Abstract
Alveolins are a recently described class of proteins common to all members of the superphylum Alveolata that are characterized by conserved charged repeat motifs (CRMs) but whose exact function remains unknown. We have analyzed the smaller of the two alveolins of Tetrahymena thermophila, TtALV2. The protein localizes to dispersed, broken patches arranged between the rows of the longitudinal microtubules. Macronuclear knockdown of Ttalv2 leads to multinuclear cells with no apparent cell polarity and randomly occurring cell protrusions, either by interrupting pellicle integrity or by disturbing cytokinesis. Correct association of TtALV2 with the alveoli or the pellicle is complex and depends on both the termini as well as the charged repeat motifs of the protein. Proteins containing similar CRMs are a dominant part of the ciliate membrane cytoskeleton, suggesting that these motifs may play a more general role in mediating membrane attachment and/or cytoskeletal association. To better understand their integration into the cytoskeleton, we localized a range of CRM-based fusion proteins, which suggested there is an inherent tendency for proteins with CRMs to be located in the peripheral cytoskeleton, some nucleating as filaments at the basal bodies. Even a synthetic protein, mimicking the charge and repeat pattern of these proteins, directed a reporter protein to a variety of peripheral cytoskeletal structures in Tetrahymena. These motifs might provide a blueprint for membrane and cytoskeleton affiliation in the complex pellicles of Alveolata.
INTRODUCTION
The vast majority of eukaryotic life is unicellular. Although protists display all of the characteristics that define a eukaryote, such as a nucleus, organelles, and sophisticated cytoskeleton, the general assumption is that these cells are less complex than those of a metazoan. However, at the level of cytoskeletal organization, the contrary would appear to be true. Advances from sequencing and genomic analysis have revealed that many of the components and their accompanying complexity that are necessary to build a multicellular organism are also present in protists. Meanwhile, intermediate filament proteins, which together with actin-based microfilaments and microtubules form one of the three pillars of the metazoan cytoskeleton, have not been clearly identified in the many known genomes of single-cell eukaryotes to the same degree. In some cases, this might be due to the poor sequence conservation of intermediate filament proteins, but the sparse phylogenetic distribution of recognizable intermediate filament proteins in protists suggests that other proteins have evolved in these lineages to assume cytoskeletal functions.
Poor sequence conservation is one reason why BLAST-based searches may fail to identify reliable homologs; another can be the presence of low-sequence-complexity regions, such as repeat motifs with a biased amino acid composition. These repeat regions hamper the definition of a single “seed region” from which the alignment commences. Moreover, the primary sequence of repetitive regions is prone to rapid evolution, so despite the location of the repeat within a homologous protein being conserved, the primary sequence is often not (1). Many structural proteins of the eukaryotic cytoskeleton are high-molecular-mass proteins with long repetitive elements, and a large number of them are predicted to form coiled coils (2, 3). There are two general classes of coiled-coil domains: (i) short ones of six or seven heptad repeats, common, for example, among transcription factors; and (ii) long ones, comprised of several hundred amino acids (4). These long coiled-coil domains are also often enriched with charged amino acids and are a core feature of intermediate filament proteins (5). They are, for example Golgi, kinetochore/centromere, and spindle apparatus associated and are involved in spindle-pole and centrosome formation. In general, long coiled coils are predominantly found among cytoskeletal or cytoskeleton-associated proteins and have been figuratively described to act as “cellular Velcro” (2, 3).
A recent proteomic profiling of the detergent-resistant membrane skeleton (epiplasm, or subpellicular network) of Tetrahymena thermophila identified a wealth of proteins that are united by their possession of repeat motifs with characteristic amino acid biases, referred to as charged repeat motif proteins (CRMPs) (1). These repeats are enriched in charged and polar amino acids, such as K, E, D, and Q, and unpolar amino acids, in particular I, L, and V. About half of the identified CRMPs are of unknown function, and many consistently recover database matches to proteins with “structural maintenance of chromosomes” (SMC) domains or “viral A-type” domains, both of which contain similar repetitive regions to those of CRMPs and appear to be the basis of their BLAST recovery (1, 6). Prominent CRMPs identified in the ciliate proteome data and known to be proteins of the pellicle are tetrins, basal body proteins 39 and 53, EPC1 (epiplasm gene 1), and the alveolins (7–11). The alveolins represent a family of proteins encoded in all alveolate genomes currently analyzed (11). In common with many pellicle proteins, the alveolins are encoded by multigene families and are apparently abundant proteins in apicomplexans, dinoflagellates, and ciliates. They appear to share conserved cytoskeletal functions associated with the alveolar sacs (12–16). The tight alliance of alveolins, alveolar membranes, and proteins involved in motility emphasizes the key role of alveolins in cellular scaffolding and organization.
These previous findings suggest a more universal, but cytoskeleton-associated function for the charged repeat motifs (CRMs) of alveolins and other proteins with similar characteristics. Here we have characterized T. thermophila ALV2 (TtALV2) and demonstrated this alveolin protein to be an essential component of the pellicular membrane skeleton of Tetrahymena, the epiplasm. Association with the alveoli and integration of TtALV2 into the pellicle are complex and partially mirror the results we obtained for the Toxoplasma gondii alveolin homolog TgALV1. Testing of other charged repeat motifs, including a synthetic repeat created to match the characteristics of Tetrahymena CRMPs, shows these proteins often have an inherent tendency to associate with the ciliate membrane skeleton.
MATERIALS AND METHODS
Both Tetrahymena thermophila strains (CU522 for fusion protein localization and CU428 for the knockdown) were cultured in SPP medium (1% proteose peptone, 0.1% yeast extract, 0.2% glucose, 0.003% Fe-EDTA) routinely at 15°C and transferred to 30°C for optimal overnight growth before the cells were harvested for all downstream experiments. The pellicle was isolated according to Williams and colleagues (17), with slight modifications. Cells were washed three times in 1 M phosphate buffer (pH 7) containing Complete-Midi tablets (Roche Applied Science) against protease degradation. RNA, DNA, and total protein were isolated using TRIzol (Invitrogen, Australia) following the manufacturer's protocol.
For green fluorescent protein (GFP) fusion constructs, we designed a new plasmid, pTtag (accession no. FJ789658), which was based on a previous GFP-tagging plasmid described by Shang and colleagues (18). The four main modifications of pTtag were (i) a Tetrahymena codon-optimized GFP, (ii) a 1-kb-shorter 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT)-expressing promoter, (iii) fewer doublet restriction sites, and (iv) an extended multiple cloning site. For the C-terminal GFP fusion, the target genes were cloned into pTtag via the HindIII and XhoI restriction sites. The primers used for the full-length Ttalv2 gene were TtA2GFPf and TtA2GFPr. The primers used for the TtALV2-Rep-GFP fusion constructs were Alv2RepeatsHindIIIF and Alv2RepeatsXhoIR. All primers are listed in Table S2 in the supplemental material. For the TtALV2-N-terminus–GFP-TtALV2-C-terminus fusion construct, the N terminus was cloned via the SphI and MluI restriction sites, and the C terminus was cloned via NsiI and BamHI. The two sets of primers used for this construct were AGA1-SphI-F and AGA1-MluI-R and AGA2-NsiI-F and AGA2BamHI-R. The knockdown of TtALV2 was based on homologous recombination and utilized plasmid pNeo4 (19). A sequence ∼800 bp upstream (restriction sites ApaI and SalI) and downstream (restriction sites PstI and SacII) of TtALV2 was used to flank the Neo4 resistance cassette. T. thermophila was transfected using a PDS-1000 Gene Gun (Bio-Rad) with 900-lb/in2 rupture discs and 20 μg of linearized plasmid DNA following a protocol established by Cassidy-Hanley and colleagues (20). Cells transfected with GFP constructs were selected for 10 days in SPP medium supplemented with 30 μg/ml paclitaxel (LC Labs). GFP fusion protein expression was induced using 2.5 μg/ml CdCl2. Cells transfected with the knockdown plasmid were initially selected with 50 μg/ml paromomycin and treated with 2 μg/ml CdCl2.
The correct substitution of the macronuclear Ttalv2 gene through the NeoR cassette in the clones analyzed was verified using the primer pairs tKD1f and tKD1r and tKD2f and tKD2r to screen for the correct 5′-upstream and 3′-downstream integrations, respectively (see Fig. S3B in the supplemental material). Real-time quantitative PCRs (RT-qPCRs) were run on a StepOnePlus using Power SYBR green master mix (both from Applied Biosystems). RNA was isolated using TRIzol and treated with DNase (Thermo Scientific) following the manufacturers' protocols. One microgram of total RNA was transcribed into cDNA using the iScript cDNA synthesis kit (Bio-Rad), and 1 μl thereof was used as a template for RT-qPCR. For each RT-qPCR, three technical replicates were run with the primers TtAlv2_qFOR and TtAlv2_qREV. For comparative quantification, inverted median threshold cycle (ΔCT) values were calculated using the wild-type CT value as a reference.
Polyclonal rabbit peptide antiserum was generated against an epitope (EKVIEVPQTQVMEKV; α74) found within the repeat motif region of alveolin family proteins from the dinoflagellate Karlodinium veneficum (11). In Western blot analysis, these primary antisera were used at concentrations between 1:50 and 1:500. The anti-GFP antibody (mouse; Sigma-Aldrich; rabbit, Abcam) was used at 1:1,000 in Western blot analyses and 1:200 for immunofluorescence assays (IFAs). The anti-tubulin antibody (mouse; Developmental Studies Hybridoma Bank) was used at 1:200 for IFAs. Secondary horseradish peroxidase (HRP)-coupled anti-rabbit or anti-mouse antibodies (Thermo Scientific) were used at 1:20,000. For signal detection, Hyperfilm ECL (GE Healthcare) and the chemifluorescence SuperSignal West Pico kit (Pierce Biotechnology) were used. For immunofluorescence labeling, ∼105 cells were concentrated at 1,100 × g for 3 min and gently resuspended in 1.5 ml PHEM buffer (60 mM PIPES, 25 mM HEPES, 10 mM EDTA, 2 mM MgCl2) containing 4% paraformaldehyde and 0.5% Triton X-100. Cells were fixed at room temperature for 30 min and then concentrated at 500 × g for 5 min, and the remaining pellet was resuspended in 1 ml phosphate-buffered saline (PBS) containing 0.1 M glycine and left for 20 min. Concentrated cells were then resuspended in 1 ml PBS with 0.2% bovine serum albumin (BSA) and blocked for 30 min at room temperature before incubation with the primary antibody at a concentration of 1:50 to 1:200, diluted in PBS containing 0.1% BSA and 0.1% Triton X-100. Secondary Alexa Fluor 488 and 633 antibodies (Invitrogen, Australia) were used at 1:1,000. All washes (three times for 10 min each after each antibody incubation) were performed using 1.5 ml PBS containing 0.2% Triton X-100. For DNA staining, the second-to-last wash additionally contained Hoechst 33342 at 5 μg/ml, and cells were mounted on Superfrost slides (Menzel) in a 2.5% DABCO [1,4-diazabicyclo(2,2,2)octane] solution and sealed with wax. The mounting medium Vectashield (Vector Laboratories), which includes DAPI (4′,6-diamidino-2-phenylindole), was also used for DNA.
CRMPs were identified using Xstream (http://jimcooperlab.mcdb.ucsb.edu/xstream/) and RADAR (www.ebi.ac.uk/Tools/pfa/radar/), as recently described (1). RADAR was also used to generate the sequence motifs shown in Fig. 4 and Fig. S1 in the supplemental material. Coiled coils were predicted using Multicoil [http://theory.lcs.mit.edu/multicoil].
Fig 4.
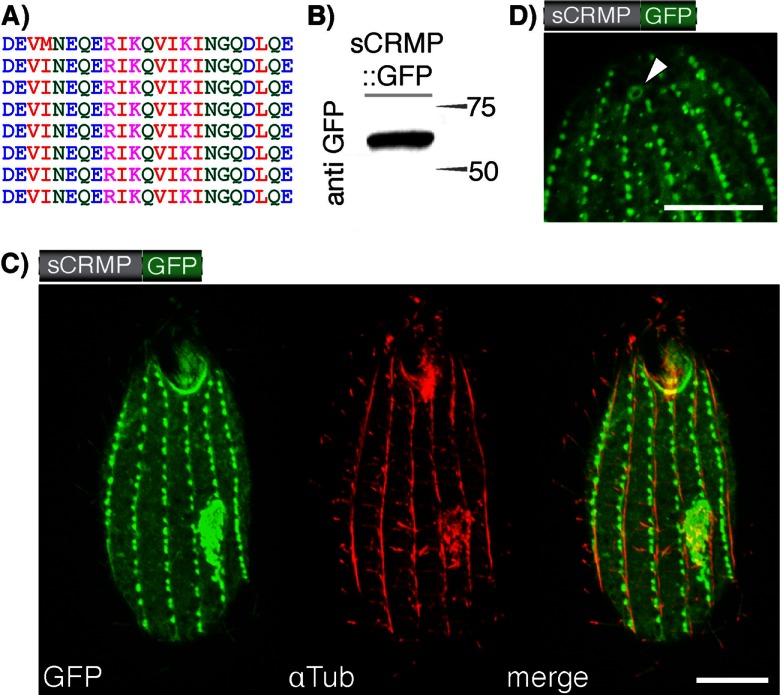
Targeting properties of a synthetic charged repeat motif. (A) We generated a synthetic charged repeat motif and fused it to a GFP reporter generating a 55-kDa fusion protein (sCRMP::GFP; Western blot in panel B). (C) The protein associates with tubulin-associated structures, such as microtubules and basal bodies, and is further targeted to the cilia. αTub, anti-tubulin. (D) Detail showing the localization of sCRMP to the contractile vacuolar pore. Scale, 10 μm.
RESULTS
Individual domains of TtALV2 localize to different cytoskeletal structures.
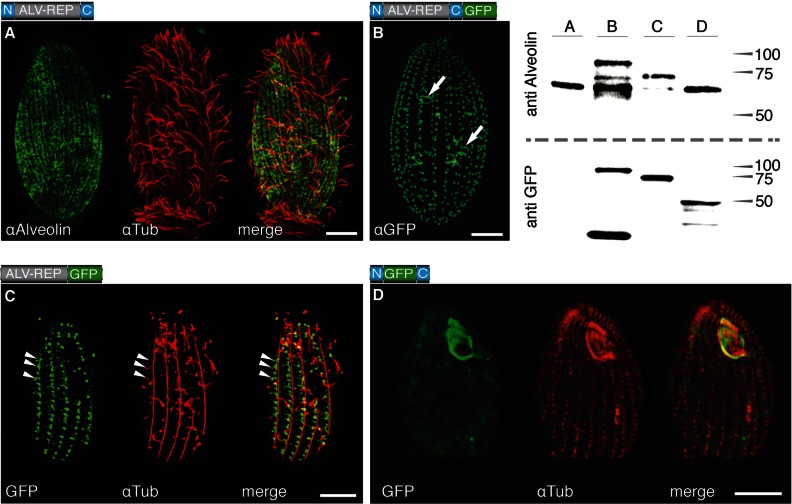
Ttalv2 coding sequence was generated from T. thermophila cDNA to verify the predicted protein sequence. This cDNA sequence (GenBank ID no. FJ789659) revealed an incorrect exon-intron boundary in the previous gene model at http://ciliate.org for TTHERM_00088030, and a predicted protein size of 67.8 kDa. To determine the location and analyze the expression of TtALV2 in T. thermophila, we used an antiserum raised against a peptide found within the characteristic repeat region of the alveolin family proteins (anti-alveolin, or α74) (11). The antibody reacts with a dominant protein band on Western blots of ∼65 kDa, consistent with the predicted mass for TtALV2 (Fig. 1, Western blot, lane A, preimmune serum nonreactive). To further verify the specificity of anti-alveolin for TtALV2 and to analyze the maturation of the TtALV2 protein (see below), we expressed two TtALV2 reporter protein fusions: one with the green fluorescent protein (GFP) and a second with a hemagglutinin (HA) tag (ALV2::GFP and ALV2::HA, respectively). The use of anti-alveolin on total protein extract from cells expressing either ALV2::GFP or ALV2::HA fusion constructs resulted in labeling of the endogenous TtALV2 plus a band of the corresponding fusion proteins of 95 and 70 kDa, respectively (Fig. 1, Western blot, lane B; see Fig. S2D in the supplemental material). These data verify the specificity of anti-alveolin for TtALV2. In immunofluorescence analysis using fixed T. thermophila cells, the anti-alveolin antibody labels a dispersed system of broken patches, arranged in longitudinal strips running along the whole cell surface. This pattern is reminiscent of military-type camouflage and is a pattern not previously observed in T. thermophila cells (Fig. 1A; see Fig. S2A) but similar to that of plateins—which are also CRMPs—from Euplotes aediculatus (21). Colabeling with tubulin demonstrates that TtALV2 is excluded from the circumciliary ring region and absent from the oral apparatus (Fig. 1A; see Fig. S2A), and preimmune anti-alveolin serum did not label fixed T. thermophila cells.
Fig 1.
Localization of TtALV2 and fragments thereof. (A) Endogenous TtALV2 (68 kDa) localizes to patches between the longitudinal microtubules and along the entire inner cell surface. αAlveolin, anti-alveolin; αTub, anti-tubulin. (B) In contrast, the TtALV2::GFP fusion protein (94 kDa) localizes predominantly to the basal bodies and forms fiber-like structures inside the cytosol (arrows). Only in this case was a GFP antibody (αGFP) used; in all other cases, autofluorescence of GFP is shown. (C) Repeat motifs alone fused to GFP (ALV-REP::GFP; 75 kDa) associate with microtubular structures, including transverse microtubules (arrowheads) and the basal bodies. (D) Replacement of the repeat motifs with GFP (N::GFP::C; 46 kDa) targets the fusion protein predominantly to the oral apparatus. The schematics of the constructs are shown at the top left corner of each panel, where N and C represent the nonrepetitive N- and C-terminal domains of TtALV2, respectively. Scale bars, 10 μm. The corresponding Western blots of the protein extracts of the individual strains are shown at the top right. Markers are in kilodaltons.
To examine the targeting properties of the different TtALV2 domains, including the CRMs, we generated three inducible GFP fusion constructs: (i) a full-length ALV2::GFP, (ii) one in which only the repeat elements were fused to GFP (ALV-REP::GFP), and (iii) another in which GFP replaces the repeat motifs (N::GFP::C). In the latter construct, the GFP is flanked by the nonrepetitive terminal regions, which have no homology to any known domain of function—a composition typical for intermediate filament proteins (5). Western blot analysis of individual cell extracts using anti-alveolin and anti-GFP confirmed expression of fusion proteins of the correct molecular mass, verifying that full-length reporters were expressed (Fig. 1). For fusion protein N::GFP::C, anti-alveolin is nonreactive, as this antibody reacts exclusively with the repeat region; however, anti-GFP verifies expression of this protein. (Note that some GFP degradation species are also evident in the GFP Western blots.) Localization of the GFP fusion proteins, intriguingly, revealed that none of the three GFP fusion constructs displayed an identical pattern to that observed for the endogenous TtALV2 identified by the anti-alveolin antibody. Nevertheless, all reporter constructs were associated with peripheral, cytoskeletal structures. ALV2::GFP localizes to the cilia-bearing basal bodies and fibrillar structures several micrometers in length (Fig. 1B) and, to a lesser extent, the oral apparatus (see Fig. S2B in the supplemental material). To visualize both endogenous TtALV2 and the ALV2::GFP fusion protein together, immunofluorescence with anti-alveolin antiserum was performed on induced cells. The pattern of labeling observed consisted of both the dispersed patchwork seen for native TtALV2 and foci at the basal bodies, consistent with the GFP localization (see Fig. S2B). ALV-REP::GFP associates with microtubular structures, including transverse microtubules around the basal bodies, and other fibrous structures (Fig. 1C). Surprisingly, when the repeats of TtALV2 were replaced by GFP (N::GFP::C), the construct predominantly localized to complex structures of the oral apparatus framing the outline of the membranelles (Fig. 1D). The GFP marker is apparently not responsible for the altered localization of ALV2::GFP, as the HA-tagged TtALV2 showed an identical localization to TtALV2::GFP (see Fig. S2D). It is also evident from the literature that tagging of proteins in Tetrahymena does not predispose those constructs to associate with cytoskeletal structures and ciliary basal bodies (22–24). Furthermore, expression of GFP alone from the same plasmid under the control of same promoter results in accumulation of GFP in the cytosol and not the cytoskeleton (see Fig. S2E). We observed similar targeting behaviors for individual domains of the Toxoplasma gondii homolog TgALV1, in which the repeat domain apparently mediates localization to the membrane skeleton (TGME49_231640; also called TgIMC1, for inner membrane complex) (see Fig. S3 in the supplemental material).
Knockdown of Ttalv2.
To explore the function of TtALV2, we first tried to generate a micronuclear, germ line knockout of the gene in Tetrahymena. Several attempts failed to produce viable offspring of a homozygous Ttalv2 knockout, suggesting that the gene is essential. At this point, we decided to generate a macronuclear, somatic knockdown of the gene instead. From one successful experiment, we isolated 48 single cells from cultures that were resistant to 50 μg/ml paromomycin (ParR) posttransfection with the knockout plasmid-based pNeo4 (19) and increased the selection pressure to 1 mg/ml of paromomycin over 2 weeks, resulting in 26 viable clones, of which 12 were phenotypic. The remaining 22 clones also had a phenotype, but were not viable at higher paromomycin concentrations. Two clones with a phenotype were chosen (KD-C1 and -C2) for further analysis, for which we first confirmed the correct integration of the NeoR cassette into the Ttalv2 gene locus by PCR (Fig. 2A). Quantitative real-time PCR was next performed on RNA isolated from (i) wild-type cells, (ii) ParR cells without a phenotype, and (iii) ParR cells with a phenotype, the latter two with identical paromomycin treatments. Ttalv2 was downregulated with respect to wild-type expression only among the clones displaying a phenotype (Fig. 2B). Increasing the drug concentration even more, to further downregulate Ttalv2, was lethal. Generally, clones with a phenotype were very unstable, but identical phenotypes were observed after four independent rounds of transfection and selection. Knockdown of Ttalv2 led to cells with undefined surface protrusions and which appear to have lost the integrity of the pellicle and cell polarity, ultimately leading to multimacronuclear, as well as multimicronuclear, cells (Fig. 2C and D).
Fig 2.
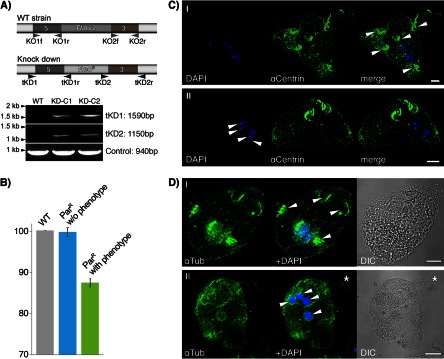
Macronuclear knockdown of TtALV2. (A) Genotypic profiling PCR of DNA of the wild type (WT) and two knockdown lines displaying a phenotype (KD-C1 and KD-C2) demonstrates the correct integration of the NeoR cassette into the Ttalv2 locus. Specific primer pairs (tKD1f and tKD1r and tKD2f and tKD2r) were used that only amplify a product upon correct 5′ and 3′ integration of the NeoR cassette into the Ttalv2 locus. A 940-bp-long fragment of TTHERM_00127110 served as a PCR control. The primer pairs KO1f and KO1r and KO2f and KO2r were used to amplify the upstream and downstream flanking regions of the Ttalv2 gene to generate the NeoR cassette for homologous recombination. αCentrin, anti-centrin; αTub, anti-tubulin. (B) Quantitative real-time PCR on RNA isolated from WT cells and paromomycin-resistant cells with and without a phenotype demonstrates the downregulation of TtALV2 to occur only in clones with a phenotype. (C and D) The ciliate appears to loose pellicle integrity and with it cell polarity and the ability to divide properly. No single cell axis can be defined, and multiple oral apparatuses, multinuclei, and macronuclei can be observed for each cell cluster. DIC, differential interference contrast. Scale bars, 10 μm.
The targeting properties of the charged repeat motifs are not limited to alveolins.
We selected three proteins from the list of predicted T. thermophila CRMPs and analyzed the targeting properties of their CRM domains. TTHERM_00777250 is a protein for which homologous proteins are only found among ciliates, and according to the Tetrahymena Functional Genomic Database (25), this protein is expressed at levels equivalent to the most abundant proteins of T. thermophila. The 311-amino-acid-long protein is entirely composed of highly conserved charged repeat motifs. During the early phase of expression, the GFP fusion protein localizes to ciliary basal bodies (see below), but after approximately 2 h, it begins to display an intricate association with the peripheral cytoskeleton of the ciliate. The protein then localizes to the cilia, the deep fiber bundle, and a row adjacent to the longitudinal microtubules (Fig. 3A; see Fig. S2F in the supplemental material). The second protein, TTHERM_00578520, is predicted to be 311 kDa and has a 20% global sequence identity (E value, 1.6e−58) to a 762-kDa protein of Toxoplasma gondii (TGME49_212880). Both proteins retrieve hits from the databases of proteins containing the domains annotated “SMC” and “viral A-type” but are otherwise of unknown function. The C-terminal domain of TTHERM_00578520 (amino acids 2026 to 2645 encoding CRMs) was sufficient to target GFP to the ciliary basal bodies (see Fig. S2C). A third CRMP tested, TTHERM_00678010, localized only to the cytosol (not shown).
Fig 3.
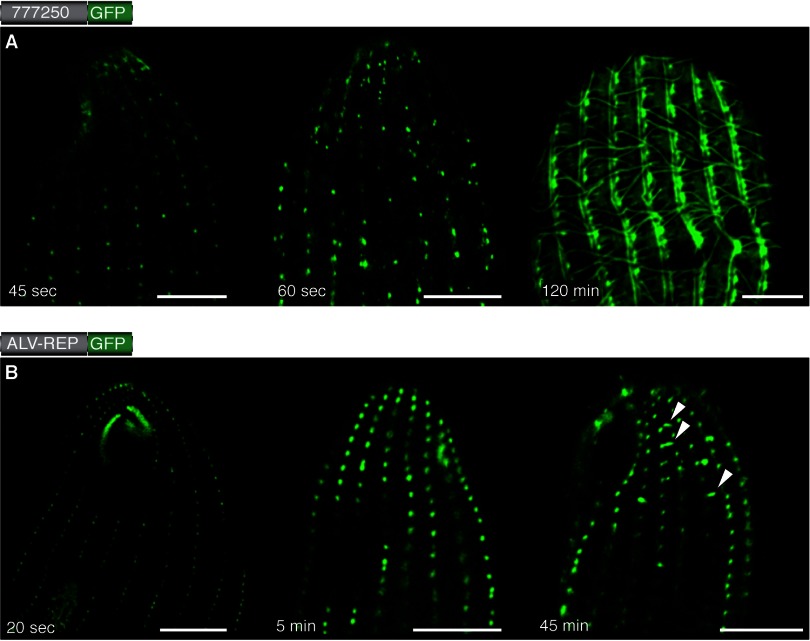
The basal bodies of Tetrahymena serve as nucleation centers. The expression of GFP fusion constructs was induced with CdCl2, and cells were fixed at different time points (given at the bottom of each image). For both exemplary constructs, the full-length TTHERM_00777250 (A) and the repeats of TtALV2 (B) fused to GFP, the first fluorescence was observed to occur at the basal bodies, from which the different patterns, depending on the individual constructs, emerged. Scale bars, 10 μm.
The integration of some CRMPs appears associated with ciliary basal bodies.
The failure of both the GFP- and HA-tagged TtALV2 to sort to the location of the endogenous protein was intriguing, particularly as they associated with the ciliary basal bodies, a core peripheral cytoskeletal structure of ciliates. We observed that many of the cytoskeletal proteins, or fragments thereof, also localize to the ciliary basal bodies after induction of expression. This prompted us to analyze whether this peculiarity may be associated with the integration of de novo-synthesized protein, which commences at the basal bodies. We induced expression of the GFP fusion constructs and fixed the cells at individual time points to analyze the progressive localization of the proteins. For ALV-REP::GFP, for example, fluorescence was observed almost immediately upon induction and initially at the basal bodies, especially around the oral apparatus, where basal bodies densely align to form the undulating membrane (26) and the three membranelles (Fig. 3B). Filamentous structures appeared to extend from the basal bodies ∼40 to 50 min after expression was induced. Similar results were obtained for TTHERM_00777250, where the final intricate localization was observed about 2 h after induction (Fig. 3A). These data imply a common propensity for CRMPs to initially associate in the regions of the basal bodies and then either develop or associate with various filamentous structures in this cell region.
A synthetic charged repeat associates with cytoskeletal structures.
The above collected information shows that some CRMPs, including the charged repeat motif unique to the alveolin family, can confer cytoskeletal association to a fusion protein. This, together with the enrichment of CRMPs in the Tetrahymena pellicle proteome, suggests some kind of “targeting function” for the motifs. To further test this hypothesis, we generated a synthetic charged repeat motif protein (sCRMP). The designed protein contains eight repetitions of a 26-amino-acid motif reading DEVINEQERIKQVIKINGQDLQERKE (Fig. 4A), which obeys the general amino acid composition and definition of CRMPs derived from the Tetrahymena pellicle proteome (E ≥ 6%, K ≥ 6%, Q ≥ 3%, and I + L + V ≥ 10%) (1). The repeats were fused between an N-terminal HA tag and a C-terminal GFP. sCRMP localized to a broad range of cytoskeletal structures in T. thermophila, which included the ciliary basal bodies, the cilia themselves, defined structures in close proximity to the basal bodies of the mature and newly forming oral apparatus (Fig. 4C), and the contractile vacuolar pores (Fig. 4D).
Charged repeat motifs might have a more general relevance in broader systems.
Given that CRMPs in the ciliate T. thermophila consistently display function or relevance to the cell cytoskeleton, we considered whether such motifs might pertain to a broader relevance in eukaryotic cytoskeletons. We screened a defined set of prominent cytoskeleton proteins from metazoans—and for which empirical evidence of an association with cytoskeletal structures exists—for charged repeat motifs and coiled coils. Genes coding for proteins such as keratin, kinesin, spectrin, epiplasmin, or tropomyosin, for example, were all found to encode such motifs, and these motifs were generally widespread across major cytoskeletal components (Table 1; see Fig. S1 in the supplemental material). Two-thirds of those proteins are furthermore predicted to form either dimeric or trimeric coiled-coil domains, including the repeat regions of the two Tetrahymena alveolin proteins, TtALV1 and TtALV2 (TTHERM_00945250 and TTHERM_00088030, respectively) (11). Intriguingly, half of the cytoskeletal proteins screened possess at least one SMC domain and one-fifth possess a viral A-like domain according to a simple BLASTP search (see Table S1 in the supplemental material).
Table 1.
Cytoskeletal proteins with charged repeat motifsa
| Organism | Gene product name | Motif |
ID no. | |||
|---|---|---|---|---|---|---|
| CRMP | Coiled coil | SMC | Viral A | |||
| Tetrahymena thermophila | TtEpiC | Yes | Dimeric | Yes | Yes | EAR95236.2 |
| TtBBC39 | Yes | Trimeric | Yes | No | EAS06694.1 | |
| Tetrin C | Yes | Trimeric | Yes | Yes | EAR86044.1 | |
| TtDFB1 | Yes | Dimeric | Yes | Yes | EAR96064.1 | |
| TtALV1 | Yes | Trimeric | No | No | EAS06387.1 | |
| TtALV2 | Yes | Dimeric | No | No | EAR92507.1 | |
| Toxoplasma gondii | TgICAMP1 | Yes | Dimeric | Yes | Yes | EEA99429.1 |
| TgSPM1 | Yes | None | No | No | EEA98311.1 | |
| TgIMC15 | Yes | None | No | No | EEB03770.1 | |
| Plasmodium falciparum | PfGAP45 | Yes | None | No | Yes | AAN36304.1 |
| PfALV1 | Yes | None | No | No | CAD51621.1 | |
| Homo sapiens | Keratin | Yes | Dimeric | Yes | Yes | EAW96642.1 |
| Spectrin | Yes | Dimeric | Yes | No | EAX00140.1 | |
| Kinesin light chain | Yes | Dimeric | Yes | No | BAB14039.1 | |
| Tropomyosin alpha chain | Yes | Dimeric | Yes | No | EAW77635.1 | |
Listed are proteins with known cytoskeletal functions that all contain charged repeat motifs. These were additionally screened for the predicted presence of coiled coils and whether a BLASTP search against Tetrahymena retrieved the SMC or viral A-type domain.
DISCUSSION
Ciliates are remarkable for their complex cell architecture, large cell size, and sophisticated taxis. These features are underpinned by a highly complex system of alveolar sacs just beneath the plasma membrane and a directly adjacent membrane cytoskeleton, of which TtALV2 is an important structural component. The alveolin protein family is best characterized in apicomplexan parasites, where this family also seems the most expanded (11). T. gondii, for instance, contains genes that encode at least 14 members, and it appears that no two alveolins have the exact same function, despite the fact that they all localize to the inner membrane complex (the alveolar sacs) of the parasite (12). Here we show that the ciliate protein TtALV2 displays a patchy localization that follows the longitudinal strip pattern known from the alveoli in Tetrahymena, which stretch from the apical to the basal end of the cell between the longitudinal microtubules (27). The epiplasmic band proteins A to C show a similar but more continuous association with the pellicular membranes (10, 28), indicating the TtALV2 localization might add a new layer of complexity to the pellicular membrane skeleton.
Our results further demonstrate that the correct integration of the alveolin protein into higher-order structures in the ciliate T. thermophila is multifarious, which was also observed for the apicomplexan parasite T. gondii (12). None of the TtALV2 fusion constructs showed identical localization, and none displayed localization identical to that of the endogenous protein, albeit all were associated with the peripheral membrane skeleton or oral apparatus. It is intriguing that ALV2::GFP and REP::GFP—which both include the coiled-coil domains—localize to the basal bodies and oral apparatus and also build filamentous structures that seem to associate with transversal microtubules (Fig. 1A to C), whereas N::GFP::C, devoid of CRMs and coiled-coil domains, mainly localizes to structures framing the membranelles of the oral apparatus, but not the basal bodies (Fig. 1D). This suggests an interdependency between (i) CRMs or coiled coils and the localization to the basal bodies and possible further membrane skeleton or alveolar associations from there on and (ii) the termini in combination with the repeats for the ultimate correct localization, in the case of TtALV2. It might be that the termini of TtALV2 need to be free to interact with one another to form alveolin filaments that associate with the alveolar membrane or into the membrane integrated “docking stations,” which would confirm a previous model suggested in reference 16. This means that the tags (both GFP and the smaller HA) might mask necessary terminal interaction sites. If that hypothesis is correct, then the N::GFP::C fusion protein, in which the termini are free to interact, might have been expected to localize correctly. However, the lack of the centrally charged repeats also perturbs the ability of the fusion constructs to associate with the alveolar membrane. Two constructs (N::GFP::ALV-C or N-ALV::GFP::C) could potentially resolve this question, but unfortunately they have resisted all cloning efforts so far. A membrane-specific interaction of coiled coils was also suggested to be essential for the correct function of the yeast (Saccharomyces cerevisiae) septin protein ScShs1 (29). It is unlikely that the localization of, for example, the full-length GFP-tagged TtALV2 is the result of overexpression. The Western blot (Fig. 1) indicates that the endogenous copy is actually more abundant than the tagged copy, and by far not all constructs expressed under the same promoter target to the basal bodies.
In Toxoplasma gondii, only the TgALV1 repeats localize to the cytoskeleton, while other fusion constructs localize to the cytosol (see Fig. S3 in the supplemental material). Similarly complex localization patterns were observed for other members of alveolin proteins in T. gondii (12), where individual domains of TgALV3 (TgIMC3) and TgALV8 (TgIMC8) targeted fusion constructs to different structures of the peripheral cytoskeleton of the parasite. As in our case, localization comparable to that of full-length proteins was not always achieved with just the domains of the proteins (12). Interestingly, the observed fluorescence distribution in cells expressing ALV-REP::mCherry appears to be similar to that previously reported for TgALV3, another CRMP (30).
The importance of TtALV2 is reflected by the severe phenotype induced through the knockdown of the gene. The protein appears indispensable for Tetrahymena, and its downregulation ultimately leads to either the loss of cell polarity through the interruption of the integrity of the cell's pellicle or somehow blocks cytokinesis, which can lead to similar phenotypes (31, 32). Several attempts to produce germ line knockouts failed, and the downregulation of the gene by macronuclear gene replacement induced massive multinuclear cells with unstructured cellular protrusions (Fig. 2C and D). Ttalv2 gene replacement through the NeoR cassette was confirmed through PCR, and only in cultures with a phenotype did we detect a downregulation of Ttalv2, albeit of only 13% on average. This suggests that Tetrahymena will only tolerate replacement of a small number of Ttalv2 gene copies in the macronucleus by the NeoR cassette. The observed phenotype is reminiscent of that of a Mob1 knockdown (TTHERM_00716080), a protein that localizes to the basal bodies and was the first reported cell polarity marker for Tetrahymena (33). It has been speculated that in apicomplexan parasites, alveolins form filaments that run along the alveolar sacs and are attached to intermembrane particles (16). Perhaps the knockdown of Ttalv2 leads to unstructured and collapsing alveolar sacs and parts of the membrane skeleton, which simultaneously induces the loss of cell polarity.
The C-terminal tags disrupted targeting to the endogenous localization of TtALV2, yet the fusion construct still associated with cytoskeletal structures. In general, we noticed an unusual number of tagged CRMPs, CRM-containing fragments thereof, and the sCRMP, to also localize to the ciliary basal body region, even when other structures were simultaneously labeled. This appears connected with the de novo synthesis of the proteins, as some of the CRMPs first localize to the basal bodies after their expression is induced and only then subsequently migrate or extend to proximate destinations (Fig. 3), altogether suggesting a two-step targeting mechanism. It might be that the basal bodies serve as some kind of “default destination” or “transfer station” for some peripheral cytoskeleton proteins in the ciliate. Maybe the ciliary basal bodies of Tetrahymena are not only microtubule organizing centers (MTOCs) but also can be considered intermediate filament-organizing centers (IFOCs). This suggestion is consistent with the previous observation that the ciliary basal bodies are pivotal cytoskeletal nucleation centers (7) that fulfill important recruitment and assembly functions for cytoskeletal structures. It reflects the complexity of the system that is best represented by the elaborate maturation of the ciliary basal bodies themselves (34), which contain up to a hundred proteins (7). However, how exactly this is achieved and to what set of proteins this applies to remain elusive and warrant further investigation. Future analysis, including detailed transmission electron microscopy of an individual candidate, might help us to better understand the individual steps underlying the complex integration of certain cytoskeletal components.
It is possible that CRMs could represent a general cytoskeleton-targeting motif, and other sequence elements—such as the N and C termini of alveolins, alone or in combination—can confer time- or cell-cycle-dependent localizations. Our results indicate that the repeats themselves are, in several cases, sufficient for cytoskeletal association (although not necessarily the correct localization). The ability of the designed sCRMP to target to similar structures as the alveolin fusions underpins this hypothesis and shows CRMs might not necessarily require a cognate partner for integration into the cell's cytoskeleton. In this respect, the localization of sCRMP within the cilia of Tetrahymena is particularly interesting. GFP expressed on its own in the ciliate does not localize to the cilia (see Fig. S2E in the supplemental material), and Kee and colleagues showed that a size-dependent barrier exists at the base of cilia (35); only proteins ≤41 kDa were able to pass the ciliary pore complex. At 55 kDa, the fusion construct sCRMP::GFP is larger than this, again supporting the independent role of CRMs being involved in a defined targeting process.
The membrane skeleton and cytoskeleton of an average eukaryote are likely composed of hundreds of proteins. One characteristic we found present in a broad range of cytoskeletal proteins is charged repeat motifs, which are also frequently predicted to form coiled-coil-based domains (see Table S1 in the supplemental material). This observation is in line with the general makeup of an intermediate filament protein that consists of a repetitive, core α-helical rod and variable head and tail domains (5, 36), which is also congruent with our model for TtALV2. Coiled coils and charged repeat motifs are obviously not limited to cytoskeletal proteins (2, 3), yet they do appear to be enriched among this set of proteins. A wealth of potentially unrecognized cytoskeleton-associated (maybe intermediate filament-like) proteins might exist not only in protists but perhaps even metazoan genomes. It will, however, be necessary to perform in-depth, genome-wide screens and examine the exact distribution of these motifs among all encoded proteins with known function. In the process, one also needs to consider that the characteristics of the CRMPs investigated here were originally defined from proteins identified in the pellicle proteome of T. thermophila. Although recently characterized cytoskeletal proteins of Toxoplasma were found to match these characteristics (37, 38), both of these organisms represent alveolates, and it might be required to fine-tune the predictions on sequences from a group of related organisms of interest. From machine-learning approaches, we know that these kinds of predictions tend to improve as the learning set of proteins and parameters are refined (39–42). We think that the identification of unrecognized cytoskeletal proteins will improve as definitions of the observed patterns are tailored toward the genomes of the organism being investigated and as the number of defining parameters—such as repeat length, frequency, charge distribution, and so forth—are increased. While we are beginning to see common patterns among cytoskeleton-associated proteins, understanding their core architecture and integration remains a challenge.
Supplementary Material
ACKNOWLEDGMENTS
S.B.G. thanks the Heinrich-Heine-University Düsseldorf (SFF starter grant), and the DFG (1825/3-1) for financial support. A Discovery grant (DP0664097) to R.F.W. and G.I.M. from the Australian Research Council is gratefully acknowledged.
12G10 was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biology, Iowa City, IA.
Footnotes
Published ahead of print 19 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00050-13.
REFERENCES
- 1. Gould SB, Kraft LGK, van Dooren GG, Goodman CD, Ford KL, Cassin AM, Bacic A, McFadden GI, Waller RF. 2011. Ciliate pellicular proteome identifies novel protein families with characteristic repeat motifs that are common to alveolates. Mol. Biol. Evol. 28:1319–1331 [DOI] [PubMed] [Google Scholar]
- 2. Rose A, Schraegle SJ, Stahlberg EA, Meier I. 2005. Coiled-coil protein composition of 22 proteomes—differences and common themes in subcellular infrastructure and traffic control. BMC Evol. Biol. 5:66 doi: 10.1186/1471-2148-5-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rose A, Meier I. 2004. Scaffolds, levers, rods and springs: diverse cellular functions of long coiled-coil proteins. Cell. Mol. Life Sci. 61:1996–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bornberg-Bauer E, Rivals E, Vingron M. 1998. Computational approaches to identify leucine zippers. Nucleic Acids Res. 26:2740–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Herrmann H, Aebi U. 2004. Intermediate filaments: molecular structure, assembly mechanism, and integration into functionally distinct intracellular scaffolds. Annu. Rev. Biochem. 73:749–789 [DOI] [PubMed] [Google Scholar]
- 6. Funahashi S, Sato T, Shida H. 1988. Cloning and characterization of the gene encoding the major protein of the A-type inclusion body of cowpox virus. J. Gen. Virol. 69:35–47 [DOI] [PubMed] [Google Scholar]
- 7. Kilburn CL, Pearson CG, Romijn EP, Meehl JB, Giddings TH, Jr, Culver BP, Yates JR, III, Winey M. 2007. New Tetrahymena basal body protein components identify basal body domain structure. J. Cell Biol. 178:905–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Honts JE, Williams NE. 1990. Tetrins: polypeptides that form bundled filaments in Tetrahymena. J. Cell Sci. 96:293–302 [DOI] [PubMed] [Google Scholar]
- 9. Williams NE, Honts JE. 1995. Isolation and fractionation of the Tetrahymena cytoskeleton and oral apparatus. Cilia Flagella 47:301–306 [DOI] [PubMed] [Google Scholar]
- 10. Williams NE, Honts JE, Dress VM, Nelsen EM, Frankel J. 1995. Monoclonal antibodies reveal complex structure in the membrane skeleton of Tetrahymena. J. Eukaryot. Microbiol. 42:422–427 [DOI] [PubMed] [Google Scholar]
- 11. Gould SB, Tham W-H, Cowman AF, McFadden GI, Waller RF. 2008. Alveolins, a new family of cortical proteins that define the protist infrakingdom Alveolata. Mol. Biol. Evol. 25:1219–1230 [DOI] [PubMed] [Google Scholar]
- 12. Anderson-White B, Ivey F, Cheng K. 2010. A family of intermediate filament-like proteins is sequentially assembled into the cytoskeleton of Toxoplasma gondii. Cell. Microbiol. 13:18–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mann T, Beckers C. 2001. Characterization of the subpellicular network, a filamentous membrane skeletal component in the parasite Toxoplasma gondii. Mol. Biochem. Parasitol. 115:257–268 [DOI] [PubMed] [Google Scholar]
- 14. Tremp AZ, Khater EI, Dessens JT. 2008. IMC1b is a putative membrane skeleton protein involved in cell shape, mechanical strength, motility, and infectivity of malaria ookinetes. J. Biol. Chem. 283:27604–27611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kono M, Herrmann S, Loughran NB, Cabrera A, Engelberg K, Lehmann C, Sinha D, Prinz B, Ruch U, Heussler V, Spielmann T, Parkinson J, Gilberger TW. 2012. Evolution and architecture of the inner membrane complex in asexual and sexual stages of the malaria parasite. Mol. Biol. Evol. 29:2113–2132 [DOI] [PubMed] [Google Scholar]
- 16. Bullen HE, Tonkin CJ, O'Donnell RA, Tham WH, Papenfuss AT, Gould S, Cowman AF, Crabb BS, Gilson PR. 2009. A novel family of apicomplexan glideosome-associated proteins with an inner membrane-anchoring role. J. Biol. Chem. 284:25353–25363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Williams NE, Honts JE, Jaeckel-Williams RF. 1987. Regional differentiation of the membrane skeleton in Tetrahymena. J. Cell Sci. 87:457–463 [DOI] [PubMed] [Google Scholar]
- 18. Shang Y, Song X, Bowen J, Corstanje R, Gao Y, Gaertig J, Gorovsky M. 2002. A robust inducible-repressible promoter greatly facilitates gene knockouts, conditional expression, and overexpression of homologous and heterologous genes in Tetrahymena thermophila. Proc. Natl. Acad. Sci. U. S. A. 99:3734–3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mochizuki K. 2008. High efficiency transformation of Tetrahymena using a codon-optimized neomycin resistance gene. Gene 425:79–83 [DOI] [PubMed] [Google Scholar]
- 20. Cassidy-Hanley D, Bowen J, Lee JH, Cole E, VerPlank LA, Gaertig J, Gorovsky MA, Bruns PJ. 1997. Germline and somatic transformation of mating Tetrahymena thermophila by particle bombardment. Genetics 146:135–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kloetzel JA, Baroin-Tourancheau A, Miceli C, Barchetta S, Farmar J, Banerjee D, Fleury-Aubusson A. 2003. Plateins: a novel family of signal peptide-containing articulins in euplotid ciliates. J. Eukaryot. Microbiol. 50:19–33 [DOI] [PubMed] [Google Scholar]
- 22. Gotesman M, Hosein RE, Gavin RH. 2011. MyTH4, independent of its companion FERM domain, affects the organization of an intramacronuclear microtubule array and is involved in elongation of the macronucleus in Tetrahymena thermophila. Cytoskeleton (Hoboken) 68:220–236 [DOI] [PubMed] [Google Scholar]
- 23. Poklepovich TJ, Rinaldi MA, Tomazic ML, Favale NO, Turkewitz AP, Nudel CB, Nusblat AD. 2012. The cytochrome b5 dependent C-5(6) sterol desaturase DES5A from the endoplasmic reticulum of Tetrahymena thermophila complements ergosterol biosynthesis mutants in Saccharomyces cerevisiae. Steroids 77:1313–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yao MC, Yao CH, Halasz LM, Fuller P, Rexer CH, Wang SH, Jain R, Coyne RS, Chalker DL. 2007. Identification of novel chromatin-associated proteins involved in programmed genome rearrangements in Tetrahymena. J. Cell Sci. 120:1978–1989 [DOI] [PubMed] [Google Scholar]
- 25. Miao W, Xiong J, Bowen J, Wang W, Liu Y, Braguinets O, Grigull J, Pearlman RE, Orias E, Gorovsky MA. 2009. Microarray analyses of gene expression during the Tetrahymena thermophila life cycle. PLoS One 4:e4429 doi: 10.1371/journal.pone.0004429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bakowska J, Frankel J, Nelsen EM. 1982. Regulation of the pattern of basal bodies within the oral apparatus of Tetrahymena thermophila. J. Embryol. Exp. Morphol. 69:83–105 [PubMed] [Google Scholar]
- 27. Satir BH, Wissig SL. 1982. Alveolar sacs of Tetrahymena: ultrastructural characteristics and similarities to subsurface cisterns of muscle and nerve. J. Cell Sci. 55:13–33 [DOI] [PubMed] [Google Scholar]
- 28. Williams NE. 2004. The epiplasm gene EPC1 influences cell shape and cortical pattern in Tetrahymena thermophila. J. Eukaryot. Microbiol. 51:201–206 [DOI] [PubMed] [Google Scholar]
- 29. Meseroll RA, Howard L, Gladfelter AS. 2012. Septin ring size scaling and dynamics require the coiled-coil region of Shs1p. Mol. Biol. Cell 23:3391–3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gubbels MJ, Wieffer M, Striepen B. 2004. Fluorescent protein tagging in Toxoplasma gondii: identification of a novel inner membrane complex component conserved among Apicomplexa. Mol. Biochem. Parasitol. 137:99–110 [DOI] [PubMed] [Google Scholar]
- 31. Jerka-Dziadosz M, Frankel J. 1979. A mutant of Tetrahymena thermophila with a partial mirror-image duplication of cell surface pattern. I. Analysis of the phenotype. J. Embryol. Exp. Morphol. 49:167–202 [PubMed] [Google Scholar]
- 32. Jerka-Dziadosz M, Jenkins LM, Nelsen EM, Williams NE, Jaeckel-Williams R, Frankel J. 1995. Cellular polarity in ciliates: persistence of global polarity in a disorganized mutant of Tetrahymena thermophila that disrupts cytoskeletal organization. Dev. Biol. 169:644–661 [DOI] [PubMed] [Google Scholar]
- 33. Tavares A, Goncalves J, Florindo C, Tavares AA, Soares H. 2012. Mob1: defining cell polarity for proper cell division. J. Cell Sci. 125:516–527 [DOI] [PubMed] [Google Scholar]
- 34. Pearson CG, Winey M. 2009. Basal body assembly in ciliates: the power of numbers. Traffic 10:461–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kee HL, Dishinger JF, Blasius TL, Liu CJ, Margolis B, Verhey KJ. 2012. A size-exclusion permeability barrier and nucleoporins characterize a ciliary pore complex that regulates transport into cilia. Nat. Cell Biol. 14:431–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chernyatina AA, Nicolet S, Aebi U, Herrmann H, Strelkov SV. 2012. Atomic structure of the vimentin central alpha-helical domain and its implications for intermediate filament assembly. Proc. Natl. Acad. Sci. U. S. A. 109:13620–13625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tran JQ, de Leon JC, Li C, Huynh M-H, Beatty W, Morrissette NS. 2010. RNG1 is a late marker of the apical polar ring in Toxoplasma gondii. Cytoskeleton (Hoboken, NJ) 67:586–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tran JQ, Li C, Chyan A, Chung L, Morrissette NS. 2012. SPM1 stabilizes subpellicular microtubules in Toxoplasma gondii. Eukaryot. Cell 11:206–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Burstein D, Zusman T, Degtyar E, Viner R, Segal G, Pupko T. 2009. Genome-scale identification of Legionella pneumophila effectors using a machine learning approach. PLoS Pathog. 5:e1000508 doi: 10.1371/journal.ppat.1000508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Burstein D, Gould SB, Zimorski V, Kloesges T, Kiosse F, Major P, Martin WF, Pupko T, Dagan T. 2012. A machine learning approach to identify hydrogenosomal proteins in Trichomonas vaginalis. Eukaryot. Cell 11:217–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Verma R, Melcher U. 2012. A support vector machine based method to distinguish proteobacterial proteins from eukaryotic plant proteins. BMC Bioinformatics 13(Suppl. 15):S9 doi: 10.1186/1471-2105-13-S15-S9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Woodcroft BJ, McMillan PJ, Dekiwadia C, Tilley L, Ralph SA. 2012. Determination of protein subcellular localization in apicomplexan parasites. Trends Parasitol. 28:546–554 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.