Abstract
Human commensal bacteria do not normally cause any diseases. However, in certain pathological conditions, they exhibit a number of curious behaviors. In HIV infection, these bacteria exhibit bidirectional relationships: whereas they cause opportunistic infections based on immunological deterioration, they also augment HIV replication, in particular, viral replication from latently infected cells, which is attributable to the effect of butyric acid produced by certain anaerobic bacteria by modifying the state of chromatin. Here, we review recent evidence supporting the contributory role of such endogenous microbes in disrupting HIV latency and its potential link to the clinical progression of AIDS.
INTRODUCTION
We begin this minireview by discussing microbes and their products in relation to human immunodeficiency virus (HIV) latency. HIV attacks the immune system, the body's defense against infectious organisms or other illnesses, creating vulnerability to various infections. Thus, microbial coinfection contributes to the course of disease progression of HIV infection and the development of AIDS-related deaths (1–4).
A number of bacteria are normal residents in body cavities surfaced by mucous membranes, including the oral cavity, gut, and vagina. As they endogenously colonize such niches, they seldom cause illness, except when the host's immunity is impaired. Recent evidence indicates that the mucosal surfaces of both the gut and vaginal cavities are predominant sites of HIV replication (5–7). These mucosal sites are densely populated with CD4+ T cells, the primary target of the virus (1, 6). While mucosal sites contain as many CD4 T cells as other sites (for example, lymph nodes) or fewer of them, they are enriched with activated CD4 T cells that express HIV coreceptors such as CCR5 and α4β7 (8). Also, Th17 cells, a subset of CD4 T cells producing interleukin-17 (IL-17), home to the gut and have been shown to be preferentially infected with HIV (9, 10). The profound loss of these cells has been associated with disease progression in both simian immunodeficiency virus (SIV) and HIV infections (11–14). Estes et al. demonstrated the presence of not only lipopolysaccharide but also Escherichia coli in the colonic lamina propria and lymph nodes of chronically infected rhesus macaques (15). In addition, Dillon et al. (16) presented evidence suggesting the preferential infection of IL-17-producing intestinal CD T cells by HIV and the enhancement of HIV productive infection in the presence of E. coli. Meanwhile, Ahmed et al. reported that certain commensal bacteria that preferentially stimulate Toll-like receptor 4 (TLR4) suppressed HIV-1 expression, whereas some with enhancing effects stimulated TLR2 (17). These findings indicate that HIV-associated impairment of epithelial barrier integrity and CD4 T cell depletion are most likely involved in a systemic microbial translocation that may give rise to an immune activation that could drive HIV either further or out of latency.
As in the gut, commensal organisms thrive in the oral cavity. Candida albicans, a polymorphic fungus that is a commensal microbe in the healthy individual, could express its pathogenic potential as HIV infection progresses because of the decay of fungal containment on the oral epithelium associated with the loss of Th17 cells (18). We have also explored the effects of certain anaerobes that are part of the oral or gut flora in reactivating latent HIV, as most of them produce butyric acid, the oldest known histone deacetylase (HDAC) inhibitor (HDACi), under anaerobic conditions. Initially, we demonstrated that the culture supernatant of Porphyromonas gingivalis, a periodontogenic bacterium, could induce the expression of quiescent HIV from latently infected T and macrophage cell lines accompanied by the induction of a hyperacetylation of histones H3 and H4 (19). We found that among the various virulence factors produced by this Gram-negative anaerobe, a high concentration of butyric acid in the culture supernatant and the augmenting effect of HIV replication were abolished upon its removal. To confirm these observations, we attempted to comprehensively examine the human resident butyric acid-producing bacteria from various tissues (20). We found that bacterial culture supernatants of P. gingivalis, Fusobacterium nucleatum, Clostridium cochlearium, and Anaerococcus tetradius increased histone acetylation and efficiently induced HIV gene expression from the latent state. Interestingly, these organisms (except P. gingivalis) produced the largest amounts of butyric acid in culture supernatants collected from among representative anaerobic organisms found in the oral, oral and gut, gut, and vaginal cavities, respectively (20). Furthermore, chromatin immunoprecipitation analysis revealed loss of HDAC1–AP-4 (transcriptional repressor complex of the HIV-1 provirus) occupancy at the long terminal repeat (LTR), whereas RNA polymerase II recruitment was increased. These effects were correlated with the presence of elevated levels of butyric acid and were not observed in culture supernatants from non-butyrate-producing bacteria. In addition, González et al. (21) reported that extracts of P. gingivalis and F. nucleatum enhanced HIV reactivation in monocytes/macrophages via TLR2 and TLR9 activation. The same group of investigators recently demonstrated that HIV reactivation in monocytes/macrophages by the oral commensal Streptococcus gordonii is Tat dependent and that it appears to involve NF-κB activation (22). Moreover, TLR5 stimulation could sufficiently induce reactivation of latent HIV in CD4+ T lymphoid cells. It was also reported earlier that P. gingivalis could upregulate CCR5 expression in oral keratinocytes, thus facilitating the transfer of infectious HIV-1 to permissive cells such as macrophages (23). Taken together, these findings support the hypothesis that periodontal and other commensal pathogens, such as butyrate-producing anaerobes, play contributory roles in the clinical progression of AIDS.
The vaginal microflora consists of different bacterial species and some anaerobic bacteria, such as A. tetradius, Anaerococcus vaginalis, Peptoniphilus asaccharolyticus, and Anaerococcus lactolyticus, have been reported to induce HIV replication. Prior studies have shown the correlation of HIV infection with an abnormal vaginal flora morphology and bacterial vaginosis (BV), with the latter considered a risk factor for further acquisition of the virus (24–27). Interestingly, these bacteria produce significant amounts of butyric acid under anaerobic conditions. In vivo, as well as in vitro, studies have demonstrated that BV-associated conditions (e.g., Candida vaginitis and herpes simplex virus infection) could also influence HIV replication and genital tract shedding (24, 28, 29). Interestingly, Spiegel et al. (30) noted the increased levels of butyrate, succinate, acetate, and propionate and decreased lactic acid levels in nonspecific vaginitis. Butyric acid production was caused by Peptococcus (Anaerococcus) bacterial species. It is noted that butyric acid, but not other short-chain fatty acids, is responsible for these effects. Moreover, others have shown that genital mycoplasmas stimulate tumor necrosis factor alpha (TNF-α) production by a murine macrophage cell line. Jiang et al. (31) recently reported the association of HIV infection with impaired regulation of innate defenses (e.g., human beta defensins) at mucosal sites and increased bacterial colonization of the female genital tract.
HIV LATENCY AS A CRITICAL OBSTACLE TO THE ERADICATION OF HIV INFECTION
Although antiretroviral (ARV) therapy (ART) has been efficient in reducing the morbidity and mortality rates of people living with HIV/AIDS, eradication of the virus has not yet been achieved. The major roadblock in the treatment of HIV is the persistence of latent HIV-1 reservoirs that are constant sources of rebound virus upon cessation of ART or treatment failure and the emergence of drug-resistant viral clones.
Maintenance of HIV latency is a multifactorial event involving several factors, including (i) the chromatin environment at the site of integration, (ii) transcriptional interference, (iii) a lack of host transcription factors needed for viral gene expression, (iv) the presence of host transcriptional repressors, and (v) epigenetic silencing of viral transcription (32–36). The elucidation of molecular mechanisms by which HIV type 1 (HIV-1) persists within infected cells provides a basis for a new therapeutic approach aimed toward combining HIV gene expression therapy and an ARV regimen. In general, it involves the use of agents that will bring latent HIV out of hiding in cells with the hope that ARV or the adaptive immune response will block new infection events (32, 37–39). Disruption of chromatin organization at the HIV-1 LTR promoter sets a threshold for transcription factor activation and eventually reversal of the state of chromatin that is responsible for the repression of HIV proviral DNA within the host genome. Interestingly, proof-of-concept studies using various combinations of HDAC or histone methyltransferase (HMT) inhibitors showed that purging of latent proviruses from latently infected cells is practically attainable (40, 41).
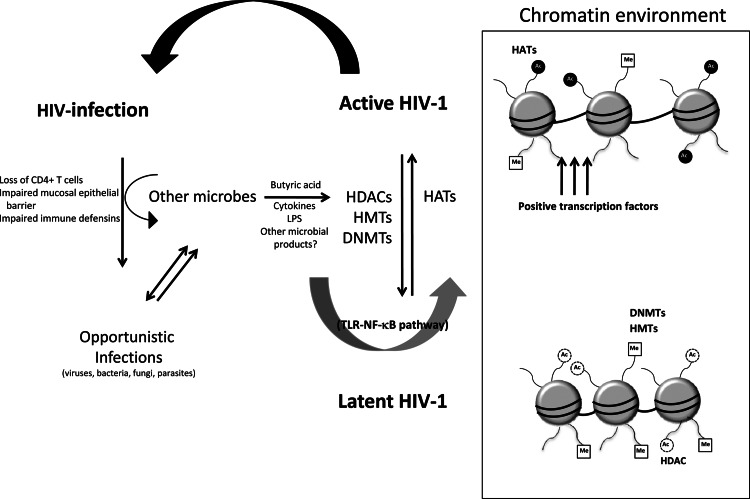
The progression to AIDS is influenced by host inflammatory responses and coinfection with other pathogens such as viruses, fungi, parasites, and bacteria (3, 20, 42–45). In particular, opportunistic infection frequently sets in when an HIV-infected individual is immunocompromised. AIDS progression is usually accompanied by the action of proinflammatory cytokines associated with inflammatory responses that are thought to be perpetuated by the cycle of immune activation brought about by opportunistic infections. AIDS-defining events are most likely consequences of cyclical host-microbe interactions within HIV-1-infected individuals. In Fig. 1, we depict one such condition caused by commensal bacteria of gastrointestinal and vaginal tissues. Interestingly, the gastrointestinal and vaginal mucosal tissues are major sites of HIV replication and amplification (6, 7). Impaired mucosal integrity and innate mechanisms of defense against gut microbes result from HIV infection because of immune activation brought about by, but not limited to, the leakage of microbial products from the gut or skewing of homeostatic responses to inflammatory stimuli (11, 46–48). A recent study of SIV-infected rhesus macaques and African green monkeys revealed the importance of the microbiome's composition during inflammation and AIDS progression and indicated a significant contribution of the endogenous microbial flora to the course of HIV infection (49).
Fig 1.
Causal association of microbial interaction with HIV-1 latency and AIDS progression. HIV-1 infection weakens the immune system, making the body susceptible to opportunistic pathogens. CD4+ T cell depletion leads to impaired mucosal epithelial barrier integrity, allowing microbial translocation. The influx of circulating microbial products is associated with a systemic hyperimmune activation (e.g., through the TLR–NF-κB pathway) that may aggravate HIV-1 disease and enhance its progression. Recently, the bacterial metabolite butyric acid, which is produced under anaerobic conditions, has been shown to reactivate latent HIV-1 by promoting dissociation of the HDAC1–AP-4 repressor complex and hyperacetylation of histones, indicating its potential involvement in the progression of AIDS (see text for details). HATs, histone acetyltransferases; Me, methyl; Ac, acetyl; LPS, lipopolysaccharide.
In the context of HIV latency, it has been clearly shown that microbial interactions can regulate the epigenetic status of HIV-1 proviral DNA within the genome of infected cells and its transcriptional competence. We previously reported that human resident butyric acid-producing bacteria from various tissues could reactivate latent HIV-1 proviruses (19, 20). These observations indicate that microbial products or changes in the composition of the microbiota could influence the progression of the disease. This review provides an overview of the interaction between some endogenous bacterial flora occupying niches in the human body and HIV persistence that may have serious implications for the pathogenesis of HIV infection.
EPIGENETIC REGULATION OF HIV GENE EXPRESSION
The chromatin organization and epigenetic regulation of the HIV-1 promoter are critical in establishing and maintaining HIV latency. Immediately downstream of the transcription start site of the HIV-1 LTR, a repressive nucleosome (nuc-1) exists in its hypoacetylated state during latency. Transcriptional reactivation can be facilitated by enzyme complexes that covalently modify tails of the core histones in nucleosomes, promoting changes in chromatin structure and allowing the recruitment of positive transcription factors to the LTR for full transcription (Fig. 1, box) (50–53).
HDACs and HMTs are two classes of enzymes closely linked to the transcriptional activation and repression of HIV. (i) HDACs.
HDACs act to repress transcription by catalyzing the hydrolytic removal of acetyl groups from histone lysine residues (54). Moreover, HDACs can interact with nonhistone proteins such as p53 and NF-κB and form multiprotein complexes whose other components help HDAC carry out its functions (55, 56). Currently, there are 18 known mammalian HDACs, which are phylogenetically divided into four groups. Class I HDACs (HDAC1, -2, -3, and -8) are related to yeast HDAC Rpd3 and are often present in multiprotein complexes harboring Nurd and SIN3 corepressors. Class II HDACs are homologous to the yeast HDAC HDA1 and are subdivided into classes IIa (HDAC4, -5, -7, and -9) and IIb (HDAC6 and -10); Class III HDACs represent the NAD-dependent sirtuins and are homologs of yeast Sir2. Class IV includes HDAC11, a constitutively nuclear protein displaying properties of both classes I and II (35).
Apart from acting on histones, HDACs can mediate HIV silencing through its physical recruitment to the HIV-1 LTR. In fact, we previously reported the negative regulation of HIV transcription by AP-4 through the recruitment of HDAC1 to the promoter, as well as by masking the binding of TATA-binding protein to the TATA box (57). Other HIV-1 LTR-bound transcription factors found to directly recruit HDAC1 and then accomplish gene silencing include the YY-1/LBP-1 complex, the NF-κB (p50) homodimer, the Sp1/Myc complex, and C promoter binding factor 1 (58–61). HDAC3 has also been reported to be present at the HIV promoter and cause transcriptional repression (62–64). Previously, we showed that the HIV-1-reactivating potential of our novel HDACi NCH-51 was abolished when the Sp1 sites at the LTR were mutated or when Sp1 expression was knocked down by small interfering RNA (siRNA) or by treatment with the Sp1 inhibitor mithramycin A (65). These data indicate that Sp1 is responsible for the recruitment of HDAC1 to the LTR. It is possible that posttranscriptional modification of Sp1 is involved. As Sp1 is widely believed to be a transcriptional activator, it is to be further elucidated in what biochemical or biological context Sp1 recruits HDAC1 and is converted to a transcriptional repressor. Further studies are needed to depict the molecular mechanism of this phenotypic transcription factor conversion.
(ii) HMTs.
Generally, HMTs catalyze the lysine methylation of histones, which can be linked to either transcriptional activation or repression (66). Methylation of histone 3 at lysine 4 (H3K4), H3K36, and H3K79 has been associated with gene activation, whereas H3K9 and H3K27 methylation has been correlated with gene repression (67). So far, latent HIV-1 proviruses have been reported to carry histones that are either trimethylated or dimethylated at Lys 9/27 or Lys 9, respectively (68–72). These repressive marks do not affect DNA or histone interactions but serve to recruit effector proteins that influence the transcriptional state of chromatin.
SUV39H1 trimethylates histone H3 at Lys 9 and mediates repression of the HIV-1 LTR in microglial cells by interacting with HP1γ (68). Chicken ovalbumin upstream promoter transcription factor-interacting protein 2 (CTIP2) forms a multienzymatic chromatin-modifying complex containing SUV39H1, HP1, HDAC1, and HDAC2 to establish a repressive heterochromatin environment that leads to HIV-1 silencing (69). Recently, it was shown that recruitment of the histone demethylase LSD1 at the HIV-1 promoter was associated with both epigenetic marks H3K4me3 and H3K9me3 and acted synergistically with CTIP2 to repress HIV transcription and viral replication (73). Meanwhile, we previously found the involvement of G9a in the maintenance of proviral latency by promoting repressive dimethylation at H3K9 in cell lines where HIV-1 proviral DNA is latently present (71). Either knockdown of G9a with siRNA or G9a inhibition with the compound BIX01294 could successfully induce activation of transcription from latent HIV provirus. Furthermore, Friedman et al. reported the contribution of EZH2, the enzyme that catalyzed the trimethylation of H3K27, in silencing HIV proviruses (74). EZH2 is part of multimeric protein complex PRC2, which serves as a recruiting platform for DNA methyltransferase 1 (DNMT1), the SWI/SNF component bromodomain-containing protein Brd7, and HDACs (75, 76). These additional components are known to be associated with the maintenance of proviral latency. For example, DNMT1 mediates the methylation of the HIV-1 LTR and reinforces HIV-1 latency (75, 77, 78). Although considered weak stimulators of the HIV-1 LTR, DNA methylation inhibitors such as 5-aza-2′-deoxycytidine could reactivate latent HIV-1 provirus and had synergistic reactivation effects with NF-κB activators prostratin and TNF-α (77, 78).
POTENTIAL THERAPEUTIC INTERVENTIONS AIMED TO DECREASE HIV RESERVOIRS OR ERADICATE HIV
The disruption of HIV latency has been proposed as part of a strategy to eradicate HIV infection. This is carried out by producing an environment permissive for transcription by altering the degree of acetylation and methylation of histones and nonhistone molecules. HDACi compounds such as trichostatin A, trapoxin, valproic acid, sodium butyrate, or vorinostat (VOR; also known as suberoylanilide hydroxamic acid [SAHA]) have the ability to disrupt latent HIV infection in both cell culture models and ex vivo assays using cells from HIV-1-infected patients or latently infected cell lines (36, 79–85). Archin et al. (83) demonstrated that a single dose of VOR increased biomarkers of cellular acetylation and simultaneously increased HIV RNA expression in resting CD4+ cells from HIV-1-infected patients. Also, we have previously demonstrated that a novel HDACi compound, NCH-51, that has better pharmacological properties than SAHA could activate latent HIV-1 gene expression with minimal cytotoxicity through Sp1 sites (65). Meanwhile, methylation inhibitors like adenosine periodate could be employed to globally inhibit protein methyltransferase activity and induce virus production (86). The EZH2-specific HKMT inhibitor 3-deazaneplanocin A (74) and the SUV39H1 inhibitor chaetocin could reactivate latent proviruses and could act cooperatively with HDACi compounds to activate HIV transcription, indicating that combination therapy reverses epigenetic silencing more efficiently (41, 74). Moreover, Bouchat et al. (40), for the first time, demonstrated the recovery of HIV from ex vivo cultures of resting CD4+ T cells isolated from HIV-1-infected individuals undergoing highly active ART by chaetocin or the G9a inhibitor BIX01294. Likewise, they observed that the reactivation activity of one HMT inhibitor was intensified when it was combined with either SAHA or prostratin. Although these findings strongly indicate the feasibility of this therapeutic approach, it has not been clearly observed that the use of such compounds led to a substantial reduction in the frequency of replication-competent cells among the resting CD4+ T cells examined (83, 87, 88).
CONCLUDING REMARKS
The commensal microbiota populating all mucosal surfaces of the body exerts its beneficial effect by offering nutritional and physiological advantages in exchange for a nutrient-rich habitat within the host. There appears to exist an interesting interplay between the host and these microbes. Just as these microorganisms help shape the mucosal immune responses, the host also shapes the microbial community by modulating both the innate and adaptive immune responses (89).
HIV infection is accompanied by functional immunodeficiency and loss of mucosal barrier integrity, allowing microbial translocation and driving disease progression (1, 90, 91). A number of HIV-related opportunistic infections have been documented while other potential microbial factors promoting AIDS progression have slowly been unraveled in the past years. It is evident that a delicate balance between commensal microbiota and immune homeostasis is critical in the persistence and progression of HIV infection. The treatment of infections associated with AIDS should conceptually slow down AIDS progression, suggesting that the prevention and treatment of such non-HIV infections might be subsidiary but significant targets for AIDS therapy (1, 92). Further identification of the components of the commensal microbiota and elucidation of the underlying mechanisms through which these microbes/microbial products potentiate or interfere with HIV-1 pathogenesis could be very important in the design of interventions against HIV/AIDS.
Biographies

Ann Florence B. Victoriano obtained her Master of Science in Public Health (Medical Microbiology) at the University of the Philippines Manila, where she was exposed to HIV studies while working as a research assistant. In 2008, she completed her Ph.D. (Doctor of Medical Sciences) on a Monbukagakusho scholarship at the Nagoya City University Graduate School of Medical Sciences (NCU-GSMS) in Japan under the supervision of Takashi Okamoto. In 2009, she got a three-year Japan Foundation for AIDS Prevention postdoctoral fellowship and continued her work in Dr. Okamoto's laboratory investigating the epigenetic mechanisms that regulate HIV latency, screening of small-molecule drugs directed at novel HIV targets, and transcriptional regulation of HIV expression. She currently holds a position as an Assistant Professor at the same institution.

Kenichi Imai received his Bachelor's and Ph.D. degrees in Dental Science from the Asahi University School of Dentistry and the Mekai University School of Dentistry, respectively. He then joined the group of Takashi Okamoto as a postdoctoral fellow to pursue his interest in the transcriptional regulation and silencing of HIV. He was appointed an Assistant Professor at NCU-GSMS and the Nihon University School of Dentistry (NUSD) in 2008 and 2010, respectively. Dr. Imai has been a recipient of a Young Investigator award from the Society for Microbial Ecology and Disease and the ECC Yamaguchi Memorial AIDS Award from the Japanese AIDS Society for his work on HIV. Currently, he holds a position as an Associate professor at NUSD and studies host-microbe interactions and infectious diseases with a focus on periodontitis-associated/other oral bacteria, Epstein-Barr virus, and HIV latency.

Takashi Okamoto obtained his degrees of Medical Doctor (MD) and Doctor of Medical Sciences (equivalent to a Ph.D.) at the School of Medicine of Keio University, Tokyo, Japan. He then became a clinical associate at the Department of Internal Medicine at Keio University and was a postdoctoral (Fogarty) fellow at the National Cancer Institute, NIH, Bethesda, MD, from 1983 to 1986 under the supervision of Dr. Flossie Wong-Staal and Dr. Robert C. Gallo. From 1986 to 1993, he joined the Virology Division of the National Cancer Center Research Institute in Tokyo, Japan. From 1993 to the present, he has been a professor and chairman of the Department of Molecular and Cellular Biology at the Nagoya City University Graduate School of Medical Sciences. He received a Tamiya Memorial Award for Cancer Research and an Award from the Japanese Rheumatology Association. His current scientific interests include transcriptional control of HIV replication with specific focuses on Tat and NF-κB. He is also interested in other pathologies where NF-κB plays major roles, such as cancer, leukemia, rheumatoid arthritis, and autoimmunity.
Footnotes
Published ahead of print 24 April 2013
REFERENCES
- 1. Sandler NG, Douek DC. 2012. Microbial translocation in HIV infection: causes, consequences and treatment opportunities. Nat. Rev. Microbiol. 10:655–666 [DOI] [PubMed] [Google Scholar]
- 2. Hunt PW. 2012. HIV and inflammation: mechanisms and consequences. Curr. HIV/AIDS Rep. 9:139–147 [DOI] [PubMed] [Google Scholar]
- 3. Báfica A, Scanga CA, Schito M, Chaussabel D, Sher A. 2004. Influence of coinfecting pathogens on HIV expression: evidence for a role of Toll-like receptors. J. Immunol. 172:7229–7234 [DOI] [PubMed] [Google Scholar]
- 4. Blanchard A, Montagnier L, Gougeon ML. 1997. Influence of microbial infections on the progression of HIV disease. Trends Microbiol. 5:326–331 [DOI] [PubMed] [Google Scholar]
- 5. Kotler DP. 2005. HIV infection and the gastrointestinal tract. AIDS 19:107–117 [DOI] [PubMed] [Google Scholar]
- 6. Belyakov IM, Ahlers JD. 2012. Mucosal immunity and HIV-1 infection: applications for mucosal AIDS vaccine development. Curr. Top. Microbiol. Immunol. 354:157–179 [DOI] [PubMed] [Google Scholar]
- 7. Brenchley JM, Douek DC. 2008. HIV infection and the gastrointestinal immune system. Mucosal Immunol. 1:23–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kader M, Wang X, Piatak M, Lifson J, Roederer M, Veazey R, Mattapallil JJ. 2009. Alpha4(+)beta7(hi)CD4(+) memory T cells harbor most Th-17 cells and are preferentially infected during acute SIV infection. Mucosal Immunol. 2:439–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Elhed A, Unutmaz D. 2010. Th17 cells and HIV infection. Curr. Opin. HIV AIDS 5:146–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dandekar S, George MD, Baumler AJ. 2010. Th17 cells, HIV and the gut mucosal barrier. Curr. Opin. HIV AIDS 5:173–178 [DOI] [PubMed] [Google Scholar]
- 11. Cecchinato V, Trindade CJ, Laurence A, Heraud JM, Brenchley JM, Ferrari MG, Zaffiri L, Tryniszewska E, Tsai WP, Vaccari M, Parks RW, Venzon D, Douek DC, O'Shea JJ, Franchini G. 2008. Altered balance between Th17 and Th1 cells at mucosal sites predicts AIDS progression in simian immunodeficiency virus-infected macaques. Mucosal Immunol. 1:279–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chege D, Sheth PM, Kain T, Kim CJ, Kovacs C, Loutfy M, Halpenny R, Kandel G, Chun TW, Ostrowski M, Kaul R. 2011. Sigmoid Th17 populations, the HIV latent reservoir, and microbial translocation in men on long-term antiretroviral therapy. AIDS 25:741–749 [DOI] [PubMed] [Google Scholar]
- 13. Favre D, Lederer S, Kanwar B, Ma ZM, Proll S, Kasakow Z, Mold J, Swainson L, Barbour JD, Baskin CR, Palermo R, Pandrea I, Miller CJ, Katze MG, McCune JM. 2009. Critical loss of the balance between Th17 and T regulatory cell populations in pathogenic SIV infection. PLoS Pathog. 5:e1000295 doi: 10.1371/journal.ppat.1000295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Macal M, Sankaran S, Chun TW, Reay E, Flamm J, Prindiville TJ, Dandekar S. 2008. Effective CD4+ T-cell restoration in gut-associated lymphoid tissue of HIV-infected patients is associated with enhanced Th17 cells and polyfunctional HIV-specific T-cell responses. Mucosal Immunol. 1:475–488 [DOI] [PubMed] [Google Scholar]
- 15. Estes JD, Harris LD, Klatt NR, Tabb B, Pittaluga S, Paiardini M, Barclay GR, Smedley J, Pung R, Oliveira KM, Hirsch VM, Silvestri G, Douek DC, Miller CJ, Haase AT, Lifson J, Brenchley JM. 2010. Damaged intestinal epithelial integrity linked to microbial translocation in pathogenic simian immunodeficiency virus infections. PLoS Pathog. 6:e1001052 doi: 10.1371/journal.ppat.1001052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dillon SM, Manuzak JA, Leone AK, Lee EJ, Rogers LM, McCarter MD, Wilson CC. 2012. HIV-1 infection of human intestinal lamina propria CD4+ T cells in vitro is enhanced by exposure to commensal Escherichia coli. J. Immunol. 189:885–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ahmed N, Hayashi T, Hasegawa A, Furukawa H, Okamura N, Chida T, Masuda T, Kannagi M. 2010. Suppression of human immunodeficiency virus type 1 replication in macrophages by commensal bacteria preferentially stimulating Toll-like receptor 4. J. Gen. Virol. 91:2804–2813 [DOI] [PubMed] [Google Scholar]
- 18. Cassone A, Cauda R. 2012. Candida and candidiasis in HIV-infected patients: where commensalism, opportunistic behavior and frank pathogenicity lose their borders. AIDS 26:1457–1472 [DOI] [PubMed] [Google Scholar]
- 19. Imai K, Ochiai K, Okamoto T. 2009. Reactivation of latent HIV-1 infection by the periodontopathic bacterium Porphyromonas gingivalis involves histone modification. J. Immunol. 182:3688–3695 [DOI] [PubMed] [Google Scholar]
- 20. Imai K, Yamada K, Tamura M, Ochiai K, Okamoto T. 2012. Reactivation of latent HIV-1 by a wide variety of butyric acid-producing bacteria. Cell. Mol. Life Sci. 69:2583–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. González OA, Li M, Ebersole JL, Huang CB. 2010. HIV-1 reactivation induced by the periodontal pathogens Fusobacterium nucleatum and Porphyromonas gingivalis involves Toll-like receptor 2 [corrected] and 9 activation in monocytes/macrophages. Clin. Vaccine Immunol. 17:1417–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. González OA, Ebersole JL, Huang CB. 2011. The oral commensal, Streptococcus gordonii, synergizes with Tat protein to induce HIV-1 promoter activation in monocytes/macrophages. Cell. Immunol. 269:38–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Giacaman RA, Asrani AC, Gebhard KH, Dietrich EA, Vacharaksa A, Ross KF, Herzberg MC. 2008. Porphyromonas gingivalis induces CCR5-dependent transfer of infectious HIV-1 from oral keratinocytes to permissive cells. Retrovirology 5:29 doi: 10.1186/1742-4690-5-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sha BE, Zariffard MR, Wang QJ, Chen HY, Bremer J, Cohen MH, Spear GT. 2005. Female genital-tract HIV load correlates inversely with Lactobacillus species but positively with bacterial vaginosis and Mycoplasma hominis. J. Infect. Dis. 191:25–32 [DOI] [PubMed] [Google Scholar]
- 25. Sewankambo N, Gray RH, Wawer MJ, Paxton L, McNaim D, Wabwire-Mangen F, Serwadda D, Li C, Kiwanuka N, Hillier SL, Rabe L, Gaydos CA, Quinn TC, Konde-Lule J. 1997. HIV-1 infection associated with abnormal vaginal flora morphology and bacterial vaginosis. Lancet 350:546–550 [DOI] [PubMed] [Google Scholar]
- 26. Mirmonsef P, Krass L, Landay A, Spear GT. 2012. The role of bacterial vaginosis and Trichomonas in HIV transmission across the female genital tract. Curr. HIV Res. 10:202–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Taha TE, Hoover DR, Dallabetta GA, Kumwenda NI, Mtimavalye LA, Yang LP, Liomba GN, Broadhead RL, Chiphangwi JD, Miotti PG. 1998. Bacterial vaginosis and disturbances of vaginal flora: association with increased acquisition of HIV. AIDS 12:1699–1706 [DOI] [PubMed] [Google Scholar]
- 28. Al-Harthi L, Roebuck KA, Olinger GG, Landay A, Sha BE, Hashemi FB, Spear GT. 1999. Bacterial vaginosis-associated microflora isolated from the female genital tract activates HIV-1 expression. J. Acquir. Immune Defic. Syndr. 21:194–202 [DOI] [PubMed] [Google Scholar]
- 29. Hashemi FB, Ghassemi M, Faro S, Aroutcheva A, Spear GT. 2000. Induction of human immunodeficiency virus type 1 expression by anaerobes associated with bacterial vaginosis. J. Infect. Dis. 181:1574–1580 [DOI] [PubMed] [Google Scholar]
- 30. Spiegel CA, Amsel R, Eschenbach D, Schoenknecht F, Holmes KK. 1980. Anaerobic bacteria in nonspecific vaginitis. N. Engl. J. Med. 303:601–607 [DOI] [PubMed] [Google Scholar]
- 31. Jiang W, Ghosh SK, Flyckt R, Kalinowska M, Starks D, Jurevic R, Weinberg A, Lederman MM, Rodriguez B. 2012. Bacterial colonization and beta defensins in the female genital tract in HIV infection. Curr. HIV Res. 10:504–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Siliciano RF, Greene WC. 2011. HIV latency. Cold Spring Harbor Perspect. Med. 1:a007096 doi: 10.1101/cshperspect.a007096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Margolis DM. 2010. Mechanisms of HIV latency: an emerging picture of complexity. Curr. HIV/AIDS Rep. 7:37–43 [DOI] [PubMed] [Google Scholar]
- 34. Hakre S, Chavez L, Shirakawa K, Verdin E. 2011. Epigenetic regulation of HIV latency. Curr. Opin. HIV AIDS 6:19–24 [DOI] [PubMed] [Google Scholar]
- 35. Colin L, Van Lint C. 2009. Molecular control of HIV-1 postintegration latency: implications for the development of new therapeutic strategies. Retrovirology 6:111 doi: 10.1186/1742-4690-6-111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Victoriano AF, Okamoto T. 2012. Transcriptional control of HIV replication by multiple modulators and their implication for a novel antiviral therapy. AIDS Res. Hum. Retroviruses 28:125–138 [DOI] [PubMed] [Google Scholar]
- 37. Choudhary SK, Margolis DM. 2011. Curing HIV: pharmacologic approaches to target HIV-1 latency. Annu. Rev. Pharmacol. Toxicol. 51:397–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Burnett JC, Lim KI, Calafi A, Rossi JJ, Schaffer DV, Arkin AP. 2010. Combinatorial latency reactivation for HIV-1 subtypes and variants. J. Virol. 84:5958–5974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Savarino A, Mai A, Norelli S, El Daker S, Valente S, Rotili D, Altucci L, Palamara AT, Garaci E. 2009. “Shock and kill” effects of class I-selective histone deacetylase inhibitors in combination with the glutathione synthesis inhibitor buthionine sulfoximine in cell line models for HIV-1 quiescence. Retrovirology 6:52 doi: 10.1186/1742-4690-6-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bouchat S, Gatot JS, Kabeya K, Cardona C, Colin L, Herbein G, De Wit S, Clumeck N, Lambotte O, Rouzioux C, Rohr O, Van Lint C. 2012. Histone methyltransferase inhibitors induce HIV-1 recovery in resting CD4(+) T cells from HIV-1-infected HAART-treated patients. AIDS 26:1473–1482 [DOI] [PubMed] [Google Scholar]
- 41. Bernhard W, Barreto K, Saunders A, Dahabieh MS, Johnson P, Sadowski I. 2011. The Suv39H1 methyltransferase inhibitor chaetocin causes induction of integrated HIV-1 without producing a T cell response. FEBS Lett. 585:3549–3554 [DOI] [PubMed] [Google Scholar]
- 42. Okamoto T, Akagi T, Shima H, Miwa M, Shimotohno K. 1987. Superinduction of trans-activation accounts for augmented human immunodeficiency virus replication in HTLV-I-transformed cells. Jpn. J. Cancer Res. 78:1297–1301 [PubMed] [Google Scholar]
- 43. Okamoto T, Matsuyama T, Mori S, Hamamoto Y, Kobayashi N, Yamamoto N, Josephs SF, Wong-Staal F, Shimotohno K. 1989. Augmentation of human immunodeficiency virus type 1 gene expression by tumor necrosis factor alpha. AIDS Res. Hum. Retroviruses 5:131–138 [DOI] [PubMed] [Google Scholar]
- 44. Mbopi-Kéou FX, Belec L, Teo CG, Scully C, Porter SR. 2002. Synergism between HIV and other viruses in the mouth. Lancet Infect. Dis. 2:416–424 [DOI] [PubMed] [Google Scholar]
- 45. Andreani G, Lodge R, Richard D, Tremblay MJ. 2012. Mechanisms of interaction between protozoan parasites and HIV. Curr. Opin. HIV AIDS 7:276–282 [DOI] [PubMed] [Google Scholar]
- 46. Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, Kazzaz Z, Bornstein E, Lambotte O, Altmann D, Blazar BR, Rodriguez B, Teixeira-Johnson L, Landay A, Martin JN, Hecht FM, Picker LJ, Lederman MM, Deeks SG, Douek DC. 2006. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat. Med. 12:1365–1371 [DOI] [PubMed] [Google Scholar]
- 47. Haynes BF. 2006. Gut microbes out of control in HIV infection. Nat. Med. 12:1351–1352 [DOI] [PubMed] [Google Scholar]
- 48. Klatt NR, Funderburg NT, Brenchley JM. 2013. Microbial translocation, immune activation, and HIV disease. Trends Microbiol. 21:6–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Handley SA, Thackray LB, Zhao G, Presti R, Miller AD, Droit L, Abbink P, Maxfield LF, Kambal A, Duan E, Stanley K, Kramer J, Macri SC, Permar SR, Schmitz JE, Mansfield K, Brenchley JM, Veazey RS, Stappenbeck TS, Wang D, Barouch DH, Virgin HW. 2012. Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell 151:253–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Verdin E, Paras P, Jr, Van Lint C. 1993. Chromatin disruption in the promoter of human immunodeficiency virus type 1 during transcriptional activation. EMBO J. 12:3249–3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jordan A, Bisgrove D, Verdin E. 2003. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 22:1868–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Laughlin MA, Zeichner S, Kolson D, Alwine JC, Seshamma T, Pomerantz RJ, Gonzalez-Scarano F. 1993. Sodium butyrate treatment of cells latently infected with HIV-1 results in the expression of unspliced viral RNA. Virology 196:496–505 [DOI] [PubMed] [Google Scholar]
- 53. Van Lint C, Emiliani S, Ott M, Verdin E. 1996. Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J. 15:1112–1120 [PMC free article] [PubMed] [Google Scholar]
- 54. Grunstein M. 1997. Histone acetylation in chromatin structure and transcription. Nature 389:349–352 [DOI] [PubMed] [Google Scholar]
- 55. Murphy M, Ahn J, Walker KK, Hoffman WH, Evans RM, Levine AJ, George DL. 1999. Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3a. Genes Dev. 13:2490–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ashburner BP, Westerheide SD, Baldwin AS., Jr 2001. The p65 (RelA) subunit of NF-kappaB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression. Mol. Cell. Biol. 21:7065–7077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Imai K, Okamoto T. 2006. Transcriptional repression of human immunodeficiency virus type 1 by AP-4. J. Biol. Chem. 281:12495–12505 [DOI] [PubMed] [Google Scholar]
- 58. Coull JJ, Romerio F, Sun JM, Volker JL, Galvin KM, Davie JR, Shi Y, Hansen U, Margolis DM. 2000. The human factors YY1 and LSF repress the human immunodeficiency virus type 1 long terminal repeat via recruitment of histone deacetylase 1. J. Virol. 74:6790–6799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Williams SA, Chen LF, Kwon H, Ruiz-Jarabo CM, Verdin E, Greene WC. 2006. NF-kappaB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J. 25:139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jiang G, Espeseth A, Hazuda DJ, Margolis DM. 2007. c-Myc and Sp1 contribute to proviral latency by recruiting histone deacetylase 1 to the human immunodeficiency virus type 1 promoter. J. Virol. 81:10914–10923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tyagi M, Karn J. 2007. CBF-1 promotes transcriptional silencing during the establishment of HIV-1 latency. EMBO J. 26:4985–4995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Keedy KS, Archin NM, Gates AT, Espeseth A, Hazuda DJ, Margolis DM. 2009. A limited group of class I histone deacetylases acts to repress human immunodeficiency virus type 1 expression. J. Virol. 83:4749–4756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Huber K, Doyon G, Plaks J, Fyne E, Mellors JW, Sluis-Cremer N. 2011. Inhibitors of histone deacetylases: correlation between isoform specificity and reactivation of HIV type 1 (HIV-1) from latently infected cells. J. Biol. Chem. 286:22211–22218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Barton K, Margolis D. 2013. Selective Targeting of the repressive transcription factors YY1 and cMyc to disrupt quiescent human immunodeficiency viruses. AIDS Res. Hum. Retroviruses 29:289–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Victoriano AF, Imai K, Togami H, Ueno T, Asamitsu K, Suzuki T, Miyata N, Ochiai K, Okamoto T. 2011. Novel histone deacetylase inhibitor NCH-51 activates latent HIV-1 gene expression. FEBS Lett. 585:1103–1111 [DOI] [PubMed] [Google Scholar]
- 66. Pinskaya M, Morillon A. 2009. Histone H3 lysine 4 di-methylation: a novel mark for transcriptional fidelity? Epigenetics 4:302–306 [DOI] [PubMed] [Google Scholar]
- 67. Kouzarides T. 2007. Chromatin modifications and their function. Cell 128:693–705 [DOI] [PubMed] [Google Scholar]
- 68. du Chéné I, Basyuk E, Lin YL, Triboulet R, Knezevich A, Chable-Bessia C, Mettling C, Baillat V, Reynes J, Corbeau P, Bertrand E, Marcello A, Emiliani S, Kiernan R, Benkirane M. 2007. Suv39H1 and HP1gamma are responsible for chromatin-mediated HIV-1 transcriptional silencing and post-integration latency. EMBO J. 26:424–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Marban C, Suzanne S, Dequiedt F, de Walque S, Redel L, Van Lint C, Aunis D, Rohr O. 2007. Recruitment of chromatin-modifying enzymes by CTIP2 promotes HIV-1 transcriptional silencing. EMBO J. 26:412–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Pearson R, Kim YK, Hokello J, Lassen K, Friedman J, Tyagi M, Karn J. 2008. Epigenetic silencing of human immunodeficiency virus (HIV) transcription by formation of restrictive chromatin structures at the viral long terminal repeat drives the progressive entry of HIV into latency. J. Virol. 82:12291–12303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Imai K, Togami H, Okamoto T. 2010. Involvement of histone H3 lysine 9 (H3K9) methyltransferase G9a in the maintenance of HIV-1 latency and its reactivation by BIX01294. J. Biol. Chem. 285:16538–16545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Easley R, Van Duyne R, Coley W, Guendel I, Dadgar S, Kehn-Hall K, Kashanchi F. 2010. Chromatin dynamics associated with HIV-1 Tat-activated transcription. Biochim. Biophys. Acta 1799:275–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Le Douce V, Colin L, Redel L, Cherrier T, Herbein G, Aunis D, Rohr O, Van Lint C, Schwartz C. 2012. LSD1 cooperates with CTIP2 to promote HIV-1 transcriptional silencing. Nucleic Acids Res. 40:1904–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Friedman J, Cho WK, Chu CK, Keedy KS, Archin NM, Margolis DM, Karn J. 2011. Epigenetic silencing of HIV-1 by the histone H3 lysine 27 methyltransferase enhancer of Zeste 2. J. Virol. 85:9078–9089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Viré E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, Bollen M, Esteller M, Di Croce L, de Launoit Y, Fuks F. 2006. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 439:871–874 [DOI] [PubMed] [Google Scholar]
- 76. Tae S, Karkhanis V, Velasco K, Yaneva M, Erdjument-Bromage H, Tempst P, Sif S. 2011. Bromodomain protein 7 interacts with PRMT5 and PRC2, and is involved in transcriptional repression of their target genes. Nucleic Acids Res. 39:5424–5438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kauder SE, Bosque A, Lindqvist A, Planelles V, Verdin E. 2009. Epigenetic regulation of HIV-1 latency by cytosine methylation. PLoS Pathog. 5:e1000495 doi: 10.1371/journal.ppat.1000495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Blazkova J, Trejbalova K, Gondois-Rey F, Halfon P, Philibert P, Guiguen A, Verdin E, Olive D, Van Lint C, Hejnar J, Hirsch I. 2009. CpG methylation controls reactivation of HIV from latency. PLoS Pathog. 5:e1000554 doi: 10.1371/journal.ppat.1000554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Van Lint C, Emiliani S, Verdin E. 1996. The expression of a small fraction of cellular genes is changed in response to histone hyperacetylation. Gene Expression 5:245–253 [PMC free article] [PubMed] [Google Scholar]
- 80. Moog C, Kuntz-Simon G, Caussin-Schwemling C, Obert G. 1996. Sodium valproate, an anticonvulsant drug, stimulates human immunodeficiency virus type 1 replication independently of glutathione levels. J. Gen. Virol. 77(Pt 9):1993–1999 [DOI] [PubMed] [Google Scholar]
- 81. Quivy V, Adam E, Collette Y, Demonte D, Chariot A, Vanhulle C, Berkhout B, Castellano R, de Launoit Y, Burny A, Piette J, Bours V, Van Lint C. 2002. Synergistic activation of human immunodeficiency virus type 1 promoter activity by NF-kappaB and inhibitors of deacetylases: potential perspectives for the development of therapeutic strategies. J. Virol. 76:11091–11103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Archin NM, Espeseth A, Parker D, Cheema M, Hazuda D, Margolis DM. 2009. Expression of latent HIV induced by the potent HDAC inhibitor suberoylanilide hydroxamic acid. AIDS Res. Hum. Retroviruses 25:207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Archin NM, Liberty AL, Kashuba AD, Choudhary SK, Kuruc JD, Crooks AM, Parker DC, Anderson EM, Kearney MF, Strain MC, Richman DD, Hudgens MG, Bosch RJ, Coffin JM, Eron JJ, Hazuda DJ, Margolis DM. 2012. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 487:482–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lehrman G, Hogue IB, Palmer S, Jennings C, Spina CA, Wiegand A, Landay AL, Coombs RW, Richman DD, Mellors JW, Coffin JM, Bosch RJ, Margolis DM. 2005. Depletion of latent HIV-1 infection in vivo: a proof-of-concept study. Lancet 366:549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Margolis DM. 2011. Histone deacetylase inhibitors and HIV latency. Curr. Opin. HIV AIDS 6:25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Willemsen NM, Hitchen EM, Bodetti TJ, Apolloni A, Warrilow D, Piller SC, Harrich D. 2006. Protein methylation is required to maintain optimal HIV-1 infectivity. Retrovirology 3:92 doi: 10.1186/1742-4690-3-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Archin NM, Cheema M, Parker D, Wiegand A, Bosch RJ, Coffin JM, Eron J, Cohen M, Margolis DM. 2010. Antiretroviral intensification and valproic acid lack sustained effect on residual HIV-1 viremia or resting CD4+ cell infection. PLoS One 5:e9390 doi: 10.1371/journal.pone.0009390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Sagot-Lerolle N, Lamine A, Chaix ML, Boufassa F, Aboulker JP, Costagliola D, Goujard C, Pallier C, Delfraissy JF, Lambotte O. 2008. Prolonged valproic acid treatment does not reduce the size of latent HIV reservoir. AIDS 22:1125–1129 [DOI] [PubMed] [Google Scholar]
- 89. Arrieta MC, Finlay BB. 2012. The commensal microbiota drives immune homeostasis. Front. Immunol. 3:33 doi: 10.3389/fimmu.2012.00033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Brenchley JM, Douek DC. 2008. The mucosal barrier and immune activation in HIV pathogenesis. Curr. Opin. HIV AIDS 3:356–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Marchetti G, Cozzi-Lepri A, Merlini E, Bellistri GM, Castagna A, Galli M, Verucchi G, Antinori A, Costantini A, Giacometti A, di Caro A, D'Arminio Monforte A. 2011. Microbial translocation predicts disease progression of HIV-infected antiretroviral-naive patients with high CD4+ cell count. AIDS 25:1385–1394 [DOI] [PubMed] [Google Scholar]
- 92. Saxena D, Li Y, Yang L, Pei Z, Poles M, Abrams WR, Malamud D. 2012. Human microbiome and HIV/AIDS. Curr. HIV/AIDS Rep. 9:44–51 [DOI] [PMC free article] [PubMed] [Google Scholar]