Abstract
Mycobacterium avium subsp. paratuberculosis causes Johne's disease, an enteric infection in cattle and other ruminants, greatly afflicting the dairy industry worldwide. Once inside the cell, M. avium subsp. paratuberculosis is known to survive harsh microenvironments, especially those inside activated macrophages. To improve our understanding of M. avium subsp. paratuberculosis pathogenesis, we examined phagosome maturation associated with transcriptional responses of M. avium subsp. paratuberculosis during macrophage infection. Monitoring cellular markers, only live M. avium subsp. paratuberculosis bacilli were able to prevent phagosome maturation and reduce its acidification. On the transcriptional level, over 300 M. avium subsp. paratuberculosis genes were significantly and differentially regulated in both naive and IFN-γ-activated macrophages. These genes include the sigma factor H (sigH) that was shown to be important for M. avium subsp. paratuberculosis survival inside gamma interferon (IFN-γ)-activated bovine macrophages. Interestingly, an sigH-knockout mutant showed increased sensitivity to a sustained level of thiol-specific oxidative stress. Large-scale RNA sequence analysis revealed that a large number of genes belong to the sigH regulon, especially following diamide stress. Genes involved in oxidative stress and virulence were among the induced genes in the sigH regulon with a putative consensus sequence for SigH binding that was recognized in a subset of these genes (n = 30), suggesting direct regulation by SigH. Finally, mice infections showed a significant attenuation of the ΔsigH mutant compared to its parental strain, suggesting a role for sigH in M. avium subsp. paratuberculosis virulence. Such analysis could identify potential targets for further testing as vaccine candidates against Johne's disease.
INTRODUCTION
Mycobacterium avium subsp. paratuberculosis is the etiological agent of Johne's disease (JD), a chronic enteritis of domestic and wild animals, especially ruminants. JD has been reported on every continent (1–3) and is considered one of the greatest causes of economic hardship to the dairy herd operations (4). More than two thirds of the U.S. dairy herds are infected with JD (5), and this wide distribution of the disease, reduced milk production, and premature culling of infected animals together cause severe economic losses estimated to be over $200 million a year for the dairy industry (6). The majority of M. avium subsp. paratuberculosis infection occurs through the fecal-oral route, and the mycobacteria are endocytosed by enterocytes and M cells in the Peyer's patches of the ileum (7). After subsequent internalization by subepithelial and intraepithelial macrophages, M. avium subsp. paratuberculosis is able to survive and persist within macrophages (8) using mechanisms that are not completely understood. To better understand mechanisms for M. avium subsp. paratuberculosis survival inside macrophages, several studies examined gene expression patterns of host defenses in bovine macrophages from naturally infected cows (9), peripheral blood mononuclear cells (PBMC) (10), or monocyte-derived macrophages (MDMs) (11). Alternatively, our group characterized the M. avium subsp. paratuberculosis responses under various in vitro stress conditions or in fecal samples from diseased cows (12). The main goal of the current study is to gain insights into how M. avium subsp. paratuberculosis responds to the intracellular microenvironments of macrophages, the primary site for mycobacterial persistence, with emphasis on mycobacterial global gene regulators.
Survival of M. avium subsp. paratuberculosis in environmental samples (13), macrophages (14), and animal models (15, 16) is well documented; however, the genetic basis for this survival remains unknown. Reports employing a large-scale screening of M. avium subsp. paratuberculosis mutants in relevant animal models (17, 18) provided some insights into virulence of this organism, with the identification of novel virulence factors associated with biofilm formation (19) and epithelial cell invasion (20). Recently, Zhu et al. analyzed intracellular M. avium subsp. paratuberculosis gene expression patterns in bovine MDMs using selective capture of transcribed sequences (SCOTS), identifying similar patterns of responses to oxidative stress, metabolic activity, and cell survival among M. avium subsp. paratuberculosis with distinct host origins (21). Further analysis of the expression profiles of M. avium subsp. paratuberculosis isolated from naturally infected bovine tissues identified tissue-specific pathways (22). Recently, characterizing the signaling network of M. avium subsp. paratuberculosis inside monocytes suggested its ability to block gamma interferon (IFN-γ) activation (23). During infection of Caco-2 cells, 7 sigma factors out of 19 encoded in the M. avium subsp. paratuberculosis genome were activated in intestinal epithelial cells (24). However, no comprehensive study has been conducted to clarify the relationship between bacterial gene expression and specific host microenvironments following macrophage infection with M. avium subsp. paratuberculosis.
In this study, we took advantage of analytical microscopy to define the phagosome environment of M. avium subsp. paratuberculosis-containing macrophages in association with the expression profile of mycobacterial bacilli using DNA microarrays. Our analysis suggested key changes in the metabolic pathways of M. avium subsp. paratuberculosis once the bacteria encounter active macrophages and the activation of various alternative sigma factors that could help M. avium subsp. paratuberculosis survive the hostile intracellular environment of macrophages. One such alternative sigma factor, sigH, has been shown to contribute to the resistance during variable environmental stress conditions, such as temperature and oxidative stress in Mycobacterium tuberculosis (25). However, the basis of transcriptional regulation of sigH remains elusive in M. avium subsp. paratuberculosis. We therefore sought to define the gene regulatory network under the control of sigH in M. avium subsp. paratuberculosis. Genetic and transcriptome analyses confirmed a role for sigH in defending M. avium subsp. paratuberculosis against thiol-specific oxidative stress and characterized the effect of sigH on global transcriptome of M. avium subsp. paratuberculosis. Further analysis of the ΔsigH mutant in bovine macrophages and murine model of paratuberculosis suggested that sigH could play an important role in mycobacterial persistence and virulence.
MATERIALS AND METHODS
Bacteria.
Escherichia coli DH5α and HB101 were used as host cells for cloning purposes in all experiments presented here. M. avium subsp. paratuberculosis K10 and Mycobacterium smegmatis mc2155 strains were grown in Middlebrook 7H9 broth and on Middlebrook 7H10 plates as previously described (1).
Construction of the ΔsigH mutant.
A specialized transduction protocol was adopted with a few modifications to delete the sigH/MAP3324c gene using the M. avium subsp. paratuberculosis strain (12). Briefly, two ∼900-bp PCR fragments flanking each end of the sigH coding region were amplified and cloned into the pYUB854 shuttle vector. The resulting pYUB854::sigH allelic-exchange substrate (AES) was then digested with PacI and ligated to the PacI-digested concatemers of a temperature-sensitive phasmid, phAE87. The ligation mixture was then packaged into phage particles with an in vitro lambda-packaging system (GIGAPackIII; Stratagene, La Jolla, CA). Mid-log-phase Escherichia coli culture was transduced with the packaged phage particles, resulting in hygromycin-resistant colonies. From the mixture of these colonies, shuttle phasmid DNA was extracted and then electroporated into M. smegmatis competent cells. Lysate of plaques formed at 30°C from the transformants was collected, propagated, and titrated in M. smegmatis to produce a high-titer recombinant phage stock. A mid-log-phase culture of M. avium subsp. paratuberculosis was transduced with the phage stock at nonpermissive temperature (37°C) with a multiplicity of infection of 10. Individual hygromycin-resistant colonies were picked and grown in broth medium following gDNA isolation. The genotype of sigH-deletion mutants was confirmed with PCR and sequence analysis as outlined before (12).
Stress treatments of M. avium subsp. paratuberculosis.
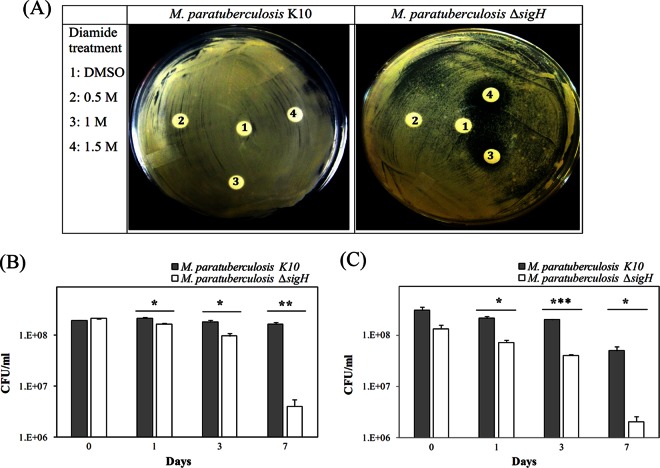
Wild-type and the ΔsigH mutant of M. avium subsp. paratuberculosis were grown to late-log phase (optical density at 600 nm [OD600] = 1.0), and 200 μl was spread on 7H10 agar plates (Difco, Sparks, MD) supplemented with 0.5% glycerol, 2 μg/ml mycobactin J, and 10% ADC (2% glucose, 5% bovine serum albumin [BSA] fraction V, and 0.85% NaCl). For disc diffusion assay (DDA), 20 μl of diamide solution (0.5 M, 1 M, or 1.5 M) and H2O2 solution (50 mM, 100 mM, or 0.5 M) was impregnated onto each 6-mm disc (Whatman, Piscataway, NJ), and discs were placed on each of the spread plates. As a positive control, ethambutol discs (5 μg/disc, Sensi-Disc; BD Diagnostics) were used. Plates were incubated at 37°C until a thick confluent lawn developed. The sustained effect of stressors (diamide and heat shock) on the viability of the wild type and ΔsigH mutant was monitored by determining their CFU counts. Aliquots of M. avium subsp. paratuberculosis cultures 1, 3, and 7 days following continuous exposure to 10 mM diamide or a 45°C water bath were serially diluted and plated. In another experiment, M. avium subsp. paratuberculosis cultures from mid-log phase were exposed to 10 mM diamide for 3 h. The cultures were centrifuged (3,000 × g, 4°C, 10 min), and pellets were immediately stored at −80°C until RNA extraction.
Mouse infections.
For the animal infections with M. avium subsp. paratuberculosis strains, female BALB/c mice (Harlan Laboratories, Indianapolis, IN) were purchased at 4 weeks of age and housed in a pathogen-free environment according to the protocol approved by the Institutional Animal Care and Use Committee, University of Wisconsin-Madison. Two groups of mice (n = 15 per group) were challenged intraperitoneally with the wild-type and ΔsigH mutant strains of M. avium subsp. paratuberculosis. Actual infection inoculum sizes (∼2 × 108 CFU per mouse) of these two strains were similar, as determined by plate count on the day of infection. Mouse groups (n = 5) were sacrificed at 3, 6, and 12 weeks postinfection (wpi), and samples from livers, intestines, and spleens were collected for bacterial CFU enumeration and histopathological examinations as described before (17). Portions of livers, spleens, and intestines were fixed in 10% neutral buffered formalin before being sectioned and stained with hematoxylin and eosin (H&E) and Ziehl-Neelsen stain. Student's t test and Mann-Whitney test were used to statistically evaluate differences in CFU counts among mouse groups infected with the wild-type and ΔsigH mutant strains of M. avium subsp. paratuberculosis.
Bovine blood monocyte isolation and infection.
Blood was collected from a Johne's disease-free herd that we maintain at the University of Wisconsin-Madison. Three cows (36-month-old Holstein, designated animals 5695, 5970, and 6117) were bled by jugular venipuncture using blood collection bags (Teruflex, Somerset, NJ) containing citrate phosphate dextrose adenine as an anticoagulant. Blood was transferred to 50-ml polypropylene tubes and centrifuged at 1,400 × g for 20 min at 25°C. Buffy coat containing white blood cells was isolated and mixed with phosphate-buffered saline (PBS) (Ca2+- and Mg2+-free) to a final volume of 30 ml. The cell suspension was layered onto 58% isotonic Percoll (Sigma) at a 1:1 ratio and centrifuged at 2,000 × g for 30 min at 25°C. Peripheral blood mononuclear cells (PBMC) were collected from the Percoll-PBS interface and washed three times with PBS to remove residual Percoll. To isolate bovine monocyte-derived macrophages (MDMs), PBMC were resuspended in RPMI 1640 (Sigma-Aldrich, St. Louis, MO) with 20% autologous serum and transferred to Teflon jars followed by incubation for 4 days at 37°C and 5% CO2. MDM cells were harvested, washed, and seeded with 2 × 106 cells/well in 24-well plates with 5% autologous serum. Immediately before MDM cell infection, M. avium subsp. paratuberculosis cultures grown to mid-log phase (OD600 of 0.4 to 0.6) were pelleted and resuspended in an appropriate volume of cell culture medium to achieve a 50:1 multiplicity of infection (MOI). The cells were incubated at 37°C with 5% CO2 for 3 h, and, subsequently, the monolayers were washed two times with warm PBS to remove extracellular bacteria, and RPMI 1640 medium containing 5% autologous serum was added. The plates were incubated at 37°C for up to 8 days, and the culture medium was replaced with fresh medium at 4 days after infection. In another set of experiments, MDM cells were pretreated overnight (18 h) with 0.01 μg/ml recombinant bovine IFN-γ (Kingfisher Biotech, St. Paul, MN) before infection with M. avium subsp. paratuberculosis strains. This concentration of IFN-γ was adequate to activate bovine monocytes (23). Bovine MDM cells were lysed at 1, 4, and 8 days postinfection for CFU plating with serial dilutions. Student's t test was used for statistical analysis, where P values of <0.05 were considered to be significant to evaluate differences in CFU counts.
J744A.1 cell culture infection.
The mouse macrophage cells (J774A.1) were maintained in RPMI 1640 (Sigma-Aldrich, St. Louis, MO) supplemented with 5 to 10% heat-inactivated fetal bovine serum (FBS) (Sigma-Aldrich) in 75-cm2 filter-cap tissue culture flasks (Techno Plastic Products, Trasadingen, Switzerland) in a water-jacketed incubator (Thermo Scientific, Waltham, MA) at 37°C with 5% CO2. When confluent, cells were detached with a cell scraper and resuspended, and 10% of the cell suspension was replenished with fresh culture medium every 3 to 4 days.
Macrophages were seeded at 1.5 × 107 cells per 15-mm cell culture dish (Techno Plastic Products) in 15 ml of culture medium as described above, 30 to 36 h prior to infection, and were incubated at 37°C with 5% CO2. At least 5 dishes were seeded for each time point. For IFN-γ activation experiments, old culture medium was discarded and 15 ml of fresh medium with 100 U/ml recombinant murine IFN-γ (Pepro Tech, Rocky Hill, NJ) was added to each dish, 16 to 20 h prior to infection. An approximate 109 CFU bacterial suspension from mid-log phase was mixed with 12 ml of cell culture medium (RPMI 1640-10% FBS, mycobactin J-free) and added to each decanted dish (MOI = 50). The cells were incubated at 37°C with 5% CO2 for 2 or 24 h before intracellular bacteria isolation. For the 24-h-time-point experiments, extracellular bacteria were washed away at 2 h postinfection with 15 ml of warm PBS at least five times or until no visible bacterial particles were observed under an inverted microscope at ×400 magnification. The washed cells were replenished with 15 ml of fresh cell culture medium and incubated until 24 h postinfection. Each condition was replicated at least twice until the quality of extracted RNA passed the criteria described below.
Immunofluorescent staining for LAMP-1 expression.
Culture cells grown on a circular coverslip were fixed in 2.5% paraformaldehyde for 10 min and permeated with cold methanol-acetone (1:1) at −20°C for 5 min. A few drops of TB Auramine M (BD Diagnostics, Franklin Lakes, NJ) were added and incubated at room temperature for 10 min to stain mycobacteria. The coverslip was washed with 95% EtOH three times and rinsed with PBS containing 0.2% saponin and 2% goat serum. Rat monoclonal antibody 1D4B against mouse LAMP-1 purchased from the Developmental Studies Hybridoma Bank at the University of Iowa was diluted to 20 μg/ml in PBS-saponin-goat serum and incubated with the fixed cells at room temperature for 1 h. The cells were washed with PBS-saponin-goat serum three times, each for 10 min. Goat antibody conjugated with Alexa Fluor 633 against rat IgG (Invitrogen, Carlsbad, CA) was diluted to 10 μg/ml in PBS-saponin-goat serum and incubated with the cells for 1 h in the dark at room temperature. The cells were then washed in the same way as described in the last step. Finally, the coverslip was mounted on a microscope slide in Vectashield mounting medium (Vector Laboratories, Burlingame, CA) and observed with a Nikon C1 confocal microscope system.
Phagosome pH measurements.
Phagosome pH measurement was slightly modified from previous studies (26, 27) based on ratiometric measurements. J774A.1 cells were seeded at 2 × 105 cells per well on a 24-well cell culture plate (Techno Plastic Products) in 0.5 ml of culture medium with or without 100 U/ml murine IFN-γ. A poly-l-lysine (Electron Microscopy Sciences, Hatfield, PA)-coated 12-mm circular coverslip was placed in each well before seeding. After overnight incubation, culture medium was replaced with 0.3 ml of prewarmed fresh medium with 5 μM LysoSensor Yellow/Blue DND-160 (Invitrogen), and the cells were incubated for 5 min at 37°C. To generate an in situ pH gradient standard curve, each coverslip was then incubated with morpholineethanesulfonic acid (MES) buffer (25 mM MES, 5 mM NaCl, 115 mM KCl, and 1.2 mM MgSO4) of known pH (from 3.5 to 7.0 at a 0.5 interval), in the presence of 10 μM nigericin and 10 μM monensin, for 2 min. The coverslip was immediately mounted on a glass slide and observed under an Olympus BX51 microscope with a reflected fluorescence system. Sixteen-bit grayscale images of two separate channels (excitation of 365/10 nm, emission of 460/50 nm, dichroic of 400 nm; excitation of 365/10 nm, emission of 540/20 nm, dichroic of 400 nm; Chroma, Bellows Falls, VT) from each field were taken.
The processing time from sample mounting to image acquisition was controlled so it took no longer than 10 min for each coverslip. Image processing was done with ImageJ 1.44k (28). For each pH standard, at least 20 individual regions of interest (ROIs) were randomly chosen, and mean intensities of each ROI from both channels were recorded. Ratios of intensities of green (540 nm) to blue (460 nm) from the same pH standard were then averaged, excluding values of ≥2 standard deviations (SD) from the mean. A standard curve of ratios was plotted against pH (see Fig. S1 in the supplemental material) by applying a Boltzmann equation, y = A2 + (A1 − A2)/{1 + exp[(x − x0)/dx]}, where A1 and A2 represent the limits of the fluorescent ratio at infinitely low and high pHs, respectively, x0 is the pH midpoint at (A1 + A2)/2, x is the observed pH, and dx is the slope of the curve. When needed, cells were infected with M. avium subsp. paratuberculosis as described above, except the bacteria were prestained with 5 μM Vybrant DiD cell-labeling solutions (Invitrogen) for 10 min (29). Intracellular bacteria could be observed with a third filter set (excitation of 535/50 nm, emission of 675/20 nm, dichroic of 565 nm). ROIs were chosen where the bacteria were colocalized with LysoSensor-stained phagosomes. The average 540/460 ratio of ROIs was plugged into the equation to calculate phagosome pH.
Intracellular bacterial isolation and RNA extraction.
Intracellular bacteria were isolated by a protocol described before (30) with modifications. At 2 or 24 h postinfection, infected cells were washed with 15 ml ice-cold PBS at least five times or until no visible bacterial particles were observed under an inverted microscope. The washed cells on each dish were then lysed with 10 ml cell lysis buffer (4 M guanidine thiocyanate, 0.5% sodium N-lauryl sarcosine, 25 mM sodium citrate, 0.5% Tween 80, and 0.1 M β-mercaptoethanol) and collected with a rubber cell scraper. To reduce viscosity and help dissolve cell debris, cell lysates from all dishes were pooled and passed through a 23-gauge needle five times. The lysate was then split into four 14-ml polypropylene centrifuge tubes (Falcon 352059; BD Biosciences, San Jose, CA) and centrifuged at 3,200 × g and 4°C for 25 min. Each pellet was washed in 1 ml of TRIzol regent (Invitrogen) twice and subjected to RNA extraction.
Total RNA was extracted by a protocol described before (12, 31). Briefly, bacterial pellets were resuspended in 2 ml of TRIzol reagent and split into two 2-ml screw-cap tubes, each with 3.0 g of 0.1-mm zirconia/silica beads (BioSpect Products, Bartlesville, OK) and disrupted in a Mini-BeadBeater-8 (BioSpect Products) at top speed for three pulses of 60 s with 30-s intervals on ice. Following a 5-min incubation at room temperature, the supernatant was transferred to RNase-free tubes and centrifuged at 12,000 × g for 15 min. RNA was then isolated according to the manufacturer's instruction. To remove genomic DNA contamination, RNA samples were treated with 10 U of Turbo DNase (Ambion, Austin, TX) at 37°C for 30 min. An IS900 241-bp PCR was performed to confirm that no genomic DNA was detectable in the RNA samples (1). DNase treatments were repeated if needed. Quality of the extracted RNA was examined with a NanoDrop 1000 (Thermo Scientific). The ratios of A260/A280 and A260/A230 must be higher than 1.8 and 1.5, respectively, before proceeding to cDNA synthesis for transcriptome studies.
Transcriptome studies.
The NimbleGen (NimbleGen System Inc., Madison, WI) M. avium subsp. paratuberculosis microarray was designed from the 4,350 open reading frame sequences in the genome of M. avium subsp. paratuberculosis (32). The whole genome was represented three times on each chip. In addition, each gene was represented by 20 probes of 60-mer oligonucleotides. As a result, each gene was represented by a total of 60 probes. Double-stranded cDNA synthesis from isolated RNA samples, microarray hybridization, and data analysis were performed as previously described (12, 33). Significantly expressed genes were selected by ±2-fold of change and P values of <0.05 by Student's t test. The intensities were also exported to GeneSpring GX (Agilent Technologies, Santa Clara CA) for principal component analysis (PCA) on treatment conditions, which is a method to reduce dimensionality in multicondition microarray experiments and to find relevant patterns across conditions (34). Two biological replicates were included for each condition.
For RNA-seq studies, purified RNA samples were used for depletion of rRNA sequences to enrich mRNA using the MICROBExpress bacterial mRNA enrichment kit (Ambion, Austin, TX). Approximately, 10 μg of total RNA for each sample was processed according to the manufacturer's instructions. For cDNA library preparation and sequencing, samples containing at least 1 μg of enriched mRNA were sent to the DNA Sequencing Facility at the University of Wisconsin-Madison Biotechnology Center. An Illumina HiSeq 2000 platform using one flow cell lane with 100-cycle paired-end chemistry (Illumina, San Diego, CA) was used to sequence the cDNA library clusters. Raw RNA-seq data files in FASTQ format were assembled against the M. avium subsp. paratuberculosis (35) using the CLC Genomics Workbench 4.8 (CLC bio, Aarhus, Denmark). Gene expression for each of the different sample conditions was calculated using “reads per kilobase million” (RPKM) expression values (36). The following formula was used to determine the RPKM values: RPKM = number of reads/(kilobase length of gene × millions of mapped reads).
This RPKM metric enables comparisons between data sets with a varying number of total reads. All reads mapping to annotated noncoding RNA (ncRNA) were removed from the data sets before determining RPKM values. Data sets were quantile normalized, and Kal's Z-test (37) was used for the comparative gene expression analysis. Genes were considered significantly, differentially expressed if they showed a ±2.5-fold change with FDR P values of <0.05.
Quantitative real-time PCR.
Quantitative real-time PCR (qRT-PCR) was previously described (12, 38) for confirmation of transcript levels. A SYBR green-based reagent with ROX (Bio-Rad, Hercules, CA) was used with 50 ng of double-stranded cDNA in each reaction. Double-stranded cDNA synthesis is described in the microarray sample preparation session. No gDNA was detected from the RNA samples for cDNA synthesis. qRT-PCRs were performed with a 7300 real-time PCR system (Applied Biosystems, Foster City, CA). The threshold cycle (CT) of each gene was normalized to the CT of the 16S rRNA gene from the same cDNA sample. The expression fold changes were calculated by comparing the normalized CT of treated samples to the control sample as detailed before (39, 40).
Microarray data accession number.
Data sets discussed in this report were deposited in NCBI's Gene Expression Omnibus (41) and are accessible through GEO Series accession number GSE43645.
RESULTS
Characterization of M. avium subsp. paratuberculosis-containing phagosomes.
In our previous study, we defined the stressome of M. avium subsp. paratuberculosis under various in vitro conditions that mimicked the hostile host microenvironments, including low pH and oxidative stress (12). In the present study, we further examined the bacterial responses in the early stage of cell infection using a murine macrophage infection model. Both naive and IFN-γ-activated cells were used in our study. We monitored the expression of inducible nitric oxide synthase (iNOS), a marker for macrophage activation, with quantitative real-time PCR following IFN-γ treatment of J774A.1 cells. The transcription activity of iNOS in IFN-γ-treated cells was over 1,000 times higher than naive cells (data not shown). The temporal profile of iNOS expression indicated that naive macrophages were activated by 2 h postinfection and throughout the course of infection, with comparable mRNA levels regardless of the viability of M. avium subsp. paratuberculosis bacilli (Fig. 1A). Activated macrophages had a similar profile, but the iNOS expression levels were between 1.6 to 2.6 times greater than those of infected naive macrophages at each time point.
Fig 1.
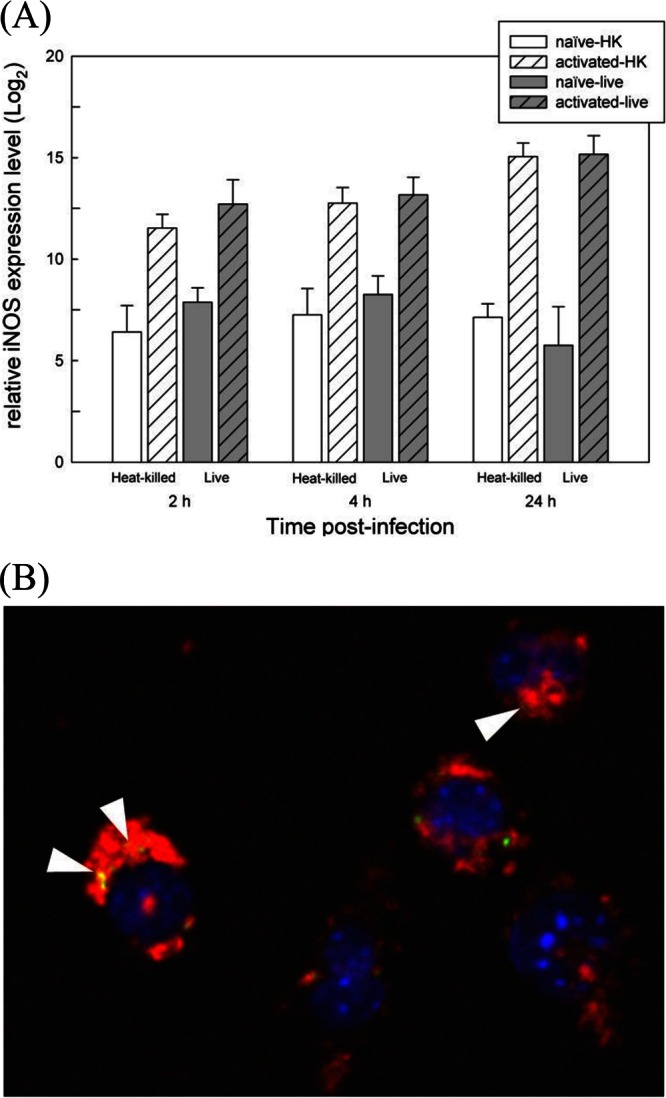
Characterization of M. avium subsp. paratuberculosis-infected murine macrophages. (A) Expression profile of iNOS in infected naive or preactivated macrophages. Naive or IFN-γ-treated J774A.1 cells were infected with either heat-killed or live M. avium subsp. paratuberculosis bacilli, and iNOS expression levels were measured with qRT-PCR. All relative expression levels were compared to the uninfected naive macrophage control sample. Error bars represent standard deviation of biological replicates under each condition. (B) Confocal microscopic examination of M. avium subsp. paratuberculosis-lysosome colocalization. A representative merged image of three wavelength channels is shown. Blue, DAPI-stained nuclei; red, LAMP-1; green, auramine-stained M. avium subsp. paratuberculosis particles. Yellow spots are results of superimposed signals of red and green, indicating colocalization of M. avium subsp. paratuberculosis and LAMP-1 (arrowheads).
Additionally, we measured the phagosomal pH in both naive and activated macrophages using a dual-emission dye LysoSensor DN-160 that emits fluorescent signals in a pH-dependent manner. Before infection, naive and activated macrophages had similar lysosomal pH levels ranging from 5.1 to 5.3 (see Fig. S2A in the supplemental material). At 2 h postinfection, the pH in phagosomes containing heat-killed M. avium subsp. paratuberculosis decreased below 4.0 regardless of cell activation status. However, the pH in phagosomes containing live M. avium subsp. paratuberculosis bacilli decreased just below the preinfection level (i.e., 4.8 to 5.0), suggesting the ability of live bacteria to prevent phagosome acidification. Activated macrophages, but not naive ones, were able to continuously decrease the pH of phagosomes containing live bacilli up to 4 h of postinfection. As the infection progressed (24 h), activated macrophages exhibited a better ability to maintain a lower pH level than naive macrophages.
To examine the role of M. avium subsp. paratuberculosis on phagosome maturation, we examined the percentage of colocalization between intracellular M. avium subsp. paratuberculosis and the lysosome marker LAMP-1 (Fig. 1B). While heat-killed bacteria showed over 85% colocalization with LAMP-1, live M. avium subsp. paratuberculosis significantly reduced the percentage of colocalization with LAMP-1 at 2 h postinfection (67.6% ± 5.5) in naive macrophages, suggesting live M. avium subsp. paratuberculosis is able to rapidly circumvent the hostile environment and to delay phagosome maturation (see Fig. S2B in the supplemental material). The percentage of colocalization did not significantly change in activated macrophages over the course of the experiment (87.92% ± 5.32 and 83.7% ± 9.5 at 2 h and 24 h, respectively), suggesting that preactivated host cells have a better ability to control invading intracellular pathogens by means of phagosome maturation. The reduced phagolysosome fusion of naive macrophages was restored to a level (78.4% ± 6.8) similar to that of the preactivated phagosome (81.6% ± 8.8) at 24 h postinfection, as also evidenced by the increased iNOS expression level of the naive macrophage infection compared to uninfected cells (data not shown). In general, both phagosomal pH and LAMP-1 colocalization indicated the ability of live, virulent M. avium subsp. paratuberculosis to prevent phagosome acidification and to delay lysosomal fusion by 2 h postinfection.
Transcriptional profiling of M. avium subsp. paratuberculosis isolated from infected macrophages.
To profile changes in the levels of M. avium subsp. paratuberculosis transcripts within macrophages, we isolated intracellular bacilli at 2 and 24 h postinfection, with or without IFN-γ activation. Because the bacteria must stay in the cell culture medium (RPMI 1640-10% FBS, mycobactin J-free) before they can infect host macrophages, we compared the transcriptomes of intracellular bacteria to those incubated in vitro in cell culture medium for 2 h. Under all conditions tested, the correlation coefficients (r) between biological replicates ranged between 0.92 and 0.99. To examine the statistical distance between each biological replicate and among treatments, a three-dimensional principal component analysis plot was generated, indicating high correlations between biological replicates (see Fig. S3 in the supplemental material). Cluster analysis identified groups of genes active only during macrophage infection (Fig. 2; see also Fig. S4 in the supplemental material). Compared to the RPMI-incubated control sample, expression levels of 136 and 333 M. avium subsp. paratuberculosis genes were significantly changed in naive macrophages at 2 and 24 h postinfection, respectively. On the other hand, in IFN-γ-activated macrophages, the numbers of genes with significantly changed expression levels were 284 and 328, respectively (see Table S1 in the supplemental material). Among those genes, 47 were common in all of the 4 examined macrophage conditions (see Table S2 in the supplemental material), representing a core set of genes responsible for interacting with the macrophage microenvironment (see Table S3 in the supplemental material).
Fig 2.
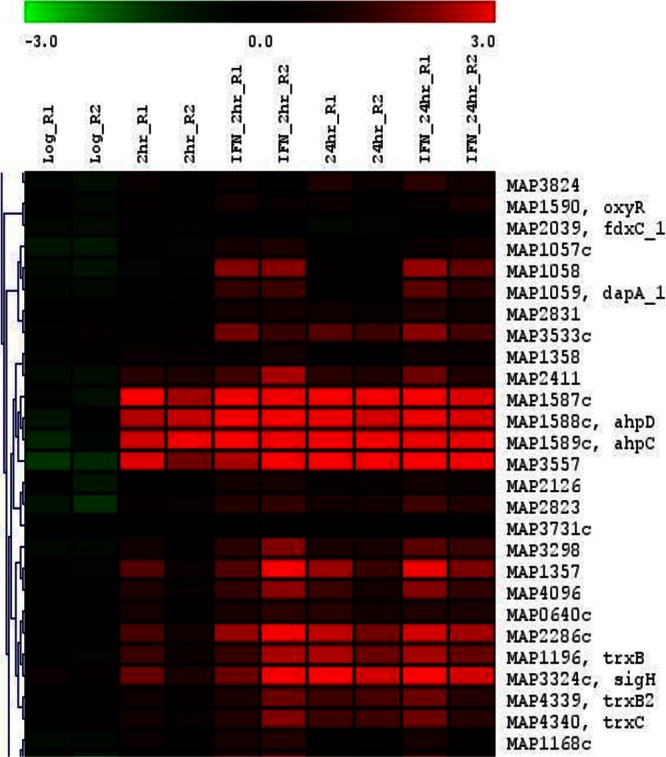
Transcriptional profiling of M. avium subsp. paratuberculosis infecting naive and activated macrophages. Clustering analysis of M. avium subsp. paratuberculosis transcriptome following DNA microarrays analysis of various times following macrophage infection. The heat map of gene transcripts shows an example of genes that are highly expressed within macrophages, compared to the control sample (2 h of incubation with cell culture medium).
In general, M. avium subsp. paratuberculosis transcriptomes in infected macrophages preactivated with IFN-γ were more similar to those observed under in vitro stressors reported earlier (12) (see Fig. S5 in the supplemental material). Also, IFN-γ activation of macrophages resulted in significant induction of a group of genes (n = 48) (see Table S4 in the supplemental material), mostly those involved in energy production and conversion (e.g., icl, fdxA, sdhABCD, and ndh) or nutrient transport and metabolism (e.g., fad genes, dapA_1, and cysH_2), at 2 h postinfection in IFN-γ-activated macrophages compared to naive macrophages.
At 24 h postinfection, we started to see a significant change in M. avium subsp. paratuberculosis transcriptomes indicative of change of their microenvironment, especially in activated macrophages. For example, the mbt operon (mbtA to mbtE and MAP2172c) involved in iron metabolism was significantly upregulated at 24 h postinfection in IFN-γ-treated macrophages compared to the RPMI-incubated control samples. A similar observation was reported for 120 h postinfection of bovine monocyte-derived macrophages (MDMs) (21). MAP2172c was shown to be repressed in M. avium subsp. paratuberculosis cultures grown in mycobactin J-depleted medium over time, where expression levels of other mbt genes remained constant (42). This paradigm could suggest that an intracellular environment is not iron exochelin-free, or there are other intracellular factors that stimulate alternative iron metabolic pathways, at least in the examined times. On the other hand, a gene involved in iron storage, bfrA (43), was significantly downregulated at the same time point, while the mbt operon was activated, suggesting the lack of access to iron inside the mycobacterial phagosome. Interestingly, the expression of the iron-dependent regulator ideR (43) remained unchanged during the examined time course regardless of macrophage activation status, suggesting a lesser role of ideR in early stages of macrophage infection. Overall, a significant time-dependent shift in M. avium subsp. paratuberculosis transcriptomes was evident from examining M. avium subsp. paratuberculosis collected at 2 and 24 h after macrophage infection.
Microenvironment of M. avium subsp. paratuberculosis.
One of our goals is to gain more insights into the intracellular environment of M. avium subsp. paratuberculosis using transcriptome analysis. Schnappinger et al. reported that M. tuberculosis upregulates β-oxidation genes by 4 h postinfection in murine bone marrow macrophages, suggesting a transition of carbon source from carbohydrates to fatty acids (44). Similarly, our study indicated activation of M. avium subsp. paratuberculosis orthologues (fadA6_3, fadB_1, fadD9, fadE17, fadE21, fadE3_1, and fadE5) in the β-oxidation pathway starting from as early as 2 h postinfection, suggesting the transition of carbon source utilization is a common bacterial strategy between M. tuberculosis and M. avium subsp. paratuberculosis. The icl gene, previously known as aceA, was also among the highly upregulated genes involved in carbon metabolism. The gene product, an isocitrate lyase, bridges the β-oxidation pathway to glyoxylate cycle, an anabolic pathway with a net product of glucose. The contribution of icl to M. avium subsp. paratuberculosis survival in macrophages remains to be analyzed.
Once entering host cell compartments, intracellular bacteria encounter host defense mechanisms such as reactive oxygen intermediates (ROIs), reactive nitrogen intermediates (RNIs), digestive enzymes, and, most importantly, lower pH. In addition, the phagosome is also a nutrient-depleted environment. Accordingly, we examined genes that are associated with stress response and intracellular bacterial survival. Several known oxidative stress-induced genes, such as oxyR, trxB, trxC, tpx, ahpC, and ahpD, were significantly upregulated in all intracellular conditions (Fig. 2). OxyR is a redox sensor protein that, when oxidized, positively regulates a group of genes, including ahpC, katG, gorA, and furA (45). Among those genes, the ahpCD operon was highly upregulated (6.2- to 11.0-fold) in samples taken from naive or activated macrophages. In addition, TrxB, TrxC, and Tpx, proteins involved in reduction of thio-disulfide and resistance of hydroperoxide processes (46), were upregulated, suggesting active machinery for counteracting oxidative stress within host cells. However, other known oxidative responsive genes, such as katG, gorA, furA, sodA, and sodC, were not activated in these samples, possibly because those genes are indirectly regulated by oxyR or also under the control of other stress-response regulators. Overall, M. avium subsp. paratuberculosis deployed specific gene products to defend against the hostile microenvironment during this early stage of infection.
Changes in global gene regulators.
Sigma factors play a central role in bacterial gene regulation (47) and pathogenesis as reviewed in M. tuberculosis (48) and other pathogens (49). Nineteen sigma factors were identified in M. avium subsp. paratuberculosis by sequence analysis (24), 13 of which are homologous to M. tuberculosis sigma factors. The sigA gene, though considered a constitutively expressed sigma factor gene in M. tuberculosis (50), was found downregulated to nearly 2-fold at 24 h in activated macrophages (see Table S5 in the supplemental material). The sigB gene, which is a dispensable sigma factor and partially responsive to some oxidative and heat shock stresses in M. tuberculosis (51), showed a slight increase (∼1.6-fold) at 24 h postinfection under both activated and naive states of macrophages. Genes of sigC, sigG, sigJ, sigM, and other extracytoplasmic sigma factors (ECF-2 through ECF-6) remained constantly expressed in all examined conditions. However, as shown in Fig. 3, sigH was the most induced among other sigma factors of M. avium subsp. paratuberculosis, and the activation seems to be augmented by macrophage activation over time. sigH transcripts were upregulated under in vitro heat shock and oxidative stress treatments in M. avium subsp. paratuberculosis (12). Also, sigL was upregulated at the 2-h time point but downregulated by 24 h postinfection, suggesting a potential role for sigL in the very early stage of infection. On the contrary, sigE expression was significantly upregulated after 2 h postinfection and remained high at 24 h postinfection, suggesting a prolonged role during M. avium subsp. paratuberculosis persistence. Overall, a few sigma factors showed a dynamic and active gene regulation transition during the first 24 h postinfection within macrophages. It is possible that other regulators could play a similar role during later times of infection or in different host cells.
Fig 3.
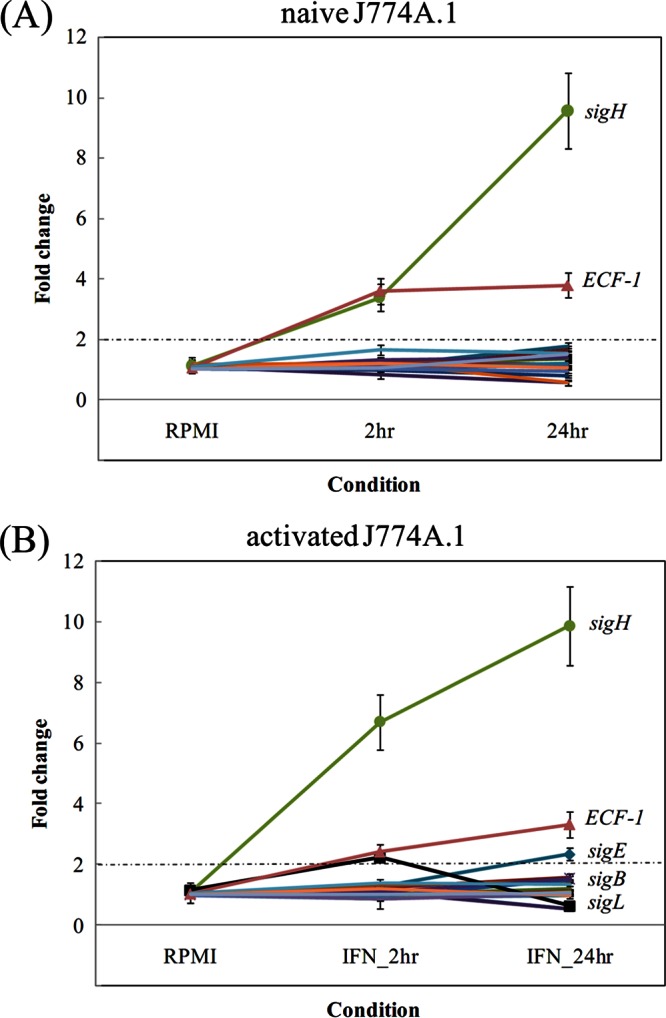
Temporal expression of M. avium subsp. paratuberculosis sigma factors within macrophages using DNA microarrays. A selected list of the M. avium subsp. paratuberculosis sigma factor genes are shown under naive (A) and activated (B) macrophages. Note that sigH and other ECF-1 (ECF-1 on the chart for clarity) were upregulated immediately after infection, followed by expression of sigE and sigB.
Role of sigH in M. avium subsp. paratuberculosis to variable stress conditions.
The sigH gene in M. tuberculosis has been shown to be upregulated upon heat shock, upon diamide treatment (25), and during survival in human macrophages (52), suggesting its importance in responding to extracellular stimuli and intracellular survival. To test the hypothesis that sigH could play an important role in M. avium subsp. paratuberculosis stress responses, we employed a specialized transduction protocol (53) to generate a sigH isogenic knockout mutant in the M. avium subsp. paratuberculosis K10 genetic background (see Fig. S6A and B in the supplemental material). Because sigH and its anti-sigma factor (MAP3323c) are likely encoded in an operon (54), the ΔsigH mutant was examined for possible polar effects on the downstream gene, MAP3323c. Using reverse transcriptase PCR, the presence of the MAP3323c transcript was confirmed in the ΔsigH mutant strain (see Fig. S6C).
After construction, the resistance of the ΔsigH mutant was evaluated against various stressors. Analysis of the disc diffusion assay revealed that the ΔsigH mutant does not tolerate thiol-specific oxidation compared to the wild-type strain, as evidenced by the observed halo zones on plates (Fig. 4A). However, no other differential resistance was observed when a cell wall stressor (sodium dodecyl sulfate) or ethambutol discs were used (data not shown). To measure viability of the ΔsigH mutant after sustained exposure to diamide or heat stress, we cultured both wild-type and the mutant strains for an extended time period in the presence of diamide or a 45°C water bath. In both stress conditions, there was significant reduction at each time point in the viability of the ΔsigH mutant compared to that of the wild-type strain (Fig. 4B and C). At day 7, the viability of the ΔsigH mutant was reduced by almost 2-log orders and more than 1-log order in CFU counts relative to the wild-type strain for diamide and heat stress, respectively. Unfortunately, replicative plasmid complementation of the ΔsigH mutant with a wild-type sigH under the control of the hsp65 promoter did not restore the diamide resistance phenotype (data not shown), most likely due to inefficient complementation in mycobacterial strains (25, 55, 56).
Fig 4.
Phenotypic differences between M. avium subsp. paratuberculosis K10 and the M. avium subsp. paratuberculosis ΔsigH mutant in responding to stress. (A) Disc diffusion assay of K10 and the ΔsigH mutant with various concentrations of diamide. Halo zones indicate that growth of the ΔsigH mutant is inhibited by thiol-specific oxidation. Sustained effect of exposure to diamide (B) or to 45°C heat shock (C) on the M. avium subsp. paratuberculosis survival. Both wild-type and ΔsigH strains were cultured in the presence of 10 mM diamide or a 45°C water bath up to 7 days. Viability of the ΔsigH mutant compared to that of the wild-type strain is shown at different times. Error bars represent the standard deviations calculated from the means of colony counts estimated from two independent experiments with variable significance levels in Student's t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Intracellular survival of the ΔsigH mutant in bovine MDM cells.
Intracellular growth kinetics of M. avium subsp. paratuberculosis strains were analyzed using bovine MDM cells. MDM cells were infected with the ΔsigH mutant and its parental strain for a prolonged time up to 8 days after infection. The MDM monolayer in the culture wells was checked at a regular interval for cell confluence under an inverted light microscope (see Fig. S7 in the supplemental material). We first determined intracellular viability of both the ΔsigH mutant and wild-type strains within the naive MDM cells. The numbers of wild-type strain of M. avium subsp. paratuberculosis bacilli increased 2-fold, whereas the growth of the ΔsigH mutant was not supported within naive MDM cells as determined by CFU plating at 8 days after infection (Fig. 5A). Next, we examined whether the ΔsigH mutant would be able to survive inside activated MDM cells pretreated with recombinant IFN-γ. At 8 days postinfection, there was more than a 2-fold increase in the number of wild-type M. avium subsp. paratuberculosis bacilli, whereas in the IFN-γ pretreated MDM cells, viability of the ΔsigH mutant was significantly reduced almost by 50% (Fig. 5B). These observations suggested an important role for sigH in defending M. avium subsp. paratuberculosis against IFN-γ activation.
Fig 5.

Survival of M. avium subsp. paratuberculosis strains in bovine macrophages. Naive (A) and IFN-γ-pretreated (B) MDM cells were infected with ΔsigH mutant and wild-type M. avium subsp. paratuberculosis strains. Cells were lysed at 1, 4, and 8 days postinfection, and numbers of viable bacilli were determined by CFU plating. The survival levels at 4 and 8 days were relative to the viable counts of bacterial strains at day 1. Survival data represent the average of macrophage infections collected from three different donor animals with significance levels in Student's t test (*, P < 0.05). Error bars represent the standard deviations.
Virulence analysis of the M. avium subsp. paratuberculosis ΔsigH mutant.
To assess the role of SigH in M. avium subsp. paratuberculosis virulence, we investigated the persistence of the M. avium subsp. paratuberculosis ΔsigH mutant using the mouse model of paratuberculosis. The initial growth kinetics of the wild-type and ΔsigH mutant strains were similar, with an equal burden of bacteria in both intestine and spleen up to 6 wpi (Fig. 6A and B). However, the colonization levels of the ΔsigH mutant compared to its parental strain were significantly reduced in spleen and intestine at 12 wpi, suggesting a role for sigH in the long-term survival of M. avium subsp. paratuberculosis in mice. Interestingly, when M. tuberculosis ΔsigH was used to challenge mice, no differences in bacterial load were observed in mouse organs compared to that of the wild-type strain (57).
Fig 6.
Virulence of M. avium subsp. paratuberculosis K10 and the ΔsigH mutant in mice. Mice groups (n = 15) were inoculated with ∼2 × 108 CFU/mouse of M. avium subsp. paratuberculosis wild-type strain or the ΔsigH mutant via intraperitoneal injection. Intestines (A) and spleens (B) were collected at 3, 6, and 12 wpi (n = 5 mice/group/time point) and cultured for bacterial counts. Colony counts for each group are represented by scattered plots accompanied with a median line. Organs with significant difference in bacterial load were denoted with * for P values of <0.05 or ** for P values of <0.01). (C) Pathology of spleen collected from mice infected with M. avium subsp. paratuberculosis K10 (a) and its isogenic ΔsigH mutant (b). H&E-stained sections with 100× magnification (scale bar = 200 μm) are shown. Inset images (1,000× magnification, scale bar = 10 μm) show the M. avium subsp. paratuberculosis bacilli in purple color (arrows). WP, white pulp; RP, red pulp.
Evaluation of the hematoxylin and eosin-stained spleen, liver, and intestine organs at 3, 6, and 12 wpi showed moderately similar tissue pathology when infected with the ΔsigH mutant or wild-type M. avium subsp. paratuberculosis. Granulomatous inflammation was evident in the liver tissues by 12 wpi, with no visual differences in mycobacterial colonization among the mouse groups infected with wild-type or mutant strains (see Fig. S8 in the supplemental material). However, mouse spleen tissues infected with the wild-type strain displayed higher follicular atrophy than the spleen tissues infected with the ΔsigH mutant. Consistent with the bacterial colonization data, Ziehl-Neelsen staining showed higher numbers of acid-fast bacilli in the mouse spleen infected with the wild-type strain than the ΔsigH mutant at 12 wpi (Fig. 6C). Taken together, our data indicated that the ΔsigH mutant was attenuated in the murine model of paratuberculosis compared to the wild-type M. avium subsp. paratuberculosis strain.
Transcriptional regulation of sigH in M. avium subsp. paratuberculosis.
Our stress experiments showed that the ΔsigH mutant was hypersensitive to elevated temperature and diamide exposure, each resulting in impaired growth. On the basis of these findings, we hypothesized that sigH may play an important role in directing transcriptional control under unfavorable environmental conditions. To identify gene regulatory networks under the control of sigH, both wild-type M. avium subsp. paratuberculosis and ΔsigH mutant transcriptomes were profiled before and after diamide exposure using the next-generation RNA sequencing (RNA-seq). When the wild-type strain was compared to the ΔsigH mutant, approximately 15% of the M. avium subsp. paratuberculosis genes (∼307 induced and ∼344 repressed; Fig. 7A) were found to be differentially regulated at 3 h postexposure to diamide stress. This large number of gene perturbation was likely orchestrated by additional sigma factors (e.g., sigB, sigD, sigE) along with various transcriptional regulators that were differentially expressed in examined samples. Genes were grouped into different functional categories (Tables 1 and 2), and a large number of genes (e.g., hsp, clpB) belonging to the chaperonin functional category (Table 1) were significantly upregulated. Many induced genes were involved in detoxification and maintaining cellular redox homeostasis (e.g., trxB2, adhE) during oxidative stress as detailed before (46, 58). Many transcriptional factors and two-component systems were found to be upregulated (e.g., sigB, sigE, whiB4, MAPK_0206, mtrA) under the control of SigH. The expression of mycobacterial sigB and sigE was known to be linked with the presence of the sigH gene (59). WhiB-like proteins are redox-responsive DNA binding factors and could play a protective role against oxidative stress (60). In mycobacteria, the role of the MtrB-MtrA two-component system is not entirely understood, but it was found to be essential for bacterial viability, particularly involved in the regulation of cell wall permeability (61, 62).
Fig 7.
Identification of the SigH regulon. RNA-seq analysis was performed on cultures of M. avium subsp. paratuberculosis and compared to those from its isogenic ΔsigH mutant following 3 h of diamide stress. (A) A circular genomic map of genes differentially expressed between the wild type and its isogenic sigH mutant strains based on significant fold expression ratios (>±2.5-fold). The inner circle shows the genome size scale. The second circle shows the location of the open reading frames in the M. avium subsp. paratuberculosis genome (35). Genes (magenta) on the forward strand are shown outside the baseline; genes (olive green) on the reverse strand are shown inside the baseline. Outer arrows (red) show induced genes, and inner arrows (green) show repressed genes in M. avium subsp. paratuberculosis relative to the levels for the ΔsigH mutant following diamide stress. Arrows indicate direction of gene transcription, and only genes with known function are listed (complete list is shown in Table S6 in the supplemental material). The figure was generated with GenVision software (DNAStar, Madison, WI). (B) Identification of the SigH-specific promoter recognition sequence. Putative M. avium subsp. paratuberculosis SigH promoter consensus elements identified in the upstream sequences of genes whose expression was downregulated in the ΔsigH mutant after exposure to thiol oxidative stress. Consensus sequence motif was determined by weblogo software (http://weblogo.berkeley.edu/), and the height of the letters is proportional to their frequency.
Table 1.
Genes induced in wild-type M. avium subsp. paratuberculosis, relative to the levels for the M. avium subsp. paratuberculosis ΔsigH mutant following diamide exposurea
| Functional classification | Geneb | Fold expression | FDR P value correction | Old locus tagc | M. tuberculosis ortholog | Protein similarity (%) |
|---|---|---|---|---|---|---|
| Chaperone/protein turnover | hsp | 11.01 | 0.00 | MAP3701c | Rv0251c | 78.61 |
| dnaJ2 | 3.57 | 0.00 | MAP2162c | Rv2373c | 84.81 | |
| clpB | 3.49 | 0.00 | MAP3853 | Rv0384c | 91.98 | |
| hflX | 7.01 | 0.00 | MAP2839c | Rv2725c | 82.88 | |
| clpC | 4.54 | 0.00 | MAP0461 | Rv3596c | 92.38 | |
| MAPK_2184 | 2.53 | 0.00 | MAP1584c | Rv2897c | 44.96 | |
| Oxidative stress scavenger/detoxification | trxB2 | 3.77 | 0.00 | MAP4339 | Rv3913 | 77.5 |
| trxC | 2.89 | 0.00 | MAP4340 | Rv3914 | 94.82 | |
| ephA | 3.06 | 0.00 | MAP0446c | Rv3617 | 85.89 | |
| adhE | 4.71 | 0.00 | MAP3596c | Rv0162c | 67.31 | |
| adhB | 2.51 | 0.00 | MAP0595c | Rv0761c | 94.66 | |
| aldB | 3.72 | 0.00 | MAP3413 | Rv3293 | 79.79 | |
| Regulatory/transcription | MAPK_0206 | 8.46 | 0.00 | MAP3562 | Rv0144 | 76.77 |
| MAPK_1421 | 5.25 | 0.00 | MAP2347 | Rv3060c | 40.61 | |
| whiB4 | 5.14 | 0.00 | MAP0393 | Rv3681c | 96.87 | |
| MAPK_0357 | 4.76 | 0.00 | MAP3411c | Rv3291c | 92 | |
| MAPK_0097 | 3.95 | 0.00 | MAP3671 | Rv0232 | 73.36 | |
| MAPK_1985 | 3.32 | 0.00 | MAP1783 | Rv0067c | 45.86 | |
| MAPK_0788 | 3.08 | 0.00 | MAP2980c | Rv2912c | 87.69 | |
| whiB2 | 3.04 | 0.00 | MAP3372c | Rv3260c | 98.59 | |
| MAPK_3589 | 2.87 | 0.00 | MAP0179c | Rv3855 | 73.48 | |
| MAPK_1209 | 2.84 | 0.00 | MAP2559 | Rv1219c | 89.95 | |
| MAPK_1920 | 2.75 | 0.01 | MAP1848c | Rv3830c | 44.38 | |
| MAPK_4083 | 2.75 | 0.02 | MAP4081 | Rv0586 | 76.39 | |
| MAPK_3159 | 2.62 | 0.01 | MAP0609 | Rv0775 | 81.77 | |
| MAPK_0193 | 2.52 | 0.00 | MAP3575 | Rv0158 | 93.8 | |
| sigH | 5.60 | 0.00 | MAP3324c | Rv3223c | 90.9 | |
| sigE | 5.04 | 0.00 | MAP2557c | Rv1221 | 88.42 | |
| sigB | 3.5 | 0.00 | MAP2826 | Rv2710 | 98.12 | |
| sigD | 2.94 | 0.00 | MAP4275 | Rv3414c | 78.28 | |
| Two-component system | MAPK_2766 | 9.89 | 0.00 | MAP1002c | Rv1033c | 85.21 |
| MAPK_2767 | 6.34 | 0.00 | MAP1001c | Rv1032c | 65.46 | |
| mtrA | 3.70 | 0.00 | MAP3360c | Rv3246c | 87.28 | |
| MAPK_2852 | 3.17 | 0.00 | MAP0916 | Rv0981 | 91.66 | |
| senX3 | 2.51 | 0.00 | MAP3982 | Rv0490 | 82.68 | |
| Virulence | MAPK_0165 | 5.01 | 0.00 | MAP3603 | Rv0168 | 66.53 |
| MAPK_1919 | 4.95 | 0.00 | MAP1849 | Rv0587 | 59.9 | |
| mceA1 | 3.79 | 0.00 | MAP3604 | Rv0169 | 77.3 | |
| MAPK_1913 | 3.76 | 0.00 | MAP1855 | Rv0593 | 48.19 | |
| MAPK_0166 | 3.71 | 0.00 | MAP3602 | Rv0587 | 89.18 | |
| MAPK_1918 | 3.66 | 0.00 | MAP1850 | Rv3500c | 50.72 | |
| mceC | 3.02 | 0.00 | MAP3606 | Rv0171 | 87.47 | |
| MAPK_2565 | 2.96 | 0.00 | MAP1203 | Rv1477 | 73.31 | |
| mce5A | 2.94 | 0.03 | MAP1851 | Rv3494c | 44.97 | |
| mceB2 | 2.81 | 0.00 | MAP3605 | Rv0170 | 89.59 | |
| MAPK_1916 | 2.80 | 0.02 | MAP1852 | Rv1967 | 54.33 | |
| MAPK_1914 | 2.59 | 0.04 | MAP1854 | Rv3496c | 50.17 | |
| mceD | 2.52 | 0.00 | MAP3607 | Rv0172 | 82.78 | |
| Central intermediary/sulfate metabolism | ppdK | 2.93 | 0.00 | MAP2664 | Rv1127c | 76.79 |
| rmlB | 5.32 | 0.00 | MAP4225c | Rv3464 | 95.46 | |
| rmlC | 4.18 | 0.00 | MAP4224c | Rv3465 | 89.28 | |
| MAPK_1169 | 10.79 | 0.00 | MAP2599c | Rv1286 | 62.76 | |
| cysQ_2 | 2.81 | 0.00 | MAP2058c | Rv2131c | 69.33 | |
| cysD | 2.52 | 0.00 | MAP2485c | Rv1285 | 82.37 | |
| Energy metabolism | icl | 2.57 | 0.05 | MAP3961 | Rv0467 | 97.36 |
| rpi | 6.53 | 0.00 | MAP2285c | Rv2465c | 96.17 | |
| tpi | 6.44 | 0.00 | MAP1166 | Rv1438 | 91.57 | |
| ccsB | 7.10 | 0.00 | MAP4025 | Rv0529 | 76.92 | |
| nuoA | 4.95 | 0.00 | MAP3201 | Rv3145 | 73.43 | |
| nuoB | 3.95 | 0.00 | MAP3202 | Rv3146 | 91.3 | |
| nuoD | 3.66 | 0.00 | MAP3204 | Rv3148 | 95.21 | |
| nuoC | 3.57 | 0.00 | MAP3203 | Rv3147 | 86.01 | |
| Cell envelope associated | mmpL4_1 | 4.60 | 0.00 | MAP0076 | Rv0402c | 68.15 |
| MAPK_4322 | 4.10 | 0.00 | MAP4320 | Rv0402c | 53.19 | |
| mmpS3 | 4.02 | 0.00 | MAP1938c | Rv2198c | 98.88 | |
| MAPK_3948 | 3.16 | 0.00 | MAP3946 | Rv0677c | 50.33 | |
| mmpS4 | 2.78 | 0.00 | MAP1241c | Rv0451c | 73.52 | |
| lpqD | 3.39 | 0.00 | MAP3481 | Rv3390 | 77.02 | |
| MAPK_0832 | 2.95 | 0.00 | MAP2936c | Rv2864c | 89.72 | |
| Cell processes/transport | MAPK_3150 | 2.66 | 0.00 | MAP0618c | Rv0783c | 70.65 |
| efpA_2 | 2.60 | 0.00 | MAP2915c | Rv2846c | 79.92 | |
| MAPK_4062 | 4.28 | 0.00 | MAP4060c | Rv3273 | 64.47 | |
| fdxC_2 | 9.70 | 0.00 | MAP2607c | Rv1177 | 96.22 |
The functional categorization of M. avium subsp. paratuberculosis genes was based on their known functions as assigned in the Integrated Microbial Genomes system (http://http://img.jgi.doe.gov).
Improved annotations according to the revised genome sequence of M. avium subsp. paratuberculosis (35).
Annotations according to Li et al. (32).
Table 2.
Genes repressed in wild-type M. avium subsp. paratuberculosis, relative to the levels for the M. paratuberculosis ΔsigH mutant following diamide exposurea
| Functional classification | Geneb | Fold expression | FDR P value correction | Old locus tagc | M. tuberculosis ortholog | Protein similarity |
|---|---|---|---|---|---|---|
| Lipid metabolism | fadE3_2 | −2.64 | 0.00 | MAP3651c | Rv0215c | 90.65 |
| fadE26_2 | −2.72 | 0.00 | MAP2585 | Rv3504 | 65.58 | |
| fadD36 | −3.15 | 0.01 | MAP2580c | Rv1193 | 75.89 | |
| echA14 | −3.60 | 0.00 | MAP2306 | Rv2486 | 75.39 | |
| MAPK_2175 | −3.81 | 0.00 | MAP1593 | Rv1136 | 55.55 | |
| fadA6_4 | −5.15 | 0.00 | MAP3337 | Rv3556c | 61.05 | |
| fadE14 | −5.42 | 0.00 | MAP1553c | Rv1346 | 84.55 | |
| fadD33_2 | −5.60 | 0.00 | MAP1554c | Rv1345 | 75.09 | |
| mbtC | −3.29 | 0.00 | MAP2175c | Rv2382c | 78.87 | |
| MAPK_1126 | −4.10 | 0.00 | MAP2642 | Rv1665 | 60.23 | |
| papA3_2 | −4.22 | 0.00 | MAP3763c | Rv3820c | 66.73 | |
| mbtD | −4.43 | 0.00 | MAP2174c | Rv2381c | 52.68 | |
| mbtA | −4.51 | 0.00 | MAP2178 | Rv2384 | 76.87 | |
| MAPK_0028 | −4.86 | 0.00 | MAP3740 | Rv0035 | 51.42 | |
| pks2 | −5.62 | 0.00 | MAP3764c | Rv1180 | 79.08 | |
| MAPK_1764 | −2.63 | 0.00 | MAP2004 | Rv2251 | 83.94 | |
| MAPK_0895 | −4.36 | 0.00 | MAP2873c | Rv2247 | 47.36 | |
| MAPK_2213 | −7.33 | 0.00 | MAP1555c | Rv1344 | 97.01 | |
| Amino acid biosynthesis | trpE2 | −2.70 | 0.00 | MAP2205c | Rv2386c | 79.33 |
| metA | −2.70 | 0.00 | MAP3458 | Rv3341 | 75.53 | |
| metC | −2.73 | 0.00 | MAP3457 | Rv3340 | 90.13 | |
| leuD | −3.79 | 0.00 | MAP3025c | Rv2987c | 97.19 | |
| leuC | −4.10 | 0.00 | MAP3026c | Rv2988c | 92.96 | |
| hacA | −11.23 | 0.00 | MAP2255 | Rv2988c | 50.28 | |
| argC | −2.80 | 0.00 | MAP1361 | Rv1652 | 83.58 | |
| Detoxification | ahpC | −12.47 | 0.00 | MAP1589c | Rv2428 | 93.33 |
| ahpD | −15.40 | 0.00 | MAP1588c | Rv2429 | 78.85 | |
| Cell envelope associated | MAPK_0029 | −5.69 | 0.00 | MAP3739c | Rv2846c | 46.68 |
| MAPK_0769 | −5.93 | 0.00 | MAP2999 | Rv2963 | 79.31 | |
| MAPK_0033 | −6.79 | 0.00 | MAP3735c | Rv1348 | 54 | |
| MAPK_3788 | −10.35 | 0.00 | MAP3786 | Rv0290 | 55.43 | |
| MAPK_3789 | −10.73 | 0.00 | MAP3787 | Rv0291 | 79.95 |
The functional categorization of M. avium subsp. paratuberculosis genes was based on their known functions as assigned in the Integrated Microbial Genomes system (http://img.jgi.doe.gov).
Improved annotations according to the revised genome sequence of M. avium subsp. paratuberculosis (35).
Annotations according to Li et al. (32).
A number of induced genes in the SigH regulon were related to virulence (Table 1), and many of them were included in the mce gene family (e.g., mceA1, mceC, mceD). Several mce genes were shown to be upregulated during phagocytosis and oxidative stress exposure (52, 63), indicating that they are active during infection. Other key functional gene categories were associated with central intermediary/sulfate metabolism (e.g., rmlB, rmlC, cysQ_2, cysD), energy metabolism (e.g., rpi, tpi, nuoA), and cell processes/transport (e.g., fdxC_2, MAPK_4062) or were cell envelope related (e.g., mmpL4_1, mmpS3). Our results indicate that many of these genes were induced under intraphagosomal stresses inside macrophages (see Table S1 in the supplemental material). Genes belonging to functional categories, including lipid metabolism (e.g., fadE14, fadD33_2, MAPK_2213), polyketides (e.g., pks2, papA3_2), and biosynthesis of amino acids (e.g., leuC, metA, trpE2), were among the downregulated genes in M. avium subsp. paratuberculosis relative to the ΔsigH mutant following diamide stress (Table 2). We also examined the differential expression profile of M. avium subsp. paratuberculosis in the absence of sigH during standard physiological growth conditions (mid-log phase). In this case, gene categories belong to lipid metabolism (e.g., fadD29), cell processes (e.g., kdpA, pstS), transcriptional regulation (e.g., MAPK_0788), and electron transport (e.g., fdxC_2). Additionally, we found that a large number of genes belong to the hypothetical functional category (∼30%). The complete lists of genes differentially expressed during diamide stress and normal physiological growth are shown in Table S6 and Table S7 in the supplemental material, respectively. To verify the transcriptome results, a few upregulated genes were randomly selected, and qRT-PCR was employed using the SYBR green method (see Fig. S9 and Table S8 in the supplemental material). The transcript levels of these genes analyzed by RNA-seq and qRT-PCR were in good agreement and corroborated with the transcriptome data.
Identification of sigH-regulated promoters in M. avium subsp. paratuberculosis.
For identification of promoters that were likely to be directly controlled by sigH, we analyzed a list of candidate transcripts with higher expression ratios (wild type/ΔsigH mutant). Since SigH of M. avium subsp. paratuberculosis is a very close homologue of M. tuberculosis SigH (59), we searched for the presence of the consensus sequence of M. tuberculosis SigH-dependent nondegenerate promoter motifs GGAA-N18–20-GTT in the 250-bp region upstream of start codons of M. avium subsp. paratuberculosis genes using the Genolist webserver (http://genodb.pasteur.fr/cgi-bin/WebObjects/GenoList). A total of 30 genes were found to be directly upregulated by SigH with GGAA and GTT core motifs at the −35 and −10 regions, respectively, in their promoter regions (see Table S9 in the supplemental material). Many of these targets of SigH in M. avium subsp. paratuberculosis were also found to be controlled in Corynebacterium glutamicum (64), Streptomyces coelicolor (65), and M. tuberculosis (59), suggesting a conserved regulon directly controlled by SigH across the high-percentage GC Gram-positive actinobacteria. The alignment of SigH-dependent consensus promoters in M. avium subsp. paratuberculosis is shown in Fig. 7B.
DISCUSSION
The intracellular pathogen M. avium subsp. paratuberculosis is known to infect and persist within host macrophages with unclear mechanisms. To examine how M. avium subsp. paratuberculosis responds to intracellular environments, especially during the early stages of infection, we used a macrophage cell line coupled with DNA microarrays to profile macrophage-induced changes in M. avium subsp. paratuberculosis transcriptome. A clear advantage of this infection model is the flexibility to control the activation status of the host cells in addition to the availability of reagents and protocols for manipulation. By comparing the results of phagosome pH and phagosome colocalization markers, we found significant differences in intracellular environments of naive versus active macrophages consistent with earlier studies (44, 66). Activated macrophages, at the time of infection, showed much higher iNOS gene expression than naive macrophages. At 2 or 24 h postinfection, they showed higher phagosome colocalization with ingested M. avium subsp. paratuberculosis particles, which clearly exhibited a better cell defense mechanism than naive macrophages. However, during the course of infection up to 24 h, overall survival of intracellular M. avium subsp. paratuberculosis did not differ in either naive or activated macrophages. This phenotype could change later during persistent infection which we did not address in this study. Most of the differentially expressed genes between these states are core stress responsive genes involved in energy production, indicating M. avium subsp. paratuberculosis initiates stress responses to a higher level more rapidly in activated intracellular environments. On the host side, once activated, the host cells maintained their activation status throughout the course of infection. This suggests that virulent M. avium subsp. paratuberculosis has the ability to prevent phagosome maturation and subsequently circumvent detrimental low pH and oxidative stresses during the very early stages of infection, possibly without interfering with the host early signal transduction pathways responsible for macrophage activation.
When M. avium subsp. paratuberculosis transcriptomes in macrophages were compared to our previous study of the in vitro stressome (12), there were more common genes with the 24-h than 2-h-postinfection samples, indicating that during the early stage of infection, M. avium subsp. paratuberculosis is adjusting to more acidic and oxidizing environments. We also observed the metabolic shift of M. avium subsp. paratuberculosis to utilize fatty acids as the major carbon source, which has already been observed in M. tuberculosis (44) and M. avium subsp. paratuberculosis (21). The shift of metabolic activity at early infection may be a common theme employed by mycobacterial pathogens under nutrient-depleted conditions. By 24 h postinfection, securing iron for M. avium subsp. paratuberculosis became a significant quest, especially in activated macrophages, as suggested by the activation of the mbt operon. It is well established that the phagosome is an iron-depleted compartment (44) and intracellular pathogens have evolved ways to scavenge iron within mammalian cells. However, iron acquisition mechanisms of M. avium subsp. paratuberculosis remain unknown given that M. avium subsp. paratuberculosis possesses a truncated mtbA gene and thus is unable to produce mycobactin (32).
Because of the important role played by global gene regulators in bacterial pathogenesis, we focused our analysis on the expression profile of the 19 sigma factors encoded in the M. avium subsp. paratuberculosis genome (32). Accordingly, in the experiments reported here, we were able to capture active gene regulation of a set of sigma factors (e.g., sigH, ECF-1) during early macrophage infection with M. avium subsp. paratuberculosis. Consistent with the M. tuberculosis infection studies, we found an immediate upregulation of sigH within macrophages. This trend continued through 24 h postinfection and indicated a crucial role of sigH to regulate stress-responsive genes, especially those activated during exposure to thiol oxidation, as indicated by the disc diffusion assay. Moreover, we have demonstrated that the ΔsigH mutant is very sensitive to sustained exposure to diamide or heat stress compared to the wild-type strain. The sigE gene, another stress-induced sigma factor, did not show higher expression levels until 24 h postinfection in either naive or activated macrophages. This delayed response of sigE as well as modest induction of sigB may indicate an indirect regulation by other immediate stress-responsive genes and support the important role played by sigma factors in mycobacterial pathogenesis (25, 59). The other sigma factor that was upregulated throughout the examined time course was ECF-1. This sigma factor may also play an important role immediately upon infection, which was not reported before.
We have also profiled the regulatory network under the control of sigH by studying the relative abundance of gene transcripts using RNA-seq. We found that a large number of M. avium subsp. paratuberculosis genes were directly or indirectly regulated by sigH after exposure to diamide stress. In fact, analysis of the upstream sequence of the upregulated genes revealed a set of genes that could be directly controlled by SigH. Among them, many genes are involved in the functional category of heat shock and protein processing. Heat shock proteins (e.g., Hsp, DnaJ2, ClpB) are found widely on prokaryotic cells and act as molecular chaperones helping to configure proteins correctly upon encountering an unfavorable milieu (67, 68). Such environments, i.e., oxidative stress, could result in nonrepairable protein structures which may necessitate full degradation by the ClpC protease (63). Oxidative stress scavengers induced following diamide stress include TrxB2, TrxC, and AdhE (46, 58). All of these genes likely play important roles in redox homeostasis under thiol oxidation and are found in high levels inside the macrophages (44, 63). Interestingly, the effect of diamide stress in M. tuberculosis also resulted in a transcriptional profile similar to that of M. avium subsp. paratuberculosis (63), indicating the pivotal role of sigH across mycobacterial species.
Consistent with the estimated large regulon of SigH, a significant difference in survival rates between the ΔsigH and wild-type strains was observed inside activated bovine macrophages. Intracellular growth of the wild-type M. avium subsp. paratuberculosis strain was not inhibited regardless of the activation state up to 8 days postinfection. This observation was in corroboration with the earlier findings which showed that activated bovine monocytes were inadequate to inhibit intracellular growth of M. avium subsp. paratuberculosis up to 9 days after infection as determined by the CFU method (69). In contrast, viability of the ΔsigH mutant was significantly impaired, indicating an important function of sigH for the intracellular growth of M. avium subsp. paratuberculosis, possibly by blocking IFN-γ activity as suggested earlier (23). In recent studies, clues have been obtained on macrophage interaction with M. avium subsp. paratuberculosis that indicate the capacity of this pathogen to subvert host immune responses by blocking the ability of mononuclear phagocyte maturation (23, 70, 71). Although it is tempting to speculate that the ΔsigH mutant failed to interfere with macrophage maturation, especially when preactivated with IFN-γ, more experiments are needed to fully understand the mechanisms that sigH could play during macrophage infection. The survival profile of M. avium subsp. paratuberculosis constructs in MDM was further supported by the inability of the ΔsigH mutant to survive in mice. Both bacteriological and histological analyses displayed impaired organ colonization of the ΔsigH mutant with a low inflammatory response. However, we did not find any comparative differences in liver organs infected with the wild-type and ΔsigH mutant strains. A recent study showed that the M. tuberculosis ΔsigH mutant was completely attenuated in nonhuman primates (72), a better experimental model than mice for studying human tuberculosis (57). It will be interesting and important to examine the survival of the M. avium subsp. paratuberculosis ΔsigH mutant in a ruminant model of paratuberculosis (e.g., goat).
In conclusion, our analyses indicated significant changes in mycobacterial gene expression during macrophage survival, most likely under the control of sigH and other sigma factors. The activation status of macrophages also directs the mycobacterial response to a specific stress-responsive profile. We demonstrated that sigH offers a massive temporal response on the M. avium subsp. paratuberculosis transcriptome to cope with the adverse effects of oxidative stress. Our data indicate that sigH could play a critical role during infection, and activation of its regulon is required for replication and full virulence of M. avium subsp. paratuberculosis. Further interrogation of these sigma factors and their regulatory networks should ultimately furnish a greater understanding of M. avium subsp. paratuberculosis pathogenesis and help design a better approach for controlling Johne's disease.
Supplementary Material
ACKNOWLEDGMENTS
We thank Matyas Sandor for providing murine IFN-γ and Erik Settles and Sarah Marcus for reading the manuscript. We also acknowledge the assistance of Joseph Heintz and Jim Windelborn with confocal microscopy and Kay Nelson and Meagan A. Cooney at the University of Wisconsin-Madison for the MDM isolation.
This work was supported by NRI 2007-35204-18400 and JDIP-Q6286224301 grants from the USDA.
Footnotes
Published ahead of print 8 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01273-12.
REFERENCES
- 1. Ghosh P, Hsu C, Alyamani EJ, Shehata MM, Al-Dubaib MA, Al-Naeem A, Hashad M, Mahmoud OM, Alharbi KB, Al-Busadah K, Al-Swailem AM, Talaat AM. 2012. Genome-wide analysis of the emerging infection with Mycobacterium avium subspecies paratuberculosis in the Arabian camels (Camelus dromedarius). PLoS One. 7:e31947 doi:10.1371/journal.pone.0031947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yalo Ayele W, Machackova M, Pavlik I. 2001. The transmission and impact of paratuberculosis infection in domestic and wild ruminants. Vet. Med. 46:205–224 [Google Scholar]
- 3. Nielsen S, Sr, Toft N. 2009. A review of prevalences of paratuberculosis in farmed animals in Europe. Prev. Vet. Med. 88:1–14 [DOI] [PubMed] [Google Scholar]
- 4. Lombard JE. 2011. Epidemiology and economics of paratuberculosis. Vet. Clin. North Am. Food Anim. Pract. 27:525–535 [DOI] [PubMed] [Google Scholar]
- 5. Lombard JE, Gardner IA, Jafarzadeh SR, Fossler CP, Harris B, Capsel RT, Wagner BA, Johnson WO. 2013. Herd-level prevalence of Mycobacterium avium subsp. paratuberculosis infection in United States dairy herds in 2007. Prev. Vet. Med. 108:234–238 [DOI] [PubMed] [Google Scholar]
- 6. Ott SL, Wells SJ, Wagner BA. 1999. Herd-level economic losses associated with Johne's disease on US dairy operations. Prev. Vet. Med. 40:179–192 [DOI] [PubMed] [Google Scholar]
- 7. Bermudez LE, Petrofsky M, Sommer S, Barletta RG. 2010. Peyer's patch-deficient mice demonstrate that Mycobacterium avium subsp. paratuberculosis translocates across the mucosal barrier via both M cells and enterocytes but has inefficient dissemination. Infect. Immun. 78:3570–3577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chacon O, Bermudez LE, Barletta RG. 2004. Johne's disease, inflammatory bowel disease, and Mycobacterium paratuberculosis. Annu. Rev. Microbiol. 58:329–363 [DOI] [PubMed] [Google Scholar]
- 9. Coussens PM, Colvin CJ, Wiersma K, Abouzied A, Sipkovsky S. 2002. Gene expression profiling of peripheral blood mononuclear cells from cattle infected with Mycobacterium paratuberculosis. Infect. Immun. 70:5494–5502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Coussens PM, Jeffers A, Colvin C. 2004. Rapid and transient activation of gene expression in peripheral blood mononuclear cells from Johne's disease positive cows exposed to Mycobacterium paratuberculosis in vitro. Microb. Pathog. 36:93–108 [DOI] [PubMed] [Google Scholar]
- 11. Weiss DJ, Evanson OA, Deng M, Abrahamsen MS. 2004. Gene expression and antimicrobial activity of bovine macrophages in response to Mycobacterium avium subsp. paratuberculosis. Vet. Pathol. 41:326–337 [DOI] [PubMed] [Google Scholar]
- 12. Wu CW, Schmoller SK, Shin SJ, Talaat AM. 2007. Defining the stressome of Mycobacterium avium subsp. paratuberculosis in vitro and in naturally infected cows. J. Bacteriol. 189:7877–7886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Whittington RJ, Marshall DJ, Nicholls PJ, Marsh IB, Reddacliff LA. 2004. Survival and dormancy of Mycobacterium avium subsp. paratuberculosis in the environment. Appl. Environ. Microbiol. 70:2989–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bannantine JP, Stabel JR. 2002. Killing of Mycobacterium avium subspecies paratuberculosis within macrophages. BMC Microbiol. 2:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stabel JR, Palmer MV, Harris B, Plattner B, Hostetter J, Robbe-Austerman S. 2009. Pathogenesis of Mycobacterium avium subsp. paratuberculosis in neonatal calves after oral or intraperitoneal experimental infection. Vet. Microbiol. 136:306–313 [DOI] [PubMed] [Google Scholar]
- 16. Wu CW, Livesey M, Schmoller SK, Manning EJ, Steinberg H, Davis WC, Hamilton MJ, Talaat AM. 2007. Invasion and persistence of Mycobacterium avium subsp. paratuberculosis during early stages of Johne's disease in calves. Infect. Immun. 75:2110–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shin SJ, Wu C-W, Steinberg H, Talaat AM. 2006. Identification of novel virulence determinants in Mycobacterium paratuberculosis by screening a library of insertional mutants. Infect. Immun. 7:3825–3833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Alonso-Hearn M, Patel D, Danelishvili L, Meunier-Goddik L, Bermudez LE. 2008. The Mycobacterium avium subsp. paratuberculosis MAP3464 gene encodes an oxidoreductase involved in invasion of bovine epithelial cells through the activation of host cell Cdc42. Infect. Immun. 76:170–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wu CW, Schmoller SK, Bannantine JP, Eckstein TM, Inamine JM, Livesey M, Albrecht R, Talaat AM. 2009. A novel cell wall lipopeptide is important for biofilm formation and pathogenicity of Mycobacterium avium subspecies paratuberculosis. Microb. Pathog. 46:222–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Alonso-Hearn M, Eckstein TM, Sommer S, Bermudez LE. 2010. A Mycobacterium avium subsp. paratuberculosis LuxR regulates cell envelope and virulence. Innate Immun. 16:235–247 [DOI] [PubMed] [Google Scholar]
- 21. Zhu X, Tu ZJ, Coussens PM, Kapur V, Janagama H, Naser S, Sreevatsan S. 2008. Transcriptional analysis of diverse strains Mycobacterium avium subspecies paratuberculosis in primary bovine monocyte derived macrophages. Microbes Infect. 10:1274–1282 [DOI] [PubMed] [Google Scholar]
- 22. Janagama HK, Lamont EA, George S, Bannantine JP, Xu WW, Tu ZJ, Wells SJ, Schefers J, Sreevatsan S. 2010. Primary transcriptomes of Mycobacterium avium subspecies paratuberculosis reveal proprietary pathways in tissue and macrophages. BMC Genomics 11:561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Arsenault RJ, Li Y, Bell K, Doig K, Potter A, Griebel PJ, Kusalik A, Napper S. 2012. Mycobacterium avium subspecies paratuberculosis inhibits gamma interferon-induced signaling in bovine monocytes: insights into the cellular mechanisms of Johne's Dis. Infect. Immun. 80:3039–3048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sechi LA, Felis GE, Ahmed N, Paccagnini D, Usai D, Ortu S, Molicotti P, Zanetti S. 2007. Genome and transcriptome scale portrait of sigma factors in Mycobacterium avium subsp. paratuberculosis. Infect. Genet. Evol. 7:424–432 [DOI] [PubMed] [Google Scholar]
- 25. Raman S, Song T, Puyang X, Bardarov S, Jacobs WR, Jr, Husson RN. 2001. The alternative sigma factor SigH regulates major components of oxidative and heat stress responses in Mycobacterium tuberculosis. J. Bacteriol. 183:6119–6125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Diwu Z, Chen CS, Zhang C, Klaubert DH, Haugland RP. 1999. A novel acidotropic pH indicator and its potential application in labeling acidic organelles of live cells. Chem. Biol. 6:411–418 [DOI] [PubMed] [Google Scholar]
- 27. Wozniak AL, Griffin S, Rowlands D, Harris M, Yi M, Lemon SM, Weinman SA. 2010. Intracellular proton conductance of the hepatitis C virus p7 protein and its contribution to infectious virus production. PLoS Pathog. 6:e1001087 doi:10.1371/journal.ppat.1001087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Abramoff MD, Magelhaes PJ, Ram SJ. 2004. Image processing with ImageJ. Biophoton. Int. 11:36–42 [Google Scholar]
- 29. Grode L, Seiler P, Baumann S, Hess J, Brinkmann V, Nasser EA, Mann P, Goosmann C, Bandermann S, Smith D, Bancroft GJ, Reyrat JM, SDvan Raupach B, Kaufmann SH. 2005. Increased vaccine efficacy against tuberculosis of recombinant Mycobacterium bovis bacille Calmette-Guerin mutants that secrete listeriolysin. J. Clin. Invest. 115:2472–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rohde KH, Abramovitch RB, Russell DG. 2007. Mycobacterium tuberculosis invasion of macrophages: linking bacterial gene expression to environmental cues. Cell Host Microbe 2:352–364 [DOI] [PubMed] [Google Scholar]
- 31. Talaat AM, Howard ST, Hale W, Lyons R, Garner H, Johnston SA. 2002. Genomic DNA standards for gene expression profiling in Mycobacterium tuberculosis. Nucleic Acids Res. 30:e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li L, Bannantine JP, Zhang Q, Amonsin A, May BJ, Alt D, Banerji N, Kanjilal S, Kapur V. 2005. The complete genome sequence of Mycobacterium avium subspecies paratuberculosis. Proc. Natl. Acad. Sci. U. S. A. 102:12344–12349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Talaat AM, Hunter P, Johnston SA. 2000. Genome-directed primers for selective labeling of bacterial transcripts for DNA microarray analysis. Nat. Biotechnol. 18:679–682 [DOI] [PubMed] [Google Scholar]
- 34. Raychaudhuri S, Stuart JM, Altman RB. 2000. Principal components analysis to summarize microarray experiments: application to sporulation time series. Pac. Symp. Biocomput. 2000:455–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wynne JW, Seemann T, Bulach DM, Coutts SA, Talaat AM, Michalski WP. 2010. Resequencing the Mycobacterium avium subsp. paratuberculosis K10 genome: improved annotation and revised genome sequence. J. Bacteriol. 192:6319–6320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5:621–628 [DOI] [PubMed] [Google Scholar]
- 37. Kal AJ, van Zonneveld AJ, Benes V, van den Berg M, Koerkamp MG, Albermann K, Strack N, Ruijter JM, Richter A, Dujon B, Ansorge W, Tabak HF. 1999. Dynamics of gene expression revealed by comparison of serial analysis of gene expression transcript profiles from yeast grown on two different carbon sources. Mol. Biol. Cell 10:1859–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ward SK, Hoye EA, Talaat AM. 2008. The global responses of Mycobacterium tuberculosis to physiological levels of copper. J. Bacteriol. 190:2939–2946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Talaat AM, Ward SK, Wu C-W, Rondon E, Tavano C, Bannantine JP, Lyons R, Johnston SA. 2007. Mycobacterial bacilli are metabolically active during chronic tuberculosis in murine lungs: insights from genome-wide transcriptional profiling. J. Bacteriol. 189:4265–4274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Edgar R, Domrachev M, Lash AE. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bannantine JP, Talaat AM. 2010. Genomic and transcriptomic studies in Mycobacterium avium subspecies paratuberculosis. Vet. Immunol. Immunopathol. 138:303–311 [DOI] [PubMed] [Google Scholar]
- 43. Janagama HK, Senthilkumar TM, Bannantine JP, Rodriguez GM, Smith I, Paustian ML, McGarvey JA, Sreevatsan S. 2009. Identification and functional characterization of the iron-dependent regulator (IdeR) of Mycobacterium avium subsp. paratuberculosis. Microbiology 155:3683–3690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schnappinger D, Ehrt S, Voskuil MI, Liu Y, Mangan JA, Monahan IM, Dolganov G, Efron B, Butcher PD, Nathan C, Schoolnik GK. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198:693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pagan-Ramos E, Master SS, Pritchett CL, Reimschuessel R, Trucksis M, Timmins GS, Deretic V. 2006. Molecular and physiological effects of mycobacterial oxyR inactivation. J. Bacteriol. 188:2674–2680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. den Hengst CD, Buttner MJ. 2008. Redox control in actinobacteria. Biochim. Biophys. Acta 1780:1201–1216 [DOI] [PubMed] [Google Scholar]
- 47. Bashyam MD, Hasnain SE. 2004. The extracytoplasmic function sigma factors: role in bacterial pathogenesis. Infect. Genet. Evol. 4:301–308 [DOI] [PubMed] [Google Scholar]
- 48. Sachdeva P, Misra R, Tyagi AK, Singh Y. 2010. The sigma factors of Mycobacterium tuberculosis: regulation of the regulators. FEBS J. 277:605–626 [DOI] [PubMed] [Google Scholar]
- 49. Kazmierczak MJ, Wiedmann M, Boor KJ. 2005. Alternative sigma factors and their roles in bacterial virulence. Microbiol. Mol. Biol. Rev. 69:527–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dubnau E, Fontan P, Manganelli R, Soares-Appel S, Smith I. 2002. Mycobacterium tuberculosis genes induced during infection of human macrophages. Infect. Immun. 70:2787–2795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Manganelli R, Provvedi R, Rodrigue S, Beaucher J, Gaudreau L, Smith I. 2004. Sigma factors and global gene regulation in Mycobacterium tuberculosis. J. Bacteriol. 186:895–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Graham JE, Clark-Curtiss JE. 1999. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS). Proc. Natl. Acad. Sci. U. S. A. 96:11554–11559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bardarov S, Bardarov S, Jr, Pavelka JM, Jr, Sambandamurthy V, Larsen M, Tufariello J, Chan J, Hatfull G, Jacobs JW., Jr 2002. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology 148:3007–3017 [DOI] [PubMed] [Google Scholar]
- 54. Song TS, Dove SL, Lee KH, Husson RN. 2003. RshA, an anti-sigma factor that regulates the activity of the mycobacterial stress response sigma factor SigH. Mol. Microbiol. 50:949–959 [DOI] [PubMed] [Google Scholar]
- 55. Karls RK, Guarner J, McMurray DN, Birkness KA, Quinn FD. 2006. Examination of Mycobacterium tuberculosis sigma factor mutants using low-dose aerosol infection of guinea pigs suggests a role for SigC in pathogenesis. Microbiology 152:1591–1600 [DOI] [PubMed] [Google Scholar]
- 56. Raman S, Hazra R, Dascher CC, Husson RN. 2004. Transcription regulation by the Mycobacterium tuberculosis alternative sigma factor SigD and its role in virulence. J. Bacteriol. 186:6605–6616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kaushal D, Schroeder BG, Tyagi S, Yoshimatsu T, Scott C, Ko C, Carpenter L, Mehrotra J, Manabe YC, Fleischmann RD, Bishai WR. 2002. Reduced immunopathology and mortality despite tissue persistence in a Mycobacterium tuberculosis mutant lacking alternative sigma factor, SigH. Proc. Natl. Acad. Sci. U. S. A. 99:8330–8335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Echave P, Tamarit J, Cabiscol E, Ros J. 2003. Novel antioxidant role of alcohol dehydrogenase E from Escherichia coli. J. Biol. Chem. 278:30193–30198 [DOI] [PubMed] [Google Scholar]
- 59. Manganelli R, Voskuil MI, Schoolnik GK, Dubnau E, Gomez M, Smith I. 2002. Role of the extracytoplasmic-function sigma Factor sigma(H) in Mycobacterium tuberculosis global gene expression. Mol. Microbiol. 45:365–374 [DOI] [PubMed] [Google Scholar]
- 60. Singh A, Crossman DK, Mai D, Guidry L, Voskuil MI, Renfrow MB, Steyn AJ. 2009. Mycobacterium tuberculosis WhiB3 maintains redox homeostasis by regulating virulence lipid anabolism to modulate macrophage response. PLoS Pathog. 5:e1000545 doi:10.1371/journal.ppat.1000545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cangelosi GA, Do JS, Freeman R, Bennett JG, Semret M, Behr MA. 2006. The two-component regulatory system mtrAB is required for morphotypic multidrug resistance in Mycobacterium avium. Antimicrob. Agents Chemother. 50:461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zahrt TC, Deretic V. 2000. An essential two-component signal transduction system in Mycobacterium tuberculosis. J. Bacteriol. 182:3832–3838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mehra S, Kaushal D. 2009. Functional genomics reveals extended roles of the Mycobacterium tuberculosis stress response factor sigmaH. J. Bacteriol. 191:3965–3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Busche T, Silar R, Picmanova M, Patek M, Kalinowski J. 2012. Transcriptional regulation of the operon encoding stress-responsive ECF sigma factor SigH and its anti-sigma factor RshA, and control of its regulatory network in Corynebacterium glutamicum. BMC Genomics 13:445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Paget MS, Kang JG, Roe JH, Buttner MJ. 1998. sigmaR, an RNA polymerase sigma factor that modulates expression of the thioredoxin system in response to oxidative stress in Streptomyces coelicolor A3(2). EMBO J. 17:5776–5782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kuehnel MP, Goethe R, Habermann A, Mueller E, Rohde M, Griffiths G, Valentin-Weigand P. 2001. Characterization of the intracellular survival of Mycobacterium avium ssp. paratuberculosis: phagosomal pH and fusogenicity in J774 macrophages compared with other mycobacteria. Cell Microbiol. 3:551–566 [DOI] [PubMed] [Google Scholar]
- 67. Stewart GR, Wernisch L, Stabler R, Mangan JA, Hinds J, Laing KG, Young DB, Butcher PD. 2002. Dissection of the heat-shock response in Mycobacterium tuberculosis using mutants and microarrays. Microbiology 148:3129–3138 [DOI] [PubMed] [Google Scholar]
- 68. Yang H, Huang S, Dai H, Gong Y, Zheng C, Chang Z. 1999. The Mycobacterium tuberculosis small heat shock protein Hsp16.3 exposes hydrophobic surfaces at mild conditions: conformational flexibility and molecular chaperone activity. Protein Sci. 8:174–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhao B, Collins MT, Czuprynski CJ. 1997. Effects of gamma interferon and nitric oxide on the interaction of Mycobacterium avium subsp. paratuberculosis with bovine monocytes. Infect. Immun. 65:1761–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Basler T, Brumshagen C, Beineke A, Goethe R, Baumer W. 2 January 2013, posting date Mycobacterium avium subspecies impair dendritic cell maturation. Innate Immun. [Epub ahead of print.] doi:10.1177/1753425912470291 [DOI] [PubMed] [Google Scholar]
- 71. Kabara E, Coussens PM. 2012. Infection of primary bovine macrophages with Mycobacterium avium subspecies paratuberculosis suppresses host cell apoptosis. Front. Microbiol. 3:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Mehra S, Golden NA, Stuckey K, Didier PJ, Doyle LA, Russell-Lodrigue KE, Sugimoto C, Hasegawa A, Sivasubramani SK, Roy CJ, Alvarez X, Kuroda MJ, Blanchard JL, Lackner AA, Kaushal D. 2012. The Mycobacterium tuberculosis stress response factor SigH is required for bacterial burden as well as immunopathology in primate lungs. J. Infect. Dis. 205:1203–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.