Abstract
When the bacterium Legionella pneumophila, the causative agent of Legionnaires' disease, is phagocytosed by alveolar macrophages, it delivers a large number of effector proteins through its Dot/Icm type IV secretion system into the host cell cytosol. Among those proteins is LidA, an effector that interacts with several host GTPases of the Rab family, including Rab6A′, a regulator of retrograde vesicle trafficking within eukaryotic cells. The effect of LidA on Rab6A′ function and the role of Rab6A′ for L. pneumophila growth within host cells has been unclear. Here, we show that LidA preferentially binds Rab6A′ in the active GTP-bound conformation. Rab6 binding occurred through the central region of LidA and followed a stoichiometry for LidA and Rab6A′ of 1:2. LidA maintained Rab6A′ in the active conformation by efficiently blocking the hydrolysis of GTP by Rab6A′, even in the presence of cellular GTPase-activating proteins, suggesting that the function of Rab6A′ must be important for efficient intracellular replication of L. pneumophila. Accordingly, we found that production of constitutively inactive Rab6A′(T27N) but not constitutively active Rab6A′(Q72L) significantly reduced the ability of L. pneumophila to initiate intracellular replication in human macrophages. Thus, the presence of an active pool of Rab6 within host cells early during infection is required to support efficient intracellular growth of L. pneumophila.
INTRODUCTION
Many microbial pathogens reside within a membrane-enclosed compartment within infected host cells. Their intravacuolar life style requires pathogens to produce and deliver signaling molecules from their own cytosol across the vacuolar membrane to the host cell in order to accomplish critical tasks, such as the acquisition of nutrients and the interference with immune signaling cascades. Among Gram-negative microbes, this is often accomplished by effector proteins that are delivered into the host cell through specialized translocation machines, such as type IV secretion systems (T4SSs). Although many pathogens take advantage of host cell vesicle transport, surprisingly few effectors have been characterized to date that directly target Rab proteins controlling these pathways. One of the better-understood examples of Rab GTPase exploitation by a pathogen occurs during infection with Legionella pneumophila, an opportunistic human pathogen that causes a potentially life-threatening pneumonia called Legionnaires' disease (1, 2). Upon phagocytosis by alveolar macrophages, the microbe delivers its effector proteins through the Dot/Icm T4SS into the host cell. These effectors help L. pneumophila intercept proteins and transport vesicles of the infected cell. By incorporating this material into the Legionella-containing vacuole (LCV), the bacterium gradually transforms its surrounding membrane into a specialized compartment that supports bacterial replication, a process called replication vacuole formation (3). L. pneumophila mutants with a nonfunctional Dot/Icm T4SS are avirulent, and their vacuoles gradually mature along the endolysosomal route into a microbicidal compartment, highlighting the critical role of T4 effectors for L. pneumophila infection (4, 5).
Marker proteins from the early secretory pathway, such as the soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) protein Sec22b, the luminal endoplasmic reticulum (ER) protein BiP, and the small GTPase Rab1, have been found to colocalize with LCVs (6–8), indicating that at least some of the material that contributes to replication vacuole formation is derived from the ER-to-Golgi complex transport route. Consistent with this observation, it has been shown that GTPases of the Arf and Rab family are direct targets of L. pneumophila effector proteins. RalF recruits Arf1 to LCVs and activates it by catalyzing exchange of GDP against GTP (9). SidM (also known as DrrA) possesses similar activities toward Rab1 (10, 11), but in addition to recruiting and activating this GTPase, SidM also catalyzes Rab1 AMPylation, a posttranslational modification that interferes with Rab1 inactivation (12). The effector SidD catalyzes de-AMPylation of Rab1 later during infection, enabling Rab1 inactivation by the L. pneumophila GTPase-activating protein (GAP) LepB and release of Rab1 from the LCV (13, 14).
The cascade of Rab1 manipulation events by L. pneumophila effectors is further complicated by the existence of another Rab1 binding protein, the L. pneumophila effector LidA (15). LidA is translocated by the Dot/Icm system throughout the duration of the infection cycle (16). Early on, the protein is detectable primarily on the LCV surface, where it is believed to assist in the recruitment of Rab1 and/or secretory transport vesicles (15, 17). As the infection cycle progresses, LidA accumulates at other locations throughout the infected host cell and associates with membranes of a yet-unknown origin (16). These findings suggest that LidA could play multiple roles in different cellular locations during the infection process.
In addition to binding Rab1, LidA has been shown to interact with other Rab GTPases of mammalian cells, including Rab6 and Rab8 (17, 18). Rab8 is involved in the transport of cargo to the apical membrane of polarized epithelial cells and has recently been found on the LCV during growth within the amoebean host Dictyostelium discoideum (19). Rab6 exists in three isoforms: Rab6A/A′, which are ubiquitously expressed, and Rab6B, which is found primarily in the brain. Rab6A/A′ are the products of alternative splicing and differ in only three amino acid residues near the GTP binding site (20, 21). Both isoforms localize to trans-Golgi apparatus cisternae and have been ascribed various functions, such as coat protein I-independent retrograde vesicle flow (22), cytokinesis (23, 24) and, more recently, delivery of exocytotic membrane carriers to plasma membrane domains enriched for Rab6-interacting protein 1A (R6IP1A) (25), as well as fission of tubular membrane extensions from the Golgi complex (26). The critical role of Rab6A/A′ in cellular trafficking to various organelles might explain why this GTPase is a valuable target of human pathogens. For instance, the intravacuolar pathogen Chlamydia trachomatis shows reduced proliferation in host cells depleted of Rab6 (29), and vacuoles or “inclusions” containing this pathogen colocalize with Rab6 (30). Unlike Chlamydia inclusions, the vacuoles containing L. pneumophila are not decorated with Rab6 during infection of mammalian cells (reference 7 and unpublished data), indicating that different pathogens exploit Rab6 for distinct purposes. In the present study, we characterized the effect of L. pneumophila LidA on the activity of Rab6A′ in vitro, and we demonstrate the importance of this GTPase for replication of the bacterium within infected cells.
MATERIALS AND METHODS
Strains, media, plasmids, and antibodies.
L. pneumophila strains were grown and maintained in medium containing thymidine as previously described (31, 32). L. pneumophila strain Lp02 (thyA hsdR rpsL) is a thymidine auxotroph derivative of the isolate Philadelphia-1 (33, 34). Escherichia coli strains for cloning and production of recombinant proteins were GC5 (Genesee) and BL21(DE3) (Novagen), respectively. Plasmids for production of recombinant tagged proteins in E. coli and of fluorescently tagged proteins in tissue culture cells were generated as described in Table 1. The oligonucleotides used to PCR amplify the desired DNA fragments and the restriction sites used to clone the open reading frames are listed in Table 2. Rab6A antibodies were purchased from Santa Cruz Biotechnology. Rabbit antibodies directed against L. pneumophila were generated using formalin-killed bacteria (Genscript USA, Inc. standard immunization protocol for polyclonal antibody). Anti-LidA serum was a kind gift of R. Isberg (Boston, MA).
Table 1.
Plasmids used in this study
| Name | Insert | Oligonucleotidesa | Source or reference |
|---|---|---|---|
| pGEX-6p-1 Rab6A′ WT | Human Rab6A′ WTb | P1, P2 | This study |
| pGEX-6p-1 Rab6A′ T27N | Human Rab6A′ T27N | P3, P4 | This study |
| pGEX-6p-1 Rab6A′ Q72L | Human Rab6A′ Q72L | P5, P6 | This study |
| pQE80L-Rab6A′ WT | Human Rab6A′ WT | P1, P2 | This study |
| pQE80L- Rab6A′ T27N | Human Rab6A′ T27N | P3, P4 | This study |
| pQE80L-Rab6A′ Q72L | Human Rab6A′ Q72L | P5, P6 | This study |
| pEGFP-Rab6A′ WT | Human Rab6A′ WT | P1, P2 | This study |
| pEGFP-Rab6A′ T27N | Human Rab6A′ T27N | P3, P4 | This study |
| pEGFP-Rab6A′ Q72L | Human Rab6A′ Q72L | P5, P6 | This study |
| pGEX-6p-1-LidA | L. pneumophila LidA (aa 1–729) | 35 | |
| pGEX-6p-1-N-LidA | L. pneumophila LidA fragment (aa 1–189); | 35 | |
| pGEX-6p-1-M-LidA | L. pneumophila LidA fragment (aa 190–600) | 35 | |
| pGEX-6p-1-C-LidA | L. pneumophila LidA fragment (aa 601–729) | 35 | |
| pQE80L-LidA | L. pneumophila LidA (aa 1–729) | 35 | |
| pQE80-GapCenA | Human TBC1D11 | P7, P8 | This study |
See Table 2 for oligonucleotide sequences and names.
WT, wild type.
Table 2.
Oligonucleotides used in this study
| Oligonucleotide no. | Name | Sequencea |
|---|---|---|
| P1 | 5′Rab6A′_BamHI | gatcGGATCCatgtccacgggcggag |
| P2 | 3′Rab6A′_SalI | gatGTCGACttagcaggaacagcctc |
| P3 | 5'′ab6A′ T27N | caaagcgttggaaagaattctttgatcaccaga |
| P4 | 3′Rab6A′ T27N | tctggtgatcaaagaattctttccaacgctttg |
| P5 | 5′Rab6A′Q72L | tgggatactgcgggtctagagcgtttccgtagc |
| P6 | 3′Rab6A′Q72L | gctacggaaacgctctagacccgcagtatccca |
| P7 | 5′GapCenA_BamHI | gatcGGATCCatggacgaccagccagggg |
| P8 | 3′GapCenA_SalI | gatcGTCGACtcagcaagtctctttccc |
Uppercase letters indicate restriction sites; nucleotide exchanges are underlined.
Production and purification of recombinant proteins.
Recombinant proteins were purified as previously described (13). Briefly, plasmids encoding GST-Rab6A′, GST-Rab1, 6×His-LidA, or 6×His-GapCenA were expressed in E. coli BL21 at 20°C overnight after induction with 0.2 mM isopropyl-β-dithiogalactopyranoside (IPTG). Harvested cells were mechanically lysed, and the soluble fraction of the lysate was incubated overnight at 4°C with either glutathione-Sepharose 4B (GE Healthcare) for glutathione S-transferase (GST)-tagged proteins or with Talon metal affinity resin (Clontech) for 6×His-tagged proteins. Beads were washed with phosphate-buffered saline, 1 mM MgCl2, and 1 mM β-mercaptoethanol (PBS-MM), and GST-tagged proteins were incubated overnight with PreScission protease to cleave the GST tag. Untagged proteins were eluted off the resin, and aliquots were stored at −80°C. 6×His-tagged proteins were eluted in PBS-MM supplemented with 125 mM imidazole. Proteins were dialyzed overnight in PBS-MM and stored in aliquots at −80°C.
Protein interaction assays.
For pulldown experiments, constitutively inactive Rab6A′(T27N) or active Rab6A′(Q72L) (10 μg each) was immobilized on Affigel beads (Bio-Rad) according to the manufacturer's recommendations. Beads were incubated for 2 h at 4°C in PBS containing a molar excess of full-length LidA, LidA variants (N-LidA, M-LidA, and C-LidA), or GapCenA. Beads were washed 5 times with cold PBS-MM and resuspended in SDS sample buffer. Proteins retained on the beads were resolved by SDS-PAGE and visualized by Coomassie staining. For pulldown from tissue culture cell lysate, 293T cells transiently transfected with plasmids encoding green fluorescent protein (GFP)-tagged R6IP1A or R6IP2B were mechanically lysed by Dounce homogenization, and postnuclear supernatant was generated by spinning the lysate at 10,000 × g and incubated with protein-coated beads. Beads were washed five times in PBS-MM, and proteins retained by the beads were detected by immunoblotting using rabbit anti-GFP antibody (Invitrogen). Isothermal titration calorimetry assays were carried out at 25°C with an ITC200 microcalorimeter (GE Healthcare). LidA (10 mg/ml) dialyzed against PBS-MM buffer was titrated into the sample cell containing 5 mg/ml wild-type Rab1. For Rab competition experiments, LidA was preincubated with Rab6A′ for 1 h prior to titration into the sample cell containing Rab1. Data were analyzed using Origin 7 software.
Fast protein liquid chromatography (FPLC) for the analysis of protein complex formation between LidA and Rab6A′ was performed by using an AektaPurifier 10 apparatus (GE Healthcare). Samples of 100 μl with purified recombinant proteins at a concentration of 1 μg/μl were loaded onto a Superdex 75 10/300 column (GE Healthcare) equilibrated with PBS-MM. The protein elution profile was monitored in real time by measuring the absorbance of light at a 280-nm wavelength. Proteins within each fraction (0.5 ml) were analyzed by SDS-PAGE followed by immunoblotting with the indicated antibody. The molecular weights of proteins/complexes were determined based on a calibration curve of a protein standard kit after calculating their partition coefficients (KAV), as follows: KAV = (elution volume of protein − void volume)/(column bed volume − void volume).
GTP hydrolysis assay.
All nucleotide hydrolysis experiments were performed at room temperature. Rab6A′ (40 μM) was incubated with 5 mM EDTA for 10 min to extract any unlabeled nucleotide. Nucleotide-free Rab6A′ was incubated for 30 min with a molar excess of [α-32P]GTP in PBS, followed by the addition of 20 mM MgCl2. Unbound [α-32P]GTP was removed by using a desalting column. GTP hydrolysis reactions were carried out in a total volume of 30 μl. To initiate GTP hydrolysis, [α-32P]GTP-loaded Rab6A′ was incubated with postnuclear supernatant (PNS) or with PNS pretreated with LidA-coated beads. To determine the protective effect of LidA on Rab6A′ inactivation, [α-32P]GTP-loaded Rab6A′ was preincubated with LidA or LidA variants (with or without proteinase K treatment) at the molar ratios indicated in the figure legends. Samples (5 μl) were removed at the indicated time points, mixed with 5 μl sample buffer (2% SDS, 20 mM EDTA), and boiled at 95°C for 10 min to release any protein-bound guanine nucleotides. Samples (6 μl) were spotted onto Polygram Cel 300 polyethylenimine cellulose paper (Macherey-Nagel) approximately 0.6 to 1.5 cm apart at a distance of 0.7 to 2.5 cm from the edge of the plate. The thin-layer chromatography (TLC) plates were run vertically in a sealed chamber filled with a 1-cm column of solvent (1 M acetic acid and 0.5 M lithium chloride). Once the solvent reached the top, the plates were dried and exposed to a film or phosphorimager screen for 12 h. The radioactive signals were quantified and normalized using ImageJ (http://rsb.info.nih.gov/ij).
Infectious center assay.
Differentiated U937 cells were transiently transfected with plasmids encoding GFP-Rab6A′ wild type or the active (Q72L) or inactive (T27N) Rab6A′ mutant protein by using the Neon transfection system (Invitrogen). U937 cell monolayers were challenged with L. pneumophila wild-type strains for 1 h at a multiplicity of infection (MOI) of 5, washed three times to remove extracellular bacteria, and incubated for an additional 12 h at 37°C to allow bacterial replication to proceed. Cells were fixed in PBS (3.7% formaldehyde) for 20 min at 37°C, blocked with PBS supplemented with 10% goat serum, and stained for extracellular bacteria by using rabbit anti-Legionella antibody and Cascade Blue-conjugated goat anti-rabbit antibody. Cells were permeabilized through incubation with −20°C methanol for 10 s and then blocked, and intracellular bacteria were labeled using rabbit anti-Legionella primary antibody and Texas Red-conjugated goat anti-rabbit antibody (Invitrogen). Coverslips were mounted using antifade reagent (Invitrogen). The number of bacteria per vacuole at 13 h postinfection was visually determined using an AxioObserver inverted microscope (Zeiss).
Coimmunoprecipitation assay.
Differentiated U937 cells were challenged with L. pneumophila wild type or a ΔlidA deletion strain at an MOI of 10. Extracellular bacteria were removed by washing the cell monolayer with RPMI. Cells were harvested either 2 h or 6 h after the initial challenge and lysed in PBS-MM containing Complete protease inhibitor cocktail (Roche) by using a Dounce homogenizer (BC Scientific). After centrifugation at 10,000 × g for 10 min, the PNS was incubated for 1 h at 4°C with Affigel beads (Bio-Rad) coated with anti-LidA antibody. Beads were washed five times in PBS-MM and boiled in SDS-PAGE loading buffer, and Rab6A′ or LidA that eluted off the beads was detected by immunoblotting with a protein-specific antibody.
RESULTS
Interaction of LidA with Rab6A′ is direct and nucleotide dependent.
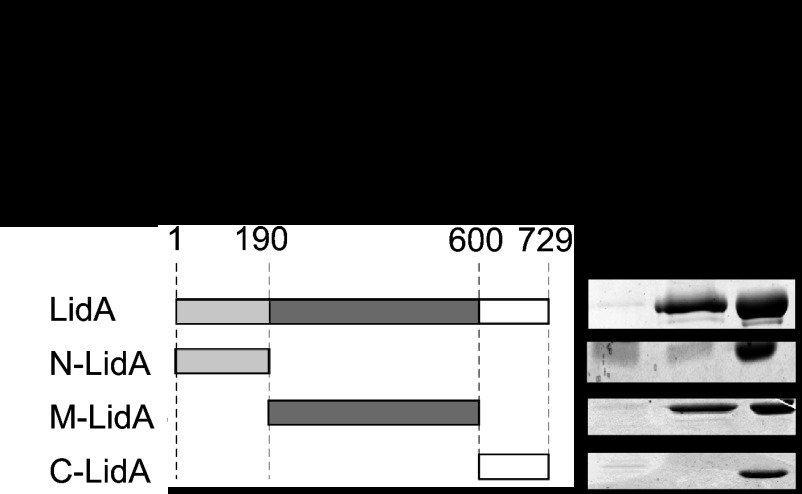
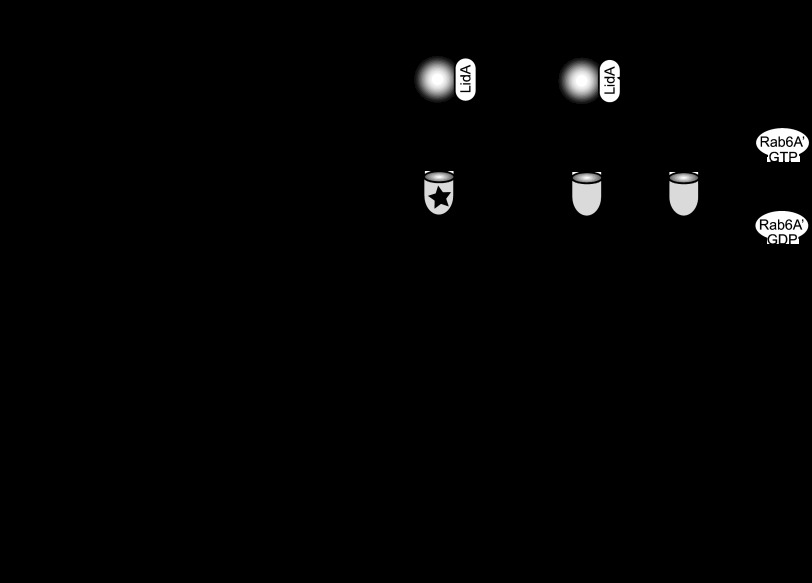
Rab6A/A′ was identified previously as a potential LidA ligand in pulldown studies from mammalian cell lysate (17). To exclude the possibility that this interaction was mediated by another protein/factor within the lysate, we probed for a direct interaction between LidA and Rab6A′. Using purified recombinant Rab6A′ variants locked in either the active [Rab6A′(Q72L)] or inactive [Rab6A′(T27N)] conformation, we found that LidA showed a strong preference for binding to the active GTP-locked form (Fig. 1). Binding of LidA to either Rab6A′(T27N)-coated beads or uncoated control beads was undetectable. Similar results were obtained using Rab6A as bait (data not shown), consistent with the idea that LidA targets both Rab6 isoforms. Any attempt to validate these results using wild-type Rab6A′ were hindered by the fact that nucleotide loading of purified Rab6A′ with either GDP or GTPγS, a nonhydrolyzable GTP analog, was incomplete (data not shown). To determine the region of LidA that mediates the Rab6A′ interaction, we performed binding experiments using truncated forms of LidA that had previously been determined, based on proteolytic degradation analyses (35): the N-terminal region (N-LidA) spanning amino acids (aa) 1 to 189, a middle region (M-LidA; aa 190 to 600) enriched in coiled-coils (a typical Rab GTPase binding domain), and a C-terminal region (C-LidA; aa 601 to 729) (Fig. 1). Upon incubation of purified LidA variants with bead-immobilized Rab6A′(Q72L), we found that M-LidA but not N-LidA or C-LidA was efficiently retained by the beads (Fig. 1). Taken together, these studies revealed that LidA binding to Rab6A and -A′ is direct, nucleotide dependent, and mediated by the central region of LidA.
Fig 1.
The central region of LidA binds Rab6A′ in a nucleotide-dependent manner. (Left) Schematic representation of LidA and the truncated variants used in this study. The numbers indicate amino acid residues. (Right) Pulldown of LidA variants by Rab6A′. Purified recombinant full-length LidA or LidA variants were incubated with beads coated with either constitutively active Rab6A′(Q72L) or inactive Rab6A′(T27N), and proteins pelleted by the beads were visualized by SDS-PAGE and Coomassie staining. The gel images are representatives of two repetitions.
Several attempts to identify additional Rab6A/A′ binding partners other than LidA from lysates of L. pneumophila strain Lp02 in pulldown studies were unsuccessful (data not shown). We therefore focused our effort on characterizing the interaction between LidA and Rab6A′ in more detail.
Rab6A′ and Rab1 compete for LidA binding.
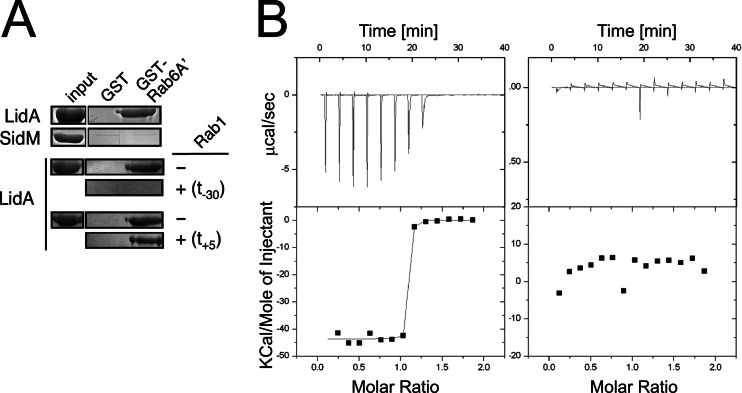
In addition to binding Rab6A′, the central region of LidA also mediates interaction with Rab1 (35). The LidA-Rab1 crystal structure reveals that the middle region of LidA (aa 224 to 559) assumes an extended fold reminiscent of a hand with four fingers that tightly grasp Rab1, explaining the exceptionally high binding affinity between the two proteins (18). To investigate whether Rab6A′ and Rab1 compete for LidA binding, we performed pulldown studies using purified recombinant proteins (Fig. 2A). LidA, but not SidM, was efficiently precipitated by beads coated with GST-Rab6A′(Q72L) but not by beads coated with GST alone. Incubation of full-length LidA with an equimolar amount of Rab1 prior to the pulldown reaction abolished its ability to bind GST-Rab6A′(Q72L)-coated beads, suggesting that Rab1 occupies an epitope on the LidA surface that overlaps with the Rab6A′-LidA interface. When Rab1 was added to a preformed complex of GST-Rab6A′(Q72L) and LidA, it did not reduce coprecipitation of LidA with GST-Rab6A′(Q72L)-coated beads, suggesting that Rab1 cannot efficiently displace Rab6A′ once it is bound by LidA. Similar results were obtained in binding studies using isothermal titration calorimetry (Fig. 2B). Whereas in the absence of Rab6A′ LidA efficiently bound Rab1 in a 1:1 complex (KD, 89.6 ± 1.6 nM [mean ± standard deviation]), preincubation of LidA with a 2-fold molar excess of Rab6A′ blocked any detectable LidA-Rab1 complex formation. Together, these results indicate that LidA interacts either with Rab1 or with Rab6A′ but not with both GTPases simultaneously, most likely because the LidA interface for Rab6A′ binding overlaps with that for Rab1.
Fig 2.
Rab6A′ and Rab1 compete for LidA binding. (A) Rab1 interferes with LidA precipitation by Rab6A′. Agarose beads coated with either GST or GST-Rab6A′(Q72L) were incubated with LidA, and LidA binding to protein-immobilized beads was determined by SDS-PAGE and Coomassie staining. Rab1 was added to LidA either 30 min prior (t = −30 min) or 5 min after (t = +5 min) addition of Rab6A′(Q72L)-coated beads. SidM served as negative control. (B) Rab6A′ prevents Rab1 from binding to LidA. Complex formation between LidA and Rab1 was analyzed by isothermal titration calorimetry in either the absence (left graph) or presence (right graph) of a 2-fold molar excess of Rab6A′. (Top) Raw data for 14 injections of full-length LidA into the isothermal titration calorimetry cell containing wild-type Rab1. Each injection peak corresponds to the heat released during that injection. (Bottom) Scatter plot showing the binding isotherm for the Rab1-LidA interaction, with the integrated heat plotted against the stoichiometry.
LidA interacts with two Rab6A′ molecules.
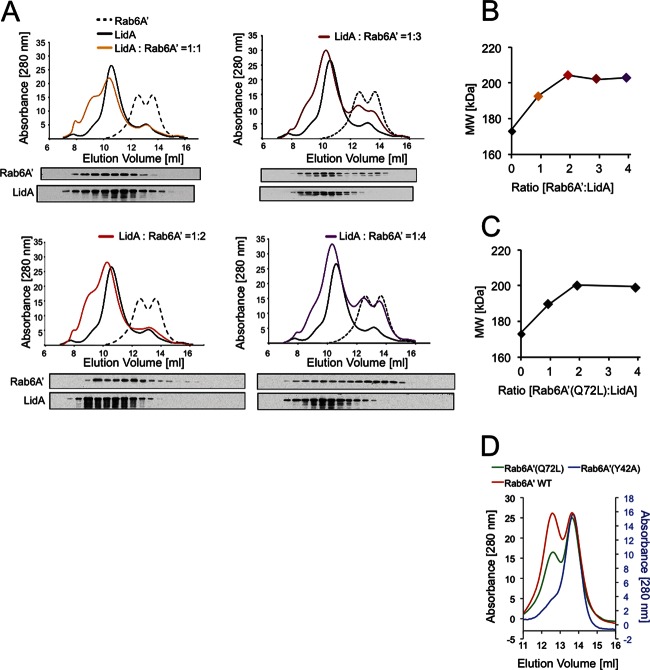
To further analyze the complex formed by LidA and Rab6A′, we performed gel permeation chromatography, which separates proteins and protein complexes based on their molecular masses as well as their topologies. Purified Rab6A′ was separated into two protein fractions with retention coefficients analogous to those of globular proteins of 54.2 kDa and 29.1 kDa, respectively, corresponding to a mixture of Rab6A′ monomer and dimer (Fig. 3A, dashed line). LidA, which has an estimated molecular mass of 83.2 kDa, eluted as a single peak with a retention coefficient equivalent to a 172-kDa protein (Fig. 3A, solid black line), corresponding to either an elongated LidA monomer or a globular LidA dimer. When LidA and Rab6 were incubated at a molar ratio of 1:1, both proteins comigrated in a single peak with a molecular mass of 192 kDa (Fig. 3A), equivalent to a heterodimeric complex of LidA and Rab6A′. No uncomplexed Rab6A′ was detected under these conditions, either by UV absorption or by immunoblotting. Similar results were obtained for a 1:2 molar ratio of LidA to Rab6A′, with no uncomplexed Rab6A′ detectable and a further increase in the size of the LidA-Rab1 complex to 204 kDa (Fig. 3A and B). Only upon addition of a 3-fold or 4-fold molar excess of Rab6A′ relative to LidA did we detect increasing amounts of the uncomplexed form of Rab6A′, both as monomer and dimer (Fig. 3A), while no further increase in the molecular mass of the LidA-Rab6A′ complex was noticeable (Fig. 3B). Densitometry analysis of the protein bands detected by immunoblotting at LidA:Rab6A′ molar ratios of 1:3 and 1:4 confirmed that 21.5% and 43.5% of the total amount of Rab6A′ was present as uncomplexed protein (Fig. 3A), which corresponded well with the 33% and 50% free Rab6A′ anticipated for a 1:2 complex of LidA and Rab6A′. Similar results were obtained when we analyzed complex formation between LidA and constitutively active Rab6A′(Q72L) (Fig. 3C) or Rab6A′(Y42A), a mutant protein that was exclusively monomeric (Fig. 3D), demonstrating that the oligomerization state of Rab6A′ (monomer versus dimer) did not affect the stoichiometry of the complex with LidA.
Fig 3.
LidA binds Rab6A′ with a 1:2 stoichiometry. (A) Gel permeation chromatogram showing LidA-Rab6A′ complex formation. Purified recombinant Rab6A′ and LidA were mixed at the indicated molar ratios and separated by gel filtration. The protein amount detected by absorbance (at 280 nm) is plotted against the elution volume. The amount of LidA and Rab6A′ within each fraction (0.5 ml) was determined by immunoblotting using protein-specific antibodies. The elution profiles of uncomplexed LidA and Rab6A′ are shown in each chromatogram. (B) Summary of the molecular mass of the LidA-Rab6A′ complex shown in panel A, plotted against the molar ratio of LidA to Rab6A′. (C) Results of the same experiment as shown in panel B, showing LidA-Rab6A′(Q72L) complex formation. (D) Gel permeation chromatogram of Rab6A′ variants. Purified recombinant Rab6A′ wild type (WT) or the indicated Rab6A′ point mutants were analyzed by gel permeation chromatography. Proteins within the eluted fractions were detected by absorbance (at a 280-nm wavelength).
LidA interferes with GTP hydrolysis by directly binding to Rab6A′.
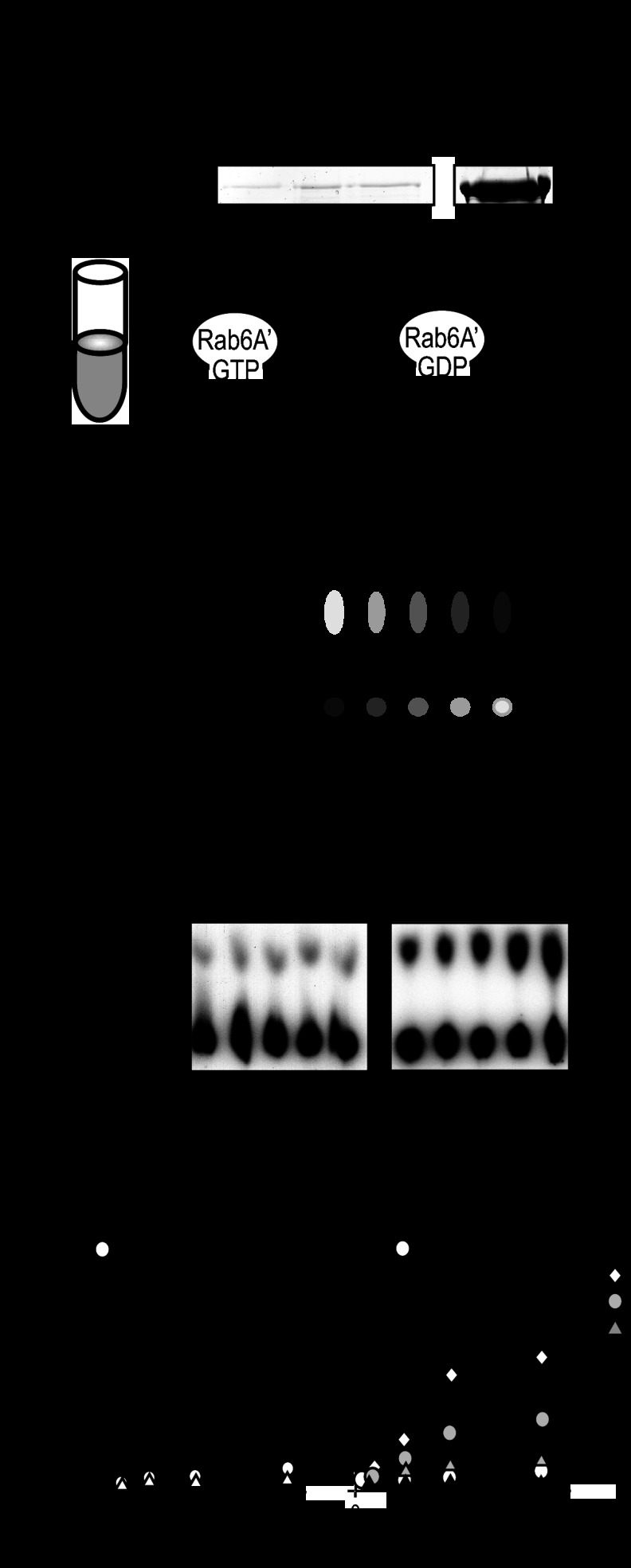
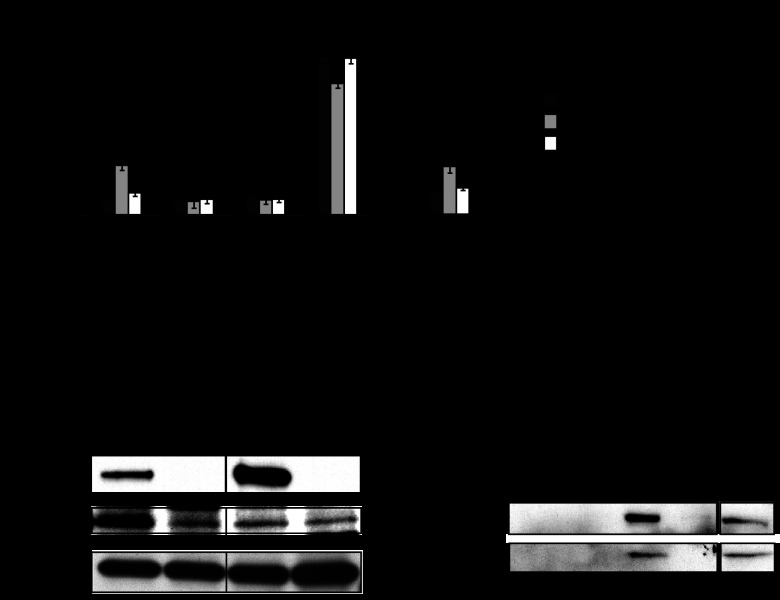
We recently showed that LidA, by binding to active Rab1, interferes with GTP hydrolysis even in the presence of bacterial or host cell GAPs (35). Given that LidA showed a strong preference for the active form of Rab6A′ (Fig. 1A), we investigated if LidA exhibits a similar protective effect toward GTP-Rab6A′. These studies were complicated by the existence of conflicting data about the identity of a mammalian Rab6-specific GAP. While the Goud lab has proposed that TBC1D11 (also known as GapCenA) possesses Rab6-GAP activity (36), Barr and colleagues reported that they found no evidence for GapCenA triggering GTP hydrolysis by Rab6 (37). We amplified the open reading frame encoding GapCenA from human cDNA and purified GapCenA from E. coli. Consistent with the findings of the Barr group, we were unable to detect any stimulating effect of GapCenA on [γ-32P]GTP hydrolysis by Rab6A′ (data not shown). In fact, when we used purified recombinant proteins in pulldown studies, we found no evidence for a direct interaction between His6-GapCenA and Rab6A′ [active (Q72L) or inactive (T27N)] (Fig. 4A), suggesting that GapCenA is not a GAP for Rab6A′ under any of the conditions tested here.
Fig 4.
LidA protects Rab6A′ from GAP-mediated inactivation. (A) GapCenA does not interact with Rab6A′. Beads coated with the indicated Rab6A′ proteins or with GST (control) were incubated with purified recombinant GapCenA, and retention of GapCenA by the beads was determined by SDS-PAGE and Coomassie staining. (B) Schematic representation of the GTP hydrolysis assay developed in this study. Rab6A′ loaded with [α-32P]GTP was incubated with PNS from 293T cell lysate. Samples taken at the indicated time points were denatured in 1% SDS (final concentration), and radiolabeled nucleotides were separated by TLC. (C) Example of TLC plates, showing that PNS but not buffer triggered conversion of [α-32P]GTP to [α-32P]GDP by Rab6A′. (D) Quantification of [α-32P]GDP formation by Rab6A′ incubated with PNS but not by Rab6A′ incubated with buffer or LidA alone. The amount of [α-32P]GTP generated by Rab6A′ in the presence of PNS after 60 min was arbitrarily set to 100%. (E) LidA blocks GTP hydrolysis by Rab6A′ in a dose-dependent manner. [α-32P]GTP-loaded Rab6 was incubated with LidA at the indicated molar ratios and with PNS, and generation of [α-32P]GDP over time was monitored by TLC. The graphs shown in panels D and E are representatives of two independent experiments.
In order to determine the effect of LidA on Rab6A′ inactivation, we developed an alternative GAP assay that utilized PNS from mammalian tissue culture cells rather than purified GAP to stimulate inactivation of Rab6A′. Rab6A′ loaded with [α-32P]GTP (not [γ-32P]GTP) was mixed with either buffer or PNS, and generation of [α-32P]GDP by Rab6A′ was monitored by thin-layer chromatography (Fig. 4B). Incubation of [α-32P]GTP-Rab6A′ with buffer alone showed no significant generation of [α-32P]GDP over time, consistent with Rab6A′ exhibiting extremely low intrinsic GTP hydrolysis activity (38). In contrast, the presence of PNS in the reaction mixture resulted in increased formation of [α-32P]GDP over time (Fig. 4C), demonstrating that the Rab6GAP protein(s) within the lysate triggered conversion of [α-32P]GTP to [α-32P]GDP by Rab6A′. Notably, LidA alone had no effect on GTP hydrolysis by Rab6A′, consistent with LidA lacking Rab6GAP activity (Fig. 4D). We subsequently analyzed the effect of LidA on GAP-mediated inactivation of Rab6A′. [α-32P]GTP-loaded Rab6A′ was incubated with PNS in the absence or presence of LidA, and [α-32P]GTP hydrolysis was monitored over time. Whereas [α-32P]GTP was efficiently hydrolyzed in the absence of LidA, increasing amounts of LidA efficiently blocked [α-32P]GTP hydrolysis by Rab6A′, with complete protection at a LidA to Rab6A′ molar ratio of 1:2 or higher (Fig. 4E), in accordance with LidA forming a 1:2 complex with Rab6A′ (Fig. 3). Thus, the presence of LidA interfered with Rab6A′ inactivation through host cell GAP(s) present in the PNS.
We wanted to determine the molecular mechanism underlying the prevention of [α-32P]GTP hydrolysis by Rab6A′ in the presence of LidA. First, we tested if the inhibitory effect of LidA on [α-32P]GTP hydrolysis by Rab6A′ was caused by a nonproteinaceous factor that may have been copurified along with recombinant LidA from the E. coli lysate. Prior to its incubation with [α-32P]GTP-loaded Rab6A′, we incubated LidA with proteinase K to degrade LidA and any other polypeptide present in the sample. Proteinase K treatment resulted in a complete loss of the inhibitory effect from the LidA sample (Fig. 5A), arguing that the block in [α-32P]GTP hydrolysis by Rab6A′ was caused by LidA rather than a small molecule.
Fig 5.
LidA targets Rab6A′ but not its GAP(s). (A) The inhibitory effect of LidA on Rab6A′ GTP hydrolysis is proteinase K sensitive. LidA pretreated with proteinase K (PK) was tested for its ability to prevent GTP hydrolysis by Rab6A′ in the presence of PNS. Note that pretreatment of PNS with the same amount of proteinase K did not affect its GAP activity. (B) Schematic representation of the experiment quantified in panel C, showing that pretreatment of PNS with LidA-coated beads did not affect the ability of PNS to trigger [α-32P]GTP hydrolysis by Rab6A′. (D) The central region of LidA protects Rab6A′ from inactivation. [α-32P]GTP-loaded Rab6A′ was incubated with PNS and the indicated LidA variants. Generation of [α-32P]GDP after 30 min was determined by TLC. Each graph is representative of two independent experiments, and the amount of [α-32P]GDP is displayed relative to the positive control, which was arbitrarily set to 100%.
We proceeded to further dissect the mechanism of LidA-mediated inhibition of GTP hydrolysis by Rab6A′. We envisioned two possible scenarios: one in which LidA binding to Rab6A′ sterically hinders potential GAPs from gaining access to the nucleotide binding pocket of Rab6A′, and another scenario where LidA directly binds to or modifies the GAP(s) present in the PNS to prevent Rab6A′ inactivation. To distinguish between these two possibilities, we designed an experiment in which PNS, prior to its incubation with [α-32P]GTP-loaded Rab6A′, was preincubated with LidA-coated beads (Fig. 5B). If LidA directly targets and inactivats the GAP(s), pretreatment with LidA should result in a reduction or loss of the Rab6A′ inactivation activity from PNS. We found that neither incubation with control beads nor treatment with LidA-coated beads had a detectable effect on the ability of PNS to trigger conversion of [α-32P]GTP to [α-32P]GDP by Rab6A′ (Fig. 5C), showing that the Rab6GAP activity had not been depleted from PNS through the exposure to LidA. We also analyzed the truncated variants of LidA for their abilities to interfere with [α-32P]GTP hydrolysis by Rab6A′. We found that preincubation of Rab6A′ with M-LidA, the region sufficient for Rab6A′ binding, inhibited [α-32P]GTP hydrolysis by Rab6A′ to a degree similar to that of the full-length form of LidA (Fig. 5D). N-LidA or C-LidA, on the other hand, had no effect on [α-32P]GDP generation by Rab6A′ in the presence of PNS, consistent with their inabilities to interact with this GTPase (Fig. 1). Thus, our findings strongly favor the hypothesis that the inhibitory effect on GTP hydrolysis is caused by direct binding of the central region of LidA to Rab6A′.
Rab6 is required for efficient intracellular replication of L. pneumophila within human macrophages.
The ability of LidA to interfere with the inactivation of Rab6A′ in vitro suggested that active Rab6 plays an important role for L. pneumophila intracellular replication. To address this question, we analyzed the ability of L. pneumophila to form large replication vacuoles in transiently transfected U937 macrophages producing GFP-tagged versions of Rab6A′ (Fig. 6A). Quantitative analysis of the number of bacteria per vacuole by immunofluorescence microscopy revealed that in the presence of either GFP-tagged Rab6A′ or GFP alone, L. pneumophila Lp02 intracellular replication appeared normal, with more than 70% large replication vacuoles and only 8.6% ± 3% (GFP; mean ± standard deviation) and 7.8% ± 3% (Rab6A′ wild type) vacuoles containing one to two bacteria, respectively. Similar results were observed in cells overproducing constitutively active Rab6A′(Q72L). In contrast, growth of L. pneumophila Lp02 in cells overproducing constitutively inactive GFP-Rab6A′(T27N) was significantly attenuated, with 23% ± 3% of vacuoles containing a single bacterium. This effect of Rab6A′(T27N) could not be attributed to secondary effects, because the previously described growth defect of a ΔlidA mutant (17) was not further exacerbated by the presence of Rab6A′(T27N) (Fig. 6A). In fact, growth of the ΔlidA strain was slightly improved in cells producing either the wild-type or active form of Rab6A′, compared to growth in control cells producing GFP (Fig. 6A), indicating that the availability of an excess amount of Rab6A′ can compensate for the absence of LidA. Notably, the number of intermediate-sized vacuoles containing 3 to 11 Lp02 bacteria did not significantly change in cells producing GFP-Rab6A′(T27N) versus GFP, indicating that the lack of Rab6A′ function affected the ability of bacteria to initiate rather than maintain replication within the LCV. Thus, active Rab6A′ plays an important role for L. pneumophila during the early stage of its intracellular replication cycle.
Fig 6.
Rab6 is required for efficient intracellular replication of L. pneumophila. (A) U937 cells transiently transfected with plasmids carrying the indicated Rab6A′ protein gene were challenged for 1 h with either L. pneumophila Lp02 or a lidA deletion strain at an MOI of 5. Extracellular bacteria were removed, and cells were incubated for an additional 12 h at 37°C. The efficiency of L. pneumophila replication vacuole formation was determined by counting the number of bacteria in at least 100 vacuoles. Results represent the means ± standard deviations of three independent experiments. ***, P < 0.001; **, P < 0.01 (t test). (B) Coprecipitation of Rab6A/A′ and LidA from L. pneumophila-infected cells. Human U937 macrophages were challenged with L. pneumophila Lp02 (WT) or a lidA deletion strain (ΔlidA). After 2 h or 6 h, the cells were mechanically lysed, LidA was precipitated using anti-LidA antibody-coated beads, and the amount of Rab6A/A′ bound to the beads was detected by immunoblotting using antibody directed against Rab6A/A′. The bottom panel shows the total amount of Rab6A/A′ within the PNS. Images are representative of two independent experiments. (C) LidA interferes with binding of cellular ligands to Rab6A′. Uncoated control beads or beads coated with either GST-Rab6A′(T27N) or GST-Rab6A′(Q72L) were incubated with postnuclear supernatant from transiently transfected 293T cells producing GFP-tagged R6IP1A or R6IP2B. Beads were washed, and proteins retained by the beads were detected by immunoblotting using anti-GFP antibody. Each panel is representative of three independent experiments.
To confirm that LidA and Rab6A′ interact during host cell infection, we tested if endogenous Rab6A′ could be coimmunoprecipitated with LidA from infected host cells. Human U937 macrophages challenged with L. pneumophila Lp02 or a ΔlidA strain were mechanically lysed, LidA was pelleted using beads coated with anti-LidA antibody, and the amount of Rab6A′ coprecipitated by the beads was determined by immunoblotting (Fig. 6B). Rab6A′ was enriched in the pulldown assay from the lysate of cells infected with L. pneumophila Lp02 but not that from cells challenged with the ΔlidA strain, consistent with LidA directly targeting host cell Rab6A′. At 6 h postinfection, Rab6A′ was not enriched in either pulldown fraction, even though the amount of LidA precipitated from Lp02-infected cell lysate had further increased relative to that at the 2-h time point, demonstrating that complex formation was not a consequence of the cell lysis procedure. These results demonstrate that LidA interacts with host cell Rab6A′ preferentially at the early stage of infection.
We recently found that LidA competes with cellular Rab1 ligands, such as p115 or GM130, for Rab1 binding, arguing for a role of LidA in restricting Rab1 interactions with host proteins (35). Given that LidA binds active Rab6A′, we determined if LidA also blocked interaction of Rab6A′ with its downstream effectors. As expected, beads coated with Rab6A′(Q72L) efficiently precipitated GFP-tagged R6IP1A and R6IP2B from lysates of transiently transfected 293T cells, whereas Rab6A′(T27N) did not (Fig. 6C). Upon preincubation of Rab6A′(Q72L) with a molar excess of LidA, neither R6IP1A nor R6IP2B was detectable in the bead fraction, demonstrating that LidA blocked Rab6A′ binding to these cellular interaction partners. Taken together, these results suggest that LidA is important for maintaining an active pool of Rab6A′ within infected cells and for controlling the interactions of Rab6A′ with cellular ligands.
DISCUSSION
LidA was among the first effector proteins reported to be delivered into the host cell by the L. pneumophila Dot/Icm T4SS (15). Subsequent studies revealed that it targets host vesicle transport GTPases of the Rab family, such as Rab1, Rab6, and Rab8 (17, 18). We and others recently described the interaction between LidA and Rab1, a GTPase that regulates ER-to-Golgi complex vesicle trafficking (18, 35). In the present study, we focused on the analysis of Rab6A′ binding to LidA and on determining the role of Rab6 for replication vacuole formation by L. pneumophila. Not surprisingly, we discovered several similarities in the interplay of LidA with either Rab6A′ or with Rab1, but also significant differences, as discussed below.
Using purified recombinant proteins in pulldown studies, we found that the interaction between LidA and Rab6A′ was direct and nucleotide dependent, with a strong preference of LidA for the constitutively active form of Rab6A′ (Fig. 1). The ability of LidA to distinguish the active from the inactive form of Rab6A′ is a distinct feature, compared to the interaction with Rab1, which is recognized by LidA in a nucleotide-independent manner (17). A recent study using a truncated version of Rab6A (not A′) found results similar to ours (39), although those authors reported that binding of LidA to the inactive form of Rab6A was reduced but not undetectable. It has been described that the isoforms Rab6A and Rab6A′, although closely related, exhibit several functional differences and that their roles are not entirely redundant. For instance, Rab6A but not Rab6A′ was found to interact with Rabkinesin-6, a Golgi complex-associated Rab6 ligand, and only Rab6A causes redistribution of Golgi complex-resident proteins into the ER when overproduced as constitutively active form in HeLa cells (21). Another study reported that depletion of Rab6A′, but not Rab6A, through RNA interference disturbed retrograde trafficking as well as cell cycle progression (20), thus emphasizing the functional specificity of each isoform. Although LidA interacts with both isoforms in vitro, it is currently unknown whether Rab6A′, Rab6A, or both represent a target for LidA during L. pneumophila infection and which of the processes that they regulate might contribute to L. pneumophila replication vacuole formation.
Our gel permeation chromatography analysis found that LidA forms a 1:2 complex with Rab6A′ (Fig. 3). To our knowledge, this has not been observed for any other known bacterial or mammalian Rab ligand, including the complex of LidA with Rab1 or Rab8, and it is a somewhat surprising finding. Rab6IP1 has been shown to bind GTP-Rab6A with a 1:1 stoichiometry (40), whereas GCC185 forms a 2:2 complex with Rab6A(Q72L) (41). A 1:2 stoichiometry suggests either that LidA possesses two separate Rab6A′ binding surfaces or that LidA interacts with a Rab6A′ dimer or stimulates its formation. Although wild-type Rab6A′ appeared to exist as a mixture of monomer and dimer in solution (Fig. 3A and D), we never observed a LidA-Rab6A′ stoichiometry above 1:2 (Fig. 3B), arguing that LidA binds either a dimer or two monomers of Rab6A′ but not two dimers (which would result in a stoichiometry of 1:4). Although the structure of Rab6A′ in complex with LidA is not yet known, it is reasonable to assume that one Rab6A′ molecule is bound in a manner very similar to that described for Rab1 and Rab8 (18, 39). In support of this hypothesis, we found that Rab6A′ competed with Rab1 for binding to LidA in vitro (Fig. 2), suggesting that both GTPases have overlapping binding sites on LidA. Given that only Rab1 and not Rab6A′ localizes to LCVs, we do not anticipate that these two GTPases compete for LidA binding in vivo. Also, we can only speculate about the mechanism of how the second Rab6A′ monomer is bound by LidA. A truncated version of Rab6A comprising only the GTPase domain (residues 13 to 174) was reported to exist as a dimer of dimers within the asymmetric unit of protein crystals (38). Although protein contacts within crystals do not necessarily represent interactions that are relevant for the function of a protein, it cannot be excluded that under certain conditions or upon binding to a particular ligand, in this case L. pneumophila LidA, Rab6A′ forms dimers. We also found that LidA interfered with inactivation of not just one but two Rab6A′ molecules simultaneously, further indicating that binding of the second Rab6A′ molecule is not random but rather coordinated to achieve the same protective effect. It is noteworthy that the crystal structures of the central region of LidA in complex with either Rab8 or Rab1 offered no definite clues about the in vivo function of LidA beyond what had already been reported (15–17). It remains to be seen if the structure and function of the N- and C-terminal domains of LidA can provide a more detailed insight into the activity of this effector and their contribution to Rab GTPase manipulation by L. pneumophila.
Interference with Rab6A′ function by overproduction of GFP-Rab6A′(T27N) in transiently transfected U937 macrophages attenuated L. pneumophila's capability to efficiently initiate intracellular replication (Fig. 6), pointing toward an important role of the Rab6A′-regulated pathway(s) for bacterial virulence. It is unlikely that this phenomenon was caused by Rab6A′(T27N) indirectly affecting interconnecting pathways, such as the early secretory trafficking route. Although Rab6(T27N) reduced retrograde cargo flow from the Golgi compartment to the ER, this pathway is not considered essential in eukaryotic cells. Rab6(Q72L), on the other hand, reduced vesicle flow through the secretory pathway, which is considered essential in eukaryotic cells, yet this Rab6A′ mutant did not cause a measurable reduction in L. pneumophila growth, suggesting that the inability of L. pneumophila to efficiently initiate replication vacuole formation was a direct consequence of the absence of active Rab6A′ rather than an indirect one due to compromised host cell viability. Another possibility is that overproduction of a recombinant Rab protein in eukaryotic cells affects the steady state of endogenous Rab proteins by competing with them for binding to the chaperone GDI. A recent report described a similar phenomenon for Rho GTPases (42). Like Rab proteins, Rho GTPases require a chaperone (RhoGDI) to cycle from the membrane-bound form to the cytosolic pool. Overproduction of an exogenous Rho protein depleted the pool of RhoGDI and caused competition among endogenous Rho GTPases for RhoGDI binding, which resulted in misfolding and increased degradation. Although we cannot exclude the possibility that overproduction of Rab6A′(T27N) altered the homeostasis of other Rab proteins within transfected cells by depleting RabGDI, we noticed no correlation between the level of GFP-Rab6A′(T27N) protein and the frequency in failure of L. pneumophila to initiate replication within those cells (data not shown), arguing against a role for Rab protein cross talk in the growth experiment (Fig. 6A).
Given that Rab6A′ is involved in several trafficking processes, we can only speculate about how and where LidA may exploit Rab6A′ in order to promote efficient intracellular multiplication of L. pneumophila. Based on the results of both the infectious center and coprecipitation assays (Fig. 6B and C), we hypothesize that LidA targets Rab6A′ early during infection. It is, however, unclear what portion of the cellular pool of Rab6A′ is affected by LidA and at what subcellular location. LidA has been reported to be most abundant at the LCV early during infection, yet it cannot be excluded that its dissemination to other locations within the infected cell occurs immediately after bacterial uptake but that its levels distal to the LCV remain below the microscopic detection limit for several hours. We do believe that the function of Rab6A′ is not redundant with that of Rab1, since we found no evidence for Rab6A′ accumulation on the LCV (data not shown), making a role for this GTPase in the recruitment of host cell vesicles to the LCV unlikely. It is also unclear if LidA exhibits a stimulating or attenuating effect on the pathway(s) regulated by Rab6A′. On one hand, LidA seems to play a promoting role in maintaining an active pool of Rab6A′ by preventing its inactivation by GAPs (Fig. 4). On the other hand, LidA binds Rab6A′ with unusually high affinity (18, 39) and competes with cellular Rab6A′ ligands for binding (Fig. 6B), which would argue for an inhibitory role of LidA on Rab6A′-controlled processes. Further studies will be needed to determine the precise time and intracellular localization at which LidA interacts with Rab6A′ during infection and how the Rab6A′-regulated cellular pathway(s) supports L. pneumophila infection.
ACKNOWLEDGMENTS
We thank H. Levin, G. Storz, and R. Hegde for insightful discussion and M. R. Neunuebel, K. Decker, and Y. Lin for comments on the manuscript.
Work in the laboratory of M. Machner is supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
Footnotes
Published ahead of print 8 April 2013
REFERENCES
- 1. Fraser DW, Tsai TR, Orenstein W, Parkin WE, Beecham HJ, Sharrar RG, Harris J, Mallison GF, Martin SM, McDade JE, Shepard CC, Brachman PS. 1977. Legionnaires' disease: description of an epidemic of pneumonia. N. Engl. J. Med. 297:1189–1197 [DOI] [PubMed] [Google Scholar]
- 2. McDade JE, Shepard CC, Fraser DW, Tsai TR, Redus MA, Dowdle WR. 1977. Legionnaires' disease: isolation of a bacterium and demonstration of its role in other respiratory disease. N. Engl. J. Med. 297:1197–1203 [DOI] [PubMed] [Google Scholar]
- 3. Horwitz MA. 1983. Formation of a novel phagosome by the Legionnaires' disease bacterium (Legionella pneumophila) in human monocytes. J. Exp. Med. 158:1319–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Segal G, Shuman HA. 1998. Intracellular multiplication and human macrophage killing by Legionella pneumophila are inhibited by conjugal components of IncQ plasmid RSF1010. Mol. Microbiol. 30:197–208 [DOI] [PubMed] [Google Scholar]
- 5. Vogel JP, Andrews HL, Wong SK, Isberg RR. 1998. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279:873–876 [DOI] [PubMed] [Google Scholar]
- 6. Derre I, Isberg RR. 2004. Legionella pneumophila replication vacuole formation involves rapid recruitment of proteins of the early secretory system. Infect. Immun. 72:3048–3053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kagan JC, Stein MP, Pypaert M, Roy CR. 2004. Legionella subvert the functions of Rab1 and Sec22b to create a replicative organelle. J. Exp. Med. 199:1201–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Swanson MS, Isberg RR. 1995. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect. Immun. 63:3609–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nagai H, Kagan JC, Zhu X, Kahn RA, Roy CR. 2002. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 295:679–682 [DOI] [PubMed] [Google Scholar]
- 10. Machner MP, Isberg RR. 2007. A bifunctional bacterial protein links GDI displacement to Rab1 activation. Science 318:974–977 [DOI] [PubMed] [Google Scholar]
- 11. Murata T, Delprato A, Ingmundson A, Toomre DK, Lambright DG, Roy CR. 2006. The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nat. Cell Biol. 8:971–977 [DOI] [PubMed] [Google Scholar]
- 12. Muller MP, Peters H, Blumer J, Blankenfeldt W, Goody RS, Itzen A. 2010. The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science 329:946–949 [DOI] [PubMed] [Google Scholar]
- 13. Neunuebel MR, Chen Y, Gaspar AH, Backlund PS, Jr, Yergey A, Machner MP. 2011. De-AMPylation of the small GTPase Rab1 by the pathogen Legionella pneumophila. Science 333:453–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tan Y, Luo ZQ. 2011. Legionella pneumophila SidD is a deAMPylase that modifies Rab1. Nature 475:506–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Conover GM, Derre I, Vogel JP, Isberg RR. 2003. The Legionella pneumophila LidA protein: a translocated substrate of the Dot/Icm system associated with maintenance of bacterial integrity. Mol. Microbiol. 48:305–321 [DOI] [PubMed] [Google Scholar]
- 16. Derre I, Isberg RR. 2005. LidA, a translocated substrate of the Legionella pneumophila type IV secretion system, interferes with the early secretory pathway. Infect. Immun. 73:4370–4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Machner MP, Isberg RR. 2006. Targeting of host Rab GTPase function by the intravacuolar pathogen Legionella pneumophila. Dev. Cell 11:47–56 [DOI] [PubMed] [Google Scholar]
- 18. Cheng W, Yin K, Lu D, Li B, Zhu D, Chen Y, Zhang H, Xu S, Chai J, Gu L. 2012. Structural insights into a unique Legionella pneumophila effector LidA recognizing both GDP and GTP bound Rab1 in their active state. PLoS Pathog. 8:e1002528 doi:10.1371/journal.ppat.1002528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Urwyler S, Nyfeler Y, Ragaz C, Lee H, Mueller LN, Aebersold R, Hilbi H. 2009. Proteome analysis of Legionella vacuoles purified by magnetic immunoseparation reveals secretory and endosomal GTPases. Traffic 10:76–87 [DOI] [PubMed] [Google Scholar]
- 20. Del Nery E, Miserey-Lenkei S, Falguieres T, Nizak C, Johannes L, Perez F, Goud B. 2006. Rab6A and Rab6A′ GTPases play non-overlapping roles in membrane trafficking. Traffic 7:394–407 [DOI] [PubMed] [Google Scholar]
- 21. Echard A, Opdam FJ, de Leeuw HJ, Jollivet F, Savelkoul P, Hendriks W, Voorberg J, Goud B, Fransen JA. 2000. Alternative splicing of the human Rab6A gene generates two close but functionally different isoforms. Mol. Biol. Cell 11:3819–3833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Girod A, Storrie B, Simpson JC, Johannes L, Goud B, Roberts LM, Lord JM, Nilsson T, Pepperkok R. 1999. Evidence for a COP-I-independent transport route from the Golgi complex to the endoplasmic reticulum. Nat. Cell Biol. 1:423–430 [DOI] [PubMed] [Google Scholar]
- 23. Hill E, Clarke M, Barr FA. 2000. The Rab6-binding kinesin, Rab6-KIFL, is required for cytokinesis. EMBO J. 19:5711–5719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Miserey-Lenkei S, Couedel-Courteille A, Del Nery E, Bardin S, Piel M, Racine V, Sibarita JB, Perez F, Bornens M, Goud B. 2006. A role for the Rab6A′ GTPase in the inactivation of the Mad2-spindle checkpoint. EMBO J. 25:278–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Grigoriev I, Splinter D, Keijzer N, Wulf PS, Demmers J, Ohtsuka T, Modesti M, Maly IV, Grosveld F, Hoogenraad CC, Akhmanova A. 2007. Rab6 regulates transport and targeting of exocytotic carriers. Dev. Cell 13:305–314 [DOI] [PubMed] [Google Scholar]
- 26. Miserey-Lenkei S, Chalancon G, Bardin S, Formstecher E, Goud B, Echard A. 2010. Rab and actomyosin-dependent fission of transport vesicles at the Golgi complex. Nat. Cell Biol. 12:645–654 [DOI] [PubMed] [Google Scholar]
- 27. Reference deleted.
- 28. Reference deleted.
- 29. Rejman Lipinski A, Heymann J, Meissner C, Karlas A, Brinkmann V, Meyer TF, Heuer D. 2009. Rab6 and Rab11 regulate Chlamydia trachomatis development and golgin-84-dependent Golgi fragmentation. PLoS Pathog. 5:e1000615 doi:10.1371/journal.ppat.1000615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rzomp KA, Scholtes LD, Briggs BJ, Whittaker GR, Scidmore MA. 2003. Rab GTPases are recruited to chlamydial inclusions in both a species-dependent and species-independent manner. Infect. Immun. 71:5855–5870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Feeley JC, Gibson RJ, Gorman GW, Langford NC, Rasheed JK, Mackel DC, Baine WB. 1979. Charcoal-yeast extract agar: primary isolation medium for Legionella pneumophila. J. Clin. Microbiol. 10:437–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gabay JE, Horwitz MA. 1985. Isolation and characterization of the cytoplasmic and outer membranes of the Legionnaires' disease bacterium (Legionella pneumophila). J. Exp. Med. 161:409–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Berger KH, Isberg RR. 1993. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol. Microbiol. 7:7–19 [DOI] [PubMed] [Google Scholar]
- 34. Berger KH, Isberg RR. 1994. Intracellular survival by Legionella. Methods Cell Biol. 45:247–259 [DOI] [PubMed] [Google Scholar]
- 35. Neunuebel MR, Mohammadi S, Jarnik M, Machner MP. 2012. Legionella pneumophila LidA affects nucleotide binding and activity of the host GTPase Rab1. J. Bacteriol. 194:1389–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cuif MH, Possmayer F, Zander H, Bordes N, Jollivet F, Couedel-Courteille A, Janoueix-Lerosey I, Langsley G, Bornens M, Goud B. 1999. Characterization of GAPCenA, a GTPase activating protein for Rab6, part of which associates with the centrosome. EMBO J. 18:1772–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fuchs E, Haas AK, Spooner RA, Yoshimura S, Lord JM, Barr FA. 2007. Specific Rab GTPase-activating proteins define the Shiga toxin and epidermal growth factor uptake pathways. J. Cell Biol. 177:1133–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bergbrede T, Pylypenko O, Rak A, Alexandrov K. 2005. Structure of the extremely slow GTPase Rab6A in the GTP bound form at 1.8 Å resolution. J. Struct. Biol. 152:235–238 [DOI] [PubMed] [Google Scholar]
- 39. Schoebel S, Cichy AL, Goody RS, Itzen A. 2011. Protein LidA from Legionella is a Rab GTPase supereffector. Proc. Natl. Acad. Sci. U. S. A. 108:17945–17950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Recacha R, Boulet A, Jollivet F, Monier S, Houdusse A, Goud B, Khan AR. 2009. Structural basis for recruitment of Rab6-interacting protein 1 to Golgi via a RUN domain. Structure 17:21–30 [DOI] [PubMed] [Google Scholar]
- 41. Burguete AS, Fenn TD, Brunger AT, Pfeffer SR. 2008. Rab and Arl GTPase family members cooperate in the localization of the golgin GCC185. Cell 132:286–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Boulter E, Garcia-Mata R, Guilluy C, Dubash A, Rossi G, Brennwald PJ, Burridge K. 2010. Regulation of Rho GTPase crosstalk, degradation and activity by RhoGDI1. Nat. Cell Biol. 12:477–483 [DOI] [PMC free article] [PubMed] [Google Scholar]