Abstract
Interactions with immune responses or exposure to certain antibiotics can remove the peptidoglycan wall of many Gram-negative bacteria. Though the spheroplasts thus created usually lyse, some may survive by resynthesizing their walls and shapes. Normally, bacterial morphology is generated by synthetic complexes directed by FtsZ and MreBCD or their homologues, but whether these classic systems can recreate morphology in the absence of a preexisting template is unknown. To address this question, we treated Escherichia coli with lysozyme to remove the peptidoglycan wall while leaving intact the inner and outer membranes and periplasm. The resulting lysozyme-induced (LI) spheroplasts recovered a rod shape after four to six generations. Recovery proceeded via a series of cell divisions that produced misshapen and branched intermediates before later progeny assumed a normal rod shape. Importantly, mutants defective in mounting the Rcs stress response and those lacking penicillin binding protein 1B (PBP1B) or LpoB could not divide or recover their cell shape but instead enlarged until they lysed. LI spheroplasts from mutants lacking the Lpp lipoprotein or PBP6 produced spherical daughter cells that did not recover a normal rod shape or that did so only after a significant delay. Thus, to regenerate normal morphology de novo, E. coli must supplement the classic FtsZ- and MreBCD-directed cell wall systems with activities that are otherwise dispensable for growth under normal laboratory conditions. The existence of these auxiliary mechanisms implies that they may be required for survival in natural environments, where bacterial walls can be damaged extensively or removed altogether.
INTRODUCTION
Bacterial cells come in an exceptionally wide variety of shapes and sizes, ranging from the well-known rods, cocci, and spirals to more complex, sometimes dramatically different morphologies, including those that are square, star shaped, multilobed, segmented, or fluted (1–3). While the reasons for this diversity are not entirely clear, it seems certain that cell shape contributes to bacterial survival by influencing nutrient uptake, surface attachment, motility, and susceptibility to predation or host immune systems, among other traits (1, 2). There is also increasing evidence that morphology affects the course of pathogenesis in those few bacteria in which this has been examined (1, 4–10). Thus, cell shape is of fundamental biological importance and has practical physiological effects.
In the eubacteria, morphology is determined and stabilized by the overall architecture of the peptidoglycan (PG) cell wall, and the organization of this structure is dictated by transmembrane mechanisms that direct when and where new PG is inserted (11–13). In particular, in E. coli, the MreBCD-penicillin binding protein 2 (PBP2)-RodA complex directs PG synthesis that results in the elongation of rod-shaped cells (11, 14, 15), and during cell division, the FtsZ protein initiates the formation of a multiprotein machine that directs septal wall synthesis and influences gross morphology (12, 13, 16–19). In other organisms, additional accessory proteins modify this baseline bacillary form so that the cells become curved or helical (8, 20, 21). Mutating or deleting proteins that affect these activities can alter cell shape, sometimes in drastic ways (11, 17, 19, 22–24). However, so far as is known, all of these systems influence cellular geometry by reorienting how or where new PG is inserted into the wall, hinting that these processes may be, in the main, maintenance mechanisms that preserve or modify preexisting bacterial shapes.
Interestingly, bacteria that have lost their cell wall can sometimes resynthesize this structure and recover a wild-type shape, indicating that cells are able to generate a defined morphology without the aid of a previously completed external template (25–28). What is not clear is whether the currently known shape maintenance pathways are sufficient for directing the recovery of cell shape in wall-less cells. Here, we show that E. coli requires additional mechanisms to recreate a normal morphology de novo. These include the Rcs envelope stress response; PBP1B and LpoB, proteins involved in PG synthesis and cell division; the Lpp lipoprotein, which connects the outer membrane (OM) to the PG layer; and PBP5 and PBP6, which modify PG. The existence of these auxiliary mechanisms implies that they may be required for the survival of E. coli and related organisms in natural environments, where their cell walls may be damaged extensively or removed altogether.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
For the E. coli strains, plasmids, and growth conditions used in this study, see the supplemental material (29, 30). Routine cultures were grown in Luria-Bertani (LB) medium, and when appropriate, antibiotics were added at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; tetracycline, 10 μg/ml; spectinomycin, 50 μg/ml. Plasmid pDEV is pLP8 (31) without the lacZ gene. The Rcs system reporter plasmid pDKR1 carries the PrprA::sfgfp gene, which places the expression of superfolder green fluorescent protein (sfGFP) (32, 33) under the control of the Rcs-specific promoter PrprA. Plasmid pDKR2 encodes the periplasmic version of sfGFP, expressed from the dsbAss-sfgfp gene. Plasmids carrying wild-type versions of rcsF (pRcsF), lpoB (pLpoB), and lpp (pLpp) were constructed by cloning each gene from E. coli MG1655. For the complete strain list and the primers used for genetic constructions, see Tables S1 and S2 in the supplemental material, respectively.
LI spheroplast preparation and shape recovery assay.
Lysozyme-induced (LI) spheroplasts of E. coli were created by adapting previous protocols (34, 35). A freshly isolated colony was inoculated into 3 ml LB medium and grown overnight, after which 100 μl was transferred into 10 ml LB broth and incubated at 37°C until the culture reached an optical density at 600 nm of 0.2. A 1-ml aliquot was washed with 1 ml of phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 8.0), and 5 μl was transferred into 500 μl of PBS containing 0.5 M sucrose and lysozyme (20 μg/ml) and incubated for 10 min at 37°C. At that time, 500 μl of PBS containing lysozyme (20 μg/ml) was added to the suspension, which was incubated at 37°C for an additional 10 min. Whole cells and LI spheroplasts were harvested by centrifugation at 500 × g for 15 min and washed with 1 ml of sucrose recovery medium (2% tryptone, 0.5% yeast extract, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 10 mM MgSO4, 20 mM glucose, 0.23 M sucrose, pH 7.0), and the resulting cell pellet was gently resuspended in 10 μl of this medium. Cells (2 μl) were placed onto a sucrose recovery soft agar pad (sucrose recovery medium plus 0.7% agar) solidified in the chambers of a Lab-Tek chamber slide (Nalge Nunc International, Rochester, NY.; catalog no. 177402). Slides were placed onto the stage of a Zeiss Axio Imager.Z1 microscope enclosed in a 37°C incubation chamber, and cells were visualized as described previously (31).
Fluorescent labeling of PG and the OM.
PG was immunolabeled by a procedure that was described previously (36) and is described fully in the supplemental material. As a control, normal rod-shaped E. coli cells were labeled and a spherical cell control was created by treating cells with 2 μg/ml amdinocillin for 1 h before labeling. In live cells, PG was labeled directly with the fluorescent compound HADA (hydroxycoumarin-carbonyl-amino-d-alanine), a gift from E. Kuru and M. S. VanNieuwenhze, Indiana University (37), and then observed in a Zeiss Axio Imager.Z1 microscope fitted with 4′,6-diamidino-2-phenylindole (DAPI) filters (358-nm excitation and 461-nm emission wavelengths). The outer surface of E. coli was labeled with N-hydroxysuccinimide (NHS) ester conjugated to Alexa Fluor 488 (Invitrogen; catalog no. A20000) before conversion into spheroplasts and then observed by microscopy with an enhanced GFP filter set (495-nm excitation and 519-nm emission wavelengths).
RESULTS
Characteristics of LI spheroplasts.
By subjecting E. coli to a mild osmotic shock in the presence of lysozyme, we created a situation in which cell shape was resynthesized from a starting point as close to de novo as possible. About 70% of the population was converted into spheres with an average volume ∼1.5 times greater than that of the original rod-shaped cells (see Fig. S1 in the supplemental material). In accordance with the original terminology of Hurwitz et al. and Birdsell and Cota-Robles, we refer to these cells as LI spheroplasts (34, 38). Spheroplasts thus prepared have no visible PG (38), and we confirmed that PG was absent from these cells by immunolabeling the sacculus (Fig. 1A to C). Also, spheroplasts derived from cells prelabeled with a fluorescent d-alanine derivative (HADA) (37) had little or no PG, while rod-shaped cells that escaped damage retained this material (Fig. 1D). In addition, a periplasmic version of sfGFP was confined to the periplasmic space bounded by an intact inner membrane (IM) and OM (Fig. 1E), and prestained OM proteins were retained (Fig. 1F). In short, LI spheroplasts lacked a detectable cell wall but possessed an intact periplasm and envelope.
Fig 1.
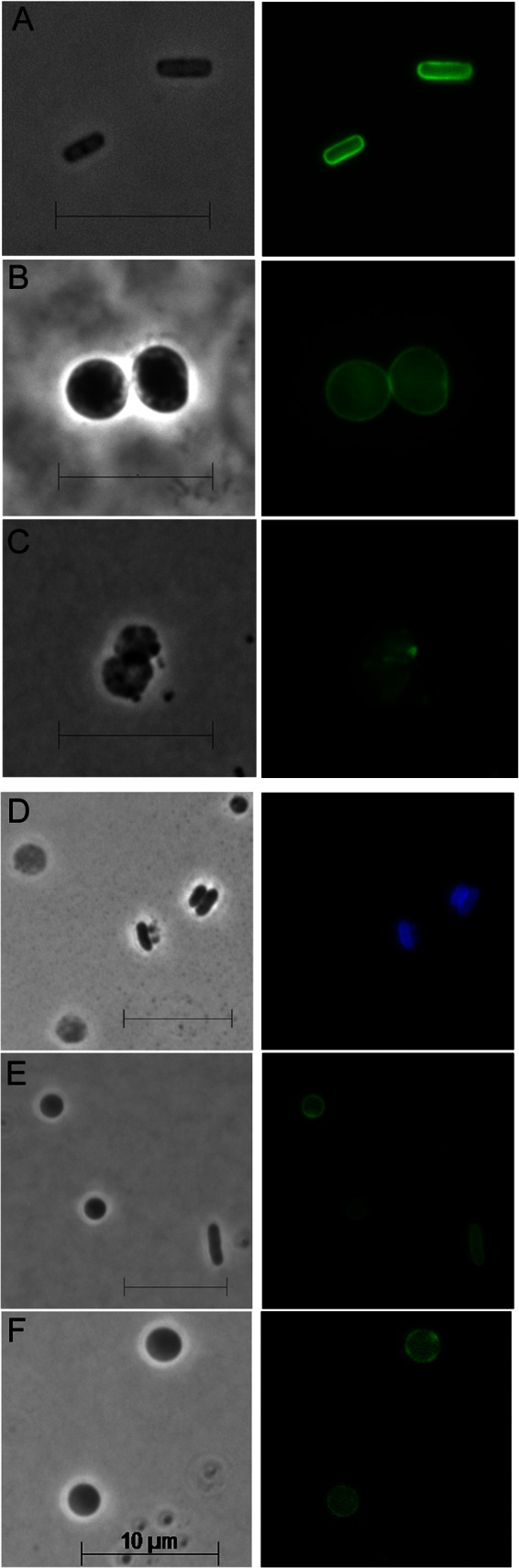
Characterization of LI spheroplasts. (A to C) PG of E. coli cells was immunolabeled with anti-murein antibody conjugated to Alexa Fluor 488. Left column, phase contrast images; right column, fluorescence images. The scale bar equals 10 μm. (A) Rod-shaped E. coli. (B) Spherical cells containing PG were prepared by growing E. coli in amdinocillin (2 μg/ml). (C) Spheroplasts created by treatment with lysozyme. (D) PG was labeled by incorporation of the fluorescent compound HADA, after which spheroplasts were generated. Rod-shaped cells (center right) that were unaffected by lysozyme retained the label, but spheroplasts (cells at the left and upper right) lost the label. (E) Periplasmic sfGFP was expressed in a population of spheroplasts. One lysozyme-unaffected rod-shaped cell is also shown (lower right). All retained sfGFP in the periplasm. (F) OM proteins were labeled with NHS-Alexa Fluor 488, after which spheroplasts were generated. See also Fig. S1 in the supplemental material.
LI spheroplasts revert via aberrant division intermediates.
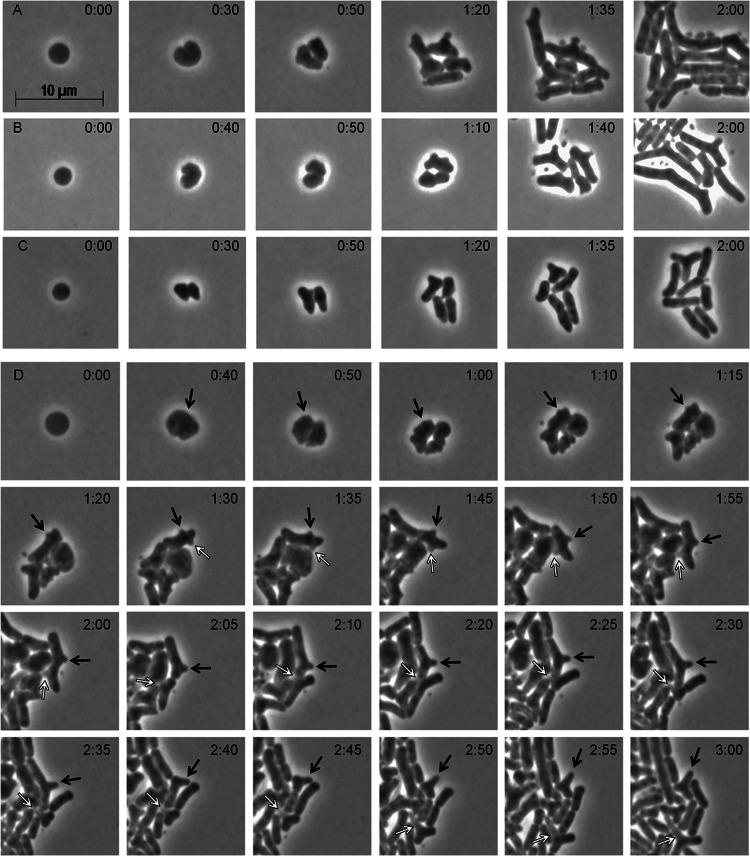
To determine if E. coli lacking its cell wall could recover its original rod-shaped morphology, we created spheroplasts from strain MG1655, mounted them on isosmotic soft agar pads, and monitored their fates by time-lapse microscopy. About 90% of the spheroplasts in isosmotic soft agar grew and developed into microcolonies of rod-shaped cells (Fig. 2A and Table 1, strain MG1655; see Movie M1 in the supplemental material). Newly created spheroplasts were smooth but rapidly became undulate before dividing, and the resulting daughter cells elongated before dividing again (Fig. 2A, time 0:30; see Movie M1). Rodlike cells appeared after 3 to 4 divisions, and after 4 to 6 divisions, the cells approached wild-type dimensions (Fig. 2A, time 2:00; see Movie M1). Spheroplasts derived from independent E. coli strains behaved similarly (Fig. 2B and C), including those from clinically derived strain 2443, which has a complete O antigen (39, 40). In every case, each spheroplast produced malformed offspring before giving rise to cells with uniform dimensions (Fig. 2A to C). Early cells exhibited aberrations (buds or bulges), some of which elongated to form branches that persisted throughout the course of the experiment (Fig. 2D). It seemed likely that aberrant cells arising from spheroplasts might originate by producing asymmetric FtsZ rings, as in other branching mutants (19), so we observed the orientation of FtsZ-GFP during shape recovery. Spheroplasts produced roughly ovoid daughter cells, with the next division occurring approximately perpendicular to the first (see Fig. S2, time 1:20 to 1:40). However, prior to the first division, randomly localized FtsZ-GFP foci and arcs formed before a single FtsZ ring coalesced to initiate invagination (see Fig. S2, time 0:00, black arrows). In all cases that could be followed for a sufficient length of time, the locations of transient foci correlated with the later development of bulges and branches (see Fig. S2, blue arrows at times 1:40 and 3:00), suggesting a probable causative connection.
Fig 2.
Regeneration of rod-shaped cells from wild-type E. coli. Spheroplasts were grown on osmotically protected sucrose recovery medium, and the recovery process was monitored by time-lapse microscopy; phase-contrast images are shown. The time after plating is displayed at the upper right of each panel (hours:minutes). (A) E. coli MG1655. (B) E. coli K-12. (C) E. coli 2443. (D) The origin and development of branches in E. coli MG1655. Black and white arrows trace protruding buds or bulges that developed into branches (also see Movie M1 in the supplemental material). The phase-contrast images in panels A and D were extracted from Movie M1 in the supplemental material. The scale bar equals 10 μm. See also Fig. S2.
Table 1.
Fates of LI spheroplasts lacking various proteins
| Strain | Protein(s) deleted | No. of spheroplasts |
|||||
|---|---|---|---|---|---|---|---|
| Totala | Dividedb | Recoveredc | Enlarged and lysedd | Lysed earlye | Inactivef | ||
| MG1655 | None | 63 | 57 | 57 | 0 | 1 | 5 |
| DR1 | RcsB | 20 | 0 | 0 | 19 | 1 | 0 |
| DR2 | CpxR | 8 | 4 | 4 | 0 | 1 | 3 |
| DR3 | OmpR | 5 | 3 | 3 | 0 | 0 | 2 |
| DR4 | RcsC | 7 | 0 | 0 | 5 | 1 | 1 |
| DR5 | RcsF | 24 | 0 | 0 | 19 | 2 | 3 |
| DR6 | RcsA | 15 | 7 | 7 | 0 | 5 | 3 |
| DR7 | PBP1B | 70 | 0 | 0 | 54 | 11 | 5 |
| DR8 | LpoB | 10 | 0 | 0 | 9 | 0 | 1 |
| DR9 | PBP1A | 10 | 9 | 9 | 0 | 0 | 1 |
| DR10 | LpoA | 12 | 9 | 9 | 0 | 0 | 3 |
| DR11 | Lpp | 31 | 24 | 0g | 0 | 7 | 0 |
| DR12m | MliC | 11 | 8 | 8 | 0 | 3 | 0 |
| DR12i | Ivy | 25 | 20 | 20 | 0 | 5 | 0 |
| DR12im | Ivy, MliC | 5 | 5 | 5 | 0 | 0 | 0 |
| DR13 | MltA | 5 | 4 | 4 | 0 | 0 | 1 |
| DR14 | MipA | 15 | 15 | 15 | 0 | 0 | 0 |
| DR15 | Pal | 36 | 23 | 23 | 0 | 13 | 0 |
| DR16 | AmiC | 6 | 6 | 6 | 0 | 0 | 0 |
| AV21-1 | PBP5 | 11 | 7 | 7h | 0 | 3 | 1 |
| AV76-1 | PBP6 | 8 | 8 | 8h | 0 | 0 | 0 |
The number of spheroplasts examined is from at least two independent experiments.
Spheroplasts that carried out cell division.
Spheroplasts that underwent cell division and produced progeny with normal rod shapes.
Spheroplasts that developed into large spheroids and lysed.
Spheroplasts that lysed without any development or physical change.
Inactive cells did not grow or lyse, remaining unchanged during the experiment.
Almost all of the cells remained spherical (see Movie M6 in the supplemental material).
Cells recovered but had abnormal or defective shapes.
The Rcs response is required for de novo shape recovery.
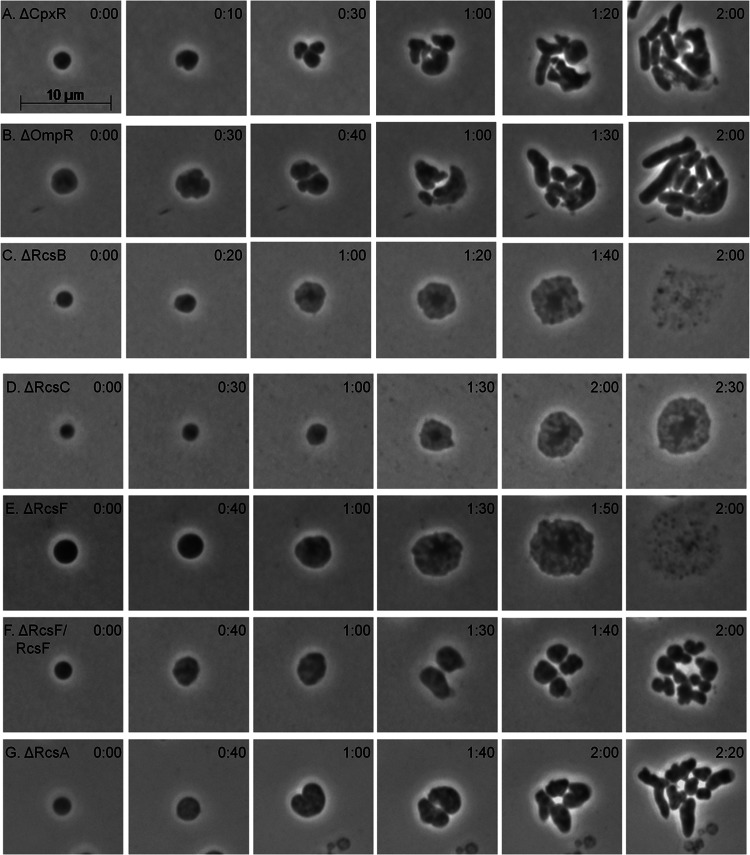
Envelope stress responses are induced by perturbations of the bacterial cell surface, including damage to the PG, the IM, or the OM, or events that affect the structure, integrity, or osmolarity of the periplasmic space (41–44). We therefore examined the behavior of spheroplasts unable to mount the Cpx, Omp, and Rcs responses. Spheroplasts prepared from ΔcpxR (Fig. 3A) or ΔompR (Fig. 3B) mutant cells divided and recovered, but a ΔrcsB mutant did not (Fig. 3C; see Movie M2 in the supplemental material). Instead, ΔrcsB spheroplasts gradually inflated to form giant spheroidal cells (∼2.7 times their original diameter) with a phase-dense center and reticulate cytoplasm (Fig. 3C; see Fig. S3 in the supplemental material). Of 20 such cells viewed for 2 to 3 h, 19 developed into large spheroids and all lysed (Fig. 3C, time 02:00; Table 1, strain DR1). A static scan of hundreds of other cells showed this to be universal (not shown). Lysis was not caused by the experimental conditions because the few rod-shaped cells that escaped lysozyme treatment grew normally (see Movie M2). Spheroplasts lacking either rcsC (Fig. 3D) or rcsF (Fig. 3E; see Movie M3) also lysed, and expression of wild-type rcsF in trans complemented the latter mutant (Fig. 3F). In contrast, mutants lacking the rcsA gene recovered normally (Fig. 3G). The lysozyme inhibitors Ivy and MliC are produced as part of the Rcs response (42, 45–47), but mutants lacking ivy and mliC recovered normally (Table 1), indicating that some other mechanism was at work. In sum, de novo shape recovery of LI spheroplasts required an RcsF- and RcsC-mediated response that proceeded by an RcsB-dependent but RcsA-independent pathway.
Fig 3.
Shape recovery of LI spheroplasts lacking different envelope stress responses. Spheroplasts were plated on soft agar overlays, and their development was monitored by time-lapse microscopy; phase-contrast images are shown. (A) E. coli DR2 (ΔCpxR). (B) E. coli DR3 (ΔOmpR). (C) E. coli DR1 (ΔRcsB). (D) E. coli DR4 (ΔRcsC). (E) E. coli DR5 (ΔRcsF). (F) E. coli DR5C (E. coli DR5 plus plasmid pRcsF, expressing wild-type rcsF). (G) E. coli DR6 (ΔRcsA). The time after plating is displayed in the upper right corner of each panel (hours:minutes). The scale bar represents 10 μm. The phase-contrast images in panels C and E were extracted from Movies M2 and M3 in the supplemental material, respectively. See also Fig. S3.
PBP1B and LpoB are required for shape recovery.
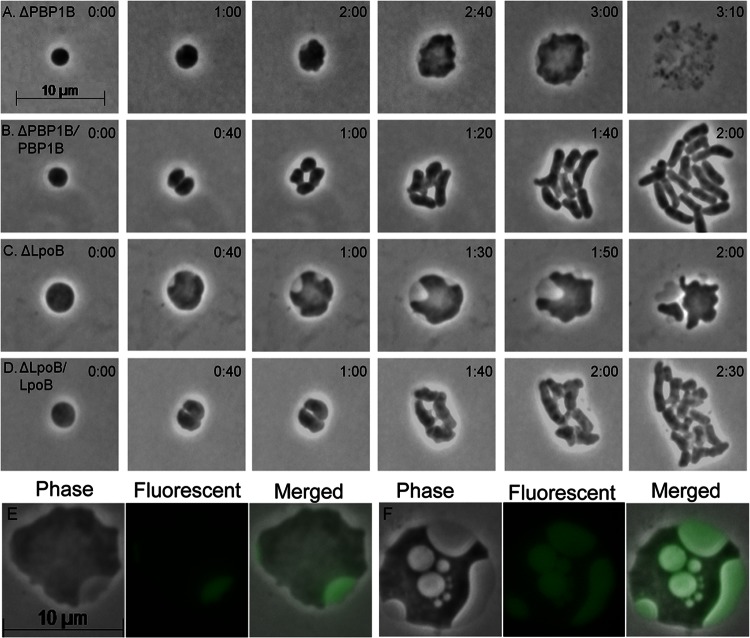
PBP1A and PBP1B polymerize and cross-link PG in E. coli (48), so it was likely that one or both were required for de novo shape recovery. Spheroplasts lacking PBP1A recovered normally (Table 1, strain DR9), but ΔmrcB spheroplasts lacking PBP1B enlarged to up to 1.6 times their original diameter and then lysed (Fig. 4A; see Fig. S3 and Movie M4 in the supplemental material). Of 70 ΔmrcB spheroplasts observed for 3 h or more, 65 lysed and 5 did not grow (Table 1, strain DR7). Expression of PBP1B in trans restored the wild-type phenotype (Fig. 4B). Lipoproteins LpoA and LpoB are essential for the proper functioning of PBP1A and PBP1B, respectively (49, 50), so we determined if these lipoproteins are required for shape recovery, as well. The lpoA mutant recovered normally (Table 1, strain DR10), whereas spheroplasts lacking lpoB did not recover but instead grew to up to two times their original diameter before lysing (Fig. 4C and Table 1, strain DR8; see Fig. S3 and Movie M5). Again, production of LpoB in trans complemented this defect (Fig. 4D). Thus, shape recovery required PBP1B and LpoB but not PBP1A or LpoA.
Fig 4.
PBP1B and LpoB are essential for the regeneration of rod-shaped cells from LI spheroplasts. (A to D) Spheroplasts were prepared from E. coli mutants and plated on sucrose soft agar overlays, and their development was monitored by time-lapse microscopy; phase-contrast images are shown. (A) E. coli DR7 (ΔPBP1B). (B) E. coli DR7C (DR7 containing plasmid pSK12, producing wild-type PBP1B). (C) E. coli DR8 (ΔLpoB). (D) E. coli DR8C (DR8 containing plasmid pLpoB, producing wild-type LpoB). (E and F) Periplasmic sfGFP (encoded by plasmid pDKR2) was produced by adding 100 μM isopropyl-β-d-thiogalactopyranoside (IPTG) to cells and incubating them for 1 h to induce gene expression, after which the cells were converted into spheroplasts and incubated for 2 h on a soft agar overlay medium containing IPTG. (E) E. coli DR7PG [ΔmrcB/dsbA(SS)-sfgfp]. (F) E. coli DR8PG [ΔlpoB/dsbA(SS)-sfgfp]. Note the presence of peripheral vesicles in the large spheroids derived from stains lacking PBP1B (A and E) and LpoB (C and F). The time after plating is displayed in the upper right corner of each panel (hours:minutes). The scale bar represents 10 μm. The phase-contrast images in panels A and C were extracted from Movies M4 and M5 in the supplemental material, respectively. See also Fig. S3.
Rcs and PBP1B affect separate processes.
The Rcs system and PBP1B could affect different recovery steps, or one system might regulate the other. Arguing for the two-pathway interpretation was the fact that the two types of spheroplasts differed visually. The cytoplasm of Δrcs spheroplasts was reticulate (Fig. 3C, D, and E; see Fig. S3 in the supplemental material), whereas that of spheroplasts lacking PBP1B-LpoB was uniform (Fig. 4A and C; see Fig. S3). Also, cells lacking PBP1B-LpoB developed peripheral vacuoles (Fig. 4A, C, E, and F) that were absent from spheroplasts lacking Rcs components (Fig. 3C, D, and E; see Fig. S3). Localization of the fluorescent protein DsbA(SS)-sfGFP marked these vacuoles as originating from the periplasm (Fig. 4E and F).
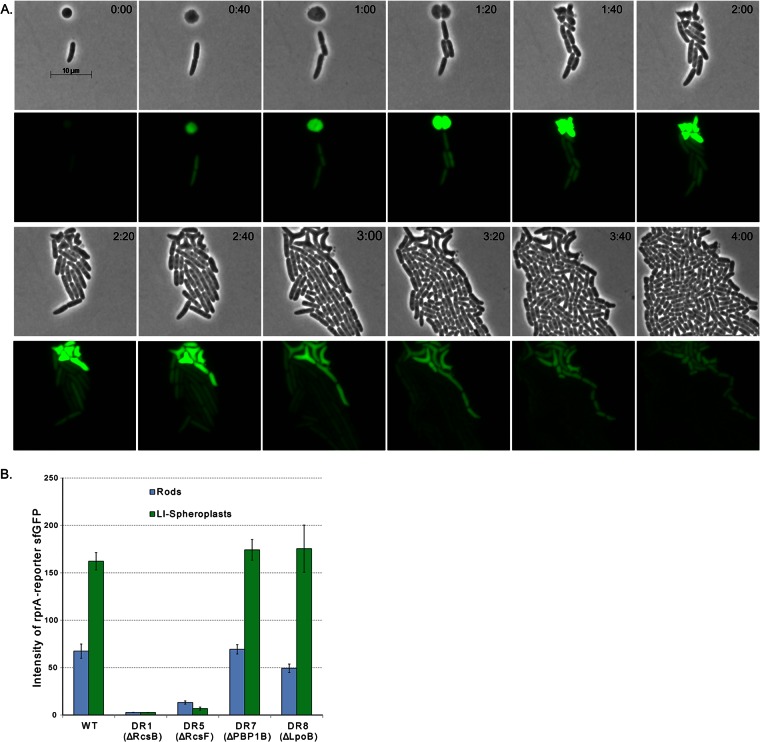
Notwithstanding these visual differences, the loss of PBP1B-LpoB might itself trigger the Rcs response. To test this, we observed the expression of sfgfp placed under the control of the Rcs-specific rprA promoter (51). As expected, sfGFP increased in spheroplasts derived from strain MG1655 (Fig. 5A and B), indicating that the Rcs response was induced by spheroplast formation. Spheroplasts lacking RcsB or RcsF did not express sfGFP, indicating that the assay was Rcs specific (Fig. 5B, ΔRcsB and ΔRcsF). Interestingly, this marker allowed us to identify and trace rod-shaped progeny that were derived from LI spheroplasts because they retained the fluorescent signal for several generations, whereas progeny descended from cells that began as rods rapidly lost their already low background fluorescence (Fig. 5A, times 1:00 and later). In rod-shaped cells lacking PBP1B-LpoB, sfGFP accumulated to the same level as in wild-type cells (Fig. 5B, ΔPBP1 and ΔLpoB, blue bars), proving that removal of these proteins did not trigger Rcs. Alternately, the absence of PBP1B-LpoB might prevent induction of the Rcs response. However, the response was just as great in spheroplasts lacking PBP1B or LpoB (Fig. 5B, ΔPBP1B and ΔLpoB, green bars). Thus, the absence of PBP1B-LpoB neither induced nor inhibited the Rcs response. Alternately, the Rcs response might reduce the expression of PBP1b or LpoB and thereby prevent spheroplast recovery. However, the inability of an rcsF or rcsB mutant to recover was not rescued when PBP1B and LpoB were produced simultaneously from separate plasmids (not shown), indicating that the Rcs response did not operate solely via an effect on these two proteins. Overall, the results suggest that the Rcs system and the PBP1b-LpoB proteins affect different stages of the recovery pathway.
Fig 5.
The Rcs system is induced in LI spheroplasts and their progeny. (A) Induction of the Rcs response was monitored by measuring sfGFP from rprA reporter plasmid pDKR1. In the typical example shown, sfGFP production was induced in the spheroplast and its rod-shaped progeny. Background amounts of sfGFP were low in rod-shaped cells that escaped lysozyme treatment (at the center of each panel, times 0:00 to 1:20) and disappeared from these cells during prolonged growth (times 1:40 to 4:00). Rows 1 and 3, phase-contrast images; rows 2 and 4, corresponding fluorescent images. The time after plating is displayed at the upper right of each panel (hours:minutes). The scale bar represents 10 μm. (B) Relative Rcs induction (sfGFP production from the rprA promoter) in rod-shaped cells (blue bars) and spheroplasts (green bars). Images were captured at 20 min after plating on sucrose recovery medium, and the relative amounts of sfGFP produced were determined by MicrobeTracker software. E. coli strains MG1655 (wild type [WT]), DR1 (ΔRcsB), DR5 (ΔRcsF), DR7 (ΔPBP1B), and DR8 (ΔLpoB) were assayed. The data were collected from 8 to 12 cells in each of two independent experiments. Error bars show the standard deviation of the mean.
PBP1B divisome partners are not required for recovery.
We tested the shape recovery ability of mutants lacking proteins that had some relationship with PBP1B or that were dispensable for cell division. PBP1B interacts with MltA when MipA is present (52), and AmiC and the Tol-Pal system play roles in daughter cell separation and in OM invagination, respectively (53–55). However, spheroplasts lacking any of these proteins recovered normally by producing aberrantly shaped and then regularly shaped rod cells (Table 1, strains DR13 to DR16). We note, though, that of 36 spheroplasts lacking Pal, 23 recovered but 13 lysed early (Table 1, strain DR15), for reasons that are not clear.
LI spheroplasts lacking Lpp grow as spheres.
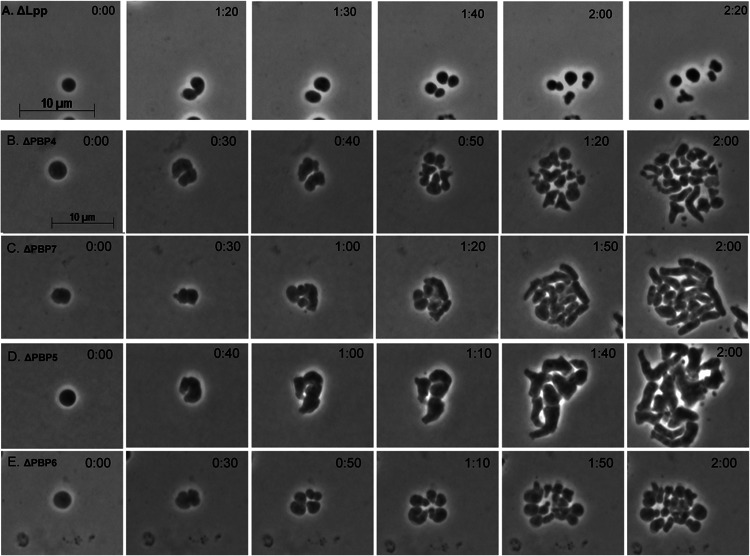
The major E. coli lipoprotein Lpp helps stabilize the Gram-negative cell envelope (56) and therefore might affect de novo shape recovery. Unlike all of the strains examined thus far, spheroplasts lacking Lpp divided into relatively equally sized spherical cells (Fig. 6A). Although ∼20% of these spheroplasts succumbed to early lysis (Table 1, strain DR11; see Movie M6 in the supplemental material), those daughter cells that survived produced spheroidal progeny (Fig. 6A). After four generations, lpp mutants produced microcolonies consisting primarily of spherical or nearly spherical cells (see Movie M6, time 2:30) with an occasion cell having a short branch (Fig. 6A, time 2:20; see Movie M6 in the supplemental material). Providing wild-type Lpp in trans reversed this phenotype (not shown).
Fig 6.
Lpp, PBP5, and PBP6 affect the trajectory by which LI spheroplasts regenerate rod-shaped cells. LI spheroplasts were prepared from E. coli mutants and placed on sucrose soft agar overlays, and their development was monitored by time-lapse microscopy; phase-contrast images are shown. (A) E. coli DR11 (ΔLpp). (B) E. coli AV14-1 (ΔPBP4). (C) E. coli AV15-1 (ΔPBP7). (D) E. coli AV21-1 (ΔPBP5). (E) E. coli AV76-1 (ΔPBP6). The time after plating is displayed in the upper right corner of each panel (hours:minutes). The scale bar represents 10 μm.
The absence of PBP5 or PBP6 alters the progression of shape recovery.
In E. coli, the low-molecular-weight PBPs modify PG, help cells maintain a uniform shape, and aid in daughter cell separation (18, 19, 29, 57–60). Spheroplasts lacking PBP4 or PBP7 recovered their normal rod shapes, though PBP4 mutants may have produced more aberrant intermediates (Fig. 6B and C). Spheroplasts lacking PBP5 produced large filamentous cells, with many having bends and branches (Fig. 6D, times 1:10 to 2:00). Most of these eventually regained nearly normal rod shapes (not shown). Although PBP6 is closely related to PBP5, spheroplasts lacking PBP6 produced microcolonies of mostly spherical cells (Fig. 6E). Rodlike protrusions erupted from a few of these and eventually elongated to form cells with normal rod shapes (Fig. 6E, time 2:00; data not shown). Thus, spheroplasts lacking PBP5 or PBP6 altered the trajectory of shape recovery in different ways.
Shape recovery does not depend on residual PG content.
One straightforward explanation for the observed differences in spheroplast recovery was that the process might depend on the overall degree of PG digestion. That is, some mutants might admit more lysozyme, which would degrade more cell wall and lead to an inability to recover, perhaps because the remaining PG seeded the rebuilding of a normal morphology. However, at least one strain, the pal mutant, has an OM defect that would be expected to admit more lysozyme (55), but these pal spheroplasts still recovered normally. Also, spheroplasts created by using lysozyme concentrations of 20 to 100 μg/ml exhibited the same course of shape recovery (not shown). Thus, the important factor did not seem to be how much lysozyme entered cells during spheroplast formation. However, to test this possibility more quantitatively, we labeled wild-type and mutant cells with fluorescent d-alanine, produced LI spheroplasts by adding lysozyme, and quantified the remaining fluorescent signal in rod-shaped cells and in spheroplasts of the parent strain and in mutants lacking RcsB, LpoB, PBP1B, or Lpp. Spheroplasts had much less PG than rods that had escaped lysozyme treatment (see Fig. S4 in the supplemental material), indicating that most of the PG in spheroplasts had been degraded. Specifically, the PG signals in spheroplasts (compared to rods) were as follows: wild-type, 2%; ΔRcsB, 6%; ΔLpoB, 13%; ΔPBP1B, 14%; ΔLpp, 4% (see Fig. S4). None of the values were significantly different from one another, suggesting that there was no direct correlation between the amount of leftover PG and the ability to recover. Thus, differences in lysozyme access or PG content did not seem to explain why some mutants recovered and others did not. In any case, all of the spheroplasts that could do so recreated their original shapes without having an explicitly rodlike template.
DISCUSSION
Virtually all of the previous work on bacterial morphology has focused on conditions that interfere with a cell's ability to retain its wild-type shape. Here, we developed a system to investigate how bacteria can reconstruct a properly shaped cell wall de novo, i.e., beginning with little to no PG or, at the very least, in the absence of a rodlike template. Recovery progresses via a series of markedly aberrant cells that probably arise because septa are placed inaccurately, consistent with earlier observations (19). Most importantly, we find that at least one stress response system (Rcs) and three accessory proteins (PBP1B, LpoB, and Lpp) are required for shape recovery even though all are dispensable in rod-shaped cells (Fig. 7). These results suggest that in the absence of a preexisting PG template, E. coli must supplement its normal cell wall synthetic machinery if it is to regenerate a wild-type morphology.
Fig 7.
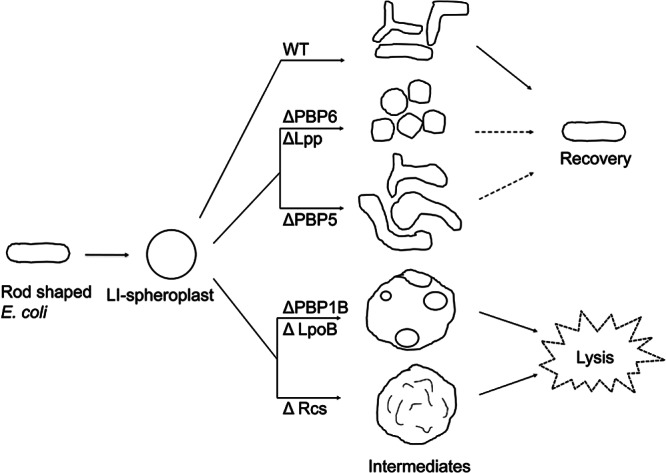
Schematic showing how different mutations affect the shape regeneration of E. coli LI spheroplasts. Through a series of reductive division events and periods of elongation, LI spheroplasts derived from wild-type (WT) cells produce branched intermediates that eventually give rise to normally shaped rod cells (far right). LI spheroplasts lacking Lpp or PBP6 produce mostly coccoidal cells, and only a few may eventually become rod shaped. Cells lacking PBP6 that become rod shaped do so by generating an elongated protrusion instead of by reductive division (not shown). LI spheroplasts lacking PBP5 produce a very high proportion of extremely aberrantly shaped and branched cells before reproducing their normal rod-shaped dimensions. LI spheroplasts lacking PBP1B, LpoB, or components of the Rcs stress pathway do not recover but instead enlarge and lyse. The enlarged spheroidal cells of mutants lacking PBP1B or LpoB contain peripheral vesicles derived by partial invagination of the IM, but such vesicles are absent from spheroidal cells derived from cells that cannot mount an Rcs response.
That lysozyme itself can produce spheroplasts seems unlikely to most contemporary investigators because it is assumed that the OM of Gram-negative bacteria prevents the enzyme from reaching the periplasmic PG. However, analogous wall-less cells were generated in just this way over 40 years ago (26, 34). For historical and semantic consistency, we have adopted the terminology of Birdsell and Cota-Robles and refer to these as LI spheroplasts. Similarly, we refer to spherical cells created in the presence of EDTA as EDTA LI spheroplasts. This terminology can be expanded to include other ways of generating spheroidal Gram-negative cells, thus maintaining the structural concepts now associated with such cells while distinguishing among entities created by different protocols. The lysozyme treatment is simple and transient and leaves most cells viable, and the resulting spheroplasts are similar to wild-type cells in size. Other model systems (e.g., β-lactam treatment) produce cells that are 4 to 30 times larger (61–63) and are therefore less useful for examining morphological recovery.
The requirements for de novo shape recovery are similar to those for the survival of E coli L-form-like cells (which require PBP1B) (61) and for the survival of L-form bacteria (which require RcsC, RcsB, RcsF, and PBP1B) (64). Here, the behavior of LI spheroplasts suggests that deficiencies in the PBP1B or Rcs system interfere with division in cells lacking PG. A plausible reason for the PBP1B-LpoB requirement is that these proteins may constitute an alternative to the Tol-Pal invagination mechanism (55). Tol-Pal is thought to link PG to the IM and OM (55), a connection that may not occur in the absence of PG. In fact, spheroplasts lacking pal recovered normally (not shown), suggesting that an alternate invagination system must be at work. In addition, spheroplasts lacking PBP1B-LpoB contained large periplasmic bays, implying that these cells could initiate IM cytokinesis but could not coinvaginate the OM. Thus, PBP1B-LpoB may mediate invagination until sufficient PG is available for the Tol-Pal system to resume its function or perhaps, because PBP1B is the major PG synthase, other enzymes are unable to make enough PG for spheroplasts to recover. The existence of two mechanisms that perform this function begs the question of whether E. coli regularly encounters situations in which all or most of its PG is degraded.
Rcs stress response proteins are present throughout the family Enterobacteriaceae (65), suggesting that they are an important survival mechanism for common normal flora and intestinal pathogens. Damage to the cell envelope or wall triggers the response, which controls the expression of over 150 genes (42, 66, 67). We do not yet know which proteins in this regulon are required for shape recovery, but the proteins responsible are not the lysozyme inhibitors Ivy and MliC, nor are the agents responsible synthesized by the RcsA branch of the pathway. This rules out colanic acid as a candidate (41, 68), even though this compound is essential for the survival of L-form bacteria (61, 64). Because the Rcs regulon includes no other PG-related genes (67), it seems that novel cell wall functionaries remain to be discovered.
The lipoprotein Lpp affects shape recovery differently than do the Rcs proteins or PBP1B. LI spheroplasts lacking Lpp divide but grow as spherical cells, and only a few recover normal rod shapes after a significant delay. Thus, Lpp has a heretofore unappreciated effect on cell shape. A complete O antigen in the outer leaflet of the OM decreases the frequency of shape abnormalities in PBP mutants, suggesting that OM integrity contributes to cellular architecture (69). Lpp is thought to stabilize the envelope by linking PG to the OM (56, 70), and so, Lpp may contribute to shape recovery by providing additional physical support or by creating the proper milieu in which relevant OM proteins can function. Recovering spheroplasts lacking PBP6 bear some similarity to Lpp mutants in that they grow as spheroidal cells. A difference is that the few cells that eventually regain a normal morphology do so by elaborating rodlike protrusions. Curiously, this morphological trajectory is unlike that of spheroplasts lacking the closely related protein PBP5 and highlights yet another physiological difference between these two proteins. Overall, the behavior of cells lacking Lpp or PBP6 suggests that de novo shape generation requires novel accessory pathways that probably assist the PG-synthesizing systems directed by FtsZ or MreB.
Finally, the present results have intriguing implications for the survival of commensal and pathogenic Gram-negative bacteria in a host environment. Most revealing is that, in the lab, E. coli grows normally and with wild-type morphology in the absence of the Rcs response, PBP1B, LpoB, Lpp, or PBP6. There is some periplasmic leakiness in lpp mutants (56, 71) and a slight β-lactam sensitivity in cells lacking PBP1B (72), but we show that these proteins play a critical role when the cell wall is removed or severely damaged. In the host, lysozyme and cationic antimicrobial peptides can produce either of these outcomes and both can trigger the Rcs stress response (42, 43, 45, 67). Lysozyme is a fundamental, innate defense that is prevalent in numerous tissues and secretions (73, 74–76), and it is present at quite high concentrations in several fluids, including tears (at ∼1.5 mg/ml) (77), saliva (0.09 mg/ml) (78), milk (0.24 to 0.89 mg/ml) (79), stomach fluid (80), and respiratory and nasal fluids (0.5 mg/ml) (81). Because lysozyme is confined to the mucus lining and crypts of the intestine (82), its true concentration is probably higher than that measured in bulk samples. In view of its widespread distribution and high concentrations, interactions between lysozyme and bacteria must be extensive and intense. Thus, it is likely that, in their natural habitat, E. coli and other intestinal flora are exposed frequently to concentrations of lysozyme that can remove or seriously damage the cell wall. Proteins required for de novo shape recovery may be essential under these dire circumstances, allowing cells to recover after they find themselves in a more congenial situation. In sum, not only does the behavior of spheroplasts represent a new way to examine the mechanics of shape generation in Gram-negative bacteria, it also hints that commensal organisms and pathogens may survive some host defenses by executing a similar spheroplast-to-rod transformation in their natural environments.
Supplementary Material
ACKNOWLEDGMENTS
We thank Suresh Kannan for plasmid pSK12 and assistance and Mary Laubacher for insights regarding the Rcs phosphorelay system. We are very grateful to Erkin Kuru and Michael S. VanNieuwenhze for providing HADA and to Miguel de Pedro for anti-murein antibody.
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award R01-GM61019 and by the Arkansas Biosciences Institute, the major research component of the Arkansas Tobacco Settlement Proceeds Act of 2000.
Footnotes
Published ahead of print 29 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00160-13.
REFERENCES
- 1. Young KD. 2006. The selective value of bacterial shape. Microbiol. Mol. Biol. Rev. 70:660–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Young KD. 2007. Bacterial morphology: why have different shapes? Curr. Opin. Microbiol. 10:596–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wanger G, Onstott TC, Southam G. 2008. Stars of the terrestrial deep subsurface: a novel ‘star-shaped’ bacterial morphotype from a South African platinum mine. Geobiology 6:325–330 [DOI] [PubMed] [Google Scholar]
- 4. Justice SS, Hunstad DA, Cegelski L, Hultgren SJ. 2008. Morphological plasticity as a bacterial survival strategy. Nat. Rev. Microbiol. 6:162–168 [DOI] [PubMed] [Google Scholar]
- 5. Rosen DA, Hooton TM, Stamm WE, Humphrey PA, Hultgren SJ. 2007. Detection of intracellular bacterial communities in human urinary tract infection. PLoS Med. 4:e329 doi:10.1371/journal.pmed.0040329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Horvath DJ, Jr, Li B, Casper T, Partida-Sanchez S, Hunstad DA, Hultgren SJ, Justice SS. 2011. Morphological plasticity promotes resistance to phagocyte killing of uropathogenic Escherichia coli. Microbes Infect. 13:426–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sycuro LK, Pincus Z, Gutierrez KD, Biboy J, Stern CA, Vollmer W, Salama NR. 2010. Peptidoglycan crosslinking relaxation promotes Helicobacter pylori's helical shape and stomach colonization. Cell 141:822–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sycuro LK, Wyckoff TJ, Biboy J, Born P, Pincus Z, Vollmer W, Salama NR. 2012. Multiple peptidoglycan modification networks modulate Helicobacter pylori's cell shape, motility, and colonization potential. PLoS Pathog. 8:e1002603 doi:10.1371/journal.ppat.1002603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Frirdich E, Biboy J, Adams C, Lee J, Ellermeier J, Gielda LD, Dirita VJ, Girardin SE, Vollmer W, Gaynor EC. 2012. Peptidoglycan-modifying enzyme Pgp1 is required for helical cell shape and pathogenicity traits in Campylobacter jejuni. PLoS Pathog. 8:e1002602 doi:10.1371/journal.ppat.1002602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rowan NJ, Kirf D, Tomkins P. 2009. Studies on the susceptibility of different culture morphotypes of Listeria monocytogenes to uptake and survival in human polymorphonuclear leukocytes. FEMS Immunol. Med. Microbiol. 57:183–192 [DOI] [PubMed] [Google Scholar]
- 11. Young KD. 2010. Bacterial shape: two-dimensional questions and possibilities. Annu. Rev. Microbiol. 64:223–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. den Blaauwen T, de Pedro MA, Nguyen-Disteche M, Ayala JA. 2008. Morphogenesis of rod-shaped sacculi. FEMS Microbiol. Rev. 32:321–344 [DOI] [PubMed] [Google Scholar]
- 13. Margolin W. 2009. Sculpting the bacterial cell. Curr. Biol. 19:R812–R822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Carballido-López R. 2006. Orchestrating bacterial cell morphogenesis. Mol. Microbiol. 60:815–819 [DOI] [PubMed] [Google Scholar]
- 15. Takacs CN, Poggio S, Charbon G, Pucheault M, Vollmer W, Jacobs-Wagner C. 2010. MreB drives de novo rod morphogenesis in Caulobacter crescentus via remodeling of the cell wall. J. Bacteriol. 192:1671–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. de Boer PAJ. 2010. Advances in understanding E. coli cell fission. Curr. Opin. Microbiol. 13:730–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Young KD. 2003. Bacterial shape. Mol. Microbiol. 49:571–580 [DOI] [PubMed] [Google Scholar]
- 18. Varma A, Young KD. 2004. FtsZ collaborates with penicillin binding proteins to generate bacterial cell shape in Escherichia coli. J. Bacteriol. 186:6768–6774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Potluri L-P, de Pedro MA, Young KD. 2012. Escherichia coli low-molecular-weight penicillin-binding proteins help orient septal FtsZ, and their absence leads to asymmetric cell division and branching. Mol. Microbiol. 84:203–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Waidner B, Specht M, Dempwolff F, Haeberer K, Schaetzle S, Speth V, Kist M, Graumann PL. 2009. A novel system of cytoskeletal elements in the human pathogen Helicobacter pylori. PLoS Pathog. 5:e1000669 doi:10.1371/journal.ppat.1000669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ausmees N, Kuhn JR, Jacobs-Wagner C. 2003. The bacterial cytoskeleton: an intermediate filament-like function in cell shape. Cell 115:705–713 [DOI] [PubMed] [Google Scholar]
- 22. Jones LJ, Carballido-Lopez R, Errington J. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104:913–922 [DOI] [PubMed] [Google Scholar]
- 23. Kruse T, Moller-Jensen J, Lobner-Olesen A, Gerdes K. 2003. Dysfunctional MreB inhibits chromosome segregation in Escherichia coli. EMBO J. 22:5283–5292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kawai Y, Asai K, Errington J. 2009. Partial functional redundancy of MreB isoforms, MreB, Mbl and MreBH, in cell morphogenesis of Bacillus subtilis. Mol. Microbiol. 73:719–731 [DOI] [PubMed] [Google Scholar]
- 25. Lederberg J, St Clair J. 1958. Protoplasts and L-type growth of Escherichia coli. J. Bacteriol. 75:143–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zinder ND, Arndt WF. 1956. Production of protoplasts of Escherichia coli by lysozyme treatment. Proc. Natl. Acad. Sci. U. S. A. 42:586–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Makemson JC, Darwish RZ. 1972. Calcium requirement and magnesium stimulation of Escherichia coli L-form induction. Infect. Immun. 6:880–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ruthe HJ, Adler J. 1985. Fusion of bacterial spheroplasts by electric fields. Biochim. Biophys. Acta 819:105–113 [DOI] [PubMed] [Google Scholar]
- 29. Denome SA, Elf PK, Henderson TA, Nelson DE, Young KD. 1999. Escherichia coli mutants lacking all possible combinations of eight penicillin binding proteins: viability, characteristics, and implications for peptidoglycan synthesis. J. Bacteriol. 181:3981–3993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Meberg BM, Sailer FC, Nelson DE, Young KD. 2001. Reconstruction of Escherichia coli mrcA (PBP 1a) mutants lacking multiple combinations of penicillin binding proteins. J. Bacteriol. 183:6148–6149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Potluri L, Karczmarek A, Verheul J, Piette A, Wilkin JM, Werth N, Banzhaf M, Vollmer W, Young KD, Nguyen-Disteche M, den Blaauwen T. 2010. Septal and lateral wall localization of PBP5, the major d,d-carboxypeptidase of Escherichia coli, requires substrate recognition and membrane attachment. Mol. Microbiol. 77:300–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dinh T, Bernhardt TG. 2011. Using superfolder green fluorescent protein for periplasmic protein localization studies. J. Bacteriol. 193:4984–4987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pédelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. 2006. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24:79–88 [DOI] [PubMed] [Google Scholar]
- 34. Birdsell DC, Cota-Robles EH. 1967. Production and ultrastructure of lysozyme and ethylenediaminetetraacetate-lysozyme spheroplasts of Escherichia coli. J. Bacteriol. 93:427–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sullivan CJ, Morrell JL, Allison DP, Doktycz MJ. 2005. Mounting of Escherichia coli spheroplasts for AFM imaging. Ultramicroscopy 105:96–102 [DOI] [PubMed] [Google Scholar]
- 36. Li G, Young KD. 2012. Isolation and identification of new inner membrane-associated proteins that localize to cell poles in Escherichia coli. Mol. Microbiol. 84:276–295 [DOI] [PubMed] [Google Scholar]
- 37. Kuru E, Hughes HV, Brown PJ, Hall E, Tekkam S, Cava F, de Pedro MA, Brun YV, VanNieuwenhze MS. 2012. In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent d-amino acids. Angew. Chem. Int. Ed. Engl. 51:12519–12523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hurwitz C, Reiner JM, Landau JV. 1958. Studies in the physiology and biochemistry of penicillin-induced spheroplasts of Escherichia coli. J. Bacteriol. 76:612–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sandlin RC, Goldberg MB, Maurelli AT. 1996. Effect of O side-chain length and composition on the virulence of Shigella flexneri 2a. Mol. Microbiol. 22:63–73 [DOI] [PubMed] [Google Scholar]
- 40. Rick PD, Hubbard GL, Barr K. 1994. Role of the rfe gene in the synthesis of the O8 antigen in Escherichia coli K-12. J. Bacteriol. 176:2877–2884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Clarke DJ. 2010. The Rcs phosphorelay: more than just a two-component pathway. Future Microbiol. 5:1173–1184 [DOI] [PubMed] [Google Scholar]
- 42. Laubacher ME, Ades SE. 2008. The Rcs phosphorelay is a cell envelope stress response activated by peptidoglycan stress and contributes to intrinsic antibiotic resistance. J. Bacteriol. 190:2065–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Farris C, Sanowar S, Bader MW, Pfuetzner R, Miller SI. 2010. Antimicrobial peptides activate the Rcs regulon through the outer membrane lipoprotein RcsF. J. Bacteriol. 192:4894–4903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ize B, Porcelli I, Lucchini S, Hinton JC, Berks BC, Palmer T. 2004. Novel phenotypes of Escherichia coli tat mutants revealed by global gene expression and phenotypic analysis. J. Biol. Chem. 279:47543–47554 [DOI] [PubMed] [Google Scholar]
- 45. Callewaert L, Vanoirbeek KG, Lurquin I, Michiels CW, Aertsen A. 2009. The Rcs two-component system regulates expression of lysozyme inhibitors and is induced by exposure to lysozyme. J. Bacteriol. 191:1979–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Abergel C, Monchois V, Byrne D, Chenivesse S, Lembo F, Lazzaroni JC, Claverie JM. 2007. Structure and evolution of the Ivy protein family, unexpected lysozyme inhibitors in Gram-negative bacteria. Proc. Natl. Acad. Sci. U. S. A. 104:6394–6399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yum S, Kim MJ, Xu Y, Jin XL, Yoo HY, Park JW, Gong JH, Choe KM, Lee BL, Ha NC. 2009. Structural basis for the recognition of lysozyme by MliC, a periplasmic lysozyme inhibitor in Gram-negative bacteria. Biochem. Biophys. Res. Commun. 378:244–248 [DOI] [PubMed] [Google Scholar]
- 48. Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. 2008. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 32:234–258 [DOI] [PubMed] [Google Scholar]
- 49. Typas A, Banzhaf M, van den Berg van Saparoea B, Verheul J, Biboy J, Nichols RJ, Zietek M, Beilharz K, Kannenberg K, von Rechenberg M, Breukink E, den Blaauwen T, Gross CA, Vollmer W. 2010. Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell 143:1097–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Paradis-Bleau C, Markovski M, Uehara T, Lupoli TJ, Walker S, Kahne DE, Bernhardt TG. 2010. Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell 143:1110–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Majdalani N, Hernandez D, Gottesman S. 2002. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol. Microbiol. 46:813–826 [DOI] [PubMed] [Google Scholar]
- 52. Vollmer W, von Rechenberg M, Holtje JV. 1999. Demonstration of molecular interactions between the murein polymerase PBP1B, the lytic transglycosylase MltA, and the scaffolding protein MipA of Escherichia coli. J. Biol. Chem. 274:6726–6734 [DOI] [PubMed] [Google Scholar]
- 53. Heidrich C, Templin MF, Ursinus A, Merdanovic M, Berger J, Schwarz H, de Pedro MA, Höltje JV. 2001. Involvement of N-acetylmuramyl-l-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol. Microbiol. 41:167–178 [DOI] [PubMed] [Google Scholar]
- 54. Bernhardt TG, de Boer PA. 2003. The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol. Microbiol. 48:1171–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gerding MA, Ogata Y, Pecora ND, Niki H, de Boer PA. 2007. The trans-envelope Tol-Pal complex is part of the cell division machinery and required for proper outer-membrane invagination during cell constriction in E. coli. Mol. Microbiol. 63:1008–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Suzuki H, Nishimura Y, Yasuda S, Nishimura A, Yamada M, Hirota Y. 1978. Murein-lipoprotein of Escherichia coli: a protein involved in the stabilization of bacterial cell envelope. Mol. Gen. Genet. 167:1–9 [DOI] [PubMed] [Google Scholar]
- 57. Ghosh AS, Young KD. 2003. Sequences near the active site in chimeric penicillin binding proteins 5 and 6 affect uniform morphology of Escherichia coli. J. Bacteriol. 185:2178–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nelson DE, Young KD. 2001. Contributions of PBP 5 and dd-carboxypeptidase penicillin binding proteins to maintenance of cell shape in Escherichia coli. J. Bacteriol. 183:3055–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Nilsen T, Ghosh AS, Goldberg MB, Young KD. 2004. Branching sites and morphological abnormalities behave as ectopic poles in shape-defective Escherichia coli. Mol. Microbiol. 52:1045–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Priyadarshini R, Popham DL, Young KD. 2006. Daughter cell separation by penicillin-binding proteins and peptidoglycan amidases in Escherichia coli. J. Bacteriol. 188:5345–5355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Joseleau-Petit D, Liebart JC, Ayala JA, D'Ari R. 2007. Unstable Escherichia coli L forms revisited: growth requires peptidoglycan synthesis. J. Bacteriol. 189:6512–6520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Domínguez-Cuevas P, Mercier R, Leaver M, Kawai Y, Errington J. 2012. The rod to L-form transition of Bacillus subtilis is limited by a requirement for the protoplast to escape from the cell wall sacculus. Mol. Microbiol. 83:52–66 [DOI] [PubMed] [Google Scholar]
- 63. Dell'Era S, Buchrieser C, Couve E, Schnell B, Briers Y, Schuppler M, Loessner MJ. 2009. Listeria monocytogenes L-forms respond to cell wall deficiency by modifying gene expression and the mode of division. Mol. Microbiol. 73:306–322 [DOI] [PubMed] [Google Scholar]
- 64. Glover WA, Yang Y, Zhang Y. 2009. Insights into the molecular basis of L-form formation and survival in Escherichia coli. PLoS One 4:e7316 doi:10.1371/journal.pone.0007316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Huang YH, Ferrieres L, Clarke DJ. 2006. The role of the Rcs phosphorelay in Enterobacteriaceae. Res. Microbiol. 157:206–212 [DOI] [PubMed] [Google Scholar]
- 66. Ferrières L, Clarke DJ. 2003. The RcsC sensor kinase is required for normal biofilm formation in Escherichia coli K-12 and controls the expression of a regulon in response to growth on a solid surface. Mol. Microbiol. 50:1665–1682 [DOI] [PubMed] [Google Scholar]
- 67. Ades SE, Hayden JD, Laubacher ME. 2011. Envelope stress, p 115–131 In Storz G, Hengge R. (ed), Bacterial stress responses, 2nd ed ASM Press, Washington, DC [Google Scholar]
- 68. Majdalani N, Gottesman S. 2005. The Rcs phosphorelay: a complex signal transduction system. Annu. Rev. Microbiol. 59:379–405 [DOI] [PubMed] [Google Scholar]
- 69. Ghosh AS, Melquist AL, Young KD. 2006. Loss of O-antigen increases cell shape abnormalities in penicillin-binding protein mutants of Escherichia coli. FEMS Microbiol. Lett. 263:252–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Nikaido H. 1996. Outer membrane, p 1549 In Neidhardt FC, Curtiss R, III, Ingraham JL, Lin CC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE. (ed), Escherichia coli and Salmonella: cellular and molecular biology, vol 1 American Society for Microbiology, Washington, DC [Google Scholar]
- 71. Hirota Y, Suzuki H, Nishimura Y, Yasuda S. 1977. On the process of cellular division in Escherichia coli: a mutant of E. coli lacking a murein-lipoprotein. Proc. Natl. Acad. Sci. U. S. A. 74:1417–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. García del Portillo F, de Pedro MA. 1990. Differential effect of mutational impairment of penicillin-binding proteins 1A and 1B on Escherichia coli strains harboring thermosensitive mutations in the cell division genes ftsA, ftsQ, ftsZ, and pbpB. J. Bacteriol. 172:5863–5870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Müller CA, Autenrieth IB, Peschel A. 2005. Innate defenses of the intestinal epithelial barrier. Cell. Mol. Life Sci. 62:1297–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Fleming A. 1922. On a remarkable bacteriolytic element found in tissues and secretions. Proc. R. Soc. Lond. B 93:306–317 [Google Scholar]
- 75. Mason DY, Taylor CR. 1975. The distribution of muramidase (lysozyme) in human tissues. J. Clin. Pathol. 28:124–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Klockars M, Reitamo S. 1975. Tissue distribution of lysozyme in man. J. Histochem. Cytochem. 23:932–940 [DOI] [PubMed] [Google Scholar]
- 77. Aine E, Morsky P. 1984. Lysozyme concentration in tears—assessment of reference values in normal subjects. Acta Ophthalmol. (Copenh.) 62:932–938 [DOI] [PubMed] [Google Scholar]
- 78. Perera S, Uddin M, Hayes JA. 1997. Salivary lysozyme: a noninvasive marker for the study of the effects of stress of natural immunity. Int. J. Behav. Med. 4:170–178 [DOI] [PubMed] [Google Scholar]
- 79. Montagne P, Cuilliere ML, Mole C, Bene MC, Faure G. 2001. Changes in lactoferrin and lysozyme levels in human milk during the first twelve weeks of lactation. Adv. Exp. Med. Biol. 501:241–247 [DOI] [PubMed] [Google Scholar]
- 80. Jollès J, Prager EM, Alnemri ES, Jolles P, Ibrahimi IM, Wilson AC. 1990. Amino acid sequences of stomach and nonstomach lysozymes of ruminants. J. Mol. Evol. 30:370–382 [DOI] [PubMed] [Google Scholar]
- 81. Cole AM, Liao HI, Stuchlik O, Tilan J, Pohl J, Ganz T. 2002. Cationic polypeptides are required for antibacterial activity of human airway fluid. J. Immunol. 169:6985–6991 [DOI] [PubMed] [Google Scholar]
- 82. Meyer-Hoffert U, Hornef MW, Henriques-Normark B, Axelsson LG, Midtvedt T, Putsep K, Andersson M. 2008. Secreted enteric antimicrobial activity localises to the mucus surface layer. Gut 57:764–771 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.