Abstract
The resurgence of invasive disease caused by Streptococcus pyogenes (group A Streptococcus [GAS]) in the past 30 years has paralleled the emergence and global dissemination of the highly virulent M1T1 clone. The GAS M1T1 clone has diverged from the ancestral M1 serotype by horizontal acquisition of two unique bacteriophages, encoding the potent DNase Sda1/SdaD2 and the superantigen SpeA, respectively. The phage-encoded DNase promotes escape from neutrophil extracellular traps and is linked to enhanced virulence of the M1T1 clone. In this study, we successfully used in vitro lysogenic conversion to transfer the Sda1-encoding phage from the M1T1 clonal strain 5448 to the nonclonal M1 isolate SF370 and determined the impact of this horizontal gene transfer event on virulence. Although Sda1 was expressed in SF370 lysogens, no capacity of the phage-converted strain to survive human neutrophil killing, switch to a hyperinvasive covRS mutant form, or cause invasive lethal infection in a humanized plasminogen mouse model was observed. This work suggests that the hypervirulence of the M1T1 clone is due to the unique synergic effect of the M1T1 clone bacteriophage-specific virulence factor Sda1 acting in concert with the M1T1 clone-specific genetic scaffold.
INTRODUCTION
Streptococcus pyogenes (group A Streptococcus [GAS]) is a strictly human pathogen, able to colonize the skin and throat asymptomatically or to trigger mild, localized superficial infections, such as impetigo and pharyngitis. Less frequently, GAS invades normally sterile body sites to cause systemic, severe, and often life-threatening pathologies, including necrotizing fasciitis and toxic shock syndrome. The virulence mechanisms that confer this invasive capacity on GAS, allowing evasion of the host immune system and penetration of deeper tissues, are complex and only partially elucidated (1, 2, 3). Although disease incidence decreased markedly in the first part of the 20th century, the past 3 decades have witnessed a resurgence in invasive GAS pathologies (4–7). A nonrandom association between invasive disease and a number of GAS serotypes (M1, M3, M18, M28) has been reported, linked particularly to the emergence of highly virulent subtypes, such as the globally disseminated M1T1 clone (3, 7, 8), defined here as GAS M1T1 strains containing the bacteriophage-encoded virulence factors Sda1 and SpeA (5, 9, 10).
The diversification of GAS serotypes has been attributed to molecular and genetic variations often related to the acquisition of phage-associated virulence factors (7, 9, 11–13). Comparative genomic analysis of the M1T1 clonal isolate MGAS5005 (14) and the nonclonal M1 isolate SF370 (15) identified the main differences between these two GAS types as the carriage of different prophages containing specific toxins (SpeC, MF2, and SpeI/SpeH within SF370; SpeA and Sda1 within MGAS5005) and the presence in MGAS5005 of an M12-derived 36-kb genomic recombination region that is absent in SF370 (6, 14, 16). The evolutionary steps that have led to the emergence of the hypervirulent M1T1 clone have been elucidated recently and confirm the major role played by horizontal gene transfer mechanisms, which include unique prophage acquisition events, in the epidemiology of GAS (17).
Much research has focused on determining the impact of individual virulence determinants on the M1T1 clone virulence phenotype through “loss-of-function” gene knockout studies. Sda1, encoded on an M1T1 prophage, is a potent streptococcal DNase (18). Targeted mutagenesis of the sda1 gene sensitizes GAS to killing within DNA-based neutrophil extracellular traps (NETs) (19). For GAS M1T1, the onset of invasive disease is associated with an in vivo switch to a SpeB-negative phenotype, where the expression of the broad-spectrum cysteine protease SpeB is abolished as a consequence of point mutations in the two-gene regulatory sensor kinase operon covRS (2, 20). The transcriptional shift that results in the loss of SpeB and increased expression of Sda1 and other streptococcal virulence factors triggers systemic infection (1). In turn, it has been shown by gene knockout studies that Sda1 expression provides selective pressure for the switch to the hypervirulent covRS mutant M1T1 genotype (1). Other gene products that also contribute to this selective pressure include the virulence determinants M1 protein and the hyaluronic acid capsule (21).
In this study, we investigated the impact of Sda1-encoding phage carriage on virulence and invasive disease propensity by a “gain-of-function” approach, via lysogenic conversion of the M1 GAS strain SF370 with the M1T1 Sda1-encoding phage. We demonstrate that the enhanced virulence of the M1T1 clone results from the unique combination of the bacteriophage-specific virulence factor Sda1 with the M1T1 clone-specific genetic backbone.
MATERIALS AND METHODS
Comparative bioinformatic analysis of M1 SF370 and M1T1 5448 GAS sequences.
Whole-genome sequencing of the M1T1 strain 5448, a strain representative of the globally disseminated M1T1 clone encoding the DNase Sda1 (16), was carried out using the Illumina (San Diego, CA) HiSeq 2000 system as described previously (17) and yielded a total of 8,879,651 paired-end 100-bp reads, corresponding to an estimated coverage of 987×. De novo assembly was performed using Velvet, version 1.1.05 (22), to produce a draft genome sequence composed of 36 scaffolds for a total of 1,800,853 assembled bases. To optimize the assembly process (which tends to counterperform with very high read coverage) and achieve optimal performance, the data set was sampled down to 1 million read pairs for an estimated 100× coverage assembly. The genome of the M1T1 strain MGAS5005 (RefSeq accession no. NC_007297) (14) was then used as a reference for reordering the 36 scaffolds of strain 5448, using ABACAS (23), and for transferring annotation, using RATT (24). To investigate variations in its genetic structure, the draft genome sequence of the M1T1 strain 5448 was compared with the completed genome sequences of MGAS5005 and the GAS M1 strain SF370 (RefSeq accession no. NC_002737) (15) by using Easyfig (25). Reads were deposited in the Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra). The SRA sample accession number is ERS123209.
GAS strains and culture.
The GAS isolates used in this study are SF370-Smr, a spontaneous streptomycin-resistant (Smr) mutant of the sequenced M1 isolate SF370 (15), obtained by subculturing on selective Todd-Hewitt (Difco Laboratories) agar supplemented with 1% (wt/vol) yeast extract (THY agar) and 150 μg/ml streptomycin; GAS strain 5448 (see above); and 5448Δsda-Cmr, an isogenic mutant of 5448 in which the sda1 gene has been replaced with a chloramphenicol resistance (Cmr) marker (19). Bacterial strains were routinely cultured on solid commercial horse blood agar (bioMérieux, Australia) or in Todd-Hewitt broth (Difco Laboratories) supplemented with 1% (wt/vol) yeast extract (THY broth) at 37°C without shaking.
In vitro lysogenic conversion of GAS SF370-Smr.
GAS 5448Δsda-Cmr was used as the phage donor strain, and SF370-Smr was used as the phage recipient strain. Briefly, overnight bacterial cultures were diluted 1:20 in prewarmed THY broth and were grown statically to an A600 of 0.25. The 5448Δsda-Cmr culture was treated with mitomycin C (0.2 mg/ml) and was grown statically for 3 h at 37°C. In order to determine successful phage induction, a 5-ml aliquot of this induced culture was retained for the detection of phage DNA in the culture supernatant. Phage DNA was purified as described previously (26). Briefly, a filter-sterilized supernatant was used for phage DNA isolation via Benzonase treatment (Novagen, USA) to eliminate genomic DNA contamination, followed by phage extraction using phenol-chloroform-isoamyl alcohol (25:24:1) (Sigma-Aldrich) and ethanol precipitation of DNA. Phage induction was verified by PCR amplification of specific M1T1 phage-encoded toxins (see Table S1 in the supplemental material). Lysogeny was performed by the addition of 100 μl of induced 5448Δsda-Cmr culture to 900 μl of uninduced SF370-Smr recipient culture, followed by static incubation at 37°C for a further 3 h. This mixed culture was used to inoculate selective THY broth (Cm, 2 μg/ml; Sm, 150 μg/ml) at a 1:10 dilution, and the mixture was then incubated statically at 37°C for 48 h for the selection and amplification of lysogenized bacteria. Culture aliquots were plated onto selective THY agar (Cm, 2 μg/ml; Sm, 150 μg/ml) and were incubated overnight at 37°C. Single colonies grown with double selection were tested by PCR for the presence of M1T1-specific and M1-specific gene sequences. A single Smr Cmr colony was selected and was designated SF370-Smr(ϕ5448.3 Cmr).
Allelic replacement to reconstruct the Sda1-encoding phage in the SF370 background.
Allelic exchange was used to replace the chloramphenicol resistance gene in SF370-Smr(ϕ5448.3 Cmr) with the wild-type sda1 gene from 5448. The methodology followed was that described previously (1). Briefly, the temperature-sensitive pHY304-sda vector (pSda) carrying the 5448 sda1 gene and an erythromycin resistance marker was employed. For selection of the plasmid, erythromycin was used at a concentration of 2 μg/ml in GAS and 500 μg/ml in Escherichia coli, and bacteria were cultured at 30°C. Plasmid pSda was introduced into SF370-Smr(ϕ5448.3 Cmr) by electroporation (27), and chromosomal integration was achieved by double crossover after the removal of selective antibiotics as described previously (28), resulting in GAS strain SF370-Smr(ϕ5448.3 Sda+).
Lysogen characterization.
PCR amplification and Sanger DNA sequencing were used to confirm the identities of the SF370-Smr lysogens and to determine phage and sda1 gene insertion sites in order to exclude the occurrence of deletion events or other gene alterations during lysogeny. Genomic DNA was isolated from overnight liquid cultures by using the DNeasy blood and tissue kit (Qiagen, CA, USA) according to the manufacturer's instructions. The primers and cycling conditions used for PCR screening are listed in Table S1 in the supplemental material. The hyaluronic acid capsule content in GAS cultures at an A600 of 0.4 was determined by the Stains-All method as described previously (29). Both SpeB in stationary-phase (overnight) supernatants and SpeC and Sda1 in mid-logarithmic-phase (A600, 0.4) supernatants were detected by Western blotting using standard protocols (30).
DNase activity and neutrophil resistance.
DNase activity was assayed essentially as described previously (18). Briefly, calf thymus DNA (Sigma) was incubated at 37°C with 2.5 μl of GAS supernatant (A600, 0.4) in activation buffer (1 mM CaCl2, 1 mM MgCl2, and 100 mM Tris buffer [pH 7.6]). A sample consisting of THY broth only was included in the experiment as a negative control. After 5 min, EDTA was added to a final concentration of 66 mM to stop the reaction. The reaction products were then loaded onto a 1.0% (wt/vol) agarose gel and were stained with ethidium bromide for DNA visualization. GAS survival following incubation with human neutrophils in vitro was assayed as described previously (1, 31). Experiments were performed in biological triplicate using mid-logarithmic-phase (A600, 0.4) GAS at a multiplicity of infection (MOI) of 10:1 (GAS to neutrophils).
Visualization and quantification of NETs.
NETs were visualized and quantified as described previously (21).
In vivo SpeB-switching assays and virulence.
To determine the rate of switching to a SpeB-negative phenotype in vivo, C57BL/J6 mice were challenged subcutaneously with Gas at 1 × 107 CFU/dose, and SpeB-switching assays were undertaken as described previously (1, 31). A modified azocaseinolytic assay (32) and culturing on Columbia skim milk agar (33) were used to assess the expression of SpeB cysteine protease. To determine the virulence potential of SF370-Smr lysogens, the humanized plasminogen transgenic AlbPLG1 mouse model (34) was used for subcutaneous GAS infection as described previously (1, 17, 28, 30, 31).
RESULTS
Comparative genomic analysis of GAS strains 5448, MGAS5005, and SF370.
A de novo-assembled draft genome of M1T1 strain 5448 (17) was used here for comparative analysis with the completed genome sequences of the M1T1 strain MGAS5005 (14) and the M1 strain SF370 (15) to confirm and visualize the nature and position of genetic differences (Fig. 1). M1T1 strains MGAS5005 and 5488 are virtually identical across their entire lengths, with no detectable sequence variation in the DNase Sda1 prophages that promote GAS virulence and invasive disease capacity (ϕ5005.3 and ϕ5448.3, respectively) (1). Most of the observed disruptions of synteny between MGAS5005 and strain 5488 were caused by gaps in the draft assembly due to repeat regions, such as insertion sequences and ribosomal operons (Fig. 1). Alignment of the three genomes confirms that the main differences between SF370 and M1T1 GAS are attributable to the presence of distinct exogenous genetic elements. These comparative analyses highlight the role of bacteriophages in GAS genetic diversity as reported previously (9, 14).
Fig 1.

Comparison of the GAS SF370 and M1T1 genomes. The linear diagrams show pairwise comparisons between the draft genome sequence of GAS M1T1 strain 5448 (top) and the complete genome sequences of the GAS M1T1 reference strain MGAS5005 (RefSeq accession no. NC_007297) and GAS M1 strain SF370 (RefSeq accession no. NC_002737). In each genome, prophages are shown as red (M1T1) or green (M1) boxes. Insertion sequences and ribosomal operons in MGAS5005 are shown as orange triangles and black boxes, respectively. The M12-derived 36-kb recombination region is indicated by gray boxes labeled M12. The colored blocks between each genome represent regions of conserved syntenic gene content, ranging in pairwise nucleotide identity (according to BLASTn) from 100% (dark blue) to 99% (yellow). The diagrams were prepared using the Easyfig genome comparison visualization tool (25).
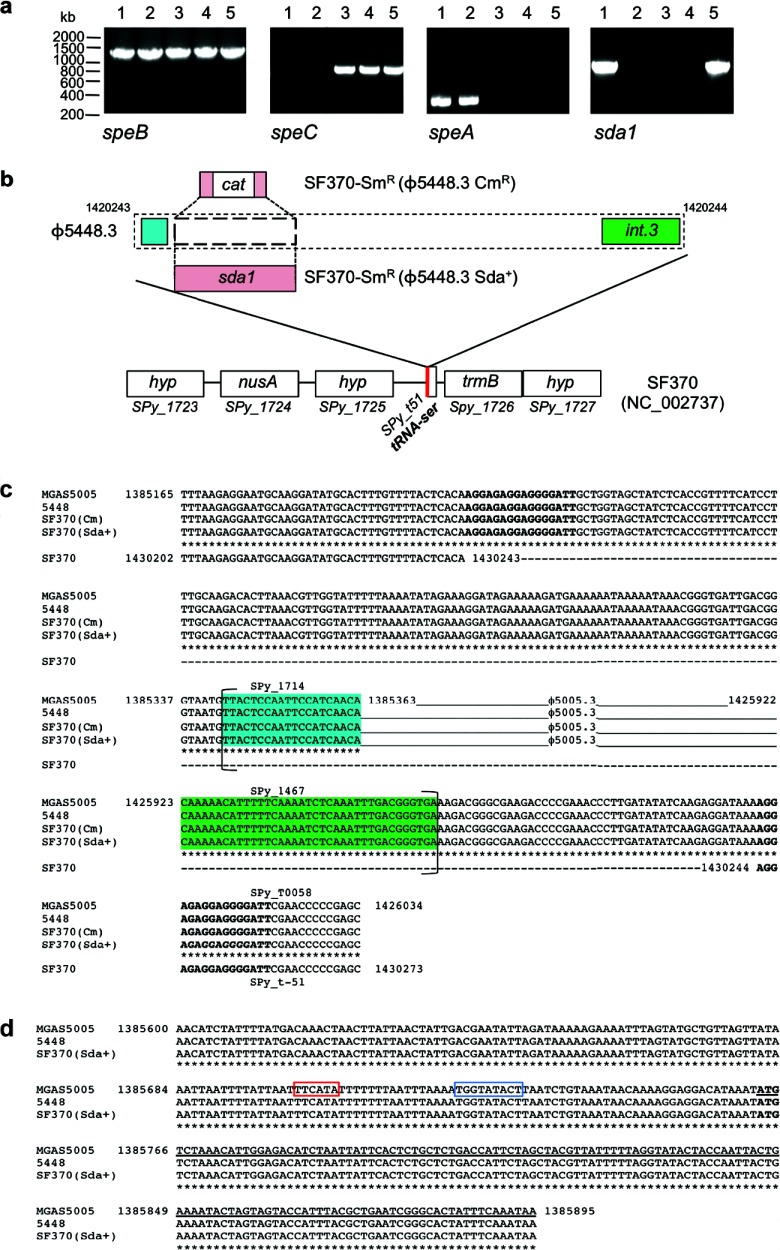
Characterization of SF370-Smr lysogens.
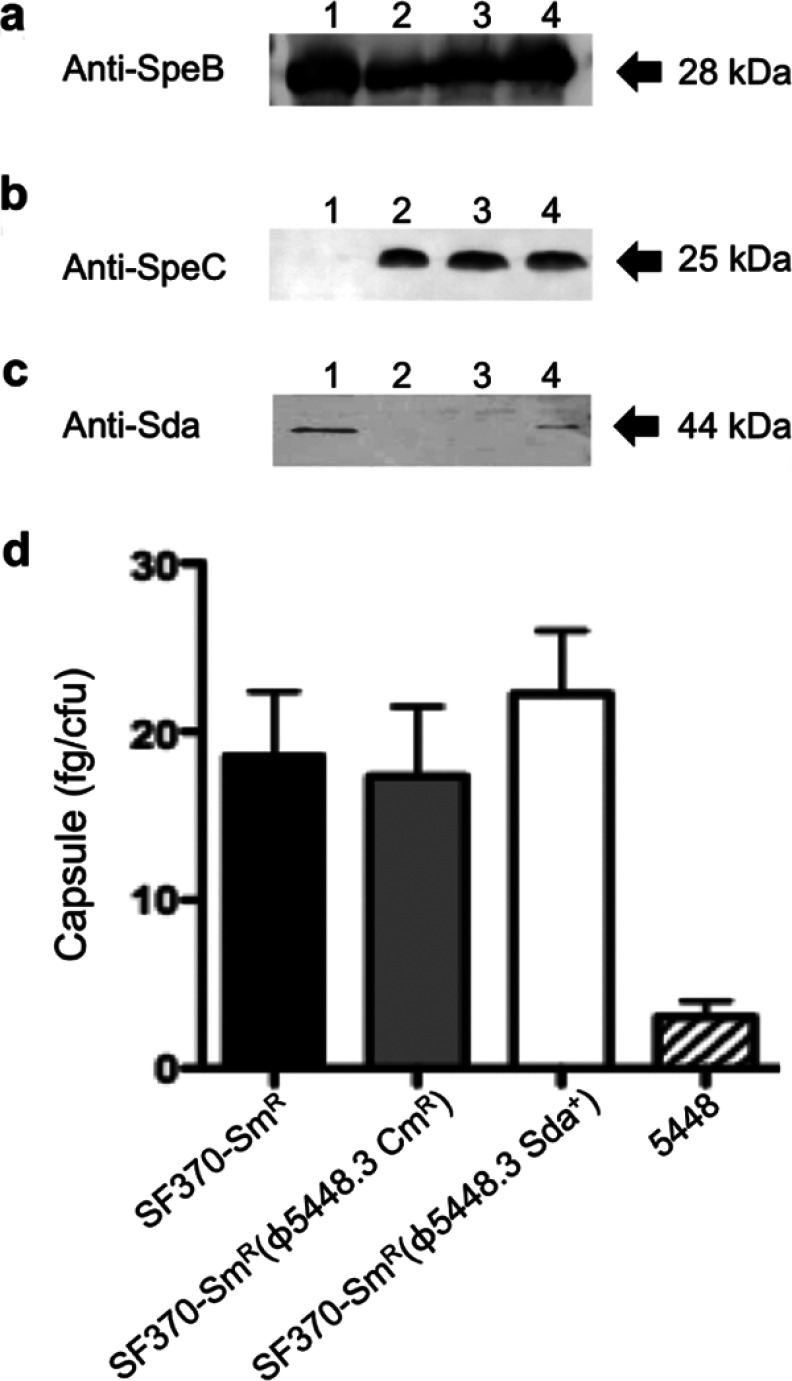
In vitro lysogenic conversion of SF370-Smr to SF370-Smr(ϕ5448.3 Cmr) was followed by allelic replacement of the chloramphenicol resistance marker with the 5448 sda1 gene to produce SF370-Smr(ϕ5448.3 Sda+). To characterize the lysogens produced by these experimental procedures, the virulence genes speC (ϕ370.1; strain SF370), speA (ϕ5448.1; strain 5448), and sda1 (ϕ5448.3; strain 5448), and the ubiquitous chromosomally located gene speB, were amplified by PCR (Fig. 2a). As expected, all GAS strains were speB positive; 5448 was positive for speA; and SF370 isogenic isolates were positive for speC. Lysogens SF370-Smr(ϕ5448.3 Sda+) and 5448 were positive for sda1 (Fig. 2a). The ϕ5448.3 phage attachment sites and the sda1 reinsertion boundaries were also PCR amplified and sequenced to confirm that phage and sda1 insertion had occurred in SF370-Smr(ϕ5448.3 Sda+) in the same genetic location as in GAS M1T1 strains (MGAS5005 and 5448) (Fig. 2b). Comparative sequence analysis showed that insertion of ϕ5448.3 and of the sda1 gene had not caused any deletions or duplications in the SF370-Smr nucleotide sequence (Fig. 2c and d). Superantigen and Sda1 DNase expression was examined in culture supernatants. The Western blot data correlated with the PCR screening results, with all isolates expressing SpeB in stationary-phase culture supernatants (Fig. 3a) and only SF370 isogenic isolates expressing SpeC (Fig. 3b). SF370-Smr(ϕ5448.3 Sda+) also expressed Sda1 in mid-log-phase culture supernatants, albeit at levels lower than those in the M1T1 isolate 5448 (Fig. 3c). Confirming previous studies (35, 36), SpeA expression was undetectable in supernatants of in vitro-grown GAS (data not shown). Hyaluronic acid capsule assays were performed on SF370-Smr, SF370-Smr(ϕ5448.3 Cmr), and SF370-Smr(ϕ5448.3 Sda+), and no significant difference in capsule production between the wild type and the lysogens was detected (Fig. 3d). All SF370 derivatives expressed higher capsule levels than the M1T1 strain 5448, in agreement with previous studies (17).
Fig 2.
Genetic characterization of SF370-Smr lysogens. (a) PCR amplification of GAS virulence genes speB, speC, speA, and sda1. Lanes 1, 5448; lanes 2, 5448Δsda; lanes 3, SF370-Smr; lanes 4, SF370-Smr(ϕ5448.3 Cmr); lanes 5, SF370-Smr(ϕ5448.3 Sda+). (b) Schematic illustrating the M1T1 ϕ5448.3 phage insertion site within the M1 SF370 genome backbone. In SF370-Smr(ϕ5448.3 Cmr), the chloramphenicol resistance gene replaces the sda1 open reading frame of phage ϕ5448.3. The red line indicates the phage insertion point immediately adjacent to a tRNA-Ser sequence. (c) Alignment of the Sda1-encoding bacteriophage attachment sites in wild-type M1T1 strains (MGAS5005 and 5448) and M1 lysogens SF370-Smr(ϕ5448.3 Cmr) and SF370-Smr(ϕ5448.3 Sda+). The sequences of the wild-type strains and the lysogens are identical. Phage insertion causes duplication of the sequence at the insertion point, resulting in a set of direct repeats (boldface). Brackets indicate the boundaries of open reading frames. The highlighted sequences represent the first (blue) and last (green) open reading frames of phages ϕ5005.3 and 5448.3. (d) Alignment of the insertion site of the sda1 gene within the Sda1-encoding bacteriophage in wild-type M1T1 strains (MGAS5005 and 5448) and the M1 lysogen SF370-Smr(ϕ5448.3 Sda+). The sequences are identical. sda1 insertion by double crossover has occurred without disruption of the sda1 open reading frame or of the 5′ sequence containing the promoter region (the −35 box is framed in red and the −10 box in blue). The starting codon for the sda1 gene is shown in boldface, and the sda1 open reading frame is underlined.
Fig 3.
Expression of virulence proteins and the hyaluronic acid capsule. (a) SpeB production was detected in stationary-phase culture supernatants. Lanes: 1, 5448; 2, SF370-Smr; 3, SF370-Smr(ϕ5448.3 Cmr); 4, SF370-Smr(ϕ5448.3 Sda+). (b) SpeC expression in mid-log-phase (A600, 0.4) culture supernatants. All lysogens expressed both SpeB and SpeC, as expected. (c) Sda1 expression in mid-log-phase (A600, 0.4) culture supernatants. (d) Hyaluronic acid capsule production was examined using the Stains-All method (29). The three M1 strains SF370-Smr, SF370-Smr(ϕ5448.3 Cmr), and SF370-Smr(ϕ5448.3 Sda+) produced comparable amounts of hyaluronic acid capsule, greater than the amount produced by the M1T1 strain 5448. Bars indicate means; error bars, standard deviations (n = 3).
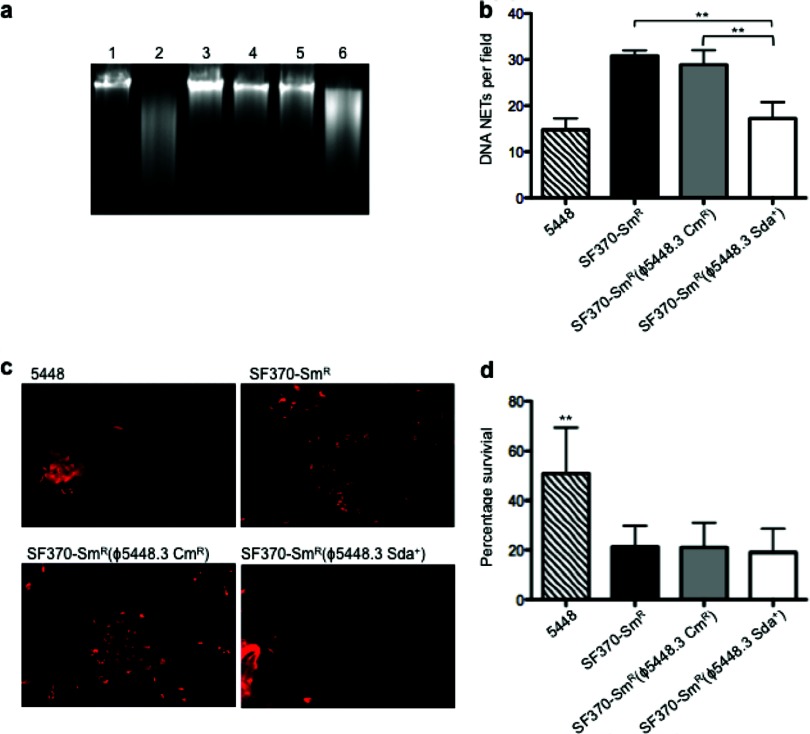
Resistance of GAS lysogens to killing by human neutrophils.
DNase activity was determined by coincubation of GAS culture supernatants with calf thymus DNA. The levels of DNA degradation by SF370-Smr(ϕ5448.3 Sda+) were greater than those for SF370-Smr and SF370-Smr(ϕ5448.3 Cmr) and were comparable to that for the M1T1 strain 5448 (Fig. 4a). Since neutrophil killing via DNA NETs plays a central role in bacterial clearance during GAS infection, and since Sda1 has been directly linked to NET degradation (19), we compared both the capacity to clear NETs and the degree of survival of SF370-Smr lysogens with that of wild-type SF370-Smr and the M1T1 strain 5448 when these strains were coincubated with human neutrophils. Even though a significant increase in the ability to degrade DNA NETs, comparable to that of M1T1 strain 5448, was observed for SF370-Smr(ϕ5448.3 Sda+) (Fig. 4b and c), this lysogenized strain did not show increased resistance to killing by neutrophils (Fig. 4d).
Fig 4.
Sda1 activities and resistance to neutrophil killing of lysogenized SF370 isolates. (a) DNase activities of stationary-phase GAS culture supernatants were determined by coincubation with calf thymus DNA. Lane 1, calf thymus DNA only (negative control); lane 2, 5448; lane 3, 5448Δsda; lane 4, SF370-Smr; lane 5, SF370-Smr(ϕ5448.3 Cmr); lane 6, SF370-Smr(ϕ5448.3 Sda+). Only 5448 and SF370-Smr(ϕ5448.3 Sda+) supernatants, containing Sda1, degraded DNA. (b) The bar graph shows the quantification of DNA NET induction per field of view after exposure to Sytox orange in wild-type GAS M1T1 (5448) and GAS M1 (SF370-Smr) and in the lysogens SF370-Smr(ϕ5448.3 Cmr) and SF370-Smr(ϕ5448.3 Sda+). In SF370-Smr(ϕ5448.3 Sda+), NET clearance was significantly greater than that for M1 GAS strains SF370-Smr and SF370-Smr(ϕ5448.3 Cmr) (P, <0.0007 for both comparisons) and comparable to that for the wild-type strain 5448. Values are arithmetic means plus standard errors (error bars) and are representative of three independent experiments performed in triplicate. Statistical 1-way analysis of variance with Dunnett's multiple-comparison posttest (P, <0.05) was performed using GraphPad Prism software. (c) Visualization of Sytox orange-stained DNA NETs (red) induced by coincubation of GAS serotype M1T1 strain 5448, M1 strain SF370-Smr, SF370-Smr(ϕ5448.3 Cmr), and SF370-Smr(ϕ5448.3 Sda+) with human neutrophils (MOI, 0.1). Fewer NETs can be observed for the SF370-Smr(ϕ5448.3 Sda+) lysogen, expressing Sda1, than for strains SF370-Smr and SF370-Smr(ϕ5448.3 Cmr). (d) Percentage of survival following coculture with human neutrophils in vitro. No significant difference was observed between the SF370-Smr parent strain and the lysogens, all of which were less neutrophil resistant than the M1T1 strain 5448 (P, <0.0008). Bars indicate means; error bars, standard deviations (n = 3). Statistical 1-way analysis of variance with Bonferroni's multiple-comparison posttest (P, <0.05) was performed using GraphPad Prism software.
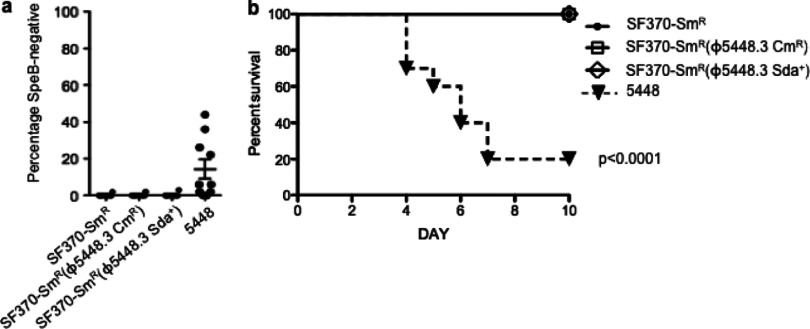
Invasive capacity and virulence of GAS lysogens.
Carriage of the sda1 gene has been characterized as a necessary selection trigger for the in vivo phase switch of GAS M1T1 to a hypervirulent phenotype (1, 17). We therefore tested the SF370-Smr lysogenized strains for a propensity to switch to the hypervirulent covRS mutant form. There was no evidence of an increased capacity to switch to the SpeB-negative hypervirulent form for any of the three SF370 derivatives characterized in this study (Fig. 5a). In order to examine virulence potential, AlbPLG1 mice were infected subcutaneously with GAS strains 5448, SF370-Smr, SF370-Smr(ϕ5448.3 Cmr), and SF370-Smr(ϕ5448.3 Sda+). SF370 is known to be avirulent in this mouse model (17), while the M1T1 isolate 5448 was hypervirulent (1, 17), with only 20% of the infected mice (at a dose of 1.3 × 107 CFU) surviving by day 7 (Fig. 5b). At comparable infection doses, SF370-Smr, SF370-Smr(ϕ5448.3 Cmr), and SF370-Smr(ϕ5448.3 Sda+) were avirulent (Fig. 5b).
Fig 5.
Characterization of SF370-Smr lysogen virulence. (a) Percentage of SpeB-positive isolates that switch to a SpeB-negative phenotype following a 3-day subcutaneous passage in C57BL/J6 mice. Each data point represents a single infected mouse (n, 10 mice per strain). (b) Percentage of survival of humanized plasminogen transgenic mice after subcutaneous infection with SF370-Smr (1.6 × 107 CFU/dose), SF370-Smr(ϕ5448.3 Cmr) (1.2 × 107 CFU/dose), SF370-Smr(ϕ5448.3 Sda+) (1.1 × 107 CFU/dose), and 5448 (1.3 × 107 CFU/dose). While 5448 infection caused the death of 80% of the mice, all SF370 strains were avirulent when an equivalent infection dose was used. Comparative analysis of survival curves (Mantel-Cox test) was performed using GraphPad Prism software.
DISCUSSION
The resurgence of GAS invasive disease in the past 30 years has coincided with the emergence and global dissemination of the hypervirulent M1T1 clone. The M1T1 serotype evolved from the ancestral M1 lineage via sequential horizontal gene transfer events, involving homologous recombination and prophage transduction, that led to the stable acquisition of two unique virulence determinants, the superantigen SpeA and the streptodornase Sda1, and increased expression of the extracellular toxins NAD+-glycohydrolase (Nga) and streptolysin O (SLO) (2, 14, 17). Our sequencing efforts confirmed the accumulation of the majority of the M1T1 serotype-specific single nucleotide polymorphisms (SNPs), in comparison to SF370, within these acquired regions (14). In the GAS M1T1 genome, the prophage encoding sda1 contains all the modules necessary for transfer and lysogeny, promotes invasive disease (1, 17), and therefore is potentially readily mobilizable to other GAS strains under appropriate conditions. In this study, we set out to determine whether the acquisition of this sda1-encoding phage was sufficient to confer enhanced virulence in a non-M1T1 GAS background.
The induced sda1-encoding phage was transferred by a combination of lysogeny and allelic replacement into the M1 strain SF370 background. Previous studies using the knockout strain 5448Δsda showed that in M1T1 strains, the Sda1 DNase plays an essential role in the clearance of NETs and is necessary for the switch to the hyperinvasive SpeB-negative covRS mutant form (1, 19). In our study, the SF370-Smr(ϕ5448.3 Sda+) lysogen, which expresses functional DNase Sda1, failed to display either enhanced resistance to neutrophil killing, a capacity to switch to a hyperinvasive phenotype, or increased virulence. The moderate difference in levels of Sda1 expression between the M1 and M1T1 strains observed in this study may be due to differing regulation of sda1 gene expression. The M1 and M1T1 genomes differ by a number of SNPs that affect regulators and modify the expression of a selected number of genes, including the has operon, slo, and nga (17, 21).
The killing of microbes by neutrophils is a multifaceted process involving DNA NETs as well as phagocytosis and the release of proteolytic enzymes, cationic antimicrobial peptides, and reactive oxygen and nitrogen species (37, 38). The evasive response of GAS is in turn complex and requires the combined action of several virulence factors (2). Protection against killing by human neutrophils is achieved not only via the production of DNases, such as Sda1 (19), but also via the inhibitory activity of the M1 protein (39) and the hyaluronic acid capsule (21) against the antimicrobial peptides embedded in NETs, as well as the activities of the streptococcal complement inhibitor protein (40) and streptolysin S (41).
Through the use of knockout strains, it was demonstrated previously that the presence of Sda1, M1, and the hyaluronic acid capsule is essential for induction of the switch to a SpeB-negative phenotype (1, 21). The proposed model for invasive disease development sees the degradation of NETs by the Sda1 DNase as the trigger for this phenotype switch in a subpopulation of M1T1 cells due to spontaneous mutations in the covRS regulatory operon, causing the upregulation of several virulence factors (the Sda1 DNase, the immunogenic M1 protein, the HasA hyaluronic acid capsule, the endopeptidase IdeS, the cytolysin SLO, and the protease SPyCEP) and the downregulation of the cysteine protease SpeB (2). This transcriptional shift promotes subversion of the human plasminogen activation system and systemic dissemination to sterile sites of the body (1, 30).
In invasive M1T1 isolates, SLO and Nga expression is higher than that in the ancestral strain SF370. Increased SLO and Nga expression is due to the presence of the M12-derived 36-kb recombination fragment found in the M1T1 clone (14, 17). Utilizing M1T1 slo and nga deletion mutants, Cole et al. showed that these factors do not contribute directly to SpeB switching and selection pressure for increased virulence in a mouse model (21). SLO nonetheless remains an important virulence factor implicated in GAS virulence and the clearance of immune system components at the site of infection (2, 42). In ancestral GAS M1 strains, such as SF370, hyaluronic acid capsule expression is higher than that of M1T1 strains (17). Capsule expression also contributes to SpeB switching and virulence (21), and thus, differing levels of capsule expression in SF370 may also potentially reduce selection for covRS mutants in vivo. The SpeA superantigen is absent in the ancestral GAS M1 strain SF370 (14). SpeA was first associated with severe cases of streptococcal shock syndrome in the late 1980s (4, 5). Recent gene knockout studies indicate that SpeA is not directly involved in the mechanism of invasive disease initiation, but its acquisition may aid in defense against the host immune system and the subsequent dissemination of the hypervirulent M1T1 clone (17).
Given that a combination of GAS virulence factors promotes neutrophil resistance, it is perhaps not surprising that the introduction of the single gene sda1 into the SF370 background did not enhance neutrophil resistance and virulence, despite improved clearance of DNA NETs. Our experimental observations may be explained by a requirement for a cohort of virulence determinants expressed at a specific level to promote virulence, including Sda1, the M1 protein, the hyaluronic acid capsule, and SLO (21, 42). In this context, it is pertinent to this hypothesis that SF370 expresses much higher levels of the hyaluronic acid capsule than GAS M1T1 strains (17), which may tip the balance away from the selection of covRS mutants in the presence of neutrophils, resulting in failure to promote selection for the hyperinvasive phenotype.
We conclude that the hypervirulence of the M1T1 clone is due to the synergic effect of the M1T1 clone bacteriophage-specific virulence factor Sda1 acting in concert with the M1T1 clone-specific genetic scaffold. This work suggests that the emergence of hypervirulent GAS clones is dependent on a fine balance of virulence determinants acting in unison.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the National Health and Medical Research Council of Australia, the Australian Research Council, and the U.S. National Institutes of Health. S.A.B. is the recipient of an Australian Research Council Australian Research Fellowship (DP0881347), and M.J.W. is the recipient of a National Health and Medical Research Council of Australia fellowship (631386).
All animal experiments conducted for this study conform to National Health and Medical Research Council of Australia guidelines for the use of animals in research and were approved by The University of Queensland Animal Ethics Committee.
All authors report no conflicts of interest.
Footnotes
Published ahead of print 25 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00192-13.
REFERENCES
- 1. Walker MJ, Hollands A, Sanderson-Smith M, Cole JN, Kirk JK, Henningham A, McArthur JD, Dinkla K, Aziz RK, Kansal RG, Simpson AJ, Buchanan JT, Chhatwal GS, Kotb M, Nizet V. 2007. DNase Sda1 provides selection pressure for a genetic and phenotypic switch promoting invasive group A streptococcal infection. Nat. Med. 13:981–985 [DOI] [PubMed] [Google Scholar]
- 2. Cole JN, Barnett TC, Nizet V, Walker MJ. 2011. Molecular insight into invasive group A streptococcal disease. Nat. Rev. Microbiol. 9:724–736 [DOI] [PubMed] [Google Scholar]
- 3. Olsen RJ, Musser JM. 2010. Molecular pathogenesis of necrotizing fasciitis. Annu. Rev. Pathol. 5:1–31 [DOI] [PubMed] [Google Scholar]
- 4. Cone LA, Woodard DR, Schlievert PM, Tomory GS. 1987. Clinical and bacteriologic observations of a toxic shock-like syndrome due to Streptococcus pyogenes. N. Engl. J. Med. 317:146–149 [DOI] [PubMed] [Google Scholar]
- 5. Cleary PP, Schlievert PM, Handley JP, Kim MH, Hauser AR, Kaplan EL, Wlazlo A. 1992. Clonal basis for resurgence of serious Streptococcus pyogenes disease in the 1980s. Lancet 339:518–521 [DOI] [PubMed] [Google Scholar]
- 6. Musser JM, Kapur V, Szeto J, Pan X, Swanson DS, Martin DR. 1995. Genetic diversity and relationships among Streptococcus pyogenes strains expressing serotype M1 protein: recent intercontinental spread of a subclone causing episodes of invasive disease. Infect. Immun. 63:994–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Aziz RK, Kotb M. 2008. Rise and persistence of global M1T1 clone of Streptococcus pyogenes. Emerg. Infect. Dis. 14:1511–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cunningham MW. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aziz RK, Edwards RA, Taylor WW, Low DE, McGeer A, Kotb M. 2005. Mosaic prophages with horizontally acquired genes account for the emergence and diversification of the globally disseminated M1T1 clone of Streptococcus pyogenes. J. Bacteriol. 187:3311–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sumby P, Barbian KD, Gardner DJ, Whitney AR, Welty DM, Long RD, Bailey JR, Parnell MJ, Hoe NP, Adams GG, DeLeo FR, Musser JM. 2005. Extracellular deoxyribonuclease made by group A Streptococcus assists pathogenesis by enhancing evasion of the innate immune response. Proc. Natl. Acad. Sci. U. S. A. 102:1679–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Banks DJ, Beres SB, Musser JM. 2002. The fundamental contribution of phages to GAS evolution, genome diversification and strain emergence. Trends Microbiol. 10:515–521 [DOI] [PubMed] [Google Scholar]
- 12. Canchaya C, Fournous G, Chibani-Chennoufi S, Dillmann ML, Brüssow H. 2003. Phage as agents of lateral gene transfer. Curr. Opin. Microbiol. 6:417–424 [DOI] [PubMed] [Google Scholar]
- 13. Beres SB, Richter EW, Nagiec MJ, Sumby P, Porcella SF, DeLeo FR, Musser JM. 2006. Molecular genetic anatomy of inter- and intraserotype variation in the human bacterial pathogen group A Streptococcus. Proc. Natl. Acad. Sci. U. S. A. 103:7059–7064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sumby P, Porcella SF, Madrigalm AG, Barbian KD, Virtaneva K, Ricklefs SM, Sturdevant DE, Graham MR, Vuopio-Varkila J, Hoe NP, Musser JM. 2005. Evolutionary origin and emergence of a highly successful clone of serotype M1 group A Streptococcus involved multiple horizontal gene transfer events. J. Infect. Dis. 192:771–782 [DOI] [PubMed] [Google Scholar]
- 15. Ferretti JJ, McShan WM, Ajdic D, Savic DJ, Savic G, Lyon K, Primeaux C, Sezate S, Suvorov AN, Kenton S, Lai HS, Lin SP, Qian Y, Jia HG, Najar FZ, Ren Q, Zhu H, Song L, White J, Yuan X, Clifton SW, Roe BA, McLaughlin R. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. U. S. A. 98:4658–4663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chatellier S, Ihendyane N, Kansal RG, Khambaty F, Basma H, Norrby-Teglund A, Low DE, McGeer A, Kotb M. 2000. Genetic relatedness and superantigen expression in group A Streptococcus serotype M1 isolates from patients with severe and nonsevere invasive diseases. Infect. Immun. 68:3523–3534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Maamary PG, Ben Zakour NL, Cole JN, Hollands A, Aziz RK, Barnett TC, Cork AJ, Henningham A, Sanderson-Smith M, McArthur JD, Venturini C, Gillen CM, Kirk JK, Johnson DR, Taylor WM, Kaplan EL, Kotb M, Nizet V, Beatson SA, Walker MJ. 9 August 2012, posting date Tracing the evolutionary history of the pandemic group A streptococcal M1T1 clone. FASEB J. doi:10.1096/fj.12-212142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aziz RK, Ismail SA, Park WH, Kotb M. 2004. Post-proteomic identification of a novel phage-encoded streptodornase, Sda1, in invasive M1T1 Streptococcus pyogenes. Mol. Microbiol. 54:184–197 [DOI] [PubMed] [Google Scholar]
- 19. Buchanan JT, Simpson AJ, Aziz RK, Liu GY, Kristian SA, Kotb M, Feramisco J, Nizet V. 2006. DNase expression allows the pathogen group A Streptococcus to escape killing in neutrophil extracellular traps. Curr. Biol. 16:396–400 [DOI] [PubMed] [Google Scholar]
- 20. Sumby P, Whitney AR, Graviss EA, DeLeo FR, Musser JM. 2006. Genome-wide analysis of group A streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog. 2:e5 doi:10.1371/journal.ppat.0020005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cole JN, Pence MA, von Köckritz-Blickwede M, Hollands A, Gallo RL, Walker MJ, Nizet V. 2010. M protein and hyaluronic acid capsule are essential for in vivo selection of covRS mutations characteristic of invasive serotype M1T1 group A streptococcus. mBio 1:e00191–10 doi:10.1128/mBio.00191-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Assefa S, Keane TM, Otto TD, Newbold C, Berriman M. 2009. ABACAS: algorithm-based automatic contiguation of assembled sequences. Bioinformatics 25:1968–1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Otto TD, Dillon GP, Degrave WS, Berriman M. 2011. RATT: Rapid Annotation Transfer Tool. Nucleic Acids Res. 39(9):e57 doi:10.1093/nar/gkq1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Banks DJ, Lei B, Musser JM. 2003. Prophage induction and expression of prophage-encoded virulence factors in group A Streptococcus serotype M3 strain MGAS315. Infect. Immun. 71:7079–7086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Simon D, Ferretti JJ. 1991. Electrotransformation of Streptococcus pyogenes with plasmid and linear DNA. FEMS Microbiol. Lett. 66:219–224 [DOI] [PubMed] [Google Scholar]
- 28. Sanderson-Smith ML, Dinkla K, Cole JN, Cork AJ, Maamary PG, McArthur JD, Chhatwal GS, Walker MJ. 2008. M protein-mediated plasminogen binding is essential for the virulence of an invasive Streptococcus pyogenes isolate. FASEB J. 22:2715–2722 [DOI] [PubMed] [Google Scholar]
- 29. Ashbaugh CD, Wessels MR. 2001. Absence of a cysteine protease effect on bacterial virulence in two murine models of human invasive group A streptococcal infection. Infect. Immun. 69:6683–6688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cole JN, McArthur JD, McKay FC, Sanderson-Smith ML, Cork AJ, Ranson M, Rohde M, Itzek A, Sun H, Ginsburg D, Kotb M, Nizet V, Chhatwal GS, Walker MJ. 2006. Trigger for group A streptococcal M1T1 invasive disease. FASEB J. 20:1745–1747 [DOI] [PubMed] [Google Scholar]
- 31. Maamary PG, Sanderson-Smith ML, Aziz RK, Hollands A, Cole JN, McKay FC, McArthur JD, Kirk JK, Cork AJ, Keefe RJ, Kansal RG, Sun H, Taylor WL, Chhatwal GS, Ginsburg D, Nizet V, Kotb M, Walker MJ. 2010. Parameters governing invasive disease propensity of non-M1 serotype group A streptococci. J. Innate Immun. 2:596–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Collin M, Olsen A. 2000. Generation of a mature streptococcal cysteine proteinase is dependent on cell wall anchored M1 protein. Mol. Microbiol. 36:1306–1318 [DOI] [PubMed] [Google Scholar]
- 33. Ashbaugh CD, Warren HB, Carey VJ, Wessels MR. 1998. Molecular analysis of the role of the group A streptococcal cysteine protease, hyaluronic acid capsule, and M protein in a murine model of human invasive soft-tissue infection. J. Clin. Invest. 102:550–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sun H, Ringdahl U, Homeister JW, Fay WP, Engleberg NC, Yang AY, Rozek LS, Wang X, Sjöbring U, Ginsburg D. 2004. Plasminogen is a critical host pathogenicity factor for group A streptococcal infection. Science 305:1283–1286 [DOI] [PubMed] [Google Scholar]
- 35. Kazmi SU, Kansal R, Aziz RK, Hooshdaran M, Norrby-Teglund A, Low DE, Halim AB, Kotb M. 2001. Reciprocal, temporal expression of SpeA and SpeB by invasive M1T1 group A streptococcal isolates in vivo. Infect. Immun. 69:4988–4995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kansal RG, Nizet V, Jeng A, Chuang WJ, Kotb M. 2003. Selective modulation of superantigen-induced responses by streptococcal cysteine protease. J. Infect. Dis. 187:398–407 [DOI] [PubMed] [Google Scholar]
- 37. Nauseef WM. 2007. How human neutrophils kill and degrade microbes: an integrated view. Immunol. Rev. 219:88–102 [DOI] [PubMed] [Google Scholar]
- 38. Nizet V. 2010. Bacteria and phagocytes: mortal enemies. J. Innate Immun. 2:505–507 [DOI] [PubMed] [Google Scholar]
- 39. Lauth X, von Kockritz-Blickwede M, McNamara CW, Myskowski S, Zinkernagel AS, Beall B, Ghosh P, Gallo RL, Nizet V. 2009. M1 protein allows group A streptococcal survival in phagocyte extracellular traps through cathelicidin inhibition. J. Innate Immun. 1:202–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pence MA, Rooijakkers SHM, Cogen AL, Cole JN, Hollands A, Gallo RL, Nizet V. 2010. Streptococcal inhibitor of complement promotes innate immune resistance phenotypes of invasive M1T1 group A Streptococcus. J. Innate Immun. 2:587–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Miyoshi-Akiyama T, Takamatsu D, Koyanagi M, Zhao J, Imanishi K, Uchiyama T. 2005. Cytocidal effect of Streptococcus pyogenes on mouse neutrophils in vivo and the critical role of streptolysin S. J. Infect. Dis. 192:107–116 [DOI] [PubMed] [Google Scholar]
- 42. Chiarot E, Faralla C, Chiappini N, Tuscano G, Falugi F, Gambellini G, Taddei A, Capo S, Cartocci E, Veggi D, Corrado C, Mangiavacchi S, Tavarini S, Scarselli M, Janulczyk R, Grandi G, Margarit I, Bensi G. 2013. Targeted amino acid substitutions impair streptolysin O toxicity and group A Streptococcus virulence. mBio 4(1):e00387–12 doi:10.1128/mBio.00387-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.