minitans was performed with strain EHA105 of Agrobacterium tumefaciens as described by Li et al. (33) and Qin et al. (10), with minor modifications. To complement the mutation of CmPEX6 in ZS-1TN22803, ZS-1TN22803 was transformed with vector pCPPE. Complemented transformants, named CmPEX6-com, were rescued on PDA containing 80 μg/ml of G418. To obtain the CmPEX6 replacement transformants, candidate transformants, termed DCmPEX6, were uncovered on PDA supplemented with 50 μg/ml hygromycin B and then subcultured on PDA with 80 μg/ml of G418. All CmPEX6-com and DCmPEX6 transformants were confirmed primarily by RT-PCR with primer pair 22803-L and 22803-R (Table 1) and further by Southern blotting.
Assay on carbon utilization.
To assay the effect of oleic acid as a carbon source on the growth of the mutants, strains ZS-1, ZS-1TN22803, CmPEX6-com, and DCmPEX6-98 were investigated for growth both on modified Czapek-Dox (MCD) medium containing oleic acid (Sigma-Aldrich) as the sole carbon source and on PDA containing 1% oleic acid and 0.5% Tween 80 at 20 to 22°C for 10 days, with MCD and PDA media as controls, according to Qin et al. (10). Mycelial growth rate and conidial production were measured. Each experiment was repeated 3 times.
To test if acetyl-coenzyme A (CoA) and glyoxylic acid could restore conidiation of the mutants, ZS-1 and DCmPEX6-98 were cultured on PDA amended with different concentrations (0.25 mM to 0.1 M) of acetyl-CoA (Sigma-Aldrich) and glyoxylic acid (Sinopharm Chemical Reagent Co., Ltd.), respectively. PDA was used as the control. The colony morphology, growth rate, and conidial production by each treatment was examined using the method described above. Three replicates were conducted for each treatment, and each experiment was repeated twice.
H2O2 and NO generation in mycelia.
To examine if there was any change for generating hydrogen peroxide (H2O2) and nitric oxide (NO) in mutants during growth on PDA, CmPEX6 mutants and ZS-1 were grown on PDA for 3 to 4 days. Mycelia (0.05 g) were scraped off, ground in liquid nitrogen, and soaked in 500 μl of lysis buffer supplied by a hydrogen peroxide assay kit or by a nitrate/nitrite colorimetric assay kit (Beyotime Institute of Biotechnology, China). Fifty microliters of the supernatant, gathered by centrifuging at 12,000 × g for 5 min, was used to determine the amount of H2O2 and NO. The measurements were carried out by following the kit protocols. There were three replicates in each treatment, and the experiment was repeated three times.
Data analyses.
SAS, version 8.1 (SAS Institute, Inc., Cary, NC), was used to analyze the variation between treatments for each experiment using analysis of variance (ANOVA). The data of conidial production were log transformed prior to analysis. When a significant treatment effect was found (P < 0.05), the mean values (whether numerical or log transforms) for different treatments in the experiments were compared with the least-significant-difference test.
Nucleotide sequence accession number.
The sequence of the polypeptide determined in the course of this work has been deposited in GenBank under accession number JN391412.
RESULTS
Characterization of conidiation-deficient mutant ZS-1TN22803 in C. minitans.
The tDNA insertion mutant ZS-1TN22803 grew well on PDA plates, and its hyphal growth rate was not significantly different from that of the wild-type strain ZS-1 (Fig. 1A and B). ZS-1TN22803 formed a light-colored colony with mycelial strands that developed into pycnidial primordia. However, only a few immature pycnidia from mycelial strands were produced; most pycnidia showed a light color without melanization, and no conidia were produced within the immature pycnidia as revealed under light microscopy. At 7 days postincubation, very few immature pycnidia turned dark, and some conidia were produced. The wild-type strain ZS-1 formed dark-colored colonies with production of mature pycnidia and conidiation, beginning at 3 days postinoculation on PDA plates, and numerous mature dark pycnidia produced at 7 days postincubation. Compared to that of the wild-type strain, conidiation of ZS-1TN22803 on PDA plates was significantly delayed. Furthermore, conidial production by ZS-1TN22803 (3.3 × 104 conidia cm−2) was considerably less than that by ZS-1 (2.6 × 107 conidia cm−2) by about 1,000-fold at 15 days postincubation (Fig. 1C and 2).
Fig 1.
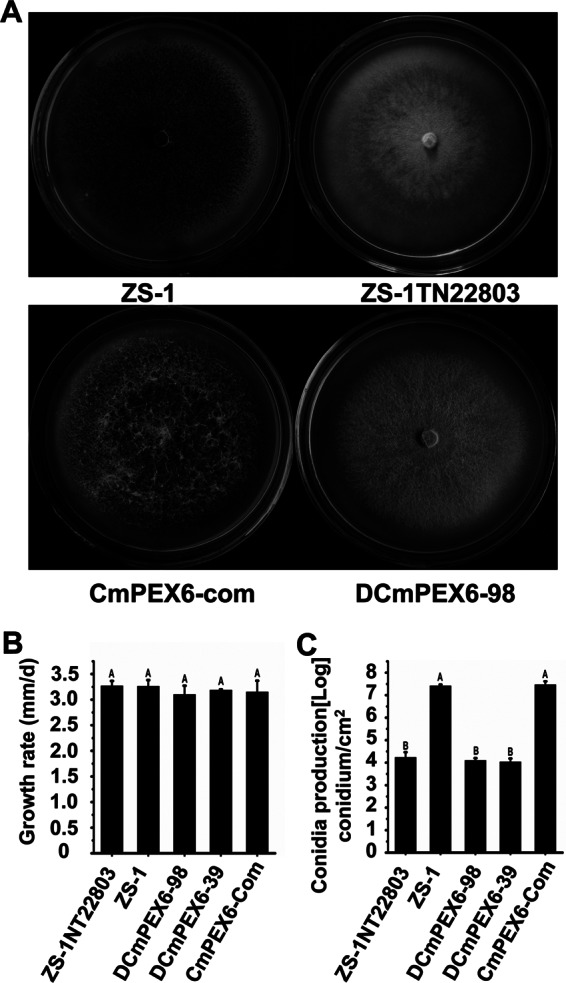
Comparison of colony morphology and conidiation by C. minitans among isolates ZS-1, ZS-1TN22803, DCmPEX6-39, DCmPEX6-98, and CmPEX6-com on PDA at 20°C for 15 days. (A) Colony morphology of C. minitans grown on PDA at 20°C for 15 days. (B) Mycelial growth rate based on colony diameter of cultures incubated at 20°C for 7 days. (C) Conidial production on PDA at 20°C for 15 days, determined with a hematocytometer. The vertical bars represent standard errors based on three replicates. Within columns, means followed by the same lowercase letter within each chart are not significantly different (P < 0.05) according to the least-significant-difference test.
Fig 2.
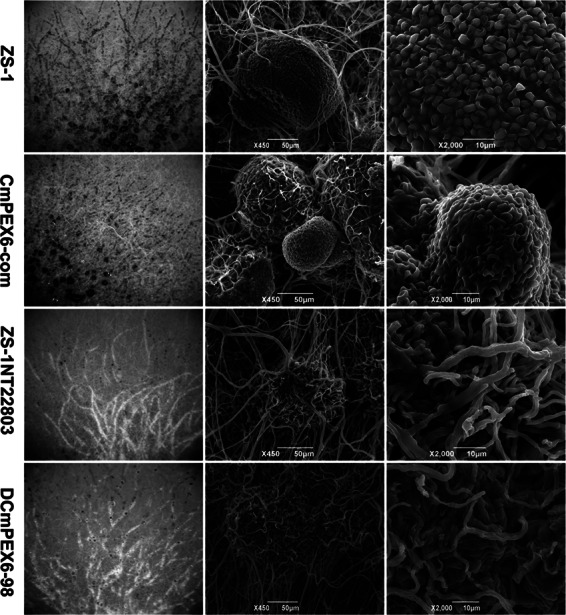
Microscopic observation of colony morphology and pycnidia formed by nonmutants first and then mutants ZS-1TN22803, DCmPEX6-98, CmPEX6-com, and ZS-1 with SEM. All strains were cultured at 20°C on PDA for 7 days. Column I shows the colony morphology. Column II shows pycnidia near the margin of the colony. Column III shows the conidia formed in the pycnidia. There were no pycnidia or conidia formed in colonies of ZS-1TN22803 or DCmPEX6-98.
ZS-1TN22803 unable to parasitize S. sclerotiorum.
When 106 conidia/ml of ZS-1TN22803 was inoculated on sclerotia and incubated at 20°C for 30 days, neither pycnidia nor conidia of C. minitans were formed on the surface or the inner part of the sclerotia (Fig. 3A and C), suggesting that ZS-1TN22803 could not parasitize sclerotia of S. sclerotiorum. To further investigate whether the mutant could parasitize the inner part of sclerotia, sclerotia were surface sterilized and then seeded on PDA amended with 50 μg/ml hygromycin for additional incubation. No colonies of ZS-1TN22803 were formed around sclerotia (Fig. 3B).
Fig 3.
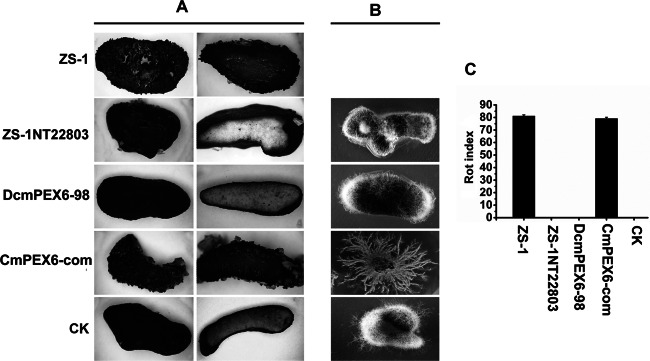
Parasitic ability of C. minitans mutants on sclerotia of S. sclerotiorum. (A) Sclerotia were parasitized by ZS-1 or CmPEX6-com, dark pycnidia were observed on the surface of sclerotia, and the inner parts of sclerotia were soft/rotted; sclerotia were not parasitized by mutants ZS-1TN22803 and DCmPEX6-98, and no pycnidia were observed on the surface of the sclerotia, just like the uninoculated sclerotia (CK). (B) Inoculated sclerotia were grown on PDA amended with hygromycin (50 μg/ml) for 7 days after surface sterilization. New colonies of C. minitans were observed only from sclerotia treated with CmPEX6-com, and mycelia of S. sclerotiorum could be observed from sclerotia treated with ZS-1TN22803 or DCmPEX6-98. (C) The sclerotial rot index induced by C. minitans mutants. Sclerotia with a similar size were surface sterilized and inoculated with the conidia of ZS-1TN22803, DcmPEX6-98, CmPEX6-com, or ZS-1, respectively, and incubated in wet sand at 20°C for 30 days.
We established a dual culture system to further investigate the interaction between the mutant ZS-1TN22803 and S. sclerotiorum. When ZS-1TN22803 was dual cultured with S. sclerotiorum for 20 days, the two colonies intermingled without a significant inhibition zone. Mycelial discs excised from the region between S. sclerotiorum and ZS-1TN22803 were placed on fresh PDA plates and formed new colonies always resembling S. sclerotiorum. Furthermore, the sclerotia formed around the colony border of S. sclerotiorum were not parasitized by the mutant, and no pycnidia of C. minitans were observed. In contrast, when the wild-type strain ZS-1 was dual cultured with S. sclerotiorum, ZS-1 continued to grow and spread over the colony of S. sclerotiorum, and mycelia discs excised from the interaction zone constantly formed colonies of C. minitans but not S. sclerotiorum. Furthermore, many mature dark pycnidia were produced on the surface of deteriorated sclerotia of S. sclerotiorum (Fig. 4). These results demonstrate that the mutant ZS-1TN22803 lost the ability to parasitize hyphae or sclerotia of S. sclerotiorum; furthermore, conidiation by ZS-1TN22803 was not restored in dual cultures with its natural living host, S. sclerotiorum.
Fig 4.
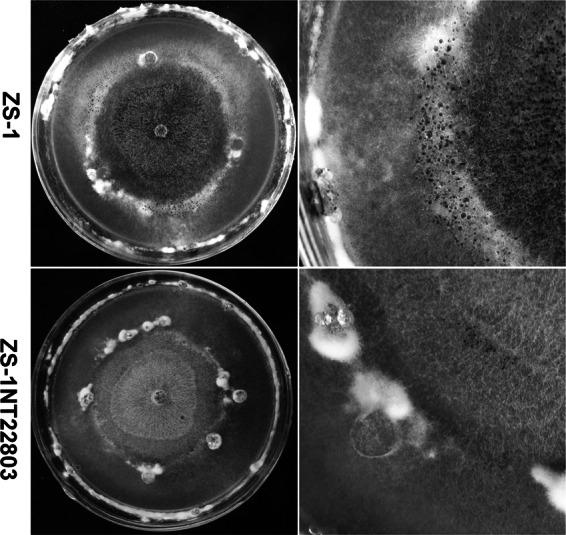
Dual cultures of ZS-1TN22803 or ZS-1 of C. minitans with host S. sclerotiorum Ep-1PNA367. The mycelial disc of mutant ZS-1TN22803 or ZS-1 was placed at the center of a PDA plate and grown for 5 days, and then four mycelial discs of S. sclerotiorum were placed on the same plate, equally spaced at ∼3.0 cm from the center, and the plates were incubated at 20°C for a further 20 days. ZS-1TN22803 did not produce dark pycnidia in the colony of S. sclerotiorum, and it did not parasitize the sclerotia (the white fuzzy material) at the interface of the two colonies.
Cloning and analysis of CmPEX6 in mutant ZS-1TN22803.
Inverse PCR was used to amplify the tDNA flanking genomic DNA sequence in ZS-1TN22803, and an 850-bp DNA fragment was obtained. The incomplete open reading frame (ORF) was predicted to encode a partial peptide of a peroxisomal biogenesis factor 6, a homolog to the PEX6 of Saccharomyces cerevisiae. Thus, we named it CmPEX6. As shown in the Southern blot analysis, only one copy of tDNA was inserted into the genome of mutant ZS-1TN22803, and CmPEX6 was likely a single-copy gene in C. minitans (see Fig. S1A and B in the supplemental material).
The coding region of CmPEX6 was obtained by amplifying C. minitans genomic DNA with specific primers. It was predicted that CmPEX6 in ZS-1 consisted of three exons and two introns (60 and 69 bp, respectively) and encoded a polypeptide with 1,399 amino acids (GenBank under accession number JN391412). The gene CmPEX6 in the ZS-1TN22803 mutant was disrupted by tDNA insertion at nucleotide position 1582 after the translational start code (ATG) (see Fig. S1C in the supplemental material). The amino acid sequence showed high homology to S. cerevisiae PEX6 (53% identity) and to genes in filamentous fungi, including A. nidulans (92% identity) and C. lagenarium (93% identity), with the conserved motifs of the AAA protein family signature at the C terminus (see Fig. S2A). Phylogenetic analysis revealed that this gene is clustered with other identified fungal PEX6 homologs (see Fig. S2B).
RT-PCR analysis showed that CmPEX6 was expressed at a high level over the time course of C. minitans growth from 48 to 96 h growing on PDA plates (see Fig. S1D in the supplemental material).
Replacement of CmPEX6 and complementation of ZS-1TN22803 mutant.
To test whether the disruption of CmPEX6 by tDNA insertion is responsible for the phenotypic changes in mutant ZS-1TN22803, replacement vector pCMPEX-3300 was transformed into ZS-1. A total of 158 transformants without resistance to G418 among 400 hygromycin-resistant transformants were obtained, and two transformants, DCmPEX6-39 and DCmPEX6-98, were selected for further analysis.
For complementation of the ZS-1TN22803 mutant, CmPEX6 with a PtrpC promoter was integrated into ZS-1TN22803 (see Fig. S3A in the supplemental material). A total of 120 putative complemented transformants were obtained, and one transformant, CmPEX6-com, was chosen for further characterization. RT-PCR analysis showed an expected 536-bp fragment of CmPEX6 in ZS-1 and CmPEX6-com but not in ZS-1TN22803, DCmPEX6-39, or DCmPEX6-98 (see Fig. S1D and S3B) when fungal RNA was extracted from mycelia grown on PDA plates at 96 h postincubation. The integration event of the CmPEX6 gene in the complemented transformant CmPEX6-com was further confirmed by Southern blotting in which two hybridization bands appeared when probed by the CmPEX6 fragment: one band represented the native CmPEX6 integrated with the tDNA insertion, and the other was the newly inserted CmPEX6 with a PtrpC promoter from the complementation vector (see Fig. S3C and D). Furthermore, Southern blot analysis demonstrated a targeted replacement of CmPEX6 in transformants DCmPEX6-39 and DCmPEX6-98, since there was one hybridization band when hph was used as a probe and no band when probed with a CmPEX6 fragment. These results confirm successful creations of both the replacement and the complemented mutants for CmPEX6 in C. minitans.
Involvement of CmPEX6 in conidiation and parasitism by C. minitans.
To decipher the roles of CmPEX6 in conidiation and parasitism to S. sclerotiorum, the phenotypic characteristics of the C. minitans strains were studied. There was no significant difference in colony morphology (Fig. 1A), mycelial growth rate (Fig. 1B), conidiation (Fig. 1C), or the ability to parasitize the sclerotia of S. sclerotiorum among the three CmPEX6-deficient mutants, DCmPEX6-39, DCmPEX6-98, and ZS-1TN22803 (Fig. 3). The conidial production of all three mutants was less than that of the wild-type strain ZS-1 by 1,000-fold (Fig. 1C and 2), and none of the mutants could parasitize the sclerotia of S. sclerotiorum (Fig. 3). Notably, the complemented mutant CmPEX6-com recovered all the defective characteristics of the mutant ZS-1TN22803 to an extent similar to that of wild-type ZS-1 (Fig. 1, 2, and 3). Taken together, these results demonstrated that CmPEX6 plays important roles in conidiation and parasitism by C. minitans.
Growth deficiency of CmPEX6 mutants on media amended with oleic acid.
Peroxisomes are the sites where lipids are broken down through fatty acid β-oxidation. Disruption of CmPEX6 is likely to cause dysfunction for fatty acid β-oxidation. To determine if CmPEX6 disruption mutants could use oleic acid, mutants were inoculated on MCD and PDA media amended with oleic acid. Results showed that the ZS-1TN22803 and CmPEX6 replacement mutants could grow on neither MCD medium, in which oleic acid was used as the sole carbon source, nor oleic acid-amended PDA, in which glucose was also available as a carbon source. In contrast, wild-type ZS-1 and the complemented mutants grew well on the same media (Fig. 5).
Fig 5.
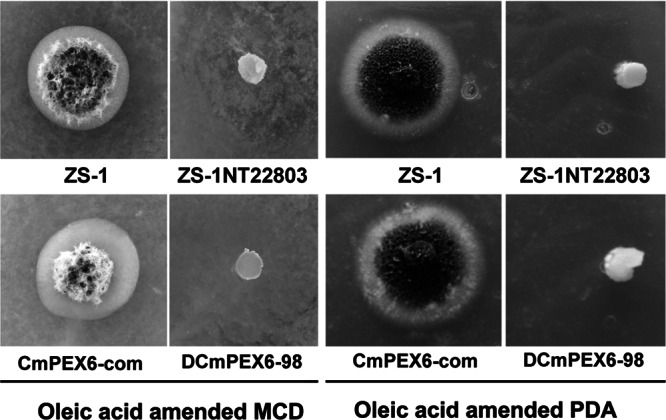
CmPEX6 mutants of C. minitans could not grow on media amended with oleic acid. ZS-1, ZS-1TN22803, DCmPEX6-98, or CmPEX6-com was placed on MCD and PDA media amended with 1% oleic acid at 20°C for 10 days. MCD medium was amended with oleic acid instead of dextrose as the sole carbon source, while PDA medium was amended with oleic acid.
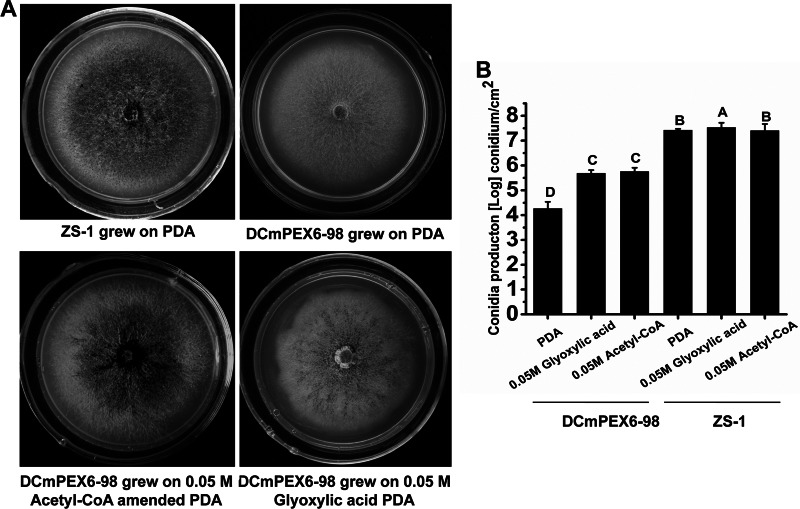
Conidiation of the CmPEX6 mutants partially restored by acetyl-CoA and glyoxylic acid.
It is well known that the β-oxidation of fatty acids producing acetyl-CoA is one of the main metabolic pathways in peroxisomes, and the acetyl-CoA-fueled glyoxylate pathway is a central component of peroxisomal function in plants and fungi (37). To confirm that this pathway is involved in conidiation by C. minitans, mutant DCmPEX6-98 was transferred onto PDA amended with acetyl-CoA or glyoxylic acid. At 15 days postincubation, conidiation by mutant DCmPEX6-98 was partially restored when the exogenous acetyl-CoA or glyoxylic acid was added to PDA medium at a final concentration of 50 mM (Fig. 6A). The conidial production by DCmPEX6-98 was increased from 1.7 × 104 conidia cm−2 on PDA to 4.6 × 105 or 5.3 × 105 conidia cm−2 on PDA amended with acetyl-CoA or glyoxylic acid, respectively (Fig. 6B). These results suggest that the CmPEX6 mutants are defective in utilization of fatty acids because of impaired β-oxidation.
Fig 6.
Partial restoration of conidiation by acetyl-CoA and glyoxylic acid. (A) Colony morphologies of ZS-1 and DCmPEX6-98 grown on PDA with or without exogenous 0.05 M acetyl-CoA or 0.05 M glyoxylic acid at 20°C for 15 days. (B) Conidial production of DCmPEX6-98 was restored partially. The vertical bars represent the standard errors based on three replicates. Columns noted by the same lowercase letter within each chart are not significantly different (P < 0.05) according to the least-significant-difference test.
Decreased production of H2O2 and NO in CmPEX6 mutants.
Peroxisomes are also the site for generating H2O2 and NO. To determine if the generation of H2O2 and NO was influenced by the disruption of the CmPEX6 gene, the amounts of H2O2 and NO in 3- and 4-day-old mycelia of mutants were monitored. Compared to levels for ZS-1, the amounts of H2O2 and NO in CmPEX6-defective mutants ZS-1TN22803, DCmPEX6-39, and DCmPEX6-98 at the two time points during incubation on PDA plates were significantly decreased. The production of H2O2 and NO in the mycelia of CmPEX6-com (3.2 and 22.7 μM) was slightly higher than that produced by ZS-1 (2.9 and 18.7 μM) (Fig. 7A and B), which correlates with possibly enhanced expression of the CmPEX6 gene in this complemented mutant, since expression of the CmPEX6 gene was driven by a trpC promoter. The results indicated that altered activities of CmPEX6 in C. minitans mutants affected the generation of H2O2 and NO during fungal growth.
Fig 7.
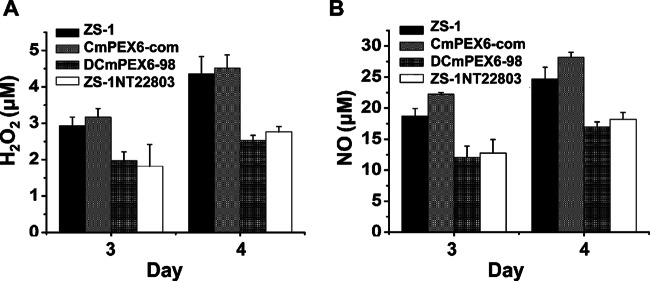
Generation of H2O2 (A) and NO (B) in mycelia of CmPEX6 mutants. ZS-1TN22803, DCmPEX6-98, CmPEX6-com, or ZS-1 was incubated on PDA at 20°C for 3 days. A hydrogen peroxide assay kit and nitrate/nitrite colorimetric assay kit (Beyotime, China) were used to measure the amount of H2O2 and NO per gram of fresh mycelia by following the kit protocol. The vertical bars represent the standard errors based on three replicates. Columns noted by the same lowercase letter within each chart are not significantly different (P < 0.05) according to the least-significant-difference test.
DISCUSSION
In this research, we cloned CmPEX6, encoding peroxisomal biogenesis factor 6, from C. minitans and demonstrated its essential role in conidiation and parasitism by C. minitans. Both tDNA insertional disruption and targeted replacement mutants of CmPEX6 were defective in conidiation and lost the ability to parasitize S. sclerotiorum. The CmPEX6 null mutants could produce pycnidial primordia on PDA plates, but most pycnidia were immature and defective in conidiation. Furthermore, the CmPEX6-disrupted mutants could not grow on media amended with oleic acid. The conidiation by the CmPEX6-disrupted mutants could be partially restored by adding acetyl-CoA or glyoxylic acid to the media, suggesting that β-oxidation of fatty acids in the mutants was impaired and that this function plays a crucial role in conidiation and parasitism.
Loss of pathogenicity caused by disruption of the PEX6 gene in plant-pathogenic fungi was reported in C. lagenarium, M. oryzae, and A. alternata (27, 30, 38). In C. lagenarium, clapex6 mutants produced small appressoria with severely reduced melanization that failed to form infectious hyphae. In M. oryzae, the Mgpex6 deletion mutant lacked appressorial melanin and was defective in host penetration and therefore was completely nonpathogenic. In A. alternata, the ΔAaPEX6 strains completely lost host-selective AK-toxin production and pathogenicity on susceptible Japanese pear leaves. Although the CmPEX6-disrupted mutants of C. minitans showed phenotypes associated with the impairment of melanization, the loss of parasitism by the CmPEX6-disrupted mutants ZS-1TN22803 and DCmPEX6 may not be due to the lack of melanization in the mutants, since other mutants with disruption of the melanin biosynthesis-associated polyketide synthase gene in C. minitans still maintain their pathogenicity to parasitize sclerotia of S. sclerotiorum (data not shown). In addition, we observed that the CmPEX6-disrupted mutants can live well on dead sclerotia and produce abundant conidia there (data not shown), suggesting that the CmPEX6 mutants are able to produce parasitism-associated fCWDEs that degrade sclerotia tissues for nutrient supply. Loss of pathogenicity for the CmPEX6-disrupted mutants on the host S. sclerotiorum suggests that the mutants are not able to overcome live host defenses during parasitism. Alternatively, C. minitans may require other virulence/pathogenicity factors to parasitize S. sclerotiorum. Notably, the generation of H2O2 and NO was significantly reduced in the CmPEX6 mutants. It is possible that these two reactive agents serve as virulence factors for C. minitans to parasitize its host. Further work is necessary to test this hypothesis.
Mutations in PEX genes can result in the absence of peroxisomes, abnormal peroxisomal structures, mistargeting of matrix proteins, and/or inability to respond to stimuli that cause increased numbers of peroxisomes (39). Peroxisomes are associated with β-oxidation of fatty acids (22, 23). It was believed that the disruption of PEX6 in C. lagenarium destroyed the β-oxidation pathway of fatty acids so that Δpex6 mutants could not utilize oleic acid as the sole carbon source (28). In this study, we showed that CmPEX6-disrupted mutants ZS-1TN22803, DCmPEX6-39, and DCmPEX6-98 could not grow on media amended with oleic acid whether or not the oleic acid was used as the sole carbon source, and the conidiation deficiency of the CmPEX6-disrupted mutants was partially restored by adding acetyl-CoA or glyoxylic acid to culture media. The results suggest that the β-oxidation of fatty acids associated with peroxisomes, along with other possible factors, plays an important role in conidiation. However, when growing on dead sclerotia or on carrot juice agar medium, the CmPEX6-disrupted mutants could produce many more conidia than on PDA amended with acetyl-CoA or glyoxylic acid (data not shown), suggesting that the dysfunction of peroxisomes in the CmPEX6-disrupted mutants can be compensated for by some unknown factor(s).
The NADPH oxidase-dependent superoxide generation by plants in response to microbial pathogen colonization is a well-known defense mechanism in plants (40). NADPH oxidases also play essential roles in development of filamentous fungi (41–43). In N. crassa, reactive oxygen species (ROS) are generated at the beginning of each morphogenetic step that occurs during asexual development (41). NoxA-generated ROS played an essential role in A. nidulans sexual differentiation but had no effect on asexual hyphal growth (42). Nox1 is required for sexual development, whereas Nox2 is involved in ascospore germination in P. anserina (43). Recently, several reports demonstrated essential roles of NO in conidiation of fungal species, including Blastocladiella emersonii (44), N. crassa (45), and C. minitans (9, 11). It is likely that NO signaling mediates fungal conidiation via a cGMP-dependent pathway and another still-unknown pathway (9, 11). Our results showed correlations between reduced generation of NO and H2O2 and conidiation deficiency in the CmPEX6-disrupted mutants, suggesting that cellular homeostasis of ROS and NO regulates conidiation in C. minitans.
PEX6 is essential for β-oxidation of fatty acids in fungi. As expected, the CmPEX6-disrupted DCmPEX6 mutants were unable to grow in MCD medium amended with oleic acid as the sole carbon source. However, we also found that the CmPEX6 mutants could not grow in oleic acid-amended PDA medium in which glucose is already available. Similarly, growth deficiency of yeast Δpex6 mutants was observed in media amended with oleic acid media containing other available carbon sources (46). The growth deficiency of both CmPEX6 mutants and yeast Δpex6 mutants in the presence of oleic acid could not be attributed to the shortage of carbon sources in the media but likely is due to the possible production of some inhibitory metabolites during β-oxidation of oleic acid or the direct cellular damage triggered by oleic acid metabolism in the CmPEX6-disrupted mutants.
Many studies have recently shown that NO is a widespread signaling molecule involved in regulation of many important physiological processes in fungi as well as in animals and plants (47). However, little is known about the origin of NO production at the cellular and subcellular levels in fungi (48). In plants, peroxisomes are organelles contributing to generation of superoxide (O2 · −) and nitric oxide radicals (24, 25). In this study, we found that the disruption of a peroxisomal CmPEX6 in C. minitans resulted in a significant decrease of NO generation in the CmPEX6-disrupted mutants, and complementation of the CmPEX6 mutation could completely restore NO generation. Our results established a potential involvement of peroxisomes in generation of NO in filamentous fungi.
In summary, we have identified a PEX6 homolog from C. minitans and have found that the CmPEX6 gene possesses critical functions in conidiation and parasitism by C. minitans. Our data suggest that the metabolism via β-oxidation of fatty acids and the production of NO and H2O2 are involved in fungal conidiation and parasitism.
Supplementary Material
ACKNOWLEDGMENTS
The research was financially supported by the National Natural Science Foundation of China (grant 309718833), the Special Fund for Agro-scientific Research in the Public Interest (grant 201103016), and the earmarked fund for the China Agriculture Research System (nycytx-00514).
We thank the anonymous reviewers for their constructive and helpful comments. We are grateful to Tom Hsiang, School of Environmental Sciences, University of Guelph, and Yangdou Wei, College of Art & Science, University of Saskatchewan, for editorial advice.
Footnotes
Published ahead of print 5 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00375-13.
REFERENCES
- 1. Bennett AJ, Leifer C, Whipps JM. 2006. Survival of Coniothyrium minitans associated with sclerotia of Sclerotinia sclerotiorum in soil. Soil Biol. Biochem. 38(1):164–172 [Google Scholar]
- 2. Boland GJ, Hall R. 1994. Index of plant hosts of Sclerotinia sclerotiorum. Can. J. Plant Pathol. 16:93–108 [Google Scholar]
- 3. Li GQ, Huang HC, Miao HJ, Erickson RS, Jiang DH, Xiao YN. 2006. Biological control of Sclerotinia diseases of rapeseed by aerial applications of the mycoparasite Coniothyrium minitans. Can. J. Plant Pathol. 114:345–355 [Google Scholar]
- 4. Smith SN, Prince M, Whipps JM. 2008. Characterization of Sclerotinia and mycoparasites Coniothyrium minitans interaction by microscale co-culture. Lett. Appl. Microbiol. 47:128–133 [DOI] [PubMed] [Google Scholar]
- 5. Whipps JM, Gerlagh M. 1992. Biology of Coniothyrium minitans and its potential for use in disease biocontrol. Mycol. Res. 96:897–907 [Google Scholar]
- 6. Yang L, Li GQ, Long YQ, Hong GP, Jiang DH, Huang HC. 2010. Effects of soil temperature and moisture on survival of Coniothyrium minitans conidia in central China. Biol. Control 55:27–33 [Google Scholar]
- 7. Adams TH, Wieser JK, Yu JH. 1998. Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 62:35–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lengeler KB, Davidson RC, D'Souza C, Ashima T, Shen WC, Wang P, Pan X, Waugh M, Heitman J. 2000. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64:746–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li B, Fu YP, Jiang DH, Xie JT, Cheng JS, Li GQ, Hamid MI, Yi XH. 2010. Cyclic GMP as a second messenger in the nitric oxide-mediated conidiation of the mycoparasite Coniothyrium minitans. Appl. Environ. Microbiol. 76:2830–2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Qin L, Gong XY, Xie JT, Jiang DH, Cheng JS, Li GQ, Huang JB, Fu YP. 2011. Phosphoribosylamidotransferase, the first enzyme for purine de novo synthesis, is required for conidiation in the sclerotial mycoparasite Coniothyrium minitans. Fungal Genet. Biol. 48:956–965 [DOI] [PubMed] [Google Scholar]
- 11. Gong XY, Fu YP, Jiang DH, Li GQ, Yi XH, Peng YL. 2007. L-arginine is essential for conidiation in the filamentous fungus Coniothyrium minitans. Fungal Genet. Biol. 44:1368–1379 [DOI] [PubMed] [Google Scholar]
- 12. Muthumeenakshi S, Sreenivasaprasad S, Rogers CW, Challen MP, Whipps JM. 2007. Analysis of cDNA transcripts from Coniothyrium minitans reveals a diverse array of genes involved in key processes during sclerotial mycoparasitism. Fungal Genet. Biol. 44:1262–1284 [DOI] [PubMed] [Google Scholar]
- 13. Rogers CW, Challen MP, Muthumeenakshi S, Sreenivasaprasad S, Whipps JM. 2008. Disruption of the Coniothyrium minitans PIF1 DNA helicase gene impairs growth and capacity for sclerotial mycoparasitism. Microbiology 154:1628–1636 [DOI] [PubMed] [Google Scholar]
- 14. Zeng FY, Gong XY, Hamid MI, Fu YP, Xie JT, Cheng JS, Li GQ, Jiang DH. 2012. A fungal cell wall integrity-associated MAP kinase cascade in Coniothyrium minitans is required for conidiation and mycoparasitism. Fungal Genet. Biol. 49:347–357 [DOI] [PubMed] [Google Scholar]
- 15. Han YC, Li GQ, Yang L, Jiang DH. 2011. Molecular cloning, characterization and expression analysis of a pacC homolog in the mycoparasite Conithyrium minitans. World J. Microbiol. Biotechnol. 27:381–391 [Google Scholar]
- 16. Ren L, Li GQ, Han YC, Jiang DH, Huang HC. 2007. Degradation of oxalic acid by Coniothyrium minitans and its effects on production and activity of β-1,3-glucanase of this mycoparasite. Biol. Control 43:1–11 [Google Scholar]
- 17. Yang R, Han YC, Li GQ, Jiang DH, Huang HC. 2008. Effects of ambient pH and nutritional factors on antifungal activity of the mycoparasite Coniothyrium minitans. Biol. Control 44:116–127 [Google Scholar]
- 18. Lazarow PB, Fujiki Y. 1985. Biogenesis of peroxisomes. Annu. Rev. Cell Biol. 1:489–530 [DOI] [PubMed] [Google Scholar]
- 19. Lin Y, Sun L, Nguyen LV, Rachubinski RA, Goodman HM. 1999. The Pex16p homolog SSE1 and storage organelle formation in Arabidopsis seeds. Science 284:328–330 [DOI] [PubMed] [Google Scholar]
- 20. Muller WH, van der Krift TP, Krouwer AJ, Wosten HA, van der Voort LH, Smaal EB, Verkleij AJ. 1991. Localization of the pathway of the penicillin biosynthesis in Penicillium chrysogenum. EMBO J. 10:489–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Petriv OI, Tang L, Titorenko VI, Rachubinski RA. 2004. A new definition for the consensus sequence of the peroxisome targeting signal type 2. J. Mol. Biol. 341:119–134 [DOI] [PubMed] [Google Scholar]
- 22. Titorenko VI, Rachubinski RA. 2001. The life cycle of the peroxisome. Nat. Rev. Mol. Cell Biol. 2:357–368 [DOI] [PubMed] [Google Scholar]
- 23. van den Bosch H, Schutgens RBH, Wanders RJA, Tager JM. 1992. Biochemistry of peroxisomes. Annu. Rev. Biochem. 61:157–197 [DOI] [PubMed] [Google Scholar]
- 24. Corpas FJ, Barroso JB, Carreras A, Quiros M, Leon AM, Romero-Puertas MC, Esteban FJ, Valderrama R, Palma JM, Sandalio LM, Gomez M, del Rio LA. 2004. Cellular and subcellular localization of endogenous nitric oxide in young and senescent pea plants. Plant Physiol. 136:2722–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Corpas FJ, Barroso JB, del Río LA. 2001. Peroxisomes as a source of reactive oxygen species and nitric oxide signal molecules in plant cells. Trends Plant Sci. 1:145–150 [DOI] [PubMed] [Google Scholar]
- 26. Peraza-Reyes L, Zickler D, Berteaux-Lecellier V. 2008. The peroxisome RING-finger complex is required for meiocyte formation in the fungus Podospora anserine. Traffic 9:1998–2009 [DOI] [PubMed] [Google Scholar]
- 27. Asakura M, Okuno T, Takano Y. 2006. Multiple contributions of peroxisomal metabolic function to fungal pathogenicity in Colletotrichum lagenarium. Appl. Environ. Microbiol. 72:6345–6354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kimura A, Takano Y, Furusawa I, Okuno T. 2001. Peroxisomal metabolic function is required for appressorium-mediated plant infection by Colletotrichum lagenarium. Plant Cell 13:1945–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang ZY, Soanes DM, Kershaw MJ, Talbot NJ. 2007. Functional analysis of lipid metabolism in Magnaporthe grisea reveals a requirement for peroxisomal fatty acid β-oxidation during appressorium-mediated plant infection. Mol. Plant Microbe Interact. 20:475–491 [DOI] [PubMed] [Google Scholar]
- 30. Imazaki A, Tanaka A, Harimoto Y, Yamamoto M, Akimitsu K, Park P, Tsuge T. 2010. Contribution of peroxisomes to secondary metabolism and pathogenicity in the fungal plant pathogen Alternaria alternata. Eukaryot. Cell 9:682–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Veenhuis M, Nordbring-Hertz B, Harde W. 1984. Occurrence, characterization and development of two different types of microbodies in the nematophagous fungus Arthrobotrys oligospora. FEMS Microbiol. Lett. 24:31–38 [Google Scholar]
- 32. Wolf J, Schliebs W, Erdmann R. 2010. Peroxisomes as dynamic organelles: peroxisomal matrix protein import. FEBS J. 277:3268–3278 [DOI] [PubMed] [Google Scholar]
- 33. Li M, Gong XY, Zheng J, Jiang DH, Fu YP, Hou MS. 2005. Transformation of Coniothyrium minitans, a parasite of Sclerotinia sclerotiorum, with Agrobacterium tumefaciens. FEMS Microbiol. Lett. 243:323–329 [DOI] [PubMed] [Google Scholar]
- 34. Cheng JS, Jiang DH, Fu YP, Li GQ, Whipps JM. 2003. Production, survival and efficacy of Coniothyrium minitans conidia produced in shaken liquid culture. FEMS Microbiol. Lett. 227:127–131 [DOI] [PubMed] [Google Scholar]
- 35. Xie J, Wei DM, Jiang DH, Fu YP, Li GQ, Ghabrial SA, Peng YL. 2006. Characterization of debilitation-associated mycovirus infecting the plant-pathogenic fungus Sclerotinia sclerotiorum. J. Gen. Virol. 87:241–249 [DOI] [PubMed] [Google Scholar]
- 36. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 37. Zimmerman R, Neupert W. 1980. Biogenesis of glyoxysomes. Synthesis and intracellular transfer of isocitrate lyase. Eur. J. Biochem. 112:225–233 [DOI] [PubMed] [Google Scholar]
- 38. Ramos-Pamplona M, Naqvi NI. 2006. Host invasion during rice-blast disease requires carnitine-dependent transport of peroxisomal acetyl-CoA. Mol. Microbiol. 61:61–75 [DOI] [PubMed] [Google Scholar]
- 39. Hynes MJ, Murray SL, Khew GS, Davis MA. 2008. Genetic analysis of the role of peroxisomes in the utilization of acetate and fatty acids in Aspergillus nidulans. Genetics 178:1355–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lamb C, Dixon RA. 1997. The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48:251–275 [DOI] [PubMed] [Google Scholar]
- 41. Hansberg W, de Groot H, Sies H. 1993. Reactive oxygen species associated with cell differentiation in Neurospora crassa. Free Radic. Biol. Med. 14:287–293 [DOI] [PubMed] [Google Scholar]
- 42. Lara-Ortiz T, Riveros-Rosas H, Aguirre J. 2003. Reactive oxygen species generated by microbial NADPH oxidase NoxA regulate sexual development in Aspergillus nidulans. Mol. Microbiol. 50:1241–1255 [DOI] [PubMed] [Google Scholar]
- 43. Malagnac F, Lalucque H, Lepere G, Silar P. 2004. Two NADPH oxidase isoforms are required for sexual reproduction and ascospore germination in the filamentous fungus Podospora anserina. Fungal Genet. Biol. 41:982–997 [DOI] [PubMed] [Google Scholar]
- 44. Vieira AL, Linares E, Augusto O, Gomes SL. 2009. Evidence of a Ca2+-NO-cGMP signaling pathway controlling zoospore biogenesis in the aquatic fungus Blastocladiella emersonii. Fungal Genet. Biol. 46:575–584 [DOI] [PubMed] [Google Scholar]
- 45. Cano-Domínguez N, Álvarez-Delfín K, Hansberg W, Aguirre J. 2008. NADPH oxidases NOX-1 and NOX-2 require the regulatory subunit NOR-1 to control cell differentiation and growth in Neurospora crassa. Eukaryot. Cell 7:1352–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lockshon D, Surface LF, Kerr EO, Kaeberlein M, Kennedy BK. 2007. The sensitivity of yeast mutants to oleic acid implicates the peroxisome and other processes in membrane function. Genetics 175:77–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Durner J, Andrew JG, Jonathan SS, Glazebrook J. 1999. Ancient origins of nitric oxide signaling in biological systems. Proc. Natl. Acad. Sci. U. S. A. 96:14206–14207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Song NK, Jeong CS, Choi HS. 2000. Identification of nitric oxide synthase in Flammulina velutipes. Mycologia 92:1027–1032 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.