Abstract
Although the plasma membrane is the terminal destination for glycosylphosphatidylinositol (GPI) proteins in higher eukaryotes, cell wall-attached GPI proteins (GPI-CWPs) are found in many fungal species. In yeast, some of the cis-requirements directing localization of GPI proteins to the plasma membrane or cell wall are now understood. However, it remains to be determined how Aspergillus fumigatus, an opportunistic fungal pathogen, signals, and sorts GPI proteins to either the plasma membrane or the cell wall. In this study, chimeric green fluorescent proteins (GFPs) were constructed as fusions with putative C-terminal GPI signal sequences from A. fumigatus Mp1p, Gel1p, and Ecm33p, as well as site-directed mutations thereof. By analyzing cellular localization of chimeric GFPs using Western blotting, electron microscopy, and fluorescence microscopy, we showed that, in contrast to yeast, a single Lys residue at the ω-1 or ω-2 site alone could retain GPI-anchored GFP in the plasma membrane. Although the signal for cell wall distribution has not been identified yet, it appeared that the threonine/serine-rich region at the C-terminal half of AfMp1 was not required for cell wall distribution. Based on our results, the cis-requirements directing localization of GPI proteins in A. fumigatus are different from those in yeast.
INTRODUCTION
Glycosylphosphatidylinositol (GPI) anchoring is a conserved posttranslational modification in eukaryotes and acts as a membrane anchor for a number of proteins. It also has been shown to be important in subcellular trafficking (1). Generally, proteins that are modified by GPI contain a hydrophobic signal sequence at the N terminus and a second hydrophobic sequence at the C terminus that signals GPI anchor addition to the so-called ω amino acid, located between 17 and 25 amino acids from the C terminus of the protein (2–6). After the precursor protein is synthesized and translocated into the endoplasmic reticulum, the GPI anchor is covalently added by a transamidase complex to the ω-site (7).
In the model yeast Saccharomyces cerevisiae, GPI proteins have been extensively studied. Two types of functions have been assigned to GPI proteins depending on their localization (8). By an as-yet-uncharacterized mechanism, plasma membrane-attached GPI proteins (GPI-PMPs) are liberated by a cleavage between the glucosamine (GlcN) moiety and the first mannose residue. GPI-PMPs possess enzymatic activities able to modify cell wall polymers and involved in cell morphology, such as β-glucanase and β-glucanosyltransferase. Although the plasma membrane is the terminal destination for GPI proteins in higher eukaryotes, cell wall-attached GPI proteins (GPI-CWPs) are found in many fungal species (8–12). A remnant of the GPI anchor reacts with β-1,6-glucan and results in cross-linking of the GPI protein into the cell wall (13–15). GPI-CWPs play a biological function in filamentation, mating, flocculation, or adhesion to the external matrix (16–20). In the fungal pathogens Candida albicans and Candida glabrata, some GPI-CWPs are adhesion molecules mediating adherence to host cells, such as Epa1p, Hwp1p, Als1p, and Als5p (21, 22).
Based on in silico analysis of GPI-anchored proteins in S. cerevisiae, it is proposed that a signal of two basic amino acids in the four amino acids upstream of the ω-site acts to retain the protein at the plasma membrane (4). Analysis of various point mutations in specific GPI anchor signal sequences also supported the importance of the dibasic motif in GPI protein localization. If basic residues are absent or replaced with hydrophobic amino acids, the protein is directed to the cell wall (23–25). However, the dibasic motif alone is not the sole determinant of plasma membrane or cell wall localization (26, 27). Indeed, the presence of a long serine- and threonine-rich stretch of amino acids also results in cell wall anchorage (28). These results suggest that more than one signal in the protein may impact on the ultimate distribution of GPI proteins in the cell. On the other hand, the localization of GPI proteins is somewhat controversial since some GPI proteins show a spectrum of distribution between cell wall and membrane. Therefore, although some of the cis-requirements directing localization of GPI proteins to the plasma membrane or cell wall are now understood, it remains to be determined how the cell actually interprets those sequence signals and sorts the two classes of GPI proteins.
In Aspergillus fumigatus several GPI-anchored proteins have been shown to play a role in cell wall morphogenesis (8). Among them, AfGel1 and AfEcm33 are identified as GPI-PMPs (29–32). However, in contrast to the case in S. cerevisiae, only one basic amino acid is present at the ω-1 and ω-2 sites of AfGel1 and AfEcm33, respectively. AfMp1, a cell wall galactomannoprotein, is thought to be GPI-CWP and contains a Ser/Thr-rich region in the C-terminal half (33). Its ω-4/-5 is neither Val nor Ile, but Asn. These observations suggest a different signal for plasma membrane and cell wall localization in A. fumigatus.
To investigate the determinant factor that signals and sorts GPI-PMPs and GPI-CWPs in A. fumigatus, we constructed chimeric green fluorescent proteins (GFPs) by fusing its C terminus with the putative GPI signal sequence of AfMp1, AfGel1, and AfEcm33, respectively, or with mutated versions of these sequences. The localizations of these chimeric GFPs were investigated in this report.
MATERIALS AND METHODS
Strains and plasmids.
A. fumigatus strain YJ-407 (China General Microbiological Culture Collection Center, CGMCC0386) was maintained on potato glucose (2%) agar slant (34). A. fumigatus strain CEA17, a pyrG mutant strain (35), was kindly provided by C. D'Enfert, Institut Pasteur, France. The bacterial strain used for transformation and amplification of recombinant DNA was E. coli DH5.
The synthetic gfp(2-5) gene encoding the S65T, V163A, I167T, and S175G variants of GFP was used in all constructs. Plasmid pGT21 contains the A. niger gla (glucoamylase) promoter and gla gene, A. nidulans trpC terminator sequence (EMBLZ32690), E. coli origin, and Amp antibiotic sequences. Plasmid pCDA14 (35) was a gift from the Institut Pasteur and contains the pyrG gene.
For the construction of plasmid pchiGFP, a full-length of gfp(2-5) fragment was digested from plasmid pMCB17 (36) with the restriction enzymes BamHI and KpnI, and a 1.8-kb A. fumigatus AfchiB1 promoter and a AfChiB1 signal peptide sequence (37) were amplified from A. fumigatus YJ407, cloned into the EcoRI/KpnI site, and ligated to the EcoRI/BamHI site of plasmid pGT21. pGFP/Mp1, pGFP/Gel1, and pGFP/Ecm33 were constructed with the C-terminal 147 bp of the coding region of the Afmp1(XM741417), 159 bp of the Afgel1 (XM744160), and 138 bp of the Afecm33 (XM 001271570), respectively, fused to the C-terminal of the gfp(2-5) gene using overlap-extension PCR, and then the fragment was cloned into the BamHI and KpnI site of pchiGFP. The primers used for generating Afmp1 were GFP-N-1, Mp1-mid-5, Mp1-mid-3, and Mp1-C; the primers used for generating Afgel1 were GFP-N-1, Gel1-mid-5, Gel1-mid-3, and Gel1-C; and the primers used for generating Afecm33 were GFP-N-1, Ecm33-mid-5, Ecm33-mid-3, and Ecm33-C (Table 1), respectively. All constructs were verified by sequencing (Sangon, China).
Table 1.
Primers used in this study
| Oligonucleotide primer | Sequence (5′–3′) |
|---|---|
| GFP-N-1 | GGGTACCATGAGTAAAGGAGAAGAACTTTTCAC |
| Mp1-C | CGGGATCC TTAGAGAGCGACGGCGATGGC |
| Mp1-mid-5 | ATGGATGAACTATACAAAGGCGGCTCCGGCTCC |
| Mp1-mid-3 | GGAGCCGGAGCCGCCTTTGTATAGTTCATCCAT |
| Ecm33-C | CGGGATTCTTACAAAACGTACTGCAC |
| Ecm33-mid-5 | GGCATGGATGAACTATACAAAGGATCTTCTGGCACCACCAC |
| Ecm33-mid-3 | GTGGTGGTGCCAGAAGATCCTTTGTATAGTTCATCCATGCC |
| Gel1-C | CGGGATCC TCACAAGAGGACGAGGCCAGCG |
| Gel1-mid-5 | GGCATGGATGAACTATACAAAGGATCTGGCTCTGCCACTGG |
| Gel1-mid-3 | CCAGTGGCAGAGCCAGATCCTTTGTATAGTTCATCCATGCC |
| Gel1(S2→K1)-N | GGCACCTCCACCTCT AAGTCCGGCGCTGCAG |
| Gel1(S2→K1)-C | AGAGGTGGAGGTGCCGCTGCTGCTGCTTCC |
| MP1(S2→K2)-N | CACCAGCACCAACCTCCTC AAGACTGGCGCCGC |
| MP1(S2→K2)-C | GAGGAGGTTGGTGCTGGTGGAGGCAGTGGC |
| Ecm(K2→S1)-N | CTCTGCTTCCGCTTCC TCCAAGAATGCTGCTGAC |
| Ecm(K2→L1)-N | CTCTGCTTCCGCTTCC CTCTCCAATGCTGCTGAC |
| Ecm(K2→SL)-C | GGAAGCGGAAGCAGAGCTGCCGCTGCTAGTG |
The site-directed mutagenesis was carried out using GeneTailor (Invitrogen) according to the manufacturer's instructions. The PCR was performed with Platinum Taq DNA polymerase high fidelity (Invitrogen) for 20 cycles (94°C for 30 s, 56°C for 1 min, 68°C for 7 min) and further incubated at 68°C for 10 min. pGFP/Mp1, pGFP/Ecm33, and pGFP/Gel1 were used as templates. The primer pairs used for generating Mp1(S2K) were MP1(S2K)-N and MP1(S2K)-C; the primer pairs used for generating Ecm(K2SS1K) were Ecm(K2SS1K)-N and Ecm(K2S/L)-C; the primer pairs used for generating Ecm(K2L) were Ecm(K2L)-N and Ecm(K2S/L)-C; and the primer pairs used for generating Gel(S2KK1S) were Gel1(S2KK1S)-N and Gel1(S2KK1S)-C (Table 1), respectively. pGFP/Mp1(S2K), pGFP/Ecm33(K2SS1K), pGFP/Ecm33(K2L), and pGFP/Gel1(S2KK1S) were also verified by sequencing (Sangon, China).
Growth and culture conditions.
A. fumigatus strain YJ-407 was grown at 37°C on YG (0.5% yeast extract, 2% glucose), complete medium (CM) (38), or minimal medium (MM) with 0.5 mM sodium glutamate as a nitrogen source (38) and solidified with 1.5% (wt/vol) agar when required. Uridine and uracil were added at a concentration of 5 mM for strain CEA17. Mycelia were harvested from strains grown in complete liquid medium at 37°C with shaking at 250 rpm. Conidia for spore inocula were prepared by growing A. fumigatus strains on solid complete medium with uridine and uracil (CMU) for 48 h at 37°C, harvested after 3 to 4 days with 0.1% Tween 20, and washed twice with distilled water. The concentration was confirmed by hemocytometer counting and viable counting. All cultures were grown at 37°C and agitated at 250 rpm when necessary.
Computer analysis.
Computer analysis and multiple sequence alignments were performed using VectorNTI 9.0.
A. fumigatus transformation.
Transformation was performed as described by Yelton et al. (39).
Screening of transformants and fluorescence microscopy.
For fluorescence microscopic analysis, mycelia were arranged upon a glass slide surface, covered with a coverslip and analyzed under fluorescent light, using Zeiss microscope equipped with a 460- to 480-nm excitation filter set, captured with a charge-coupled device [CCD] camera, and edited with the image analyzer program Image (AxioVision Rel.4.6).
To plasmolyse the mycelial cells, the mycelia were washed twice with phosphate-buffered saline (PBS) buffer (pH 7.4) and then transferred to plasmolytic solution (0.5 M sorbitol). For analysis of the first stages of plasmolysis, nonplasmolysed hyphae were mounted in water. Directly under the fluorescence microscopy, the plasmolytic solution was added on one side of the coverslip with a pipette and drawn from the other side with filter paper.
Immunogold staining and electron microscopy.
A. fumigatus YJ-407 were grown in CM at 37°C for 56 h with shaking at 250 rpm. Mycelia were harvested and washed twice in PBS. The cells were fixed with 4% (wt/vol) paraformaldehyde plus 0.5% (vol/vol) glutaraldehyde for 2 h at 4°C, followed by being embedded in 10% (wt/vol) gelatin for 20 min at room temperature and 20 min at 4°C. They were then fixed for about 30 min and cut into blocks (1 by 1 mm). The blocks were infused with 20% polyvinylpyrrolidone (PVP) in 2.3 M sucrose-PBS solution overnight at 4°C and frozen in liquid N2. Ultrathin cryosections obtained by using a Leica EM UC6+FC6 (Leica Microsystems, Austria) were incubated with anti-GFP polyclonal antibodies (Clontech) at a 1:50 dilution in PBS containing 1% bovine serum albumin and 0.15% glycine and then revealed with specific goat anti-mouse 10-nm gold conjugates at a 1:10 dilution (Life-Holder). Samples were examined with Tecnai Spirit (120 kV) transmission electron microscope (FEI Company, Netherlands). Negative controls were performed by reacting samples with an irrelevant murine IgG or with the immunoconjugates alone.
PCR and Southern blot analysis.
Genomic DNA was prepared from fresh mycelium. A total of 30 cycles (94°C for 30 s, 60°C for 30 s, and 72°C for 45 s) of PCR amplification were carried out by using Taq DNA polymerase (Tiangen, China). The primers used were GFP-N-1 and Mp1-C, Ecm33-C, and Gel-C, respectively (Table 1). The concentration of DNA was determined by using a NanoDrop ND100 spectrometer (Japan). Ten micrograms of genomic DNA digested with KpnI/BamHI was separated by electrophoresis on 0.8% agarose gel and transferred to Hybond-N nylon membrane (Amersham, USA). Hybridization was carried out with a digoxigenin (DIG)-labeled KpnI/BamHI fragment of the GFP as a probe using a DIG-labeled detection kit (Roche, USA).
Membrane and cell wall protein preparation.
A total of 108 conidia of A. fumigatus YJ-407 were inoculated into CM and cultured at 37°C for 56 h with shaking at 250 rpm. Mycelia were collected by paper filtration under vacuum, extensively washed with distilled water, and then saved at −80°C for further use. Extracellular proteins were precipitated from the culture supernatant, and intracellular proteins were precipitated from the cell lysate with 4 volumes of ethanol. The membrane fraction was prepared using the method described by Fontaine et al. (40). The cell wall was isolated as described by Damveld et al. (41). Ground mycelia were lyophilized, weighed, and resuspended in 25 μl of Tris buffer (0.05 M Tris-HCl [pH 7.8]) per mg (dry weight). The cytosolic fraction was separated from the cell wall and membrane by centrifugation (13,000 rpm) at 4°C for 10 min. To remove residual cytosolic contaminants, membrane proteins, and disulfide-linked cell wall proteins, the pellets were boiled three times in 25 μl of sodium dodecyl sulfate (SDS) extraction buffer (50 mM Tris-HCl [pH 7.8], 2% [wt/vol] SDS, 20 mM sodium EDTA, and 40 mM β-mercaptoethanol) per mg (dry weight). The supernatants were stored as SDS fractions (SDS1 to SDS3) (42). The cell wall pellet was recovered by centrifugation (12,000 rpm) at 4°C for 10 min, washed three times with deionized water, and then freeze-dried.
Freeze-dried SDS/β-mercaptoethanol-extracted cell walls were treated in two ways. (i) The cell-wall sample was subjected to Quantazyme digestion (recombinant endo-1,3-β-glucanase; Quantum Biogene). First, 10 mg of lyophilized cell wall was resuspended in 300 μl of buffer (10 mM Na3PO4 [pH 7.5], 150 mM NaCl, 5 mM EDTA, and 50 U of Quantazyme), followed by incubation at 37°C for overnight (43). Enzymatic hydrolysis was stopped by boiling the samples for 10 min in the extraction buffer. Solubilized proteins were recovered in the supernatant after centrifugation at 4,000 rpm for 10 min. (ii) The cell wall sample was incubated with 10 μl of hydrofluoride (HF)-pyridine per mg (dry weight) for 3 h at 0°C (44). After centrifugation, the supernatant containing the HF-extracted proteins was collected (in 100-μl aliquots), and proteins were precipitated by the addition of 9 volumes of 100% methanol buffer (100% [vol/vol] methanol, 50 mM Tris-HCl [pH 7.8]) and subsequently incubated at 0°C for 2 h. Precipitated proteins were collected by centrifugation (13,000 rpm, 10 min, at 4°C). The pellet was washed three times with 90% methanol buffer (90% [vol/vol] methanol, 50 mM Tris-HCl [pH 7.8]) and lyophilized. The Quantazyme-/HF-extracted proteins were detected by SDS-PAGE and Western blotting.
Deglycosylation of glycoproteins.
Enzymatic deglycosylation kit (PROzyme) was used to remove N-linked carbohydrates from glycoproteins. A total of 100 μg of HF-extracted cell wall proteins (HF treated for 3 h) was dissolved in 30 μl of deionized water and subjected to denaturation and deglycosylation according to the manufacturer's instructions. Deglycosylated proteins were analyzed by Western blotting.
TFMS (trifluoromethanesulfonate; Sigma) was also used to remove N-/O-linked carbohydrates from glycoproteins. A total of 40 mg of cell wall proteins were treated with HF for 3 h to release GPI proteins. The cell wall glycoproteins released by HF were freeze-dried, treated with precooled, anhydrous TFMS on ice for 2 h, neutralized with pyridine solution (pyridine-methanol-water [3:1:1]) and ammonium bicarbonate (0.5% [wt/vol]), and then washed three times with 80% ethanol (45, 46). Deglycosylated proteins were analyzed by SDS-PAGE and Western blotting.
Western blotting.
Prepared proteins were loaded to 12% polyacrylamide gel. After separation, the proteins were electrotransformed to nitrocellular membrane. After blocking in 5% defatted dried milk in Tris-buffered saline (TBS; 10 mM Tris-HCl, 150 mM NaCl [pH 8.0]), the membrane was probed with antibodies at 1:1,000 dilution in TBS containing 1% dried milk. The membrane was then washed for 15 min each in TBS plus 0.05% Triton X-100 (TBST), TBST plus 0.5 M NaCl, and TBST before incubating with alkaline phosphatase-conjugated anti-rabbit IgG 1:5,000 in TBS with 1% dried milk. After washing as described above, immunolabeling was visualized by using NCIP/NBT solution (Amersco). Anti-GFP polyclonal antibody was from Clontech, and customer-designed antibody against the AfEcm33p, AfGel1p, or AfMp1p, which were developed in specific-pathogen-free rabbit antiserum using synthesized peptide (AfEcm33p, TITISSQSDADGYSSC; AfGel1p, CPAKDAPNWDVDNDALPA; AfMp1p, DKFVAANAGGTVYEDLK), were obtained from B&M.
RESULTS
Analysis of location of GPI-anchored proteins.
AfMp1p is a GPI-CWP (33), while AfGel1p and AfEcm33p are recognized as GPI-PMPs (8, 40). In order to verify their location, Gel1p antibody, Ecm33p antibody, and Mp1p antibody were used to detect the distribution of AfGel1p, AfEcm33p, and AfMp1p in cell membrane and cell wall separately. Both AfGel1p and AfEcm33p were detected in the cell wall and membrane fraction of A. fumigatus, while AfMp1p was only detected in the cell wall (Fig. 1). Although both AfGel1p and AfEcm33p could be found in the SDS-extracted and β-1,3-glucanase-released fraction. It is interesting to notice that AfMp1p was only detected as a smear band in the fraction released by β-1,3-glucanase, which might be due to different lengths of β-glucans attached to AfMp1p. Based on these observations, it can be concluded that AfMp1p is a strict GPI-CWP covalently linked to the cell wall β-glucan, whereas AfGel1p and AfEcm33p are continuously distributed from the membrane to the cell wall of A. fumigatus.
Fig 1.
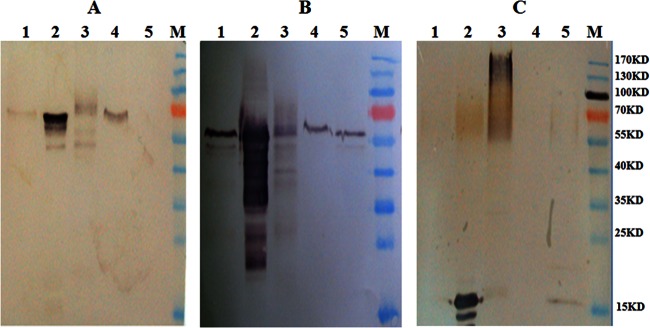
Distribution of Ecm33p, Gel1p, and Mp1p in A. fumigatus. Proteins extracted from the cell wall or membrane fraction were detected by Western blotting with anti-Gel1p (A), anti-Ecm33p (B), or anti-Mp1p (C) antibody. Lane 1, SDS-extracted supernatant of the cell wall; lane 2, extracellular proteins; lane 3, cell wall proteins extracted by β-1,3-glucanase; lane 4, membrane proteins; lane 5, intracellular proteins.
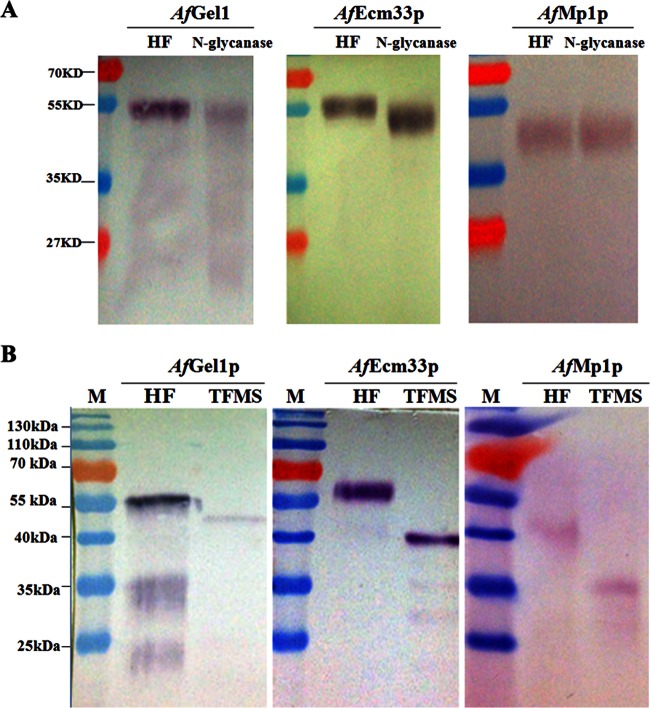
AfGel1p consists of 452 amino acids with 2 potential N-glycosylation sites and some O-glycosylation sites, and its predicted ω-site lies at the 418 amino acid of the C terminus signal peptides. The putative molecular mass of AfGel1p is ∼49.7 kDa, whereas glycosylated AfGel1p is 56 kDa (Fig. 2A, left panel). AfEcm33p is made up of 398 amino acids with nine potential N-glycosylation sites and some O-glycosylation sites, and amino acid 372 is its predicted ω-site. The putative molecular mass of AfEcm33p is ∼43.8 kDa, whereas the glycosylated AfEcm33p is 61 kDa (Fig. 2A, middle panel). AfMp1p is composed of 284 amino acids with some potential O-glycosylation sites, its predicted ω-site is at amino acid 259 of the C terminus signal peptides, and its putative molecular mass is ∼31.2 kDa, whereas the glycosylated AfMp1p is 42 kDa (Fig. 2A, right panel).
Fig 2.
Detection of cell wall proteins in A. fumigatus. Western blotting was carried out with anti-Gel1p, anti-Ecm33p, or anti-Mp1p antibody. (A) Cell wall proteins released by HF-pyridine (HF) were de-N-glycosylated with N-glycanase. (B) Cell wall proteins released by HF-pyridine (HF) were de-N/O-glycosylated with TFMS.
In fact, many studies have mentioned that proteins in the cell wall were found bigger than their predicted sizes, it may most likely due to protein glycosylation (41, 47). In order to confirm this hypothesis, the cell wall proteins were firstly extracted by HF-pyridine to release the GPI anchor. Subsequently, N-glycans were released by N-glycanase (Fig. 2A). After de-N-glycosylation, both AfGel1p and AfEcm33p became smaller than their untreated forms, but they were still bigger than their theoretical sizes, whereas AfMp1p remained unchanged although the corresponding band was confirmed to be AfMp1p by liquid chromatography-mass spectrometry. We further removed N-/O-glycans on AfGel1p, AfEcm33p, and AfMp1p with TFMS. As shown in Fig. 2B, AfGel1p, AfEcm33p, and AfMp1 moved to the position corresponding to their theoretical sizes. These results demonstrate that both AfGel1p and AfEcm33p are N and O glycosylated, whereas AfMp1p is O glycosylated. Moreover, our results also indicate that AfMp1p is covalently linked to the cell wall β-glucans via O-glycans.
Analysis of GPI signal sequences in A. fumigatus.
When these three proteins were analyzed using the Big-Pi prediction program (http://mendel.imp.ac.at/gpi/cgi-bin/gpipred.cgi), their putative GPI signals were identified. As listed in Table 2, featured hydrophobic amino acid tails are present at the C terminus of these putative GPI signals. The up- and downstream of ω-site regions are nonpolar amino acids and the upstream sequences of ω-site are rich in Ser/Thr at a similar level. It is interesting that AfGel1p or AfEcm33p only contains one single Lys residue at ω-1 or ω-2 site, which is different from that in yeast. In terms of AfMp1p, although the ω-4 of is Leu, the ω-5 site is Asn, not Val or Ile, as in yeast GPI-CWP.
Table 2.
GPI signal sequences identified by the Big-Pi program
| Protein | Amino acid sequence (5′–3′)a | Predicted location |
|---|---|---|
| AfMp1p | GGSGSGSGSSTGTATASTSTNLLSTGAASKEHFSYSLGGAVVAAAIAVAL | Cell wall |
| AfGel1p | GSGSATGSSSSGTSTSSKGAAAGLTVPSLTMAPVVVGAVTLLSTVFGGLVLL | Plasma membrane |
| AfEcm33p | GSSGTTTSSGSSASASKSNAADLNAANLPALGFAAVFGALVQYVL | Plasma membrane |
GPI proteins were analyzed using the Big-Pi prediction program (http://mendel.imp.ac.at/gpi/cgi-bin/gpipred.cgi). The ω amino acid is indicated in boldface. The shaded amino acids are up-and downstream nonpolar regions of the ω-site. The underlined amino acid sequences are hydrophobic tails at the C terminus.
In yeast, dibasic residues (Lys and Arg or Lys alone) in the ω-minus region (ω-4 to ω-1) were found to be a determinant factor for the plasma membrane location (4, 23, 24, 43). However, in A. fumigatus, AfGel1p and AfEcm33p only have one basic residue (K) at its ω-1 or ω-2 site, while AfMp1p has no basic residue. Therefore, we hypothesized that one single basic residue at ω-minus site is sufficient to retain A. fumigatus GPI proteins in the plasma membrane.
Construction and expression of GPI-anchored GFPs.
In order to study the feature of GPI-anchor attachment signal sequence in A. fumigatus, the putative GPI signal sequences of AfMp1p, AfGel1p, and AfEcm33p were fused to C terminus of GFP to construct chimeric GFP-Mp1, GFP-Gel1, and GFP-Ecm33, respectively (Fig. 3). To ensure the proper targeting of these chimeric proteins, an AfChiB1 N-terminal signal peptide sequence was fused to the N terminus of GFPs. All three chimeric proteins were under the control of the AfchiB1 (AY271350) promoter (37).
Fig 3.
Constructs of chimeric GFPs. Chimeric GFPs were constructed by fusing the N-terminal signal peptide sequence of AfChiB1 and different GPI-anchor signals derived from A. fumigatus GPI proteins with the N and C termini of GFP, respectively. The ω-site amino acid is shown in boldface. The ω-minus sites are underlined. White and gray bars indicate N-terminal signal sequences from AfChiB and GFP, respectively.
The vectors harboring the encoding regions for GFP-Mp1, GFP-Gel1, and GFP-Ecm33 were constructed and transformed into A. fumigatus as described in Materials and Methods. The transformants of pGFP/Mp1, pGFP/Gel1, and pGFP/Ecm33 were screened for GFP expression by fluorescence microscopy. Three transformants were obtained, and these were named MP1-21, GEL1-6, and ECM33-1, respectively. As shown in Fig. 4, the genes encoding for chimeric GFPs could be detected in genome by PCR (Fig. 4A) or Southern blotting (Fig. 4B).
Fig 4.
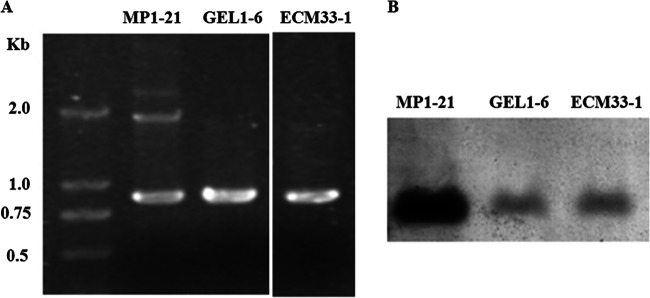
Confirmation of transformants by PCR (A) and Southern blotting (B). Transformants expressing GFP-Mp1, GFP-Gel1, and GFP-Ecm33 were screened and named as MP1-21, GEL1-6, and ECM33-1, respectively. The genomic DNA extracted from the transformant MP1-21, GEL1-6, or ECM33-1 was used for PCR and Southern blotting.
Distribution of chimeric GFPs in A. fumigatus.
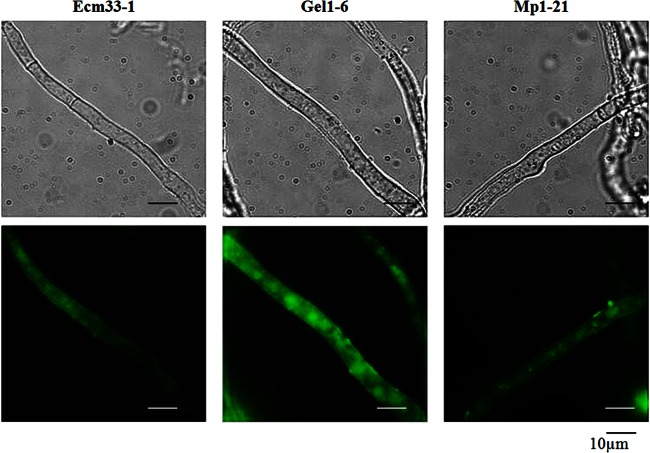
Strains MP1-21, GEL1-6 and ECM33-1 were cultivated in CM at 37°C for 24 to 56 h. At intervals, mycelia were collected and analyzed by fluorescence microscopy. The chimeric GFPs were synthesized inside the cells and transported to the surfaces of mycelia within 24 h (Fig. 5). These results demonstrate that all chimeric GFPs can be expressed and targeted correctly inside A. fumigatus.
Fig 5.
Expression of chimeric GFP proteins. After cultivation at 37°C for 24 h, the mycelia were harvested and analyzed under fluorescent light, using a Zeiss microscope equipped with a 460- to 480-nm excitation filter set, captured with a charge-coupled device (CCD) camera and edited with the image analyzer program Image (AxioVision Rel.4.6). Panels: left, strain expressing chimeric GFP-ECM33; middle, strain expressing chimeric GFP-Gel1; right, strain expressing chimeric GFP-Mp1.
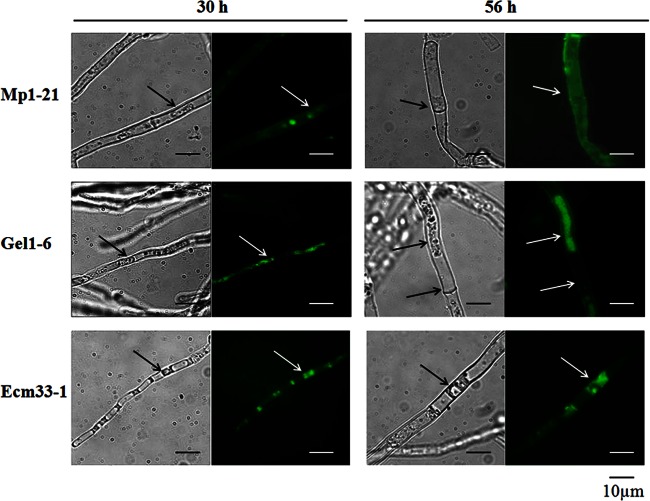
When the mycelia are treated with 0.5 M sorbitol, a high osmotic environment, the mycelial cell shrinks and its plasma membrane can be separated away from its cell wall. Taking advantage of this simple treatment, we were able to visualize the location of chimeric GFPs under fluorescence microscopy. As shown in Fig. 6, after 30 to 56 h of incubation, sorbitol treatment did not change the fluorescent distribution of the GFP-Mp1, suggesting its cell wall localization. On the other hand, a condensed signal of GFP-Gel1 was observed mainly associated with the plasma membrane and slightly associated with the cell wall, suggesting a spectrum of distribution between plasma membrane and cell wall (Fig. 6). As for strain ECM33-1, plasmolyzing of mycelia caused a shrunk and condensed signal inside the mycelia cell wall, and the signal on mycelia surface was completely disappeared after 116 h, which suggests a terminal destination of GFP-Ecm33 to plasma membrane.
Fig 6.
Distribution of chimeric GFP-Mp1, GFP-Gel1, and GFP-Ecm33. After cultivation at 37°C for 24 or 56 h, the mycelia were harvested, treated with 0.5 M sorbitol, and then analyzed under fluorescent light, using a Zeiss microscope equipped with a 460- to 480-nm excitation filter set, captured with a CCD camera, and edited with the image analyzer program Image (AxioVision Rel.4.6). The cell wall is marked with an arrow.
To further confirm the cell wall localization of GFP-Mp1 and GFP-Gel1, gold immunolabeling of hyphae with anti-GFP antibody was carried out. As shown in Fig. 7, gold particles were only present in the mannoprotein layer of cell wall in strain MP1-21 and present both in mannoprotein layer and plasma membrane of strain GEL1-6; the number of gold particles in cell wall of strain MP1-1 was greater than that in strain GEL1-6. In contrast, gold particles were found in the plasma membrane or cytosol of strain ECM33-1. These results confirmed a localization of GFP-Mp1 in the cell wall, a spectrum distribution of GFP-Gel1 between the plasma membrane and cell wall, and a final location of GFP-Ecm33 in the plasma membrane.
Fig 7.
Distribution of GFP-Mp1, GFP-Gel1, and GFP-Ecm33. A. fumigatus strains were cultured in CM at 37°C for 56 h with shaking at 250 rpm. Immunogold labeling with anti-GFP polyclonal antibodies (Clontech) was carried out as described in Materials and Methods. Gold particles are marked with arrows.
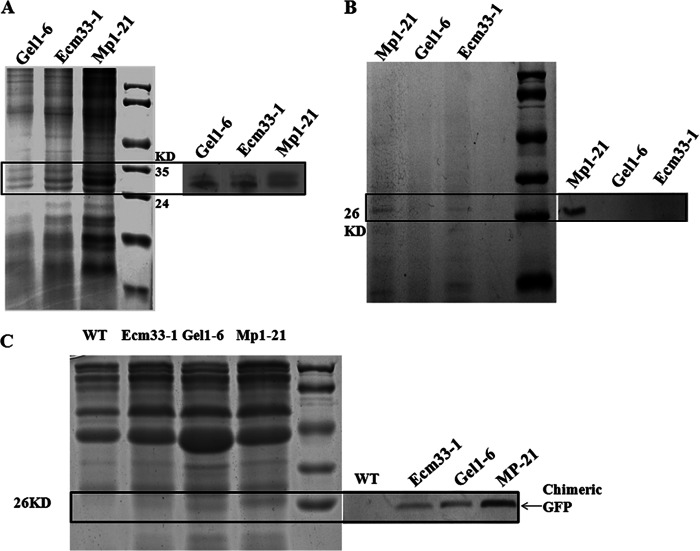
Using anti-GFP antibody, we verified the locations of chimeric GFPs in the cell membrane (Fig. 8A), cell wall (Fig. 8B), and culture supernatant (Fig. 8C) of strains MP1-21, GEL1-6, and ECM33-1, respectively. As a result, all chimeric GFP-Mp1, GFP-Gel1, and GFP-Ecm33 were visualized in membrane extracts after 36 h of incubation, whereas only GFP-Mp1 appeared attached to the cell wall (Fig. 8B). These results indicate that signals contained in the GPI signal of AfEcm33 or AfGel1 can retain GFP in the membrane, while the signals contained in AfMp1 directs a localization of GFP in the cell wall. In addition, all three chimeric GFPs were detected in the culture supernatant (Fig. 8C). The amount of secreted GFP-Gel1, which was mainly distributed in the plasma membrane and slightly in the cell wall (Fig. 6 and Fig. 7), was lower than GFP-Mp1 and higher than GFP-Ecm33 in the culture supernatant. These results suggest that all three chimeric GFPs can be secreted into culture medium. Although we were not able to detect GFP-Gel1 in cell wall by Western blotting, the amount of GFP-Gel1 in culture supernatant was higher than GFP-Ecm33 and lower than GFP-Mp1. It appears that GPI-CWP and cell wall localized GPI protein can be easily released compared to GPI-PMP.
Fig 8.
Examination of chimeric GFPs in membrane (A), cell wall (B), and culture medium (C) of A. fumigatus. The same amount of proteins in the membrane, cell wall, or culture supernatant of strain MP1-21, GEL1-6, or ECM33-1 was analyzed by SDS-PAGE (left side of each panel) and Western blotting (right side of each panel). Anti-GFP polyclonal antibodies (Clontech) at a 1:1,000 dilution were used for Western blotting.
Identification of the determinant factor for membrane distribution.
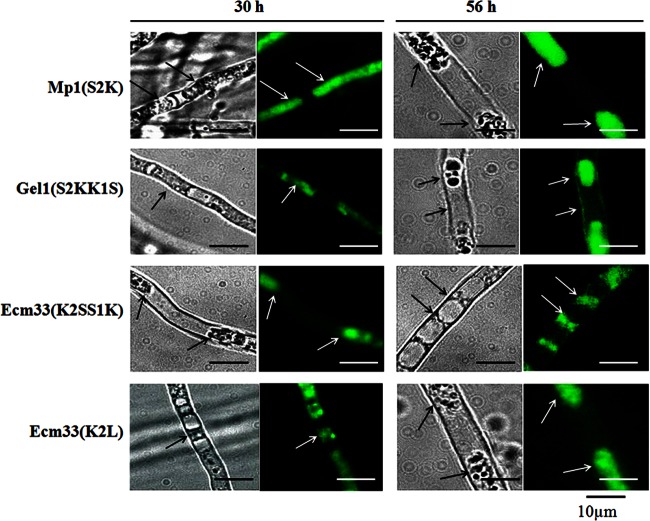
To confirm whether one single basic residue alone at ω-minus region could direct the plasma membrane localization, the mutant chimeric GFPs were constructed as described in Materials and Methods (Fig. 2). The only basic residue Lys in the ω-minus region of GFP-Ecm33 was replaced by Leu, and a basic residue Lys was introduced into GFP-Mp1 to replace Ser, to construct the mutant chimeric GFP-Ecm33 (K2L) and GFP-Mp1 (S2K). To further explore whether the site of basic residue at ω-minus region could influence the locations of proteins, the Lys residue was translocated from ω-2 to ω-1 in GFP-Ecm33 and from ω-1 to ω-2 in GFP-Gel1, which generated the mutant chimeric GFP-Ecm33(K2SS1K) and GFP-Gel1(S2KK1S), respectively. As shown in Fig. 9, when Ser at ω-2 position was changed into Lys in GFP-Mp1(S2K), the chimeric GFP changed its localization from cell wall to cell membrane. The switch of two amino acids at ω-1 and ω-2 site in GFP-Gel1(S2KK1S) did not change a spectrum of distribution of GFP-Gel1 between plasma membrane and cell wall (Fig. 9). Similarly, the exchange of ω-1 Lys and ω-2 Ser in GFP-Ecm33(K2SS1K) had no effect on localization of GFP. On the other hand, replacement of Lys at ω-1 with Leu in GFP-Ecm33(K2L) led to a loss of basic residue at ω-minus region; However, the mutated chimeric GFP-Ecm33 was still retained in plasma membrane. These results demonstrate that one single basic amino acid at the ω-1 or ω-2 site is a signal to retain GPI-proteins in plasma membrane. However, it should be pointed out that basic residue alone may not be the sole determinant to retain GPI protein in plasma membrane. Indeed, although an introduction of one basic residue into ω-2 site could shift the final localization of GFP-Mp1 from cell wall to plasma membrane, the absence of Lys at ω-2 site in GPI signal of AfEcm33 did not lead to a final distribution in cell wall, which suggests that some other unknown factors would contribute to the final distribution of GPI protein in cell wall.
Fig 9.
Distribution of GFP-Mp1(S2K), GFP-Gel1(S2KK1S), GFP-Ecm33(K2SS1K), and GFP-Ecm33(K2L). After cultivation at 37°C for 50 h, the mycelia were harvested, treated with 0.5 M sorbitol, and examined under fluorescent light using Zeiss microscope equipped with a 460- to 480-nm excitation filter set, captured with CCD camera, and edited with the image analyzer program Image (AxioVision Rel.4.6).
DISCUSSION
In general, the GPI attachment signal of S. cerevisiae contains either an Asn or Glu at the anchoring site which was named ω-site, a spacer region consisting of 8 to 12 amino acids, and a hydrophobic tail with 10 to 15 amino acids. In yeast, the final destination of GPI proteins is the plasma membrane or the cell wall. The GPI-CWPs show some preferences at the ω-2, ω-4, and ω-5 sites, i.e., Tyr, Val, or Asn at the ω-2 site and Val, Ile, or Leu at ω-4 or ω-5 sites. On the other hand, dibasic residues (Lys and Arg or Lys alone) in the ω-minus region (ω-4 to ω-1) are a determining factor, but not the sole, for plasma membrane location (4, 23, 24, 26, 27, 43). For instance, the presence of 200 to 300 amino acids rich in serine and threonine residues can result in cell wall anchorage (11, 28). Indeed, ca. 70% of yeast GPI-CWPs have a Ser/Thr content above 30% (28). These results suggest that more than one signal in the protein may impact on the ultimate distribution of GPI proteins in the cell. Nevertheless, the signaling for GPI protein localization is controversial and De Sampaio et al. (26) showed that Gas1p and fusions of the Gas1p to α-galactosidase, although they both have a dibasic motif at ω-minus site, are both cell wall localized. Neither the Gas1p nor α-galactosidase is particularly rich in serine and threonine residues. Moreover, the finding that some GPI proteins show a spectrum of distribution between cell wall and membrane makes the mechanism for signaling and sorting of the GPI proteins even more complicated.
In A. fumigatus, both AfGel1p and AfEcm33p are identified as GPI-PMPs. AfGel1p, a β-1,3-glucanosyltransferase involved in the elongation of β-1,3-glucan side chain (30), is homologous to Gas1p in S. cerevisiae, as well as Phr1p and Phr2p in C. albicans. AfGel1p is attached to the membrane through a GPI anchor in a manner similar to that of the yeast homologous proteins (30). AfEcm33p is a homolog of a putative yeast GPI-anchored protein (4) and shares 30% identity to the S. cerevisiae Ecm33p. However, both AfGel1p and AfEcm33p do not have dibasic amino acids at their ω-minus region, which is a typical feature for GPI-anchored membrane protein in S. cerevisiae. Indeed, they have only one basic residue (K) at either the ω-1 or the ω-2 site. Based on this observation, we hypothesized that one single basic residue would be enough for retaining GPI protein on the plasma membrane in A. fumigatus. To verify this hypothesis, we used GFP as an indicator molecule by fusing its C terminus to a putative GPI signal sequence of AfGel1p or AfEcm33p. Our results showed that both GFP-Ecm33 and GFP-Gel1 were localized to the plasma membrane. Shifting of Lys from the ω-1 to the ω-2 site or from the ω-2 to the ω-1 site did not affect the membrane localization of GFP-Ecm33 or GFP-Gel1. These results confirm that a single basic residue at the ω-1 or ω-2 site can retain GPI protein in cell membrane of A. fumigatus.
It should be pointed out that we also found some GFP-Gel1 proteins were present in cell wall and shifting of Lys from ω-2 to ω-1 site did not affect its mixed distribution from membrane to cell wall. In yeast, it has been observed that some GPI-PMPs can be liberated by a cleavage between its GlcN moiety and the first mannose residue. Although the mechanism is not yet known, it is reasonable to propose that the presence of GFP-Gel1 in the cell wall is due to the cleavage and release of membrane-localized GFP-Gel1 via a similar manner observed in yeast.
On the other hand, A. fumigatus Mp1p is a cell wall galactomannoprotein (33). Ultrastructural analysis by immune-gold staining indicated that AfMp1p is present in cell wall of A. fumigatus. Sequence analysis of AfMp1p shows that, although the ω-4 is Leu, the ω-2 is neither Val nor Tyr, but Ser, which is not a typical cell wall localization signal found in yeast. Considering that the AfMp1p has a Ser/Thr-rich region in the C-terminal half (29.9% of Thr/Ser content), its cell wall localization may be due to the presence of Ser/Thr-rich region. However, ProFASTA analysis revealed that only 30% of A. fumigatus putative GPI-CWPs contains above 30% Ser/Thr residues (48), suggesting an existence of some unknown determinants except Ser/Thr-rich region. It remains unknown whether its GPI attachment signal makes contribution to cell wall localization or not. We therefore fused the GPI signal of AfMp1p to GFP, which does not contain the Ser/Thr-rich region in the C-terminal half of AfMp1p, indeed the content of Ser/Thr in the AfMp1p GPI signal is similar to that in the AfEcm33p or AfGel1p GPI signal. Our results showed that the ω-minus region of AfMp1p alone, rather than the Ser/Thr-rich region in the C-terminal half of this protein, could direct the protein to the cell wall. When Ser at ω-2 site was changed into Lys, the localization of chimeric GFP was shifted from cell wall to membrane. These results clearly demonstrate that one single basic residue at the ω-2 site can signal a retention of GFP in cell membrane of A. fumigatus. However, when we replaced Lys at the ω-1 with Leu in GFP-Ecm33(K2L), the mutated GFP-Ecm33 was still present in plasma membrane, and no chimeric GFP was found in the cell wall. Taken together, we conclude that the presence or absence of a basic residue at the ω-minus site is not the sole determinant of plasma membrane or cell wall localization and that an unknown signal in the GPI signal sequence of AfMp1p may contribute to cell wall localization.
Interestingly, GFP-Mp1, GFP-Gel1, and GFP-Ecm33 were detected in culture supernatant. Although GPI anchoring is also thought to be as an important secretory pathway, AfMp1p, AfEcm33p, and AfGel1p are usually not supposed to be secreted by A. fumigatus. Therefore, a plausible explanation for the presence of GFP-Mp1 or GFP-Gel1 in supernatant is due to a leakage of cell wall components during cell wall remodeling in growing cells and during formation of new branches or autolysis of aged cells. As for membrane-located GFP-Ecm33, the presence of GFP-Ecm33 in culture supernatant might be caused by autolysis of aged cells.
Using the newly developed software ProFASTA (48), 86 GPI-anchored proteins are predicted with positive Big-PI, signal P, and TMHMM domain in A. fumigatus. Among these predicted GPI proteins, 23 of them are found to have one basic residue at their ω-minus sites (see Table S1 in the supplemental material) and therefore, based on our results, can be predicted as GPI-PMPs.
In conclusion, we have shown that one basic residue at the ω-1 or ω-2 site signals retention of GPI proteins in plasma membrane in A. fumigatus, which is different from that in yeast. Although the signal for cell wall distribution has not been identified yet, it is clear that the signal for the cell wall distribution in A. fumigatus is different from that in yeast. Furthermore, GPI anchoring cannot limit the distribution of GPI-proteins strictly in the plasma membrane or cell wall of A. fumigatus. GPI-CWPs can be secreted into culture medium by an unknown mechanism, while GPI-PMPs also can be found in culture medium.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (31011130030, 31030025, and 31100113) and partly by the Austrian Science Fund FWF (I391 to I.B.H.W.).
Footnotes
Published ahead of print 12 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00351-12.
REFERENCES
- 1. Muniz M, Riezman H. 2000. Intracellular transport of GPI-anchored proteins. EMBO J. 19:10–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Caras IW, Weddell GN. 1989. Signal peptide for protein secretion directing glycophospholipid membrane anchor attachment. Science 243:1196–1198 [DOI] [PubMed] [Google Scholar]
- 3. Caras IW, Weddell GN, Williams SR. 1989. Analysis of the signal for attachment of a glycophospholipid membrane anchor. J. Cell Biol. 108:1387–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Caro LH, Tettelin H, Vossen JH, Ram AF, van den Ende H, Klis FM. 1997. In silico identification of glycosylphosphatidylinositol- anchored plasma-membrane and cell wall proteins of Saccharomyces cerevisiae. Yeast 13:1477–1489 [DOI] [PubMed] [Google Scholar]
- 5. Orlean P, Menon AK. 2007. GPI anchoring of protein in yeast and mammalian cells, or: how we learned to stop worrying and love glycophospholipids. J. Lipid Res. 48:993–1011 [DOI] [PubMed] [Google Scholar]
- 6. Moran P, Caras IW. 1991. A nonfunctional sequence converted to a signal for glycophosphatidylinositol membrane anchor attachment. J. Cell Biol. 115:329–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fraering P, Imhof I, Meyer U, Strub JM, van Dorsselaer A, Vionnet C, Conzelmann A. 2001. The GPI transamidase complex of Saccharomyces cerevisiae contains Gaa1p, Gpi8p, and Gpi16p. Mol. Biol. Cell 12:3295–3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bruneau J-M, Magnin T, Tagat E, Legrand R, Bernard M, Diaquin M, Fudali C, Latgé J-P. 2001. Proteome analysis of Aspergillus fumigatus identifies glycosylphosphatidylinositol-anchored proteins associated to the cell wall biosynthesis. Electrophoresis 22:2812–2823 [DOI] [PubMed] [Google Scholar]
- 9. Lu CF, Kurjan J, Lipke PN. 1994. A pathway for cell wall anchorage of Saccharomyces cerevisiae alpha-agglutinin. Mol. Cell. Biol. 14:4825–4833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kapteyn JC, Hoyer LL, Hech JE, Mülle WH, Andel A, Verkleij AJ, Makarow M, Van Den Ende H, Klis FM. 2000. The cell wall architecture of Candida albicans wild-type cells and cell wall-defective mutants. Mol. Microbiol. 35:601–611 [DOI] [PubMed] [Google Scholar]
- 11. Frieman MB, McCaffery JM, Cormack BP. 2002. Modular domain structure in the Candida glabrata adhesion Epa1p, a β1,6 glucan-cross-linked cell wall protein. Mol. Microbiol. 46:479–492 [DOI] [PubMed] [Google Scholar]
- 12. Brul S, King A, van der Vaart JM, Chapman J, Klis F, Verrips CT. 1997. The incorporation of mannoproteins in the cell wall of Saccharomyces cerevisiae and filamentous Ascomycetes. Antonie Van Leeuwenhoek 72:229–237 [DOI] [PubMed] [Google Scholar]
- 13. Fujii T, Shimoi H, Iimura Y. 1999. Structure of the glucan-binding sugar chain of Tip1p, a cell wall protein of Saccharomyces cerevisiae. Biochim. Biophys. Acta 1427:133–144 [DOI] [PubMed] [Google Scholar]
- 14. Kollar R, Reinhold BB, Petrakova E, Yeh HJ, Ashwell G, Drgonova J, Kapteyn JC, Klis FM, Cabib E. 1997. Architecture of the yeast cell wall: β(1-6)-glucan interconnects mannoprotein, β(1-)3-glucan, and chitin. J. Biol. Chem. 272:17762–17775 [DOI] [PubMed] [Google Scholar]
- 15. Van Der Vaart JM, Rte Biesebeke Chapman JW, Klis FM, Verrips CT. 1996. The β-1,6-glucan containing side chain of cell wall proteins of Saccharomyces cerevisiae is bound to the glycan core of the GPI moiety. FEMS Microbiol. Lett. 145:401–407 [DOI] [PubMed] [Google Scholar]
- 16. Lussier M, White AM, Sheraton J, di Paolo T, Treadwell J, Southard SB, Horenstein CI, Chen-Weiner J, Ram AF, Kapteyn JC, Roemer TW, Vo DH, Bondoc DC, Hall J, Zhong WW, Sdicu AM, Davies J, Klis FM, Robbins OW, Bussey H. 1997. Large-scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics 147:435–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rodriguez-Penz JM, Cid VJ, Arroyo J, Nomvela C. 2000. A novel family of cell wall-related proteins regulated differently during the yeast life cycle. Mol. Cell. Biol. 20:3245–3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guo B, Styles CA, Feng Q, Fink GR. 2000. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc. Natl. Acad. Sci. U. S. A. 97:12158–12163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Martinez-Lopez R, Monteoliva L, Diez-Orejas R, Nombela C, Gil C. 2004. The GPI-anchored protein CaEcm33p is required for cell wall integrity, morphogenesis and virulence in Candida albicans. Microbiology 150:3341–3354 [DOI] [PubMed] [Google Scholar]
- 20. De Groot PW, Ram AF, Klis FM. 2005. Features and functions of covalently linked proteins in fungal cell walls. Fungal Genet. Biol. 42:657–675 [DOI] [PubMed] [Google Scholar]
- 21. Cormak BP, Ghori N, Falkow S. 1999. An adhesin of the yeast pathogen Candida glabrata mediating adherence to human epithelial cells. Science 285:578–582 [DOI] [PubMed] [Google Scholar]
- 22. Hoyer LL. 2001. The ALS gene family of Candida albicans. Trends Microbiol. 9:176–180 [DOI] [PubMed] [Google Scholar]
- 23. Hamada K, Terashima H, Arisawa M, Kitada K. 1998. Amino acid sequence requirement for efficient incorporation of glycosylphosphatidyl-inositol-associated proteins into the cell wall of Saccharomyces cerevisiae. J. Biol. Chem. 273:26946–26953 [DOI] [PubMed] [Google Scholar]
- 24. Hamada K, Terashima H, Arisawa M, Yabuki N, Kitada K. 1999. Amino acid residues in the omega-minus region participate in cellular localization of yeast glycosylphosphatidylinositol-attached proteins. J. Bacteriol. 181:3886–3889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Frieman MB, Cormack BP. 2003. The omega-site sequence of glycosylphosphatidylinositol-anchored proteins in Saccharomyces cerevisiae can determine distribution between the membrane and the cell wall. Mol. Microbiol. 50:883–896 [DOI] [PubMed] [Google Scholar]
- 26. De Sampaio G, Bourdineaud JP, Lauquin GJ. 1999. A constitutive role for GPI anchors in Saccharomyces cerevisiae: cell wall targeting. Mol. Microbiol. 34:247–256 [DOI] [PubMed] [Google Scholar]
- 27. Meyer U, Fraering P, Bosson R, Imhof I, Benghezal M, Vionnet C, Conzelmann A. 2002. The glycosylphosphatidylinositol (GPI) signal sequence of human placental alkaline phosphatase is not recognized by human Gpi8p in the context of the yeast GPI anchoring machinery. Mol. Microbiol. 46:745–748 [DOI] [PubMed] [Google Scholar]
- 28. Frieman MB, Cormack BP. 2004. Multiple sequence signals determine the distribution of glycosylphosphatidylinositol proteins between the plasma membrane and cell wall in Saccharomyces cerevisiae. Microbiology 150:3105–3114 [DOI] [PubMed] [Google Scholar]
- 29. Bernard M, Mouyna I, Dubreucq G, Debeaupuis JP, Fontaine T, Vorgias C, Fuglsang C, Latgé J-P. 2002. Characterization of a cell-wall acid phosphatase (PhoAp) in Aspergillus fumigatus. Microbiology 148:2819–2829 [DOI] [PubMed] [Google Scholar]
- 30. Mouyna I, Fontaine T, Vai M, Monod M, Fonzi WA, Diaquin M, Popolo L, Hartland RP, Latgé J-P. 2000. Glycosyl-phosphatidylinositol-anchored glucanosyltransferases play an active role in the biosynthesis of the fungal cell wall. J. Biol. Chem. 275:14882–14889 [DOI] [PubMed] [Google Scholar]
- 31. Romano J, Nimrod G, Ben-Tal N, Shadkchan Y, Baruch K, Sharon H, Osherov N. 2006. Disruption of the Aspergillus fumigatus ECM33 homologue results in rapid conidial germination, antifungal resistance, and hypervirulence. Microbiology 152:1919–1928 [DOI] [PubMed] [Google Scholar]
- 32. Chabane S, Sarfati J, Ibrahim-Granet O, Du C, Schimidt C, Mouyna I, Prevost M-C, Calderone R, Latgé J-P. 2006. Glycosylphosphatidyl-inositol-anchored Ecm33p influences conidial cell wall biosynthesis in Aspergillus fumigatus. Appl. Environ. Microbiol. 72:3259–3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yuen K-Y, Chan C-M, Chan K-M, Woo PCY, Che X-Y, Leung ASP, Cao L. 2001. Characterization of AFMP1: a novel target for serodiagnosis of aspergillosis. J. Clin. Microbiol. 39:3830–3837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xia G, Jin C, Zhou J, Yang S, Zhang S, Jin C. 2001. A novel chitinase having a unique mode of action from Aspergillus fumigatus YJ-407. Eur. J. Biochem. 268:4079–4085 [DOI] [PubMed] [Google Scholar]
- 35. Weidner G, D'Enfert C, Koch A, Mol PC, Brakhage AA. 1998. Development of a homologous transformation system for the human pathogenic fungus Aspergillus fumigatus based on the pyrG gene encoding orotidine 5′-monophosphate decarboxylase. Curr. Genet. 33:378–385 [DOI] [PubMed] [Google Scholar]
- 36. Fernández-Ábalos JM, Fox H, Pitt C, Wells B, Doonan JH. 1998. Plant-adapted green fluorescent protein is a versatile vital reporter for gene expression, protein localization and mitosis in the filamentous fungus, Aspergillus nidulans. Mol. Microbiol. 27:121–130 [DOI] [PubMed] [Google Scholar]
- 37. Wang Y, Wang J, Hu H, Yang X, Yang S, Yu X, Jin C. 2004. Cloning and expression of chitinase gene from Aspergillus fumigatus YJ-407. Chinese J. Biotech. 20:843–850 [Google Scholar]
- 38. Cove DJ. 1966. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim. Biophys. Acta 113:51–56 [DOI] [PubMed] [Google Scholar]
- 39. Yelton M, Hamer J, Timerberlake W. 1984. Transformation of Aspergillus nidulans by using a trpC plasmid. Proc. Natl. Acad. Sci. U. S. A. 81:1470–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fontaine T, Magnin T, Melhert A, Lamont D, Latgé J-P, Ferguson MAJ. 2003. Structures of the glycosylphosphatidylinositol membrane anchors from Aspergillus fumigatus membrane proteins. Glycobiology 13:169–177 [DOI] [PubMed] [Google Scholar]
- 41. Damveld RA, Arentshorst M, VanKuyk PA, Klis FM, van den Hondel CAMJJ, Ram AFJ. 2005. Characterisation of CwpA, a putative glycosylphosphatidylinositol-anchored cell wall mannoprotein in the filamentous fungus Aspergillus niger. Fungal Genet. Biol. 42:873–885 [DOI] [PubMed] [Google Scholar]
- 42. Montijn RC, van Rinsum J, van Schagen FA, Klis FM. 1994. Glucomannoproteins in the cell wall of Saccharomyces cerevisiae contain a novel type of carbohydrate side chain. J. Biol. Chem. 269:19338–19342 [PubMed] [Google Scholar]
- 43. Terashima H, Hamada K, Kitada K. 2003. The localization change of Ybr078w/Ecm33, a yeast GPI-associated protein, from the plasma membrane to the cell wall, affecting the cellular function. FEMS Microbiol. Lett. 218:175–180 [DOI] [PubMed] [Google Scholar]
- 44. De Groot PW, de Boer AD, Cunningham J, Dekker HL, de Jong L, Hellingwerf KJ, de Koster C, Klis FM. 2004. Proteomic analysis of Candida albicans cell walls reveals covalently bound carbohydrate-active enzymes and adhesins. Eukaryot. Cell 3:955–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Edge ASB, Faltynek CR, Hof L, Jr, Reichert LE, Weber P. 1981. Deglycosylation of glycoproteins by trifluoromethanesulfonic acid. Anal. Biochem. 118:131–137 [DOI] [PubMed] [Google Scholar]
- 46. Edge ASB. 2003. Deglycosylation of glycoproteins with trifluoromethanesulfonic acid: elucidation of molecular structure and function. Biochem. J. 376:339–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Van Der Vaart JM, Caro LHP, Chapman JW, Klis FM, Verrips CT. 1995. Identification of three mannoproteins in the cell wall of Saccharomyces cerevisiae. J. Bacteriol. 177:3104–3110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. de Groot PW, Brandt BW. 2012. ProFASTA: a pipeline web server for fungal protein scanning with integration of cell surface prediction software. Fungal Genet. Biol. 49:173–179 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.