Abstract
Natural genetic transformation is common among many species of the genus Streptococcus, but it has never, or rarely, been reported for the Streptococcus pyogenes and S. bovis groups of species, even though many streptococcal competence genes and the competence regulators SigX, ComR, and ComS are well conserved in both groups. To explore the incidence of competence in the S. bovis group, 25 isolates of S. infantarius and S. macedonicus were surveyed by employing culture in chemically defined media devoid of peptide nutrients and treatment with synthetic candidate pheromone peptides predicted from the sequence of the gene comS. Approximately half of strains examined were transformable, many transforming at high rates comparable to those for the well-characterized streptococcal natural transformation systems. In S. infantarius, nanomolar amounts of the synthetic pheromone LTAWWGL induced robust but transient competence in high-density cultures, but mutation of the ComRS locus abolished transformation. We conclude that at least these two species of the S. bovis group retain a robust system of natural transformation regulated by a ComRS pheromone circuit and the alternative sigma factor SigX and infer that transformation is even more common among the streptococci than has been recognized. The tools presented here will facilitate targeted genetic manipulation in this group of streptococci.
INTRODUCTION
Horizontal gene transfer is widely recognized as an important driver of bacterial evolution. Natural genetic transformation provides a pathway of horizontal gene transfer in many groups of bacteria, including the streptococci, in which it has been known for over 70 years (1, 2). Even among the streptococci, however, the proportion of species exhibiting a capacity for transformation remains uncertain (3). Interestingly, the incidence of reported natural transformation varies greatly among the six recognized groups of species within the genus Streptococcus. Many species in the Streptococcus anginosus, S. mitis, and S. salivarius groups are recognized as naturally transformable (4). At the other extreme, no species of the S. pyogenes group has been reported to transform in laboratory culture (5). For the S. bovis group of species, the literature provides only a single case of natural genetic transformation, reported for S. bovis strain JB1. Although two groups reported competence in this strain (6, 7), no further characterization of S. bovis competence has been reported, and natural genetic transformation has not yet been employed as a tool for genetic analysis in any of the six species of the S. bovis group.
Members of the S. bovis group are common commensal bacteria of the gut microbiotas of humans, birds, and mammals, but they also are associated with products of milk fermentation. After recent revisions of the classification of the S. bovis group, most isolates can be assigned to one of six species, S. equinus, S. gallolyticus, S. infantarius, S. lutetiensis, S. macedonicus, or S. pasteurianus (8). While S. equinus, S. pasteurianus, S. gallolyticus, and S. lutetiensis are mainly isolated as commensal or clinical isolates, and S. macedonicus is isolated mainly in food, S. infantarius seems to share both commensal and food habitats (8–15). Interestingly, the recent publication of the genome of the food strain S. infantarius CJ18 (16, 17) indicates recent acquisitions of new genes facilitating adaptation of this strain to milk. The extent of these transfers suggests descent from an ancestor with an active mechanism of gene acquisition, such as natural competence.
In the streptococci, as in several other bacterial genera, competence for natural genetic transformation is not constitutive but depends on a developmental switch to coordinated expression of a complex array of effector genes that enable DNA acquisition and genetic recombination. As the natural cues that lead to this switch are poorly understood, or simply unknown, it has not been possible to design a definitive test of whether a given species is naturally transformable. Rather, naturally transformable species have been identified by adventitious discovery of laboratory culture conditions that sufficiently mimic, or substitute for, the circumstances of natural development of competence to achieve a detectable level of transformation (for examples, see references 18–21).
A shortcut to revealing the potential for natural competence for genetic transformation in many streptococci was opened by the discovery that species in the S. mitis and S. anginosus groups share a conserved regulatory circuit through which a peptide pheromone, competence-stimulating peptide (CSP), coordinates the switch leading to development of competence among nearby conspecifics (22). Elements of this circuit, including a dedicated peptide export machine, ComA/ComB, and a two-component signal transduction pathway, ComD/ComE, that senses CSP are so well conserved that synthetic preparations of candidate CSPs designed directly from genomic sequences often allow artificial induction of competence development (23). For example, in S. pneumoniae, of the S. mitis group, synthetic CSP peptide short-circuits the (unknown) upstream regulators of pheromone production and eliminates the need for strain-by-strain optimization of culture media and protocols for eliciting natural development of competence, making genetic manipulation of fresh isolates of this species a routine matter (24). However, the ComD/ComE circuit appears to be restricted to the S. mitis and S. anginosus streptococcal groups (4). Recently, another shortcut has emerged in streptococci lacking the CSP/ComD/ComE circuit. In the S. salivarius and S. mutans groups, a different class of peptide signal, encoded by genes designated comS, act as an intercellular pheromone to coordinate competence development (25, 26). The mature ComS peptides (designated XIP, for sigX-inducing peptide) are smaller than the CSP signal and are sensed after uptake by an oligopeptide permease transporter. This dependence on a nutritional peptide permease may explain why sensitivity to ComS signals is lower in rich culture media prepared from protein digests than in chemically defined media devoid of peptides. In S. thermophilus, of the S. salivarius group, and in S. mutans, of the S. mutans group, this approach has allowed discovery of conditions for robust endogenous competence development and identification of new peptide pheromones that are readily available in synthetic form (25, 27–29).
To explore the incidence of competence in species of the S. bovis group more broadly, we have examined an expanded set of genomic sequences and looked directly for genetic transformation in type strains of S. macedonicus and S. infantarius and in two dozen additional strains assigned to these species. Natural competence was frequent in both taxa.
MATERIALS AND METHODS
Bacterial strains and culture media.
Table 1 lists wild-type isolates of S. infantarius and S. macedonicus used in this study, as well as derivatives made during this work (17, 30–35; C. Jans, D. W. M. Kaindi, D. Böck, P. M. K. Njage, M. Kouamé, B. Bonfoh, C. Lacroix, and L. Meile, unpublished data). Samples were generously provided by Christoph Jans, Christophe Lacroix, and Leo Meile (Eidgenössische Technische Hochschule [ETH] Zürich); a single colony isolated from each was assigned a Jouy/INRA/Micalis (JIM) serial number and stocked in M17 medium (36) with 10% glycerol at −80°C. Routine culture was done at 37°C either in capped tubes or in open multiwell plates containing 150 to 300 μl of medium per well. Two chemically defined culture media (CDM) were used: CDMT, as described by Sissler et al. (37) but containing 1% (wt/vol) glucose as a carbon source and supplemented with morpholinepropanesulfonic acid (MOPS) (8 g/liter), urea (1.2 g/liter), and ascorbic acid (0.5 g/liter), or CDMK, described previously (38). The former was prepared from concentrated stock solutions of components, while the latter was prepared from a mixture of dry components obtained from JRH Biosciences (Lenexa, KS). Genomic donor DNA was prepared by the method of Lin Tao, as described in previously (26). Plates containing M17 medium solidified with 1.2% agar were incubated at 37°C or 30°C in ambient air. Peptides were obtained as custom syntheses from NeoPeptide or from RS Synthesis and stored at −20°C in dimethyl sulfoxide (DMSO) as 1 mM solutions. Peptide names are assigned to indicate cognate species source and residues of ComS as follows: SinComS9-15, LTAWWGL; SgaComS9-15, ITGWWGL; SeqComS9-15, LTSWWGL; and SpaComS9-15, LTGWWGV.
Table 1.
Strains used in this study
| Straina | JIM no. | Source type and country of isolation or description | Source or reference | Transformation (log Rifr/ml)b |
|---|---|---|---|---|
| S. macedonicus strains | ||||
| DSM15879T | 9421 | Greek cheese, Greece | 30 | 6 |
| 139C | 9423 | Suusac garoor, Somalia | 31 | 6 |
| 34I | 9425 | Fènè, Mali | 32 | <3 |
| AV3B(1) | 9427 | Sour milk, Ivory Coast | Jans et al., unpublished | <3 |
| S. infantarius strains | ||||
| ATCC BAA-102 | 9376 | Human infant feces, not available | 33 | 5 |
| 11FA-1 | 9430 | Fènè, Mali | 32 | 7 |
| 13AF | 9375 | Fènè, Mali | 32 | 7 |
| 3AG-1 | 9431 | Fènè, Mali | 32 | 6 |
| 6C | 9433 | Fènè, Mali | 32 | 4 |
| AB2VB1-2 | 9386 | Sour milk, Ivory Coast | Jans et al., unpublished | 5 |
| AB2VB1-3 | 9387 | Sour milk, Ivory Coast | Jans et al., unpublished | 4 |
| AV2A(1) | 9394 | Sour milk, Ivory Coast | Jans et al., unpublished | 7 |
| AV3A(3) | 9395 | Sour milk, Ivory Coast | Jans et al., unpublished | 7 |
| AV3B(3) | 9396 | Sour milk, Ivory Coast | Jans et al., unpublished | 7 |
| P2VC1-1 | 9383 | Sour milk, Ivory Coast | Jans et al., unpublished | 6 |
| 13DW | 9374 | Fènè, Mali | 32 | <3 |
| BEV4C(1) | 9392 | Sour milk, Ivory Coast | Jans et al., unpublished | <3 |
| CJ18 | 9377 | Suusac, Kenya | 34 | <3 |
| CJ246 | 9381 | Suusac, Kenya | 17 | <3 |
| CJ249 | 9379 | Suusac garoor, Somalia | 35 | <3 |
| CJ251 | 9378 | Suusac, Kenya | 17 | <3 |
| L2VB3-3 | 9389 | Sour milk, Ivory Coast | Jans et al., unpublished | <3 |
| LV1A(1) | 9390 | Sour milk, Ivory Coast | Jans et al., unpublished | <3 |
| LV1A(2) | 9391 | Sour milk, Ivory Coast | Jans et al., unpublished | <3 |
| P2VC1-4 | 9385 | Sour milk, Ivory Coast | Jans et al., unpublished | <3 |
| Derivative strains | ||||
| JIM9435 | JIM9431 (3AG-1) but ΔcomS::erm; Ermr | This study | 7 | |
| JIM9436 | JIM9431 (3AG-1) but ΔcomR comS::erm; Ermr | This study | <2 | |
| JIM9437 | JIM 9394 [AV2A(1)] but ΔcomR comS::erm; Ermr | This study | <2 | |
| JIM9438 | JIM9377 (CJ18) but Rifr | This study | ||
| JIM9439 | JIM9421 (DSM15879T) but Rifr | This study |
The species designations S. macedonicus and S. infantarius are used according to the recommendation of Poyart et al. (8). They are also known as S. gallolyticus subsp. macedonicus and S. infantarius subsp. infantarius, respectively.
Transformation reported as the highest yield of Rifr recombinants observed with cognate ComS peptide.
Transformation assay.
In routine assays, aliquots of a growing culture were mixed with DNA and XIP and held at 37°C for 40 min in a 96-well plate (300 μl/well) or in Eppendorf tubes (200 to 500 μl/tube). The transformed cultures were then diluted 3-fold into M17 broth containing 2 μg/ml of DNase I (Boehringer Mannheim) and held for 40 min further at 37°C to allow expression of new genes, before dilution and plating on or in selective agar containing 5 μg/ml of rifampin (Rif) or erythromycin (Erm).
Construction of new strains.
Portions of the comR comS locus were deleted from strain 3AG-1 and replaced with an Ermr gene, using overlapping PCR to prepare donor DNA fragments bounded by sequences flanking the deleted material, as described previously (39). The Erm cassette, derived from pAMβ-1, was amplified from pGh9:ISS1 (40) with primers EG1576/7 (5′-ACGGTTCGTGTTCGTGCTGACTTGCACCAT-3′/5-AGCTCCTTGGAAGCTGTCAGTAGTATACCT-3′). The downstream flanking fragment was amplified from strain 3AG-1 with primers EG1570/1 (5′-AGGTATACTACTGACAGCTTCCAAGGAGCTTTGTAGACTGTTATCAAAACAATTCACTGA-3′/5′-AGATGATCATATGCTTTAGCTGTA-3′). Upstream fragments were amplified from strain 3AG-1 with primers EG1568/9 (ΔcomR comS) (5′-TCCACGTGACCTTCACGCAGAATA-3′/5′-ATGGTGCAAGTCAGCACGAACACGAACCGTCATCAAGCGGATCTTCTCCCCAAATTTTTC-3′) or EG1574/5 (ΔcomS) (5′-TCACGTGAAGAATTCTGTGGGGAT-3′/5′-ATGGTGCAAGTCAGCACGAACACGAACCGTTCCCTTTCTCTAATTTCTAATATTATTGTA-3′). For assembly, three fragments were mixed and amplified by overlapping PCR with primers EG1571 and either EG1574 (for ΔcomS) or EG1568 (for ΔcomR comS). After transformation of 3AG-1, isolated Ermr clones were named JIM9435 or JIM9436, respectively. Verification of the mutant structure in each one was done by PCR using primers EG1572/1573(5′-TGTTGAAGCTTATGTGTGTGAA-3′/5′-ATAGTTATAGTATATAAAGACTTGC-3′) and other pairs internal and external to the planned insertion, with the expected presence (or absence) of fragments observed in all cases. The ΔcomR comS deletion in JIM9436 was transferred to strain AV2A (1) by XIP-induced transformation, with Erm selection, to create strain JIM9437. Spontaneous Rifr mutants were isolated from colonies appearing on M17-Rif agar spread with 100 μl of overnight cultures of S. infantarius strain CJ18 (JIM9377) or S. macedonicus strain DSM15879T (JIM9421). The mutations increased the rifampin MIC from <1 to >100 μg/ml.
Genome analysis of strains.
Partial or complete genomic sequences were collected from published and INRA sources. Other sequences were newly determined (see below).
Nucleotide sequence accession numbers.
Sequences determined at INRA/Jouy are assigned BioProject numbers PRJEB239, PRJEB257, PRJEB268, and PRJNA189563.
RESULTS
Conserved competence regulons in S. bovis genomes.
In the naturally transformable streptococci, development of competence is regulated through tightly controlled expression of the alternative sigma factor SigX (also designated ComX [41]), which, in turn, drives transcription of a few dozen effector genes, known as “late” genes (42, 43). Regulation of sigX expression is coordinated by peptide pheromone quorum-sensing circuits of three types, which are distributed in a group-specific pattern. In the S. mitis and S. anginosus groups, the ∼17-amino-acid (aa) comC pheromone product CSP is sensed outside the cell by a two-component signal transduction system comprising a membrane receptor, ComD, and cognate response regulator, ComE. The two other types of pheromone circuit are encoded by bicistronic loci designated comR comS. In the S. salivarius group, a 7-aa pheromone is sensed by an intracellular receptor, ComR, an Rgg-like DNA binding protein (25, 44). In the S. mutans and S. pyogenes groups, a distinct 7-aa pheromone is sensed by a paralogous class of receptors designated type II ComR (45). ComS products associated with type I ComR receptors have the sequences PYFAGCL and VPFFMIYY, whereas the type II ComR receptors are associated with ComS products that share a subterminal WW motif (26).
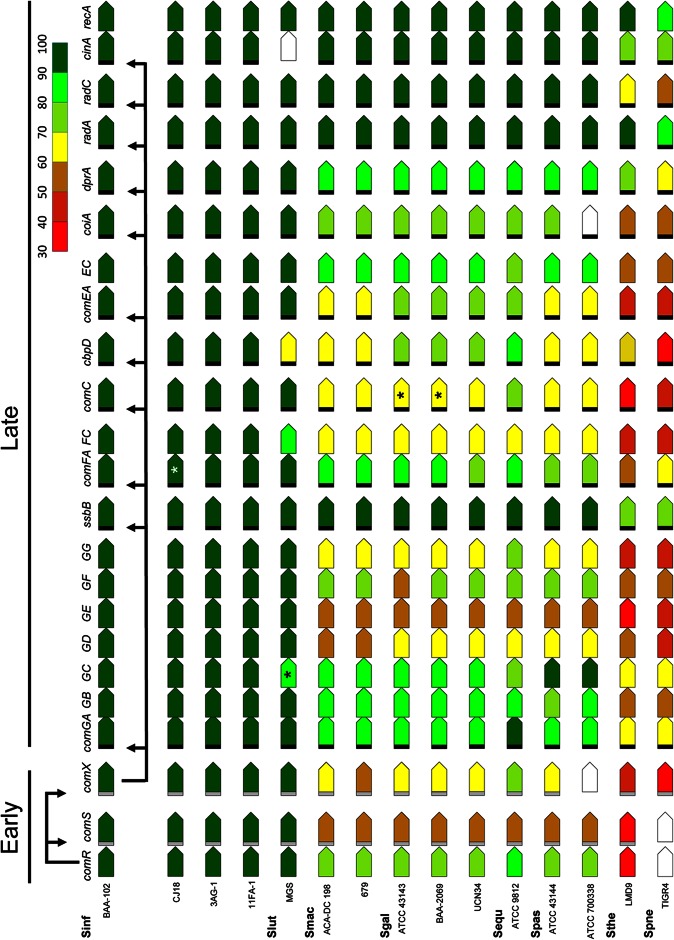
To explore the genetic potential for natural competence in the S. bovis group more thoroughly, we determined four new draft genome sequences, to supplement the information from the 12 genome sequences currently available for the group. This collection afforded a comprehensive view of genome compositions in six S. bovis group species. The genomes were searched for homologs of genes known to be required for genetic transformation in other streptococcal groups. Remarkably, both sigX and a full set of orthologs of all late effector genes required for transformation in S. pneumoniae were identified in each genome examined (Fig. 1). Furthermore, each such effector gene was associated with the cinbox noncanonical promoter (TACGAATA) known as the target of SigX. Finally, the S. bovis genomes all carried orthologs of the type II comR genes, as well as two copies of the conserved promoter and 9-bp inverted repeat (termed ComR box [44]) that are recognized by type II ComR proteins to control transcription of comS and sigX (25, 26). The exceptions to this pattern among the genomes analyzed are an inactive allele of comFA in the genome of S. infantarius CJ18, a comGC pseudogene in S. lutetiensis MGS, and two S. gallolyticus genomes that share an inactive allele of comC, which encodes a peptidase required for assembly of the DNA transport apparatus. Finally, inspection of the sequences immediately downstream of comR revealed a type II ComR box linked to a small unannotated open reading frame (ORF) encoding an ∼15-aa peptide containing the WW subterminal motif characteristic of type II comR comS loci. The sequences of the latter ORFs, which we designate ComS, are highly conserved, as shown in Table 2, which lists the four similar C-terminal heptapeptides that can be predicted as sigX-inducing peptides (XIPs) by analogy with known type II ComS products.
Fig 1.
Conservation of competence regulons in the S. bovis group. The presence of full-length gene orthologs of sigX (comX), of its target late competence genes, and of the comRS pheromone circuit genes was determined by analysis of complete or draft genomes, for which species and strains are indicated at the left. Genes are not drawn to scale. Black bar, cinbox promoter; gray bar, ComR box promoter; empty pentagon, missing or incomplete gene sequence; *, truncated gene; ▽, presence of other genes. The radA cinbox is located in front of the upstream gene, dut. Colors indicate percent sequence match of protein products to the translation products of genes in S. infantarius ATCC BAA-102: comR (STRINF_00393), comS (not annotated), comX (STRINF_00080), comGA (STRINF_00857), comGB (STRINF_00856), comGC (STRINF_00855), comGD (STRINF_00854), comGE (STRINF_00853), comGF (STRINF_00852), comGG (STRINF_00851), ssbB (STRINF_00838), comFA (STRINF_00254), comFC (STRINF_00255), comC (STRINF_00665), cbpD (lytF) (STRINF_01688), comEA (STRINF_00591), comEC (STRINF_00590), coiA (STRINF_01410), dprA (STRINF_01270), radA (STRINF_00885), radC (STRINF_01176), cinA (STRINF_00511), and recA (STRINF_00510). Sources of sequences analyzed: S. infantarius (Sinf) ATCC BAA-102 (PRJNA54885), CJ18 (PRJNA87033), 3AG-1 (PRJEB257), and 11FA-1 (PRJEB239); S. lutetiensis (Slut) metagenomic assembly (MGS) (PRJNA189563); S. macedonicus (Smac) ACA-DC-198 (PRJNA81631) and 679 (PRJEB268); S. gallolyticus (Sgal) ATCC 43143 (PRJDA162103), ATCC BAA-2069 (PRJNA63617), and UCN34 (PRJNA46061); S. equinus (Sequ) ATCC 9812 (PRJNA62297); S. pasteurianus (Spas) ATCC 43144 (PRJDA62519) and ATCC 700338 (designated in GenBank as S. bovis but assigned here as an S. pasteurianus strain based on its sodA gene sequence) (PRJNA52359); S. thermophilus (Sthe) LMD9 (PRJNA58327); and S. pneumoniae (Spne) TIGR4 (PRJNA57857).
Table 2.
ComS peptides encoded in genomes of S. bovis group species
| Species | Strain | ComS | Putative XIP |
|---|---|---|---|
| S. equinus | ATCC 9812 | MKVFSILLTSWWGL | LTSWWGL (SeqComS8–15) |
| S. gallolyticus | ATCC BAA-2069 | MLNIFSIVITGWWGL | ITGWWGL (SgaComS9–15) |
| UCN34 | MLNIFSIVITGWWGL | ITGWWGL (SgaComS9–15) | |
| ATCC 43143 | MLNIFSIVITGWWGL | ITGWWGL (SgaComS9–15) | |
| S. infantarius | 3AG-1 | MLKGFTVLLTAWWGL | LTAWWGL (SinComS9–15) |
| 11FA-1 | MLKGFTVLLTAWWGL | LTAWWGL (SinComS9–15) | |
| ATCC BAA-102 | MLKGFTVLLTAWWGL | LTAWWGL (SinComS9–15) | |
| CJ18 | MLRGFTVLLTAWWGL | LTAWWGL (SinComS9–15) | |
| S. lutetiensis | MGS | MLKGFTVLLTAWWGL | LTAWWGL (SluComS9–15) |
| S. macedonicus | ACA-DC-198 | MLKFFSIVITGWWGL | ITGWWGL (SmaComS9–15) |
| 679 | MLKFFSIVITGWWGL | ITGWWGL (SmaComS9–15) | |
| S. pasteurianus | ATCC 43144 | MLNIFSIVLTGWWGV | LTGWWGV (SpaComS9–15) |
| ATCC 700338 | MLNIFSIVLTGWWGV | LTGWWGV (SpaComS9–15) |
Altogether, homologs of all genetic elements of the SigX regulon of the natural competence systems identified in the other streptococcal groups appeared to be present in the genomes of all six S. bovis group species examined, with apparent pseudogenes in only 4 among 13 strains examined. In addition, a type II ComR ComS pheromone circuit orthologous to those known in S. mutans (26, 28) and S. pyogenes (5) appeared to be linked to expression of sigX in each case by means of a ComR box element at the sigX gene. Taken together, this pattern of conservation of effector proteins, regulators, and cis regulatory sites that are linked to competence in other groups of streptococci suggests that species of the S. bovis group often retain a capacity for natural genetic transformation.
Widespread competence in S. bovis group food species.
The sequenced S. bovis group genomes available to date represent only a few strains per species. To test the hypothesis of widespread competence in the S. bovis group more broadly, we examined multiple S. infantarius strains by direct experiment, using the type strain ATCC BAA-102 and a collection of 20 recent isolates from the camel milk food chain in Africa (Table 1). In initial surveys, cultures growing in CDMT were exposed at a range of densities to either the Ermr plasmid pFED756 or Rifr genomic donor DNA, with or without a supplement of the candidate XIP peptide, SinComS9–15 (LTAWWGL), predicted from genome analysis of S. infantarius strains (Table 2). Many of the strains yielded transformants upon such XIP treatment, including the type strain ATCC BAA-102, and some were competent even without addition of the peptide. Ten strains never yielded any transformants, while 11 yielded various levels of recombinants after exposure to XIP. Table 1 identifies both sets of strains and presents the highest yield of Rifr transformants observed for each. Endogenous competence development varied among strains but was typically absent or detected only at high cell densities. We also explored the potential for competence of four representatives of S. macedonicus in the same way, but using CDMK. The type strain, DSM15879, readily exhibited endogenous competence in this medium, and one among three recent food isolates also transformed upon treatment with the predicted S. macedonicus XIP peptide, SmaComS9–15 (ITGWWGL) (Table 1). We regard the observed levels of natural competence in these two species as a minimum estimate, because the failure to observe transformants applies only to the culture conditions used in our survey, and we did not attempt to optimize culture conditions or choice of peptide for individual strains. Overall, competent strains in these two species originated from five countries, suggesting that natural transformation is common and widespread in both species.
Competence development by S. infantarius in CDM and enhancement by XIP.
To begin to define better the conditions for competence development in S. infantarius, we examined in more detail competence development in strain AV2A (1), which often exhibited endogenous competence in the preliminary surveys described above. As Fig. 2A shows, endogenous competence development by this strain during growth in CDMT was tightly regulated; competence appeared suddenly in late log phase and then disappeared in less than 30 min, effectively limiting transformation to culture densities (optical densities [OD]) between 0.3 and 0.5. In contrast to this limited window for endogenous development of competence, the same strain became highly competent over a broad range of culture densities (0.05 to 1) if treated with the XIP peptide, as illustrated in Fig. 2B. Similar patterns were observed for strains 11FA-1 and 3AG-1 (data not shown). We conclude that S. infantarius can develop a high level of competence in this CDM and that the C-terminal heptapeptide fragment of ComS stimulates the switch to competence gene expression. While we have not tested other C-terminal derivatives of ComS, and the identity of the native ComS product is not yet known, activity of the candidate C-7 ComS peptide is consistent with activity of the C-7 form of ComS peptides in several other type II ComR ComS systems, including S. mutans (28, 29) and S. pyogenes (5), as well as S. parauberis, S. porcinus, and S. agalactiae (46).
Fig 2.
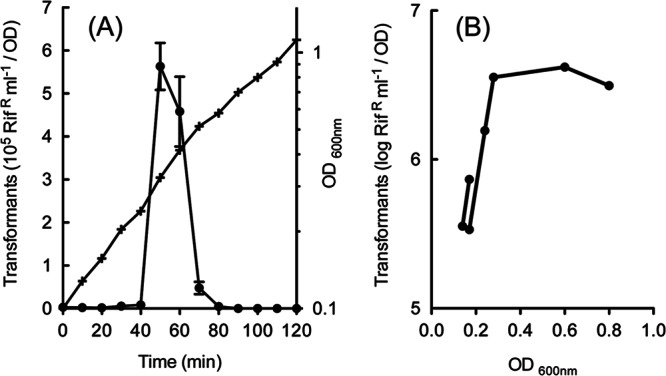
Development of competence in S. infantarius. (A) Endogenous development of competence. A culture of strain AV2A (1) in CDM was initiated by 1:100 dilution of a log-phase stock. After reaching an OD of 0.1, samples (300 μl) were treated with 2 μg/ml of DNA for 8 min, diluted 1/30 in M17 medium with DNase for expression, and then plated after further 1/30 dilution in M17-Rif agar to determine transformants. Error bars represent standard deviations among triplicate determinations. (B) XIP-induced development of competence. After growth of multiple cultures of strain AV2A (1) in CDM to the indicated densities, samples (300 μl) were treated with 0.4 μg/ml of DNA and 1 μM SinComS9–15 in microwell plates for 40 min and diluted 1/3 in M17 medium with DNase for expression. After further serial dilution in M17 medium, 6-μl drops were spotted on Rif agar to determine transformants. Values are the average of two determinations.
Regulation of S. infantarius competence by ComRS.
To examine if, as could be predicted from the activity of the candidate XIP, comRS is required for the response to synthetic XIP, a deletion of the comR comS locus was constructed by gene replacement in the transformable strain 3AG-1, using our draft genome sequence of this strain as a guide for targeted mutagenesis. To examine its effect on endogenous competence development, this mutation was transferred to strain AV2A (1), which has displayed endogenous competence development more consistently than strain 3AG-1. Deletion of comR comS eliminated both endogenous expression of competence and response to synthetic peptide in this strain (Fig. 3). In contrast, a deletion of comS alone, made in strain 3AG-1 in the same way, did not prevent a response to XIP (see below). We conclude that ComR is a key regulator of competence gene expression in S. infantarius and that it is required for sensing a ComS product. We hypothesize further, on the basis of conserved ComR box sites in S. bovis genomes at both sigX and comS genes, that ComR regulates expression of sigX and comS directly in all species of this group.
Fig 3.
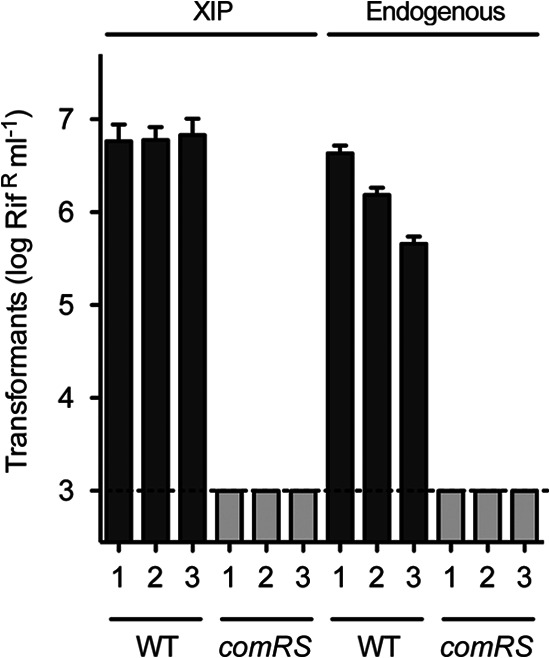
Linkage of transformation-defective phenotype to comRS mutation. Transformation was determined for three parallel AV2A (1) cultures (wild type [WT]), and parallel cultures of three independent ΔcomR comS::erm transformants of strain AV2A (1) (comRS). XIP-induced transformation (left) was determined at OD of 0.4 to 0.6 (left), by 40-min exposures to 1 μg/ml of DNA and 4 μM SinComS9–15. Endogenous development of competence (right) was determined as the maximum among three successive 40-min exposures to 1 μg/ml of DNA during growth from OD of 0.6 to 2. The dashed line indicates the limit of detection. Error bars represent standard deviations among triplicate determinations for each culture.
Kinetics of XIP-induced competence development.
To characterize additional fundamental properties of transformation in S. infantarius, we used the 3AG-1ΔcomS strain to ensure absence of both endogenous competence development and the refractory period that may follow it. As Fig. 4 shows, competent 3AG-1ΔcomS cells yielded transformants in direct proportion to the amount of added DNA, displaying a linear dose response which saturated above 1 μg/ml of donor genomic DNA. In the linear response range, the yield was approximately 2 million transformants per microgram of input DNA. Peptide titration revealed a high sensitivity to XIP, with substantial levels of competence elicited by as little as 1 nM peptide, with little or no inhibition resulting from a 10,000-fold excess (Fig. 5A). The time course of the response to addition of XIP was monitored by determining the yield of transformants arising from short exposures to DNA at various times after XIP addition to a growing CDM culture of 3AG-1ΔcomS. Competence development followed a delay of 10 min, reached a maximum by 20 min, and then declined rapidly, recapitulating the temporal pattern seen in endogenous competence development in this medium (Fig. 5B). Together, these patterns describe a typical streptococcal competence regime, operating transiently at high cell density and responding to unusually low levels of pheromone peptide.
Fig 4.
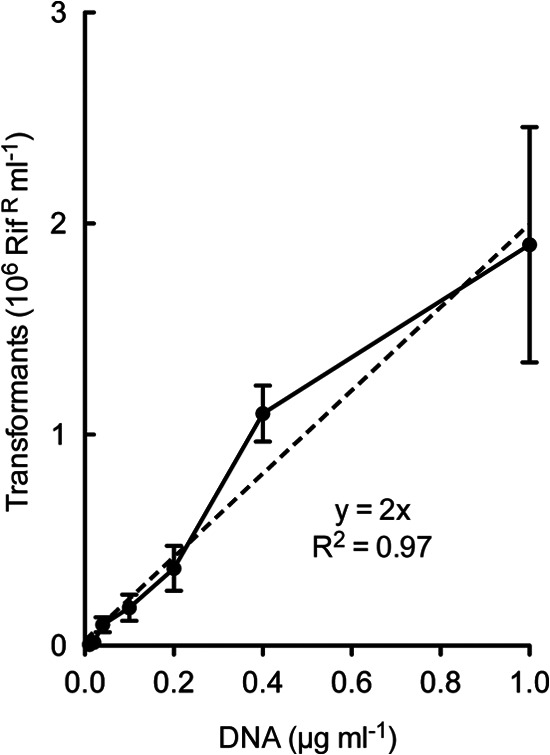
Linear DNA dose dependence of transformation of S. infantarius strain 3AG-1 by a genomic marker. Strain 3AG-1ΔcomS growing in CDM at an OD of 0.5 was exposed to SinComS9–15 (1 μM) and the indicated amounts of donor DNA in 250-μl volumes. After 40 min at 37°C, samples were diluted in M17 medium for expression and plating. The dashed line indicates linear regression fit to data between 0 and 1 μg of DNA/ml. Error bars represent standard deviations among triplicate determinations.
Fig 5.
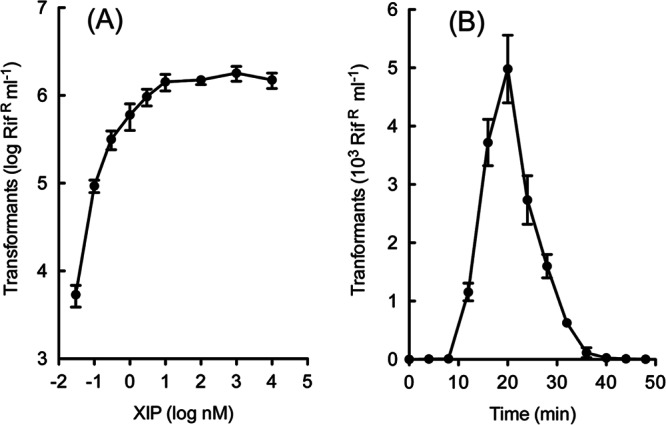
Kinetics of competence development in S. infantarius. (A) Pheromone dependence of competence development. Aliquots of a CDM culture of strain 3AG-1ΔcomS at an OD of 0.5 were mixed with DNA (0.5 μg/ml) and the indicated amounts of SinComS9–15 peptide. After 40 min at 37°C, samples were diluted in M17 medium for expression and plating. Error bars represent standard deviations among triplicate determinations. (B) Temporal pattern of competence induction by XIP. After addition of 10 μM SinComS9–15 to a culture of 3AG-1ΔcomS in CDM at an OD of 0.3, 250-μl samples were exposed to DNA (2 μg/ml) for 3 min at the indicated times and then diluted 30-fold into M17 medium with 2 μg/ml of DNase. After expression, further dilutions were plated in M17-Rif agar to obtain an estimate of relative competence level. Error bars represent standard deviations among triplicate determinations.
Peptide cross talk.
To examine whether the XIP signal recognition was highly specific, strain 3AG-1ΔcomS was treated with synthetic peptides that differed by 1 or 2 residues from the S. infantarius sequence but matched the ComS C terminus of other species. As Fig. 6 shows, the A3S and L1I/A3G substitutions in S. equinus and S. macedonicus peptides hardly affected their activity in inducing competence development in S. infantarius, indicating efficient cross-species communication within the S. bovis group. To explore sensitivity to type II ComS peptide signals from a more distantly related species, we also tested the ComS signal from S. mutans, which differs by four substitutions from the S. infantarius peptide. Its effectiveness was about 100-fold lower but was not abolished completely.
Fig 6.
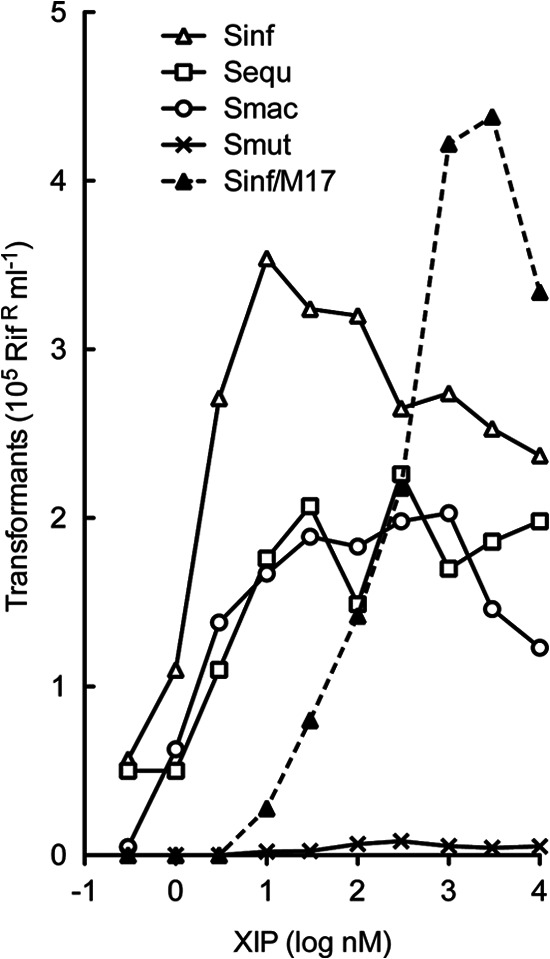
Competence development in S. infantarius with variant peptides. Transformation of strain 3AG-1ΔcomS treated with various peptides was determined by incubation of 250-μl aliquots of a culture at an OD of 0.2 with 2 μg/ml of Rifr DNA for 40 min at 37°C in microtiter wells, followed by expression after 1:3 dilution into M17 medium with 2 μg/ml of DNase. Seven further serial 3× dilutions were made from the DNase well in the same plate, and 6-μl samples were spotted onto M17-Rif agar in triplicate. Values are calculated from colonies counted in three successive dilution spots and standard deviations of the triplicate estimates. Competence induction by SinComS9-15 in M17 medium was determined in the same way, using a culture growing in M17 medium at an OD of 0.2 (▲). Peptides were SinComS9-15 (△), SeqComS9-15 (□), SmaComS9-15 (○), or SmuComS11-17 (GLDWWSL) from S. mutans (×).
Peptide competition.
ComS signaling is commonly absent or reduced in rich culture media, an effect attributed to competition for access to the peptide transporters by peptides in the protein hydrolysates used in formulating these media (27, 28, 45, 47). To examine if this pattern holds for the S. bovis competence regulators, we compared XIP titration curves for 3AG-1ΔcomS in CDMT versus the rich M17 medium. The profiles, shown in Fig. 6, revealed a strong (hundredfold) reduction of effectiveness of XIP in M17 medium but also showed that the apparent competition could be overwhelmed with a large enough dose of the pheromone.
DISCUSSION
Natural genetic transformation and control of expression of competence regulons have been characterized in some detail for species of the S. mitis, S. anginosis, S. mutans, S. salivarius, and S. pyogenes groups. However, until now, competence in species of the S. bovis group was not characterized in such detail. The results presented here begin to fill this gap, revealing a transient state of competence regulated by SigX and a type II ComR ComS pheromone circuit in S. infantarius. Competence in S. infantarius shares features with that in other groups of streptococci, but in a new combination. Endogenous competence development occurred at high cell densities, similar to the case for S. mutans and contrasting with the patterns in S. thermophilus and S. pneumoniae, where low densities are optimal. On the other hand, competence was transient, similar to the pattern in S. pneumoniae and S. thermophilus but different from that seen with S. mutans, where competence is maintained for hours. S. infantarius offers a potential readily accessible model of competence regulation for the S. bovis group. The development of competence in CDM at high density (optical density [OD] of 0.5, 109/ml) for S. infantarius contrasts to that reported by Mercer et al. (6) for S. bovis strain JB1 in brain heart infusion (BHI), for which competence was reported as developing very early during exponential growth, at a density of less than 2 × 107 CFU/ml. This broad description of S. bovis competence raises, of course, many questions about its biology. While the robust competence seen in CDM in laboratory culture establishes that competence is available to this species, it does not reveal the nature of native cues affecting comRS activation, or even whether competence develops in milk during fermentation. Nor is it known at present whether the readily available competence in fresh isolates of S. infantarius and S. macedonicus is typical for other S. bovis group species or exceptional. However, the recent demonstration that the ComR protein of S. gallolyticus strain UCN34 exhibits ComS-dependent binding to its cognate ComR box (44) suggests that this species may also possess natural competence and strengthens the inference that competence is common throughout the S. bovis group.
An aspect of competence we did not investigate in this study is its common association with expression of bacteriocins and other lytic proteins (48, 49). As suggested by Håvarstein (3), the variety of regulators of sigX may reflect different evolutionary solutions to a need to coordinate attack on neighboring cells with expression of systems for DNA uptake for effective horizontal gene transfer. Although the food S. infantarius strains can produce potent bacteriocins, which likely play a role in its dominance of camel milk fermentations (17), the relation of such bacteriocins to competence is unknown.
The present observations provide useful tools for future studies of the S. bovis group. First, the similar behaviors of two S. bovis species suggest that the CDM/XIP strategy will be useful in beginning to explore competence in the remaining species of this group. Second, the large number of additional food S. bovis strains available from collection campaigns in Africa offer a rich resource for exploring the natural distribution and variation of competence in this group in more detail. Third, the highly synchronous response to XIP suggests that direct gene expression profiling could be used to define quite precisely the ComR and the SigX regulons of this group. Finally, readily available conditions for robust natural transformation can now be exploited for direct molecular genetic analysis of at least two species of the S. bovis group.
Identification of ComR, ComS, and SigX as regulators of competence in S. bovis group species strengthens the experimental foundation for a unified general description of competence for genetic transformation in the streptococci. All major groups of species possess functional competence genes regulated by the alternative sigma factor SigX, but each group has evolved a specific arrangement for the control of expression of sigX. In two groups, it is controlled by an extracellular receptor, a two-component signal transduction system, and a signaling peptide. In the four other groups, two paralogous families of ComR proteins control sigX expression directly, type I in the S. salivarius group and type II in the three others. In this view, the Gram-positive streptococci have followed a pattern of evolution like that described for competence in the Gram-negative Pasteurellaceae by Redfield et al. (50), where an ancestral competence system has occasionally been lost by strains or taxa within the group but is maintained in most extant lineages. The geographical range of competent S. bovis isolates described here suggests directly that competence in these species is widespread and that there is selective pressure for its maintenance in the artisanal dairy environment. Indeed, the widespread maintenance of such a complex trait as competence in all streptococcal groups argues for such selective pressure in many streptococcal niches.
ACKNOWLEDGMENTS
We are grateful to Christoph Jans, Christophe Lacroix, and Leo Meile for providing strains from their collections. The experiments reported here were carried out during a sabbatical research visit at MICALIS by D.A.M. Analytical support from the MG-RAST server is gratefully acknowledged.
This material is based upon work supported in part by the National Science Foundation under grant no. MCB-1020863.
Footnotes
Published ahead of print 29 March 2013
REFERENCES
- 1. Avery OT, Macleod CM, McCarty M. 1944. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J. Exp. Med. 79:137–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Johnsborg O, Eldholm V, Håvarstein LS. 2007. Natural genetic transformation: prevalence, mechanisms and function. Res. Microbiol. 158:767–778 [DOI] [PubMed] [Google Scholar]
- 3. Håvarstein LS. 2010. Increasing competence in the genus Streptococcus. Mol. Microbiol. 78:541–544 [DOI] [PubMed] [Google Scholar]
- 4. Martin B, Quentin Y, Fichant G, Claverys JP. 2006. Independent evolution of competence regulatory cascades in streptococci? Trends Microbiol. 14:339–345 [DOI] [PubMed] [Google Scholar]
- 5. Mashburn-Warren L, Morrison DA, Federle MJ. 2012. The cryptic competence pathway in Streptococcus pyogenes is controlled by a peptide pheromone. J. Bacteriol. 194:4589–4600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mercer DK, Melville CM, Scott KP, Flint HJ. 1999. Natural genetic transformation in the rumen bacterium Streptococcus bovis JB1. FEMS Microbiol. Lett. 179:485–490 [DOI] [PubMed] [Google Scholar]
- 7. Asanuma N, Yoshii T, Kanada K, Yoshizawa K, Arai Y, Ichikawa T, Kawamura A, Hino T. 2010. Involvement of two-component signal transduction system, ComDE, in the regulation of growth and genetic transformation, in the ruminal bacterium Streptococcus bovis. Anaerobe 16:405–411 [DOI] [PubMed] [Google Scholar]
- 8. Poyart C, Quesne G, Trieu-Cuot P. 2002. Taxonomic dissection of the Streptococcus bovis group by analysis of manganese-dependent superoxide dismutase gene (sodA) sequences: reclassification of ‘Streptococcus infantarius subsp. coli’ as Streptococcus lutetiensis sp. nov. and of Streptococcus bovis biotype 11.2 as Streptococcus pasteurianus sp. nov. Int. J. Syst. Evol. Microbiol. 52:1247–1255 [DOI] [PubMed] [Google Scholar]
- 9. Beck M, Frodl R, Funke G. 2008. Comprehensive study of strains previously designated Streptococcus bovis consecutively isolated from human blood cultures and emended description of Streptococcus gallolyticus and Streptococcus infantarius subsp. coli. J. Clin. Microbiol. 46:2966–2972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kok H, Jureen R, Soon CY, Tey BH. 2007. Colon cancer presenting as Streptococcus gallolyticus infective endocarditis. Singapore Med. J. 48:e43–e45 [PubMed] [Google Scholar]
- 11. Leclercq R, Huet C, Picherot M, Trieu-Cuot P, Poyart C. 2005. Genetic basis of antibiotic resistance in clinical isolates of Streptococcus gallolyticus (Streptococcus bovis). Antimicrob. Agents Chemother. 49:1646–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Onoyama S, Ogata R, Wada A, Saito M, Okada K, Harada T. 2009. Neonatal bacterial meningitis caused by Streptococcus gallolyticus subsp. pasteurianus. J. Med. Microbiol. 58:1252–1254 [DOI] [PubMed] [Google Scholar]
- 13. Romero B, Morosini MI, Loza E, Rodriguez-Banos M, Navas E, Canton R, Campo RD. 2011. Reidentification of Streptococcus bovis isolates causing bacteremia according to the new taxonomy criteria: still an issue? J. Clin. Microbiol. 49:3228–3233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rusniok C, Couve E, Da Cunha V, El Gana R, Zidane N, Bouchier C, Poyart C, Leclercq R, Trieu-Cuot P, Glaser P. 2010. Genome sequence of Streptococcus gallolyticus: insights into its adaptation to the bovine rumen and its ability to cause endocarditis. J. Bacteriol. 192:2266–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schlegel L, Grimont F, Ageron E, Grimont PA, Bouvet A. 2003. Reappraisal of the taxonomy of the Streptococcus bovis/Streptococcus equinus complex and related species: description of Streptococcus gallolyticus subsp. gallolyticus subsp. nov., S. gallolyticus subsp. macedonicus subsp. nov. and S. gallolyticus subsp. pasteurianus subsp. nov. Int. J. Syst. Evol. Microbiol. 53:631–645 [DOI] [PubMed] [Google Scholar]
- 16. Jans C, Follador R, Hochstrasser M, Lacroix C, Meile L, Stevens MJ. 2013. Comparative genome analysis of Streptococcus infantarius subsp. infantarius CJ18, an African fermented camel milk isolate with adaptations to dairy environment. BMC Genomics 14(1):200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jans C. 2011. Biodiversity of lactic acid bacteria in raw camel milk products of East Africa including genomic and functional characterization of predominant lactose-adapted Streptococcus infantarius subsp. infantarius. Ph.D. thesis Eidgenössische Technische Hochschule Zürich, Zürich, Switzerland [Google Scholar]
- 18. Dawson MH, Sia RH. 1931. In vitro transformation of pneumococcal types: I. A technique for inducing transformation of pneumococcal types in vitro. J. Exp. Med. 54:681–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Alexander HE, Leidy G. 1953. Induction of streptomycin resistance in sensitive Hemophilus influenzae by extracts containing desoxyribonucleic acid from resistant Hemophilus influenzae. J. Exp. Med. 97:17–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Spizizen J. 1958. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc. Natl. Acad. Sci. U. S. A. 44:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Meibom KL, Blokesch M, Dolganov NA, Wu CY, Schoolnik GK. 2005. Chitin induces natural competence in Vibrio cholerae. Science 310:1824–1827 [DOI] [PubMed] [Google Scholar]
- 22. Håvarstein LS, Coomaraswamy G, Morrison DA. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. U. S. A. 92:11140–11144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Håvarstein LS, Hakenbeck R, Gaustad P. 1997. Natural competence in the genus Streptococcus: evidence that streptococci can change pherotype by interspecies recombinational exchanges. J. Bacteriol. 179:6589–6594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ramirez M, Morrison DA, Tomasz A. 1997. Ubiquitous distribution of the competence related genes comA and comC among isolates of Streptococcus pneumoniae. Microb. Drug Resist. 3:39–52 [DOI] [PubMed] [Google Scholar]
- 25. Fontaine L, Boutry C, de Frahan MH, Delplace B, Fremaux C, Horvath P, Boyaval P, Hols P. 2010. A novel pheromone quorum-sensing system controls the development of natural competence in Streptococcus thermophilus and Streptococcus salivarius. J. Bacteriol. 192:1444–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mashburn-Warren L, Morrison DA, Federle MJ. 2010. A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp via an Rgg regulator. Mol. Microbiol. 78:589–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gardan R, Besset C, Guillot A, Gitton C, Monnet V. 2009. The oligopeptide transport system is essential for the development of natural competence in Streptococcus thermophilus strain LMD-9. J. Bacteriol. 191:4647–4655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Desai K, Mashburn-Warren L, Federle MJ, Morrison DA. 2012. Development of competence for genetic transformation of Streptococcus mutans in a chemically defined medium. J. Bacteriol. 194:3774–3780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Khan R, Rukke HV, Ricomini Filho AP, Fimland G, Arntzen MO, Thiede B, Petersen FC. 2012. Extracellular identification of a processed type II ComR/ComS pheromone of Streptococcus mutans. J. Bacteriol. 194:3781–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tsakalidou E, Zoidou E, Pot B, Wassill L, Ludwig W, Devriese LA, Kalantzopoulos G, Schleifer KH, Kersters K. 1998. Identification of streptococci from Greek Kasseri cheese and description of Streptococcus macedonicus sp. nov. Int. J. Syst. Bacteriol. 48:519–527 [DOI] [PubMed] [Google Scholar]
- 31. Wullschleger S. 2009. Biodiversity and microbial safety of artisanal Malian sour milk fènè and development of adapted starter cultures for controlled production. Ph.D. thesis Eidgenössische Technische Hochschule Zürich, Zürich, Switzerland [Google Scholar]
- 32. Wullschleger S, Lacroix C, Bonfoh B, Sissoko-Thiam A, Hugenschmidt S, Romanens E, Baumgartner S, Traoré I, Yaffee M, Jans C, Meile L. 2013. Analysis of lactic acid bacteria communities and their seasonal variations in a spontaneously fermented dairy product (Malian fènè) by applying a cultivation/genotype-based binary model. Int. Dairy J. 29:28–35 [Google Scholar]
- 33. Schlegel L, Grimont F, Collins MD, Regnault B, Grimont PA, Bouvet A. 2000. Streptococcus infantarius sp. nov., Streptococcus infantarius subsp. infantarius subsp. nov. and Streptococcus infantarius subsp. coli subsp. nov., isolated from humans and food. Int. J. Syst. Evol. Microbiol. 50:1425–1434 [DOI] [PubMed] [Google Scholar]
- 34. Jans C, Bugnard J, Njage PMK, Lacroix C, Meile L. 2012. Lactic acid bacteria diversity of African raw and fermented camel milk products reveals a highly competitive, potentially health-threatening predominant microflora. LWT Food Sci. Technol. 47:371–379 [Google Scholar]
- 35. Jans C, Gerber A, Bugnard J, Njage PM, Lacroix C, Meile L. 2012. Novel Streptococcus infantarius subsp. infantarius variants harboring lactose metabolism genes homologous to Streptococcus thermophilus. Food Microbiol. 31:33–42 [DOI] [PubMed] [Google Scholar]
- 36. Terzaghi BE, Sandine WE. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sissler M, Delorme C, Bond J, Ehrlich SD, Renault P, Francklyn C. 1999. An aminoacyl-tRNA synthetase paralog with a catalytic role in histidine biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 96:8985–8990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. van de Rijn I, Kessler RE. 1980. Growth characteristics of group A streptococci in a new chemically defined medium. Infect. Immun. 27:444–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sperandio B, Gautier C, Pons N, Ehrlich DS, Renault P, Guedon E. 2010. Three paralogous LysR-type transcriptional regulators control sulfur amino acid supply in Streptococcus mutans. J. Bacteriol. 192:3464–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Maguin E, Prevost H, Ehrlich SD, Gruss A. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 178:931–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee MS, Morrison DA. 1999. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J. Bacteriol. 181:5004–5016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Peterson SN, Sung CK, Cline R, Desai BV, Snesrud EC, Luo P, Walling J, Li H, Mintz M, Tsegaye G, Burr PC, Do Y, Ahn S, Gilbert J, Fleischmann RD, Morrison DA. 2004. Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol. Microbiol. 51:1051–1070 [DOI] [PubMed] [Google Scholar]
- 43. Dagkessamanskaia A, Moscoso M, Henard V, Guiral S, Overweg K, Reuter M, Martin B, Wells J, Claverys JP. 2004. Interconnection of competence, stress and CiaR regulons in Streptococcus pneumoniae: competence triggers stationary-phase autolysis of ciaR mutant cells. Mol. Microbiol. 51:1071–1086 [DOI] [PubMed] [Google Scholar]
- 44. Fontaine L, Goffin P, Dubout H, Delplace B, Baulard A, Lecat-Guillet N, Chambellon E, Gardan R, Hols P. 2013. Mechanism of competence activation by the ComRS signalling system in streptococci. Mol. Microbiol. doi:10.1111/mmi.12157 [DOI] [PubMed] [Google Scholar]
- 45. Federle MJ, Morrison DA. 2012. One if by land, two if by sea: signalling to the ranks with CSP and XIP. Mol. Microbiol. 86:241–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wolfer M. 2011. Specific and interspecies recognition of pheromones regulates the competence regulon in pyogenic and mutans group streptococci. M.S. thesis Technical University of Kaiserslautern, Kaiserslautern, Germany [Google Scholar]
- 47. Son M, Ahn SJ, Guo Q, Burne RA, Hagen SJ. 2012. Microfluidic study of competence regulation in Streptococcus mutans: environmental inputs modulate bimodal and unimodal expression of comX. Mol. Microbiol. 86:258–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Guiral S, Mitchell TJ, Martin B, Claverys JP. 2005. Competence-programmed predation of noncompetent cells in the human pathogen Streptococcus pneumoniae: genetic requirements. Proc. Natl. Acad. Sci. U. S. A. 102:8710–8715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Claverys JP, Håvarstein LS. 2007. Cannibalism and fratricide: mechanisms and raisons d'etre. Nat. Rev. Microbiol. 5:219–229 [DOI] [PubMed] [Google Scholar]
- 50. Redfield RJ, Findlay WA, Bosse J, Kroll JS, Cameron AD, Nash JH. 2006. Evolution of competence and DNA uptake specificity in the Pasteurellaceae. BMC Evol. Biol. 6:82. [DOI] [PMC free article] [PubMed] [Google Scholar]