Abstract
The Gram-negative bacterium Gallibacterium anatis is a major cause of salpingitis and peritonitis in egg-laying chickens, leading to decreased egg production worldwide. Widespread multidrug resistance largely prevents treatment of this organism using traditional antimicrobial agents, while antigenic diversity hampers disease prevention by classical vaccines. Thus, insight into its pathogenesis and knowledge about important virulence factors is urgently required. A key event during the colonization and invasion of mucosal surfaces is adherence, and recently, at least three F17-like fimbrial gene clusters were identified in the genomes of several G. anatis strains. The objective of this study was to characterize the putative F17-like fimbrial subunit protein FlfA from G. anatis 12656-12 and determine its importance for virulence. In vitro expression and surface exposure of FlfA was demonstrated by flow cytometry and immunofluorescence microscopy. The predicted function of FlfA as a fimbrial subunit protein was confirmed by immunogold electron microscopy. An flfA deletion mutant (ΔflfA) was generated in G. anatis 12656-12, and importantly, this mutant was significantly attenuated in the natural chicken host. Furthermore, protection against G. anatis 12656-12 could be induced by immunizing chickens with recombinant FlfA. Finally, in vitro expression of FlfA homologs was observed in a genetically diverse set of G. anatis strains, suggesting the potential of FlfA as a serotype-independent vaccine candidate This is the first study describing a fimbrial subunit protein of G. anatis with a clear potential as a vaccine antigen.
INTRODUCTION
The global demand for meat and animal products is rising as the world population and income increases. Poultry meat and eggs are considered very important and highly sustainable components of the future global diet (1). Gallibacterium is a genus in the Gram-negative Pasteurellaceae family (2, 3) that is commonly associated with poultry (4, 5). Besides constituting a part of the normal microflora of the upper respiratory tract and lower genital tract in chickens (4–6), Gallibacterium anatis has recently been recognized as a major cause of lesions in the reproductive tracts of egg layers (7–10), causing a drop in egg production and increased mortality (11). Multiple-drug resistance (12) and a substantial antigenic diversity (13) make it difficult to prevent the negative effects of G. anatis using traditional antimicrobial agents and vaccines. Thus, novel prevention and treatment strategies are urgently needed. To date, very little is known about the pathogenesis of G. anatis and only a few putative virulence factors have been identified. These include a new type of RTX toxin GtxA (14), a polysaccharide capsule (15), secreted metalloproteases capable of degrading avian immunoglobulin IgG (16), and the ability of some strains to agglutinate avian erythrocytes (17).
A critical step in microbial colonization and invasion of mucosal tissues is adherence. Preventing adherence is therefore an attractive intervention strategy against further negative effects (18). Although G. anatis can adhere to plastic surfaces, form biofilms, and produce a glycoprotein-like hemagglutinin (19, 20), little is known about the importance of these factors during colonization and infection of the natural host. A variety of adhesins, including fimbriae, has been reported from other members of Pasteurellaceae (21). Fimbriae are long, hair-like, extracellular appendages on the surfaces of bacterial cells and are important virulence factors in many bacterial species (22). Recently, several F17-like fimbrial biosynthesis gene clusters were identified in the genomes of three different G. anatis strains (23). The most frequently occurring fimbrial gene cluster includes a gene predicted to encode a 20.5-kDa fimbrial subunit designated FlfA (GenBank accession no. JX855927), which showed sequence similarity to the F17-like fimbrial protein precursor identified in the extraintestinal pathogenic Escherichia coli (ExPEC) strain 536 (24). The F17 fimbria family, previously known as FY or Att25 (25), comprises many variants such as the F17, G, K99, and 20K fimbriae, which are produced by several E. coli pathotypes, including human ExPEC, animal enterotoxigenic E. coli (ETEC), and avian pathogenic E. coli strains (24–30). For all of these strains it is proposed that fimbriae mediate bacterial adhesion to glycoprotein receptors in the mucus layer (31, 32). F17 fimbriation requires expression of the entire four-gene fimbrial gene cluster (33), and subunit assembly on the cell surface occurs via a chaperone/usher pathway (34).
To assess the structure and function of the F17-like fimbriae in G. anatis, we characterized the subunit protein FlfA from G. anatis 12656-12 and studied its importance in pathogenesis. We demonstrated in vitro expression and surface exposure of FlfA in a bistable manner, and confirmed that FlfA is incorporated into surface exposed fimbrial structures by immunogold electron microscopy. Moreover, a nonfimbriated ΔflfA mutant showed decreased virulence in chickens and we showed that protection against G. anatis 12656-12 could be induced by immunization of chickens with recombinant FlfA. Finally, we demonstrated that FlfA homologs are expressed in other G. anatis strains, suggesting the cross-protective potential of FlfA. The results from this study therefore indicate that F17-like fimbriae are key factors in the pathogenesis of G. anatis in chickens and focus interest on the FlfA protein as a future vaccine candidate.
MATERIALS AND METHODS
Animal ethics statement.
All work on experimental animals was carried out in accordance with legislation of The Danish Ministry of Justice and with the approval of the Danish National Animal Ethics Committee (approval no. 2008/561-1481).
Bacterial strains and growth conditions.
G. anatis bv. haemolytica strain 12656-12 Liver (referred to here as G. anatis 12656-12) was used as a source of DNA for cloning of flfA, for the in vitro characterization of FlfA, and as the challenge strain in the chicken infection experiments. In addition, seven G. anatis strains were selected and analyzed for the expression of a FlfA protein homolog. The genomes of the strains have been annotated using Wasabi, a web-based annotation system for prokaryotic organisms developed by the Victorian Bioinfomatics Consortium, Monash University, Melbourne, Australia (36, 37). Protein similarity was determined by use of BLASTp (38), and multiple alignment was conducted using CLUSTAL 2.1 (39). The bacterial strains and plasmids used in the present study are listed in Table 1.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| G. anatis | ||
| 12656-12 | Wild-type strain; biovar 4, isolated in 1981 (Denmark) | 51 |
| ΔflfA mutant | flfA deletion mutant of G. anatis 12656-12; Kmr | This study |
| F149T | Wild-type strain; isolated in 1979 (Denmark) | 2 |
| 10672/6 | Wild-type strain; biovar 1 (Denmark) | 2 |
| 4895 | Wild-type strain; biovar 4 (Mexico) | 52 |
| 7990 | Wild-type strain; biovar 1 (Mexico) | 52 |
| Avicor | Wild-type strain; biovar 3 (Mexico) | 52 |
| CCM5995 | Wild-type strain; biovar 20, isolated in 1978 (Czech Republic) | 2 |
| IPDH 697-78 | Wild-type strain; biovar 15, isolated in 1978 (Germany) | 2 |
| E. coli | ||
| TOP10 | E. coli host strain for routine cloning | Invitrogen |
| DH5α | E. coli host strain for routine cloning | Invitrogen |
| BL21 CodonPlus | E. coli strain for expression of recombinant proteins | Stratagene |
| Plasmids | ||
| pENTR/SD/D-TOPO | Directional cloning vector for entry to the Gateway System | Invitrogen |
| pDEST17 | Gateway Destination vector (Ampr, N-terminal His6 fusion tag) | Invitrogen |
| pDEST17flfA | FlfA expression vector (Ampr, N-terminal His6 fusion tag) | This study |
| pBluescript II KS(+) | General purpose cloning vector (Ampr) | Stratagene |
| pUC4K | Vector carrying the Kmr cassette from Tn903 | 53 |
| pBSΔflfA | pBluescript II KS(+) containing flanking regions of the flfA gene (Ampr) | This study |
| pBSΔflfA-Kmr | Plasmid for flfA deletion with flanking regions of the gene cloned on either side of the Kmr cassette from pUC4K into pBluescript II KS(+) (Ampr Kmr) | This study |
Ampr, ampicillin resistance; Kmr, kanamycin resistance; His, histidine.
G. anatis 12656-12 was incubated at 37°C on brain heart infusion (BHI) agar supplemented with 5% citrated bovine blood in a closed plastic bag or in BHI broth with aeration. E. coli was incubated at 37°C on Luria-Bertani (LB) agar plates or in LB broth with aeration. The media were supplemented with 50 μg of kanamycin/ml or 100 μg of ampicillin/ml when appropriate.
Cloning, expression, and purification of recombinant FlfA.
The flfA gene was amplified by PCR using G. anatis 12656-12 genomic DNA as the template together with the primers 1162F and 1162R (Table 2). Primers were designed as described previously (40), by use of Oligo Explorer 1.2 (Gene Link). Nucleotides facilitating TOPO cloning (CACC), together with a start codon (ATG), were added to 1162F.
Table 2.
Primers used in this study
| Primer | Sequence (5′–3′)a | Function | Amplicon |
|---|---|---|---|
| 1162F | CACCATGGGTGCATTTGCGGATGATCC | PCR amplification of FlfA | flfA |
| 1162R | TATTCGTATGCGATAGTATAGTTC | PCR amplification of FlfA | flfA |
| FlfA_UF_n2 | GGCTCTAGACTCCAAAATGGAGTAGTGAT | Deletion mutant generation | Upstream fragment of FlfA |
| FlfA_UR_n2 | TATCCCGGGATCTGTTGCAGATTGACAAC | Deletion mutant generation | Upstream fragment of FlfA |
| FlfA_DF | TTACCCGGGAAGCAAGAAAACAGCCACA | Deletion mutant generation | Downstream fragment of FlfA |
| FlfA_DR | GGACTCGAGGCTCGGTGAAATGGTTAAT | Deletion mutant generation | Downstream fragment of FlfA |
| FlfA_UF2 | ACCTTATGTATGCTCCTATG | Mutant confirmation | |
| FlfA_DR2 | AAAAATCGGGCAGGAAATCT | Mutant confirmation |
The introduced TOPO cloning site (CACC) and start codon (ATG) is indicated in boldface. Introduced restriction sites are underlined.
Cloning was carried out using the Gateway System (41) with chemically competent E. coli strains and plasmids as listed in Table 2. Briefly, the PCR product was cloned into the Gateway entry vector pENTR/SD/D-TOPO and introduced by transformation into E. coli TOP10. The insert in the entry clone was verified by PCR and sequencing. To produce pDEST17galFA, the insert was transferred to pDEST17 by recombination (LR Clonase kit; Invitrogen), and the resulting expression clones were transferred to E. coli DH5α.
Recombinant FlfA was expressed using the Overnight Express System (Novagen), and purification was carried out at the Protein Production Unit, Monash University, Melbourne, Australia. Briefly, expression clones were introduced into E. coli BL21 CodonPlus. Cells were incubated overnight in 200 ml of Overnight Express System medium with 100 μg of ampicillin/ml at 28°C. Recombinant FlfA was purified by Ni-NTA affinity chromatography, followed by gel filtration on an S75 column, and the protein was solubilized in sodium phosphate buffer (100 mM sodium phosphate [pH 7.4], 0.15 M NaCl) containing 8 M urea. The molecular mass, purity, and total protein concentration were analyzed using the Agilent 2100 Bioanalyzer and Protein 230 (P230) kit (Agilent Technologies), together with SDS-PAGE and Lowry assay (DC protein assay; Bio-Rad) to further confirm size and concentration. The identity of FlfA was subsequently confirmed by matrix-assisted laser desorption ionization–time of flight mass spectrometry, as described previously (42).
Production of polyclonal antisera in rabbits.
Polyclonal antisera against intact G. anatis 12656-12 and FlfA were produced in female rabbits (University of Copenhagen, Faculty of Health Sciences, Copenhagen, Denmark). Each rabbit was immunized subcutaneously with 107 bacterial cells fixed in phosphate-buffered saline (PBS) containing 1% paraformaldehyde or with 100 μg of purified recombinant FlfA, both mixed 1:1 with Freund incomplete adjuvant. Immunization was performed at days 0, 21, and 35. Blood for serum was collected at day 0 (preimmune) and day 41 (immune).
Construction of a flfA deletion mutant in G. anatis 12656-12.
A G. anatis flfA deletion mutant was generated by replacing a part of the flfA gene with a kanamycin resistance (Kmr) cassette. The DNA construct was prepared using chemically competent E. coli DH5α and plasmids as listed in Table 1. Briefly, the 485-bp upstream and 649-bp downstream regions of flfA were amplified by PCR from G. anatis 12656-12 with the primers FlfA_UF_n2, FlfA_UR_n2, FlfA_DF, and FlfA_DR (Table 3). The upstream fragment was digested with XbaI and SmaI, while the downstream fragment was digested with XhoI and SmaI. The cloning vector, pBluescript II KS(+), was digested with XbaI and XhoI. All restriction enzymes were purchased from New England BioLabs. Digested fragments and the vector were ligated to generate pBSΔflfA, and the insert was verified by PCR and sequencing. To generate pBSΔflfA-Kmr, the Kmr cassette was purified from pUC4K using HincII, and ligated with SmaI-digested pBSΔFlfA. Linear DNA was produced by PCR amplification using the primer pair FlfA_UF_n2 and FlfA_DR. Natural competence of G. anatis 12656-12 was induced by the MIV method, and transformation with linear DNA was carried out as described previously (14). Transformants were selected on blood agar plates with 5 μg of kanamycin/ml. The size of the deletion site in flfA was confirmed by using the primer pair FlfA_UF2 and FlfA_DR2, and the PCR product was sequenced by BigDye sequencing to verify the genomic localization of the deletion (Macrogen, Korea). The flfA deletion mutant was referred to here as the ΔflfA mutant.
Table 3.
Bacterial reisolation rates of G. anatis from chickens
| Study and group | G. anatis inoculation strain | No. of chickens | Immunizationa | Reisolation (no. of chickens) of G. anatis |
||
|---|---|---|---|---|---|---|
| Ovary | Salpinx | Peritoneum | ||||
| Challenge | ||||||
| A | 12656-12 | 10 | No | 7 | 10 | 6 |
| B | ΔflfA mutant | 10 | No | 3 | 6 | 3 |
| Immunization | ||||||
| C | 12656-12 | 20 | Yes | 11 | 12 | 15 |
| D | 12656-12 | 6 | No | 6 | 4 | 6 |
Chickens in group C were immunized twice with 200 μg of recombinant FlfA, whereas those in group D were sham immunized with a placebo.
Whole-cell extraction and Western blotting.
G. anatis 12656-12 was incubated in liquid BHI to reach the mid-logarithmic growth phase. Cells were harvested by centrifugation, washed and resuspended in PBS, and lysed by three cycles of freeze-thawing. To quantify the amount protein, samples were diluted 1:1,000 in 0.1 M NaOH plus 1%SDS, and the optical density at 260 nm (OD260) was measured. Whole-cell extracts were electrophoresed in 4 to 12% NuPAGE Bis-Tris gels (Invitrogen) under reducing conditions, and transferred to a polyvinylidene difluoride (PVDF) membrane by use of the iBlot dry blotting system (Invitrogen). Membranes were incubated with preimmune or anti-FlfA immune serum. To investigate FlfA expression in G. anatis 12656-12, horseradish peroxidase (HRP)-conjugated polyclonal goat anti-rabbit immunoglobulin (DakoCytomation) was used as a secondary antibody, and blots were developed using SuperSignal West Pico chemiluminescent substrate (Pierce). For the investigation of expression of FlfA homologs, polyclonal goat anti-rabbit IgG (Fc):HRP (AbD Serotec) was used as secondary antibody, and blots were developed using a Novex ECL chemiluminescent substrate reagent kit (Invitrogen).
FACS analysis.
G. anatis 12656-12 was grown in liquid BHI to mid-logarithmic growth phase. Cells were harvested by centrifugation, fixed in 1% paraformaldehyde, and resuspended in PBS plus 1% bovine serum albumin (BSA). Samples of 2.5 × 106 cells were dispensed into a 96-multiwell round-bottom plate, followed by incubation for 1 h with anti-FlfA immune serum diluted in the range 1:100 to 1:3,200 in PBS plus 1% BSA. Antibody binding was detected by 30 min of incubation with fluorescein isothiocyanate-conjugated, goat anti-rabbit IgG (Sigma-Aldrich) diluted 1:100 in PBS plus 1% BSA. Fluorescence-activated cell sorting (FACS) acquisition was performed using the FACSCalibur flow cytometer (Becton Dickinson). The data were analyzed using CellQuest Pro software. As a negative control, anti-FlfA preimmune serum was included. As a control for cell homogeneity, bacteria were incubated with 1:200 dilutions of anti-G. anatis 12656-12 preimmune and immune serum. Finally, staining with secondary antibody alone was included to confirm the specificity of the signal.
Confocal immunofluorescence microscopy.
Bacterial cells from mid-logarithmic growth phase were prepared as described for FACS analysis. Cells were paraformaldehyde-fixed on glass slides and blocked in PBS plus 2% BSA, followed by incubation with preimmune or anti-FlfA immune serum diluted 1:100 in PBS plus 2% BSA. Antibody binding was detected using a secondary goat anti-rabbit IgG rhodamine RedX-conjugated antibody (Jackson Immunoresearch Laboratories), and slides were mounted using the ProLong Gold antifade reagent with DAPI (4′,6′-diamidino-2-phenylindole; Invitrogen). Images were acquired using the Zeiss LSM 710 laser scanning microscope and analyzed using the ZEN software. As controls, samples were prepared using 1:400 dilutions of preimmune or anti-G. anatis 12656-12 immune rabbit serum, followed by incubation with goat anti-rabbit Alexa Fluor 488-conjugated IgG (Invitrogen).
Immunogold electron microscopy.
G. anatis 12656-12 wild-type (WT) and ΔflfA strains was grown to mid-logarithmic phase and washed in PBS. Formvar-carbon-coated nickel grids were floated on 20-μl drops of G. anatis suspensions (∼109 CFU/droplet) for 2 min, and fixed in 2% paraformaldehyde for 15 min. Grids were then placed in blocking solution (PBS plus 1% BSA) for 30 min, floated on drops of anti-FlfA immune serum diluted in PBS plus 1% BSA for 30 min (room temperature), washed with six drops of blocking solution, and floated on secondary antibody conjugated to 10-nm gold particles (diluted 1:20 in PBS plus 1% BSA) for 30 min. Grids were examined by using a JEOL 1200EX II transmission electron microscope at a minimal magnification of ×15,000.
Challenge of chickens with G. anatis WT or ΔflfA strains.
Lohmann Brown layer chickens (29 weeks old) were purchased from a commercial breeder with high biosecurity standards. Chickens were kept under free indoor housing conditions and were provided with freshwater and feed ad libitum. After 1 week of acclimatization the chickens were randomly separated in two groups: A and B. Group A (n = 10) was inoculated intraperitoneally as described previously (15) with G. anatis strain 12656-12, and group B (n = 10) was inoculated intraperitoneally with the ΔflfA mutant. Primary cultures were prepared from single colonies and incubated in BHI at 37°C overnight. Approximately 10 ml of each primary culture was added to 20 ml of BHI, followed by incubation at 37°C for 3 h until the bacteria were in the late-logarithmic growth phase (∼109 CFU/ml). The bacterial concentration in the inoculum was verified by duplicate plate counts. All birds were subjected to postmortem examination 24 h after inoculation, including bacterial isolation and recording of gross lesions. Bacterial isolations from spleen, ovary, and three fixed locations in the salpinx were obtained by streaking tissue swabs. If more than 10 colonies were found on the plate the location was scored positive for reisolation. Five isolates of G. anatis from each chicken were incubated on blood agar plates with 5 μg of kanamycin/ml at 37°C overnight and examined for growth. Gross lesions in the reproductive organs and in the peritoneum were evaluated using a scale from 0 to 4 based on severity, distribution, and characteristics of the lesions. A score of 0 was assigned if no lesions were found; a score of 1 indicated lesions with only slight changes from normal state, focal distribution, and without exudation; a score of 2 was used for lesions with moderate changes from normal state, multifocal distribution, and with light exudation; a score of 3 indicated the presence of lesions with marked changes from normal state, diffuse distribution, and marked exudation or deformation; and a score of 4 was used for the presence of severe lesions with total deformation, diffuse distribution, and heavy exudation.
Immunization of chickens with recombinant FlfA.
Twenty Lohmann Brown layer chickens (group C, n = 20) were immunized twice subcutaneously, 2 weeks apart, with 200 μg of recombinant FlfA in 0.5 ml of 8 M urea sodium phosphate buffer (100 mM sodium phosphate [pH 7.4], 0.15 M NaCl) and 0.5 ml of Freund incomplete adjuvant (Sigma-Aldrich). As a control, chickens (group D, n = 6) were sham immunized with a placebo (8 M urea sodium buffer and Freund incomplete adjuvant). Three weeks after the second injection, all chickens were inoculated intraperitoneally as described in the previous section. Ten of the immunized and three of the nonimmunized chickens were euthanized 24 h postinfection, and the remaining chickens 48 h postinfection. Postmortem examination was performed as described in the previous section.
Statistical analysis.
Data analysis was performed using Mann-Whitney test for comparison of the score of gross lesions and with the Fisher exact test for comparison of bacterial reisolation rates. P values of <0.05 were considered statistically significant.
Nucleotide sequence accession number.
The sequences corresponding to G. anatis 12656-12 flfA, flfD, flfC, and flfG genes have been deposited in GenBank under accession numbers JX855927, JX855928, JX855929, and JX855930, respectively. The sequences corresponding to the flfA homologs identified in G. anatis have been deposited in GenBank under accession numbers JX915814 (G. anatis F149), JX915815 (G. anatis 7990), JX915816 (G. anatis Avicor), JX915817 (G. anatis CCM5995), and JX915818 (G. anatis IPDH 697-78).
RESULTS
The FlfA protein is expressed in vitro in G. anatis 12656-12.
The flfA gene is located in a four-gene cluster in G. anatis 12656-12 and encodes a 20.5-kDa protein with 58% similarity (identical or conserved residues) to the F17-like fimbrial protein precursor identified in the ExPEC strain 536. To produce recombinant FlfA, the flfA gene was amplified from G. anatis 12656-12 without its predicted signal sequence and cloned into the pDEST17 vector, which adds a 4.1-kDa N-terminal sequence including a His6 tag. The protein was expressed in E. coli BL21 and purified by Ni-NTA affinity with a purity of 94% (data not shown). The observed mass of ∼26 kDa (Fig. 1A) corresponded to the predicted size plus the addition of the N-terminal tag generated by pDEST17. The identity of recombinant FlfA was confirmed by mass spectrometry (data not shown). Polyclonal antiserum against recombinant FlfA was produced in rabbits and Western blot demonstrated recognition of recombinant FlfA by the immune serum but not by the preimmune serum (Fig. 1B).
Fig 1.
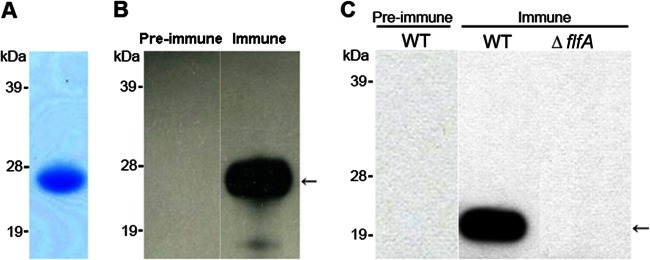
Analysis of recombinant FlfA, specificity of anti-FlfA serum, and FlfA expression within G. anatis 12656-12. (A) The purity and size of recombinant FlfA protein (∼26 kDa) were verified by SDS-PAGE. A 5-μg portion of protein was loaded into the lane. (B) The specificity of rabbit anti-FlfA antiserum was demonstrated by Western blots comparing the recognition of recombinant FlfA (arrow) by preimmune or immune sera. A 0.5-μg portion of protein was loaded into each lane. (C) Western blots demonstrating expression of FlfA in G. anatis 12656-12 (20.5 kDa, arrow). Whole-cell extract equivalent to an OD260 of 10 in 10 μl was loaded into each lane. The positions of molecular mass standards (in kDa) are indicated on the left.
To further confirm the reactivity and specificity of the rabbit antiserum and investigate the role of FlfA during infection, an flfA deletion mutant (ΔflfA) was constructed in G. anatis 12656-12. G. anatis 12656-12 is naturally competent (35), and a stable mutant was obtained by natural transformation with linear DNA. In the ΔflfA mutant, nucleotides 282 to 488 of the flfA gene were replaced by a kanamycin resistance (Kmr) cassette. The size of the deletion site was confirmed by PCR and verified by sequencing. No differences were observed in the in vitro growth of the ΔflfA mutant (data not shown).
To demonstrate expression of FlfA in G. anatis 12656-12 in vitro, a whole-cell extract was prepared from cells in the mid-logarithmic growth phase. Western blotting with anti-FlfA immune serum indicated the presence of a protein corresponding to FlfA (Fig. 1C). This band was not recognized in the whole-cell extract from the ΔflfA mutant, nor in Western blots using preimmune serum, thus confirming the deletion of flfA in the ΔflfA mutant and the specificity of the immune serum.
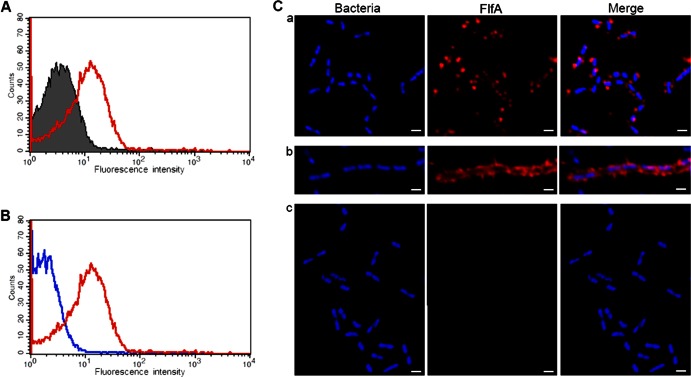
FlfA surface-exposure reveals two different cell populations.
Since exposure to the host is an important feature of any virulence factor, surface expression of FlfA on G. anatis 12656-12 during mid-logarithmic growth phase was investigated by FACS. Incubation of WT cells with anti-FlfA immune serum showed recognition of the protein on the cell surface, with a fluorescence peak significantly higher than that observed with preimmune serum (Fig. 2A). In addition, there was a clear difference in the fluorescence intensity observed between the ΔflfA mutant (Fig. 2B, blue line) and WT cells (Fig. 2B, red line) incubated with anti-FlfA immune serum, thus supporting the surface-localization of FlfA in G. anatis 12656-12. Secondary antibody alone was used as an internal control and did not result in any fluorescence for either WT or ΔflfA strains (data not shown). In summary, these results support the hypothesis that FlfA is surface exposed on G. anatis 12656-12 during in vitro growth.
Fig 2.
Analysis of FlfA surface exposure in G. anatis 12656-12. (A) FACS analysis demonstrating FlfA surface exposure on WT cells in the mid-logarithmic growth phase. Bacterial cells were incubated with preimmune serum (gray shaded) or anti-FlfA immune serum (red line). (B) The specificity of the signal in panel A was verified by incubation of WT cells (red line) or ΔflfA mutant cells (blue line) in anti-FlfA immune serum. (C) The surface exposure of FlfA was confirmed by using confocal immunofluorescence microscopy. G. anatis 12656-12 WT cells (panels a and b) or ΔflfA mutant cells (panel c) were incubated with anti-FlfA immune serum, followed by incubation with rhodamine RedX-labeled secondary antibody (red staining). DAPI (blue staining) was used to localize the bacterial cells. Staining of the WT cells from the same culture indicated the presence of a low-level (panel a) and a hyperfimbriated (panel b) population of cells. Magnification, ×63. Scale bar, 1 μm.
The spatial distribution of FlfA on the surface of G. anatis 12656-12 was also investigated by confocal immunofluorescence microscopy. Incubation of WT cells with anti-FlfA serum, followed by incubation with a red-fluorescent secondary antibody, demonstrated the presence of the FlfA protein on the cell surface (Fig. 2C, panels a and b). No reaction was observed between the anti-FlfA serum and the ΔflfA mutant (Fig. 2C, panel c). Interestingly, binding of FlfA antibodies to WT cells revealed a mixed population of cells comprising two FlfA phenotypes: a majority (95 to 98%) expressing a relatively low level of FlfA (Fig. 2C, panel a) and a few cells (2 to 5%) expressing a relatively high level of FlfA (Fig. 2C, panel b). The proposed hyperfimbriated group of cells was not identified in the FACS analysis. A likely explanation for this finding is that hyperfimbriated cells were excluded due to their large size and tendency to aggregate and form long chains. The same ratio of low-level and hyperfimbriated cells was also observed in in vitro cultures inoculated from a single colony or when samples were prepared directly from single colonies without culturing (data not shown). In general, the FlfA protein was found to extend from the surface of the cells and a fimbria-like staining pattern was observed especially with those cells showing hyperfimbriation. The preimmune serum did not show any fluorescence with either WT or ΔflfA cells, confirming the specificity of the binding (data not shown). As demonstrated by FACS, incubation of the WT or ΔflfA cells with antiserum against intact G. anatis 12656-12 bacteria confirmed the purity of the populations, since the observed staining was similar, specific, and homogenous (data not shown). Thus, confocal immunofluorescence analysis confirmed the surface localization of FlfA of G. anatis 12656-12, as demonstrated by FACS analysis, and the staining pattern is consistent with the prediction of FlfA as a fimbrial subunit protein.
FlfA is a fimbrial protein precursor in G. anatis 12656-12.
To confirm the prediction of FlfA as a fimbrial subunit protein, immunogold electron microscopy was used for a more precise and detailed localization of FlfA. This analysis clearly demonstrated the presence of FlfA in long fimbria-like structures on the cell surface (Fig. 3A and B). As observed using confocal immunofluorescence microscopy, the WT bacterial population could be divided into two separate subpopulations based on the fimbrial phenotype: a large population (95 to 98% of cells) exposing up to five long fimbriae per cell (Fig. 3A) and a small number of cells with a high number of fimbriae intermingled on the bacterial surface (Fig. 3B). Incubation of the ΔflfA mutant with anti-FlfA immune serum did not result in any immunogold labeling (Fig. 3C). In summary, the results clearly showed that G. anatis 12656-12 possesses 1- to 2-μm-long fimbrial structures on its surface, and the specific staining pattern corresponds to the prediction of FlfA as a fimbrial subunit protein in G. anatis 12656-12.
Fig 3.
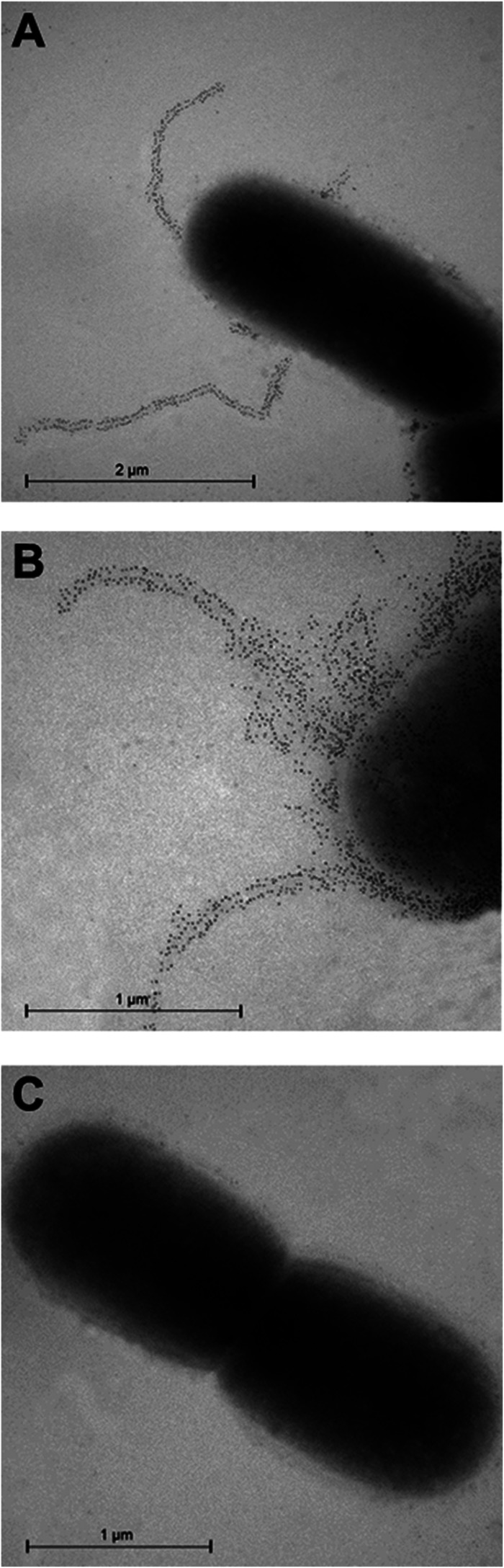
Immunogold electron microscopy with anti-FlfA immune serum. G. anatis 12656-12 WT cells (A and B) or ΔflfA mutant cells (C) were incubated with anti-FlfA immune serum and labeled with a secondary antibody conjugated to 10-nm gold particles. Two fimbrial cell populations were observed: a low-level population (A) and a hyperfimbriated population (B).
The F17-like fimbriae are important for the virulence of G. anatis 12656-12 in chickens, and immunization with recombinant FlfA induces protective immunity.
To investigate the role of the F17-like fimbriae during infection of the natural host, a G. anatis infection model in chickens was used, as described previously (15). Chickens were divided into two groups of 10 birds each and infected intraperitoneally with G. anatis WT (9.8 × 108CFU) or ΔflfA (1.04 × 109 CFU) strains. All birds were subjected to postmortem examination 24 or 48 h after inoculation. Bacterial reisolation and recording of gross lesions were also performed. In all infected birds, lesions of varying severity and dissemination were found at 24 h postinoculation. However, the lesion scores in the group infected with WT cells were significantly higher than those in the group infected with ΔflfA cells (P = 0.006, Fig. 4A). Furthermore, only two of the birds infected with the G. anatis WT were in lay, compared to six in the group infected with the ΔflfA mutant. There was no significant difference in the rate of reisolation of G. anatis between the two groups, although the reisolation rates in birds infected with ΔflfA mutant were numerically lower at all of the sites tested (Table 3, groups A and B). These results strongly indicate that the F17-like fimbriae composed of FlfA contributes to virulence during infection of the natural host.
Fig 4.
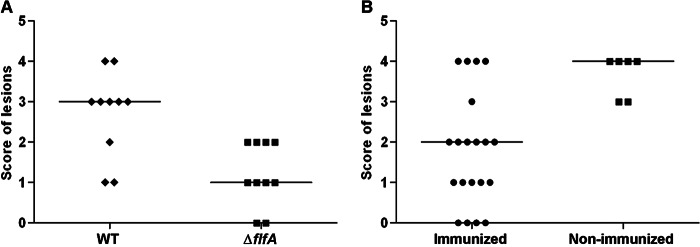
Challenge of chickens with G. anatis 12656-12 and immunization with recombinant FlfA. (A) Chickens were challenged intraperitoneally with G. anatis 12656-12 or ΔflfA mutant and subjected to postmortem examination at 24 h after inoculation. Gross lesions in the reproductive organs and in the peritoneum were evaluated using a scale from 0 to 4 based on the severity, distribution, and characteristics of the lesions. The median is given for each group (horizontal bars). A significant difference was found between the scores in the two groups (P = 0.006). (B) Chickens were immunized twice with 200 μg of recombinant FlfA (immunized) or sham immunized with placebo (nonimmunized), followed by challenge with G. anatis 12656-12. The scores are as described in panel A, showing a significant difference between the two groups (P = 0.008).
To investigate the protective potential of FlfA, recombinant FlfA was used to immunize a group of 20 chickens, followed by intraperitoneal challenge with the G. anatis 12656-12 (8 × 108 CFU). A significantly lower (P = 0.008) lesion score was found in the immunized chickens compared to the nonimmunized chickens (Fig. 4B). Again, G. anatis was reisolated from a lower number of immunized chickens compared to nonimmunized chickens (Table 3, groups C and D). No differences were found between 24 and 48 h postinfection (data not shown). Thus, FlfA has the potential of eliciting protective immunity against infections caused by G. anatis 12656-12.
FlfA homologs are expressed in vitro in other G. anatis strains.
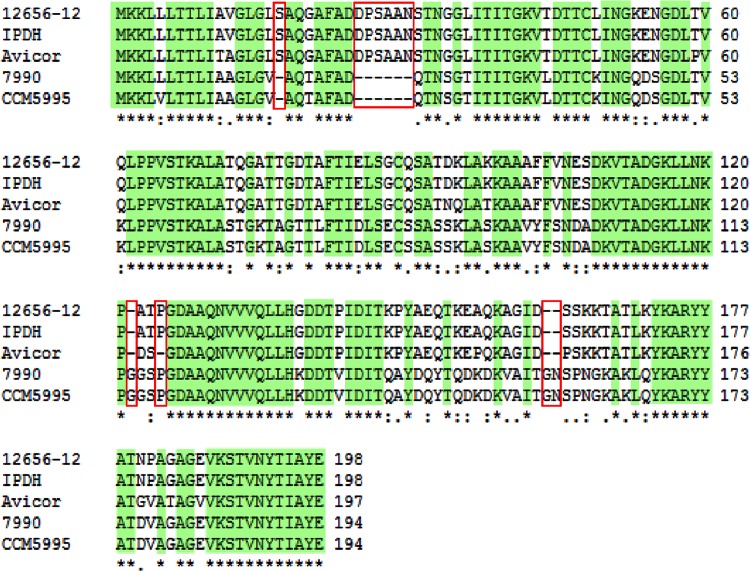
Since the substantial antigenic diversity described for G. anatis (13) is a major challenge in terms of disease prevention, the conservation of the F17-like fimbrial gene cluster was investigated across G. anatis. For this purpose, seven additional G. anatis strains were included in the study (Table 1). These strains were selected in order to provide as much variation as possible based on place and time of isolation and antigenic diversity. In general, between one and three F17-like gene clusters could be identified in each of the investigated strains (Table 4), thus supporting the notion that the F17-like fimbriae is common in G. anatis. A flfA fimbrial gene cluster was identified in six of the strains; however, the flfA gene homolog in G. anatis F149 lacks the first 17 bp including the start codon, suggesting that this strain is incapable of expressing a FlfA protein homolog. The four intact FlfA homologs were found to be 78 to 100% similar to FlfA from G. anatis 12656-12 (Table 4). Moreover, sequence alignment demonstrated several stretches of fully conserved amino acid sequences (Fig. 6).
Table 4.
Distribution of F17-like gene clusters in G. anatis
| G. anatis strain | Gene cluster identificationa |
||
|---|---|---|---|
| FlfA (% similarity)b | UMN179_295 | UMN179_750 | |
| 12656-12 | + (100) | + | + |
| F149T | +c | + | + |
| 10672/6 | – | – | + |
| 4895 | – | – | + |
| 7990 | + (78) | + | + |
| Avicor | + (93) | + | – |
| CCM5995 | + (78) | – | – |
| IPDH 697-78 | + (100) | + | + |
+, Identification of a gene cluster, –, no gene cluster identified. Data for UMN179_295 and UMN179_750 are from Johnson et al. (23).
The percent similarity (indicated in parentheses) is based on the number of identical or conserved residues.
The gene lacks the first 17 bp, and no protein product is expected to be expressed.
Fig 6.
Sequence alignment of FlfA and FlfA homologs. Sequence alignment of FlfA from G. anatis 12656-12 (JX855927) and FlfA homologs from G. anatis IPDH 697-78 (JX915818), Avicor (JX915816), 7990 (JX915815), and CCM5995 (JX915817) was conducted using CLUSTAL X 2.1 (38). Positions with identical residues (*) are highlighted in green. Highly similar and similar residues are indicated by “:” and “.”, respectively. Deletions observed in one or more of the sequences are framed in red.
To investigate whether the identified FlfA-like fimbrial gene clusters of G. anatis were expressed in the eight diverse strains, whole-cell extracts were prepared from cells in late-logarithmic growth phase. Western blotting with anti-FlfA immune serum demonstrated recognition of a protein band corresponding to FlfA in five of the eight strains, including G. anatis 12656-12, while no expression was observed in G. anatis F149, 4895, and 10672/6 (Fig. 5). The FlfA homolog expressed from G. anatis 7990 appeared smaller (∼2 kDa) in comparison to the proteins expressed from the other strains. Since this difference in migration cannot be explained by the amino acid compositions, which are highly conserved (Fig. 6), or by the calculated masses of the proteins, ranging between 20.1 and 20.5 kDa, the result suggests differences at the posttranslational level. The FlfA protein was not recognized in Western blots using anti-FlfA preimmune serum, confirming the specificity of the binding (data not shown). In conclusion, the majority of the investigated G. anatis strains express a protein antigenically similar to FlfA, suggesting that recombinant FlfA has the potential to elicit cross-protective immunity against heterologous G. anatis strains.
Fig 5.
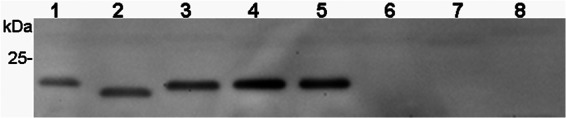
Expression of FlfA in G. anatis. Western blotting demonstrated recognition of a protein band corresponding to FlfA by anti-FlfA immune serum in whole-cell extracts prepared from five of the eight investigated G. anatis strains, including G. anatis 12656-12 (lane 1). The positions of molecular mass standards (in kDa) are shown on the left. Whole-cell extract equivalent to OD260 between 4 and 8 in 10 μl was loaded to each lane. Lane 1, G. anatis 12656-12; lane 2, G. anatis 7990; lane 3, G. anatis Avicor; lane 4, G. anatis CCM5995; lane 5, G. anatis IPDH 697-78; lane 6, G. anatis F149; lane 7, G. anatis 10672/6; lane 8, G. anatis 4895.
DISCUSSION
G. anatis is a major cause of salpingitis and peritonitis in egg-laying chickens, leading to decreased egg production and increased mortality worldwide. It has previously been suggested that different means of attachment and the ability to form biofilms are important for G. anatis to overcome the propulsive effect of peristalsis and allow colonization of the upper reproductive tract (19), and yet specific factors and their contributions to these processes have remained unknown. Here we have characterized an F17-like fimbrial subunit protein from G. anatis and demonstrated that it is expressed and surface exposed in vitro. The FlfA protein shows 59% similarity to the F17 fimbrial subunit protein described in the ExPEC strain 536 (24), where the fimbriae are encoded by a 5-kb operon composed of four genes (27, 43). Similarly, in G. anatis 12656-12 (23), flfA is located within a 5,070-bp gene cluster upstream of the three genes flfD (GenBank accession no. JX855928), flfC (GenBank accession no. JX855929), and flfG (GenBank accession no. JX855930), encoding putative F17-like chaperone, usher, and adhesion proteins, respectively.
By use of confocal immunofluorescence analysis and immunogold electron microscopy, we found that FlfA is surface exposed and that the bacterial population could be divided into two phenotypically distinct groups based on surface localization of F17-like fimbriae: a dominant group expressing only 1 to 5 fimbriae, and a minor, apparently hyperfimbriated, group. Phase variation is a well-described heterogeneity-generating process involving regulatory alterations within the genome, which has been shown to affect fimbrial expression in many bacterial species (44). However, we were not able to identify any sequence motifs or genetic elements, such as 2-bp repeats, 4-bp repeats, or methylation sites, which could support regulation by phase variation of the FlfA fimbriae in G. anatis 12656-12. Another recently defined phenomenon is bistability, which is used to describe the occurrence of a heterogeneous population of genetically identical bacteria, propagated in a homogenous environment (45). In the case of bistability, the switch is epigenetic, and bistability is considered an optimal bacterial strategy for coping with infrequent changes in the environment. A bistable fimbriation pattern has previously been described for the S-fimbriae in the ExPEC strain 536 (46), in which the in vitro occurrence of hyperfimbriated to normally fimbriated bacteria in a 1:100 ratio was described despite the lack of apparent genomic alterations. Thus, the F17-like fimbriae in G. anatis 12656-12 could be subject to bistability, which may allow the population to respond to sudden changes on the mucosal lining. However, the exact mechanisms for regulation of FlfA expression remain unknown.
Although the specific role of FlfA and the F17-like fimbriae during infection still remains to be elucidated, our results clearly demonstrate a decreased ability of the ΔflfA mutant to cause lesions in vivo. Moreover, immunization of chickens with recombinant FlfA protein resulted in a significantly lower lesion score, further supporting the role of the F17-like fimbriae as an important virulence factor in the pathogenesis of G. anatis. We propose that the F17-like fimbriae could play a role in the previously observed tissue tropism of G. anatis, by providing G. anatis with the adhesive ability that allows the bacteria to ascend from the cloaca to the upper reproductive tract and beyond, as observed frequently in egg-laying chickens (10).
In the present study, the vaccine potential of recombinant FlfA against G. anatis infection was examined. However, to overcome the substantial antigenic diversity within the G. anatis population, it is important that recombinant FlfA demonstrates a serotype-independent protective potential. Along this line, we demonstrated the presence of homologs of the flfA gene in four G. anatis strains besides G. anatis 12656-12, all encoding proteins with similarity to FlfA. However, the flfA gene of G. anatis F149 lacks the first 17 bp, including the start codon, which was reflected in the lack of expression in vitro. Thus, recombinant FlfA does show some potential at eliciting cross-protective immunity against G. anatis. As mentioned previously, the majority of the G. anatis strains possess more than one F17-like fimbrial gene cluster. The occurrence of a varying copy number of the F17 fimbrial gene cluster has previously been demonstrated in E. coli, in which the gene clusters were suggested to have arisen from duplications of the entire operon and to be either silent copies or copies encoding different types of fimbriae expressed at different time points depending on the host and type of tissue colonized (43, 47). In G. anatis 12656-12, only one type of FlfA-like fimbriae could be identified during in vitro growth in the present study, suggesting that (i) only the FlfA-like fimbrial gene cluster is expressed in G. anatis 12656-12 or (ii) the different putative F17-like fimbrial gene clusters encode fimbriae composed of subunit proteins with different antigenic specificities. The latter hypothesis is likely, since sequence alignments of the three identified subunit proteins in G. anatis 12656-12 showed that the variability is localized in the N-terminal parts of the proteins, while the C-terminal fimbrial domain, previously shown to be required for the general assembly of the fimbriae (48), is conserved in all of the subunit proteins. Thus, G. anatis might be capable of expressing antigenically different F17-like fimbria. If so, an even higher degree of protective immunity might be reached if two or three subunit proteins, adhesion proteins, and/or isolated fimbriae were combined as a fusion protein. This strategy has been used successfully in several recent studies, demonstrating the potential of adhesin-fimbria (49) or adhesin-adhesin-toxoid (50) fusion proteins as antigens in a vaccine against porcine ETEC diarrhea.
In conclusion, we have demonstrated (i) that G. anatis 12656-12 expresses surface-exposed F17-like fimbriae, composed of the FlfA protein, possibly in a bistable manner, (ii) that the F17-like fimbriae play a prominent role in the pathogenesis of G. anatis in chickens, (iii) that recombinant FlfA can elicit protective immunity against infections caused by G. anatis 12656-12 in chickens, and (iv) that FlfA is expressed in other G. anatis strains, suggesting a cross-protective potential. Taken together, these results indicate that F17-like fimbriae are a conserved and important virulence factor in G. anatis and the fimbrial subunit protein FlfA is likely to possess broad vaccine potential in chickens.
ACKNOWLEDGMENTS
This study was supported by the Danish Research Council for Technology and Production (grant 09-065909), a Ph.D. stipend from the Faculty of Life Sciences of Copenhagen University, and Novartis Vaccines, Siena, Italy.
We thank Massimiliano Biagini and Nathalie Norais from Novartis Vaccines for their help with the mass spectrometry analysis and Anna Rita Taddei from the Università degli Studi della Tuscia (Viterbo, Italy) for assistance with the electron microscopy.
Footnotes
Published ahead of print 18 March 2013
REFERENCES
- 1. AVEC 2011. Annual report 2011. Assoc. Poultry Processors Poultry Trade EU Countries 41:1–52 [Google Scholar]
- 2. Christensen H, Bisgaard M, Bojesen AM, Mutters R, Olsen JE. 2003. Genetic relationships among avian isolates classified as Pasteurella haemolytica, “Actinobacillus salpingitidis” or Pasteurella anatis with proposal of Gallibacterium anatis gen. nov., comb. nov. and description of additional genomospecies within Gallibacterium. Int. J. Syst. Evol. Microbiol. 53:275–287 [DOI] [PubMed] [Google Scholar]
- 3. Bisgaard M, Korczak BM, Busse Kuhnert H-JP, Bojesen AM, Christensen H. 2009. Classification of the taxon 2 and taxon 3 complex of Bisgaard within Gallibacterium and description of Gallibacterium melopsittaci sp. nov., Gallibacterium trehalosifermentans sp. nov. and Gallibacterium salpingitidis sp. nov. Int. J. Syst. Evol. Microbiol. 59:735–744 [DOI] [PubMed] [Google Scholar]
- 4. Mushin R, Weisman Y, Singer N. 1980. Pasteurella haemolytica found in the respiratory tract of fowl. Avian Dis. 24:162–168 [Google Scholar]
- 5. Bisgaard M. 1977. Incidence of Pasteurella haemolytica in the respiratory tract of apparently healthy chickens and chickens with infectious bronchitis: characterization of 213 strains. Avian Pathol. 6:285–292 [DOI] [PubMed] [Google Scholar]
- 6. Bojesen AM, Nielsen SS, Bisgaard M. 2003. Prevalence and transmission of haemolytic Gallibacterium species in chicken production systems with different biosecurity levels. Avian Pathol. 32:503–510 [DOI] [PubMed] [Google Scholar]
- 7. Gerlach H. 1977. Die Bedeutung von Pasteurella haemolytica in Hühnerbeständen. Prakt. Tierartz. 5:324–328 [Google Scholar]
- 8. Mirle C, Schöngarth M, Meinhart H, Olm U. 1991. Untersuchungen zu Auftreten and Bedeutung von Pasteurella haemolytica Infektionen bei Hennen unter Besonderer Berücksichtigung von Erkrankungen des Legeapparates. Mh. Vet. Med. 46:545–549 [Google Scholar]
- 9. Jordan FTW, Williams NJ, Wattret A, Jones T. 2005. Observations on salpingitis, peritonitis, and salpingoperitonitis in a layer breeder flock. Vet. Rec. 157:573–577 [DOI] [PubMed] [Google Scholar]
- 10. Neubauer C, De Souza-Pilz M, Bojesen AM, Bisgaard M, Hess M. 2009. Tissue distribution of haemolytic Gallibacterium anatis isolates in laying birds with reproductive disorders. Avian Pathol. 38:1–7 [DOI] [PubMed] [Google Scholar]
- 11. Bojesen AM, Christensen JP, Bisgaard M. 2008. Gallibacterium infections and other avian Pasteurellaceae, p 160–163 In Pattison M, McMullin PF, Bradbury JM, Alexander DJ. (ed), Poultry diseases, 6th ed Saunders/Elsevier, Edinburgh, United Kingdom [Google Scholar]
- 12. Bojesen AM, Vazquez ME, Bager RJ, Ifrah D, Gonzalez C, Aarestrup FM. 2011. Antimicrobial susceptibility and tetracycline resistance determinant genotyping of Gallibacterium anatis. Vet. Microbiol. 148:105–110 [DOI] [PubMed] [Google Scholar]
- 13. Vazquez ME, Gonzalez C, De la Mora R, Bojesen AM. 2006. Prevalence of Gallibacterium anatis in Mexico and their effect in laying hens, poster 25-29/6–2006. 4th International Veterinary and Vaccines Diagnostics Conference, Oslo, Norway [Google Scholar]
- 14. Kristensen BM, Frees D, Bojesen AM. 2010. GtxA from Gallibacterium anatis, a cytolytic RTX-toxin with a novel domain organisation. Vet. Res. 41:25 doi:10.1051/vetres/2009073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bojesen AM, Nielsen OL, Christensen JP, Bisgaard M. 2004. In vivo studies of Gallibacterium anatis infection in chickens. Avian Pathol. 33:145–152 [DOI] [PubMed] [Google Scholar]
- 16. García-Gómez E, Vaca S, Pérez-Méndez A, Ibarra-Caballero J, Pérez-Márquez V, Tenorio VR, Negrete-Abascal E. 2005. Gallibacterium anatis-secreted metalloproteases degrade chicken IgG. Avian Pathol. 34:426–429 [DOI] [PubMed] [Google Scholar]
- 17. Zepeda A, Ramirez S, Vega V, Morales V, Talavera M, Salgado-Miranda C, Simón-Martínez J, Bojesen A, Soriano-Vargas ME. 2009. Hemagglutinating activity of Gallibacterium strains. Avian Dis. 53:115–118 [DOI] [PubMed] [Google Scholar]
- 18. Klemm P, Vejborg RM, Hancock V. 2010. Prevention of bacterial adhesion. Appl. Microbiol. Biotechnol. 88:451–459 [DOI] [PubMed] [Google Scholar]
- 19. Vaca S, Monroy E, Rojas L, Vazquez C, Sanchez P, Soriano-Va E, Bojesen AM, Abascal EN. 2011. Adherence of Gallibacterium anatis to inert surfaces. J. Anim. Vet. Adv. 10:1688–1693 [Google Scholar]
- 20. Ramirez-Apolinar S, Guerra-Infante FM, Haro-Cruz M, Salgado-Miranda C, Madrid-Morales E, Kristensen BM, Bojesen AM, Negrete-Abascal E, Soriano-Vargas E. 2012. Characterization of a Gallibacterium genomospecies 2 hemagglutinin. J. Anim. Vet. Adv. 11:556–560 [Google Scholar]
- 21. Jacques M, Paradis SE. 1998. Adhesin-receptor interactions in Pasteurellaceae. FEMS Microbiol. Rev. 22:45–59 [DOI] [PubMed] [Google Scholar]
- 22. Klemm P, Schembri MA. 2000. Bacterial adhesins: function and structure. Int. J. Med. Microbiol. 290:27–35 [DOI] [PubMed] [Google Scholar]
- 23. Johnson TJ, Danzeisen JL, Trampel D, Nolan L, Seemann T, Bager R, Bojesen AM. 2013. Genome analysis and phylogenetic relatedness of Gallibacterium anatis strains from poultry. PLoS One 8:e54844 doi:10.1371/journal.pone.0054844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dobrindt U, Blum-Oehler G, Nagy G, Schneider G, Johann A, Gottschalk G, Hacker J. 2002. Genetic structure and distribution of four pathogenicity islands (PAI I(536) to PAI IV(536)) of uropathogenic Escherichia coli strain 536. Infect. Immun. 70:6365–6372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lintermans PF, Pohl P, Bertels A, Charlier G, Vandekerckhove J, Van Damme J, Schoup J, Schlicker C, Korhonen T, De Greve H. 1988. Characterization and purification of the F17 adhesin on the surface of bovine enteropathogenic and septicemic Escherichia coli. Am. J. Vet. Res. 49:1794–1799 [PubMed] [Google Scholar]
- 26. Ghanbarpour R, Sami M, Salehi M, Ouromiei M. 2011. Phylogenetic background and virulence genes of Escherichia coli isolates from colisepticemic and healthy broiler chickens in Iran. Trop. Anim. Health Prod. 43:153–157 [DOI] [PubMed] [Google Scholar]
- 27. Stordeur P, Marlier D, Blanco J, Oswald E, Biet F, Dho-Moulin M, Mainil J. 2002. Examination of Escherichia coli from poultry for selected adhesin genes important in disease caused by mammalian pathogenic E. coli. Vet. Microbiol. 84:231–241 [DOI] [PubMed] [Google Scholar]
- 28. Nguyen TD, Vo TT, Vu-Khac H. 2011. Virulence factors in Escherichia coli isolated from calves with diarrhea in Vietnam. J. Vet. Sci. 12:159–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Martin C, Rousset E, De Greve H. 1997. Human uropathogenic and bovine septicemic Escherichia coli strains carry an identical F17-related adhesin. Res. Microbiol. 148:55–64 [DOI] [PubMed] [Google Scholar]
- 30. Samadder P, Xicohtencatl-Cortes J, Saldaña Z, Jordan D, Tarr PI, Kaper JB, Girón Ja. 2009. The Escherichia coli ycbQRST operon encodes fimbriae with laminin-binding and epithelial cell adherence properties in Shiga-toxigenic Escherichia coli O157:H7. Environ. Microbiol. 11:1815–1826 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31. Le Bouguénec C, Bertin Y. 1999. AFA and F17 adhesins produced by pathogenic Escherichia coli strains in domestic animals. Vet. Res. 30:317–342 [PubMed] [Google Scholar]
- 32. Nagy B, Fekete PZ. 1999. Enterotoxigenic Escherichia coli (ETEC) in farm animals. Vet. Res. 30:259–284 [PubMed] [Google Scholar]
- 33. Lintermans PF, Bertels A, Schlicker C, Deboeck F, Charlier G, Pohl P, Norgren M, Normark S, Van Montagu M, De Greve H. 1991. Identification, characterization, and nucleotide sequence of the F17-G gene, which determines receptor binding of Escherichia coli F17 fimbriae. J. Bacteriol. 173:3366–3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thanassi DG, Saulino ET, Hultgren SJ. 1998. The chaperone/usher pathway: a major terminal branch of the general secretory pathway. Curr. Opin. Microbiol. 1:223–231 [DOI] [PubMed] [Google Scholar]
- 35. Kristensen BM, Frees D, Bojesen AM. 2011. Expression and secretion of the RTX-toxin GtxA among members of the genus Gallibacterium. Vet. Microbiol. 153:116–123 [DOI] [PubMed] [Google Scholar]
- 36. Bulach DM, Zuerner RL, Wilson P, Seemann T, McGrath A, Cullen PA, Davis J, Johnson M, Kuczek E, Alt DP, Peterson-Burch B, Coppel RL, Rood JI, Davies JK, Adler B. 2006. Genome reduction in Leptospira borgpetersenii reflects limited transmission potential. Proc. Natl. Acad. Sci. U. S. A. 103:14560–14565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Seeman T. 2006. Providing essential support for Victorian science. Bioinform. Asia Pacific 10:1400 [Google Scholar]
- 38. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 39. Larkin M a., Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. CLUSTAL W and CLUSTAL X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 40. Al-Hasani K, Boyce J, McCarl VP, Bottomley S, Wilkie I, Adler B. 2007. Identification of novel immunogens in Pasteurella multocida. Microb. Cell Fact. 6:3 doi:10.1186/1475-2859-6-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hartley JL, Temple GF, Brasch MA. 2000. DNA cloning using in vitro site-specific recombination. Genome Res. 10:1788–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Berlanda Scorza F, Doro F, Rodríguez-Ortega MJ, Stella M, Liberatori S, Taddei AR, Serino L, Gomes Moriel D, Nesta B, Fontana MR, Spagnuolo A, Pizza M, Norais N, Grandi G. 2008. Proteomics characterization of outer membrane vesicles from the extraintestinal pathogenic Escherichia coli ΔtolR IHE3034 mutant. Mol. Cell. Proteomics 7:473–485 [DOI] [PubMed] [Google Scholar]
- 43. Bertin Y, Martin C, Oswald E, Girardeau JP. 1996. Rapid and specific detection of F17-related pilin and adhesin genes in diarrheic and septicemic Escherichia coli strains by multiplex PCR. J. Clin. Microbiol. 34:2921–2928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Van der Woude MW, Bäumler AJ. 2004. Phase and antigenic variation in bacteria. Clin. Microbiol. Rev. 17:581–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dubnau D, Losick R. 2006. Bistability in bacteria. Mol. Microbiol. 61:564–572 [DOI] [PubMed] [Google Scholar]
- 46. Ott M, Hacker J. 1991. Analysis of the variability of S-fimbriae expression in an Escherichia coli pathogen. FEMS Microbiol. Lett. 63:233–238 [DOI] [PubMed] [Google Scholar]
- 47. Stordeur P, Brée A, Mainil J, Moulin-Schouleur M. 2004. Pathogenicity of pap-negative avian Escherichia coli isolated from septicemic lesions. Microbes Infect. 6:637–645 [DOI] [PubMed] [Google Scholar]
- 48. Choudhury D, Thompson A, Stojanoff V, Langermann S, Pinkner J, Hultgren SJ, Knight SD. 1999. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science 285:1061–1066 [DOI] [PubMed] [Google Scholar]
- 49. Tiels P, Verdonck F, Coddens A, Goddeeris B, Cox E. 2008. The excretion of F18+ Escherichia coli is reduced after oral immunization of pigs with a FedF and F4 fimbriae conjugate. Vaccine 26:2154–2163 [DOI] [PubMed] [Google Scholar]
- 50. Ruan X, Liu M, Casey TA, Zhang W. 2011. A tripartite fusion, FaeG-FedF-LT(192)A2:B, of enterotoxigenic Escherichia coli (ETEC) elicits antibodies that neutralize cholera toxin, inhibit adherence of K88 (F4) and F18 fimbriae, and protect pigs against K88ac/heat-labile toxin infection. Clin. Vaccine Immunol. 18:1593–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bojesen AM, Torpdahl M, Christensen H, Olsen JE, Bisgaard M. 2003. Genetic diversity of Gallibacterium anatis isolates from different chicken flocks. J. Clin. Microbiol. 41:2737–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kristensen BM, Sinha S, Boyce JD, Bojesen AM, Mell JC, Redfield RJ. 2012. Natural transformation of Gallibacterium anatis. Appl. Environ. Microbiol. 78:4914–4922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Taylor LA, Rose RE. 1988. A correction in the nucleotide sequence of the Tn903 kanamycin resistance determinant in pUC4K. Nucleic Acids Res. 16:358. [DOI] [PMC free article] [PubMed] [Google Scholar]