Abstract
Desulfovibrio species are Gram-negative anaerobic sulfate-reducing bacteria that colonize the human gut. Recently, Desulfovibrio spp. have been implicated in gastrointestinal diseases and shown to stimulate the epithelial immune response, leading to increased production of inflammatory cytokines by macrophages. Activated macrophages are key cells of the immune system that impose nitrosative stress during phagocytosis. Hence, we have analyzed the in vitro and in vivo responses of Desulfovibrio vulgaris Hildenborough to nitric oxide (NO) and the role of the hybrid cluster proteins (HCP1 and HCP2) and rubredoxin oxygen oxidoreductases (ROO1 and ROO2) in NO protection. Among the four genes, hcp2 was the gene most highly induced by NO, and the hcp2 transposon mutant exhibited the lowest viability under conditions of NO stress. Studies in murine macrophages revealed that D. vulgaris survives incubation with these phagocytes and triggers NO production at levels similar to those stimulated by the cytokine gamma interferon (IFN-γ). Furthermore, D. vulgaris hcp and roo mutants exhibited reduced viability when incubated with macrophages, revealing that these gene products contribute to the survival of D. vulgaris during macrophage infection.
INTRODUCTION
Desulfovibrio spp. are anaerobic sulfate-reducing bacteria (SRB) that occur in several environmental niches, such as marine and freshwater sediments, as well as in humans as part of the normal oral cavity and gut flora. In particular, four Desulfovibrio spp., namely, D. fairfieldensis, D. desulfuricans, D. piger, and D. vulgaris, were detected in healthy humans (1–3). Furthermore, Desulfovibrio spp. have also been implicated in gastrointestinal diseases, such as inflammatory bowel diseases and periondontitis, since Desulfovibrio strains were isolated from biopsy specimens of patients with ulcerative colitis, brain, abdominal wall, and liver abscesses, and appendicitis (4–6).
Recently, D. desulfuricans and D. fairfieldensis were shown to be able to invade nonprofessional phagocytic cells such as the oral epithelial cells and to stimulate the epithelial immune response by increasing the production of inflammatory interleukins (7). Nonetheless, the ability of Desulfovibrio spp. to survive professional phagocytes, such as macrophages, remains to be evaluated.
Two of the main weapons of the innate immune system to eradicate pathogens are the generation of reactive oxygen species (ROS) and the generation of reactive nitrogen species (RNS), which are derived from the superoxide and nitric oxide produced by the NADPH oxidase and the mammalian inducible nitric oxide synthase (iNOS), respectively (8, 9). These chemicals can inflict serious damage in bacteria, which employ the expression of several detoxification systems to avoid such damage (9). The flavodiiron proteins (FDP) constitute a large family of enzymes widespread among archaea and bacteria, including in Desulfovibrio spp. They are believed to contribute to bacterial survival under oxidative and nitrosative stress conditions (10). FDPs are homodimeric proteins, with each monomer formed by a flavodoxin-like domain, containing a FMN cofactor, and a β-lactamase-like domain, harboring a diiron center (11). The first Desulfovibrio FDP to be studied was that of D. gigas, namely, the rubredoxin oxygen oxidoreductase (ROO) (12), which was shown to reduce dioxygen to water with electrons from rubredoxin (13). Since subsequent studies reported that several prokaryotic FDPs have significant nitric oxide reductase activity, FDPs are currently believed to be either oxygen or NO reductases or even to be bifunctional (10, 14, 15).
The hybrid cluster proteins (HCPs) constitute another family of bacterial proteins proposed to protect against ROS and RNS toxicity. HCPs contain two redox-active iron-sulfur clusters, namely, a canonical [4Fe-4S]2+/1+ or [2Fe-2S]2+/1+ cluster and a hybrid iron-sulfur-oxygen cluster [4Fe-2S-2O] (16, 17). Previous work reported that an Escherichia coli strain mutated in the hcp gene has lower resistance to hydrogen peroxide and S-nitrosoglutathione (GSNO) (18–20). Furthermore, the recombinant E. coli HCP exhibited hydrogen peroxide and hydroxylamine reductase activities, the latter being also described for the HCPs of Rhodobacter capsulatus and Pyrococcus furiosus (18, 21–23). Hence, HCPs seem also to have more than one enzymatic function.
Like many other Desulfovibrio spp. with known genome sequences, D. vulgaris Hildenborough contains two homologues of the FDPs (ROO1 and ROO2) and of HCPs (HCP1 and HCP2). The genes encoding ROO1 (DVU2014) and HCP1 (DVU2013) are adjacent within a genomic island (24), while the genes encoding ROO2 (DVU3185) and HCP2 (DVU2543) are located elsewhere and are separated in the genome. In Desulfovibrio spp., these proteins are proposed to promote survival in oxygenated environments and to remove RNS generated by nitrite reduction (24–28).
Although several reports have implicated Desulfovibrio spp. in infectious processes, the behavior of these bacteria when they are contacting cells of the immune system such as macrophages has not yet been evaluated. Since infected macrophages produce NO that contributes to eradication of pathogens (9), in this work we started by analyzing the expression of the four D. vulgaris Hildenborough hcp and roo genes under conditions of in vitro NO stress. The phenotype and the NO consumption activity of the D. vulgaris wild-type strain and the transposon mutants with inactivated roo and hcp genes were evaluated in cells exposed to NO donors. Moreover, we also tested the ability of these strains to survive exposure to macrophages.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
D. vulgaris Hildenborough strains used in this study are listed in Table 1. All Desulfovibrio strains were grown anaerobically, at 37°C, in Wall LS4 medium (29). For phenotype assays, cells were grown anaerobically until the stationary phase (approximately 48 h), collected by centrifugation, diluted in fresh medium to an optical density at 600 nm (OD600) of ∼0.2, and further incubated until they reached the early exponential-growth phase (OD600, ∼0.3). At this stage, cells were left untreated or exposed to the NO releaser dipropylenetriamine NONOate (DPTA NONOate) (Cayman) (100 μM; half-life of 3 h at 37°C), and growth was monitored for 8 h.
Table 1.
D. vulgaris Hildenborough strains used in this study
| Strain | Description | Source |
|---|---|---|
| Wild type | D. vulgaris ATCC 29579 | ATCC |
| GZ6896 | hcp1-398::Tn5-RL27; insertion at bp 398/1662 for the gene; Kmra | J. D. Wall Laboratory |
| GZ11714 | hcp2-173::Tn5-RL27; insertion at bp 173/1620 for the gene; Kmr | J. D. Wall Laboratory |
| GZ2505 | roo1-164::Tn5-RL27; insertion at bp 164/1176 for the gene; Kmr | J. D. Wall Laboratory |
| GZ14874 | roo2-134::Tn5-RL27; insertion at bp 134/1209 for the gene; Kmr | J. D. Wall Laboratory |
Kmr, kanamycin resistance.
Quantitative real-time PCR analysis.
The D. vulgaris wild-type strain was grown anaerobically, at 37°C in Wall LS4 medium, until an OD600 of ∼0.3 was reached and was then treated with an NO releaser (100 μM). Cells were then exposed during 1 h to the fast releaser spermine NONOate (Sigma), which decomposes with a half-life of 39 min, at a temperature of 37°C. To analyze the gene transcription of D. vulgaris exposed to NO for 4 h, a slower NO releaser was used, namely, the DPTA NONOate. Total RNA was isolated with an RNeasy Minikit (Qiagen) and quantified in a Nanodrop ND-100 spectrophotometer (NanoDrop Technologies), and its integrity was confirmed by gel agarose electrophoresis. cDNA was synthesized from 2 μg total RNA with a Transcriptor High Fidelity cDNA synthesis kit following the manufacturer's protocol (Roche Applied Science). Quantitative real-time PCRs were performed in a LightCycler instrument according to the instructions provided with a LightCycler FastStart DNA Master SYBR green I kit (Roche Applied Science). The amplification reactions were carried out with equal amounts of cDNA (10 ng) as the initial template, together with the specific pair of oligonucleotides, which were designed to amplify an internal region of 200 to 300 bp for each target gene (Table 2). The ratio of the expression of the target gene to that of a D. vulgaris 16S rRNA reference gene whose transcription remains unchanged under all tested conditions was determined. Quantitative real-time PCR experiments were performed for two biologically independent samples that were assayed in triplicate.
Table 2.
Oligonucleotides used in this study
| Protein | Locus/gene name | Oligonucleotide sequence |
|---|---|---|
| HCP1 | DVU2013/hcp1 | Fw: 5′-GAACCCCGGCATCCTCATC |
| Rv: 5′-GGATGGGGCCGTTGAAGG | ||
| HCP2 | DVU2543/hcp2 | Fw: 5′-GGCGCTTCAGGACCTCACCATC |
| Rv: 5′-CTGTGCCACCAGCCCGTCG | ||
| ROO1 | DVU2014/roo1 | Fw: 5′-GGGTACATGAAGCGGCAAAACG |
| Rv: 5′-CGAAGGGAAAGGCCACCAGG | ||
| ROO2 | DVU3185/roo2 | Fw: 5′-CCTGCCCGAACTGATAGCCC |
| Rv: 5′-GCGTAGCGTTCGGTGGAGG | ||
| 16S rRNA | Dv16S/rrs | Fw: 5′-CCTAGGGCTACACACGTACTACAA |
| Rv: 5′-GAGCATGCTGATCTCGAATTACTA |
NO consumption assays.
Wild-type and mutant strains were grown anaerobically in Wall LS4 medium. When the cultures reached an OD600 of ∼0.3, the cells were left untreated or exposed to 100 μM DPTA NONOate for 4 h. D. vulgaris lysates were prepared by incubating the cells for 15 min with 0.1 mg lysozyme/ml and 0.01% (wt/vol) sodium deoxycholate. Assays were carried out anaerobically, at room temperature, in phosphate-buffered saline (PBS) buffer supplemented with 20 mM glucose, 130 U catalase/ml, 17 U glucose oxidase/ml, 0.2 mM NADPH, 0.2 mM NADH, and 4 to 6 μM NO. NO was added to the samples by means of injection of an appropriate volume of a 2 mM NO-saturated water solution, prepared as previously described (30). Upon addition of the cell lysates, the NO consumption rate was monitored amperometrically with an NO electrode (ISO-NOP) connected to an APOLLO-4000 free radical analyzer (WPI-Europe). Two biological samples were assayed in triplicate.
Macrophage assays and determination of nitrite.
RAW264.7 murine macrophages (ATCC Tib71) were inoculated (5 × 105 cells/ml) in 24-well plates containing Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (vol/vol) fetal bovine serum (Gibco), 70 U penicillin/ml, and 70 μg streptomycin/ml (Gibco) antibiotics and cultured for 3 h, at 37°C, in a 5% CO2 air atmosphere. Prior to infection, macrophages were activated for 12 h with 0.3 μg/ml gamma interferon (IFN-γ) (Sigma). When required, 400 μM NG-monomethyl-l-arginine acetate salt (L-NMMA; Sigma) was added to inhibit the activity of the murine iNOS. Bacterial suspensions of D. vulgaris wild-type and mutant strains were grown anaerobically in Wall LS4 medium with no antibiotics. When cultures reached the stationary-growth phase, cells were collected, washed three times with PBS and resuspended in DMEM to obtain an OD600 of ∼0.3, and the viability (CFU/ml) was evaluated before incubation in macrophages (time zero). Cells were then used to infect macrophages, at a multiplicity of infection (MOI) of 40, during 5, 8, and 24 h. Bacterial survival was evaluated by colony formation on plates loaded with serial dilutions of cultures in PBS. Briefly, 5 μl of at least two different dilutions was spread on tryptic soy agar medium supplemented (per liter) with 2.5 g sodium lactate, 2.0 g magnesium sulfate, 0.5 g ammonium iron (II) sulfate, and 20 mg sodium thioglycolate/ascorbic acid solution (31). The plates were then incubated, at 37°C, in a jar containing an anaerobic generator (GENbox anaer from bioMérieux), and after 2 days, the number of colonies was evaluated. Four independent biological samples with three replicates each were analyzed.
To determine the number of cells phagocytized by the macrophages, D. vulgaris was firstly incubated with macrophages for 2 h. Next, the macrophages were washed and the noninternalized bacteria eliminated by incubation, for 5 min, with DMEM supplemented with 70 U penicillin/ml and 70 μg streptomycin/ml. After the addition of fresh DMEM, the infection proceeded for an extra 3, 6, and 22 h, at which time macrophages were washed and lysed with saponin (2% [wt/vol]) and the intracellular bacterial content was evaluated by counting of CFU.
The amount of NO produced by macrophages was measured as the nitrite accumulated in the supernatants of murine macrophage cell cultures grown in DMEM and either activated by 0.3 μg IFN-γ/ml or infected with D. vulgaris (MOI, ∼40) in the absence and presence of the inhibitor L-NMMA (400 μM). The microtiter plate colorimetric assay (Multiskan GO; Thermo Scientific) was performed by reading the absorbance at 540 nm of 1:1 mixtures of supernatants (100 μl) and Griess reagent (1% [wt/vol] sulfanilamide, 0.1% [wt/vol] naphthylene diamine dihydrochloride, 2% [vol/vol] phosphoric acid). Sodium nitrite was used as a standard.
RESULTS
Transcriptional response of hcp and roo genes to nitrosative stress.
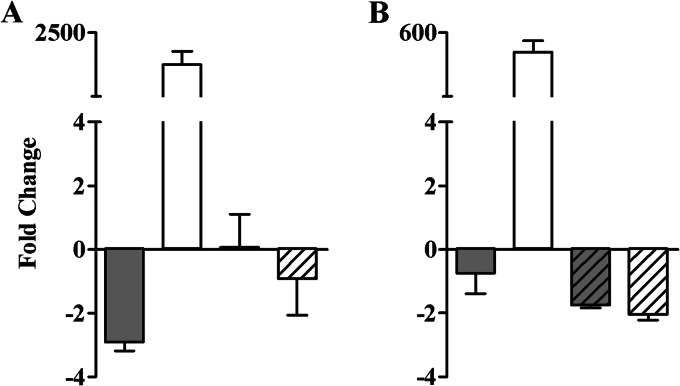
D. vulgaris Hildenborough, a model organism in the study of SRB, was used to assess the function of ROO and HCP proteins in in vitro protection against NO. To this end, we first analyzed the expression of genes DVU2013 (hcp1), DVU2543 (hcp2), DVU2014 (roo1), and DVU3185 (roo2) in D. vulgaris grown to the early exponential-growth phase and exposed to an NO donor (Fig. 1). Exposure of D. vulgaris to NONOates caused no significant change in the transcription of the roo genes but slightly lowered the hcp1 mRNA abundance after treatment for 1 h (Fig. 1). Importantly, high increases of the hcp2 gene expression of approximately 1,900-fold and 400-fold were observed in cells treated with nitrosative stress for 1 h and 4 h, respectively (Fig. 1).
Fig 1.
Effect of NO stress on the transcription of D. vulgaris hcp and roo genes. The fold variations of the expression of the genes hcp1 (gray bars), hcp2 (white bars), roo1 (gray striped bars), and roo2 (white striped bars) upon exposure of the D. vulgaris wild-type strain to 100 μM spermine NONOate for 1 h (A) and 100 μM DPTA NONOate for 4 h (B) are indicated. Fold change values represent the ratio of the expression level of treated culture to that of untreated culture and were considered significant when they exceeded 2-fold. Values are means ± standard errors (n = 6).
Sensitivity of D. vulgaris wild-type and mutant strains to nitric oxide donors.
Next, the in vitro susceptibility of D. vulgaris and transposon mutants of roo and hcp to nitrosative stress was evaluated by monitoring the anaerobic growth behavior of untreated cells and of cells exposed to 100 μM DPTA NONOate. Under these conditions, D. vulgaris transposon mutants of roo1 and roo2 were slightly more resistant to NO than the wild-type strain whereas the mutant lacking hcp2 stopped growing immediately after the introduction of the stress (Fig. 2). These results revealed that inactivation of hcp2 resulted in a D. vulgaris strain with a lower ability to cope with NO stress.
Fig 2.
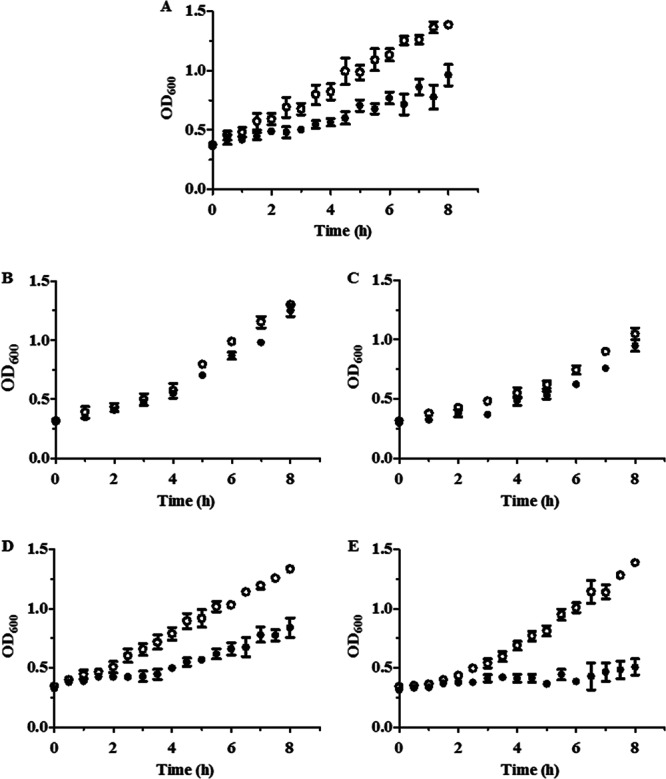
Growth of D. vulgaris wild-type and mutant strains in the presence of NO. Growth curves of the D. vulgaris wild-type strain (A) and of strains mutated in roo1 (B), roo2 (C), hcp1 (D), and hcp2 (E) which were left untreated (ο) or treated with 100 μM DPTA NONOate (•) are shown. Three biological samples were analyzed, and values are means ± standard errors.
Since the roo mutants did not show reduced susceptibility to NO compared with the wild-type strain, we further tested the effect of another NO source, GSNO, on their growth behavior. For all strains, 10 μM GSNO had a moderate inhibitory effect on growth, whereas 50 μM GSNO caused strong growth impairment (see Fig. S1 in the supplemental material). However, in all cases no significant differences were observed between the growth of the wild-type strain and that of the roo mutant strains.
NO consumption activity of D. vulgaris wild-type and mutant strains.
The cellular NO reductase activity of the D. vulgaris wild-type strain was also evaluated by measuring the NO consumption of lysates prepared from cells grown anaerobically and left untreated or exposed to 100 μM DPTA NONOate. We observed that the activity of the wild-type strain was slightly higher in NO-treated cells (91.5 ± 24.1 and 140.8 ± 21.7 pmol NO/min/mg protein total for untreated and NO-treated cells, respectively) (Fig. 3). Analysis of the mutants showed that in untreated cells, inactivation of either the roo or hcp gene did not change the NO consumption rates (Fig. 3A). However, for NO-treated cells, the strains interrupted in roo1 or roo2 exhibited NO consumption that was lower than that of the NO-treated wild-type strain at approximately 85% and 25% for the roo1 and roo2 transposon mutants, respectively (Fig. 3B). These results revealed that, among the four proteins, ROO1 is the major contributor to the NO reduction capability of D. vulgaris.
Fig 3.
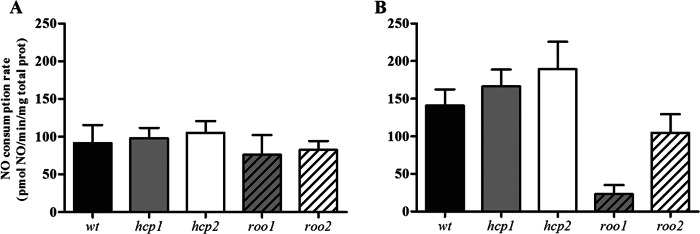
NO reduction activity. NO consumption activity was determined amperometrically in D. vulgaris cell lysates prepared from untreated cultures (A) and cultures exposed for 4 h to 100 μM DPTA NONOate (B). D. vulgaris wild-type strain (wt), black bars; hcp1 mutant, gray bars; hcp2 mutant, white bars; roo1 mutant, gray striped bars; roo2 mutant, white striped bars. Values are means ± standard errors (n = 6). Prot, protein.
Infection of macrophages with D. vulgaris.
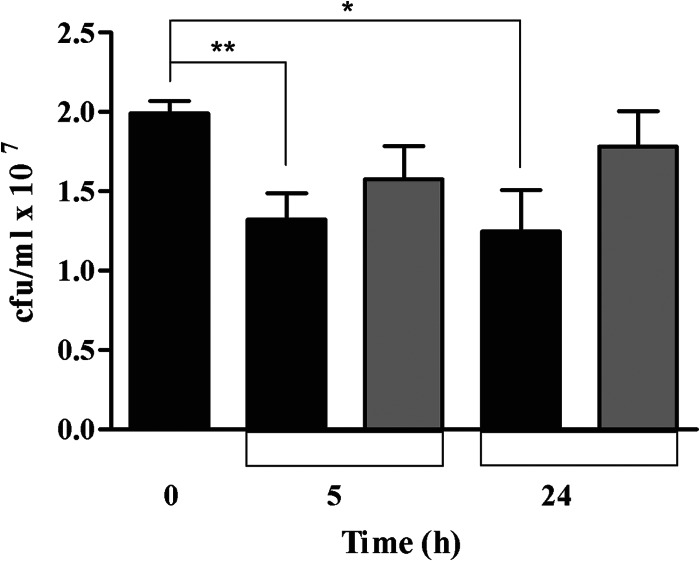
Although it has been proposed that Desulfovibrio spp. are involved in infectious processes, so far, no data have been available on the viability of Desulfovibrio within key cells of the innate immune system, such as macrophages. Hence, in this work the D. vulgaris wild-type strain was cultured anaerobically until the late exponential-growth phase and incubated with RAW264.7 murine macrophages. After 2 h of infection, the extracellular bacteria were eliminated by addition of the standard antibiotics used in macrophage culturing (see Materials and Methods). After periods of 5 h and 24 h, the macrophages were lysed and the intracellular bacterial content was determined. Under all conditions, no viable bacterial cells were detected (data not shown), suggesting that D. vulgaris is not capable of intracellular replication in macrophages. However, D. vulgaris was able to survive extracellularly in DMEM (see Fig. S2 in the supplemental material), and upon coculture with macrophages, the D. vulgaris wild-type strain suffered an approximately 30% decrease in survival (Fig. 4). Similar experiments done in the presence of L-NMMA, an inhibitor of the mammalian iNOS, showed that inhibition of macrophage NO production allows the recovery of the D. vulgaris wild-type strain, particularly after 24 h of infection (Fig. 4). At that time, the proportion of D. vulgaris viable cells in macrophages that do not produce NO is similar to that observed in DMEM, indicating an ∼100% viability recovery of the wild-type strain (Fig. 4; see also Fig. S2 in the supplemental material).
Fig 4.
Survival of D. vulgaris upon interaction with macrophages. Activated RAW264.7 murine macrophages were infected with D. vulgaris in the absence and in the presence of the iNOS inhibitor L-NMMA (black and gray bars, respectively). The bacterial survival was determined by CFU counting immediately before D. vulgaris was incubated with macrophages (time zero) and at 5 h and 24 h postinfection. Values are means ± standard errors (n = 12) determined with a t test (*, P < 0.005; **, P < 0.01).
To determine whether D. vulgaris was able to activate the production of NO by the mammalian iNOS, cultures of macrophages were infected with the D. vulgaris wild-type strain for 14 h and the nitrite content was measured in the supernatants (Fig. 5). For comparison purposes, assays that were similar but in which the activation of iNOS was achieved by addition of the macrophage activator gamma interferon (IFN-γ) (32) were also performed. The results showed that D. vulgaris activates the production of NO in macrophages to a level similar to that stimulated by IFN-γ (Fig. 5). Moreover, experiments done in the presence of L-NMMA inhibitor caused a significant decrease of the nitrite content in the supernatants (Fig. 5).
Fig 5.
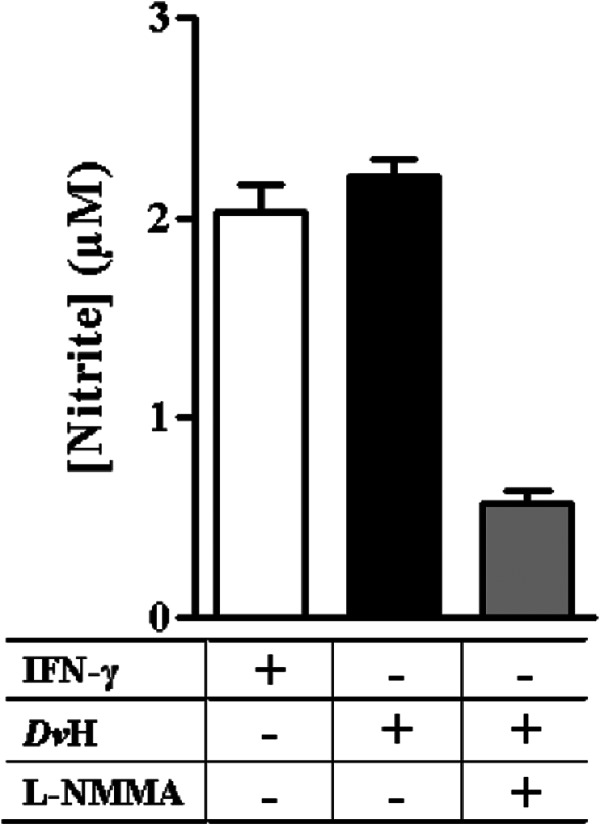
Nitrite production by D. vulgaris-infected macrophages. Nitrite concentrations accumulated, during 14 h, in macrophages preactivated with IFN-γ (white bar) or infected with D. vulgaris Hildenborough (DvH) in the absence (black bar) or in the presence (gray bar) of L-NMMA are shown. Values represent means and the corresponding standard errors (n = 8).
Altogether, these results revealed that D. vulgaris is able to trigger the induction of iNOS in macrophages.
Survival of D. vulgaris mutant strains upon contact with macrophages.
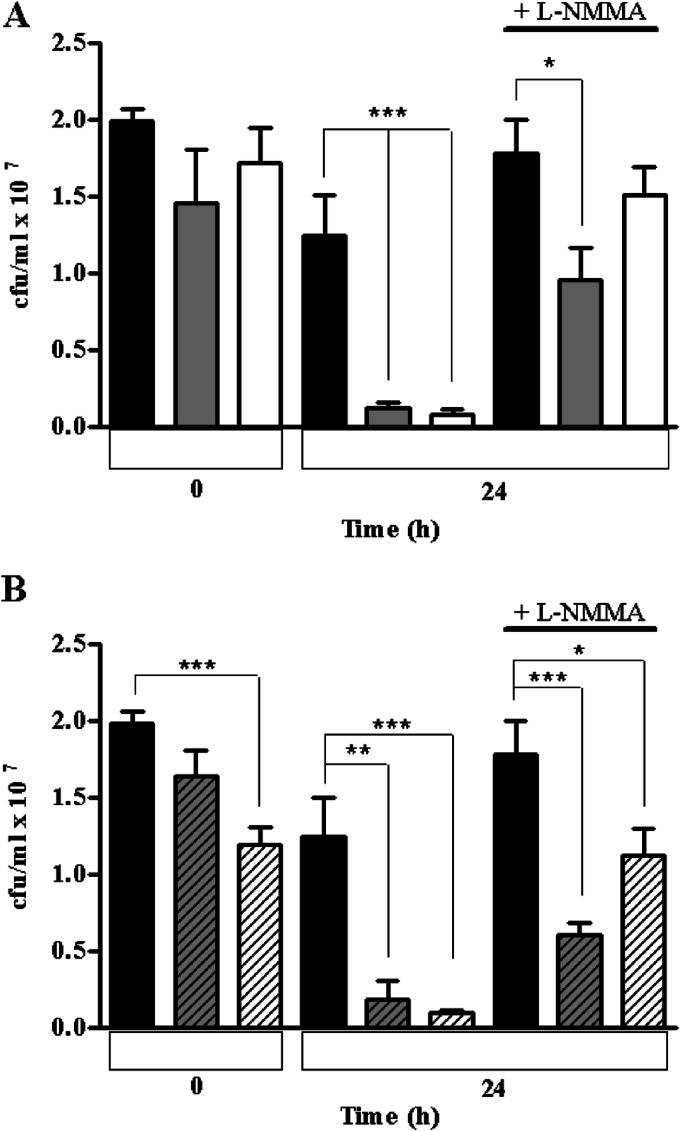
The contribution of the HCP and ROO proteins to the survival of D. vulgaris in macrophages was also investigated. For this purpose, macrophages were infected with D. vulgaris wild-type and mutant strains for 5 h, 8 h, and 24 h. While no differences between the wild-type strain and mutants were seen at up to 5 h and 8 h of incubation (data not shown), the results after 24 h of infection were distinct (Fig. 6). At this time, the survival of all mutant strains was reduced by approximately 90% relative to that of the wild-type strain, while the viability of the parental strain decreased only 30% (Fig. 6). Impairment of the macrophage NO production by L-NMMA resulted in an increase in the survival of the four mutant strains. Nevertheless, in the absence of NO production, the recovery of viability of the mutant strains was still lower than that observed in the absence of macrophages (time zero in Fig. 6). These data suggest that the compromised viability of the mutant strains upon incubation with macrophages is related, albeit partially, to the lower nitrosative stress resistance of these strains.
Fig 6.
Survival of D. vulgaris wild-type and mutant strains upon interaction with macrophages. Activated macrophages were infected with the D. vulgaris wild-type strain (black bars) and with the following transposon mutants: the hcp1 mutant (gray bars) and the hcp2 mutant (white bars) (A) and the roo1 mutant (gray striped bars) and the roo2 mutant (white striped bars) (B). The bacterial survival was determined by CFU counting immediately before D. vulgaris was incubated with macrophages or had the iNOS inhibitor L-NMMA added (time zero). Again, CFU were determined after 24 h of exposure to either macrophages or inhibitor. Values are means ± standard errors (n = 12) determined with a t test (*, P < 0.005; **, P < 0.01; ***, P < 0.001).
DISCUSSION
In this work, it was demonstrated that, among the four studied genes, the hcp2 gene is the highest upregulated by NO stress. A strong induction of hcp2 was also seen in D. vulgaris cells exposed to nitrate and nitrite stress conditions (33, 34). Moreover, only the hcp2 transposon mutant generated a D. vulgaris strain with high susceptibility to NO. Hence, HCP2 seems to primarily contribute to the in vitro survival of D. vulgaris under conditions of NO stress.
Recent data from Voordouw and coworkers showed that D. vulgaris HCP1 and HCP2 are required to maintain the high rates of nitrite reduction by the nitrite reductase NrfHA, and those authors therefore proposed a role for HCPs in protection from nitrite-derived products (28). Although our phenotypic data indicate that HCP2 participates in NO defense, the NO consumption rates of D. vulgaris remained unaltered upon deletion of hcp2. In agreement, none of the HCP proteins studied exhibited NO reductase activity (18, 21–23).
Concerning the D. vulgaris roo genes, we observed no significant transcriptional alterations under conditions of nitrosative stress, which fully agrees with earlier gene expression studies (25, 26, 33, 34). Furthermore, the growth behavior of the D. vulgaris roo1 and roo2 mutants upon exposure to the NO donor was similar to that of the wild-type strain. Since earlier work with disk diffusion assays indicated that the D. vulgaris roo2 transposon mutant was moderately more sensitive to GSNO (27), it is possible that these divergent results are related to the growth conditions and sources of NO used in each case. Nevertheless, we cannot exclude the possibility that, in the presence of other D. vulgaris RNS protecting enzymes/proteins, the contribution of roo2 to the overall growth is not discernible, a situation that recalls that of E. coli, which encodes several NO detoxifying enzymes (15, 35).
Interestingly, we observed that the NO reduction rate of the strain lacking roo1 is significantly lower, suggesting that ROO1 is the major contributor to the NO consumption activity in D. vulgaris. In agreement, the D. gigas ROO was previously shown to be able to rescue an E. coli strain deleted in the NO-reductase flavodiiron gene and to have significant in vitro NO reductase activity (36).
While the results here support the previously proposed idea of the role of ROO as a NO detoxifier (27, 36), a more complex mechanism may explain the protection conferred by HCP. So far, HCPs have been described as having peroxidase activity (E. coli and D. desulfuricans ATCC 27774) (18) and hydroxylamine reductase activity (E. coli, Pyrococcus furiosus, and Rhodobacter capsulatus E1F1) (21–23). However, it was assumed that hydroxylamine is not the natural substrate because of the low catalytic efficiency of the reaction (21–23). Here also, the direct involvement of HCP in NO detoxification could not be inferred. Although the hcp2 mutant was more sensitive to the NO donor than the wild-type strain, the NO consumption rate of the Δhcp2 mutant was similar to the consumption rate measured for the wild-type strain. Hence, other roles for HCP need to be considered. For example, Vine and Cole have proposed that HCP may be involved in the repair of damage caused by nitrosative stress (35).
Since Desulfovibrio spp. are proposed to act as opportunistic pathogens, in this study we addressed the ability of these bacteria to survive when contacting macrophages. Our results show that D. vulgaris is not capable of intracellular replication in macrophages but survives extracellularly. This is consistent with the capacity of Desulfovibrio spp. to replicate outside host cells in the gastrointestinal tract (37). Furthermore, this bacterium stimulates NO production in macrophages at levels similar to those induced by IFN-γ. The fact that D. vulgaris triggers NO production is consistent with the observed induction of IL-8 and IL-6 cytokines in HeLA cells upon infection with D. fairfieldensis and D. desulfuricans (7). Moreover, the NO released lowers Desulfovibrio survival, as the viability of the wild-type and mutant strains was inversely related to the level of NO generated.
We have also found that HCP and ROO proteins contribute to survival of D. vulgaris during infection of macrophages. Interestingly, the lack of correlation between the absence of increased susceptibility of the hcp1 and the two roo mutants upon in vitro exposure to NO and the positive contribution of all strains to survival in animal cells observed here was also seen in Salmonella enterica. Indeed, although the hcp mutant of S. enterica serovar Typhimurium (ΔnipA) displayed no defects under in vitro stress conditions, the NipA protein did contribute to the virulence in mice (38).
The increased viability of D. vulgaris mutants in macrophages that do not produce NO may be interpreted to mean that ROO and HCP proteins are related to NO defense mechanisms. However, the incomplete recovery of the mutant strains observed upon inhibition of iNOS suggests that they also participate in protection against other stresses, such as oxidative stress, imposed by the macrophages. Studies in oxygenated environments showing that the roo and hcp mutations decrease D. vulgaris survival under conditions of oxidative stress corroborate this hypothesis (18, 24, 27, 28, 39).
Another interesting issue relates to the presence in D. vulgaris of two HCPs and ROOs that share high amino acid sequence identities (ROO1 and ROO2, 29% identity; HCP1 and HCP2, 42% identity), including the conservation of the ligands for the diiron centers and iron-sulfur centers, respectively. This conservation suggests that the two pairs of homologues share similar functional roles. However, our results indicate otherwise, as the hcp2 transposon mutant is more susceptible to elimination by NO than the hcp1 mutant, and ROO1 contributes significantly more than ROO2 to cellular NO consumption. It is possible that these apparently distinct roles result from different gene regulation mechanisms. Whereas in D. vulgaris, HcpR, a Cpr/Fnr-like global regulator that responds to nitrosative stress (40), has been proposed to control hcp2 expression, the mechanism of regulation of roo2 and that of the gene cluster hcp1-roo1, present in an isolated D. vulgaris genomic island (24), remain unknown. Noteworthy, in E. coli, the hcp gene is regulated by the peroxide-sensing transcriptional factor OxyR and the recombinant protein exhibits peroxidase activity (18, 19). Therefore, we speculate that in each case the in vivo function is controlled by transcriptional factors that respond to either oxidative or nitrosative stress and that they may be involved in NO defenses as RNS/ROS detoxifiers or indirectly in a not-yet-recognized way. Hence, the finding that the four gene products independently contribute to bacterial protection against macrophages may be rationalized by considering that macrophages expose D. vulgaris to both oxidative and nitrosative stress conditions.
In summary, we have shown for the first time that D. vulgaris triggers macrophage effectors and that the HCP and ROO proteins contribute to the resistance of the bacterium during macrophage infection.
Supplementary Material
ACKNOWLEDGMENTS
We thank Grant Zane for the generation and confirmation of the transposon mutants of D. vulgaris Hildenborough used in this work.
The work was funded by PEst-OE/EQB/LA0004/2011, FCT project PTDC/BIA-PRO/098224/2008, and by the following FCT student fellowships: SFRH/BD/44140/2008 (M.C.O.F.), SFRH/BPD/63944/2009 (S.A.L.L.), SRFH/BPD/69325/2010 (L.S.N.), and BI/001/BI-BI/2012 (S.H.S.). J.D.W. was funded by ENIGMA Ecosystems and Networks Integrated with Genes and Molecular Assemblies (http://enigma.lbl.gov), a Scientific Focus Area Program at Lawrence Berkeley National Laboratory, supported by the Office of Science, Office of Biological and Environmental Research, of the U.S. Department of Energy under contract no. DE-AC02-05CH11231.
Footnotes
Published ahead of print 5 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00074-13.
REFERENCES
- 1. Jia W, Whitehead RN, Griffiths L, Dawson C, Bai H, Waring RH, Ramsden DB, Hunter JO, Cauchi M, Bessant C, Fowler DP, Walton C, Turner C, Cole JA. 2012. Diversity and distribution of sulphate-reducing bacteria in human faeces from healthy subjects and patients with inflammatory bowel disease. FEMS Immunol. Med. Microbiol. 65:55–68 [DOI] [PubMed] [Google Scholar]
- 2. Ichiishi S, Tanaka K, Nakao K, Izumi K, Mikamo H, Watanabe K. 2010. First isolation of Desulfovibrio from the human vaginal flora. Anaerobe 16:229–233 [DOI] [PubMed] [Google Scholar]
- 3. Nakao K, Tanaka K, Ichiishi S, Mikamo H, Shibata T, Watanabe K. 2009. Susceptibilities of 23 Desulfovibrio isolates from humans. Antimicrob. Agents Chemother. 53:5308–5311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lozniewski A, Maurer P, Schuhmacher H, Carlier JP, Mory F. 1999. First isolation of Desulfovibrio species as part of a polymicrobial infection from a brain abscess. Eur. J. Clin. Microbiol. Infect. Dis. 18:602–603 [DOI] [PubMed] [Google Scholar]
- 5. Rowan F, Docherty NG, Murphy M, Murphy B, Calvin Coffey J, O'Connell PR. 2010. Desulfovibrio bacterial species are increased in ulcerative colitis. Dis. Colon Rectum 53:1530–1536 [DOI] [PubMed] [Google Scholar]
- 6. Gaillard T, Pons S, Darles C, Beausset O, Monchal T, Brisou P. 2011. Desulfovibrio fairfieldensis bacteremia associated with acute sigmoiditis. Med. Mal. Infect. 41:267–268 [DOI] [PubMed] [Google Scholar]
- 7. Bisson-Boutelliez C, Massin F, Dumas D, Miller N, Lozniewski A. 2010. Desulfovibrio spp. survive within KB cells and modulate inflammatory responses. Mol. Oral Microbiol. 25:226–235 [DOI] [PubMed] [Google Scholar]
- 8. Nathan C, Shiloh MU. 2000. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc. Natl. Acad. Sci. U. S. A. 97:8841–8848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kidd SP. 2011. Stress response in pathogenic bacteria, vol 19 CAB International, Oxfordshire, United Kingdom [Google Scholar]
- 10. Vicente JB, Carrondo MA, Teixeira M, Frazão C. 2008. Flavodiiron proteins: nitric oxide and/or oxygen reductases. In Messerschmidt A. (ed), Handbook of metalloproteins. John Wiley & Sons, Ltd., Chichester, United Kingdom [Google Scholar]
- 11. Saraiva LM, Vicente JB, Teixeira M. 2004. The role of the flavodiiron proteins in microbial nitric oxide detoxification. Adv. Microb. Physiol. 49:77–129 [DOI] [PubMed] [Google Scholar]
- 12. Frazão C, Silva G, Gomes CM, Matias P, Coelho R, Sieker L, Macedo S, Liu MY, Oliveira S, Teixeira M, Xavier AV, Rodrigues-Pousada C, Carrondo MA, Le Gall J. 2000. Structure of a dioxygen reduction enzyme from Desulfovibrio gigas. Nat. Struct. Biol. 7:1041–1045 [DOI] [PubMed] [Google Scholar]
- 13. Gomes CM, Silva G, Oliveira S, LeGall J, Liu MY, Xavier AV, Rodrigues-Pousada C, Teixeira M. 1997. Studies on the redox centers of the terminal oxidase from Desulfovibrio gigas and evidence for its interaction with rubredoxin. J. Biol. Chem. 272:22502–22508 [DOI] [PubMed] [Google Scholar]
- 14. Baptista JM, Justino MC, Melo AM, Teixeira M, Saraiva LM. 2012. Oxidative stress modulates the nitric oxide defense promoted by Escherichia coli flavorubredoxin. J. Bacteriol. 194:3611–3617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Justino MC, Vicente JB, Teixeira M, Saraiva LM. 2005. New genes implicated in the protection of anaerobically grown Escherichia coli against nitric oxide. J. Biol. Chem. 280:2636–2643 [DOI] [PubMed] [Google Scholar]
- 16. Macedo S, Mitchell EP, Romão CV, Cooper SJ, Coelho R, Liu MY, Xavier AV, LeGall J, Bailey S, Garner DC, Hagen WR, Teixeira M, Carrondo MA, Lindley P. 2002. Hybrid cluster proteins (HCPs) from Desulfovibrio desulfuricans ATCC 27774 and Desulfovibrio vulgaris (Hildenborough): X-ray structures at 1.25 Å resolution using synchrotron radiation. J. Biol. Inorg. Chem. 7:514–525 [DOI] [PubMed] [Google Scholar]
- 17. Aragão D, Macedo S, Mitchell EP, Romao CV, Liu MY, Frazao C, Saraiva LM, Xavier AV, LeGall J, van Dongen WM, Hagen WR, Teixeira M, Carrondo MA, Lindley P. 2003. Reduced hybrid cluster proteins (HCP) from Desulfovibrio desulfuricans ATCC 27774 and Desulfovibrio vulgaris (Hildenborough): X-ray structures at high resolution using synchrotron radiation. J. Biol. Inorg. Chem. 8:540–548 [DOI] [PubMed] [Google Scholar]
- 18. Almeida CC, Romão CV, Lindley PF, Teixeira M, Saraiva LM. 2006. The role of the hybrid cluster protein in oxidative stress defense. J. Biol. Chem. 281:32445–32450 [DOI] [PubMed] [Google Scholar]
- 19. Seth D, Hausladen A, Wang YJ, Stamler JS. 2012. Endogenous protein S-nitrosylation in E. coli: regulation by OxyR. Science 336:470–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vine CE, Cole JA. 2011. Nitrosative stress in Escherichia coli: reduction of nitric oxide. Biochem. Soc. Trans. 39:213–215 [DOI] [PubMed] [Google Scholar]
- 21. Cabello P, Pino C, Olmo-Mira MF, Castillo F, Roldan MD, Moreno-Vivian C. 2004. Hydroxylamine assimilation by Rhodobacter capsulatus E1F1. requirement of the hcp gene (hybrid cluster protein) located in the nitrate assimilation nas gene region for hydroxylamine reduction. J. Biol. Chem. 279:45485–45494 [DOI] [PubMed] [Google Scholar]
- 22. Overeijnder ML, Hagen WR, Hagedoorn PL. 2009. A thermostable hybrid cluster protein from Pyrococcus furiosus: effects of the loss of a three helix bundle subdomain. J. Biol. Inorg. Chem. 14:703–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wolfe MT, Heo J, Garavelli JS, Ludden PW. 2002. Hydroxylamine reductase activity of the hybrid cluster protein from Escherichia coli. J. Bacteriol. 184:5898–5902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Johnston S, Lin S, Lee P, Caffrey SM, Wildschut J, Voordouw JK, da Silva SM, Pereira IA, Voordouw G. 2009. A genomic island of the sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough promotes survival under stress conditions while decreasing the efficiency of anaerobic growth. Environ. Microbiol. 11:981–991 [DOI] [PubMed] [Google Scholar]
- 25. He Q, He Z, Joyner DC, Joachimiak M, Price MN, Yang ZK, Yen HC, Hemme CL, Chen W, Fields MM, Stahl DA, Keasling JD, Keller M, Arkin AP, Hazen TC, Wall JD, Zhou J. 2010. Impact of elevated nitrate on sulfate-reducing bacteria: a comparative study of Desulfovibrio vulgaris. ISME J. 4:1386–1397 [DOI] [PubMed] [Google Scholar]
- 26. He Q, Huang KH, He Z, Alm EJ, Fields MW, Hazen TC, Arkin AP, Wall JD, Zhou J. 2006. Energetic consequences of nitrite stress in Desulfovibrio vulgaris Hildenborough, inferred from global transcriptional analysis. Appl. Environ. Microbiol. 72:4370–4381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wildschut JD, Lang RM, Voordouw JK, Voordouw G. 2006. Rubredoxin:oxygen oxidoreductase enhances survival of Desulfovibrio vulgaris Hildenborough under microaerophilic conditions. J. Bacteriol. 188:6253–6260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yurkiw MA, Voordouw J, Voordouw G. 2012. Contribution of rubredoxin:oxygen oxidoreductases and hybrid cluster proteins of Desulfovibrio vulgaris Hildenborough to survival under oxygen and nitrite stress. Environ. Microbiol. 14:2711–2725 [DOI] [PubMed] [Google Scholar]
- 29. Keller KL, Bender KS, Wall JD. 2009. Development of a markerless genetic exchange system for Desulfovibrio vulgaris Hildenborough and its use in generating a strain with increased transformation efficiency. Appl. Environ. Microbiol. 75:7682–7691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Beckman JS, Wink DA, Crow JP. 1996. Methods in nitric oxide research. John Wiley & Sons Ltd., Chichester, United Kingdom [Google Scholar]
- 31. van den Berg WAM, Stokkermans JPWG, van Dongen WMAM. 1989. Development of a plasmid transfer system for the anaerobic sulphate reducer, Desulfovibrio vulgaris. J. Biotechnol. 12:173–184 [Google Scholar]
- 32. MacMicking J, Xie QW, Nathan C. 1997. Nitric oxide and macrophage function. Annu. Rev. Immunol. 15:323–350 [DOI] [PubMed] [Google Scholar]
- 33. Haveman SA, Greene EA, Stilwell CP, Voordouw JK, Voordouw G. 2004. Physiological and gene expression analysis of inhibition of Desulfovibrio vulgaris Hildenborough by nitrite. J. Bacteriol. 186:7944–7950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Haveman SA, Greene EA, Voordouw G. 2005. Gene expression analysis of the mechanism of inhibition of Desulfovibrio vulgaris Hildenborough by nitrate-reducing, sulfide-oxidizing bacteria. Environ. Microbiol. 7:1461–1465 [DOI] [PubMed] [Google Scholar]
- 35. Vine CE, Cole JA. 2011. Unresolved sources, sinks, and pathways for the recovery of enteric bacteria from nitrosative stress. FEMS Microbiol. Lett. 325:99–107 [DOI] [PubMed] [Google Scholar]
- 36. Rodrigues R, Vicente JB, Felix R, Oliveira S, Teixeira M, Rodrigues-Pousada C. 2006. Desulfovibrio gigas flavodiiron protein affords protection against nitrosative stress in vivo. J. Bacteriol. 188:2745–2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Macfarlane GT, Cummings JH, Macfarlane S. 2007. Sulphate-reducing bacteria and the human large intestine, p 503–521 In Barton LL, Hamilton WA. (ed), Sulphate-reducing bacteria: environmental and engineered systems. Cambridge University Press, New York, NY [Google Scholar]
- 38. Kim CC, Monack D, Falkow S. 2003. Modulation of virulence by two acidified nitrite-responsive loci of Salmonella enterica serovar Typhimurium. Infect. Immun. 71:3196–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Figueiredo MC, Lobo SA, Carita JN, Nobre LS, Saraiva LM. 2012. Bacterioferritin protects the anaerobe Desulfovibrio vulgaris Hildenborough against oxygen. Anaerobe 18:454–458 [DOI] [PubMed] [Google Scholar]
- 40. Zhou A, Chen YI, Zane GM, He Z, Hemme CL, Joachimiak MP, Baumohl JK, He Q, Fields MW, Arkin AP, Wall JD, Hazen TC, Zhou J. 2012. Functional characterization of Crp/Fnr-type global transcriptional regulators in Desulfovibrio vulgaris Hildenborough. Appl. Environ. Microbiol. 78:1168–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.