Abstract
NtrYX is a sensor-histidine kinase/response regulator two-component system that has had limited characterization in a small number of Alphaproteobacteria. Phylogenetic analysis of the response regulator NtrX showed that this two-component system is extensively distributed across the bacterial domain, and it is present in a variety of Betaproteobacteria, including the human pathogen Neisseria gonorrhoeae. Microarray analysis revealed that the expression of several components of the respiratory chain was reduced in an N. gonorrhoeae ntrX mutant compared to that in the isogenic wild-type (WT) strain 1291. These included the cytochrome c oxidase subunit (ccoP), nitrite reductase (aniA), and nitric oxide reductase (norB). Enzyme activity assays showed decreased cytochrome oxidase and nitrite reductase activities in the ntrX mutant, consistent with microarray data. N. gonorrhoeae ntrX mutants had reduced capacity to survive inside primary cervical cells compared to the wild type, and although they retained the ability to form a biofilm, they exhibited reduced survival within the biofilm compared to wild-type cells, as indicated by LIVE/DEAD staining. Analyses of an ntrX mutant in a representative alphaproteobacterium, Rhodobacter capsulatus, showed that cytochrome oxidase activity was also reduced compared to that in the wild-type strain SB1003. Taken together, these data provide evidence that the NtrYX two-component system may be a key regulator in the expression of respiratory enzymes and, in particular, cytochrome c oxidase, across a wide range of proteobacteria, including a variety of bacterial pathogens.
INTRODUCTION
Respiration is a key process for cellular survival and energy generation. Specific enzymes have evolved to mediate efficient respiration under conditions of varying oxygen availability and in the absence of this electron acceptor (reviewed in reference 1). Recent work has linked cytochrome oxidases to the virulence of several bacteria, including Neisseria spp. (2, 3). The cytochrome oxidase mediating these effects is cytochrome cbb3 (cytochrome oxidase; Cco), a respiratory oxidase that is present in a wide variety of nonenteric bacteria (4). Cytochrome cbb3 accepts electrons from c-type cytochromes and terminates a respiratory chain in which electrons from ubiquinol are transferred to c-type cytochromes via a cytochrome bc1 complex. Cytochrome cbb3 can use ascorbate and N,N,N,N-tetra-methyl-p-phenylenediamine (TMPD) as an artificial electron donor in respiration (5), and its presence results in the bacteria staining positive in the classical oxidase test (6, 7). Enteric bacteria that only possess oxidases that directly oxidize ubiquinol, such as Escherichia coli, yield a negative oxidase test. An important characteristic of cytochrome cbb3 is its remarkable affinity for oxygen, often being in the nM range (5, 8–10), with activity levels being highest under microaerobic or oxygen-limited conditions (8, 11, 12).
Two-component systems (TCSs) composed of a sensor histidine kinase (SHK) and response regulator (RR) have emerged as key signal transducers and global regulators of gene expression in prokaryotes. TCSs are known to regulate metabolic and respiratory processes. For example, in enteric bacteria the ArcBA TCS regulates the expression of many metabolic genes (13), including components of the citric acid cycle (e.g., succinate dehydrogenase and malate dehydrogenase) (14, 15) and aerobic quinol oxidase complexes (16, 17). The regulation of cytochrome cbb3 expression in proteobacteria has mostly been studied in Rhodobacter capsulatus, where the RegBA TCS and FNR are involved (18–20). This suggests that TCS may play a role in modulating cytochrome cbb3 expression in other bacteria. As the RegBA TCS system, like ArcBA, is a master regulator of many cellular functions, it is plausible to expect that respiration may be controlled by an alternative TCS(s) in those bacteria possessing a cytochrome cbb3 but lacking RegBA. In contrast to the situation in metabolically versatile bacteria, such as Rhodobacter spp., the human-adapted pathogen Neisseria gonorrhoeae is known to contain cytochrome cbb3 as its only respiratory oxidase (2). Furthermore, N. gonorrhoeae contains only five TCSs (GenBank accession no. AE004969), none of which resembles RegBA. However, one of these five N. gonorrhoeae TCSs appears to be an ortholog of the NtrYX system that plays a role in the regulation of nitrogen fixation and metabolism in some bacteria (21, 22). Despite having been implicated in regulating various cellular processes, NtrYX is usually regarded as being associated with the NtrBC system in Alphaproteobacteria (23) and is typically associated with the regulation of nitrogen metabolism. However, N. gonorrhoeae (a betaproteobacterium) does not contain an NtrBC system, raising the question of the functional role of NtrYX in a bacterium that lacks metabolic versatility. We report herein the role of NtrX in two unrelated “oxidase-positive” bacteria. Characterization of an ntrX mutant in N. gonorrhoeae shows that this response regulator has a role in regulating the expression of respiratory enzymes and a key role in the adaptive ability of this bacterium. The effect of an ntrX mutation on cytochrome oxidase and respiratory chain activity in the facultative phototroph R. capsulatus is also briefly described. These results, together with recent observations on the role of the sensor histidine kinase NtrY in Brucella abortus (24), led us to propose that the NtrYX TCS is a regulator of respiratory gene expression in a diverse range of oxidase-positive bacteria that includes several bacterial pathogens.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
N. gonorrhoeae strains were grown on GC agar supplemented with IsoVitaleX (1% [vol/vol]; Becton, Dickinson) at 37°C with 5% (vol/vol) CO2, or in supplemented brain heart infusion or GC (sBHI or sGC, respectively) broth supplemented with Levinthal's base (10% [vol/vol]) and IsoVitaleX (1% [vol/vol]). R. capsulatus SB1003 was routinely grown on either RCV (25) or TYS medium (26) at 28°C. Growth media were supplemented with antibiotics as described previously (27). Escherichia coli strains DH5α, JM109 (Promega), and S 17-1 (28) were used to propagate plasmids and were routinely grown at 37°C in Luria-Bertani (LB) medium supplemented with kanamycin (100 μg/ml) or were grown as described previously (27).
Molecular biology.
Standard methods, as described by Sambrook et al. (29), were used throughout. All enzymes were sourced from New England BioLabs. An N. gonorrhoeae ntrX::kan mutant was made by insertional inactivation of the ntrX gene in strain 1291 (accession no. EEH63133.1) using a promoterless kanamycin resistance cassette inserted in the same orientation as the ntrX transcriptional unit. Briefly, the ntrX gene (1.3 kb), including upstream and downstream flanking regions (350 bp each), was amplified by PCR using primers ntrX-F (5′-GATACGACCGCCATGCGGCAG-3′) and ntrX-R (5′-CATCCTGAAGCAGCATCAG-3′). The PCR product was A-tailed and subcloned into the T-tailed pGEM-T Easy vector (Promega) to generate pGEM-T::ntrX. A kanamycin resistance cassette was obtained from pUC4k (GenBank accession no. X06404) by excision with HincII and subsequently inserted into the unique BmgBI restriction site within ntrX to generate pGEM-T::ntrX::kan. The ntrX::kan cassette was finally amplified by PCR using primers DUS-F (5′-TGCCGTCTGAAGACTTCAGACGGCGTAAAACGACGGCAGT-3′) and DUS-R (5′-GGAACAGCTATGACCATG-3′). The resulting PCR product carried the gonococcal DNA uptake sequence GCCGTCTGAA at the 5′ end and was used to transform N. gonorrhoeae strain 1291. Kanamycin-resistant colonies were visible after 1.5 to 2 days of growth on GC agar containing 1% (vol/vol) IsoVitaleX supplemented with 100 μg/ml kanamycin. Correct insertion into the chromosome was verified by PCR using combinations of cloning primers as well as primers complementary to the kanamycin resistance cassette: Km-out-F (5′-CATTTGATGCTCGATGAGTTTTCTAA-3′) and Km-out-R (5′-AGACGTTTCCCGTTGAATATGGCTGCAT-3′). This strain was designated the Ng-ntrX mutant.
The Ng-ntrX mutant was complemented in cis by double crossover of a wild-type (WT) copy of the ntrYX locus into the proB locus in the gonococcal chromosome using the complementation construct pCTS32 (30). The entire ntrYX locus, including 300-bp upstream and downstream flanking regions, was amplified from N. gonorrhoeae 1291 wild-type genomic DNA using primers COMP-F (5′-TACAAACTAAGTTTCCATCCG-3′) and COMP-R (5′-AAAGCGCGTTTTTCCGCAT-3′). The PCR product was ligated into the SmaI site of pCTS32 to generate pCTS32-ntrYX. The plasmid was linearized with ScaI and transformed into the Ng-ntrX mutant. Positive transformants were selected on GC agar supplemented with both kanamycin (100 μg/ml) and spectinomycin (50 μg/ml). The complemented strain was verified using a combination of PCR and DNA sequencing using primers specific for the ntrYX locus (check-F, 5′-CGGTAGAAACTTATGCGTAG-3′) and the spectinomycin resistance cassette (check-R, 5′-GAATGGTTACAAGAGCTTTA-3′). This strain was designated the Ng-ntrX-COMP strain.
An ntrX mutant in R. capsulatus strain SB1003 was generated by transferring the suicide plasmid pDG9-3II (31) into R. capsulatus SB1003 by conjugation. The identity of the strain, designated the Rc-ntrX mutant, was confirmed by Southern blotting using digoxigenin (DIG)-dUTP (Roche Biochemicals, NSW, Australia)-labeled DNA probes as described previously (32). Selection and propagation of Rc-ntrX mutants were carried out aerobically on antibiotic-supplemented TYS medium (26).
Preparation of cell extracts and respiratory enzyme assays.
The N. gonorrhoeae 1291 wild-type strain and the Ng-ntrX mutant strain were grown aerobically to the mid-exponential phase in 100 ml sBHI broth in 250-ml flasks with shaking (37°C, 200 rpm). Cells were harvested by centrifugation (15,000 × g for 20 min at 4°C) and resuspended in 50 mM HEPES-Na (pH 7.5). N. gonorrhoeae cytochrome oxidase activity was assayed as a measure of oxygen consumption using an OxyGraph oxygen electrode and the accompanying software (Hansatech), based on the method of Markwell and Lascelles (33). The reaction mixture contained 50 mM HEPES-Na (pH 7.5), 25 μl cell suspension, 50 mM substrate (lactic acid), 10 μM myxothiazol, 10 mM ascorbate, and 20 μM TMPD. Reactions were carried out in a total volume of 2 ml at 30°C. Protein concentrations of cell suspensions were determined by the method of Markwell et al. (34).
R. capsulatus cell extracts were prepared using late-exponential-phase cells as described previously (27). The activity of R. capsulatus cytochrome oxidase was measured using a Rank oxygen electrode, also as described previously (35). Specific activity of transhydrogenase was measured as described previously (36) with the omission of KCN as experiments were carried out in an anaerobic cabinet (<2 ppm O2). Specific activity of the NADH:quinone oxidoreductase (also performed in an anaerobic cabinet) was measured by the reduction of decylubiquinone at 340 nm (50 μM decylubiquinone, 100 μM NADH, 2 to 10 μl chromatophores in 50 mM HEPES-Na [pH 7.5] in a total volume of 1 ml).
Nitrite reductase assays.
Methyl viologen-linked nitrite reductase assays were carried out using intact N. gonorrhoeae cells in a Hitachi U3000 spectrophotometer, based on the method of Sellars et al. (37) with slight modifications. N. gonorrhoeae strains were grown aerobically to the mid-exponential phase in sBHI. Cells were harvested by centrifugation (15,000 × g for 20 min at 4°C) and resuspended in 10 mM Tris-Cl (pH 7.5). The assay was carried out in a total volume of 1 ml and consisted of 10 mM Tris-Cl (pH 7.5) and 100 μM methyl viologen contained in a screw-topped quartz cuvette (Hellma Analytics) fitted with a silicone rubber seal to prevent gas exchange. After the addition of cells and sparging with nitrogen gas for approximately 20 min to make the mixture anaerobic, aliquots of freshly prepared sodium dithionite solution were injected into the cuvette until the absorbance at 600 nm was stable at approximately 1.1 absorbance units. The assay was then started by the addition of sodium nitrite to the cuvette to yield a final concentration of 5 mM. Rates of nitrite reductase activity were calculated using an extinction coefficient for methyl viologen of 8.25 mM−1 cm−1 at 600 nm. Protein concentrations of cell suspensions were determined by the method of Markwell et al. (34).
Microarray analysis.
Triplicate cultures of the N. gonorrhoeae 1291 wild-type strain and the Ng-ntrX mutant were grown aerobically to the exponential phase (optical density at 600 nm of 0.5 to 0.6) in sGC prior to RNA extraction using the RNeasy maxikit according to manufacturer's instructions (Qiagen). The growth rates of the wild-type and mutant strain pairs used to make RNA for microarray comparison were equivalent, thereby ensuring that RNA isolated for subsequent microarray comparison was obtained from bacteria in the same growth phase. The culture medium for RNA isolation was free of antibiotics as the chromosomal ntrX::kan mutation is stable without selection. For each strain, triplicate samples (100 μg RNA each) were isolated and pooled, and the integrity and concentration of RNA were determined using a Bioanalyzer (Agilent Technologies).
All microarray analysis was performed on N. gonorrhoeae or Neisseria meningitidis genome arrays (J. Craig Venter Institute; http://www.jcvi.org/). Each microarray consists of 6,389 70-mer oligonucleotides representing open reading frames (ORFs) from N. gonorrhoeae strains FA1090 and ATCC 700825 (reference strain), as well as from N. meningitidis strains Z2491 (serogroup A) and MC58 (serogroup B). Methods and analysis were performed as previously described (38). All primary data were imported into an in-house installation of the comprehensive microarray relational database BASE.
Biofilm formation by N. gonorrhoeae.
Examination of biofilm formation was carried out via confocal microscopy using a Nikon PCM-2000 confocal microscope scanning system (Nikon, Melville, NY). The N. gonorrhoeae 1291 wild-type strain and the Ng-ntrX isogenic mutant were transformed with plasmid pCmGFP, encoding a green fluorescent protein (GFP) (39). Formation and analyses of biofilms were performed as described previously (38, 40), with the exception that cells were grown under the same aerobic conditions described for the microarray analysis, prior to their inoculation into biofilm growth chambers. The colonies used to inoculate cultures for biofilm assays were assessed for morphology to ensure equivalent levels of piliation as described previously (41). The biofilm images are three-dimensional reconstructions of stacked z-series taken at ×200 magnification and were rendered by Volocity (Improvision, Inc., Lexington, MA) as described previously (40).
N. gonorrhoeae association, invasion, and survival assays.
Primary cervical epithelial (pex) cells were procured from surgical cervical tissue and maintained as described previously (42). Cervical tissue was obtained from premenopausal women undergoing hysterectomy at The Ohio State University Medical Center for medically indicated reasons not related to our study and was provided by the Cooperative Human Tissue Network (The Research Institute at Nationwide Children's Hospital, Columbus, OH). In accordance with NIH guidelines, these tissues do not constitute human subjects. Quantitative association, invasion, and survival assays were performed as we have described previously (43). Briefly, pex cells were challenged with gonococci at a multiplicity of infection of 100 for 90 min. Gentamicin was then omitted from (association assays) or added to (invasion and intracellular survival assays) infected pex cell monolayers to kill extracellular cell-associated bacteria. The pex cell monolayers were subsequently lysed, or they were subjected to a second incubation in antibiotic-free medium before cell lysis (intracellular survival assays). For all assays, serial dilutions of the cell lysates were plated to enumerate viable CFU. The percentage of association, invasion, or survival was determined as a function of the original inoculum and the number of colonies formed with subsequent plating of the cellular lysate. Each assay was performed in triplicate on at least three separate occasions. A Kruskal-Wallis analysis of variance (ANOVA) was used to determine the statistical significance of the calculated percentage of association, invasion, or survival for each assay. Student's t test was used to determine the significance of the calculated invasion and survival indices.
Phylogenetic analyses.
Protein sequences related to the NtrX proteins (accession numbers in parentheses) from N. gonorrhoeae FA1090 (YP_208898), R. capsulatus SB1003 (YP_003577952), Pseudovibrio sp. strain JE062 (ZP_05082799.1), Nitrococcus mobilis Nb-231 (ZP_01126634.1), and Geobacter sulfurreducens PCA (NP_901868) were identified using the BLAST algorithm (44). All sequences had to have at least 350 amino acids and share the basic domain architecture of the NtrX proteins (N terminus, response regulator receiver domain; middle section, P-loop ATPase-σ54 interaction domain; C terminus, DNA binding domain). After the removal of duplicates, sequences were aligned using Mega5.0 (45) followed by construction of phylogenetic trees using the neighbor-joining, minimum evolution, unweighted-pair group method using average linkages (UPGMA) and maximum likelihood algorithms as embedded in Mega5.0 using the default settings. Robustness testing was carried out using the bootstrap method and 500 resampling cycles.
RESULTS
Disruption of the N. gonorrhoeae ntrX gene affects expression of genes involved in aerobic and anaerobic respiration.
We constructed an ntrX mutant in N. gonorrhoeae strain 1291 by disrupting the ntrX gene (GenBank accession no. EEH63133.1; annotated as ntrX in the 1291 genome) with a promoterless kanamycin antibiotic resistance cassette (Fig. 1A) and designated this strain as the Ng-ntrX mutant. To identify NtrX-regulated genes, levels of gene expression in the 1291 wild type and Ng-ntrX mutant were compared by analysis of Neisseria gonorrhoeae or Neisseria meningitidis genome microarrays (the J. Craig Venter Institute) using total RNA isolated from exponential-phase gonococci that had been grown aerobically. Overall, 11 genes were differentially regulated and showed a greater than 2-fold change in expression (P < 0.01) between the 1291 wild-type and Ng-ntrX strains (Table 1). Seven genes showed reduced expression in the Ng-ntrX mutant compared to the wild type, with four of these genes (ccoP, aniA, norB, and ccpR) encoding terminal reductases of the Neisseria respiratory chain (Fig. 1B; Table 1). ccoP (NGO1371) encodes the triheme c-type cytochrome component of the cytochrome cbb3 oxidase complex (cytochrome oxidase; GenBank accession no. AE004969) (46, 47), and ccpR (NGO1769) encodes a cytochrome c peroxidase (a diheme c-type cytochrome that uses hydrogen peroxide as an electron acceptor) (48); aniA (NGO1276) and norB (NGO1275) encode a copper-containing nitrite reductase and a nitric oxide reductase, respectively. The latter two enzymes make up the partial denitrification pathway present in N. gonorrhoeae, which converts nitrite to nitrous oxide with nitric oxide as an intermediate (49). The three remaining genes that also showed reduced expression in the Ng-ntrX strain were (i) NGO1024, which shows the highest similarity to nitroalkane oxygenases; (ii) NGO1064, which is related to a carbon starvation protein found in E. coli; and (iii) NGO0718, which is annotated as encoding an rpiR family transcription factor—most probably hexR, which encodes a repressor of genes involved in glucose catabolism via the Entner-Doudoroff pathway (50, 51) (GenBank accession no. AE004969). Of the four genes whose expression was increased in the Ng-ntrX strain, two (NGO0640 and NGO0641) encode RmsR and a putative type III restriction/modification methylase (GenBank accession no. AE004969), whereas the other two encode hypothetical proteins.
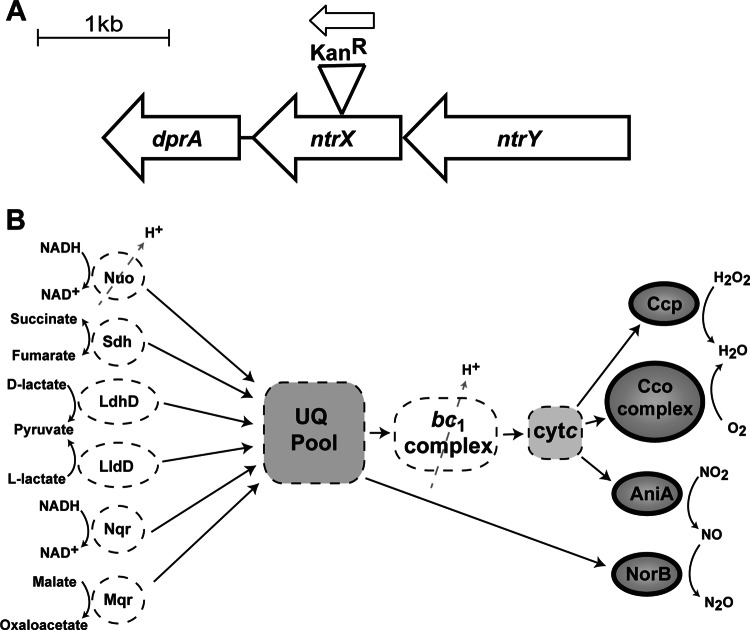
Fig 1.
(A) Construction of an ntrX::kan insertion-deletion mutant in N. gonorrhoeae strain 1291, using a nonpolar promoterless kanamycin resistance cassette as detailed in Materials and Methods. The ntrY gene is upstream of ntrX. No polar effects are expected on the gene downstream of ntrX, dprA, as there is a large intergenic region of >80 bp between these two genes. The dprA gene is annotated as DNA processing chain A (NGO1865) in the FA1090 genome. (B) Summary of the N. gonorrhoeae respiratory chain. A number of different substrates can be used to provide reducing power to the electron transport chain, which transfers electrons from the ubiquinone-ubiquinol pool (UQ pool) to cytochrome c (cyt c) via the cytochrome bc1 complex. Electrons are then transferred to a number of final electron acceptors—oxygen (cytochrome cbb3 oxidase [Cco]), hydrogen peroxide (cytochrome c peroxidase [CCP])—or to nitrite and then nitric oxide (nitrite reductase [AniA] and nitric oxide reductase [NorB], respectively). The latter two enzymes comprise the partial denitrification pathway present in the gonococcus. NorB takes electrons directly from the UQ pool and is only energy conserving if used in tandem with the Nuo NADH dehydrogenase. All four terminal reductases (Cco, CCP, AniA, and NorB) are downregulated in the Ng-ntrX mutant.
Table 1.
Genes that are differentially regulated in the Ng-ntrX mutant compared to the 1291 wild-type strain
| Gene type and no.a | Product description (gene) |
ntrX mutant/WT expression ratio by: |
|
|---|---|---|---|
| Microarray (B statistic)b | Enzyme activityc | ||
| Downregulated | |||
| NGO1276 | Nitrite reductase (aniA) | 0.33 (6.91) | 0.29 |
| NGO1275 | Nitric oxide reductase (norB) | 0.37 (4.09) | |
| NGO1769 | Cytochrome c peroxidase (ccpR) | 0.39 (4.19) | |
| NGO1371 | Cytochrome oxidase subunit (ccoP) | 0.40 (5.23) | 0.46 |
| NGO1024 | Hypothetical protein | 0.40 (5.13) | |
| NGO1064 | CstA | 0.45 (2.18) | |
| NGO0718 | Putative RpiR family transcriptional regulator | 0.50 (4.77) | |
| Upregulated | |||
| NGO0641 | Putative type III restriction/modification methylase | 2.64 (1.01) | |
| NGO0640 | RmsR | 2.62 (3.70) | |
| NGO1712 | Hypothetical protein | 2.15 (5.15) | |
| NGO1889 | Hypothetical protein | 2.07 (3.67) | |
The “NGO” gene identification numbers refer to the annotation of the Neisseria gonorrhoeae strain FA1090 genome in the Los Alamos database (http://www.stdgen.lanl.gov/stdgen/bacteria/ngon/index.html).
The ratio presented is the mean of the ratio of expression of the mutant to that of the wild type (WT) from six replicate spots on three independent microarrays, incorporating a dye swap. Thus, the expression of each gene was measured six times. Only those genes with an expression value above 2-fold (upregulated) or below 0.5-fold (downregulated) and having a B statistic value above 0.0 were considered significant and included in this study. A threshold in the B statistic of 0.0 was adopted because genes with a B score of >0 have a >50% probability of being truly differentially expressed.
Enzyme assays were performed as detailed in Materials and Methods, and the results are detailed in Fig. S1 in the supplemental material.
To confirm that a mutation in ntrX affects the activity of gonococcal respiratory complexes, the activities of cytochrome oxidase and nitrite reductase in the 1291 wild-type strain and Ng-ntrX mutant were determined (Table 1; see Fig. S1 in the supplemental material). Cytochrome oxidase activity and nitrite reductase activity were significantly lower in the Ng-ntrX mutant than in wild-type bacteria, a finding that is consistent with the microarray results. Complementation of the Ng-ntrX mutant using the entire ntrYX operon resulted in a restoration of the wild-type phenotype when cytochrome oxidase activity was used as a test for successful mutant complementation (see Fig. S1). This confirms that the downregulation of respiratory chain components in the Ng-ntrX strain was due to loss of functional NtrX. Taken together, these data showed that N. gonorrhoeae NtrX has a role in the regulation of terminal reductase expression/activity and, especially, in the regulation of cytochrome cbb3. We found no evidence that NtrX affected genes involved in assimilatory nitrogen metabolism in this bacterium.
The gonococcal ntrX mutant is compromised in biofilm formation.
The ability to generate energy using respiration or other mechanisms is a key metabolic process, and therefore the effect of the ntrX mutation on growth and survival of the Ng-ntrX mutant relative to the wild type was assessed. No difference was observed in the growth rates of wild-type or Ng-ntrX mutant gonococci during aerobic and microaerobic growth in planktonic culture, and COMSTAT analysis showed no difference in either average thickness or amount of biomass in the biofilms of wild-type versus Ng-ntrX cells (data not shown). However, when gonococcal biofilms were examined, after 48 h of growth on glass that had been stained using a LIVE/DEAD stain (Fig. 2), a substantially greater number of dead bacteria were present in the lower strata of the Ng-ntrX biofilm (Fig. 2B) compared to the biofilm formed by wild-type bacteria (Fig. 2A). It is known that gonococcal growth in a biofilm requires both nitrite reductase and nitric oxide reductase (52, 53), and both of these enzymes, as well as cytochrome cbb3, showed decreased expression in the Ng-ntrX strain. This result suggests that NtrX and the genes regulated by NtrX play an important role in gonococcal survival in the oxygen-depleted substrata of gonococcal biofilms.
Fig 2.
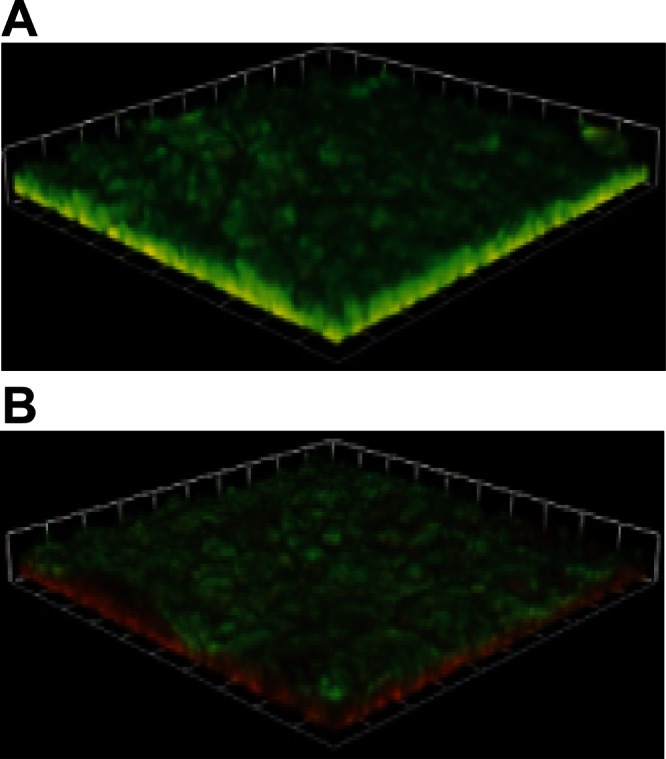
Biofilm images of the 1291 wild-type strain (A) and the Ng-ntrX isogenic mutant strain (B) using green fluorescent protein to show LIVE/DEAD cells. Wild-type biofilms contain predominantly live cells, as evidenced by the large amount of GFP produced. Ng-ntrX cells in contrast, contain predominantly dead cells, indicating the genes regulated by NtrX are important in the formation of a live, functional biofilm. Images were taken as 1-dimensional (1D) slices at ×200 magnification and stacked to produce a 3D image using Volocity software.
Gonococcal mutants lacking NtrX show a significant decrease in the ability to invade and survive within primary human cervical epithelial cells.
The use of primary human cervical epithelial (pex) cells as a model system of gonococcal cervicitis is well established and has been used to examine various aspects of the cervical tissue-gonococcus interaction (38, 42). To determine the biological significance of the ntrX mutation using this pex cell culture model, we performed quantitative association, invasion, and survival assays using Ng-ntrX mutant and wild-type gonococci (Fig. 3). The Ng-ntrX cells showed a significant (P < 0.0001) decrease in their ability to associate with, invade, and survive within pex cells compared to wild-type N. gonorrhoeae cells (Fig. 3B). Together with the biofilm data, these results suggest that NtrX plays an important role in the pathobiology of N. gonorrhoeae by aiding survival of the bacteria in the context of a cervical cell infection as well as during extracellular biofilm formation.
Fig 3.
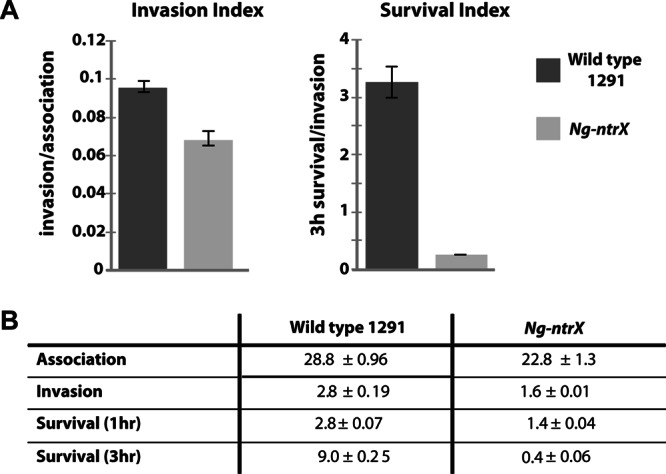
(A) Pex cell survival assays of the 1291 wild-type strain and the Ng-ntrX mutant represented as an index of invasion (invasion/association) and survival (3-h survival/invasion). For the invasion index, P = 0.0016; for the survival index, P = 0.0001. Error bars are a measure of variance from the mean; P values for the invasion and survival indices were determined using Student's t test. (B) Data from pex cell association, invasion, and survival assays used to generate the data shown in panel A, shown as a percentage of the initial inoculum. All data are averages from three separate experiments. P values for association, invasion, and the 1- and 3-h survival rates for the 1291 wild-type strain versus the Ng-ntrX mutant are all ≤0.0001. Errors represent variance from the mean; P values were calculated using a Kruskal-Wallis ANOVA. Experiments were performed in triplicate on three separate occasions.
An ntrX mutant of R. capsulatus is also affected in cytochrome cbb3 respiratory activity.
Previous studies of NtrYX and its role in the regulation of nitrogen and general metabolism were carried out in Alphaproteobacteria, including R. capsulatus (31). To determine whether NtrYX is also involved in the regulation of specific respiratory processes in R. capsulatus, an insertion-deletion mutation in the ntrX locus of wild-type R. capsulatus SB1003 (designated the Rc-ntrX mutant) was constructed. To enable comparison of the data derived from the use of the Ng-ntrX strain, respiratory activities were measured using an oxygen electrode with chromatophores (inverted membrane vesicles) prepared from R. capsulatus grown aerobically with lactate or succinate (carbon source) plus ammonium (nitrogen source). The activities of the respiratory chain complexes in the Rc-ntrX mutant were significantly lower in all cases than those of the wild-type strain (Fig. 4). Respiratory chain activities using NADH, succinate, and lactate as electron donors were all reduced by between 2- and 4-fold. Of particular significance was the low level (reduced by 80%) of respiratory activity observed with the use of ascorbate-TMPD in the Rc-ntrX mutant, which corresponds to the activity of cytochrome cbb3 and thus indicates a significant decrease in the expression of this terminal oxidase complex in the ntrX mutant (Fig. 4).
Fig 4.
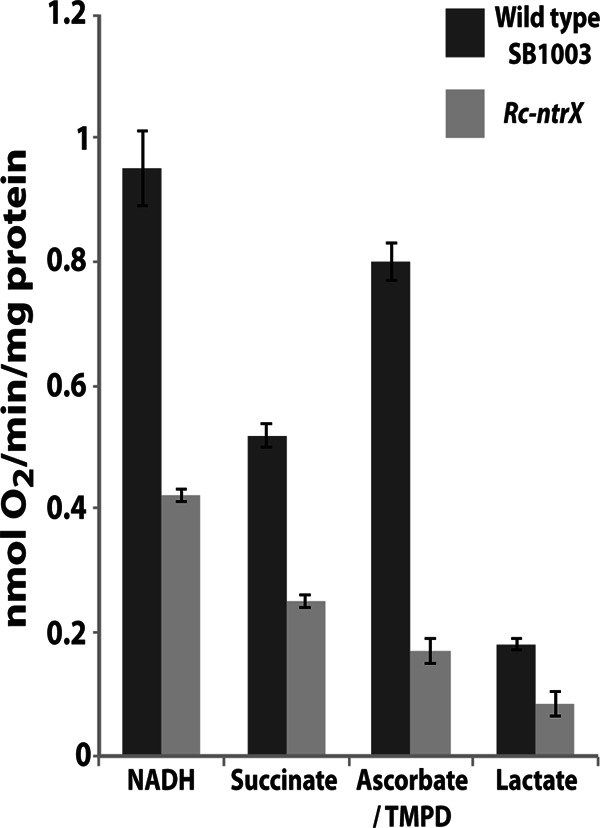
R. capsulatus SB1003 wild-type versus Rc-ntrX mutant strain respiratory assays using chromatophores, with NADH, succinate, ascorbate-TMPD, and lactate, provided as the substrates. Rates are expressed as a measure of nmol O2 consumed/min/mg protein and are the result of three separate experiments. Rates were measured using a Rank oxygen electrode as detailed in Materials and Methods. In all cases, there was a statistically significant difference between the wild-type and Rc-ntrX strains (P < 0.0001), calculated using Student's t test. Error bars represent 1 standard error of the mean.
To further confirm the effect of the ntrX mutation on the respiratory chain and its associated components, the specific activities of two enzymes, NADH:decylubiquinone oxidoreductase (NADH dehydrogenase/complex I) and pyridine nucleotide transhydrogenase (which interconverts NADH and NADPH), were also measured in membranes from wild-type R. capsulatus and the Rc-ntrX mutant strain. The activities of both enzymes were reduced in the Rc-ntrX mutant compared to those in the wild-type strain (NADH dehydrogenase, wild type, 2.6 ± 0.87; Rc-ntrX mutant, 0.80 ± 0.53; transhydrogenase, wild type, 0.14 ± 0.021; Rc-ntrX mutant, 0.075 ± 0.043 [all rates expressed as mU activity/mg protein]), indicating a possible role for NtrX in the regulation of these respiratory components as well as cytochrome cbb3 in R. capsulatus.
Comparative analysis of ntrX genes across completed bacterial genomes.
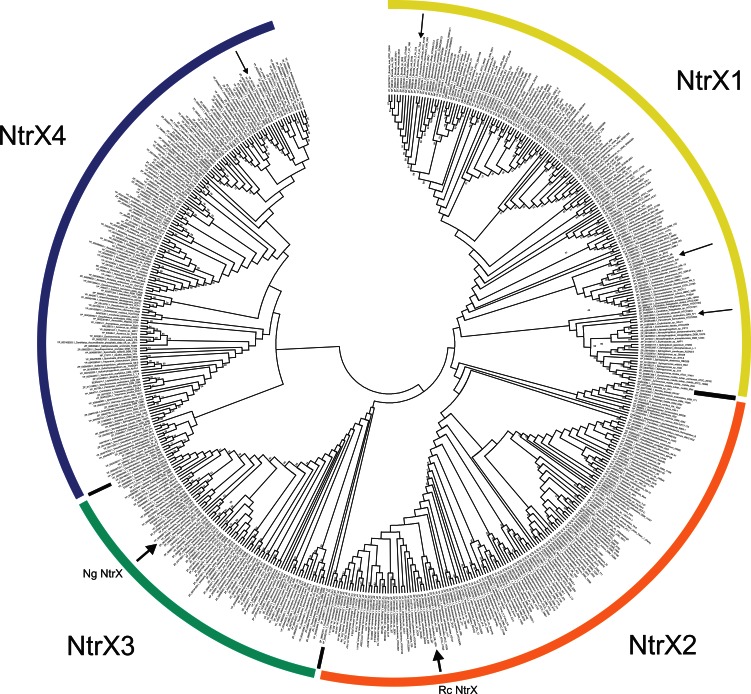
Our data show that ntrX mutations in both N. gonorrhoeae strain 1291 and R. capsulatus SB1003 affected the expression of genes involved in respiration, in spite of the fact that these NtrX proteins originated from two bacteria exhibiting completely different lifestyles and divergent phylogenetic associations. A comparison of the NtrX proteins from N. gonorrhoeae and R. capsulatus showed that these proteins have an amino acid identity of 30.4% (similarity, 45.3%) and share similar domain architectures. In both proteins the N-terminal region (∼120 amino acids [aa]) showed strong similarities to a signal receiver (Rec) domain (cd00156), followed by a P-loop ATPase domain (cl09099 and cd00009) or σ54-binding domain (pfam00158), which is less well conserved in the N. gonorrhoeae NtrX protein, and a DNA binding domain (helix-turn-helix type) in the C-terminal region. This domain architecture, which closely resembles that of the σ54-activating NtrC response regulator (54–56), together with the proximity of the ntrYX genes to the nitrogen regulatory genes ntrBC in many Alphaproteobacteria (including R. capsulatus) led to the original naming of this regulator, as well as to the suggestion that the NtrYX system is involved in nitrogen regulation (22, 31). As the basic function of this regulator appears to be conserved between Alpha- and Betaproteobacteria, we analyzed the phylogenetic distribution of ntrX genes in sequences available in public databases. A total of 534 NtrX homologues were identified and analyzed. The identified sequences were mostly of proteobacterial origin, with Alphaproteobacteria species (families Rickettsiales, Rhizobiales, and Rhodobacteriales) being the most abundant, followed by the Neisseriales (Betaproteobacteria). Other NtrX-related sequences originated from Gamma- and Deltaproteobacteria as well as from representatives of the spirochetes and Aquificales (e.g., Persephonella marina and Sulfurihydrogenibium). Phylogenetic analyses suggested the presence of four major groups of NtrX sequences (NtrX1 to -4), with NtrX1, NtrX2 and NtrX4 containing multiple subgroups (Fig. 5). Neighbor-joining and minimum evolution analyses returned identical tree topologies using UPGMA analysis to generate a phylogenetic tree, albeit some branch points were changed, leading to an altered placement of some groups. As the outer nodes were less well supported in bootstrap analyses of all trees generated, this is not unusual.
Fig 5.
Phylogenetic relationships between NtrX sequences. The four NtrX groupings are marked in color; small arrows indicate NtrX sequences for which experimental data are available, with the NtrX sequences from Rhodobacter capsulatus and Neisseria gonorrhoeae highlighted with large arrows (labeled Rc NtrX and Ng NtrX, respectively). Sequences were aligned and analyzed in Mega 5.0; the phylogenetic tree shown was constructed using the neighbor-joining algorithm and 500 bootstrap resampling cycles. Only bootstrap values above 50% are shown.
The NtrX sequences from N. gonorrhoeae and R. capsulatus are in groups NtrX3 and NtrX2, respectively, whereas NtrX sequences from other bacteria (for which functional analyses of the NtrYX system have been published) are located in NtrX1 (B. abortus, Azospirillum brasilense, Azorhizobium caulinodans) and NtrX4 (Ehrlichia chaffeensis) (21, 22, 24, 57). This indicates that the sequences identified in our database search are true NtrX homologues, although the reasons underlying the sequence divergence are unclear at present and functional analyses and comparisons between representatives of different NtrX groups will be necessary. The NtrX groups did not arise from the differential evolution of taxonomically related species as they all contain representatives of more than one taxonomic unit. For example, NtrX3 is made up of both betaproteobacterial and gammaproteobacterial sequences, and similar trends are also observed for the other groups.
We also analyzed the correlation of the occurrence of ntrYX genes together with ntrBC genes and found that, with the exception of the Neisseriales and the Rickettsiales, the majority of bacteria that contain ntrYX genes also contain copies of ntrBC genes. Moreover, in Rickettsiales and in Ehrlichia spp., both of which fall into group NtrX4, it appears that the ntrX gene encodes an orphan response regulator, with the NtrY sensor kinase being encoded elsewhere in the genome. It is possible that this is true of additional species within the NtrX4 group, and unlike most other species represented in the phylogenetic tree, cco genes appear to be absent from Rickettsiales and Ehrlichia spp. An interesting observation is that whereas most NtrX sequences contain a full-length P-loop NTPase domain, in NtrX sequences obtained from Neisseria spp., this domain, which mediates interactions with σ54, appears to be truncated (∼50 aa instead of ∼100 aa). This truncation is likely a specific adaptation that may have occurred in response to the loss of function of the σ54 sigma factor in Neisseria spp. NtrX from N. gonorrhoeae retains the same functionality as other NtrX proteins as well as the synteny of the ntrX and ntrY genes, indicating that it is a true NtrX protein.
DISCUSSION
Adaptive responses controlled by a variety of TCSs are central to the success of a number of bacterial pathogens in their interactions with the host (reviewed in reference 58). One important adaptive response is an adjustment to altered oxygen availability or redox environments, and it is established that the ArcBA TCS is required for the survival of Haemophilus influenzae in a mouse model of bacteremia (59). The ArcBA TCS, however, appears to be restricted to “oxidase-negative” bacteria such as the Enterobacteriaceae and Pasteurellaceae. Our phenotypic characterization of an ntrX mutant of N. gonorrhoeae suggests that in oxidase-positive bacteria, the NtrYX TCS may fulfill a similar adaptive role to that of ArcBA in a changing oxygen availability or redox environment. This conclusion is consistent with those of Carrica et al. (24), who found that an ntrY mutant of B. abortus exhibited reduced expression of those operons encoding enzymes of denitrification (nar, nir, nor, and nos). Our results show that in N. gonorrhoeae, the expression of the operons encoding the two respiratory complexes of the partial denitrification pathway (aniA and norB) is dependent on NtrX. However, our results also show that in N. gonorrhoeae NtrYX has a broader role in the regulation of respiratory gene expression, as both cytochrome cbb3 and cytochrome c peroxidase were also downregulated in the Ng-ntrX mutant. Although B. abortus does possess a cytochrome cbb3 (60), the effect of an ntrY mutation on its expression was not reported by Carrica et al. (24).
The NtrY sensor kinase (Conserved Domain Database [CDD] designation COG5000) is predicted to be a membrane-bound protein with cytoplasmic HAMP, PAS, and histidine kinase domains. PAS domains are critical sensory components of oxygen- and redox-sensing systems (61), and Carrica et al. (24) have shown that NtrY contains a heme prosthetic group that is involved in sensing redox changes. Our observation that the Ng-ntrX mutant biofim was necrotic beneath the surface layer is consistent with the view that the mutant was unable to adjust to the limitation in oxygen that would occur in the substrata of the biofilm. Together these results suggest that the NtrYX TCS is a system required for adaptation to oxygen limitation. Support for this idea is derived from Carrica et al. (24), who show that NtrY-dependent gene expression increases as cell culture conditions switch from aerobic to anaerobic. The modest difference that we observed in NtrX-dependent gene expression in the wild-type and Ng-ntrX mutant gonococci is similar to that observed for B. abortus NtrY-dependent gene expression occurring under microaerobic conditions.
It is increasingly clear that cytochrome oxidases with a high affinity for oxygen are required for the intracellular survival of some bacterial pathogens. For example, cytochrome bd is required for the intracellular survival and virulence of the enteric bacterium Shigella flexneri (62). In Brucella suis, cytochrome cbb3 is required for chronic infection of oxygen-limited organs in a murine model (63). Hence, one of the factors that may have contributed to the failure of the gonococcal ntrX mutant to survive within pex cells may be a lowered level of cytochrome cbb3 activity. However, the decreased expression of norB observed in the Ng-ntrX mutant also likely played a predominant role in the impaired ability of mutant gonococci to survive within pex cells, as previous data indicate that nitric oxide utilization plays a paramount role in promoting gonococcal survival within pex cells (64, 65). The human and bacterial responses driving the cervical tissue-gonococcus dynamic are complex (65). Therefore, additional, alternative adaptive responses controlled by the NtrYX TCS may have a contributory role in promoting the intracellular survival and growth of gonococci in the pex cell model of infection. Carrica et al. (24) also observed that the ntrY mutant of B. abortus showed lower survival within macrophages, lending further support to the importance of the NtrYX TCS to intracellular survival of bacterial pathogens.
Almost all studies of NtrYX to date have been conducted using bacteria in which the ntrBC genes lie upstream of the ntrYX genes. The NtrBC TCS is central to the adaptation of bacteria to changes in the quality of the nitrogen source, and it is involved in the control of the expression of nif genes in nitrogen-fixing bacteria. The NtrYX TCS was first discovered through transposon mutagenesis in Azorhizobium caulinodans, where it was described as an NtrBC-related TCS with a role in controlling nitrogen fixation (22). The authors concluded that ntrY and -X form an operon as ntrY mutations had a polar effect on ntrX. Mutants with mutations in both genes were impaired in their ability to use different nitrogen sources and to fix dinitrogen. Further, in nodulation assays, phenotypes similar to those of fixL and ntrC mutants as well as altered growth characteristics were observed for this A. caulinodans ntrYX double mutant, suggesting that symbiotic growth was affected by mutagenesis (22). Although NtrYX has been considered to be involved directly in nitrogen metabolism, the phenotype described above for A. caulinodans could have also arisen from a deficiency in the expression of cytochrome cbb3 because this oxidase is essential for symbiotic nitrogen fixation (11). Although we did not investigate rhizobial species, we did observe that in an ntrX mutant of another diazotrophic alphaproteobacterium, R. capsulatus, the activity of cytochrome cbb3 was also reduced. Thus, these data support the idea that control of cco expression is a function shared by the NtrYX TCS from a highly diverse range of bacteria. Several regulators are already known to control Cco expression in R. capsulatus, including the RegBA TCS and FNR (19). From our data we can now add the NtrYX TCS to this list, which contributes to the complexity of the regulation of Cco in this organism. Our observation that NADH dehydrogenase and pyridine nucleotide transhydrogenase activities were also affected by the mutation in R. capsulatus ntrX suggests that the NtrYX TCS may have a substantive effect on wider bioenergetic functions in R. capsulatus. FNR has been shown to also regulate cco and aniA expression in N. gonorrhoeae (66). The NarPQ TCS and the repressor NsrR are also involved in the control of aniA expression as part of the partial (AniA-NorB) denitrification pathway in N. gonorrhoeae (67). NorB, AniA, and Ccp, all of which are controlled by NtrX, are all essential for biofilm formation in N. gonorrhoeae (53). In the NtrYX-containing pathogen Ehrlichia chaffeensis, inhibitors of histidine kinase activity also reduced the ability of these bacteria to evade the cellular clearance that normally occurs through their delivery to the lysosomes (57). Clearly, further studies are required to understand the complex interplay between NtrYX and the other regulators of respiratory gene expression in N. gonorrhoeae and other bacteria, as well as the impact of an ntrX mutation on the virulence and/or ability of these bacteria to form symbiotic relationships with their respective hosts.
The activity of the NtrYX system in at least some Alphaproteobacteria is also associated with the alternative sigma factor σ54 (31, 68). Together, these regulators allow a bacterium to adjust to altered nitrogen status. However, in N. gonorrhoeae the locus encoding σ54 is a nonfunctional pseudogene (69). Collectively this is consistent with the view that adaptations made in response to altered nitrogen availability are not a primary function of the NtrYX system in this bacterium. Nevertheless, the absence of a functional gene encoding a σ54 protein is consistent with the observed truncation of the P-loop NTPase domain of NtrX in Neisseriales in that the P-loop NTPase domain is required for the efficient interaction of NtrC- or NtrX-type regulators with σ54 proteins.
Our phylogenetic analysis of NtrX showed that this response regulator is found in four distinct clades, all of which contain studied representatives of the NtrX regulator. NtrYX is present not only in the pathogenic Neisseria but also in the intracellular pathogens Brucella and Bartonella, as well as Ehrlichia and Anaplasma. It is unclear what the exact role of NtrX is in many of these species, but the conservation of this regulator in bacterial pathogens that contain very few two-component regulatory systems indicates its significance. It is possible that through the control of respiratory gene expression, the NtrYX system enables the adaptation and survival of a variety of intracellular oxidase-positive pathogens in addition to N. gonorrhoeae.
In conclusion, our results and those of Carrica et al. (24) indicate that the NtrYX TCS is a novel regulator of respiratory gene expression in a wide range of oxidase-positive bacteria and that NtrYX most likely responds to signals arising from oxygen limitation. This TCS may be critical for bacterial survival under conditions where oxygen is limiting: for example, within biofilms or intracellular growth within the eukaryotic host ranging from epithelial cells to macrophages and plant root nodules, depending on the host-microbe interaction.
ACKNOWLEDGMENTS
This work was supported by NH & MRC Program grants 284214 and 565526 to A.G.M. and M.P.J. J.P.W. was a recipient of an Australian Postgraduate Award.
Footnotes
Published ahead of print 5 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02062-12.
REFERENCES
- 1. Poole RK, Cook GM. 2000. Redundancy of aerobic respiratory chains in bacteria? Routes, reasons and regulation. Adv. Microb. Physiol. 43:165–224 [DOI] [PubMed] [Google Scholar]
- 2. Aspholm M, Aas FE, Harrison OB, Quinn D, Vik Å, Viburiene R, Tønjum T, Moir J, Maiden MCJ, Koomey M. 2010. Structural alterations in a component of cytochrome c oxidase and molecular evolution of pathogenic Neisseria in humans. PLoS Pathog. 6:e1001055 doi:10.1371/journal.ppat.1001055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Colburn-Clifford J, Allen C. 2010. A cbb3-type cytochrome c oxidase contributes to Ralstonia solanacearum R3bv2 growth in microaerobic environments and to bacterial wilt disease development in tomato. Mol. Plant Microbe Interact. 23:1042–1052 [DOI] [PubMed] [Google Scholar]
- 4. Pitcher RS, Watmough NJ. 2004. The bacterial cytochrome cbb3 oxidases. Biochim. Biophys. Acta 1655:388–399 [DOI] [PubMed] [Google Scholar]
- 5. Nagata K, Tsukita S, Tamura T, Sone N. 1996. A db-type cytochrome c oxidase terminates the respiratory chain in Helicobacter pylori. Microbiology 142:1757–1763 [DOI] [PubMed] [Google Scholar]
- 6. Gordon J, McLeod JW. 1928. The practical application of the direct oxidase reaction in bacteriology. J. Pathol. Bacteriol. 31:185–190 [Google Scholar]
- 7. Kovacs N. 1956. Identification of Pseudomonas pyocyanae by the oxidase reaction. Nature 178:703. [DOI] [PubMed] [Google Scholar]
- 8. Ekici S, Pawlik G, Lohmeyer E, Koch Daldal H-GF. 2012. Biogenesis of cbb3-type cytochrome c oxidase in Rhodobacter capsulatus. Biochim. Biophys. Acta 1817:898–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Garcia-Horsman JA, Barquera B, Rumbley J, Ma J, Gennis RB. 1994. The superfamily of heme-copper respiratory oxidases. J. Bacteriol. 176:5587–5600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Preisig O, Zufferey R, Thony-Meyer L, Appleby CA, Hennecke H. 1996. A high-affinity cbb3-type cytochrome oxidase terminates the symbiosis-specific respiratory chain of Bradyrhizobium japonicum. J. Bacteriol. 178:1532–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Preisig O, Anthamatten D, Hennecke H. 1993. Genes for a microaerobically induced oxidase complex in Bradyrhizobium japonicum are essential for a nitrogen-fixing endosymbiosis. Proc. Natl. Acad. Sci. U. S. A. 90:3309–3313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Swem LR, Elsen S, Bird TH, Swem DL, Koch Myllykallio H-GH, Daldal F, Bauer CE. 2001. The RegB/RegA two-component regulatory system controls synthesis of photosynthesis and respiratory electron transfer components in Rhodobacter capsulatus. J. Mol. Biol. 309:121–138 [DOI] [PubMed] [Google Scholar]
- 13. Partridge JD, Sanguinetti G, Dibden DP, Roberts RE, Poole RK, Green J. 2007. Transition of Escherichia coli from aerobic to micro-aerobic conditions involves fast and slow reacting regulatory components. J. Biol. Chem. 282:11230–11237 [DOI] [PubMed] [Google Scholar]
- 14. Park SJ, Chao G, Gunsalus RP. 1997. Aerobic regulation of the sucABCD genes of Escherichia coli, which encode alpha-ketoglutarate dehydrogenase and succinyl coenzyme A synthetase: roles of ArcA, Fnr, and the upstream sdhCDAB promoter. J. Bacteriol. 179:4138–4142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Park SJ, Cotter PA, Gunsalus RP. 1995. Regulation of malate dehydrogenase (mdh) gene expression in Escherichia coli in response to oxygen, carbon, and heme availability. J. Bacteriol. 177:6652–6656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cotter PA, Melville SB, Albrecht JA, Gunsalus RP. 1997. Aerobic regulation of cytochrome d oxidase (cydAB) operon expression in Escherichia coli: roles of Fnr and ArcA in repression and activation. Mol. Microbiol. 25:605–615 [DOI] [PubMed] [Google Scholar]
- 17. Govantes F, Albrecht JA, Gunsalus RP. 2000. Oxygen regulation of the Escherichia coli cytochrome d oxidase (cydAB) operon: roles of multiple promoters and the Fnr-1 and Fnr-2 binding sites. Mol. Microbiol. 37:1456–1469 [DOI] [PubMed] [Google Scholar]
- 18. Bauer C, Elsen S, Swem LR, Swem DL, Masuda S. 2003. Redox and light regulation of gene expression in photosynthetic prokaryotes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358:147–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Swem DL, Bauer CE. 2002. Coordination of ubiquinol oxidase and cytochrome cbb3 oxidase expression by multiple regulators in Rhodobacter capsulatus. J. Bacteriol. 184:2815–2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Swem LR, Kraft BJ, Swem DL, Setterdahl AT, Masuda S, Knaff DB, Zaleski JM, Bauer CE. 2003. Signal transduction by the global regulator RegB is mediated by a redox-active cysteine. EMBO J. 22:4699–4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ishida ML, Assumpcao MC, Machado HB, Benelli EM, Souza EM, Pedrosa FO. 2002. Identification and characterization of the two-component NtrY/NtrX regulatory system in Azospirillum brasilense. Braz. J. Med. Biol. Res. 35:651–661 [DOI] [PubMed] [Google Scholar]
- 22. Pawlowski K, Klosse U, de Bruijn FJ. 1991. Characterization of a novel Azorhizobium caulinodans ORS571 two-component regulatory system, NtrY/NtrX, involved in nitrogen fixation and metabolism. Mol. Gen. Genet. 231:124–138 [DOI] [PubMed] [Google Scholar]
- 23. Masepohl B, Klipp W, Puhler A. 1988. Genetic characterization and sequence analysis of the duplicated nifA/nifB gene region of Rhodobacter capsulatus. Mol. Gen. Genet. 212:27–37 [DOI] [PubMed] [Google Scholar]
- 24. Carrica MdC, Fernandez I, Martí MA, Paris G, Goldbaum FA. 2012. The NtrY/X two-component system of Brucella spp. acts as a redox sensor and regulates the expression of nitrogen respiration enzymes. Mol. Microbiol. 85:39–50 [DOI] [PubMed] [Google Scholar]
- 25. Weaver PF, Wall JD, Gest H. 1975. Characterization of Rhodopseudomonas capsulata. Arch. Microbiol. 105:207–216 [DOI] [PubMed] [Google Scholar]
- 26. Beringer JE. 1974. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84:188–198 [DOI] [PubMed] [Google Scholar]
- 27. Kappler U, Huston WM, McEwan AG. 2002. Control of dimethylsulfoxide reductase expression in Rhodobacter capsulatus: the role of carbon metabolites and the response regulators DorR and RegA. Microbiology 148:605–614 [DOI] [PubMed] [Google Scholar]
- 28. Simon R, Priefer U, Puhler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1:784–791 [Google Scholar]
- 29. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 30. Steichen CT, Shao JQ, Ketterer MR, Apicella MA. 2008. Gonococcal cervicitis: a role for biofilm in pathogenesis. J. Infect. Dis. 198:1856–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Drepper T, Wiethaus J, Giaourakis D, Gross S, Schubert B, Vogt M, Wiencek Y, McEwan AG, Masepohl B. 2006. Cross-talk towards the response regulator NtrC controlling nitrogen metabolism in Rhodobacter capsulatus. FEMS Microbiol. Lett. 258:250–256 [DOI] [PubMed] [Google Scholar]
- 32. Kappler U, Sly LI, McEwan AG. 2005. Respiratory gene clusters of Metallosphaera sedula—differential expression and transcriptional organization. Microbiology 151:35–43 [DOI] [PubMed] [Google Scholar]
- 33. Markwell JP, Lascelles J. 1978. Membrane-bound, pyridine nucleotide-independent l-lactate dehydrogenase of Rhodopseudomonas sphaeroides. J. Bacteriol. 133:593–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Markwell MAK, Haas SM, Bieber LL, Tolbert NE. 1978. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal. Biochem. 87:206–210 [DOI] [PubMed] [Google Scholar]
- 35. Dupuis A, Peinnequin A, Darrouzet E, Lunardi J. 1997. Genetic disruption of the respiratory NADH-ubiquinone reductase of Rhodobacter capsulatus leads to an unexpected photosynthesis-negative phenotype. FEMS Microbiol. Lett. 148:107–114 [Google Scholar]
- 36. Anderlund M, Nissen TL, Nielsen J, Villadsen J, Rydstram J, Hahn-Hagerdal B, Kielland-Brandt MC. 1999. Expression of the Escherichia coli pntA and pntB genes, encoding nicotinamide nucleotide transhydrogenase, in Saccharomyces cerevisiae and its effect on product formation during anaerobic glucose fermentation. Appl. Environ. Microbiol. 65:2333–2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sellars MJ, Hall SJ, Kelly DJ. 2002. Growth of Campylobacter jejuni supported by respiration of fumarate, nitrate, nitrite, trimethylamine-N-oxide, or dimethyl sulfoxide requires oxygen. J. Bacteriol. 184:4187–4196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Seib KL, Wu HJ, Srikhanta YN, Edwards JL, Falsetta ML, Hamilton AJ, Maguire TL, Grimmond SM, Apicella MA, McEwan AG, Jennings MP. 2007. Characterization of the OxyR regulon of Neisseria gonorrhoeae. Mol. Microbiol. 63:54–68 [DOI] [PubMed] [Google Scholar]
- 39. Edwards JL, Apicella MA. 2005. I-domain-containing integrins serve as pilus receptors for Neisseria gonorrhoeae adherence to human epithelial cells. Cell. Microbiol. 7:1197–1211 [DOI] [PubMed] [Google Scholar]
- 40. Falsetta ML, Bair TB, Ku SC, vanden Hoven RN, Steichen CT, McEwan AG, Jennings MP, Apicella MA. 2009. Transcriptional profiling identifies the metabolic phenotype of gonococcal biofilms. Infect. Immun. 77:3522–3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Neil RB, Apicella MA. 2009. Role of HrpA in biofilm formation of Neisseria meningitidis and regulation of the hrpBAS transcripts. Infect. Immun. 77:2285–2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Edwards JL, Shao JQ, Ault KA, Apicella MA. 2000. Neisseria gonorrhoeae elicits membrane ruffling and cytoskeletal rearrangements upon infection of primary human endocervical and ectocervical cells. Infect. Immun. 68:5354–5363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wu HJ, Seib KL, Edwards JL, Apicella MA, McEwan AG, Jennings MP. 2005. Azurin of pathogenic Neisseria spp. is involved in defense against hydrogen peroxide and survival within cervical epithelial cells. Infect. Immun. 73:8444–8448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Deeudom M, Koomey M, Moir JWB. 2008. Roles of c-type cytochromes in respiration in Neisseria meningitidis. Microbiology 154:2857–2864 [DOI] [PubMed] [Google Scholar]
- 47. Snyder LAS, Davies JK, Ryan CS, Saunders NJ. 2005. Comparative overview of the genomic and genetic differences between the pathogenic Neisseria strains and species. Plasmid 54:191–218 [DOI] [PubMed] [Google Scholar]
- 48. Atack JM, Kelly DJ. 2007. Structure, mechanism and physiological roles of bacterial cytochrome c peroxidases. Adv. Microb. Physiol. 52:73–106 [DOI] [PubMed] [Google Scholar]
- 49. Barth KR, Isabella VM, Clark VL. 2009. Biochemical and genomic analysis of the denitrification pathway within the genus Neisseria. Microbiology 155:4093–4103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Blattner FR, Plunkett G, III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462 [DOI] [PubMed] [Google Scholar]
- 51. Dempsey JA, Litaker W, Madhure A, Snodgrass TL, Cannon JG. 1991. Physical map of the chromosome of Neisseria gonorrhoeae FA1090 with locations of genetic markers, including opa and pil genes. J. Bacteriol. 173:5476–5486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Falsetta ML, McEwan AG, Jennings MP, Apicella MA. 2010. Anaerobic metabolism occurs in the substratum of gonococcal biofilms and may be sustained in part by nitric oxide. Infect. Immun. 78:2320–2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Falsetta ML, Steichen CT, McEwan AG, Cho C, Ketterer M, Shao J, Hunt J, Jennings MP, Apicella MA. 2011. The composition and metabolic phenotype of Neisseria gonorrhoeae biofilms. Front. Microbiol. 2:75 doi:10.3389/fmicb.2011.00075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kern D, Volkman BF, Luginbuhl P, Nohaile MJ, Kustu S, Wemmer DE. 1999. Structure of a transiently phosphorylated switch in bacterial signal transduction. Nature 402:894–898 [DOI] [PubMed] [Google Scholar]
- 55. Pelton JG, Kustu S, Wemmer DE. 1999. Solution structure of the DNA-binding domain of NtrC with three alanine substitutions. J. Mol. Biol. 292:1095–1110 [DOI] [PubMed] [Google Scholar]
- 56. Volkman BF, Nohaile MJ, Amy NK, Kustu S, Wemmer DE. 1995. Three-dimensional solution structure of the N-terminal receiver domain of NTRC. Biochemistry 34:1413–1424 [DOI] [PubMed] [Google Scholar]
- 57. Kumagai Y, Cheng Z, Lin M, Rikihisa Y. 2006. Biochemical activities of three pairs of Ehrlichia chaffeensis two-component regulatory system proteins involved in inhibition of lysosomal fusion. Infect. Immun. 74:5014–5022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Beier D, Gross R. 2006. Regulation of bacterial virulence by two-component systems. Curr. Opin. Microbiol. 9:143–152 [DOI] [PubMed] [Google Scholar]
- 59. Wong SMS, St. Michael F, Cox A, Ram S, Akerley BJ. 2011. ArcA-regulated glycosyltransferase Lic2B promotes complement evasion and pathogenesis of non-typeable Haemophilus influenzae. Infect. Immun. 79:1971–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Halling SM, Peterson-Burch BD, Bricker BJ, Zuerner RL, Qing Z, Li LL, Kapur V, Alt DP, Olsen SC. 2005. Completion of the genome sequence of Brucella abortus and comparison to the highly similar genomes of Brucella melitensis and Brucella suis. J. Bacteriol. 187:2715–2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Taylor BL, Zhulin IB. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Way SS, Sallustio S, Magliozzo RS, Goldberg MB. 1999. Impact of either elevated or decreased levels of cytochrome bd expression on Shigella flexneri virulence. J. Bacteriol. 181:1229–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jiménez de Bagüés MP, Loisel-Meyer S, Liautard JP, Jubier-Maurin V. 2007. Different roles of the two high-oxygen-affinity terminal oxidases of Brucella suis: cytochrome c oxidase, but not ubiquinol oxidase, is required for persistence in mice. Infect. Immun. 75:531–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Edwards JL. 2010. Neisseria gonorrhoeae survival during primary human cervical epithelial cell infection requires nitric oxide and is augmented by progesterone. Infect. Immun. 78:1202–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Edwards JL, Butler EK. 2011. The pathobiology of Neisseria gonorrhoeae lower female genital tract infection. Front. Microbiol. 2:102 doi:10.3389/fmicb.2011.00102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Whitehead RN, Overton TW, Snyder LA, McGowan SJ, Smith H, Cole JA, Saunders NJ. 2007. The small FNR regulon of Neisseria gonorrhoeae: comparison with the larger Escherichia coli FNR regulon and interaction with the NarQ-NarP regulon. BMC Genomics 8:35 doi:10.1186/1471-2164-8-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Overton TW, Whitehead R, Li Y, Snyder LAS, Saunders NJ, Smith H, Cole JA. 2006. Coordinated regulation of the Neisseria gonorrhoeae truncated denitrification pathway by the nitric oxide-sensitive repressor, NsrR, and nitrite-insensitive NarQ-NarP. J. Biol. Chem. 281:33115–33126 [DOI] [PubMed] [Google Scholar]
- 68. Masepohl B, Drepper T, Paschen A, Gross S, Pawlowski A, Raabe K, Riedel KU, Klipp W. 2002. Regulation of nitrogen fixation in the phototrophic purple bacterium Rhodobacter capsulatus. J. Mol. Microbiol. Biotechnol. 4:243–248 [PubMed] [Google Scholar]
- 69. Laskos L, Dillard JP, Seifert HS, Fyfe JA, Davies JK. 1998. The pathogenic Neisseriae contain an inactive rpoN gene and do not utilize the pilE sigma54 promoter. Gene 208:95–102 [DOI] [PubMed] [Google Scholar]