Abstract
Fungi have been used as model systems to define general processes in eukaryotes, for example, the one gene-one enzyme hypothesis, as well as to study polar growth or pathogenesis. Here, we show a central role for the regulator protein Ras in a mushroom-forming, filamentous basidiomycete linking growth, pheromone signaling, sexual development, and meiosis to different signal transduction pathways. ras1 and Ras-specific gap1 mutants were generated and used to modify the intracellular activation state of the Ras module. Transformants containing constitutive ras1 alleles (ras1G12V and ras1Q61L), as well as their compatible mating interactions, did show strong phenotypes for growth (associated with Cdc42 signaling) and mating (associated with mitogen-activated protein kinase signaling). Normal fruiting bodies with abnormal spores exhibiting a reduced germination rate were produced by outcrossing of these mutant strains. Homozygous Δgap1 primordia, expected to experience increased Ras signaling, showed overlapping phenotypes with a block in basidium development and meiosis. Investigation of cyclic AMP (cAMP)-dependent protein kinase A indicated that constitutively active ras1, as well as Δgap1 mutant strains, exhibit a strong increase in Tpk activity. Ras1-dependent, cAMP-mediated signal transduction is, in addition to the known signaling pathways, involved in fruiting body formation in Schizophyllum commune. To integrate these analyses of Ras signaling, microarray studies were performed. Mutant strains containing constitutively active Ras1, deletion of RasGap1, or constitutively active Cdc42 were characterized and compared. At the transcriptome level, specific regulation highlighting the phenotypic differences of the mutants is clearly visible.
INTRODUCTION
The homobasidiomycete white-rot fungus Schizophyllum commune Fr. grows mainly on wood of deciduous trees and less frequently on coniferous wood and shoots and roots of herbs, with a worldwide distribution (1). It is genetically tractable and has been used to study the tetrapolar mating system (2), as well as fruiting body formation (3–6).
Mating is regulated by two independent factors, where the A locus encodes homeodomain transcription factors (7) while B genes are pheromone receptor and pheromone genes (8). Compatible mating leads to the establishment of a fertile dikaryon that, under favorable environmental conditions, is able to develop fruiting bodies. Within the basidia, karyogamy and meiosis occur, linking spore production to a compatible mating interaction (9).
Little is known about the signal transduction involved in fruiting body formation by basidiomycetes (4, 9). One of the major players in intracellular signaling is the small G protein Ras. Since the 1960s, Ras proteins have been studied for their regulatory role in oncogenesis and in intracellular signaling pathways (for a review, see reference 10). Their ability to bind and hydrolyze GTP allows Ras proteins to exist either in an active, GTP-bound or an inactive, GDP-bound form. Guanine nucleotide exchange factors promote the formation of the active, GTP-bound form of Ras by exchange of GDP for GTP. GTPase-activating proteins accelerate the intrinsic GTP hydrolytic activity of Ras to promote the formation of inactive Ras. Active, GTP-bound Ras protein has been shown to mediate cellular proliferation via mitogen-activated protein kinase (MAPK), cyclic AMP (cAMP), and Cdc42 signaling (11–13).
Ras signaling and morphotypes connected to this regulatory pathway have been studied with several fungi. Two Ras-encoding genes have been identified in almost all fungal species, as in the yeast Saccharomyces cerevisiae (14). Both Ras2 and Ras1 expressed at lower levels can activate adenylate cyclase, leading to enhanced intracellular cAMP levels. The higher cAMP level, in turn, activates protein kinase A (PKA) by binding the regulatory subunits of the tetramer, which releases the active catalytic subunits (15). In addition, Ras2 activates a MAPK cascade through Cdc42 (16). Through this pathway, Ras influences the cytoskeleton (17). Differences in regulatory pathways have been shown by investigations of different fungi, e.g., with Schizosaccharomyces pombe, where the single Ras protein regulates pheromone response, sporulation, and morphogenesis (18). Ras1 of S. pombe is not involved in cAMP signaling.
The isolation of the basidiomycete Cryptococcus neoformans (19), Ustilago maydis (20), Lentinus edodes (21), Coprinopsis cinerea (NCBI: BAA02552), Suillus bovinus (NCBI: AAF65465, AAF65466), and Laccaria bicolor (22) Ras-encoding genes has also been described. The two Ras-encoding genes of S. commune and their homology to known small G proteins of other fungi have been reviewed recently (13). In the dimorphic pathogenic basidiomycete C. neoformans, two Ras proteins regulate pheromone response, filamentous growth, and the actin cytoskeleton (19, 23). The two Ras proteins of the heterobasidiomycete U. maydis have distinct roles. While Ras2 regulates pheromone response, as well as filamentous growth and virulence, via a MAPK cascade (20), Ras1 regulates the yeast-filamentous transition and enhances pheromone expression via cAMP signaling. These signal transduction pathways are connected through the transcription factor Prf1 (24).
In an attempt to identify mating type-specific, B-regulated genes, Schubert et al. (25) isolated a cDNA fragment coding for a Ras-dependent Gap from S. commune. Disruption of gap1, and hence enhanced Ras signaling, led to a reduced and disorientated growth pattern. Homozygous Δgap1/Δgap1 dikaryons showed a strong phenotype during clamp formation, where hook cells failed to fuse with the peg beside them. Instead, the hooks fused with nearby developing branches, thus re-establishing a dikaryon with subterminal, aberrant clamp-like structures. Mature fruiting bodies formed no or abnormal gills lacking spore production (25). The phenotype of absent or partially developed gills resembled the description of aberrant fruiting bodies by Schwalb (26), where extracellular cAMP had been added to dikaryons. Since deletion of gap1 leads to inferior GTP hydrolysis and hence results in sustained activation of Ras signaling, we postulated that Ras regulates intracellular cAMP levels in S. commune.
Here, we describe the influence of the small G protein Ras1 and the RasGap protein Gap1 on mushroom formation of the basidiomycete S. commune. We could show that activation of Ras1 affects polar growth, development of side branches, mating, fruiting body development, and spore formation. In order to test the involvement of intracellular cAMP signaling, we identified the gene coding for a catalytic PKA subunit and measured PKA activities in constitutively active Ras and Δgap1 mutants and performed transcriptome analyses to identify targets of signaling.
MATERIALS AND METHODS
Strains and growth conditions.
The strains of S. commune used in this study (Table 1) were grown on minimal medium or complex yeast medium (CYM) (27) with or without supplementation with 4 mM tryptophan at 28°C. For the formation of fruiting bodies, strains were grown at 30°C in darkness for 5 days. From then on, they were kept in a normal day-night rhythm at 25°C with no exposure to direct sunlight. For transcriptome studies, mycelia were grown on a cellophane membrane covering CYM agar for 3 days at 28°C in the dark, except for SccdcG12V, which was grown on carboxymethyl cellulose (28).
Table 1.
S. commune strains used in this study
| Strain | Mating type, relevant genotype | Source or reference |
|---|---|---|
| 4-40 | matA4,6 matB1,1 | JMRCa |
| 4-39 | matA1,1 matB3,2 | JMRC |
| 12-43 | matA3,5 matB2,2 ura− | JMRC |
| H4-8 | matA4,6 matB3,2 | 4 |
| T2 | matA4,1 matB2,2 ura− trp− | JMRC |
| T3 | matA8,1 matB2,2 ura− trp− | JMRC |
| T62 | matA3,5 matB7,2 ura− | JMRC |
| W22 | matA4,6 matB3,2 | JMRC |
| II-1 | matA1,1 matB3,2 ura− trp− | 25 |
| F28 | matA4,6 matB3,2 Δgap1 | 25 |
| F15 | matA1,1 matB1,1 Δgap1 | 25 |
| F20H | matA4,6 matB3,2 Δgap1 | 25 |
| 12-44 | bse mutant | 3 |
| T2G12V | matA4,1 matB2,2 ura− | This work |
| II-1Q61L | matA4,6 matB1,1 ura− | This work |
| II-1G12V | matA4,6 matB1,1 trp− | This work |
| SccdcG12V | matA4,2 matB2,3 | 28 |
JMRC, Jena Microbial Resource Center.
Diameters of at least 10 colonies of each S. commune strain were measured every 24 h for up to 7 days. Mycelia used for DNA and RNA isolation were grown in liquid CYM supplemented with tryptophan if necessary (27). The potential phenotypic differences relating to genotypic variance between strains were compensated for by using multiple strains of different mating types throughout this study (Table 1). Escherichia coli K-12 DH5α (Bethesda Research Laboratories) was used for plasmid construction.
Characterization of ras1.
A ras1 genomic gene fragment was amplified from S. commune strain 4-40 by PCR with primers Rasdown1 and Rasup2 (see Table S1 in the supplemental material), which were designed from a ras1 cDNA fragment (AF268471). For DNA and RNA isolation, standard procedures were followed (25, 29). An additional 840 bp of the 5′ region was amplified by thermal asymmetric interlaced PCR (30). The entire ras1 gene with its native promoter region was amplified with primers RasPromdown and Rasup2. Cloning of the 1.3-kb fragment into pDrive (Qiagen, Hilden, Germany) resulted in the pRasWT plasmid. DNA sequences (JenaGen, Jena, Germany) were analyzed with DNASTAR (Lasergene, Madison, WI).
For Southern blot analysis, DNA was digested with restriction enzymes, separated on an agarose gel, and blotted onto nylon membranes (Porablot NY amp; Macherey-Nagel, Düren, Germany). Labeling of the DNA probe, hybridization, and detection of DNA-DNA hybrids were performed by using the labeling and detection system of Roche Diagnostics (Mannheim, Germany) according to the manufacturer's recommendations under stringent conditions at 65°C.
For quantitative reverse transcription (qRT)-PCR, total RNA extracted with the RNeasy plant kit (Qiagen, Hilden, Germany) was treated with RNase-free DNase (Qiagen, Hilden, Germany), cDNA was synthesized (iScript; Bio-Rad, Munich, Germany), and a real-time PCR assay was performed (MiniOpticon; Bio-Rad, Munich, Germany) with Maxima SYBR green (Thermo Scientific, Waltham, MA) with two technical and two biological replicates. PCR was performed with 2 μl of cDNA in 36 cycles of denaturation for 20 s at 94°C, annealing for 20 s at 60°C, and extension for 20 s at 72°C after initial denaturation for 10 min at 94°C and followed by a final melting curve analysis from 65 to 95°C at 0.2°C/s. Gene-specific efficiencies were calculated from the slope of a standard curve obtained with different cDNA concentrations (0.5, 1, 5, 10, 100, and 150 ng cDNA/12.5 μl PCR mixture). The expression of ras1 was analyzed with primers ras-1 and ras-2 (primer efficiency, 89.9%). Primers tef-1 and tef-2, spanning an intron for the detection of contaminating genomic DNA, were used as internal standards (primer efficiency, 106%). Student's t test was used for statistical analysis.
Mutagenesis.
Plasmid pRasWT was used as the template for three site-directed mutagenesis experiments with the QuikChange site-directed mutagenesis kit (Stratagene, Groningen, The Netherlands). The mutagenic primer pairs used were G12V-1 and G12V-2, Q61L-1 and Q61L-2, and G15N-1 and G15N-2 (see Table S1 in the supplemental material). After mutagenesis, each insert was digested with EcoRI. The inserts were cloned into S. commune selection plasmids ptrp and pCHI. Plasmid ptrp is a pBluescript SKII derivate that harbors the EcoRI-HindIII fragment of cosmid pTC20 containing the trp1 gene (31). The resulting plasmids were named Raswt-trp, RasG12V-trp, RasQ61L-trp, and RasG15N-trp. For uracil as a selection marker, the pCHI vector (32) was used, creating plasmids Raswt-CHI, RasG12V-CHI, RasQ61L-CHI, and RasG15N-CHI.
S. commune transformation.
Protoplast transformation and selection of transformants were performed as reported by Schuren and Wessels (33). The resulting S. commune transformants, T2G12V, II-1Q61L, and II-1G12V, were used for immunofluorescence staining, expression analyses, and phenotypic characterization.
Protein kinase A activity.
S. commune strains were grown for 3 days in liquid CYM with shaking at 28°C. After harvesting, mycelia were ground in liquid nitrogen, suspended in extraction buffer (25 mM Tris/HCl, 1 mM dithiothreitol, 1 mM EDTA, pH 7.4), and incubated on ice for 15 min. Samples were centrifuged at 4°C and 13,000 rpm for 10 min. The supernatant with the soluble protein fraction was stored at −80°C. Protein concentrations were determined by the method of Bradford (34). The activity of PKA was measured with the PepTag Assay (Promega, Mannheim, Germany); 1.5 μg/ml protein-containing supernatant was used in the assay, in accordance with the instructions of the manufacturer. One strain (12-43) did show a prevalent retardation of migration, likely by binding of cellular components, of the nonphosphorylated kemptide, albeit with lower intensity, which is also visible with the other strains.
Immunofluorescence staining.
For immunocytochemical staining (35), small colonies of S. commune were inoculated on agar plates next to sterile coverslips. Mycelia attached to the coverslips were transferred into fixation solution, which was PME buffer containing 50 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES, pH 6.7), 25 mM EGTA (pH 8.0), 5 mM MgSO4, and 4% formaldehyde, for 1 h at room temperature and then washed three times with PME buffer. Cell walls were digested with lysing enzyme (30 mg lysozyme in 500 μl PME buffer containing 500 μl egg white) for 1 to 1.5 h at room temperature and then washed three times in PME buffer. The mycelium was incubated for 5 min at room temperature in extraction solution (100 mM PIPES [pH 6.7], 25 mM EGTA [pH 8.0], 0.1% IGEPAL [Sigma-Aldrich, Munich, Germany]) and then for 10 min at −20°C in methanol. After repeated washing with PME buffer, blocking was performed with 3% milk powder in TBS (20 mM Tris/HCl [pH 7.6], 137 mM NaCl, 0.1 vol% Tween 20) at room temperature. The mycelium was then incubated overnight at 4°C with the first antibody (mouse anti-tubulin and rabbit anti-actin; Sigma-Aldrich, Munich, Germany) diluted 1:500 and 1:250, respectively, in TBS with 3% milk powder and then washed three times with TBS. The mycelium was incubated for 1 h at room temperature with the second antibody (fluorescein isothiocyanate-conjugated goat anti-mouse and Cy3-conjugated sheep anti-rabbit; Sigma-Aldrich, Munich, Germany) diluted 1:100 in TBS with 3% milk powder. After being washed with TBS, the coverslip was mounted in 0.1 M Tris/HCl (pH 8.0)–50% glycerol–1 mg/ml phenylene diamine dihydrochloride containing, if necessary, 0.1 to 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) and observed by fluorescence microscopy (Axioplan 2; Zeiss, Jena, Germany). Images were taken with an Insight Firewire 4 image sample digital camera (Diagnostic Instruments, Sterling Heights, MI) and Spot version 4.6 (Diagnostic Instruments, Munich, Germany).
Cross sections.
Primordia and fruiting bodies at different stages of development were fixed for at least 7 days in Pfeiffer's solution (36) containing 30% methanol, 13.5% formalin, and 3.5% acetic acid. Dehydration was performed with Pfeiffer's solution in a methanol dilution series. The objects were embedded in Technovit 7100 (Kulzer-Mikrotechnik, Hanau, Germany). Deviating from the manufacturer's instructions, the time of preinfiltration was increased (1 to 2 days instead of 2 h), as was the quantity of hardener (8.3% instead of 6.7%). Microtome sections 8 to 10 μm thick were obtained with rotation microtome Mikrom HM 355 (Microm, Walldorf, Germany). Sections were attached to glass coverslips with n-propanol (37) and stained with toluidine blue O (0.1% [wt/vol] in H2O). Coverslips were attached with Merckoglas (Merck, Darmstadt, Germany). Selected unstained slides were stained with the nuclear stain DAPI in embedding solution for DAPI (35). The slides were incubated with 10 μl DAPI (1 μg/ml) in phosphate-buffered saline or embedding medium at 25°C for 30 min. This treatment was repeated once with fresh DAPI. Microscopic examination was performed with an Axioplan 2 microscope (Zeiss, Jena, Germany) with filter 02. Micrographs were taken with an Insight Firewire 4 image sample digital camera (Diagnostic Instruments, Sterling Heights, MI) and Spot version 4.6 software (Diagnostic Instruments, Sterling Heights, MI).
Microarray-based transcriptome analysis.
Microarrays of the 13,181 genes predicted were designed from the genome sequence (NCBI 67931) of S. commune strain H4-8 by febit biomed (Heidelberg, Germany). The monokaryons 4-39 and 12-43 and the dikaryon W22 × 12-43, which were used for comparison, had been deposited in NCBI Gene Expression Omnibus series GSE26401 earlier (38). The overall experimental design (total RNA extraction, microarray analysis by biotin-labeled cDNA, and processing and normalization of data) was as previously described (38). Both studies GSE26401 and GSE43965 were performed in parallel. For the study described here, a ≥3-fold change cutoff and a P value cutoff of ≤0.05 were used. The Pearson coefficient for all array experiments (overall, 16 arrays; so far, data for 11 arrays are available online) was averaged by using 0.972 for technical replicates and 0.958 for biological replicates. Two technical and at least two biological replicates were used for each analysis. Genes showing differences between two monokaryons (12-43 and 4-39) were omitted from the analysis as strain-specific differences if the change was >2- or <−2-fold. A comparison of a mutant and the wild type defined the regulated genes specific for this mutant. For example, the array signals of the monokaryotic mutant T2G12V were compared to those of monokaryotic strains 12-43 and 4-39. Genes showing differences in these comparisons were considered differentially regulated because of mutation of Ras1. Also, dikaryotic and monokaryotic expression was controlled for the subsets chosen; regulated genes of interest are found in (Ras1 versus monokaryon) versus (gap1 versus dikaryon) but not in Ras1 versus gap1. In this way, relevant changes in expression due to ras1 mutation were enriched in the sample for detailed analysis.
Microarray data accession number.
The data included in this publication were deposited in the NCBI Gene Expression Omnibus within Platform GPL11376, series GSE43965 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE43965). This holds true for mutant strains T2G12V and SccdcG12V and the F15 × F28 (Δgap1) dikaryon.
RESULTS
Isolation and characterization of ras1 and ras2 of S. commune.
A 1.9-kb genomic fragment coding for Ras1 was cloned from S. commune 4-40. The open reading frame of 576 bp codes for 192 amino acids disrupted by five introns, one of which is found in front of the translation start site. Within the 840-bp 5′ upstream genomic region, no conserved promoter elements were identified. The highest sequence similarities of the conceptual translation product observed were to Ras1 from L. bicolor, with 88% identity, and to Ras2 from S. bovinus, with 82% identity. While ras1 appeared as a single gene in the haploid genome by Southern blotting, a second Ras-encoding gene with only 44.3% amino acid identity was identified in the genome sequence. Ras2 is also expressed, as evidenced by cDNA and massively parallel signature sequencing analyses (4) and shows 88.8% identity to Ras1 of C. neoformans and 82.9% identity to Ras2 of U. maydis. Both S. commune Ras proteins contain the consensus sequences for functional GTP binding domains (see Fig. S2 in the supplemental material) and CaaX farnesylation signal consensus sequences at the C terminus indicating membrane attachment, as known also for other Ras proteins.
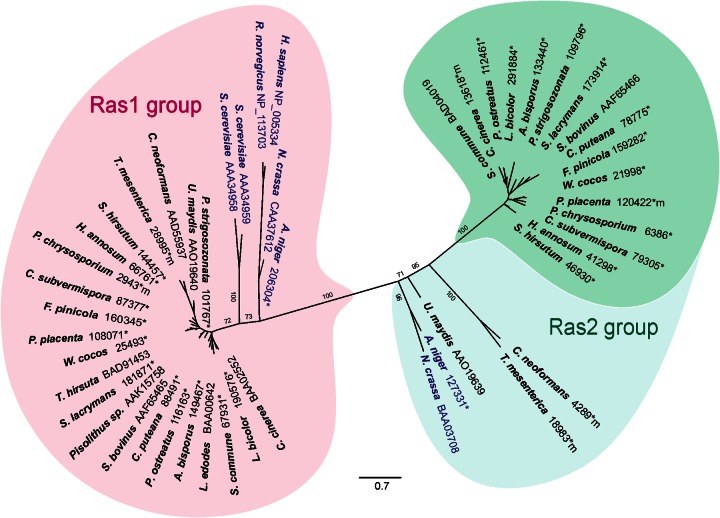
Since the similarity between Ras1 and Ras2 of S. commune was limited and two Ras proteins have been identified in most fungi, a phylogenetic analysis was performed to check for different clusters of Ras proteins. Two Ras clusters can be separated, with one cluster, the Ras1 group, containing basidiomycete sequences, two Ras proteins of S. cerevisiae and one each of Aspergillus nidulans and Neurospora crassa (Fig. 1). For the latter two filamentous ascomycetes, a second Ras protein of the Ras2 group can be found in a slightly offset phylogenetic line that separates outgroups, ascomycetes and phylogenetically distant basidiomycetes from the mushroom-forming basidiomycete group. The protein termed Ras1 of S. commune in earlier publications (4, 9, 13, 39) clusters in this Ras2 group. Ras2 of S. commune is represented in the Ras1-like group. This phylogenetic separation of two Ras protein-encoding genes for each filamentous fungus might be indicative of distinct, only partially overlapping functions for the two proteins, and was therefore investigated in detail during S. commune development.
Fig 1.
Rooted phylogenetic tree of Ras proteins based on maximum likelihood as implemented in RAxML (substitution matrix Blosum62) (60). HRas (Homo sapiens), KRas (Rattus norvegicus), and ascomycete Ras were included as an outgroup (blue lettering). NCBI and JGI (*) accession numbers are shown. The sequences compiled are those of N. crassa, A. nidulans, H. sapiens, R. norvegicus, S. cerevisiae, Punctularia strigosozonata; U. maydis, C. neoformans, Tremella mesenterica; Stereum hirsutum; Heterobasidion annosum; Phanaerochaete chrysosporium; Ceriporiopsis subvermispora; Fomitopsis pinicola; Postia placenta; Wolfiporia cocos; Trametes hirsuta; Serpula lacrymans; S. bovinus, Coniophora puteana; Pleurotus ostreatus; Agaricus bisporus; Lentinula edodes; S. commune, L. bicolor, and C. cinerea.
Expression of constitutively active Ras1-encoding alleles.
Since Ras2 in U. maydis had been seen to be involved in pheromone signaling and filamentous growth, we investigated the function of the homologous Ras1 protein of S. commune in more detail. To ascertain the role of ras1 in the sexual development of S. commune, independent mutations (G12V, Q61L) were introduced into the GTP hydrolysis domain. Both mutations abolish the intrinsic GTPase activity, causing Ras proteins to become constitutively active (10). The mutant alleles under the control of the native promoter (378-bp upstream sequence) were cloned and transformed into wild-type strains. The transformants containing the dominant active allele showed an obvious and stable phenotype exhibiting a small colony size and hyphae with a disoriented growth pattern. No comparable changes in the hyphal phenotype were observed in transformants with extra copies of the wild-type ras1 allele. Southern hybridization analysis indicated ectopic integration of one to seven copies of the modified alleles into the genome of transformants in addition to the endogenous, wild-type copy of ras1 (data not shown). A dominant negative allele containing the mutation G15N rendering the GTP-binding site nonfunctional, however, did not lead to stable integration in >2,000 transformants, potentially hinting at an essential function for Ras1 that is obliterated by the dominant negative function of an integrated mutant gene copy.
The resulting constitutively active Ras1 strains T2G12V, II-1G12V, and II-1Q61L were analyzed by qRT-PCR to score possible changes in expression levels. A Δgap1 mutant strain (F15, F28) was used for comparison, since deletion of gap1 also leads to the accumulation of active, GTP-bound Ras in the cell. The expression of ras1 in all of the mutant strains was similar to that in the wild type (Fig. 2). This indicates transcriptional regulation independent of gene copy number, controlled by the ras1 promoter fragment. Our finding of no obvious transcriptional regulation during development is in good agreement with activation at the protein level through the binding of GTP.
Fig 2.
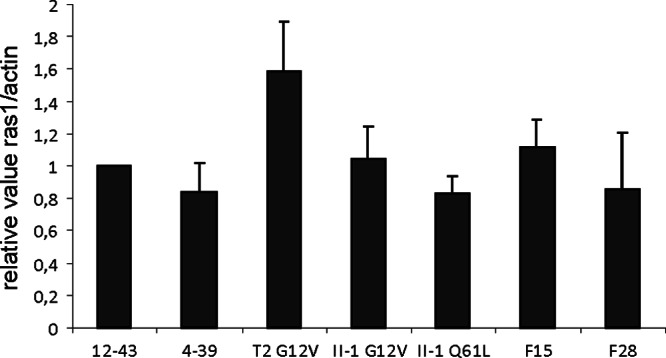
Expression of ras1 measured by qRT-PCR. No statistically significantly different expression is visible in different wild-type (12-43, 4-39), constitutively active Ras1 (T2G12V, II-1G12V, II-1Q61L), or Δgap1 mutant (F15, F28) strains.
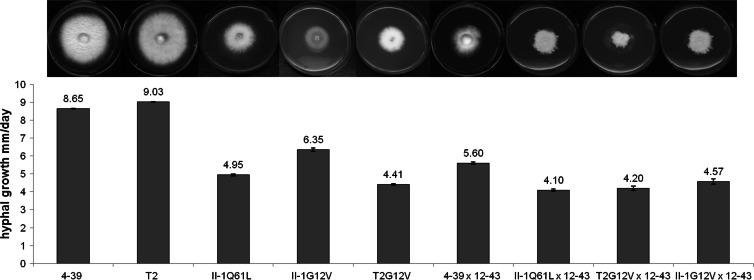
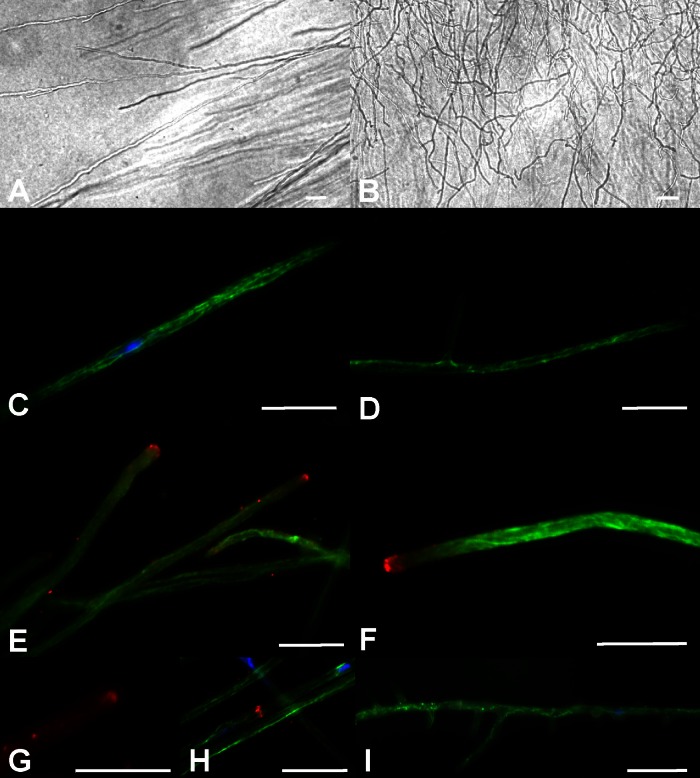
The phenotypes of the transformants carrying constitutively active Ras alleles revealed 28 to 52% growth rate reductions as measured by the diameters of developing colonies (4.41 to 6.35 mm/day versus 8.65 to 9.03 mm/day for the wild type) in haploid mycelial growth, while in heterozygous dikaryons the growth rate was reduced by 20 to 28% (Fig. 3). Similar values were seen in different genetic backgrounds, in both the control and different mutants. At the same time, microscopic examination of the hyphae of the mutants on solid medium showed a disorientated growth pattern. While hyphae of wild-type strains keep their growth axis, hyphae of the ras1 mutants grew curly, with irregular changes in the orientation of the direction of growth (Fig. 4A and B). Thus, the slower radial growth is due to both slower growth and a lack of area covered by newly grown mycelium. This phenotype implies changes in cytoskeleton organization dependent on altered signaling through Ras. Thus, we decided to analyze microtubules and the actin cytoskeleton. Immunofluorescence microscopy visualized microtubules in wild-type strains with multiple bundles parallel to the hyphal axis and in hyphal tips (Fig. 4C). Since delocalization of the vesicle supply center may induce changes in the growth axis, hyphal tips in the mutant strain were analyzed. However, no obvious changes in microtubuli were recorded (Fig. 4D). The actin cytoskeleton in wild-type mycelia showed actin patches and accumulated actin at the hyphal tips, actin rings at the site of septum formation, and actin cables, all of which could be seen with no obvious differences in the mutant strains (Fig. 4E to H).
Fig 3.
Comparison of radial growth rates of wild-type monokaryons (4-39, T2), monokaryotic Ras mutants (T2G12V, II-1G12V, II-1Q61L), a wild-type dikaryon (4-39 × 12-43), and heterozygous Rasmut dikaryons (II-1Q61L × 12-43, T2G12V × 12-43, II-1Q61L).
Fig 4.
Microscopic examination of hyphal morphology and cytoskeleton organization. Wild-type hyphae (A) keep their growth axis, while hyphae of the ras1 mutants grow curly with irregular changes in the orientation of the direction of growth (B). Microtubules in wild-type strains form multiple bundles parallel to the hyphal axis and hyphal tips (C). No obvious changes in the formation of microtubuli in the mutant strains could be observed (D). The actin cytoskeleton in wild-type mycelia (E) showed actin patches and accumulated actin at the hyphal tips with no obvious differences from the mutant strains (F). Actin cables (G) and actin rings at the site of septum formation (H) are shown here for wild-type strains. Increased formation of side branches could be observed in some subapical compartments of constitutively active Ras mutant strains (I). Bars represent 10 μm.
Increased formation of side branches could be observed in some subapical compartments of constitutively active Ras integration strains (Fig. 4I). This phenotype was in contrast to that of the Δgap1 mutant, where such hyperbranching phenotypes have not been observed (25). Usually, up to one side branch per compartment is seen with S. commune and only 10% of the cells of a wild-type strain show more than one branch (n = 100). In the Ras mutants, up to 14 branches in a single compartment with a median of 7 branches per cell could be observed (n = 100). In approximately a third of the developing side branches, septa were formed at the base of the branch which, in the absence of nuclear division, generated anucleate, small compartments visualized by nuclear staining with DAPI. Such anucleate cells subsequently ceased growth. The strong phenotypes seen already in monokaryotic strains led us to study morphogenesis in competent mating interactions.
Role of Ras in fruiting body development.
To evaluate the effects of constitutively active Ras1 on sexual development, the transformants were mated with compatible wild-type strains of S. commune. The transformants unilaterally donated nuclei to their mating partners, but the mutant strains were not able to accept nuclei. As a result, clamp formation could not be detected on the mutant's side and fruiting bodies were formed exclusively on the side where the wild-type strain had been inoculated. This indicates that the preformed mycelium with the mutant gene copy was unable to sustain the entry of a mate's nucleus and therefore was unable to develop into a dikaryon capable of inducing fruiting body formation.
The resulting heterozygous fruiting bodies appeared normal and produced spores. However, the germination rate of the spores was decreased by up to 60%, and in approximately 50% of the spores, no or only one nucleus was revealed by DAPI staining. In wild-type matings, in contrast, spores always contain two nuclei after a postmeiotic division of nuclei (data not shown). In matings of compatible Ras1 mutant strains (T2G12V × II-1Q61L and T2G12V × II-1G12V), no dikaryotization could be observed since both mutant strains were unable to accept nuclei from their mating partners.
Thus, homozygous matings could not be set up productively. However, homozygous Δgap1/Δgap1 dikaryons of S. commune had been shown to develop (aberrant) fruiting bodies (25). To investigate the role of Ras in mushroom formation, detailed micromorphological analyses of homozygous Δgap1/Δgap1 dikaryotic strains were performed in comparison to the development of fruiting bodies in the wild type.
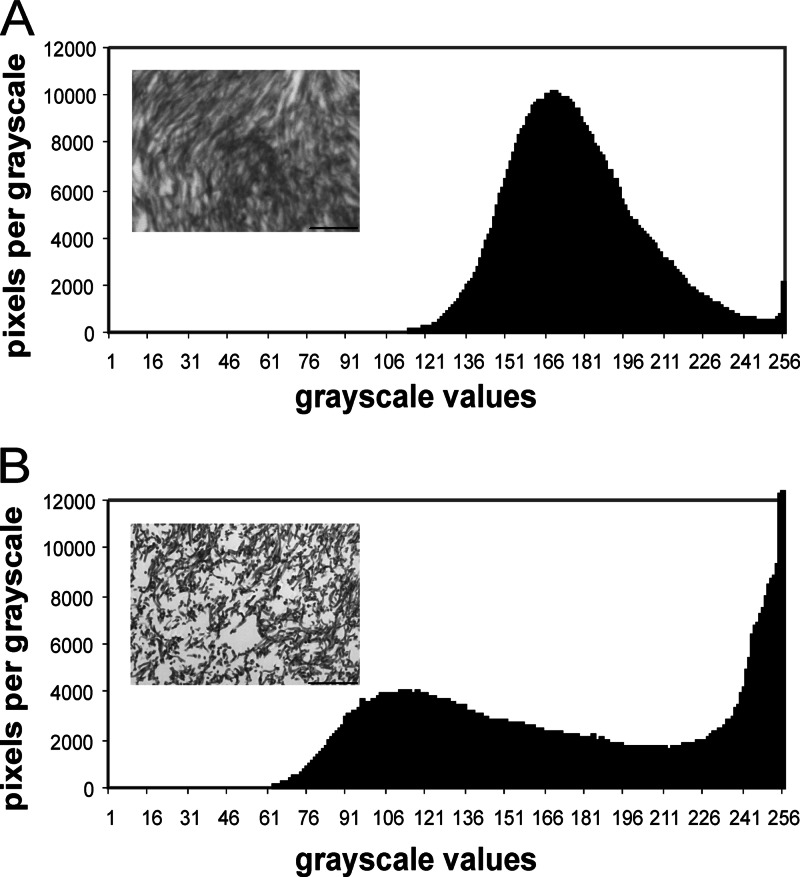
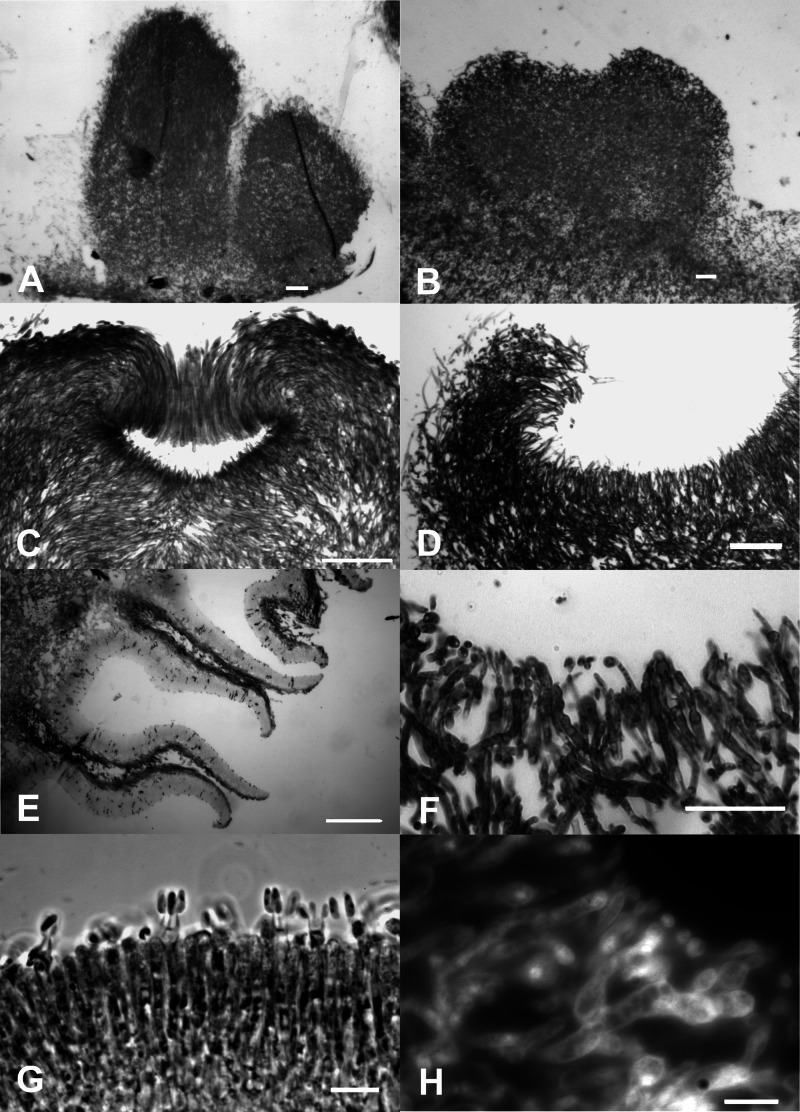
The formation of noduli, termed stage I, was not affected (for information about fruiting body formation, see references 3 and 4). Starting with the differentiation of central and peripheral hyphae in stage II primordial development, an effect of altered Ras signaling was seen with a less dense and compact plectenchyme containing numerous cavities in the Δgap1 mutant. This observation was quantitatively confirmed by an area coverage analysis that shows a dramatic increase in the number of light gray (200 to 255) to pure white (256) pixels in the histogram of the mutants (Fig. 5). Wild-type stage III primordia show elongation, formation of the apical pit, and development of the hymenial palisade (Fig. 6). This results in fruiting bodies that develop sporulating basidia late in stage III (Fig. 6). The developmental steps during primordial development up to the formation of the apical pit are not markedly affected in the homozygous Δgap1 mutant. However, because of the decreased polar growth of the hyphae, the typical bending of peripheral hyphae resulting in the formation of an apical pit is less pronounced in Δgap1 mutant primordia. At the same time, peripheral hyphae of the Δgap1 mutant do not exhibit the strong parallel growth pattern seen in wild-type primordia. In late stage III fruiting bodies, the observed differences became more prominent, with locally thickened and constricted hyphal ends as opposed to the typical thin and parallel hyphae of the wild type. The mutant subhymenium was only weakly developed. In the wild type, basidium formation and meiosis lead to spore formation. To reveal the lack of spore development in mutant fruiting bodies, we stained the club-shaped probasidia with DAPI to visualize karyogamy (Fig. 6H). However, the meiosis following karyogamy was blocked in the mutant dikaryon, which results in a complete lack of the formation of basidiospores and a lack of further development toward stages IV and V, which is necessary in the wild type for the formation of pseudolamellae. Thus, the mutant's development is arrested in stage IV.
Fig 5.
Grayscale microphotographs of microtome-sectioned fruiting body plectenchymes. Constitutively active Ras leads to a less dense and compact plectenchyme in stage II primordia with numerous cavities in the Δgap1 mutant, quantitatively confirmed by an area coverage analysis that shows a dramatic increase in the number of light gray pixels (200 to 255) in the wild type (A) toward pure white pixels (256) in the histogram of the mutants (B). Bars represent 100 μm.
Fig 6.
Micromorphological analysis of wild-type and Δgap1 mutant fruiting bodies. Wild-type stage III primordia did show elongation (A), also seen in the mutant strain (B), before the formation of the apical pit (C). Because of the decreased polar growth of the hyphae, the typical bending of peripheral hyphae resulting in the formation of an apical pit was less pronounced in Δgap1 mutant primordia (D). In late stage III fruiting bodies, the observed differences became more prominent, with typical thin and parallel hyphae in the wild type (E) and locally thickened and constricted hyphal ends in the mutant (F). The mutant subhymenium was only weakly developed. In the wild type, basidium formation and meiosis led to spore formation (G). The mutant was able to form club-shaped probasidia, and karyogamy was seen (H). Bars represent 10 μm (G, H), 50 μm (F), or 100 μm (A, B, C, D, E).
Role of Ras in cAMP signaling and PKA activation.
Investigations of Schwalb (26) had shown that cAMP has an influence on fruiting body development in S. commune. The addition of 10−3 M cAMP caused many fruiting bodies to stop morphogenesis at an early stage. Those fruiting bodies that continued to grow showed an irregular development of pseudolamellae that were unable to produce spores. This phenotype was reminiscent of Δgap1/Δgap1 mutant fruiting bodies (25). Since Ras1 mutant strains are assumed to lead to elevated concentrations of intracellular cAMP and signaling via adenylate cyclase, intracellular cAMP levels were investigated. In an earlier investigation, Δgap1 mutant strains showed no clear and reproducible increase in intracellular cAMP (25). Thus, the activity of cAMP-dependent PKA was examined by using protein extracts of wild-type, constitutively active RasG12V and Δgap1 mutant strains. While very low PKA activities were detectable in the wild-type strain, enhanced PKA activity was seen in the constitutively active Ras1 and Δgap1 mutant strains (Fig. 7). The original bse mutant strain (12-44) (26), with a reported increase in the intracellular cAMP concentration in vegetative mycelia and fruiting bodies, showed the highest activation (see Fig. S3 in the supplemental material). These results point to Ras1 signaling via cAMP-mediated signal transduction in S. commune.
Fig 7.
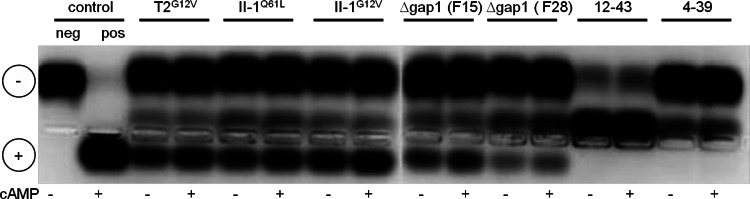
Examination of the cAMP-dependent PKA activity in protein extracts of wild-type (12-43, 4-39), constitutively active Ras (T2G12V, II-1G12V, II-1Q61L), and Δgap1 mutant (F15, F28) strains. PKA activity was assayed with a colored peptide (kemptide) that is phosphorylated by PKA. In the wild-type strains, no PKA activity was detectable even after the addition of cAMP. In contrast, the negative charge led to the migration of phosphorylated kemptide toward the positively charged electrode in all of the mutant strains, also without the addition of cAMP.
Targets of Ras1-associated signaling.
As we have shown, Ras1 and Gap1 have overlapping phenotypes. Hence, the signaling should be at least partly identical and we decided to include both mutations in our study. Similarly, an influence of Ras proteins on the Rho family protein Cdc42 has been shown for several fungi, including S. cerevisiae (16), Candida albicans (40), and C. neoformans (41). For further investigations, we therefore also included a constitutively active Cdc42 mutant of S. commune (28). For this mutant, alterations in tip growth and branching behavior had been described. Hence, again a partial overlap of phenotypes could already be established and we decided to include this mutant in our transcriptome analyses as well.
A comparison of all of the differentially regulated genes in the Ras1, ΔGap1, and Cdc42 mutants allowed us to identify genes influenced in a Ras-dependent manner. In addition, carefully chosen subsets for comparisons allowed us to differentiate among regulation exclusively through Ras, alternative signaling to Ras (via Gap1), and Cdc42-dependent or -independent Ras signaling (Table 2).
Table 2.
Identification of significant changes in gene regulation through the Ras1/Gap1/Cdc42 signaling cascadea
| Regulation through | ΔGap1 × ΔGap1 vs W22 × 12-43 | Ras1 vs 12-43 | Ras1 vs 4-39 | Cdc42 vs 12-43 | Cdc42 vs 4-39 | No. of genes | Group with greatest functional enrichment |
|---|---|---|---|---|---|---|---|
| Ras1/Gap1 | Induced | Induced | Induced | Induced | Induced | 7 | Protein turnover |
| Cdc42 | Repressed | Repressed | Repressed | Repressed | Repressed | 7 | |
| Ras1/Gap1 | Induced | Induced | Induced | 6 | Cell cycle control | ||
| Repressed | Repressed | Repressed | 7 | ||||
| Ras1/Cdc42 | Induced | Induced | Induced | Induced | 8 | Metabolism | |
| Repressed | Repressed | Repressed | Repressed | 21 | |||
| Gap1 | Induced | 80 | Signal transduction | ||||
| Repressed | 51 | ||||||
| Ras1 | Induced | Induced | 29 | Chromatin structure/dynamics | |||
| Repressed | Repressed | 70 | |||||
| Cdc42 | Induced | Induced | 64 | Cytoskeleton | |||
| Repressed | Repressed | 128 |
All of the genes mentioned are not regulated in the comparison of 12-43 versus 4-39. Functional enrichment is shown by KOG category (see legend to Fig. 8) of highest prevalence over the monokaryon group's prevalence. The statistical significance of differences was determined by t test (P ≤ 0.05) with a with a 3-fold change cutoff.
With a 3-fold regulation (induction or repression) cutoff and the use of statistical significance tests (P < 0.05), 99 genes, corresponding to 0.75% of the genome, were identified as targets of Ras signaling. In the group of genes regulated by the Ras1/Gap1/Cdc42 pathway, 14 genes did show highly statistically significant regulation. In the Ras1/Gap1 signaling pathway, we found 13 genes that were significantly changed, and in the Ras1/Cdc42 pathway, there were 29 regulated genes. Exclusive regulation through Ras1 (99 genes), Gap1 (131 genes), and Cdc42 (192 genes) adds up to 422 genes. This is equivalent to 3.2% of the genes of S. commune being regulated with some involvement of Ras. This high number underlines the importance of Ras signaling in this mushroom-forming basidiomycete.
The 29 genes induced in the constitutively active Ras1 mutant were classified according to KOG groups (Fig. 8; see Fig. S4 in the supplemental material). They contain a histone deacetylase (catalytic subunit RPD3, protein identifier [PI] 77889) involved in the condensation and inactivation of DNA, a meiotic cell division protein (Pelota/DOM34, PI 48206), and an SWI-SNF chromatin-remodeling complex (PI 53495). A histone acetyltransferase SAGA/ADA (subunit PCAF/GCN5, PI 109113) and a regulator of chromosome condensation (RCC1, PI 256079) are among the 70 specifically repressed genes. This interesting finding might point toward extensive epigenetic processes related to Ras signaling.
Fig 8.
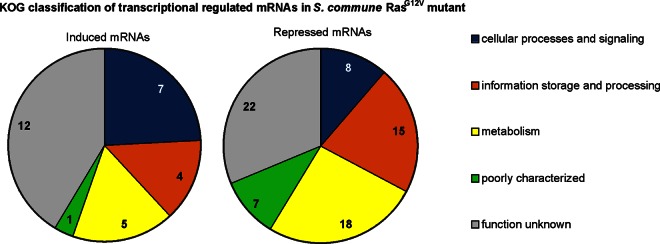
KOG classification of transcriptionally regulated mRNAs in the S. commune RasG12V mutant. The value in each wedge is the number of regulated mRNAs in that specific group. Functional groups of regulated genes (P ≤ 0.05; fold change, ≥3/≤−3) are sorted by KOG classification as follows: cellular processes and signaling (M, cell all/membrane/envelope biogenesis; N, cell motility; O, posttranslational modification, protein turnover, chaperones; T, signal transduction; U, intracellular trafficking, secretion, and vesicular transport; V, defense mechanisms; W, extracellular structures; Y, nuclear structure; Z, cytoskeleton), information storage and processing (A, RNA processing and modification; B, chromatin structure and dynamics; J, translation, ribosomal structure and biogenesis; K, transcription; L, replication, recombination, and repair), metabolism (C, energy production and conversion; D, cell cycle control, cell division, chromosome partitioning; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; G, carbohydrate transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; P, inorganic ion transport and metabolism; Q, secondary metabolite biosynthesis, transport, and catabolism), and poorly characterized (R, general function prediction only; S, function unknown).
A lipid phosphate phosphatase (PAP2 family, PI 65863), a glucan-1,4-α-glucosidase (PI 103679), a glucan endo-1,3-α-glucosidase (PI 110853), and a β-1,6-N-acetylglucosamine transferase (PI 84499), all involved in cell wall assembly and integrity, might be related to the prominent growth phenotype. Hydrophobins were not significantly and consistently changed because of Ras1 activity; instead, several hydrophobins showed repression in the Δgap1 dikaryon and in the Cdc42 mutant. Additional genes included an inositol monophosphatase (PI 47747) and a phosphatidylinositol synthase (PI 81380) referring to alternative signaling pathways. For detailed data, see Tables S5 and S6 in the supplemental material.
Similar lists were assembled for the Gap1 deletion mutant or the constitutively active Cdc42 mutant. In the former, a spindle pole body protein (Sad1), a chitinase, signal transduction components like a 3-phosphoinositide-dependent protein kinase (PDK1), several members of other families of small G proteins (Rab GTPase activator, GTP1/OBG, and several protein kinases), and a sexual differentiation protein (ISP4) were found to be regulated (see Table S7 in the supplemental material). The latter is one of the few genes related to sexual development that were statistically significantly changed because of one of the genes investigated. For Cdc42, a two-component phosphorelay intermediate involved in MAPK cascade regulation (Hpt) and repression of dynactin (subunit p27/WS-3) and a microtubule-associated protein, TAU, were identified that may relate to phenotypes overlapping with Ras signaling (see Table S8 in the supplemental material).
Our analysis allowed us also to define genes regulated by a pathway combining Ras1, Gap1, and Cdc42 signaling (see Tables S9 and S10 in the supplemental material). Induced genes common to Ras1/Gap1/Cdc42 signaling include aay4, a mating type homeodomain transcription factor (other mating type genes were not expected since mating type specificities are encoded by genes sufficiently different not to bind to the microarray sequences), and a Zn finger nucleic acid binding protein. Repression of transcription was visible for genes coding for a serine/threonine kinase (involved in autophagy) and a ubiquitin ligase, MIB2. Thus, a link to mating via direct control of a mating type protein and rearrangement of the proteome occurring during mating interactions were directly identified in our transcriptome analysis.
The larger subsets of genes regulated by Ras1/Gap1 (upregulation of a DNA helicase while phosphatases were repressed) and the Ras1/Cdc42 pathway (increased expression of a couple of cyclin regulation proteins, repression of the SWI-SNF chromatin remodeling complex and genes suggestive of metabolic shifts) show the involvement of Ras in general developmental shifts in the fungal cell. Additionally, we identified ankyrin, which is expected to be associated with cytoskeleton remodeling via Cdc42 signaling, and a LabA-like protein that is involved in circadian rhythm control and has not yet been explored in S. commune (42). For a complete overview of all of the genes identified, see Tables S5 through S8 in the supplemental material.
DISCUSSION
The investigation of Ras signaling in fungi shows a central role for the small G protein in a variety of intracellular signaling and development pathways. Ras proteins are important for cell morphology (18), asexual development (43, 44), mating (24, 41), and cell growth and polarity (17, 20, 45, 46). Two Ras-encoding genes were found in S. commune and in most fungi, with the exception of L. edodes, which codes for only one Ras protein. In accordance with the phylogeny of basidiomycetes, C. neoformans and U. maydis each contain a Ras2 protein that clusters apart from the mushroom-forming basidiomycetes and is more similar to the ascomycete A. nidulans and N. crassa Ras2 family genes. The numbering of Ras1/Ras2 is inconsistent with these phylogenetic lineages in several cases.
The cluster of the Ras2 group, including Suillus, Cryptococcus, and Ustilago, might be indicative of a similar function for these proteins. While Ras2 group members like C. neoformans or U. maydis Ras2 show a function in pheromone response, this is not seen in members of the Ras1 family. Interestingly, the S. commune targets of this Ras homolog include an array of other G protein modules (for a review of S. commune G proteins, see reference 19). RanBP1 interacts with GTP-bound Ran, and therefore it is also a negative regulator of RCC1, which is repressed in the mutant. The RCC1 homolog in yeast, Pim1 (47), prevents premature initiation of mitosis.
The Ras1 family, including both copies of S. cerevisiae (14), show signaling through MAPK and G protein pathways related to filamentous growth. An independent role or partially overlapping function of the Ras1 group protein Ras2 in S. commune thus is likely, potentially with connection to other G proteins and MAPK cascades, as seen also with other Ras1 family proteins (16, 40, 41).
Both overlapping and distinct phenotypes of Ras1 and Gap1 signaling (for Gap1, see reference 25) were observed, including growth reduction, interference with hyphal morphology, hyperbranching, and alteration of sexual development. Gap1 deletion leaves a low, intrinsic GTPase within Ras1 intact, which might explain the differences in the phenotypes, or an independent Gap protein might be involved. Signaling is expected rather through binding of GTP; an unlikely transcriptional activation could not be seen.
In ascomycetes, the deletion of ras mostly leads to decreased growth rates, e.g., with deletion of rasB in Aspergillus fumigatus (45) or ras1 of C. albicans (40), whereas the human-pathogenic fungus Penicillium marneffei displayed a decreased growth rate in dominant active rasA mutants (48). Here, the decrease in the growth rate was, however, not associated with a defect in polar growth or hyphal orientation.
Hyperbranching was seen with the constitutive ras mutants but not with deletion of gap1, and this phenotype was similar to an earlier one observed for Cdc42 signaling in S. commune (28). For constitutively active Cdc42 mutants, however, the polar growth at the tip was unchanged. In A. nidulans, the septin protein AspB was identified as a premitotic marker of branch formation (49), while in Ashbya gossipii, hyperbranching was seen when the polarisome component Spa2 was mislocalized (50). In our transcriptome analysis, inositol phosphate signaling was identified. This is known to target phospholipase C, an enzyme involved in polar tip growth (51) and in SAGA and SWI/SNF recruitment in yeast (52). Also, SAGA and SWI/SNF were found to be regulated in our study. In addition, several cell wall/membrane assembly and integrity proteins appeared as Ras1 regulated. Outstanding was the repression of the gene coding for a Wiskott-Aldrich syndrome protein-interacting protein, WIP/VRP. Vrp1 is important for polarized growth and polarization of actin filaments by interaction with myosin motor proteins for endocytosis in C. albicans (53) and S. cerevisiae (54). The Ras1 phenotype in growth direction maintenance can be linked (albeit not phenotypically in S. commune) to actin mislocation.
Another role for Ras was found in the course of this investigation in mating and nuclear distribution after hyphal anastomoses. While mating was abolished in the strains carrying a constitutive ras1 allele, this was not the case in Δgap1 mutant strains. The unilateral nuclear migration seen with matings of mutant and wild-type strains shows that nuclear migration is induced via Ras signaling.
A central role for Ras signaling was also observed during mushroom formation in S. commune. The fruiting body ontogeny was shown to be severely impacted. The nodulus or hyphal knot is formed independently of Ras, while the curly growth pattern phenotype interferes with all later stages of fruiting body development and is first visible as a failure to form the apical pit correctly. Additionally, a block in meiosis leads to arrest at stage IV of fruiting body development. This phenotype exactly matches the one observed with the bse mutant of S. commune (3). The addition of exogenous cAMP to this mutant caused a complete loss of fruiting body anatomical structures in heterozygous bse mutants (3). According to these results, the cAMP concentration would have to be slightly and temporarily decreased in order to allow the proper formation of pseudolamellae. Transcription factors involved in mushroom formation of S. commune have been published recently (5, 6). Ras1 and Ras2, as well as Gap1, were not differentially expressed in Δhom2 and Δfst4 dikaryons, and neither were these differentially regulated in our analyses, reflecting an epistasis of hom2/fst4 over gap1. This is also observed from our Ras1/Gap1 phenotypes in the formation of pseudolamellae and a block in meiosis late in mushroom formation, while the aforementioned transcription factors are involved in earlier steps of mushroom formation. Among the genes in the Ras1 mutant T2G12V, epigenetic processes were highlighted. Hence, interference with regulation exerted by epistatic genes, including the earlier transcription factors, is feasible.
Investigations in L. edodes had shown ras mRNA expression at constitutive levels during fruiting body development, although an increasing cAMP level was detected (21). In addition, spatially differentiated expression of the two ras genes in different parts of the fruiting body was shown in this mushroom-forming basidiomycete (55). Some other investigations had addressed intracellular cAMP signaling. The application of caffeine leads to increased intracellular cAMP levels (56). Interestingly, caffeine-resistant mutants were shown to be phenocopying pheromone-dependent mating reactions (56). This indicates that a change in intracellular cAMP levels is involved in pheromone-dependent signaling. As shown by intracellular cAMP measurements, cAMP levels peak just before the formation of primordia. From the drop just after peak values, an incremental increase during the rest of development is seen (57). A connection between cAMP and UV-A-dependent fruiting body development was observed for S. commune by Yli-Mattila (58). Also, a light-induced increase in cAMP by the activation of adenylate cyclase during fruiting body formation was shown in C. cinerea (59). In our study, effects of UV or daylight were not addressed.
In conclusion, by investigation of ras1 in this mushroom-forming basidiomycete, Ras signaling was shown to influence the growth rate, growth orientation, branching, nuclear migration, nuclear distribution in mating interactions and in spore production, clamp fusion, fruiting body morphology, spore production, and meiosis (Fig. 9). The downstream signaling pathways involved Cdc42 (exerting an influence on the cytoskeleton), cAMP (mainly influencing fruiting body formation), and a MAPK cascade (involved in pheromone response and clamp fusion). Target genes point to chromatin remodeling and substantial cross talk between signaling pathways. Thus, we show here for the first time that Ras exerts an influence on different processes in features that can be used to study all aspects of filamentous growth; mating reactions, including nuclear distribution and migration; and mushroom formation.
Fig 9.
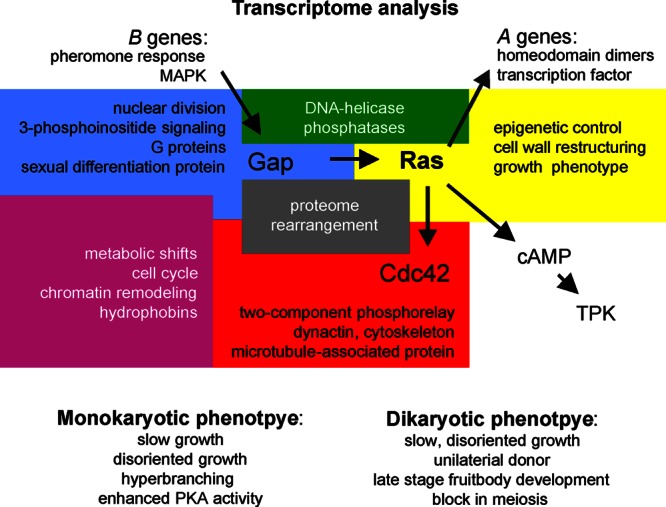
Schematic model of the interconnection of the genes addressed in this study with pathways in mating and development of S. commune. The genetic background with mating type genes and the phenotypes associated with central Ras signaling are specifically included. Yellow, functional groups of genes regulated by Ras1; blue, regulation by Gap1; red, regulation by Cdc42; green, regulation by Ras1/Gap1; purple, regulation by Gap1/Cdc42; gray, regulation by Ras1/Gap1/Cdc42. TPK, tyrosine protein kinase.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Jena School for Microbial Communication for funding.
We thank Wolfgang Schmidt-Heck of the Hans Knöll Institute, Jena, Germany, for bioinformatic analyses and Matthias Gube and Marjatta Raudaskoski for their help.
Footnotes
Published ahead of print 19 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00355-12.
REFERENCES
- 1. Raper JR, Krongelb GS, Baxter MG. 1958. The number and distribution of incompatibility factors in Schizophyllum. Am. Nat. 92:221–232 [Google Scholar]
- 2. Kothe E, Gola S, Wendland J. 2003. Evolution of multispecific mating-type alleles for pheromone perception in the homobasidiomycete fungi. Curr. Genet. 42:268–275 [DOI] [PubMed] [Google Scholar]
- 3. Schwalb MN. 1978. Regulation of fruiting, p 135–165 In Schwalb MN, Miles PG. (ed), Genetics and morphogenesis in the basidiomycetes. Academic Press, Inc., New York, NY [Google Scholar]
- 4. Ohm RA, de Jong JF, Lugones LG, Aerts A, Kothe E, Stajich JE, de Vries RP, Record E, Levasseur A, Baker SE, Bartholomew KA, Coutinho PM, Erdmann S, Fowler TJ, Gathman AC, Lombard V, Henrissat B, Knabe N, Kües U, Lilly WW, Lindquist E, Lucas S, Magnuson JK, Piumi F, Raudaskoski M, Salamov A, Schmutz J, Schwarze FWMR, vanKuyk PA, Horton JS, Grigoriev IV, Wösten HAB. 2010. Genome sequence of the model mushroom Schizophyllum commune. Nat. Biotechnol. 28:957–963 [DOI] [PubMed] [Google Scholar]
- 5. Ohm RA, de Jong JF, de Bekker C, Wösten HAB, Lugones LG. 2011. Transcription factor genes of Schizophyllum commune involved in regulation of mushroom formation. Mol. Microbiol. 81:1433–1445 [DOI] [PubMed] [Google Scholar]
- 6. Ohm RA, Aerts D, Wösten HA, Lugones LG. 2013. The blue light receptor complex WC-1/2 of Schizophyllum commune is involved in mushroom formation and protection against phototoxicity. Environ. Microbiol. 15:943–955 [DOI] [PubMed] [Google Scholar]
- 7. Specht CA, Stankis MM, Giasson L, Novotny CP, Ullrich RC. 1992. Functional analysis of the homeodomain-related proteins of the Aα locus of Schizophyllum commune. Proc. Natl. Acad. Sci. U. S. A. 89:7174–7178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vaillancourt LJ, Raudaskoski M, Specht CA, Raper CA. 1997. Multiple genes encoding pheromones and a pheromone receptor define the Bβ1 mating-type specificity in Schizophyllum commune. Genetics 146:541–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Raudaskoski M, Kothe E. 2010. Basidiomycete mating type genes and pheromone signaling. Eukaryot. Cell 9:847–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Malumbres M, Barbacid M. 2003. RAS oncogenes: the first 30 years. Nat. Rev. Cancer. 3:459–465 [DOI] [PubMed] [Google Scholar]
- 11. Lowy DR, Willumsen BM. 1993. Function and regulation of RAS. Annu. Rev. Biochem. 62:851–891 [DOI] [PubMed] [Google Scholar]
- 12. Lengeler KB, Davidson RC, D'Souza C, Harashima T, Shen WC, Wang P, Pan X, Waugh M, Heitman J. 2000. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64:746–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Raudaskoski M, Kothe E, Fowler TJ, Jung EM, Horton JS. 2012. Ras and Rho small G proteins: insights from the Schizophyllum commune genome sequence and comparisons to other fungi. Biotechnol. Genet. Eng. Rev. 28:61–100 [DOI] [PubMed] [Google Scholar]
- 14. Kataoka T, Powers S, McGill C, Fasano O, Strathern J, Broach J, Wigler M. 1984. Genetic analysis of yeast RAS1 and RAS2 genes. Cell 37:437–445 [DOI] [PubMed] [Google Scholar]
- 15. Toda T, Uno I, Ishikawa T, Powers S, Kataoka T, Broek D, Cameron S, Broach J, Matsumoto K, Wigler M. 1985. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell 40:27–36 [DOI] [PubMed] [Google Scholar]
- 16. Mösch HU, Roberts RL, Fink GR. 1996. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 93:5352–5356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ho J, Bretscher A. 2001. Ras regulates the polarity of the yeast actin cytoskeleton through the stress response pathway. Mol. Biol. Cell 12:1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fukui Y, Kozasa T, Kaziro Y, Takeda T, Yamamoto M. 1986. Role of a ras homolog in the life cycle of Schizosaccharomyces pombe. Cell 44:329–336 [DOI] [PubMed] [Google Scholar]
- 19. Alspaugh JA, Lora MC, John RP, Heitman J. 2000. RAS1 regulates filamentation, mating and growth at high temperature of Cryptococcus neoformans. Mol. Microbiol. 36:352–365 [DOI] [PubMed] [Google Scholar]
- 20. Lee N, Kronstad JW. 2002. ras2 controls morphogenesis, pheromone response, and pathogenicity in the fungal pathogen Ustilago maydis. Eukaryot. Cell 1:954–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hori K, Kajiwara S, Saito T, Miyazawa H, Katayose Y, Shishido K. 1991. Cloning, sequence analysis and transcriptional expression of a ras gene of the edible basidiomycete Lentinus edodes. Gene 105:91–96 [DOI] [PubMed] [Google Scholar]
- 22. Sundaram S, Kim SJ, Suzuki H, McQuattie CJ, Hiremah ST, Podila GK. 2001. Isolation and characterization of a symbiosis-regulated ras from the ectomycorrhizal fungus Laccaria bicolor. Mol. Plant Microbe Interact. 14:618–628 [DOI] [PubMed] [Google Scholar]
- 23. Waugh MS, Vallim MA, Heitman J, Alspaugh JA. 2003. Ras1 controls pheromone expression and response during mating in Cryptococcus neoformans. Fungal Genet. Biol. 38:110–121 [DOI] [PubMed] [Google Scholar]
- 24. Müller P, Katzenberger JD, Loubradou G, Kahmann R. 2003. Guanyl nucleotide exchange factor Sql2 and Ras2 regulate filamentous growth in Ustilago maydis. Eukaryot. Cell 2:609–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schubert D, Raudaskoski M, Knabe N, Kothe E. 2006. Ras GTPase-activating protein Gap1 of the homobasidiomycete Schizophyllum commune regulates hyphal growth orientation and sexual development. Eukaryot. Cell 5:683–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schwalb MN. 1974. Effect of adenosine 3′,5′-cyclic monophosphate on the morphogenesis of fruit bodies of Schizophyllum commune. Arch. Mikrobiol. 96:17–20 [DOI] [PubMed] [Google Scholar]
- 27. Schwalb MN, Miles PG. 1967. Morphogenesis of Schizophyllum commune. II. Effect of microaerobic growth. Mycologia 59:610–622 [PubMed] [Google Scholar]
- 28. Weber M, Salo V, Uuskallio M, Raudaskoski M. 2005. Ectopic expression of a constitutively active Cdc42 small GTPase alters the morphology of haploid and dikaryotic hyphae in the filamentous homobasidiomycete Schizophyllum commune. Fungal Genet. Biol. 42:624–637 [DOI] [PubMed] [Google Scholar]
- 29. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 30. Liu YG, Whittier RF. 1995. Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25:674–681 [DOI] [PubMed] [Google Scholar]
- 31. Muñoz-Rivas AM, Specht CA, Ullrich RC, Novotny CP. 1986. Isolation of the DNA sequence coding indole-3-glycerol phosphate synthetase and phosphoribosylanthranilate isomerase of Schizophyllum commune. Curr. Genet. 10:909–913 [DOI] [PubMed] [Google Scholar]
- 32. Lengeler K, Kothe E. 1994. Molecular characterization of ura1, a mutant allele for orotidine-5′-monophosphate decarboxylase in Schizophyllum commune. FEMS Microbiol. Lett. 119:243–247 [DOI] [PubMed] [Google Scholar]
- 33. Schuren FH, Wessels JG. 1994. Highly-efficient transformation of the homobasidiomycete Schizophyllum commune to phleomycin resistance. Curr. Genet. 26:179–183 [DOI] [PubMed] [Google Scholar]
- 34. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 35. Fischer R, Timberlake WE. 1995. Aspergillus nidulans apsA (anucleate primary sterigmata) encodes a coiled-coil protein required for nuclear positioning and completion of asexual development. J. Cell Biol. 128:485–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pfeiffer F. 1898. Beiträge zur Fixierung und Präparation der Süßwasseralgen. Österr. Bot. Ztg. 48:53–59 [Google Scholar]
- 37. Clémençon H. 1990. Fixierung, Einbettung und Schnittfärbungen für die plectologische Untersuchung von Hymenomyceten mit dem Lichtmikroskop. Mycol. Helv. 3:451–466 [Google Scholar]
- 38. Erdmann S, Freihorst D, Raudaskoski M, Schmidt-Heck W, Jung EM, Senftleben D, Kothe E. 2012. Transcriptome and functional analysis of mating in the basidiomycete Schizophyllum commune. Eukaryot. Cell 11:571–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yamagishi K, Kimura T, Suzuki M, Shinmoto H, Yamaki K. 2004. Elevation of intracellular cAMP levels by dominant active heterotrimeric G protein alpha subunits ScGP-A and ScGP-C in the homobasidiomycete Schizophyllum commune. Biosci. Biotechnol. Biochem. 68:1017–1026 [DOI] [PubMed] [Google Scholar]
- 40. Leberer E, Harcus D, Dignard D, Johnson L, Ushinsky S, Thomas DY, Schroppel K. 2001. Ras links cellular morphogenesis to virulence by regulation of the MAP kinase and cAMP signalling pathways in the pathogenic fungus Candida albicans. Mol. Microbiol. 42:673–687 [DOI] [PubMed] [Google Scholar]
- 41. Nichols CB, Perfect ZH, Alspaugh JA. 2007. A Ras1-Cdc24 signal transduction pathway mediates thermotolerance in the fungal pathogen Cryptococcus neoformans. Mol. Microbiol. 63:1118–1130 [DOI] [PubMed] [Google Scholar]
- 42. Taniguchi Y, Katayama M, Ito R, Takai N, Kondo T, Oyama T. 2007. labA: a novel gene required for negative feedback regulation of the cyanobacterial circadian clock protein KaiC. Genes Dev. 21:60–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fortwendel JR, Panepinto JC, Seitz AE, Askew DS, Rhodes JC. 2004. Aspergillus fumigatus rasA and rasB regulate the timing and morphology of asexual development. Fungal Genet. Biol. 41:129–139 [DOI] [PubMed] [Google Scholar]
- 44. Kana-uchi A, Yamashiro CT, Tanabe S, Murayama T. 1997. A ras homologue of Neurospora crassa regulates morphology. Mol. Gen. Genet. 254:427–432 [DOI] [PubMed] [Google Scholar]
- 45. Fortwendel JR, Zhao W, Bhabhra R, Park S, Perlin DS, Askew DS, Rhodes JC. 2005. A fungus-specific ras homolog contributes to the hyphal growth and virulence of Aspergillus fumigatus. Eukaryot. Cell 4:1982–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ha YS, Memmott SD, Dickman MB. 2003. Functional analysis of Ras in Colletotrichum trifolii. FEMS Microbiol. Lett. 226:315–321 [DOI] [PubMed] [Google Scholar]
- 47. Matsumoto T, Beach D. 1991. Premature initiation of mitosis in yeast lacking Rcc1 or an interacting GTPase. Cell 66:347–360 [DOI] [PubMed] [Google Scholar]
- 48. Boyce KJ, Hynes MJ, Andrianopoulos A. 2005. The Ras and Rho GTPases genetically interact to co-ordinately regulate cell polarity during development in Penicillium marneffei. Mol. Microbiol. 55:1487–1501 [DOI] [PubMed] [Google Scholar]
- 49. Westfall PJ, Momany M. 2002. Aspergillus nidulans septin AspB plays pre- and postmitotic roles in septum, branch, and conidiophore development. Mol. Biol. Cell 13:110–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bauer Y, Knechtle P, Wendland J, Helfer H, Philippsen P. 2004. A Ras-like GTPase is involved in hyphal growth guidance in the filamentous fungus Ashbya gossypii. Mol. Biol. Cell 15:4622–4632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ischebeck T, Seiler S, Heilmann I. 2010. At the poles across kingdoms: phosphoinositides and polar tip growth. Protoplasma 240:13–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Guha N, Desai P, Vancura A. 2007. Plc1p is required for SAGA recruitment and derepression of Sko1p-regulated genes. Mol. Biol. Cell 18:2419–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Borth N, Walther A, Reijnst P, Jorde S, Schaub Y, Wendland J. 2010. Candida albicans Vrp1 is required for polarized morphogenesis and interacts with Wal1 and Myo5. Microbiology 156:2962–2969 [DOI] [PubMed] [Google Scholar]
- 54. Mochida J, Yamamoto T, Fujimura-Kamada K, Tanaka K. 2002. The novel adaptor protein, Mti1p, and Vrp1p, a homolog of Wiskott-Aldrich syndrome protein-interacting protein (WIP), may antagonistically regulate type I myosins in Saccharomyces cerevisiae. Genetics 160:923–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tanaka Y, Kaneko S, Katsukawa S, Yamazaki T, Ohta A, Miyazaki Y, Shishido K. 2005. Specific distribution in homobasidiomycete hymenophores of the transcripts of Ras protein and G-protein α-subunit genes. FEMS Microbiol. Lett. 242:169–175 [DOI] [PubMed] [Google Scholar]
- 56. Klein KK, Deppe CS. 1985. Complementation and noncomplementation among nonallelic mutations altering development in Schizophyllum commune. Genetics 109:333–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kinoshita H, Sen K, Iwama H, Samadder PP, Kurosawa S, Shibai H. 2002. Effects of indole and caffeine on cAMP in the ind1 and cfn1 mutant strains of Schizophyllum commune during sexual development. FEMS Microbiol. Lett. 206:247–251 [DOI] [PubMed] [Google Scholar]
- 58. Yli-Mattila T. 1987. The effect of UV-light on cAMP level in the basidiomycete Schizophyllum commune. Physiol. Plant. 69:451–455 [Google Scholar]
- 59. Moore D. 1998. Fungal morphogenesis. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 60. Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web-servers. Syst. Biol. 75:758–771 [DOI] [PubMed] [Google Scholar]
- 61. Beadle GW, Tatum EL. 1941. Genetic control of biochemical reactions in Neurospora. Proc. Natl. Acad. Sci. U. S. A. 27:499–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wessels JGH. 1993. Wall growth, protein excretion and morphogenesis in fungi. New Phytol. 123:397–413 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.